Abstract
Purpose:
Validating artificial intelligence algorithms for clinical use in medical images is a challenging endeavor due to a lack of standard reference data (ground truth). This topic typically occupies a small portion of the discussion in research papers since most of the efforts are focused on developing novel algorithms. In this work, we present a collaboration to create a validation dataset of pathologist annotations for algorithms that process whole slide images. We focus on data collection and evaluation of algorithm performance in the context of estimating the density of stromal tumor-infiltrating lymphocytes (sTILs) in breast cancer.
Methods:
We digitized 64 glass slides of hematoxylin- and eosin-stained invasive ductal carcinoma core biopsies prepared at a single clinical site. A collaborating pathologist selected 10 regions of interest (ROIs) per slide for evaluation. We created training materials and workflows to crowdsource pathologist image annotations on two modes: an optical microscope and two digital platforms. The microscope platform allows the same ROIs to be evaluated in both modes. The workflows collect the ROI type, a decision on whether the ROI is appropriate for estimating the density of sTILs, and if appropriate, the sTIL density value for that ROI.
Results:
In total, 19 pathologists made 1645 ROI evaluations during a data collection event and the following 2 weeks. The pilot study yielded an abundant number of cases with nominal sTIL infiltration. Furthermore, we found that the sTIL densities are correlated within a case, and there is notable pathologist variability. Consequently, we outline plans to improve our ROI and case sampling methods. We also outline statistical methods to account for ROI correlations within a case and pathologist variability when validating an algorithm.
Conclusion:
We have built workflows for efficient data collection and tested them in a pilot study. As we prepare for pivotal studies, we will investigate methods to use the dataset as an external validation tool for algorithms. We will also consider what it will take for the dataset to be fit for a regulatory purpose: study size, patient population, and pathologist training and qualifications. To this end, we will elicit feedback from the Food and Drug Administration via the Medical Device Development Tool program and from the broader digital pathology and AI community. Ultimately, we intend to share the dataset, statistical methods, and lessons learned.
Keywords: Artificial intelligence validation, medical image analysis, pathology, reference standard, tumor-infiltrating lymphocytes
INTRODUCTION
Artificial intelligence (AI) is often used to describe machines or computers that mimic “cognitive” functions associated with the human mind, such as “learning” and “problem-solving.”[1] Machine learning (ML) is an AI technique that can be used to design and train software algorithms to learn from and act on data. Although AI/ML has existed for some time, recent advances in algorithm architecture, software tools, hardware infrastructure, and regulatory frameworks have enabled health-care stakeholders to harness AI/ML as a medical device. Such medical devices have the potential to offer enhanced patient care by streamlining operations, performing quality control, supporting diagnostics, and enabling novel discovery.
While AI/ML has already found utility in radiology, the role of AI/ML algorithms in pathology has been a matter of wide discussion.[2,3,4,5,6,7] Recent technological advancements and market access of systems that scan glass slides to create digital whole slide images (WSIs) have opened the door to a myriad of opportunities for AI/ML applications in digital pathology.[8,9] While pathology is new to digitization, the field is expected to extend algorithms to a broad range of clinical decision support tasks. This technology shift is reminiscent of the digitization of mammography in 2000[10] and the first computer-aided detection (CADe) device in radiology in 1998, the R2 ImageChecker.[11] The R2 CADe device marked regions of interest (ROIs) likely to contain microcalcifications or masses, initially evaluating digitized screen-film mammograms rather than digital acquisition of mammography images.
Fourteen years after the R2 ImageChecker was approved by the US Food and Drug Administration (FDA), regulatory guidance for CADe was finalized in two documents. While both guidance documents are specific to radiology, their principles are applicable to other specialties, including digital pathology. The first document generally delineates how to describe a CADe device and assess its “stand-alone” performance.[12] In the pathology space, this might be referred to as analytical validation. The second guidance document covers clinical performance assessment, or clinical validation.[13] The document was recently updated and discusses issues such as study design, study population, and the reference standard. Related issues are also discussed in a paper summarizing a meeting jointly hosted by the FDA and the Medical Imaging Perception Society.[14]
Regardless of the technology providing the data or the algorithm architecture, Software as a Medical Device (SaMD) must be analytically and clinically validated to ensure safety and effectiveness before clinical deployment.[15] One critical aspect of algorithm validation is to assess accuracy. Accuracy compares algorithm predictions to true labels using holdout validation data, data that are independent from data used in development. Validation data include patient data (images and metadata) on which the algorithm will make predictions as well as the corresponding reference standard (ground truth or label). The reference standard can be established using an independent “gold standard” modality, longitudinal patient outcomes, or when these are not available or appropriate, a reference standard established by human experts. What constitutes the “ground truth” and how to approach it is a topic of discussion even in more traditional diagnostic test paradigms, and certainly so in evolving areas such as SaMD.
In this work, we focus on the often challenging task of establishing a reference standard using pathologists. The “interpretation by a reviewing clinician” is listed as a reference standard in the radiology CADe guidance documents and acts as the reference standard (in full or in part) in many precedent-setting radiology applications.[16,17,18] In pathology, the reference standard for evaluating performance in the Philips IntelliSite Pathology Solution regulatory submission, “was based on the original sign-out diagnosis rendered at the institution, using an optical (light) microscope.”[8]
In this manuscript, we present a collaborative project to produce a validation dataset established by pathologist annotations. The project will additionally produce statistical analysis tools to evaluate algorithm performance. The context of this work is the validation of an algorithm that measures, or estimates, the density of tumor-infiltrating lymphocytes (TILs), a prognostic biomarker in breast cancer. Resulting tools and data may be used to facilitate the external validation of an algorithm within the applied context. Given the cross-disciplinary nature of the study, the volunteer effort comprises an international, multidisciplinary team working in the precompetitive space. Project participants include the FDA Center for Devices and Radiological Health's Office of Science and Engineering Laboratories, clinician-scientists from international health systems, academia, professional societies, and medical device manufacturers. By incorporating diverse stakeholders, we aim to address multiple perspectives and emphasize interoperability across platforms.
We are pursuing qualification of the final validation dataset as an FDA Medical Device Development Tool (MDDT).[19] In doing so, we have an opportunity to receive feedback from an FDA review team while building the dataset. If the dataset qualifies as an MDDT, it will be a high-value public resource that can be used in AI/ML algorithm submissions, and our work may guide others to develop their own validation datasets.
Definitions of terms in AI-based medical device development and regulation are evolving. For example, there has been inconsistent usage of “testing” versus “validation.” To avoid this confusion, we are referring to building, training, tuning, and validating algorithms, where tuning is for hyperparameter optimization, and validation is for assessing or testing the performance of AI/ML algorithms. There is also some confusion between the terms “algorithm” and “model.” In this work, we will use the term “algorithm” to refer to the SaMD, the device, the software that is or will be deployed. Some may refer to the SaMD as the “model,” but we shall use “model” to refer to the description of the algorithm (the architecture, image normalization, transfer learning, augmentation, loss function, training, hyperparameter selection, etc.).
