Abstract
Digital pathology is gaining prominence among the researchers with developments in advanced imaging modalities and new technologies. Generative adversarial networks (GANs) are a recent development in the field of artificial intelligence and since their inception, have boosted considerable interest in digital pathology. GANs and their extensions have opened several ways to tackle many challenging histopathological image processing problems such as color normalization, virtual staining, ink removal, image enhancement, automatic feature extraction, segmentation of nuclei, domain adaptation and data augmentation. This paper reviews recent advances in histopathological image processing using GANs with special emphasis on the future perspectives related to the use of such a technique. The papers included in this review were retrieved by conducting a keyword search on Google Scholar and manually selecting the papers on the subject of H&E stained digital pathology images for histopathological image processing. In the first part, we describe recent literature that use GANs in various image preprocessing tasks such as stain normalization, virtual staining, image enhancement, ink removal, and data augmentation. In the second part, we describe literature that use GANs for image analysis, such as nuclei detection, segmentation, and feature extraction. This review illustrates the role of GANs in digital pathology with the objective to trigger new research on the application of generative models in future research in digital pathology informatics.
Keywords: Artificial intelligence, deep learning, digital pathology, generative adversarial networks, histopathology, image processing, whole-slide imaging
INTRODUCTION
Digital pathology is rapidly gaining prominence with the development of modern imaging modalities and advancements in novel technologies. During histopathology assessment, which is the current gold standard for cancer diagnosis, surgical samples obtained from tumor biopsy and/or resection are typically prepared and stained using hematoxylin and eosin (H&E) stains and/or other techniques.[1] Hematoxylin stains cell nuclei blue/purple in color, whereas eosin gives a pink color to the cytoplasm and the connective tissues. Manual examination of the H&E stained slides is performed by trained pathologists for tumor diagnosis and grading, with the aim to guide further treatment and to give prognostic information. The inception and evolution of whole-slide imaging in the late 1990s has enabled complete histopathological slides to be recorded and digitally stored at high resolution using microscope-based slide scanners or whole-slide scanners.[2] A whole-slide image (WSI), also referred to as virtual slide, is the digital representation of a histopathology slide which provides visual information about the H&E and/or other staining techniques, in a range of multiple scales and different focal planes.
The increasing presence of digital pathology in contemporary medicine has motivated many researchers to explore the opportunities in digitally analyzing histopathological images for automated pathological diagnosis and decision-making.[3,4] Several novel methods in image processing, including computational fractal-based analysis and artificial intelligence (AI), have been tested against confirmed cases in their ability to extract reliable information from images to extend the limits of human interpretation beyond viewing a microscopic slide.[5,6] Several studies have applied fractal dimension analysis for the diagnosis, staging, and prognosis of different cancer types.[7,8] The application of AI methodologies in digital pathology has enabled pathologists to maximize the knowledge extracted from WSIs and has facilitated the automation of tasks that are traditionally manual and operator dependent. Niazi et al.[9] reviewed the role of AI in digital pathology by highlighting its applications, including education (training next generation of pathologists) and quality assurance (assessing interobserver and intraobserver variance). The synergetic integration of pathology with AI could lead to improvements in education and quality in the health care and medical field.
Deep learning is emerging as a powerful tool in machine vision and natural language processing and is opening new venues in AI with transformative impact on medicine, including neurosciences and digital pathology.[10,11] While traditional methods usually require “handcrafted” domain-specific features, deep learning methods can directly and automatically detect features from the data without the need for labour intensive manual feature extraction. Deep learning methods are used in digital pathology in many tasks including image preprocessing,[12] segmentation and detection of histologic primitives,[13,14] and grading and prognosis.[15,16] In 2014, Goodfellow et al.[17] introduced the generative adversarial networks (GANs) as a deep learning concept capable of learning representative distributions of data and generating new, synthetic data that can be used as real data substitutes or complements.
GANs have achieved state-of-the-art performance in many image generation tasks,[18] including text-to-image synthesis,[19] super-resolution images,[20] unpaired image-to-image translation,[21] and various applications in medical imaging such as image reconstruction,[22,23] image synthesis,[24] segmentation,[25] classification,[26] and other tasks.[27,28] Conditional GANs (cGANs), a variant of GANs, have been able to virtually stainimages of unstained specimens, to make them resemble H&E stained pathology slides.[29] This research using the GAN model has shown the possibility that digital virtual staining technology could lead to replacement of the laborious and expensive process of actual histological staining.
The aim of this paper is to provide an overview of the role of GANs in digital pathology, thereby triggering new research on the application of generative models in future research in digital pathology informatics.
METHODS
Search strategy
The papers used in this review were retrieved by conducting a search on Google Scholar. Meanwhile, as GANs are relatively new, a substantial number of articles are still in the publication process toward different journals and conferences, so we have also covered pre-prints published in arXiv. The search was limited to the English language and a year range of 2014–March 2021 using the keywords “histopathol*” and “generative adversarial network*”, “histopathol*” and “gan”, “histology” and “generative adversarial network*,” “histology” and “gan.”
Inclusion criteria
We included papers that made use ofH&E stained digital pathology images for histopathological image processing. All papers that (i) applied GANs to other imaging modalities such as ultrasound imaging, magnetic resonance imaging (MRI), computed tomography, and positron emission tomography or (ii) used digital slides which were stained with staining techniques other than hematoxylin and eosin were eliminated. Out of the 435 results listed from the search query, 48 articles were chosen for the study after applying a filter of title and abstract on the retrieved results to eliminate irrelevant papers. Figure 1 shows how the selected publications are distributed over different areas of image processing in digital histopathology.
Figure 1.
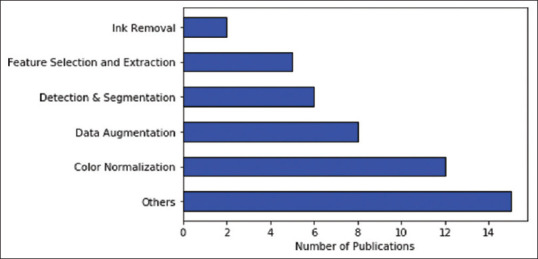
The distribution of papers selected for this review between the different areas in histopathological image processing that use generative adversarial networks, dated between 2014 and March 2021. Others here refer to tasks such as domain adaptation, image synthesis, image enhancement, and virtual staining
OVERVIEW OF GENERATIVE ADVERSARIAL NETWORKS
In this section, we provide a general description of GANs and a few extensions such as cGANs and cycle-consistent GANs (cycleGANs), which are used as the basis for further model development in histopathological image processing.
