Significance
Strawberry is a very popular fruit. The strawberry genus (Fragaria) has emerged as a model system for various fundamental and applied research in recent years. Here, by using high-throughput sequencing technologies, we provide de novo whole-genome sequences for five wild strawberry species and genome resequencing data for 128 additional accessions of key species. Our analyses resulted in robust estimates of the evolutionary history for most diploid strawberry species, the discovery of a new diploid species (Fragaria emeiensis Jia J. Lei), and the construction of a pan-genome for strawberry. We also examined the evolutionary dynamics of gene families. This study provides a powerful genomic platform and resource for future studies in strawberry.
Keywords: strawberry (Fragaria spp.), pan-genome, comparative genomics, genetic differentiation, MYB transcription factors
Abstract
Strawberry (Fragaria spp.) has emerged as a model system for various fundamental and applied research in recent years. In total, the genomes of five different species have been sequenced over the past 10 y. Here, we report chromosome-scale reference genomes for five strawberry species, including three newly sequenced species’ genomes, and genome resequencing data for 128 additional accessions to estimate the genetic diversity, structure, and demographic history of key Fragaria species. Our analyses obtained fully resolved and strongly supported phylogenies and divergence times for most diploid strawberry species. These analyses also uncovered a new diploid species (Fragaria emeiensis Jia J. Lei). Finally, we constructed a pan-genome for Fragaria and examined the evolutionary dynamics of gene families. Notably, we identified multiple independent single base mutations of the MYB10 gene associated with white pigmented fruit shared by different strawberry species. These reference genomes and datasets, combined with our phylogenetic estimates, should serve as a powerful comparative genomic platform and resource for future studies in strawberry.
Multiple genomes of closely related species provide an ideal framework for comparative and evolutionary genomics (1–3). Strawberry, Fragaria L. (Rosaceae), includes roughly 25 described species and contains diverse mating systems (self-compatible, self-incompatible, dioecious) and natural ploidy variation (2×, 4×, 5×, 6×, 8×, and 10×) (4, 5). There are currently 14 diploid species assigned to the genus Fragaria, among which 9 species endemic to East Asia, the hypothesized center area of origin for the genus (6, 7). The phenotypic diversity of important agronomic traits and relatively small genome sizes of the diploids (∼250 Mb), combined with the ease of growing and genetic manipulation, make this genus an emerging model system for various fundamental and applied research (5, 6, 8).
To elevate Fragaria as a model system for evolutionary genomics, a phylogenomic framework is needed, which requires understanding the phylogenetic relationships among species and having reference genomes available for multiple species. A previous study estimated the phylogeny of Fragaria based on sequences from the plastid genome (7). However, it has long been known that the evolutionary history of either organellar genome, mitochondrial or plastid, which are both maternally inherited, do not always reflect true species relationships. Phylogenomic methods using multiple markers distributed across the nuclear genome (referred to as phylogenomics) may provide more accurate estimates of species relationships (9–13).
Multiple genomes of Fragaria also provide a unique opportunity to investigate the variation of genomic features (e.g., gene content) at the genus level. Through genome-wide intra- and interspecies comparisons, one can uncover the shared (core) or species-specific (dispensable) gene content that was gained or lost since divergence from the most recent common ancestor (14). High-quality reference genomes are currently available for five diploid species: woodland strawberry (Fragaria vesca) (15), Nōgō strawberry (Fragaria iinumae) (16), Nilgiri strawberry (Fragaria nilgerrensis) (17), creamy strawberry (Fragaria viridis) (18), and Tibet strawberry (Fragaria nubicola) (18).
In the present study, we aimed to resolve the phylogenetic relationships of most Fragaria diploid species, date the divergence times of species, estimate the genetic diversity of several key species, uncover the evolutionary dynamics of gene families associated with important agronomic traits, and elucidate the pan-genome of the genus Fragaria. We assembled de novo chromosome-scale reference genomes of five diploid strawberry species (Fragaria mandschurica, Fragaria daltoniana, Fragaria pentaphylla, F. nilgerrensis, and F. viridis) using single-molecule sequencing technologies (Pacific Biosciences [PacBio] SMRT or Oxford Nanopore platforms). Furthermore, we generated genome resequencing data for 128 accessions spanning 10 species, including a newly described diploid species (Fragaria emeiensis Jia J. Lei) by us, and detected genome-wide polymorphisms and genetic structure in different species of Fragaria. Our study provides insights into the evolutionary history of strawberry, including a pan-genome estimate for strawberry, updated phylogenetic estimates of diploid species, and large-scale genomic resources for future studies.
Results and Discussion
Genome Assembly and Annotation.
The genomes of five diploid Fragaria species were sequenced using long-read technologies and assembled de novo using FALCON v0.3.0 (19) or wtdbg v2.4 (20). Among those, F. nilgerrensis, F. pentaphylla, and F. mandschurica were sequenced using PacBio, and F. daltoniana and F. viridis were sequenced using Oxford Nanopore (Table 1). Additional short-fragment Illumina libraries (250 bp and 450 bp, or 350 bp) were constructed and sequenced for each of the five species. These high-quality short reads were used to estimate the genome size and for correcting sequencing errors in the PacBio and Nanopore assemblies. The predicted genome sizes for the five species ranged from 229.61 Mb (F. viridis) to 305.90 Mb (F. nilgerrensis); and the assembled genome sizes ranged from 223.08 Mb (F. viridis) to 288.97 Mb (F. daltoniana) with sequencing depths ranging from 246.81X (F. daltoniana) to 425.33X (F. viridis). The contig N50 size was highest for F. viridis (9.83 Mb) and lowest for F. pentaphylla (0.91 Mb) (Table 1).
Table 1.
Genome assembly and annotation of five newly sequenced species and F. iinumae (16)
| Assembly parameters | F. iinumae | F. nilgerrensis | F. daltoniana | F. mandshurica | F. pentaphylla | F. viridis |
| Mating system | SC | SC | SC | SI | SI | SI |
| Predicted genome size (Mb) | 265.56 | 305.90 | 290.79 | 265.75 | 282.33 | 229.61 |
| Predicted heterozygous | 0.18% | 0.23% | 0.15% | 1.31% | 1.02% | 0.61% |
| Illumina reads (250 bp) | 14.21G | 13.72G | / | 15.25G | 19.41G | / |
| Illumina reads (450 bp) | 14.21G | 17.76G | / | 15.17G | 14.74G | / |
| Pacbio reads | 45.77G | 48.97G | / | 35.46G | 37.71G | / |
| Illumina reads (350 bp) | / | / | 10.06G | / | / | 46.37G |
| Nanopore reads | / | / | 61.71G | / | / | 51.29G |
| Total reads | 74.19G | 80.45G | 71.77G | 65.88G | 71.86G | 97.66G |
| Total sequence coverage depths | 279.37 | 262.99 | 246.81 | 247.9 | 256.82 | 425.33 |
| Genome coverage ≥ 4X | 99.80% | 98.13% | 98.21% | 99.78% | 89.82% | 94.48% |
| Assembled genome size (MB) | 240.58 | 288.43 | 288.97 | 239.83 | 279.04 | 223.08 |
| Total number contigs | 94 | 425 | 870 | 291 | 726 | 382 |
| Length of contig N50 (MB) | 10.67 | 3.16 | 4.29 | 1.29 | 0.908 | 9.83 |
| Number of contig N50 | 8 | 28 | 20 | 59 | 96 | 8 |
| Length of contig N90 (MB) | 3.13 | 0.68 | 0.89 | 0.46 | 0.26 | 1.96 |
| Number of contig N90 | 22 | 93 | 75 | 176 | 314 | 26 |
| Anchored chromosomes Size (MB) | 239.09 | 255.41 | 266.88 | 219.81 | 252.82 | 211.2 |
| Percent of anchored chromosomes | 99.38% | 88.55% | 92.35% | 91.96% | 90.60% | 94.67% |
| GC content | 39.70% | 39.70% | 42.76% | 38.50% | 42.76% | 38.16% |
| Gene numbers | 23,665 | 24,491 | 28,131 | 25,411 | 23,853 | 24,779 |
| BUSCO assessment | 94.80% | 94.50% | 93.30% | 90.70% | 83.40% | 94.50% |
SC: self-compatible; SI: self-incompatibility.