Herein, we present our efforts to source a pathologist-driven reference standard and apply it to algorithm validation, with an eye toward generating a fit-for-regulatory-purpose dataset. Specifically, we review the clinical association between TILs and patient outcomes in the context of accepted guidelines for estimating TIL density in tumor-associated stroma (stromal TIL [sTIL] density). We then imagine an algorithm that similarly estimates sTIL density and could use a sTIL density annotated dataset for validation. Next, we describe the breast cancer tissue samples used in our pilot study, the data collection methods and platforms, and the pathologists we recruited and trained to provide sTIL density estimates in ROIs using digital and microscope platforms. We also present some initial data and outline how we plan to account for pathologist variability when estimating algorithm performance.
TECHNICAL BACKGROUND
Tumor-infiltrating Lymphocytes
TILs are an inexpensively assessed, robust, prognostic biomarker that is a surrogate for antitumor, T-cell-mediated immunity. Clinical validity of TILs as a prognostic biomarker in early-stage, triple-negative breast cancer (TNBC), as well as in HER2+ breast cancer, has been well-established via Level 1b evidence.[20,21,22,23] Two pooled analyses of TILs, in the adjuvant setting for TNBC[21] and neoadjuvant setting across BC subtypes,[22] included studies that have evaluated TILs in archived tissue samples based on published guidelines.[24] Incorporating TILs into standard clinical practice for TNBC is endorsed by international clinical and pathology standards (St. Gallen 2019 recommendation, WHO 2019 recommendation, and ESMO2019 recommendation).[25,26,27,28] It is expected that TILs will be assessed to monitor treatment response in the future.[29,30] Further, evidence is emerging that TIL-assessment will be done in other tumor types as well, including melanoma, gastrointestinal tract carcinoma, non-small cell lung carcinoma and mesothelioma, and endometrial and ovarian carcinoma.[31,32]
Visual and Computational Tumor-infiltrating Lymphocyte Assessment
Given the recent and evolving evidence of the prognostic value of TIL assessment, there have been several efforts to create algorithms to estimate TIL density in cancer tissue. Amgad et al. provide an excellent summary of this space, including a table of algorithms from the literature, an outline with visual aids for TIL assessment, as well as a discussion on validation and training issues.[32,33] While some algorithms are leveraging details about the spatial distribution of individual TILs in different tissue compartments,[34,35,36] the guidelines for pathologists are to calculate the sTIL density[24] defined as the area of sTILs divided by the area of the corresponding tumor-associated stroma.
In this work, we imagine an algorithm that estimates the density of sTILs in pathologist-marked ROIs in WSIs of hematoxylin- and eosin-stained slides (H&E) containing breast cancer needle core biopsies. Amgad et al. refer to these quantitative values as computational TIL assessments and visual TIL assessments, respectively.[32] Such an algorithm produces quantitative values[37] that are equivalent to those proposed in the guidelines for pathologists. This provides the opportunity for using pathologist evaluations as the reference standard for such an algorithm.
We propose the following clinical workflow: (1) patient imaging finds an abnormality suspected for breast cancer. Physicians order a needle core biopsy to assess the tissue. (2) TILs will be scored during histopathologic evaluation and diagnosis. Specifically, pathologists will score the TILs in each H&E-stained breast cancer core biopsy with assistance from an algorithm. Or, depending on the algorithm intended use, the sTIL score could be created automatically, without pathologist input. (3) The sTIL density will then be reported in the patient's pathology report.
Algorithm Validation
Before it can be marketed and applied in the clinical workflow, any algorithm/SaMD should be well validated. Validation of algorithms for clinical use comes after the building, training, and tuning phases of algorithm development. There are two main categories of algorithm validation: analytical and clinical. For both categories, a reference standard is needed. For algorithms that evaluate WSIs of H & E slides, there are generally three kinds of truth: patient outcomes, evaluation of the tissue with other diagnostic methods, and evaluation of the slide by pathologists. This work focuses on truth as determined by pathologists.
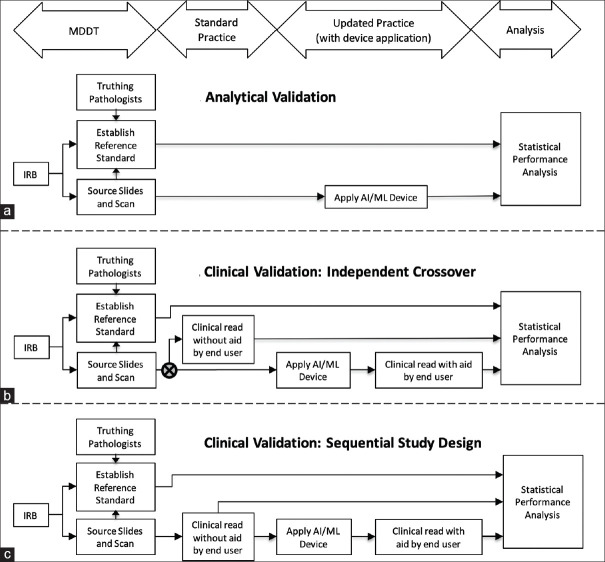
Analytical validation, or stand-alone performance assessment, focuses on the precision and accuracy of the algorithm, and compares algorithm outputs directly against the reference standard [Figure 1a]. In a clinical validation study, the algorithm end user, here a pathologist, evaluates cases without and with the algorithm outputs; Figure 1b shows an independent-crossover clinical validation study design. There is typically a washout period between the evaluations by the same pathologists evaluating the same cases without and with the algorithm outputs, where the order in which these viewing modes are executed is randomized and balanced across pathologists and batches of cases. Figure 1c shows a putative sequential clinical validation study design for an algorithm intended to be used as a decision support tool after the clinician makes their conventional evaluation. We have depicted two populations of pathologists in our proposed clinical validation studies: experts for establishing the reference standard and end users for evaluating performance without and with the algorithm outputs.
Figure 1.
(a) Study design for analytical validation of an algorithm (stand-alone performance assessment). Algorithm outputs are compared to the reference standard. (b) Independent crossover study design for clinical validation has two arms corresponding to pathologist evaluations without and with the algorithm. We compare the performance of these two evaluation modes. (c) Sequential study design for clinical validation has one arm corresponding to end user evaluations first without and then with the algorithm as an aid. A comparison is made between the performance of these two evaluation modes
The current best practice for algorithm validation is to source slides from multiple independent sites different from the algorithm development site to ensure algorithm generalizability, also known as external validation.[38,39,40,41] Developers should also be blinded to the validation data before a validation study, eliminating potential bias arising from developers' training to the test.[41,42,43,44] These practices generally assume that the algorithm is locked; the architecture, parameters, weights, and thresholds should not be changed before the algorithm is released into the field. Validation of algorithms that are not locked – algorithms that rely on “active learning” and “online” learning, or hard negative mining, where the training is done iteratively and continuously – is an area that is still evolving and not in the scope of this work.[32,45,46,47,48,49]
APPROACH: PILOT STUDY
Data – Pathology Tissue and Images
We, through a partnership with the Institute Jules Bordet, Brussels, Belgium, sourced 77 matched core biopsies and surgical resections. Of these cases, 65 were classified as invasive ductal carcinoma and 12 were invasive lobular carcinoma. There was no patient information provided with these slides, no metadata such as age, race, cancer stage, or subtype (morphologic or molecular). This study was approved by the Ethics Commission of the Institute Jules Bordet.