Generative adversarial network
GANs are a special type of neural network model consisting of two networks: a generator network and a discriminator network which are trained simultaneously when competing with each other.[17] The generative network function G (x) generates new data and the discriminative network function D (y) learns to distinguish between the real and the generated data. Both networks always compete to optimize themselves; the discriminator D tries to maximize its objective function and the generator G tries to minimize its objective function. Equation 1 mathematically expresses the objectives of D and G as:
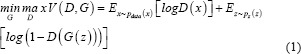
where D (x) is the discriminator, G (z) is the generator, x is the real data, and pz (z) is the input noise. Both networks train each other through multiple cycles of generation and discrimination when simultaneously trying to outwit each other. This is treated as a two-player minimax game which converges at a state known as the Nash equilibrium.[17,30] Figure 2a shows a graphical representation of the GAN model.
Figure 2.
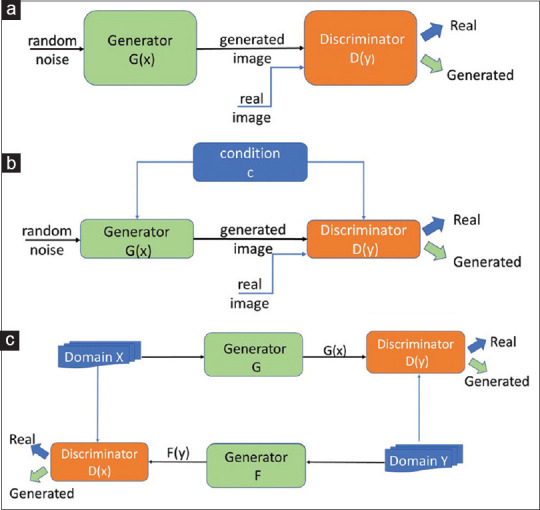
Graphical representation of (a) the generative adversarial network model, (b) the conditional generative adversarial network model, and (c) the cycle-consistent generative adversarial network model. For more details please see references 17, 21 and 31.
GANs have gained attention in many areas due to their effectiveness in generating new synthetic images which can complement or even replace real data.[32,33,34] GANs can be potentially utilized in several steps of digital pathology, including synthesizing images for augmenting the training data and processing of histopathology images. State-of-the-art GANs, such as a medical imaging GAN developed for retinal images, have recently shown impressive results in generating synthetic images and their segmented masks which can be used for the application of supervised analysis of medical images.[32] Moreover, GAN models have been proposed as useful tools to learn complex representations of cancer tissues in histopathology images.[28,35]
Conditional generative adversarial networks
cGAN are an extension of the GAN architecture, in which both the generator and discriminator are conditioned on auxiliary information such as a label associated with an image or a more detailed tag.[31] The condition information c is fed into both the generator and the discriminator as an additional input to direct the data generation process. The generator learns to generate a sample with this specific condition or characteristics rather than a generic sample from unknown noise distribution. Equation 2 mathematically represents the loss function of cGAN as:
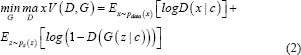
where D (xǀc) and G (zǀc) demonstrate discriminating and generating an image given a condition c, x is again the real data, and pz (z) is the input noise. Figure 2b shows a graphical representation of the cGAN model.
Cycle-consistent generative adversarial network
A CycleGAN is another GANs' variant, capable of translating images from one domain to another, for example, translating images of zebras to images of horses.[21] Image-to-image translation is traditionally performed by mapping between an input image and an output image using a training set of aligned image pairs. However, for many tasks, paired training data will not be available. The cycleGAN is capable of capturing special characteristics of one image domain and translating the image into another domain, without having paired images. The model of cycleGAN includes two generator networks G and F with mapping G : X → Y, F : Y → X and two discriminator networks D (x) and D (y). While D (x) aims to distinguish between images in domain X and translated images {F (y)}, D (y) aims to discriminate between images in domain Y and {G (x)}. Figure 2c shows a graphical representation of the cycleGAN model.
GENERATIVE ADVERSARIAL NETWORKS IN HISTOPATHOLOGICAL IMAGE PREPROCESSING
Histopathological WSIs are large-sized images that provide a great source of information. Nevertheless, the information contained in the histopathological WSIs is affected by many factors such as the type and quality of the microscope or scanner used for imaging, the size and magnification of the image, vibrations caused by external factors, physical color variation due to staining and tissue section preparation process, contamination in the sample, noise and defocusing, and artifacts. Consequently, preprocessing of histopathological WSIs is essential to improve the quality of the images prior to applying AI learning algorithms. The importance of preprocessing is illustrated by a recent study where various preprocessing techniques have been applied to histopathological WSIs before feeding them to a convolutional neural network (CNN) architecture for classification.[36] The results showed an improved classification performance with traditional preprocessing techniques such as background noise reduction and cell enhancement, as compared to the original images. Excessive additional preprocessing, however, for example, thresholding or the application of morphological operations applied on the already preprocessed images, resulted in removal of the important image features and subsequent performance reduction of the CNN.
The introduction of GANs has triggered a novel approach for histopathological image preprocessing. CycleGANs have been effectively applied to color normalization of breast histopathological images eliminating the requirement of having a reference template slide.[37] Super-resolution GANs (SRGANs) are a variant of GANs that are capable of inferring photorealistic natural images. SRGAN has been employed to increase image resolution and eliminate image noise from breast histopathological images.[38] The SRGAN-based method produced high-resolution images that are cleaner than images preprocessed by classical interpolation methods for increasing resolution. The following subsections describe various preprocessing tasks using generative models: color normalization, virtual staining of histological tissue, image enhancement, removal of ink marks from histopathological images, and data augmentation.[27,39,40,41,51,66,67]
Color normalization
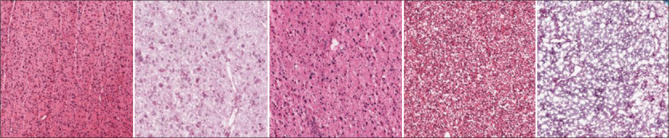
Color variations in digital slides hinder the successful performance of deep learning algorithms in digital pathology across laboratories and settings. Physical color variations can be a result of the biochemical staining process (difference in stain batches, variability in staining amount, time, thickness of sections, and staining protocol), while color variations can also be due to the imaging and digitizing parameters (variations in illumination, (de) focusing, imaging resolution and magnification, microscope or scanner model, and spectral sensitivity of the detector).[42,43] Normalization of the color represented by digital slides is thus an important preprocessing task in digital pathology. An example of observed color variations in the sample images is shown in Figure 3.
Figure 3.
Original H&E stained glioblastoma pathology slides obtained from The Cancer Genome Atlas database[44] showing diverse color variations in the sample images
Traditional image processing approaches such as histogram techniques[45,46] and color models[47,48] have been used for color normalization for a long time. The progress in artificial neural networks such as convolutional neural networks and the generative models has opened innovative paths for color normalization. Some of these GAN-based techniques and approaches for color normalization in digital pathology are highlighted in Table 1. The listed color normalization approaches are based on a style transfer method in which the style of the input image is modified based on the style image, when preserving the content of the input image.[37,39,50,51,53,54,55] The methods based on cycleGAN explore the capability of unpaired image-to-image translation which makes it a flexible architecture for stain normalization. Other approaches discussed here use alternative formulations such as self-attention models,[56] cGAN,[31] and encoder–decoder architecture.[57] The stain normalization stage can be integrated into a classification approach to eliminate bias introduced in these types of architectures.[50,58]
Table 1.