Utilizing a high-density linkage map of F. iinumae consisting of 4,173 markers (21), we successfully anchored the scaffolds of all five species into seven pseudomolecules, numbered according to the linkage group nomenclature (Fig. 1A and SI Appendix, Fig. S1). A total of 211.20 Mb (F. viridis) to 266.88 Mb (F. daltoniana) genome sequence was anchored to the linkage map, accounting for 88.55% (F. nilgerrensis) to 94.67% (F. viridis) of the total scaffolded sequence. The completeness of the five assembled genomes ranged between 83.4% and 94.8% when evaluated using Benchmarking Universal Single-Copy Orthologs (BUSCO) (22). The GC depth analysis revealed that all five genomes had a unimodal GC content distribution with an average GC content ranging between 38.16% and 42.76% (Table 1). Collectively, these results support a high-quality assembly for four species and one draft assembly for F. pentaphylla.
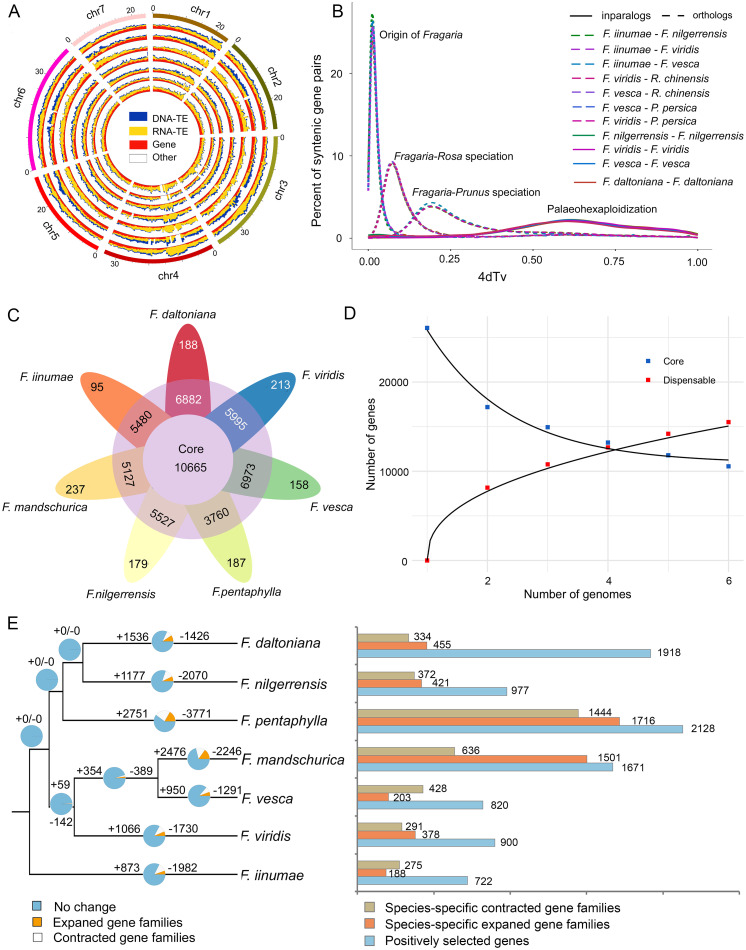
Fig. 1.
Evolution of seven diploid Fragaria genomes. (A) Circular representation of the comparative genome analysis of seven diploid Fragaria species (also see SI Appendix, Fig. S1). Collinear alignment was conducted with F. vesca as a reference. The outer layer of the colored blocks represents the seven chromosomes of Fragaria with tick marks every 5 Mb in size. Tracks displayed with seven genomes from outside to inside: F. vesca, F. daltoniana, F. iinumae, F. mandschurica, F. nilgerrensis, F. pentaphylla, F. viridis. The plots within each track exhibit densities of genes (red), RNA transposons (orange), DNA transposons (blue), and other types of genome components (white) from inside to outside, respectively. (B) 4DTv of in-paralogous (solid lines) and orthologous (dashed lines) genes of Fragaria spp. 4DTv of orthologous pairs between species are shown with different colors. (C) The flower plot displays the core orthogroups number (in the center), the orthogroups in a subset of species (in the annulus), and the species-specific orthogroups (in the petals) for the seven Fragaria species. (D) Modeling the core and dispensable Fragaria pan-genome. Square dots show the variation of shared gene content (core genome). Triangle dots show the variation of the species-specific gene content (dispensable genome). Curves are fitted separately for core and dispensable mean values shown in red. (E) Expanded and contracted gene families (Left), species-specific expanded and contracted gene families as well as positively selected genes (Right) in seven strawberry species.
Ab initio gene prediction programs, homology searches, and evidence-based (RNA-sequencing) analysis were integrated to annotate these genomes (Materials and Methods). In total, 23,665 to 28,131 protein-coding genes were predicted with the average gene length and average exons number per gene varying from 2648.36 to 2918.41 bp and from 4.88 to 5.14, respectively (SI Appendix, Table S1). In addition, 38.23 to 48.74% repetitive elements were annotated in these five assembled genomes by combining de novo and homology-based approaches. Among the annotated repeats, long-terminal repeat retrotransposons were the most abundant, accounting for 22.11 to 33.77% of the entire genome (SI Appendix, Table S1).
The synonymous substitutions at fourfold degenerate sites (4dTv) values showed that only one peak value corresponding to the paleo-hexaploidization event (Ks = ∼0.65) shared by the core eudicots was identified (23), suggesting that there have not been any additional whole-genome duplication (WGD) events in Fragaria. This was consistent with previous reports that other ancient WGDs are absent in Fragaria and that only Maloideae (apple subfamily) of Rosaceae share another ancient WGD event (24, 25). A divergence peak value (Ks = ∼0.03) was estimated for the origin of Fragaria (Fig. 1B).
Comparative Genomic Analysis of Multiple Strawberry Species.
We estimated the core and dispensable genome sizes by constructing and analyzing a pan-genome graph following the alignment of the genomes of six of the seven species using Progressive Cactus (26). The genome of F. pentaphylla was excluded from this analysis due to the overall fragmented state of the assembly. This analysis suggested that roughly 42.9% of genes (11,173 total) on average make up the core genome (Fig. 1D). When compared to the annotated gene content of individual genomes, the core genome size ranges from 10,271 genes in F. iinumae to 11,953 genes in F. mandschurica (SI Appendix, Fig. S2). Through this approach, annotated gene features present in a single reference genome were assessed if present or absent, if annotated or not, and present or not in each of the other five species genomes. We also compared the gene content of these six species in syntenic regions with SynMap (27, 28). This analysis suggests that roughly 44.4% of the genes are in the core genome. For both approaches, the largest reduction of shared genes comes from comparing any two genomes, with the slope and variance decreasing as additional genomes are added to the analysis. This estimate for the size of the core genome is similar to that previously estimated for Maize (29) and Brachypodium (30). Our analysis, together with previous studies (29, 30), thus suggests that roughly half of all genes in a particular angiosperm genome fall into either the core or dispensable portion of the genome.