The slides are 2019 recuts of formalin-fixed, paraffin-embedded tissue blocks from a single institution. Slide preparation was performed at the same institution by a single laboratory technician. Specifically, one 5 um-thick section was mounted on a glass slide and stained with H & E. The slides were scanned on a Hamamatsu Nanozoomer 2.0-RS C10730 series at ×40 equivalent magnification (scale: 0.23 um per pixel).
For our pilot study, we included eight batches of eight cases each; a case refers to the slide image pair. The remaining 13 slides were not used for the pilot study. All 64 cases were biopsies of invasive ductal carcinomas; no resection specimens were used. Batches split data collection into manageable chunks for pathologists. Each batch was expected to take about 30 min to annotate. Batches also allowed us to make assignments for pathologists that help distribute evaluations across all cases and ROIs. We targeted five pathologist evaluations per ROI for the pilot study.
Data Collection = Region of Interest Annotation
Data collection, or ROI annotation, is broken into ROI selection and ROI evaluation in this work. ROI selection is a data curation step preceding ROI evaluation. The purpose of selecting ROIs ahead of ROI evaluation is to allow multiple pathologists to evaluate the same ROIs quickly. For our pilot study, ROI selection was performed by a collaborating pathologist using the digital platforms. Subsequent ROI evaluation was performed by recruited pathologists using digital and microscope platforms. The platforms, ROI selection and evaluation, and the pathologists that participated in the pilot study are described in more detail below.
Digital Platforms
For this work, we have two digital platforms for viewing and annotating WSIs: PathPresenter[50] and caMicroscope.[51] Screenshots of the user interfaces are shown in Figure 2a and b. Pathologists can log in from anywhere in the world, and annotate images using web-based viewers.
Figure 2.
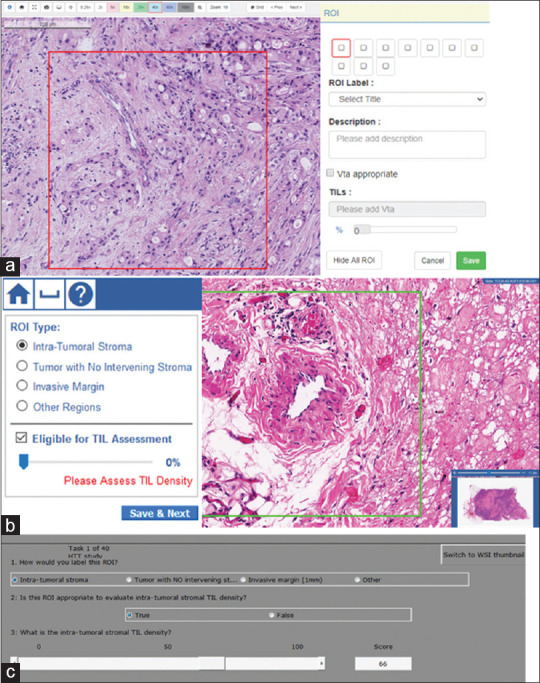
Screenshots from graphical user interfaces of three platforms used in data collection. All three collect a descriptive label of the regions of interest [Table 1], a binary evaluation of whether the regions of interest are appropriate for stromal tumor-infiltrating lymphocyte density estimation, and an estimate of stromal tumor-infiltrating lymphocyte density via slider bar or keyboard entry. (a) PathPresenter and (b) caMicroscope are digital platforms. (c) Evaluation environment for digital and analog pathology microscope platform. In data collection, the pathologist is at the microscope, while a study coordinator records evaluations through the graphical user interface
Both PathPresenter and caMicroscope leadership are collaborators in this project and supported development of controlled and standardized workflows to select ROIs and to evaluate ROIs. Both platforms can read and write annotations using the ImageScope XML format,[52] and we have used that format to share ROIs and create an identical study on both platforms. Both platforms also record the pixel width and height and the zoom setting of the WSI area being viewed. We have not yet imposed display requirements in the pilot study but that will be discussed for future phases of our project.
Using more than one platform, including the microscope platform described next, allows us to involve more partners that can provide different perspectives, build redundancy to mitigate against a collaborator leaving the team, and promote interoperability as we progress to future phases of the project. The validation dataset will be based on the microscope platform, and the digital platforms allow fast development and understanding of our study and also allow us to compare microscope mode to digital mode evaluations.
Microscope Platform
The microscope platform we use is a hardware and software system called Evaluation Environment for Digital and Analog Pathology (eeDAP).[53] The system uses a computer-controlled motorized stage and digital camera mounted to a microscope. eeDAP software registers the location of what is seen in the physical tissue through the microscope to the corresponding location in a WSI. Registration is accomplished through an interactive process that links the coordinates of the motorized stage to the coordinates of a WSI image. Registration enables the evaluation of the same ROIs in both the digital and microscope domains.
Similar to the digital platforms, the eeDAP software includes a utility to read and write ImageScope XML files, and a graphical user interface (GUI) implementing the ROI evaluation workflow [Figure 2c].[54] A research assistant supports the pathologist by entering data into the eeDAP GUI and monitoring registration accuracy. The square ROI is realized with a reticle in the eyepiece. As annotations are collected on the slide, they are scanner agnostic and may be mapped to any scanned version of the slide using the eeDAP registration feature.
Region of Interest Selection: Study Preparation
A board-certified collaborating pathologist marked 10 ROIs on each of the 64 cases using the digital platforms described above. The ROIs were 500 um × 500 um squares. The instructions were to target diverse morphology from various locations within the slide. More specific instructions were to target areas with and without tumor-associated stroma, areas where sTIL densities should and should not be evaluated. More details on selecting specific ROI types can be found in Table 1. An algorithm is expected to perform well in all these areas, so it is vital that the dataset include them.
Table 1.
Region of interest types
| Intra-tumoral stroma (aka tumor-associated stroma): Select ~3 ROIs |
|---|
| • Be sure to include regions with lymphocytes (TILs) |
| • If there are lymphocytic aggregates, make sure to capture both lymphocyte- depleted and lymphocyte-rich areas within the same ROI if possible |
| • Preferable to include some tumor in the same ROI — i.e. carcinoma cells as well |
| • If variable density within the slide, make sure to capture ROIs from different |
|
|
| Invasive margin (Tumor-stroma transition): Select ~2 ROIs |
|
|
| • If heterogeneous tumor morphology, sample from different tumor-stroma transitions for each |
|
|
| Tumor with no interveninq stroma: Select ~2 ROIs, if possible |
|
|
| • If heterogeneous tumor morphology, sample from different morphologies |
| • Be sure to sample from: vacuolated tumor cells, dying tumor cells, regions of |
| • Will be used to capture/assess intra-tumoral TILs and/or detect false positive Til- detections in purely cancerous regions. |
|
|
| Other regions: Select ~3-4 ROIs |
|
|
| • ~1 from “empty”/distant/uneventful stroma • ~1 from hyalinized stroma, if any • ~2 other regions: |
| ⁰ Necrosis transition (including comedo pattern) ⁰ Normal acini/ducts ⁰ Blood vessels ⁰ Others at patholoaist discretion |
Region of Interest Evaluation
In current project protocols, we crowdsource pathologists to participate in ROI evaluation, separate from the pathologist who completed ROI selection. These pathologists will first label the ROI by one of the four labels given in Table 1. Pathologists then mark if the ROI is appropriate for evaluating sTIL density. This question is designed to determine if the area has tumor-associated stroma or not. If there is no tumor-associated stroma, annotation is complete. If there is tumor-associated stroma, the pathologist needs to estimate the density of TILs appearing in the tumor-associated stroma. The platforms allow integers 0–100, with no binning or thresholds. The motivation is to allow for thresholds to be determined later as the role of TILs becomes more clear and patient management guidelines are developed.