Novel generative adversarial networks approaches used for color normalization of histopathological images
| Reference | Tissue type | Dataset | Architecture | Method |
|---|---|---|---|---|
| Cho et al., 2017[49] | Lymph node samples | CAMELYON16 | cGAN | Stain style transfer approach |
| Bentaieb and Hamarneh, 2018[50] | Breast histology images, colon adenocarcinoma tissue images, ovarian carcinoma images | MITOS-ATYPIA14 challenge, MICCAI16 GlaS challenge, nonpublic dataset | GAN | Stain style transfer approach using encoder-decoder architecture and skip connections |
| Zanjani et al., 2018[39] | Lymph node samples | Nonpublic dataset | GAN | Unsupervised GAN based model for stain color normalization |
| Rana et al., 2018[51] | Prostate core biopsy tissue samples | Nonpublic dataset | cGAN | Staining and de-staining models used for learning hierarchical non-linear mappings between non-stained and stained WSIs |
| Zhou et al., 2019[52] | Breast cancer samples | CAMELYON16 | cycleGAN | Enhanced cycleGAN based method using stain color matrices for translation |
| Shaban et al., 2019[37] | Breast cancer samples | MITOS-ATYPIA14 challenge, CAMELYON16 | cycleGAN | Structure preserving stain style transfer |
| Cai et al., 2019[53] | Breast cancer samples | MITOS-ATYPIA14 challenge | cycleGAN | Structure preserving stain style transfer |
| Shrivastava et al., 2019[54] | Duodenal biopsy samples | Nonpublic dataset | GAN | Stain transfer approach using self-attentive adversarial network |
| Salehi and Chalechale et al., 2020[55] | Breast cancer samples | MITOS-ATYPIA14 challenge | cGAN | Pix2Pix based stain to stain translation |
cGAN: Conditional generative adversarial networks, GAN: Generative adversarial networks, WSIs: Whole-slide image
Virtual staining of histological tissue
GANs have gained significant attention in many other medical image processing tasks, including virtual staining of tissues slides.[27,29,51,59,60] Histological staining is commonly used in pathological diagnosis to highlight the important features of tissue. These staining processes are usually lengthy, relatively costly, and laborious due to time-sensitive steps.
Virtual staining can be achieved starting from unstained slides imaged using different imaging modalities. In one study, lung tissue slides were imaged using a hyperspectral microscope where the tissue structure is imaged at many different wavelength bands (colors) of the light spectrum.[29] The most important information from this multidimensional image cube was captured by principal component analysis to reduce the cube to a three-component image which was subsequently fed to a cGAN to train the staining network model. The trained network was able to transform images from the hyperspectral domain to the H&E domain by highly nonlinear mappings, resulting in their virtually stained versions.
Virtual staining can also be achieved using conventional broadband light illumination, by training a cGAN using RGB-colored WSI pairs before and after H&E staining.[51] In addition, a “destaining” model was developed that learned the reversed mapping between an RGB image pair of an H&E stained WSI and a non-stained WSI of the same biopsy. The authors could examine their models qualitatively by comparing the original H&E stained section to their virtually destained and restained counterparts; however, further suitability of the generated images for tumor diagnosis was not pursued and further clinical validation is necessary.
A related domain transfer approach in virtual staining techniques is the transfer from H&E stained specimens to their immunohistochemistry (IHC)-stained equivalents. IHC stains target specific proteins on cells on the tissue slide and display different colors. A conditional cycleGAN was trained to achieve unpaired image-to-image translation for multiclass virtual IHC staining.[27] Optimized structure and authenticity preservation was achieved by including additional loss functions for structural and photorealistic properties of the created virtual slide.[61,62] To evaluate the results, computer vision researchers were asked to score the image quality of real and fake images, and professional pathologists were asked to score the staining quality of the virtually stained patches. Although the conditional cycleGAN had a higher authenticity score as compared to the cycleGAN, both models suffered considerably from incorrect staining. The authors suggest to improve this domain transfer issue by increasing and optimizing the training dataset.
The ability of virtually staining is also demonstrated in many other image modalities such as autofluorescence image captured by a standard fluorescence microscope,[60] quantitative phase images,[59] confocal microscopy images,[63] and holographic virtual staining of individual biological cells.[64] Virtual staining of histological tissues has the potential to bypass the disadvantages of manual histological staining, provided it is clinically validated and approved. In addition, virtual staining preserves the unlabeled and unaltered tissue sections for further analysis.
Image enhancement
Image enhancement acts as a preprocessing step that improves the quality of the images in terms of color, brightness, and contrast. Researchers in this field have explored several image enhancement techniques using AI methods.[65] Deep neural networks are employed for image quality enhancement but need a large quantity of paired training data of low-quality images and their corresponding high-quality images. To alleviate this barrier, SRGANs can generate high-resolution images, and for such a reason, they have attracted attention among the researchers in this area. For instance, SRGANs have been employed to generate super-resolution images and to eliminate noise from breast histopathological images.[38] Another study proposed a mixed-supervision GAN, based on SRGAN, that can take both high- and medium-quality images as input for training.[66] This approach leverages training data from high quality to medium quality to improve the performance when limiting the costs of data curation.[66] The method uses a sequence of generators and discriminators, dealing with increasingly higher quality training data at each level, compared to the low-quality input data. Quantitative evaluation of the images was done using metrics such as the structural similarity index and the relative root mean square error, while the images were not tested for further image analysis such as classification and segmentation tasks.
Another study employing SRGAN performed classification of super-resolution, high-resolution, and low-resolution images along with the comparison of different performance measures.[67] This study uses a wide attention SRGAN which uses wide-residual blocks and a self-attention layer. Wider channels before activation in residual blocks were shown to improve the performance of image super-resolution networks.[54] By including an in-built self-attention function that calculates the weighted mean of all pixels in an image, the model learns relationships between widely separated spatial regions and distant pixels based on patch appearance similarity. The classification results for high- and low-resolution images displayed an accuracy of 99.49% and 95.82%, respectively.[67] These studies emphasize the role of the ability of GANs to increase the resolution during the reconstruction of poor-quality images, thereby tackling the cost of hardware and storage to employ high-resolution images.
Ink mark removal
Histopathological glass slides are often annotated by pathologists using pens or other markers to indicate regions of interest, such as neoplastic areas or other specific features. Figure 4 displays some examples of ink-marked WSIs from The Cancer Genome Atlas dataset.
Figure 4.