Next, we performed ortholog clustering using OrthoFinder2 (31). A total of 25,687 orthogroups (roughly comparable with gene families) were identified across seven diploid Fragaria species: our five genomes, plus the genomes of F. vesca (15) and F. iinumae (16). A total of 10,665 orthogroups containing all species, 13,765 orthogroups containing a subset of species, and 1,257 species-specific orthogroups were identified. F. vesca and F. daltoniana shared 16,217 common orthogroups, which was the highest among paired species. All seven species had species-specific orthogroups (Fig. 1C and SI Appendix, Fig. S3). To investigate gene content changes among lineages, we examined the rates and direction of changes in orthogroup size among each of the species. Across the Fragaria phylogeny, most species have higher numbers of orthogroup contractions than expansions, except for F. daltoniana and F. mandschurica (Fig. 1E and SI Appendix, Table S2). Orthogroups in the F. pentaphylla genome exhibit the highest number of expansions and contractions. F. pentaphylla and F. mandschurica also showed a higher number of species-specific expanded orthogroups than other species (Fig. 1E).
Finally, we aimed to identify and functionally characterize genes under positive selection in each Fragaria species. Of the 19,135 orthologous genes from 10,665 orthogroups, we identified between 722 and 2,128 genes in each species that show signs of positive selection (referred to as PSGs) (Fig. 1E). These PSGs were enriched in various gene ontology functional categories. For example, F. daltoniana, which is adapted to high altitude (>3,200 m) regions of Southwest China, has a group of genes significantly enriched in categories associated with reproduction and stress response, such as the “ubiquitin-dependent ERAD pathway,” which is associated with abiotic stress response (e.g., drought, cold, and heat) (32) (SI Appendix, Table S3).
Phylogenetic Tree and Dating of the Genus Fragaria.
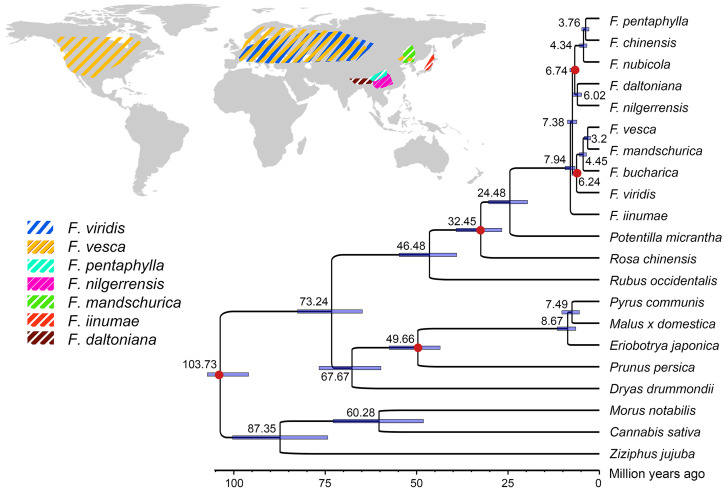
The phylogenetic relationships of species in the genus Fragaria have remained controversial. Based on 1,007 conserved single-copy nuclear genes, we estimated the relationships for the 10 diploid strawberry species, 8 additional Rosaceae species, as well as Ziziphus jujuba, Morus notabilis, and Cannabis sativa as outgroups (Fig. 2). The phylogenetic tree clearly indicated two major clades: F. pentaphylla, Fragaria chinensis, F. nubicola, F. daltoniana, and F. nilgerrensis, most of which are endemic to Southwest China, cluster together into one clade, while F. vesca, F. mandschurica, Fragaria bucharica, and then F. viridis are grouped into another clade, which roughly corresponds to south and north clades, respectively, in the phylogenetic tree established based on chloroplast genomes (33). F. iinumae, an extant relative of one of the diploid progenitors of the octoploid cultivated strawberry (16), is the sole species sister to these two clades. These phylogenetic relationships are consistent with previous studies of Fragaria except for F. viridis and F. nilgerrensis, the phylogenetic relationship of which were not well resolved previously using transcriptome and plastome sequences (6, 7). The topology of this phylogeny is also consistent with the geographic distribution of the species in each clade (Fig. 2).
Fig. 2.
Geographic distribution and phylogeny of seven diploid Fragaria species and outgroup species. (Upper Left) Geographic distribution of seven diploid Fragaria species with available reference genomes from this and our previous study (15, 16). Mapped ranges are adapted from Global Biodiversity Information Facility, Chinese Virtual Herbarium, and our own collections. (Lower Right) The phylogenetic tree of Rosales with three species (C. sativa, M. notabilis Z. jujuba) used as outgroups. The estimated divergence times are shown above branch nodes. Red dots mark the fossil calibration points used to estimate the divergence times.
We also estimated the age of divergences across the Fragaria phylogeny, using five fossils that possess synapomorphies of their respective clades as crown group calibrations (34–36), including the minimum age (2.9 Mya) for the root node of the north clade. Furthermore, a newly discovered achenes fossil of Fragaria from the late Pliocene (2.6 to 3.6 Mya) in Yunnan, Southwest China (36) suggested that the south clade, at least, had originated before 2.6 Mya (the minimum age for the root node of the south clade). Based on these fossils, our estimate of the origin of Fragaria, 7.94 (6.5 to 10.3) Mya (Fig. 2), is consistent with our dating of Fragaria (7.99 Mya) using transcriptome sequences (6). Moreover, the age of Fragaria estimated in this study is also overlapping with that estimated recently using a phylogenomic tree of Rosaceae (6.37: 5.54 to 8.38 Mya) (18). A previous evolutionary study of Rosaceae based on nuclear genomes using 124 species with 19 fossil constraints (37) indicated that the time of origin of Fragaria is ∼13 Mya. Another phylogenetic study of Rosaceae using 142 plastomes (35) showed that the divergence time between Fragaria and closely related genera (Chamaerhodos, Drymocallis, Potaninia, and Sasiphora) is ∼32 Mya. These estimated times of origin and divergence of Fragaria are older than some other estimations (from 2.12 to 3.57 Mya) using relatively few plastid genomes of Rosaceae (7, 33, 34). These differences may be caused by utilizing different fossil calibrations, choice of maximum constraints for calibrated nodes, different numbers of samples, as well as genes encoded on the plastid and nuclear genomes evolving at different rates.
Furthermore, our estimated date of the divergence between the south and north clade of Fragaria is 7.38 (6.3 to 8.8) Mya (Fig. 2). The south clade, which contains species mostly distributed throughout mountains of Southwest China, subdivided further into two subclades, ∼6.74 Mya. This age estimate coincides with dramatic elevational and climatic changes (e.g., monsoon intensification and cooling) associated with geological events in the Tibet-Himalaya-Hengduan region of Southwest China between the late Miocene and Pliocene (38, 39), suggesting that the differentiation of microhabitats associated with elevational gradients may have contributed to the diversification of Fragaria in this region.
Population Genetic Structure and a New Species Discovery in Fragaria.