Pathologist Participants in Region of Interest Evaluation
Pathologist participants were recruited at a meeting of the Alliance for Digital Pathology immediately preceding the February 2020 USCAP [United States and Canadian Academy of Pathology] annual meeting.[6] That meeting launched the in-person portion of pilot phase data collection. Board-certified anatomic pathologists and anatomic pathology residents were eligible to participate. To participate, they were asked to review the informed consent[55] and the training materials: the guidelines on sTIL evaluation[24] and a video tutorial and corresponding presentation about sTIL evaluation, the project, and using the platforms.[56] Reviewing the sTIL evaluation training was required before participating and took about 30 min. Pathologists were asked to label the ROI according to the types given in Table 1, a true-false decision about whether sTIL densities should or should not be evaluated, and if true, an estimate of the sTIL density.
In total, 19 pathologists made 1645 ROI evaluations during the February event and the 2 weeks following. The primary platform at the event was the eeDAP microscope system where 7 pathologists made 440 evaluations. Most of the evaluations made on the digital platforms were made by pathologists who could not attend in person. Data collection in digital mode took approximately 30–40 min per batch and twice that long in microscope mode. The increased time for microscope evaluation was due to the motorized stage movements.
Reference Standard (Truth) from Pathologists
The sTIL density measurements from pathologists are subject to bias and variance due to differences in pathologist expertise and training. In this work, we collected observations from multiple pathologists for each ROI, and then, we averaged over the pathologists. While the precision of these values can be estimated, averaging over pathologists ultimately ignores pathologist variability in the subsequent algorithm performance metric. As such, we also let the observations from each pathologist stand as noisy realizations of the truth. This approach is used in related research on inferring truth from the crowd for the purpose of training an algorithm.[57] For our work, however, the purpose is to properly account for pathologist variability when estimating the uncertainty of algorithm performance.
Performance Metric for Stromal Tumor-Infiltrating Lymphocyte Density Values
The primary endpoint of an algorithm that produces quantitative values needs to measure how close the values from the algorithm (Predictedi) are to the reference standard (Truthi). To evaluate “closeness,” one appropriate performance metric that we are focusing on is the root mean squared error (RMSE):
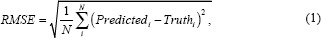
where N is the number of ROIs. Smaller values of RMSE indicate that the predicted values are closer to the truth, and thus better algorithmic performance. Equation 1 shows the RMSE estimated from a finite population (e.g., a finite sample of ROIs). As we consider a statistical analysis for our work – estimating uncertainty, confidence intervals, and hypothesis tests – we look to the infinite population quantity without the square root.[38,58,59,60]
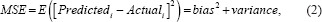
Here, we see that mean squared error measures accuracy and precision, similar to Lin's concordance correlation coefficient.[61]
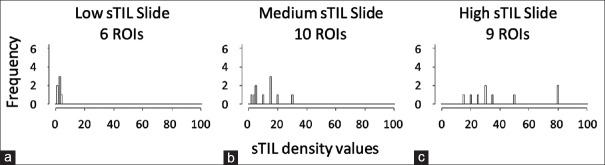
There are two main challenges to analyzing the differences between predictions and truth in our work. First, the sum in Equation 1 is really a sum over ROIs nested within cases. These values are not independent and identically distributed (iid), as is generally assumed for Equation 1. There should be a subscript for both case and ROI, and the statistical analysis needs to account for the correlation between values from ROIs within a case. In Figure 3, we show that sTIL densities are not iid across cases. The data are from one pathologist evaluating three cases that have different levels of sTIL infiltration. We see the sTIL densities are correlated within a case, and the variance is increasing with the mean. The distribution of sTIL densities is not the same for every case.
Figure 3.
The distribution of stromal tumor-infiltrating lymphocyte densities in three slides with different levels of infiltration: (a) Low, (b) Medium, (c) High. The stromal tumor-infiltrating lymphocyte densities were from one pathologist. As not all region of interest labels are appropriate for stromal tumor-infiltrating lymphocyte density evaluation, not every case will contain tumor-infiltrating lymphocyte evaluations for all 10 regions of interests
The second challenge in our work is to account for the variability from pathologist to pathologist. This variability is shown in Figure 4, which is a scatter plot showing the paired sTIL densities from two pathologists. Our strategy for addressing pathologist variability is to replace the single reference score in Equation 1 with pathologist-specific values.
Figure 4.
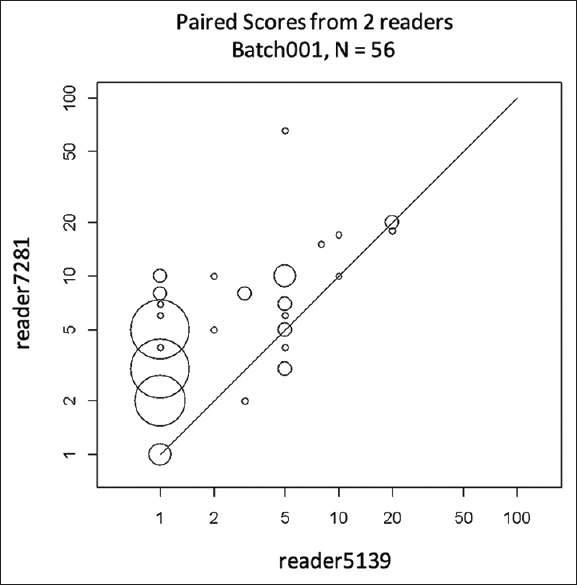
Scatter plot of stromal tumor-infiltrating lymphocyte densities from two pathologists on eight slides (one batch) that led to 56 paired observations. The plot is scaled by a log-base-10 transformation (with zero stromal tumor-infiltrating lymphocyte values changed to ones). The size of the circles is proportional to the number of observations at that point
To address these two challenges, we rewrite Equation 2 as

Where Xjkl denotes the sTIL density from pathologist j evaluating the ROI l in case k and Ykl denotes the sTIL density from the algorithm evaluating the ROI l in case k. Furthermore, the expected value averages over pathologists, cases, and ROIs. It is this quantity that we wish to estimate, and we are developing such methods to account for the correlation of ROIs within a case and pathologist variability. The estimate may take the form of a summation over readers, cases, and ROIs, or it may be the result of a model that needs to be solved by more sophisticated methods that do not permit an explicit closed-form expression. The methods build on previous work on so-called multi-reader multi-case methods[62,63,64,65] and methods to evaluate intra- and inter-reader agreement.[66]
DISCUSSION
The “high-throughput truthing” (HTT) moniker for this project reflects the data collection methods as well as the spirit of the effort. The project was inspired by perception studies that have been run at annual meetings of the Radiological Society of North America.[67] Society meetings provide an opportunity to reach a high volume of pathologists away from the workload of their day job. A similar opportunity is available at organizations with many pathologists. We have explored both of these kinds of data collection opportunities via an event at the American Society of Clinical Pathology Annual Meeting 2018,[68,69] and an event at the Memorial Sloan Kettering Cancer Center.[70,71]
In addition to live events where we can use the eeDAP microscope system, our workflows on web-based platforms (PathPresenter and caMicroscope) can crowdsource pathologists from anywhere in the world. We have found these events to be low-cost, efficient opportunities to recruit pathologists and collect data. We plan to continue the project by scaling our efforts to a pivotal phase and disseminating our final validation dataset.