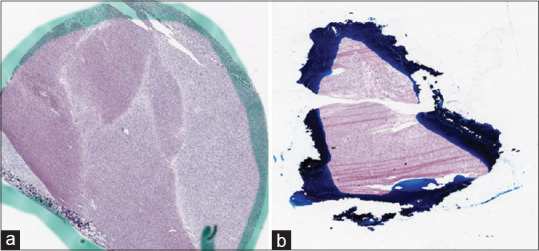
Examples of whole-slide images annotated by (a) green and (b) blue ink marks obtained from The Cancer Genome Atlas database[44]
This pragmatic approach unfortunately hinders computer-aided interpretation of marked WSIs. For example, color thresholding or other automatic selection algorithms fail to select the correct areas in the presence of ink marks. State-of-the-art color normalization methods are not sufficient in removing ink marks as they are designed to handle sensitive variations in unmarked H&E images;[68] therefore, other solutions based on generative models have been explored.
A fully automatic CNN-based approach to remove ink from H&E stained prostate WSIs consisted of three separate architectures.[40] First, a sequential classical CNN classifies the image tiles into three categories: ink-free tiles, background contaminated tiles (tiles with ink marks, but not on the tissue), and foreground contaminated tiles (tiles with ink marks on the tissue). Subsequently, the background contaminated tiles are cleaned by replacing the ink marks with a white background. A fast region detector performs the precise localization of the ink marks on the foreground contaminated tiles. Finally, a cycleGAN is applied to these detected localized marks to restore these image tiles to ink-free image tiles.
A similar study has approached the ink removal as a style transfer problem using a cycleGAN to remove ink marks from human melanoma tissues, when preserving the tissue structure underneath the marker region.[69] The melanoma WSIs were first tiled to obtain marker patches which contain full or partial ink marks and clean patches which do not contain any ink marks. The cycleGAN used, here, is composed of two generative and one discriminative CNN. The first generative CNN is trained to remove ink marks from the marker patches and to generate patches similar to clean tissue that cannot be differentiated by the discriminator model. The second generative CNN is trained to reconstruct the output of the first generative CNN to ensure the preservation of the individual visual information from each image patch. The application of cycleGAN to ink removal exploits the image-to-image translation method to produce visually coherent images and outperforms the state-of-the-art color normalization methods that are not capable of removing ink marks from the WSIs.
Data augmentation
The availability of large training sets is essential to achieve good performance in deep learning. However, due to privacy concerns, high cost, and complexity of data collection and labeling, medical datasets, especially pathological datasets, are relatively small. Data augmentation is a method to significantly increase the amount and variety of data available for training models, without collecting new data. Verified data augmentation methods can be used in the development of automated medical image analysis to mitigate the effects of data imbalance and improve overall performance of the training model.[70,71] Traditional data augmentation methods include image transformations (rotation, flipping, reflection, zooming, etc.) and color transformations (histogram matching). The success of data augmentation methods has stimulated an interest in researching advanced approaches to generate new images for training. After the initial development of GANs, generative models that synthesize completely new images have become a popular method for generating augmented data.
GAN-based data augmentation methods were able to generate new skin melanoma photographs, histopathological images, and breast MRI scans.[41] Here, the GAN style transfer method was applied to combine an original picture with other image styles to obtain a multitude of pictures with a variety in appearance. The augmented dataset was utilized for diagnostic image classification. An image translation approach using cycleGAN was used in another study to generate synthetic colorectal polyp images from normal colonic mucosa images.[72] The generated synthetic images maintained the general structure of the normal images but exhibited adenomatous features. The quality of the augmented images was then evaluated by a pretrained classifier and through clinical evaluation by pathologists, finding that three of the four pathologists could not differentiate at least half of the synthetic images from real images. In another study, cGAN was used to synthesize realistic cervical histopathology images to expand a limited training dataset.[72] The authors used a filtering mechanism to control the feature quality of augmented synthetic images and boost the performance of the classifier. A cycleGAN was proposed to augment existing nuclei segmentation datasets by generating synthetic H&E patches representing several different organs for nuclei segmentation algorithm development.[33] Another study has recently investigated the use of GAN to augment the dataset of histological specimens of gliomas with synthetic gliomas, to increase the training and accuracy of deep learning techniques to predict the IDH gene mutation status.[73] The accuracy of the deep learning model for predicting the IDH status onH&E stained histopathological images was increased from 80% (without augmentation) to 85% with GAN augmented data. Figure 5 shows some examples of GAN-synthesized glioma images.
Figure 5.
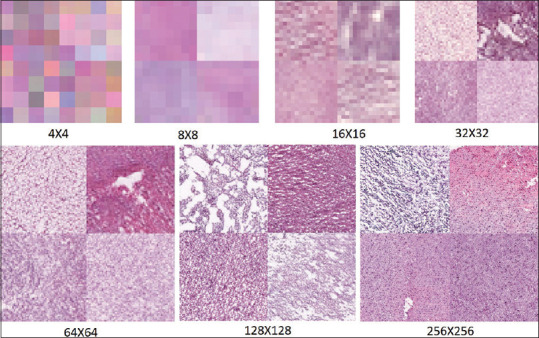
Samples of generative adversarial network synthesized glioma images from coarse to fine scales
The studies listed in this section show the capability of GANs to generate synthetic images: from image-to-image translation for normalization and cleaning of color-shaded and marked pathology images to the generation of augmented databases to solve histopathological data limitations. Further exploration of the application of GANs to WSI preprocessing will help in the workflow of digital pathology as it preserves the tissue sample structure when enabling further AI-based image analysis. These studies also show the capability of GANs to generate synthetic images that can alleviate some of the limitations related to histopathological data.
GENERATIVE ADVERSARIAL NETWORKS IN HISTOPATHOLOGICAL IMAGE ANALYSIS
Computer-aided analysis of images has been widely explored in many areas of studies for feature extraction and pattern recognition. With the recent advancement of digital pathology, histopathological image analysis has gained attention of many researchers.[74] Computer-assisted quantitative analysis of histopathological images can significantly speed up clinical analysis and research project outcomes. In addition, AI has the potential to uncover information in pathology images that results in novel insights about diseases and therapy. GANs and their extensions have opened encouraging ways to challenge long-standing image analysis problems such as segmentation, feature extraction, detection, and classification. In the following subsections, we categorized the histopathological image analysis articles into: automated detection and segmentation, other feature selection, extraction and quantification, and domain adaptation.[28,75,76]
Nuclei detection and segmentation
Automated segmentation and detection of tissue and cellular structures is an important task in histopathological image analysis. Classical methods for nuclei segmentation techniques include watershed algorithms, color-based thresholding, morphological processing, active contours, and their variants.[77] During the last few decades, numerous state-of-the-art methods have been proposed for nucleus detection and segmentation in digital pathology images.[78] GANs have also effectively enabled a range of nuclei segmentation tasks, including segmentation of nuclei from WSIs and the identification of nuclei within a specific region.