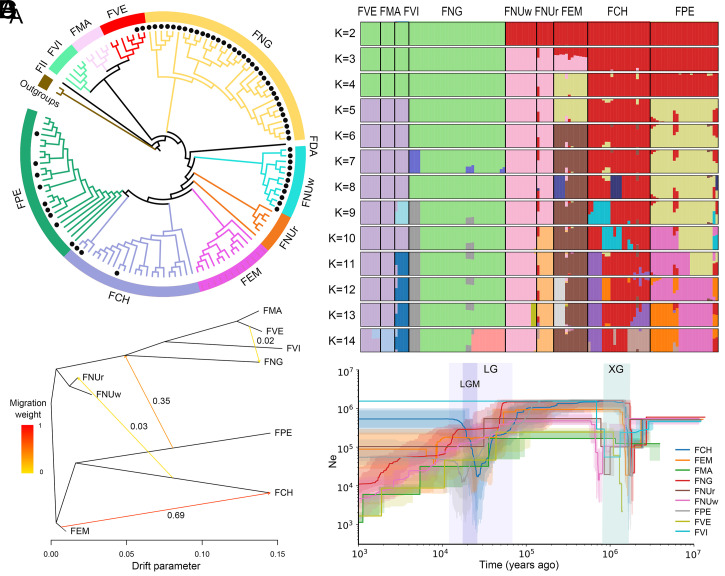
In Fragaria, the lack of many distinguishing phenotypic features between different species makes classification difficult. Furthermore, the classification of some species has been controversial due to lack of adequate genetic evidence. Here, we generated genome resequencing data for 128 individuals spanning 10 diploid species (including one candidate new species discovered in this study, F. emeiensis Jia J. Lei; see below) to investigate genome-wide nucleotide diversity (SI Appendix, Table S4). After quality-control filtering, we obtained a total of 910.5 Gb clean data and mapped these data to the F. vesca reference genome (15). After mapping, 25.50-fold average coverage depth per diploid sample was obtained. The mapping rate of individual libraries varied from 61.19 to 96.13% (average 85.31%) (SI Appendix, Table S4). As expected, seven individuals of F. vesca had the highest mapping rates (93.54 to 96.13%). The species F. nubicola, F. nilgerrensis, and F. chinensis, endemic to East Asia, had the lowest mapping rates (>61.19%). The differences in mapping rates are mainly due to genetic divergence between these species and the reference F. vesca genome. Using a set of stringent criteria (Materials and Methods), we retained 1,907,898 high-quality single nucleotide polymorphisms (SNPs) from 19,938,809 initially identified SNPs for further analysis. We estimated phylogenetic relationships of all samples based on the whole-genome polymorphisms using the maximum likelihood (ML) method (Fig. 3A and SI Appendix, Fig. S4). The SNP data grouped accessions into 10 clusters mirroring species designations, except for 3 individuals of F. chinensis, which were intermingled with F. pentaphylla. This analysis suggests that F. vesca, F. mandschurica, and F. viridis are genetically closer related to each other than to other species. Similarly, F. chinensis, F. pentaphylla, F. nubicola, and F. emeiensis clustered together and overlapped in principle components analysis (PCA) (SI Appendix, Fig. S5). The topology of major clades presented in Fig. 3A is nearly identical with our species tree estimated with conserved single-copy nuclear genes (Fig. 2) and previous estimates (6, 7, 40). However, the species distributed in Southwest China (e.g., F. pentaphylla, F. chinensis, F. nubicola, F. daltoniana, and F. nilgerrensis) formed two subclades (Fig. 2), but were successive sisters in the phylogenetic tree estimated using SNP data (Fig. 3A).
Fig. 3.
Phylogenetic tree and structure of 128 samples from key diploid species in Fragaria. (A) ML tree of diploid species in Fragaria with two outgroups based on whole-genome polymorphisms. Black dots show the white fruit type accessions. (B) Structure bar plots showing the assignment probabilities. (C) Estimated gene flow between eight species. Heat-map colors represent the migration weight for each pairwise comparison. (D) Demographic history of eight species in Fragaria. FCH: F. chinensis; FDA: F. daltoniana; FEM: F. emeiensis; FII: F. iinumae; FMA: F. mandschurica; FNG: F. nilgerrensis; FNUr: red fruit type of F. nubicola; FNUw: white fruit type of F. nubicola; FPE: F. pentaphylla; FVE: F. vesca; FVI: F. viridis; XG: Xixiabangma glaciation.
It is worth noting that, based on the whole-genome SNPs, a new species, named F. emeiensis Jia J. Lei, was identified in this study (SI Appendix, Figs. S6 and S7, Taxonomical treatment). F. emeiensis is most similar to F. nubicola, F. chinensis, and F.pentaphylla in morphology, which is also supported by the phylogenomic results (Fig. 3A and SI Appendix, Fig. S4). However, F. emeiensis has unique and stable morphological features and can be distinguished from related species. Plants of F. emeiensis are robust with thick petiole and runner, and leaves are also thick with deep veins, which separate it from its closely related species F. nubicola. Different from F. chinensis and F. viridis with trifoliate leaves, F. emeiensis has pinnately quinquefoliolate leaves. In addition, petioles, runners, and peduncles of F. emeiensis are covered with appressed hairs, while those of F. pentaphylla are covered with spreading hairs. Another trait differing F. emeiensis from F. pentaphylla is the color of abaxial leaf. The abaxial leaf of F. emeiensis is light green, while that of F. pentaphylla is green to purplish. Therefore, the identification of this new species (F. emeiensis) is well supported by both genetic and phenotypic data. F. emeiensis is diploid with the standard karyotype (2n = 2x = 14), and named F. emeiensis because the type specimen was discovered in the Emei Mountains within the Sichuan Province in Southwest China. Our study confirmed that the genome-wide phylogenomic analysis can discriminate very closely related organisms and uncover cryptic species (9, 10).
We further estimated individual ancestry assignment using ADMIXTURE (41). An optimum population K = 9 was recovered with smallest cross-validation error and increasing K values to 14 provided additional resolution by identifying intraspecific structure (Fig. 3B). The results also showed that F. chinensis, F. pentaphylla, F. nubicola, and F. emeiensis had a mixed pattern of ancestry, suggesting recent introgression or incomplete lineage sorting taking place between these closely related species (Fig. 3B). Both recent introgression and incomplete lineage sorting could be invoked to explain the relative low levels of divergence among these four species revealed by pairwise FST values (from 0.151 to 0.203) (SI Appendix, Table S5). We additionally used the TreeMix program (42) to estimate the migration direction and weight of ancestral gene flow among these species based on allele frequencies (SI Appendix, Fig. S8). Consistently, the result suggested four migration edges and supported that strong ancestral gene flow was detected from F. emeiensis to F. chinensis (migration weight = 0.69), as well as from F. pentaphylla to the root of F. nilgerrensis, F. vesca, F. mandschurica, and F. viridis (migration weight = 0.35) (Fig. 3C). The gene flow between these species may have been important in facilitating Fragaria’s rapid diversification into diverse habitats.
Genetic Diversity, Demographic History, and Natural Selection in Key Species.
We selected 126 samples that span the major eight species (excluded F. daltoniana and F. iinumae) for further genetic diversity, selective sweep, linkage disequilibrium (LD) decay, and demographic history analyses (Fig. 3D and SI Appendix, Figs. S9 and S10 and Table S5). The level of nucleotide diversity (θπ) varied across the eight species, ranging from 0.007 (F. vesca) to 0.024 (F. viridis) (SI Appendix, Table S5). The lowest θw and θπ were detected in F. vesca, which was consistent with low levels of heterozygosity previously reported using microsatellite markers (43). The low level of genetic diversity might be explained by high selfing rates of F. vesca. Furthermore, the mean Tajima’s D values were negative in F. chinensis, F. emeiensis, F. pentaphylla, and F. viridis (from −0.075 to −0.216) (SI Appendix, Table S5), indicating potential population expansions in these species. Accordingly, the LD plots of F. chinensis, F. emeiensis, and F. pentaphylla exhibited faster decay (SI Appendix, Fig. S9), suggesting a large ancestral population size or that population expansion has occurred in these species (44).