Food and Drug Administration Medical Device Development Tool Program
A key aim of this project is to pursue the qualification of this dataset as a tool through the FDA MDDT program.[19] Pursuing qualification offers an opportunity to receive feedback from an FDA review team about building the dataset to be fit for a regulatory purpose. As we disseminate our work, we believe that this feedback will be valuable for the project and more generally, for other public health stakeholders interested in the collection of validation datasets (industry, academia, health providers, patient advocates, professional societies, and government). A qualified tool has the potential to streamline the submission and review of validation data and allows the FDA to compare algorithms on the same prequalified data. In this way, the project may benefit the agency and medical device manufacturers, as well as the larger scientific community.
The MDDT program was created by the FDA as a mechanism by which any public health stakeholder may develop and submit a tool to the agency for formal review. Tools are not medical devices. Rather, tools facilitate and increase predictability in medical device development and evaluation. Each tool is qualified for a specific context of use and may be used in a manufacturer's submission without needing to reconfirm its suitability and utility.[19] Qualified tools are expected to be made publicly available, which can include a licensing arrangement. In this way, qualified tools reduce burden to both the agency and the manufacturer and ultimately increase product quality and better patient outcomes. The proposed context of use for this work is given in Table 2.
Table 2.
Proposed context of use for a stromal tumor-infiltrating lymphocyte density annotated dataset
| The sTIL-density Annotated Dataset is a tool to be used to assess the accuracy of algorithms that quantify the density of stromal tumor infiltrating lymphocytes (sTILs). It is comprised of a dataset of slides, digital whole slide images and annotations in regions of interest (ROIs) compiled by pathologists using microscopes to evaluate glass slides of tissue samples from breast cancer needle core biopsies, where the tissue sections are stained with Hematoxylin and eosin (H&E). |
The exact platform and mechanisms for sharing the dataset have yet to be determined. However, the dataset will be shared broadly at no cost with any entity, subject to applicable terms required by either the FDA or the MDDT program. Possible terms would protect against data being used to “train to the test” using strategies such as data access via containers or data governance by written agreements. We can look to public challenges[72,73,74,75] to inform our data sharing plans and educational dissemination opportunities.
An MDDT dataset has the potential to significantly reduce the burden of manufacturers, especially small companies. Validation in the commercial space tends to be siloed, with each developer using distinct, licensed, and proprietary data. Our proposed MDDT may allow manufacturers and the FDA to avoid the time- and resource-consuming back-and-forth discussions to formulate a study design and protocol. Manufacturers may also be able to skip burdensome steps such as obtaining Investigational Review Board approvals, slide sourcing, reader recruitment, and collecting the data. Instead of planning statistical analyses from scratch, manufacturers may use the analyses developed from this project as an example to guide their work. These bypassed steps are represented in the column headings of Figure 1.
Data Representativeness/Generalizability
A random set of breast cancer biopsies are naturally expected to include the different immunophenotypic subtypes of TILs (CD4+, CD8+ T-cells, and natural killer cells) and a variety of shapes, locations, colors, and clustering of TILs.[76,77,78,79] Our current strategy of selecting ROIs gathers areas for sTIL evaluation with and without tumor-associated stroma, areas where sTIL densities should and should not be evaluated [Table 1]. Despite efforts to assemble a balanced and stratified sample of ROI types, our pilot study data yielded an abundant number of cases with nominal sTIL infiltration. While this may be the true clinical distribution, for our MDDT, we want to balance and stratify the sTIL density values across the expected range. For this, we intend to realize some data curation before ROI evaluation in our future pivotal study.
The MDDT dataset should also adequately represent the variability arising from preanalytic differences (slide preparation) and the intended population (clinical subgroups). As such, for our pivotal study, we intend to source slides from at least three sites and stratify the cases across important clinical subgroups. If possible, we will also create some cases that systematically explore the H & E staining protocol (incubation time, washing time, and stain strength).
There are several clinical subgroups that are appropriate to sample, such as patient age, breast cancer subtypes and stages,[28,80,81,82] and treatment at various time intervals. Sampling from all possible subgroups is challenging if not impossible. While our inclusion and exclusion criteria limit the use of our MDDT to a selective population, we do not expect to sample all the subgroups that might be required in an algorithm submission, and we do not expect to have the same metadata for all cases. It is important to note that while TILs are known to have the most prognostic value in certain molecular (genomic) subtypes (e.g., TNBC and HER2+), a TIL algorithm is most likely to be confounded by histologic subtype and characteristics. While there is some correspondence between genomic and histologic classifications of breast tumors, the histological presentation (morphology) of, say, a ductal carcinoma does not necessarily correlate well with its genomic composition. Any data that is not part of the MDDT but is required for a regulatory submission of an algorithm will ultimately be the responsibility of the algorithm manufacturer. We do not intend to sample treatment methods or longitudinal data.
Pathologists and Pathologist Variability
In this work, our initial data shows notable variability in independent sTIL density estimates from multiple pathologists on each ROI [Figure 4], which is consistent with previous work in this area.[32] These findings further reinforce the need to collect data from multiple pathologists and the need to better understand this variability. We intend to explore the difference between averaging over pathologists and keeping them distinct when evaluating algorithm performance. In either case, we believe that a statistical analysis method should account for reader variability in addition to case variability. A final statistical analysis plan for our pivotal study, including sizing the number of pathologists and cases, will be developed based on the pilot data, simulation studies, and feedback from the FDA's MDDT review team.
As we are crowdsourcing pathologists, we have received questions regarding the expertise of the participating pathologists. Initially, we accepted any board-certified pathologist or anatomic pathology resident, but the reader variability observed in the pilot data has caused us to reconsider. As such, this is a limitation in the reliability of the pilot study data. Improving the expertise of annotating pathologists will reduce pathologist variability and allow us to reduce the number of pathologists. Therefore, for our pivotal study, we are expanding our current training materials to include testing with immediate feedback, providing the reference standard for each ROI. We are also creating a proficiency test. These training materials may be built from the pilot study dataset. A robust training program could additionally serve the community beyond our specific project need.