In an unsupervised nuclei segmentation method formulated as an image-to-image translation problem, diverse histopathological images were selected as nuclear domain and randomly generated nuclear shapes were selected as label domain.[75] Then, a cycleGAN was used to translate the histopathology images to the label domain by generating segmentation maps. Several challenges were reported in this study, including (1) reconstruction losses for segmentation resulting in the suppression of less bright nuclei with each gradient update, and (2)incorrectly generated nuclei shape annotations resulting in poorly segmented nuclei. Despite the challenges, the study showed that generative models can be a promising solution for the nuclei segmentation problem, including the separation of differently shaped nuclei. In another study, nuclei detection on colorectal adenocarcinomas histopathology images was peformed using a residual attention GAN, which is based on cGAN.[79] The residual attention mechanism is used to capture the characteristics of the nuclear regions more clearly by generating a more accurate probability map with spatial contiguity, resulting in improved nuclei detection.
The performance of deep learning algorithms for segmentation highly depends on the quality and volume of labeled histopathology data available for training. Furthermore, most of the work has focused on developing cellular and cellular feature segmentation methods for single organs. In a multiorgan study, cGANs are used to train synthetically generated data and real data to perform nuclei segmentation.[80] First, a cycleGAN model was trained with images from four different organs to synthetically generate pathology data with perfect nuclei segmentation labels. These real and synthetically generated images were subsequently used to train a cGAN to perform nuclei segmentation. The study demonstrates that this approach performs better than the state-of-the-art methods for nuclei segmentation.
The concept of the information-maximizing GAN (infoGAN) architecture,[35] which learns the visual representations of the cell nuclei by utilizing features in the image such as shape, nuclear density, and color, was explored for the classification of breast cancer histology images.[81] First, a CNN-based stain transfer method is used to normalize the H&E stained breast cancer histology images. The stain-normalized breast histology images are then trained by a neural network to segment out the nuclei. Finally, infoGAN is used to classify the segmented nuclei in an unsupervised manner based on visual representations of different types of nuclei.
Feature selection, extraction, and quantification
Selection, extraction, and quantification of relevant features from histopathological images have long been an active area of research within the image processing community. Automated cancer diagnosis relies on capturing cellular and tissue level features to quantify the changes in a given tissue.[45] Traditional methods used for feature extraction include morphological, topological, textural, fractal, and/or intensity-based methods.
Pathology GAN[28] is a framework that generates high-fidelity synthetic cancer images using GANs to capture the key tissue features such as color, texture, spatial features of cancer and normal cells, and their interaction. The proposed model uses the generated images to obtain the feature space based on cell density and morphology in cancer tissue and captures pathologically meaningful representations. The study also demonstrated that pathologists were not able to reliably find differences between the real and generated images. Another study used a GAN architecture to perform unsupervised representation learning for cell-level images segmented from bone marrow histopathology images.[35] Each cell-level image contains only one cell, enabling GAN-based learning of interpretable and disentangled cellular elements. These cell-level visual representations can be used for a variety of tasks such as cell classification based on their semantic features. The proposed pipeline used the cell-level information to calculate the cellular class proportions in the histopathological images, combining this with nuclei segmentation to perform image-level classification of histopathology images. Then, image-level classification was performed to differentiate between normal and abnormal bone marrow pathology images. The study revealed that the algorithm can classify different cell types such as myeloblasts, monocytes, granulocytes, lymphocytes, and erythrocyte precursors and quantify their presence to confidently discriminate normal from abnormal images. However, some abnormal images were wrongly classified as normal which the authors attribute to the atypical staining of erythroid precursors. The authors suggested improvements to make the model more robust to these variations and also highlighted that acquiring high-quality histopathological images is a priority for reliable results.
To improve segmentation accuracy, a GAN-based image-to-image translation method was proposed that is based on image content enrichment rather than increasing the number of samples, by overlapping virtual images with the given (original) image.[82] First, the input image to be segmented is translated to different domains (stains) by several generators to produce virtual images with stains different from the original image. All these virtual images are concatenated and merged with the original image, resulting in an enriched multichannel image. As the second step, the enriched images are used to train and test a fully connected convolutional network for segmentation. An experiment with mouse kidney pathology images showed an improved segmentation score for kidney glomeruli for enriched images compared to using the original images only. The study relies on the rationale that the two models individually trained to solve a specific subproblem are more powerful than one single model trained to solve the whole, more complex problem. The image-to-image translation approach here produces highly realistic and detailed histopathological images with added information from the virtual stains, leading to improved segmentation.
Domain adaptation
Domain variations such as differences in staining between laboratories can prevent the predictions based on a model learned from one domain being directly applicable to another domain (other laboratory). Oftentimes, paired images from the different domains (for example, the same samples imaged at different laboratories) are not available; thus, direct transformation cannot be learned. Domain adaptation is the task of transferring the image features between domains, so that classification based on the annotated source domain can be reliably applied to the unlabeled target domain. A study was proposed for classification and grading of breast cancer images by transforming the target images to the source domain and then applying a deep learning method.[76] The authors used cycleGAN for unsupervised domain adaptation to transform data from four different medical centers to data from one center to reduce the variability in the data and improve the classification results. Another study[26] addressed unsupervised domain adaptation using a different paradigm, namely the alignment of the learned image feature space of the source domain with the unlabeled target domain. The adversarial training minimized the distribution discrepancy in the feature space between the source and target domains. Asymmetric adaptation resulted in fine-tuning of the target domain network guided by the source-domain network, thereby mimicking the distribution of the source-domain feature space. Thus, the target network gets trained to extract the domain invariant features from input samples. Using a set of prostate histopathology images, the classification results showed improvement compared to baseline models, indicating that adversarial training may not only decrease color differences but also mechanical distortions, morphologic, and structural differences resulting from different processing routines.
In medical image processing, learning of distinctive patterns and feature extraction plays an important role in the diagnosis of diseases. The discussions in the above sections show the rich feature extraction capability of GANs, their power to extract semantically meaningful information from the histopathological images, and transferring the image features between different domains.
DISCUSSION AND CONCLUSION
Digital pathology is rapidly growing alongside the recent developments in AI and deep learning. The integration of AI with pathology can contribute exciting changes to health care, including precision medicine through personalized patient models and treatments.[83] In particular, the innovative development of GANs and its variants has led to an improved digital pathology workflow resulting in more informed and comprehensive cancer diagnosis.[28,82] In recent years, GANs have gained significant attention in the area of various medical image processing tasks, including virtual staining of tissues slides, which bypasses the laborious and expensive process of histological staining.[59,60] We consider the application of GANs to digital pathology as a promising upcoming research area with many opportunities to be explored. Fortunately, it is gaining the interest of the computational and medical researchers.
In this study, we have presented the application of GAN models to histopathological image processing. The above discussion shows that GAN has the power to facilitate several practical applications in digital pathology. The advantages of GANs in digital pathology can be summarized as the following.