Demographic history analysis using stairway plots revealed that all species have undergone a first bottleneck during the Xixiabangma glaciation (1.17 to 0.8 Mya). After the retreat of the Xixiabangma glaciation, the effective population size (Ne) of all eight species expanded and reached its peak. Then, a second bottleneck (∼70 Kya) (Fig. 3D) was detected during the last glaciation (LG, 11.5 to 70.0 Kya), especially during the LG maximum (LGM, 19.0 to 26.5 Kya), in all Fragaria species except F. viridis. Following the end of the LG, F. viridis, F. chinensis, F. pentaphylla, and F. emeiensis maintained a relatively high Ne, while the Ne of F. vesca, F. mandschurica, and F. nilgerrensis continued to decline. These demographic history estimations are also consistent with the negative Tajima’s D values in the former four species and the positive Tajima’s D values in the latter three species (SI Appendix, Table S5).
It is worth noting that two population substructures were detected in F. nubicola: one comprising individuals with red fruits and the other comprising individuals with white fruits (Fig. 3 A and B). The FST value between both fruit types of F. nubicola (FST = 0.177) was even higher than that of different species (e.g., F. pentaphylla vs. F. chinensis vs. F. emeiensis) (SI Appendix, Table S5), suggesting that these two fruit types of F. nubicola possibly are in the early stages of speciation. Identification of the signatures of a selective sweep with highest 5% FST values, as well as the highest 5% of θπ ratio, enable detection of loci that have undergone natural selection between these two fruit types of F. nubicola (SI Appendix, Fig. S10). The selective sweep regions contained more than 20 genes related to stress or disease resistance (e.g., pentatricopeptide repeat genes) genes were detected in white fruit type. The gene encoding “flowering time control protein FY” also has undergone selection between both types of F. nubicola. The 12 nucleotide deletion of FY that altered the original start codon ATG > GTG resulted in the subsequent 10 amino acids not being translated, which was uncovered in all white fruit individuals of F. nubicola only (SI Appendix, Fig. S11). Flowering time control protein (FCA) and FY function together in a complex that regulates flowering time in the autonomous flowering pathway by downregulating the floral repressor FLC (encoding FLOWERING LOCUS C) mRNA level (45). This is consistent with our observation that white fruited types flowered 2 wk later than red fruited types in our strawberry germplasm resources nursery in Shenyang, China. This difference in flowering time may have led to potential prezygotic reproductive isolation, which may explain the observed high genetic differentiation between these two fruit types.
Multiple Independent MYB10 Mutations Cause Fruit Color Variation in Wild Strawberries.
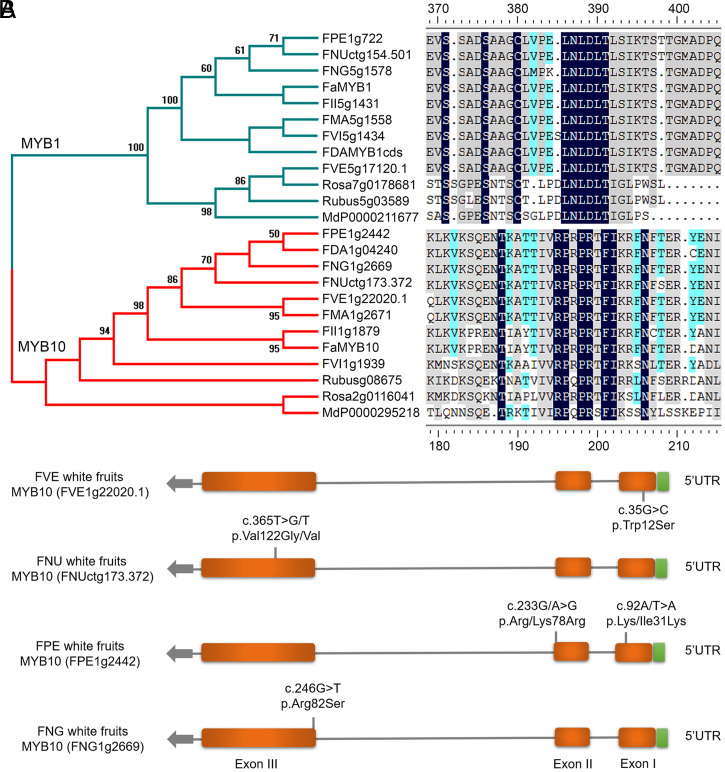
There are white fruited individuals in several wild strawberries (e.g., F. vesca, F. nubicola, F. pentaphylla, and F. nilgerrensis). The genes encoding R2R3-MYB transcription factors regulate the spatial and temporal expression of flavonoid genes in plants (46), and are important for fruit color in strawberry (47). The total gene number of the R2R3-MYB gene family was highest in F. vesca (138) and lowest in F. daltoniana (119) (SI Appendix, Fig. S12). FaMYB1, belonging to the R2R3-MYB family, has been shown to suppress the biosynthesis of flavonoids in cultivated strawberry (48). In contrast, FaMYB10 is known to promote the biosynthesis of anthocyanins in cultivated strawberry (49). To dissect the cause of white fruit phenotype in Fragaria, we first identified the candidate orthologs of FaMYB1 and FaMYB10 in the genome of the eight Fragaria species. Our phylogenetic analysis revealed that MYB1s and MYB10s from each species formed two distinct clusters (Fig. 4A), and all the MYB1s share the repressor motif “LNLDLTLS” (50), while all the MYB10s contained the promoting anthocyanin biosynthesis motif of “RPRPRTFII” (51). Previous studies have shown that fruit color mutants in strawberries were mostly associated with MYB10 (49, 52, 53). To reveal the cause of fruit color transition, we used the resequencing data to detect the polymorphisms of MYB10 between white and red fruited individuals for each species (Fig. 4B and SI Appendix, Fig. S13).
Fig. 4.
Evolution of the R2R3-MYB gene family in Fragaria. (A, Left) Phylogenetic tree of MYB1 and MYB 10. (Right) Conserved anthocyanin-promoting motif “RPRPRTF” of MYB10 and conserved anthocyanin-repressive motif “LNLDLTLSIKT” of MYB1. Similar residues are highlighted, with the homology level ranging from black (100% identity), grey (>75%), to light blue (>50%). (B) Nucleotide and amino acid variation of MYB10 between red and white fruits in four wild strawberry species. Fragaria × ananassa (Fa), F. daltoniana (FDA), F. iinumae (FII), F. mandschurica (FMA), F. nilgerrensis (FNG), F. pentaphylla (FPE), F. vesca (FVE), F. viridis (FVI), M. domestica (Md), R. chinensis (Rosa), and R. occidentalis (Rubus).
By comparing the white fruit (Hawaii4) and red fruit accessions of F. vesca, we found a single mutation (35G > C) causing an amino acid change (Trp12Ser) in the MYB10 of Hawaii4 accessions, which was also reported to be responsible for the loss of anthocyanins and the pale color of “Yellow Wonder” fruits of F. vesca (52). In F. nubicola, one nonsynonymous mutation (c.365T > G, p.Val122Gly) was exclusively detected in the third exon of the MYB10 gene of white fruited accessions, with only two individuals with white fruits harboring allelic heterozygous mutations (c.365T > G/T) (Fig. 4B). We speculate that the mutation in the MYB10 gene in white fruit accessions of F. nubicola is dominant. Unlike F. nubicola, two allelic heterozygous mutations (c.92A/T > A, p.Lys/Ile31Lys; c.233A/G > G, p.Lys/Arg78Arg) were detected in all the red fruit accessions of F. pentaphylla (Fig. 4B). In addition, F. nilgerrensis only has the white fruit type. This phenotype was recently reported to be caused by sequence variations in the promoter of MYB10 (17). In our analysis, one nonsynonymous mutation (c.246G > T, p.R82S) in the R2R3 motif of MYB10 gene was exclusively fixed in all accessions of F. nilgerrensis compared to F. vesca, suggesting this mutation also might be responsible for the loss of anthocyanins in F. nilgerrensis fruit. Although further experimental verification is needed, our results suggest that these multiple independent MYB10 mutations in wild strawberries may explain the fruit color transition from dark red and pink to fully white fruits in Fragaria (54).