As relates to the RMSE performance metric, which summarizes the bias as well as the variance of an algorithm, it is not clear whether the bias comes from the algorithm or the pathologist. Amgad et al.[83] found their algorithm to be biased low compared to the pathologists. They also found that the Spearman rank-based correlation was stronger for the algorithm-to-pathologist-consensus comparison compared to the pathologist-to-pathologist comparison (R = 0.73 vs. R = 0.66). The authors believe these results are related to pathologist bias and variability, and not the algorithm. While this may be true, it is difficult to know as only two pathologists provided sTIL density values. Furthermore, the comparison does not account for pathologist variability in either correlation result and is not an apple-to-apple comparison due to the consensus process. Still, we expect that our expanded pathologist training will improve pathologist correlation, and we will compare the correlation of our pivotal study data to that of Amgad et al. and to our pilot study results. In preparation for this comparison, we will explore Spearman's rank correlation and Kendall's tau on the pilot study data. These metrics treat pathologist sTIL density estimates as ordinal data rather than quantitative and calibrated data.[66,84,85,86]
Relaunch and Future Pivotal Study
While the live event portion of pilot phase data collection was a burdensome process, we totaled 1645 evaluations in 10 h. The live event was set up with four evaluation stations: 2 digital platforms and 2 microscope platforms. We created training materials and hosted an online training seminar before the event. We assembled recruitment materials and sent invitations to pathologists. We trained study administrators to operate eeDAP and assist pathologists with data collection at the microscope. All equipment was shipped and assembled on site. Data aggregation was completed via APIs. Not surprisingly, the data are stored quite differently on the two digital platforms, so we created scripts to clean and harmonize the raw data into common data frames. We began building a software package to analyze the clean data. In sum, the process took a lot of time and effort, but offered experiences to inform the next phase of our project.
To help pathologists improve their sTIL density estimates and collect more detailed data, we thought about what an algorithm generally would do: identify and segment the tumor, tumor-associated stroma, and sTILs. We thought that it would be worthwhile to parallel these steps. We were already asking pathologists to label ROIs by tumor, margin, and the presence of tumor-associated stroma. We decided that in our pivotal study, we would ask the pathologist to estimate the percent of the ROI area that contains tumor-associated stroma.
Data collection on the microscope system was put on hold because of the COVID-19 pandemic, but we relaunched data collection on the digital platforms in September 2020 to fill out observations across all batches of the pilot study. We invite board-certified pathologists to spend approximately 30 min on training and 30 min per batch on data collection.[87] With newly established agreements for sharing materials, we are in the process of securing more slides and images to sample the patient subgroups mentioned from multiple sites in our future pivotal study, bolstering our single-site pilot data. We welcome parties that are able and willing to share such materials to contact us through the corresponding author. Similarly, we are looking for opportunities to set up HTT events or find collaborating sites interested in hosting data collection events on their own. There are opportunities to set up their own eeDAP microscope system or borrow an existing system from us. We are willing to supervise and assist remotely.
CONCLUSION
On the volunteer efforts of many and a nominal budget, we have created a team and a protocol, administrative materials, and infrastructure for our HTT project. We have sourced breast-cancer slides and crowdsourced pathologists in a pilot study, and we are actively planning a pivotal study with more data and better pathologist training. Our goal is to create a sTIL-density annotated dataset that is fit for a regulatory purpose. We hope that this project can be a roadmap and inspiration for other stakeholders (industry, academia, health providers, patient advocates, professional societies, and government) to work together in the precompetitive space to create similar high-value, fit-for-purpose, broadly accessible datasets to support the field in bringing algorithms to market and to monitor algorithms on the market.
FINANCIAL SUPPORT AND SPONSORSHIP
R. S. is supported by a grant from the Breast Cancer Research Foundation (grant No. 17 194), and J. S. is supported by grants from the NIH (UH3CA225021 and U24CA180924). A. M. is supported via grants fromthe NCI (1U24CA199374-01, R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01), National Heart, Lung and Blood Institute (1R01HL15127701A1, R01HL15807101A1), National Institute of Biomedical Imaging and Bioengineering (1R43EB028736-01), National Center for Research Resources (1 C06 RR12463-01), VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Ohio Third Frontier Technology Validation Fund, the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. Sponsored research agreements from Bristol Myers-Squibb, Boehringer-Ingelheim, and Astrazeneca.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
CONFLICTS OF INTEREST
A. M. is an equity holder in Elucid Bioimaging and in Inspirata Inc. In addition he has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol Meyers-Squibb and Merck. Currently he serves on the advisory board of Aiforia Inc and currently consults for Caris, Roche and Aiforia. He also has sponsored research agreements with Philips, AstraZeneca, Boehringer-Ingelheim and Bristol Meyers-Squibb. His technology has been licensed to Elucid Bioimaging. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc.
ACKNOWLEDGMENTS
The HTT team acknowledges the work of collaborating pathology networks such as the International Immuno Oncology Biomarker Working Group and the Alliance for Digital Pathology in their participant pathologist recruitment (www.tilsinbreastcancer.org and https://digitalpathologyalliance. org). Ioanna Laios and Ligia Craciun contributed technical support and expertise in the preparation of glass slides. Finally, the team thanks their developers: Krushnavadan Acharya (PathPresenter), Nan Li (caMicroscope), and Qi Gong (eeDAP).
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2021/12/1/4/307702
REFERENCES
- 1.Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. 3rd ed. Upper Saddle River, New Jersey: Prentice Hall; 2009. [Google Scholar]
- 2.Fuchs TJ, Buhmann JM. Computational pathology: Challenges and promises for tissue analysis. Comput Med Imaging Graph. 2011;35:515–30. doi: 10.1016/j.compmedimag.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Chang HY, Jung CK, Woo JI, Lee S, Cho J, Kim SW, et al. Artificial Intelligence in Pathology. J Pathol Transl Med. 2019;53:1–2. doi: 10.4132/jptm.2018.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serag A, Ion-Margineanu A, Qureshi H, McMillan R, Saint Martin MJ, Diamond J, et al. Translational AI and deep learning in diagnostic pathology. Front Med (Lausanne) 2019;6:185. doi: 10.3389/fmed.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology – New tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marble HD, Huang R, Dudgeon SN, Lowe A, Herrmann MD, Blakely S, et al. A regulatory science initiative to harmonize and standardize digital pathology and machine learning processes to speed up clinical innovation to patients. J Pathol Inform. 2020;11:22. doi: 10.4103/jpi.jpi_27_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niazi MK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–61. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.FDA CDRH, “De Novo Request Evaluation of Automatic Class III Designation for Philips IntelliSite Pathology Solution (PIPS): Decision Summary;”. 2017. [Last accessed on 2018 May 17]]. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN160056.pdf .
- 9.FDA CDRH, “510(k) Summary Aperio AT2 DX System;” 2019. [Last accessed on 2020 Apr 21]]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf19/K190332.pdf .
- 10.FDA CDRH, “Summary of Safety and Effectiveness Data: GE FFDM (p990066);” 2001. [Last accessed on 2021 Sept 02]]. Available from: http://www.accessdata.fda.gov/cdrh_docs/pdf/P990066b.pdf .
- 11.FDA CDRH, “Summary of Safety and Effectiveness Data: R2 Technology, Inc. ImageChecker M1000 (p990066);” 1998. [Last accessed on 2020 Aug 06]]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf/p970058.pdf .
- 12.FDA CDRH, “Guidance for Industry and FDA Staff – Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data – Premarket Notification [510(k)] Submissions.” FDA. 2012. [Last accessed on 2020 Apr 21]]. Available from: https://www.fda.gov/media/77635/download .
- 13.FDA CDRH, “Guidance for Industry and FDA Staff – Clinical Performance Assessment: Considerations for Computer-Assisted Detection Devices Applied to Radiology Images and Radiology Device Data in Premarket Notification [510(k)] Submissions.” FDA. 2020. [Last accessed on 2020 Aug 05]]. Available from: https://www.fda.gov/media/77642/download .