Resolving scarcity of training datasets by producing realistic synthetic data
Due to privacy concerns, high cost, and complexity of data collection and labeling, medical datasets tend to be relatively small compared to the general datasets. GAN-based models can be a promising solution for the limited size of publicly available clinical datasets. The ability of GANs to mimic data distributions and produce realistic synthetic data can be employed to augment data, enlarge the medical datasets, and handle class imbalance. Furthermore, the manual labeling of WSIs is a time-consuming task that could be solved using synthetic labeled images.
Augmentation by generating data with diversity
Augmentation of data can be performed in many ways, such as image transformation and color transformation methods. However, these augmentation techniques do not provide diversity in the data being generated. GANs are additionally effective for the augmentation that is difficult to implement with standard augmentation techniques, through its capability to generate realistic and customized variations.
Preprocessing of input data
GANs have proved to be effective candidates for image preprocessing tasks, such as color normalization, ink removal, and data augmentation. We envisage that it could be further explored to use generative models for the development of universal image preprocessing steps that can be applied to all types of histopathology images irrespective of the sample types.
Virtual staining of the whole-slide images
Histological staining of tissue slides is usually a laborious and expensive process during pathological diagnosis. GANs can be a potential candidate for virtual staining of tissue slides which can generate images with reduced stain variations. This will eventually reduce the labour and cost incurred in the actual histological staining.
Effective in learning and extracting features
Feature extraction and pattern recognition play an important role in medical image analysis for the diagnosis of diseases. GANs are powerful tools in extracting and quantifying semantically meaningful information from images, with several advantages in digital pathology for automatic detection, segmentation, and classification applications.
Although the number of studies applying GANs has increased significantly in the last 3 years, it should be noted that most of the research has been focused to MRI instead of other medical imaging modalities. One reason for this might be the large number of publicly available MRI datasets compared to the pathological datasets. In the area of histopathology, most of the GAN-based methods applied to histopathological images focused on breast cancer or prostate cancer datasets. This leaves ample scope to adopt these state-of-the-art GAN-based methods to datasets representing other organs.
Despite its significant success as a generative model, the training of GAN is an expert task and the training process encounters many challenges such as vanishing gradients, mode collapse, and failure to converge. The possible solutions to handle GANs challenges are to choose appropriate network architectures, to use customized loss functions, and to apply various forms of regularization.[84] Another important challenge with GANs is that they are often focused on matching the input image to a target distribution of images, sometimes resulting in additional trivial or misleading features by adding or removing textures. A study employing cycleGAN used a structural loss to suppress these features, which the authors refer as “imaginary” ability of the GANs especially while encountering new features.[27] Many variants of GAN with diverse characteristics have been proposed for handling the drawbacks of GANs, but several issues persist to be dealt with for future research to take full advantage of the extensive capabilities of GAN-based methods to transform and advance digital pathology.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
Prof. Antonio Di Ieva received the 2019 John Mitchell Crouch Fellowship from the Royal Australasian College of Surgeons (RACS), which, along to Macquarie University co-funding, supported the opening of the Computational NeuroSurgery (CNS) Lab at Macquarie University, Sydney, Australia. Moreover, he is supported by an Australian Research Council (ARC) Future Fellowship (2019-2023, F190100623).
Dr. Sidong Liu acknowledges the support of an Australian National Health and Medical Research Council grant (NHMRC Early Career Fellowship 1160760).
Dr. Annemarie Nadort acknowledges the support of the National Health and Medical Research Council (NHMRC ECF APP1124160).
Laya Jose acknowledges the support of the ARC Centre of Excellence for Nanoscale BioPhotonics and iMQRES scholarship, Macquarie University..
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2021/12/1/4/307702
REFERENCES
- 1.Alturkistani HA, Tashkandi FM, Mohammedsaleh ZM. Histological stains: A literature review and case study. Glob J Health Sci. 2015;8:72–9. doi: 10.5539/gjhs.v8n3p72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farahani N, Parwani AV, Pantanowitz L. Whole slide imaging in pathology: Advantages, limitations, and emerging perspectives. Pathol Lab Med Int. 2015;7:4321. [Google Scholar]
- 3.Lichtblau D, Stoean C. Cancer diagnosis through a tandem of classifiers for digitized histopathological slides. PloS One. 2019;14:e0209274. doi: 10.1371/journal.pone.0209274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aeffner F, Zarella MD, Buchbinder N, Bui MM, Goodman MR, Hartman DJ, et al. Introduction to digital image analysis in whole-slide imaging: A white paper from the digital pathology association. J Pathol Inform. 2019;10:9. doi: 10.4103/jpi.jpi_82_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Ieva A, Bruner E, Widhalm G, Minchev G, Tschabitscher M, Grizzi F. Computer-assisted and fractal-based morphometric assessment of microvascularity in histological specimens of gliomas. Sci Rep. 2012;2:429. doi: 10.1038/srep00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esgiar AN, Naguib RN, Sharif BS, Bennett MK, Murray A. Fractal analysis in the detection of colonic cancer images. IEEE Trans Inf Technol Biomed. 2002;6:54–8. doi: 10.1109/4233.992163. [DOI] [PubMed] [Google Scholar]
- 8.Di Ieva A. Fractal analysis of microvascular networks in malignant brain tumors. Clin Neuropathol. 2012;31:342–51. doi: 10.5414/np300485. [DOI] [PubMed] [Google Scholar]
- 9.Niazi MK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–61. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards BA, Lillicrap TP, Beaudoin P, Bengio Y, Bogacz R, Christensen A, et al. A deep learning framework for neuroscience. Nat Neurosci. 2019;22:1761–70. doi: 10.1038/s41593-019-0520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21:4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 12.Janowczyk A, Basavanhally A, Madabhushi A. Stain normalization using sparse autoencoders (StaNoSA): Application to digital pathology. Comput Med Imaging Graph. 2017;57:50–61. doi: 10.1016/j.compmedimag.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Chang H, Barner KE, Parvin B. Nuclei segmentation via sparsity constrained convolutional regression. IEEE 12th International Symposium on Biomedical Imaging (ISBI) 2015:1284–7. doi: 10.1109/ISBI.2015.7164109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Snead DR, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016;35:1196–206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 15.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–67. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Li J, Sarma KV, Ho KC, Shen S, Knudsen BS, et al. Path R-CNN for prostate cancer diagnosis and gleason grading of histological images. IEEE Trans Med Imaging. 2019;38:945–54. doi: 10.1109/TMI.2018.2875868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative Adversarial Networks. Proceedings of the International Conference on Neural Information Processing Systems (NIPS 2014) 2014:672–80. [Google Scholar]