Conclusion
Here, we present chromosome-scale genome assemblies for five strawberry species (F. daltoniana, F. mandschurica, F. pentaphylla, F. viridis, and F. nilgerrensis) as a resource to the community. The analyses of these five genomes, combined with other available genomes and resequencing datasets, allowed us to infer the phylogenetic relationships for most diploid species in the genus Fragaria, date the divergence of various Fragaria species and the origin of Rosaceae, identify gene families associated with important agronomic traits, estimate the genetic diversity of key strawberry species, and identify selective sweeps that may be associated with certain traits. Furthermore, our analyses uncovered a new diploid species (F. emeiensis Jia J. Lei). We anticipate that these reference genomes and datasets, combined with our phylogenetic estimates of species relationships, is the phylogenomic framework needed to elevate Fragaria as a true model system for evolutionary genomics.
Materials and Methods
Sample Collection and Genome Sequencing.
Healthy young leaves of five Fragaria species collected from the strawberry germplasm resources nursery in Shenyang Agricultural University and Yunnan University were used for high-quality genomic DNA extraction. For Illumina sequencing, paired-end libraries with insert sizes of 250 bp, 450 bp, or 350 bp were constructed and sequenced on an Illumina HiSeq X Ten platform (Illumina). Genome size and heterozygosity of the five species were estimated using the k-mer statistics (55) based on the Illumina HiSeq reads.
For PacBio sequencing, a SMRTbell DNA library was prepared and sequenced according to the manufacturer’s protocols (Pacific Biosciences), and a 20-kb SMRTbell library was generated using a BluePippin DNA size-selection instrument (Sage Science) with a lower size limit of 10 kb for each species. For Nanopore sequencing, 30- to 80-kb genomic DNA fragments were selected with BluePippin (Sage Science) and processed according to the Ligation Sequencing Kit 1D (SQK‐LSK109) protocol. The final library was sequenced on different R9.4 flow cells using the PromethION DNA sequencer (Oxford Nanopore Technologies).
Total RNA was extracted from leaves, flower, and fruit tissues of each species using the Qiagen RNeasy Plant Mini Kit (Qiagen). RNA-sequencing libraries were then prepared using the Illumina TruSeq RNA Library Preparation Kit and paired-end sequencing with a read length of 150 bp was conducted on the HiSeq. 2000 platform. The transcriptome data were used for genome annotation.
Genome Assembly.
For PacBio sequences, genome assembly was performed on full PacBio long reads using FALCON v0.3.0 (19). Error correction and preassembly were carried out with the FALCON pipeline after evaluating the outcomes of using different parameters in FALCON. For the Nanopore data, reads with mean quality scores >7 were retained and further corrected with NextDenovo (https://github.com/Nextomics/NextDenovo) and then assembled into contigs by program wtdbg v2.4 (20). The draft genome was polished with Pilon v1.22 (56) using the Illumina reads under the default settings. A GC depth analysis was conducted to assess potential contamination during sequencing and to assess the coverage of the assembly. The completeness of the genome assembly was also evaluated using the BUSCO software (22).
Previously a high-density linkage map of F. iinumae was constructed by 4,173 markers, with 3,280 identified with an array and 893 from genotyping by sequencing (21). Here we anchored these markers to the genome of five Fragaria species. The assembled scaffolds of these species were aligned and oriented to the F. iinumae reference linkage map. If the scaffold mapped to more than one linkage group on the genetic map, it finally was anchored to the linkage group with most markers.
Genome Annotation.
Homology-based, de novo-based, and RNA-sequencing–based gene prediction methods were used in combination to identify the protein-coding genes. For homology-based predictions, protein sequences of five species including Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, F. vesca, and Malus domestica were used as references. Augustus v2.4 (57), GlimmerHMM v3.0.4 (58), SNAP v2006 (59), GeneID v1.4 (60), and Genscan (61) with default parameters were used for de novo-based gene prediction.
For the RNA-sequencing–based prediction, TransDecoder v2.0 (transdecoder.github.io.), GeneMarkS-T v5.1 (62), and PASA v2.0.2 (63) were used. Finally, the results from the three methods were integrated using EVM v1.1.1 (64). All the genes were annotated by aligning to the NR database, Swiss-Prot, and Kyoto Encyclopedia of Genes and Genomes (KEGG database release 84.0). Then, the InterProScan (65) package was used to annotate the predicted genes using the InterPro database.
For repeat detection, four software packages—RepeatModeler (66) (www.repeatmasker.org/RepeatModeler/), RepeatScout (67), Piler (68), and LTR-Finder (69)—were used to build a de novo repeat library based on our assembly with the default settings. To identify known transposable elements in the genomes, RepeatMasker (66) was used to screen the assembled genome against the Repbase v22.11 (70) and Mips-REdat libraries (71).
Genome Evolution.
To identify syntenic regions within and between genomes, homologous gene pairs between two pairs of seven Fragaria species were identified using BLASTP, and high-confidence collinear blocks were determined using MCScanX (72). Collinear alignment was conducted with F. vesca as a reference. For each gene pair in a syntenic block, the amino acid alignments were conducted by MUSCLE (73) to obtain the conserved protein sequence of each species, which were then reverse-translated to the corresponding codon-based nucleotide alignments by PAL2NAL (74). Finally, nonsynonymous (Ka) and synonymous (Ks) substitution values were calculated by KaKs_Calculator (75). The 4dTv (transversion substitutions at fourfold degenerate sites) distances were calculated to identify putative whole-genome duplication events as well as divergence times between species. Expansions and contractions of orthologous gene families were determined using OrthoFinder2 (31) and CAFÉ v3.0 (76) programs.
Phylogenetic Tree Construction.
To estimate the evolutionary relationships of these species, ortholog clustering analysis was performed using OrthoFinder2 (31) on all the protein-coding genes of 10 Fragaria diploid species and 11 other sequenced Rosales species (Malus x domestica, Pyrus communis, Prunus persica, Rosa chinensis, Rubus occidentalis, Potentilla micrantha, Eriobotrya japonica, Dryas drummondii, M. notabilis, C. sativa, Z. jujuba). Among these 10 Fragaria species, translated transcriptomes for F. bucharica and F. chinensis, and whole-genome sequence data for other 8 species were used. Orthogroups where at least 94.4% of the species had single-copy genes in an orthogroup were selected and sequences aligned by MAFFT (77). Phylogenetic trees based on ML were conducted by IQ-TREE (78) with the JTT+F+R3 model and 1,000 bootstraps. Finally, divergence times were estimated based on one-to-one orthologs using Bayesian inference as implemented in MCMCtree of PAML 4.9 (79) with the options “independent rates” and “GTR” model. A Markov chain Monte Carlo analysis was run for 10,000 generations, using a burn-in of 1,000 iterations. Five time-calibrated points were used based on fossil records, including the Crown Rosales (106.5 to 90 Mya), Stem Prunus (>47.8 Mya), and Stem Rosa (>47.8 Mya) (38), the root node of the north clade of Fragaria (>2.9 Mya) (34), and the root node of south clade (>2.6 Mya) (36).