- 14.Gallas BD, Chan HP, D'Orsi CJ, Dodd LE, Giger ML, Gur D, et al. Evaluating imaging and computer-aided detection and diagnosis devices at the FDA. Acad Radiol. 2012;19:463–77. doi: 10.1016/j.acra.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.FDA/CDRH, “Software as a Medical Device (SAMD): Clinical Evaluation,” FDA, Silver Spring, MD. 2017. [Last accessed on 2018 Jan 05]]. Available from: https://www.fda.gov/media/100714/download .
- 16.FDA CDRH, “Evaluation of Automatic Class III Designation for QuantX: Decision Summary;”. 2017. [Last accessed on 2020 Sep 16]]. Available from: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/denovo.cfm?id=DEN170022 .
- 17.FDA CDRH, “Evaluation of Automatic Class III Designation for ContaCT: Decision Summary;”. 2017. [Last accessed on 2020 Sep 16]]. Available from: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/denovo.cfm?id=DEN170073 .
- 18.FDA/CDRH, “Qualification of Medical Device Development Tools.” FDA. 2017. Auguest 10, [Last accessed on 2020 Aug 03]]. Available from: https://www.fda.gov/media/87134/download .
- 19.FDA/CDRH, “Qualification of Medical Device Development Tools.” FDA. 2017. Auguest 10, [Last accessed on 2020 Aug 03]]. Available from: https://www.fda.gov/media/87134/download .
- 20.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–52. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loi S, Drubay D, Adams S, Pruneri G, Francis PA, Lacroix-Triki M, et al. Tumor-infiltrating lymphocytes and prognosis: A pooled individual patient analysis of early-stage triple-negative breast cancers. J Clin Oncol. 2019;37:559–69. doi: 10.1200/JCO.18.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 23.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–72. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 24.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–71. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, et al. Early breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up†. Ann Oncol. 2019;30:1194–220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 26.Morigi C. Highlights of the 16th St Gallen International Breast Cancer Conference, Vienna, Austria, 20-23 March 2019: Personalised treatments for patients with early breast cancer. Ecancermedicalscience. 2019;13:924. doi: 10.3332/ecancer.2019.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balic M, Thomssen C, Würstlein R, Gnant M, Harbeck N. St.Gallen/Vienna 2019: A brief summary of the consensus discussion on the optimal primary breast cancer treatment. Breast Care (Basel) 2019;14:103–10. doi: 10.1159/000499931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Agency for Research on Cancer. Breast Tumours. In: WHO Classification of Tumours Series. 5th ed., Vol. 2. Lyon (France): WHO Classification of Tumours Editorial Board. 2019. [Last accessed on 2020 Jun 12]]. Available from: https://tumourclassification.iarc.who.int/chapters/32 .
- 29.Luen SJ, Salgado R, Dieci MV, Vingiani A, Curigliano G, Gould RE, et al. Prognostic implications of residual disease tumor-infiltrating lymphocytes and residual cancer burden in triple-negative breast cancer patients after neoadjuvant chemotherapy. Ann Oncol. 2019;30:236–42. doi: 10.1093/annonc/mdy547. [DOI] [PubMed] [Google Scholar]
- 30.Luen SJ, Griguolo G, Nuciforo P, Campbell C, Fasani R, Cortes J. On-treatment changes in tumor-infiltrating lymphocytes (TIL) during neoadjuvant HER2 therapy (NAT) and clinical outcome. J Clin Oncol. 2019;37(Suppl 15):574. [Google Scholar]
- 31.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the international immunooncology biomarkers working group: Part 1: Assessing the host immune response, TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv Anat Pathol. 2017;24:235–51. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amgad M, Stovgaard ES, Balslev E, Thagaard J, Chen W, Dudgeon S, et al. Report on computational assessment of tumor infiltrating lymphocytes from the international immuno-oncology biomarker working group. NPJ Breast Cancer. 2020;6:16. doi: 10.1038/s41523-020-0154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klauschen F, Müller KR, Binder A, Bockmayr M, Hägele M, Seegerer P, et al. Scoring of tumor-infiltrating lymphocytes: From visual estimation to machine learning. Semin Cancer Biol. 2018;52:151–7. doi: 10.1016/j.semcancer.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K, et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2019;25:1526–34. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–93. doi: 10.1016/j.celrep.2018.03.086. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AbdulJabbar K, Raza SE, Rosenthal R, Jamal-Hanjani M, Veeriah S, Akarca A, et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med. 2020;26:1054–62. doi: 10.1038/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.F. C. FDA/CDRH, “Technical Performance Assessment of Quantitative Imaging in Device Premarket Submissions: Draft Guidance for Industry and Food and Drug Administration Staff.” FDA. 2019. [Last accessed on 2020 Apr 28]]. Available from: https://www.fda.gov/regulatory-information/search-fda-guidancedocuments/technical-performance-assessment-quantitative-imagingdevice-premarket-submissions .
- 38.Altman DG, Royston P. What do we mean by validating a prognostic model? Stat Med. 2000;19:453–73. doi: 10.1002/(sici)1097-0258(20000229)19:4<453::aid-sim350>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann Intern Med. 2015;162:W1–73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 40.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–9. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 42.De A, Meier K, Tang R, Li M, Gwise T, Gomatam S, et al. Evaluation of heart failure biomarker tests: A survey of statistical considerations. J Cardiovasc Transl Res. 2013;6:449–57. doi: 10.1007/s12265-013-9470-3. [DOI] [PubMed] [Google Scholar]
- 43.Pennello GA. Analytical and clinical evaluation of biomarkers assays: When are biomarkers ready for prime time? Clin Trials. 2013;10:666–76. doi: 10.1177/1740774513497541. [DOI] [PubMed] [Google Scholar]
- 44.Williams BJ, Treanor D. Practical guide to training and validation for primary diagnosis with digital pathology. J Clin Pathol. 2020;73:418–22. doi: 10.1136/jclinpath-2019-206319. [DOI] [PubMed] [Google Scholar]
- 45.Doyle S, Monaco J, Feldman M, Tomaszewski J, Madabhushi A. An active learning based classification strategy for the minority class problem: Application to histopathology annotation. BMC Bioinformatics. 2011;12:424. doi: 10.1186/1471-2105-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.FDA, “Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD) – Discussion Paper and Request for Feedback.” US Food and Drug Administration. 2019. Apr 02, [Last accessed on 2020 Mar 15]]. Available from: https://www.fda.gov/media/122535/download .
- 47.Feng J, Emerson S, Simon N. Approval policies for modifications to machine learning-based software as a medical device: A study of biocreep. Biometrics. 2020;77:31–44. doi: 10.1111/biom.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pennello G, Sahiner B, Gossmann A, Petrick N. Discussion on 'Approval policies for modifications to machine learning-based software as a medical device: A study of bio-creep' by Jean Feng, Scott Emerson, and Noah Simon. Biometrics. 2021;77:45–48. doi: 10.1111/biom.13381. [DOI] [PubMed] [Google Scholar]
- 49.McShane LM, Cavenagh MM, Lively TG, Eberhard DA, Bigbee WL, Williams PM, et al. Criteria for the use of omics-based predictors in clinical trials. Nature. 2013;502:317–20. doi: 10.1038/nature12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.“PathPresenter”. [Last accessed on 2020 Sep 16]]. Available from: https://pathpresenter.net .