- 18.Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: A review. Med Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 19.Xu T, Zhang P, Huang Q, Zhang H, Gan Z, Huang X, et al. AttnGAN: Fine-Grained Text to Image Generation With Attentional Generative Adversarial Networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2018:1316–24. [Google Scholar]
- 20.Ledig C, Theis L, Huszar F, Caballero J, Cunningham A, Acosta A, et al. Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017:4681–90. [Google Scholar]
- 21.Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision (ICCV) 2017:2223–32. [Google Scholar]
- 22.Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging. 2017;36:2536–45. doi: 10.1109/TMI.2017.2708987. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Shi Y, Christodoulou F, Xie AG, Zhou Y, Li Z, et al. Efficient and Accurate MRI Super-Resolution Using a Generative Adversarial Network and 3D Multi-level Densely Connected Network. Medical Image Computing and Computer Assisted Intervention – MICCAI. 2018:91–99. [Google Scholar]
- 24.Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321:321–31. [Google Scholar]
- 25.Gadermayr M, Gupta L, Appel V, Boor P, Klinkhammer BM, Merhof D. Generative adversarial networks for facilitating stain-independent supervised and unsupervised segmentation: A study on kidney histology. IEEE Trans Med Imaging. 2019;38:2293–302. doi: 10.1109/TMI.2019.2899364. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Hacihaliloglu I, Singer EA, Foran DJ, Qi X. Unsupervised domain adaptation for classification of histopathology whole-slide images. Front Bioeng Biotechnol. 2019;7:102. doi: 10.3389/fbioe.2019.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Moro CF, Bozóky B, Zhang Q. GAN-Based Virtual Re-Staining: A Promising Solution for Whole Slide Image Analysis. arXiv. 2019 arXiv: 1901.04059. [Google Scholar]
- 28.Quiros AC, Murray-Smith R, Yuan K. PathologyGAN: Learning deep Representations of Cancer Tissue. Proceedings of the Third Conference on Medical Imaging with Deep Learning, Vol. 121. PMLR. 2020:669–95. [Google Scholar]
- 29.Bayramoglu N, Kaakinen M, Eklund L, Heikkila J. Towards virtual H&E staining of hyperspectral lung histology images using conditional generative adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision Workshops. 2017:64–71. [Google Scholar]
- 30.Nash JF. Equilibrium points in N-person games. Proc Natl Acad Sci U S A. 1950;36:48–9. doi: 10.1073/pnas.36.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirza M, Osindero S. Conditional Generative Adversarial Nets. arXiv 2014. arXiv: 1411.1784 [Google Scholar]
- 32.Iqbal T, Ali H. Generative adversarial network for medical images (MI-GAN) J Med Syst. 2018;42:231. doi: 10.1007/s10916-018-1072-9. [DOI] [PubMed] [Google Scholar]
- 33.Mahmood F, Chen R, Borders D, McKay GN, Salimian K, Baras A, et al. Adversarial U-net with spectral normalization for histopathology image segmentation using synthetic data. Proc. SPIE 10956, Medical Imaging 2019. Digit Pathol. 2019:109560N. doi: 10.1117/12.2512918. [Google Scholar]
- 34.Jin Y, Zhang J, Li M, Tian Y, Zhu H, Fang Z, et al. Towards the Automatic Anime Characters Creation with Generative Adversarial Networks. arXiv Preprint arXiv: 1708.05509. 2017 [Google Scholar]
- 35.Hu B, Tang Y, Chang EI, Fan Y, Lai M, Xu Y. Unsupervised learning for cell-level visual representation in histopathology images with generative adversarial networks. IEEE J Biomed Health Inform. 2019;23:1316–28. doi: 10.1109/JBHI.2018.2852639. [DOI] [PubMed] [Google Scholar]
- 36.Öztürk Ş, Akdemir B. Effects of histopathological image pre-processing on convolutional neural networks. Procedia Comput Sci. 2018;132:396–403. [Google Scholar]
- 37.Shaban MT, Baur C, Navab N, Albarqouni S. Staingan: Stain style transfer for digital histological images. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE. 2019:953–56. [Google Scholar]
- 38.Çelik G, Talu MF. Resizing and cleaning of histopathological images using generative adversarial networks. Physica A Stat Mech Appl. 2019;554:122652. [Google Scholar]
- 39.Zanjani FG, Zinger S, Bejnordi BE, Laak JV. Histopathology stain-color normalization using deep generative models. 1st Conference on Medical Imaging with Deep Learning (MIDL 2018) 2018:1–11. [Google Scholar]
- 40.Ali S, Alham NK, Verrill C, Rittscher J. Ink Removal from Histopathology Whole Slide Images by Combining Classification, Detection and Image Generation Models. IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) 2019:928–32. [Google Scholar]
- 41.Mikołajczyk A, Grochowski M. Data augmentation for improving deep learning in image classification problem. In 2018 International Interdisciplinary PhD Workshop (IIPhDW). IEEE. 2018:117–22. [Google Scholar]
- 42.Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kothari S, Phan JH, Stokes TH, Wang MD. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc. 2013;20:1099–108. doi: 10.1136/amiajnl-2012-001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–57. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabesh A, Teverovskiy M, Pang HY, Kumar VP, Verbel D, Kotsianti A, et al. Multifeature prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans Med Imaging. 2007;26:1366–78. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 46.Sertel O, Catalyurek UV, Shimada H, Gurcan MN. Computer-aided prognosis of neuroblastoma: Detection of mitosis and karyorrhexis cells in digitized histological images. Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:1433–6. doi: 10.1109/IEMBS.2009.5332910. [DOI] [PubMed] [Google Scholar]
- 47.Zarella MD, Breen DE, Plagov A, Garcia FU. An optimized color transformation for the analysis of digital images of hematoxylin & eosin stained slides. J Pathol Inform. 2015;6:33. doi: 10.4103/2153-3539.158910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001;21:34–41. [Google Scholar]
- 49.Cho H, Lim S, Choi G, Min H. Neural Stain-Style Transfer Learning Using Gan for Histopathological Images. arXiv 2017. arXiv: 1710.08543. [Google Scholar]
- 50.Bentaieb A, Hamarneh G. Adversarial stain transfer for histopathology image analysis. IEEE Trans Med Imaging. 2018;37:792–802. doi: 10.1109/TMI.2017.2781228. [DOI] [PubMed] [Google Scholar]
- 51.Rana A, Yauney G, Lowe A, Shah P. Computational Histological Staining and Destaining of Prostate Core Biopsy RGB Images with Generative Adversarial Neural Networks. 17th IEEE International Conference on Machine Learning and Applications (ICMLA) IEEE. 2018 [Google Scholar]