Pan-Genome Modeling.
Because the overall fragmented state of the assembly of the F. pentaphylla genome, this species was excluded from the Fragaria pan-genome analysis. Core and dispensable genomes size estimates for Fragaria based on the comparison of six species: F. vesca, F. daltoniana, F. iinumae, F. mandschurica, F. nilgerrensis, and F. viridis. These six genomes were aligned to each other using Progressive Cactus (v0.1 August 2017) (26) with default parameters and the representative Newick tree ((((vesca:1, mandschurica:1)Anc4:1,viridis:1)Anc2:1,(daltoniana:1,nilgerrensis:1)Anc3:1)Anc1:1,iinumae:1)Anc0. The resulting multiple alignment file (hal) was used with HalLiftover to convert the genome coordinates of each gene, where possible, to the corresponding location in each of the other Fragaria genomes to determine the subset of core genes without an explicit reference bias. For the F. vesca genome, the most recent annotation 4.0.a2 (80) was used, while for all other genomes the annotation accompanying each build was used. Before genome coordinate conversion, the gff3 file for each reference was filtered for the first transcript model for each gene and converted to a GenePred using gff3ToGenePred (kentUtils, https://github.com/ENCODE-DCC/kentUtils). The GenePred was then sorted and converted to a bed format using genePredToBed (https://github.com/ENCODE-DCC/kentUtils). This generated a bed-12 file that described the intron-exon structure of each gene as annotated in the gff3. These bed files were lifted over using the bed-12 compatible utility halLiftover from the Comparative Genomics Toolkit package (https://github.com/ComparativeGenomicsToolkit/hal) with options –noDupes (to retain primarily syntenic alignments) (26) and –inMemory. A gene in a reference genome was considered to have been successfully identified in the nonreference genome if it both mapped from and to a nonzero pseudomolecule (chromosome zero was used in some genomes as an aggregation of unplaced contigs) and the pseudomolecule ID assigned according to broad synteny was the same in the reference and nonreference. A successful identification also required the sum of the exon sizes of the identified gene in the nonreference to be at least 80% that of the exons in the gene-model in the reference. In conjunction with the requirement that the genes are discovered on the corresponding orthologous chromosome, these requirements removed heavily degraded genes as well as mappings to anciently diverged paralogous sequences (e.g., paleo-hexaploid event shared with Arabidopsis). Curves modeling the mean number of core and dispensable genes were fit separately in R using the ‘nls’ function. Code to reproduce this analysis can be found at: https://github.com/Aeyocca/Edger-Lab/blob/master/Fragaria_pangenome_modeling.Rmd
Signs of Positive Selection.
A total of 19,135 orthologs from seven Fragaria diploid species were aligned. Genes showing signs of positive selection were identified based on likelihood-ratio tests of nonsynonymous to synonymous substitution rates. Substitution rates were estimated using the branch-site model of the codeml program in the PAML v4.9 package (79). For genes of each species tested for positive selection, the P value was measured by likelihood-ratio tests and corrected with false-discovery rate (FDR < 0.01). The functional annotation of positively selected genes was conducted by gene ontology and the KEGG database.
Population Sample Collection, Resequencing, and SNP Calling.
We collected 128 samples of 10 species (including one candidate new species discovered in this study, F. emeiensis Jia J. Lei) with two outgroups (Duchesnea indica and Rubus corchorifolius) from our strawberry germplasm resources nursery in Shenyang and Yunnan. Sequencing libraries were prepared from each sample. DNA was sequenced by standard procedures on an Illumina HiSeq. 2500 platform. Raw data in fastq format was first processed through a series of quality-control procedures. Quality-control standards were the following: 1) removing reads with ≥10% unidentified nucleotides (N); 2) removing reads with >50% bases having phred quality <5; 3) removing reads with > 10 nt aligned to the adapter; 4) removing putative PCR duplicates generated in the library construction process.
After trimming low-quality bases, paired-end reads of each sample were mapped to the F. vesca reference genome (15) using Burrows-Wheeler Aligner (BWA) software (81) with parameters of mem -t 4 -k 32 -M -R. Alignment files were converted to BAM files using SAMtools software (82). If multiple read pairs had identical external coordinates, only the pair with the highest mapping quality was retained. Variants calling were performed for all samples using the UnifiedGenotyper function in GATK software (83). The filter parameters of high-quality SNP were used as follows: sequencing depth: dp 8; deletion rate: Miss 0.2; and minimum allele frequency: maf 0.01. ANNOVAR (84) was used to annotate SNPs based on the GFF3 files for the reference genome.
Population Genetic Structure, Genetic Differentiation, and Demographic History.
SNPs with LD were pruned (–indep-pairwise 50 10 0.0575) using PLINK software (85). We conducted the ML tree of 128 individuals with two outgroups (D. indica and R. corchorifolius) using IQ-TREE software (78) with 1,000 bootstraps. The best model was TVMe+ASC+R3 (the ASC model is specific used for SNPs data). Then, ADMIXTURE (41) was used to estimate the genetic ancestry of 126 samples (the single individual species F. daltoniana and F. iinumae were discarded for this analysis), specifying a range of K = 2 to 14 hypothetical ancestral populations. The smallest CV error value and the first inflection point occurred when K = 9. The PCA was performed with population-scale LD filtered SNPs using GCTA (86). A sliding-window approach (10-kb windows sliding in 5-kb steps) was applied to quantify polymorphism levels (θπ), genetic differentiation (FST), and selection statistics (Tajima’s D) between key species.
TreeMix (42) was also used to create a phylogeny of these species. We used the -global option and -se option to calculate SEs of migration proportions and the -noss option to prevent overcorrection for species with small sample sizes. Then, the OptM R package (https://cran.r-project.org/web/packages/OptM) was used to determine the optimal number of migration edges. LD decay analysis based on the coefficient of determination (r2) was calculated between each pair of SNPs using PopLDdecay (87).
The demographic history of each species was inferred by the trend in Ne change over time using Stairway Plot (88). We estimated the mutation rate of strawberry using fourfold degenerate sites which are considered as putatively neutral sites by MCMCTREE of PAML 4.9 (79). The results showed a mutation rate for strawberry of about 2.8 × 10−9 per site per year. For demographic history analysis with stairway plot, we only used neutral sites which excluded gene regions and upstream and downstream 2,000-bp sequences of genes.
Analysis of Transcription Factors MYB1 and MYB10 in Fragaria.
To identify members of the R2R3-MYB gene family in eight diploid Fragaria species, the hidden Markov model (HMM) profile for the MYB binding domain (PF00249) retrieved from the Pfam 3.0 database (pfam.xfam.org/) was used to search the Fragaria genomes using the hmmscan program of HMMER (hmmer.org/). First, the known MYB1s and MYB10s in strawberry-related species (48, 49) were retrieved from National Center for Biotechnology Information (NCBI) databases. Then they were used as query in BLASTP or aligned to construct the HMM profiles to search for the potential Fragaria MYB1s and MYB10s. Finally, the entire protein sequences, which were further confirmed by Pfam, were retained and aligned by the MAFFT (77) and DNAMan system (Lynnon Biosoft). The ML tree was constructed using RAxML (89) with the PROTGAMMAJTT model and estimated clade support with 100 rapid bootstrap replicates. In order to detect polymorphisms in coding regions of MYB10 between white and red fruit phenotypes, we mapped the short reads of resequencing data from both white and red fruits phenotypes accessions to the reference of each corresponding Fragaria genome, using the standard BWA-mem (81) and SAMtools (82) pipeline. The detected mutations were visualized and evaluated using the Integrative Genomics Viewer (90).