- 51.Saltz J, Sharma A, Iyer G, Bremer E, Wang F, Jasniewski A, et al. A containerized software system for generation, management, and exploration of features from whole slide tissue images. Cancer Res. 2017;77:e79–82. doi: 10.1158/0008-5472.CAN-17-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.“Aperio ImageScope.”. [Last accessed on 2020 Sep 16]]. Available from: https://www.leicabiosystems.com/digital-pathology/manage/aperio-imagescope .
- 53.Gallas BD, Gavrielides MA, Conway CM, Ivansky A, Keay TC, Cheng WC, et al. Evaluation Environment for Digital and Analog Pathology (eeDAP): A platform for validation studies. J Med Imaging. 2014;1:037501. doi: 10.1117/1.JMI.1.3.037501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallas BD. eeDAP: Evaluation environment for digital and analog histopathology. [Last accessed on 2020 Aug 13]]. Available from: https://github.com?DIDSR/eeDAP/releases .
- 55.“High-Throughput Truthing Project Informed Consent Form.”. [Last acessed on 2020 Sep 17]]. Available from: https://ncihub.org/groups/eedapstudies/wiki/HTTinformedConsent .
- 56. [Last accessed on 2020 Sep 17]];“High-Throughput Truthing Training Materials.”. Available from: https://ncihub.org/groups/eedapstudies/wiki/HTTdataCollectionTraining . [Google Scholar]
- 57.Zheng Y, Li G, Li Y, Shan C, Cheng R. Truth inference in crowdsourcing: Is the problem solved? Proc VLDB Endow. 2017;10:541–52. [Google Scholar]
- 58.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- 59.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–60. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 60.Casella G, Berger RL. Statistical Inference. 2nd ed. Pacific Grove, CA: Duxbury; 2002. [Google Scholar]
- 61.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–68. [PubMed] [Google Scholar]
- 62.Gallas BD, Bandos A, Samuelson F, Wagner RF. A framework for random-effects ROC analysis: Biases with the bootstrap and other variance estimators. Commun Stat Theory. 2009;38:2586–603. [Google Scholar]
- 63.Gallas BD, Chen W, Cole E, Ochs R, Petrick N, Pisano ED, et al. Impact of prevalence and case distribution in lab-based diagnostic imaging studies. J Med Imaging (Bellingham) 2019;6:015501. doi: 10.1117/1.JMI.6.1.015501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen W, Gong Q, Gallas BD. Paired split-plot designs of multireader multicase studies. J Med Imaging (Bellingham) 2018;5:031410. doi: 10.1117/1.JMI.5.3.031410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabata K, Uraoka N, Benhamida J, Hanna MG, Sirintrapun SJ, Gallas BD, et al. Validation of mitotic cell quantification via microscopy and multiple whole-slide scanners. Diagn Pathol. 2019;14:65. doi: 10.1186/s13000-019-0839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawid AP, Skene AM. Maximum likelihood estimation of observer error-rates using the EM algorithm. J R Stat Soc Ser C Appl Stat. 1979;28:20–8. [Google Scholar]
- 67.Toomey RJ, McEntee MF, Rainford LA. The pop-up research centre – Challenges and opportunities. Radiography (Lond) 2019;25(Suppl 1):S19–24. doi: 10.1016/j.radi.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Gallas BD, Amgad M, Chen W, Cooper LAD, Dudgeon S, Gilmore H. A collaborative project to produce regulatory-grade pathologist annotations to validate viewers and algorithms, In: Abstracts. J Pathol Inform. 2019;10:28. [Google Scholar]
- 69.Gallas BD, Amgad M, Chen W, Cooper LAD, Dudgeon S, Gilmore H. A collaborative project to produce regulatory-grade pathologist annotations to validate viewers and algorithms. In: Abstracts, Supplementary Materials. [Last accessed 2020 Aug 04]]. Available from: https://ncihub.org/groups/eedapstudies/wiki/HighthroughputTruthingYear2 . [Google Scholar]
- 70.A reader study on a 14-head microscope In: Pathology Informatics Summit 2018. J Pathol Inform. 2018;9:50. [Google Scholar]
- 71.Gallas BD. Reader Study on a 14-Head Microscope, In: Pathology Informatics Summit 2018, Supplementary Materials. 2018. Jan 01, [Last accessed on 2020 Aug 04]]. Available from: https://nciphub.org/groups/eedapstudies/wiki/Presentation: AReaderStudyona14headMicroscope .
- 72.van Ginneken B, Kerkstra S, Meakin J. “Grand Challenge.”. [Last accessed on 2020 Aug 11]]. Available from: https://grand-challenge.org/
- 73.Cha K. “Overview – Grand Challenge.”. [Last accessed on 2020 Aug 11]]. Available from: https://breastpathq.grand-challenge.org/
- 74.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Litjens G, Bandi P, Ehteshami Bejnordi B, Geessink O, Balkenhol M, Bult P, et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: The CAMELYON dataset. Gigascience. 2018;7:giy065. doi: 10.1093/gigascience/giy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zgura A, Galesa L, Bratila E, Anghel R. Relationship between tumor infiltrating lymphocytes and progression in breast cancer. Maedica (Bucur) 2018;13:317–20. doi: 10.26574/maedica.2018.13.4.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romagnoli G, Wiedermann M, Hübner F, Wenners A, Mathiak M, Röcken C, et al. Morphological evaluation of Tumor-Infiltrating Lymphocytes (TILs) to investigate invasive breast cancer immunogenicity, reveal lymphocytic networks and help relapse prediction: A retrospective study. Int J Mol Sci. 2017;18:1936. doi: 10.3390/ijms18091936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1:155–67. doi: 10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Çelebi F, Agacayak F, Ozturk A, Ilgun S, Ucuncu M, Iyigun ZE, et al. Usefulness of imaging findings in predicting tumor-infiltrating lymphocytes in patients with breast cancer. Eur Radiol. 2020;30:2049–57. doi: 10.1007/s00330-019-06516-x. [DOI] [PubMed] [Google Scholar]
- 80.Tan PH, Ellis I, Allison K, Brogi E, Fox SB, Lakhani S, et al. The 2019 World Health Organization classification of tumours of the breast. Histopathology. 2020;77:181–5. doi: 10.1111/his.14091. [DOI] [PubMed] [Google Scholar]
- 81.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.American Joint Committee on Cancer. Breast. In: AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer; 2017. [Google Scholar]
- 83.Amgad M, Elfandy H, Hussein H, Atteya LA, Elsebaie MA, Abo Elnasr LS, et al. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics. 2019;35:3461–7. doi: 10.1093/bioinformatics/btz083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stevens SS. On the theory of scales of measurement. Science. 1946;103:677. doi: 10.1126/science.103.2684.677. [DOI] [PubMed] [Google Scholar]
- 85.Kim JO. Predictive measures of ordinal association. Am J Sociol. 1971;76:891–907. [Google Scholar]
- 86.Kendall MG. A new measure of rank correlation. Biometrika. 1938;30:81–93. [Google Scholar]
- 87.“High-Throughput Truthing Project Data Collection Information.”. [Last accessed on 2020 Sep 17]]. Available from: https://ncihub.org/groups/eedapstudies/wiki/HTTStartDataCollection .