- 52.Zhou N, Cai D, Han X, Yao J. Enhanced Cycle-Consistent Generative Adversarial Network for Color Normalization of H&E Stained Images. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. :694–702. [Google Scholar]
- 53.Cai S, Xue Y, Gao Q, Du M, Chen G, Zhang H, et al. Stain style transfer using transitive adversarial networks. Machine learning for medical Image Reconstruction. MLMIR 2019. Lecture Notes in Computer Science. Vol. 11905. Springer, Cham. 2019 [Google Scholar]
- 54.Shrivastava A, Adorno W, Sharma Y, Ehsan L, Ali SA, Moore SR, et al. Self-Attentive Adversarial Stain Normalization. arXiv Preprint arXiv: 1909.01963. 2019 doi: 10.1007/978-3-030-68763-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salehi P, Chalechale A. Pix2Pix-based Stain-to-Stain Translation: A Solution for Robust Stain Normalization in Histopathology Images Analysis. International Conference on Machine Vision and Image Processing (MVIP) 2020:1–7. [Google Scholar]
- 56.Zhang H, Goodfellow I, Metaxas D, Odena A. Self-attention generative adversarial networks. In International Conference on Machine Learning. PMLR. 2019:7354–63. [Google Scholar]
- 57.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015:3431–40. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 58.Cong C, Liu S, Di Ieva A, Pagnucco M, Berkovsky S, Song Y. Texture Enhanced Generative Adversarial Network for Stain Normalisation in Histopathology Images. International Symposium on Biomedical Imaging(ISBI) IEEE. 2021 [Google Scholar]
- 59.Rivenson Y, Liu T, Wei Z, Zhang Y, de Haan K, Ozcan A. PhaseStain: The digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci Appl. 2019;8:23. doi: 10.1038/s41377-019-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y, et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng. 2019;3:466–77. doi: 10.1038/s41551-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 61.Fujun L, Sylvain P, Eli S, Kavita B. Deep Photo Style Transfer. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017:4990–8. [Google Scholar]
- 62.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: From error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–12. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 63.Combalia Escudero M, Pérez Ankar J, García Herrera A, Alos L, Vilaplana Besler V, Marqués Acosta F, et al. London, United Kingdom: proceedings of Machine Learning Research; 2019. Digitally stained confocal microscopy through deep learning. International Conference on Medical Imaging with Deep Learning: 8-10 July 2019; pp. 121–9. [Google Scholar]
- 64.Nygate YN, Levi M, Mirsky SK, Turko NA, Rubin M, Barnea I, et al. Holographic virtual staining of individual biological cells. Proc Natl Acad Sci U S A. 2020;117:9223–31. doi: 10.1073/pnas.1919569117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abraham T, Todd A, Orringer DA, Levenson R. Applications of artificial intelligence for image enhancement in pathology. Artificial Intelligence and Deep Learning in Pathology. USA: Elsevier; 2021. pp. 119–48. [Google Scholar]
- 66.Uddeshya U, Awate Suyash P. Mixed-Supervision Multilevel GAN Framework for Image Quality Enhancement. Medical Image Computing and Computer Assisted Intervention – MICCAI. 2019:556–64. [Google Scholar]
- 67.Shahidi F. Breast Cancer Histopathology Image Super-Resolution Using Wide-Attention GAN with Improved Wasserstein Gradient Penalty and Perceptual Loss. IEEE Access. 2021;9:32795–809. [Google Scholar]
- 68.Niethammer M, Borland D, Marron J, Woosley J, Thomas NE. Appearance normalization of histology slides. International Workshop on Machine Learning in Medical Imaging. Berlin, Heidelberg: Springer; 2010. pp. 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venkatesh B, Shaht T, Chen A, Ghafurian S. Restoration of Marker Occluded Hematoxylin and Eosin Stained Whole Slide Histology Images Using Generative Adversarial Networks. IEEE 17th International Symposium on Biomedical Imaging (ISBI) 2020:591–5. [Google Scholar]
- 70.Wong WS, Amer M, Maul T, Liao IY, Ahmed A. Advances in Intelligent Systems and Computing. Vol. 978. Cham: Springer; 2020. Conditional Generative Adversarial Networks for Data Augmentation in Breast Cancer Classification. Recent Advances on Soft Computing and Data Mining. SCDM 2020. [Google Scholar]
- 71.Pandey S, Singh PR, Tian J. An image augmentation approach using two-stage generative adversarial network for nuclei image segmentation. Biomed Signal Process Control. 2020;57:101782. [Google Scholar]
- 72.Xue Y, Zhou Q, Ye J Long LR, Antani S, Cornwell C. Synthetic Augmentation and Feature-Based Filtering for Improved Cervical Histopathology Image Classification. Medical Image Computing and Computer Assisted Intervention – MICCAI. 2019:387–96. [Google Scholar]
- 73.Liu S, Shah Z, Sav A, Russo C, Berkovsky S, Qian Y, et al. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci Rep. 2020;10:7733. doi: 10.1038/s41598-020-64588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: A review. IEEE Rev Biomed Eng. 2009;2:147–71. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koyun OC, Yildirim T. Adversarial Nuclei Segmentation on H&E Stained Histopathology Images. IEEE International Symposium on INnovations in Intelligent SysTems and Applications (INISTA) 2019:1–5. [Google Scholar]
- 76.Wollmann T, Eijkman CS, Rohr K. Adversarial Domain Adaptation to Improve Automatic Breast Cancer Grading in Lymph Nodes. IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 2018:582–5. [Google Scholar]
- 77.Xing F, Yang L. Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: A comprehensive review. IEEE Rev Biomed Eng. 2016;9:234–63. doi: 10.1109/RBME.2016.2515127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Höfener H, Homeyer A, Weiss N, Molin J, Lundström CF, Hahn HK. Deep learning nuclei detection: A simple approach can deliver state-of-the-art results. Comput Med Imaging Graph. 2018;70:43–52. doi: 10.1016/j.compmedimag.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 79.Li J, Shao W, Li Z, Li W, Zhang D. Residual Attention Generative Adversarial Networks for Nuclei Detection on Routine Colon Cancer Histology Images. Machine Learning in Medical Imaging. MLMI. 2019:142–50. [Google Scholar]
- 80.Mahmood F, Borders D, Chen RJ, Mckay GN, Salimian KJ, Baras A, et al. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2020;39:3257–67. doi: 10.1109/TMI.2019.2927182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan E, Suh J. Neural Stain Normalization and Unsupervised Classification of Cell Nuclei in Histopathological Breast Cancer Images. arXiv 2018; arXiv: 1811.03815 [Google Scholar]
- 82.Gupta L, Klinkhammer BM, Boor P, Merhof D, Gadermayr M. GANBased Image Enrichment in Digital Pathology Boosts Segmentation Accuracy. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI. 2019:631–9. [Google Scholar]
- 83.Schork NJ. Cancer Treatment and Research. Vol. 178. Cham: Springer; 2019. Artificial intelligence and personalized medicine. Precision Medicine in Cancer Therapy; pp. 265–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saxena D, Cao J. Generative Adversarial Networks (GANs): Challenges, Solutions, and Future Directions. ACM Computing Surveys. 2021 [Google Scholar]