Acknowledgments
We dearly cherish the memory of our respected mentor and friend Prof. Yang Zhong at Fudan/Tibet University for his full support to this project. We thank Rengang Zhang, Quanzheng Yun, and Huanchong Wang for their helpful discussion. This work is supported by National Natural Science Foundation of China Grants 32060085 and 31760082 (to Q.Q.), 32060237 and 31770408 (to T.Z.), and 31760127 (to L.Q.); National Key Research and Development Project Grant 2019YFD1000800 (to J. Lei and L.X); Michigan State University AgBioResearch (P.P.E.), US Department of Agriculture-National Institute of Food and Agriculture (USDA-NIFA) HATCH 1009804 (to P.P.E.); USDA-NIFA AFRI 2020-67013-30870 (to P.P.E.); and National Science Foundation - PGRP 2029959 (to P.P.E.). Y.V.d.P. acknowledges funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (no. 833522) and from Ghent University (Methusalem funding, BOF.MET.2021.0005.01).
Footnotes
The authors declare no competing interest.
Reviewers: V.A.A., University at Buffalo, State University of New York; and E.A.K., Donald Danforth Plant Science Center.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105431118/-/DCSupplemental.
Data Availability
The raw genomic reads generated in this study have been deposited in the NCBI Sequence Read Archive (BioProject nos. PRJNA743176 and PRJNA757203). The genome assembly and annotation files are available at the Genome Database for Rosaceae (F. daltoniana: https://www.rosaceae.org/Analysis/11885161; F. pentaphylla: https://www.rosaceae.org/Analysis/12137892; F. mandschurica: https://www.rosaceae.org/Analysis/12137893; F. nilgerrensis: https://www.rosaceae.org/Analysis/12137894; F. viridis: https://www.rosaceae.org/Analysis/12137895).
References
- 1.Clark A. G., et al., Drosophila 12 Genomes Consortium, Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Stein J. C., et al., Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Jia N., et al., Tick Genome and Microbiome Consortium (TIGMIC), Large-scale comparative analyses of tick genomes elucidate their genetic diversity and vector capacities. Cell 182, 1328–1340.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Johnson A. L., Rajanikanth G., Tia‐Lynn A., Bioclimatic evaluation of geographical range in Fragaria (Rosaceae): Consequences of variation in breeding system, ploidy and species age. Bot. J. Linn. Soc. 176, 99–114 (2014). [Google Scholar]
- 5.Liston A., Cronn R., Ashman T. L., Fragaria: A genus with deep historical roots and ripe for evolutionary and ecological insights. Am. J. Bot. 101, 1686–1699 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Qiao Q., et al., Comparative transcriptomics of strawberries (Fragaria spp.) provides insights into evolutionary patterns. Front Plant Sci 7, 1839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Njuguna W., Liston A., Cronn R., Ashman T. L., Bassil N., Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol. Phylogenet. Evol. 66, 17–29 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Vergauwen D., De Smet I., The strawberry tales: Size matters. Trends Plant Sci. 24, 1–3 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Jarvis E. D., et al., Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Árnason E., Halldórsdóttir K., Codweb: Whole-genome sequencing uncovers extensive reticulations fueling adaptation among Atlantic, Arctic, and Pacific gadids. Sci. Adv. 5, eaat8788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novikova P. Y., et al., Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 48, 1077–1082 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Sun Y., Lu Z., Zhu X., Ma H., Genomic basis of homoploid hybrid speciation within chestnut trees. Nat. Commun. 11, 3375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng L., et al., Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 5, 4956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Q., et al., Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Edger P. P., et al., Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edger P. P., et al., Reply to: Revisiting the origin of octoploid strawberry. Nat. Genet. 52, 5–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., et al., The high-quality genome of diploid strawberry (Fragaria nilgerrensis) provides new insights into anthocyanin accumulation. Plant Biotechnol. J. 18, 1908–1924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng C., et al., Tracing the diploid ancestry of the cultivated octoploid strawberry. Mol. Biol. Evol. 38, 478–485 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin C. S., et al., Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan J., Li H., Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17, 155–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahoney L. L., et al., A high-density linkage map of the ancestral diploid strawberry, F. iinumae, constructed with single nucleotide polymorphism markers from the IStraw90 array and genotyping by sequencing. Plant Genome 9, 10.3835/plantgenome2015.08.0071 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Vekemans D., et al., Gamma paleohexaploidy in the stem lineage of core eudicots: Significance for MADS-box gene and species diversification. Mol. Biol. Evol. 29, 3793–3806 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Wu J., et al., The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S., An H., Xu F., Zhang X., Chromosome-level genome assembly and annotation of the loquat (Eriobotrya japonica) genome. Gigascience 9, giaa015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong J., et al., Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587, 246–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H., et al., SynFind: Compiling syntenic regions across any set of genomes on demand. Genome Biol. Evol. 7, 3286–3298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyons E., Pedersen B., Kane J., Freeling M., The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the Rosids. Trop. Plant Biol. 1, 181–190 (2008). [Google Scholar]
- 29.Hirsch C. N., et al., Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26, 121–135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon S. P., et al., Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 8, 2184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emms D. M., Kelly S., OrthoFinder2: Fast and accurate phylogenomic orthology analysis from gene sequences. BioRxiv [Preprint] (2018). 10.1101/466201. Accessed 24 April 2019. [DOI]
- 32.Liu L., et al., The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21, 957–969 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J., et al., Complete chloroplast genome sequencing of ten wild Fragaria species in China provides evidence for phylogenetic evolution of Fragaria. Genomics 113, 1170–1179 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Dillenberger M. S., Wei N., Tennessen J. A., Ashman T. L., Liston A., Plastid genomes reveal recurrent formation of allopolyploid Fragaria. Am. J. Bot. 105, 862–874 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Zhang S. D., et al., Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Huang Y. J., Zhu H., Momohara A., Jia L. B., Zhou Z. K., Fruit fossils of Rosoideae (Rosaceae) from the late Pliocene of northwestern Yunnan, Southwest China. J. Syst. Evol. 57, 180–189 (2019). [Google Scholar]
- 37.Xiang Y., et al., Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 34, 262–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding W. N., Ree R. H., Spicer R. A., Xing Y. W., Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578–581 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Xing Y., Ree R. H., Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseau-Gueutin M., et al., Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): New insights from phylogenetic analyses of low-copy nuclear genes. Mol. Phylogenet. Evol. 51, 515–530 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilmarsson H. S., et al., Population genetic analysis of a global collection of Fragaria vesca using microsatellite markers. PLoS One 12, e0183384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez R. D., et al., Demographic histories and patterns of linkage disequilibrium in Chinese and Indian Rhesus macaques. Science 316, 240–243 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Simpson G. G., Dijkwel P. P., Quesada V., Henderson I., Dean C., FY is an RNA 3' end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Feller A., Machemer K., Braun E. L., Grotewold E., Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Labadie M., et al., Metabolite quantitative trait loci for flavonoids provide new insights into the genetic architecture of strawberry (Fragaria × ananassa) fruit quality. J. Agric. Food Chem. 68, 6927–6939 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Salvatierra A., Pimentel P., Moya-León M. A., Herrera R., Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90, 25–36 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Medina-Puche L., et al., MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J. Exp. Bot. 65, 401–417 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Dubos C., et al., MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Stracke R., Werber M., Weisshaar B., The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Hawkins C., Caruana J., Schiksnis E., Liu Z., Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep. 6, 29017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castillejo C., et al., Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 32, 3723–3749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James F. H., Peter W. C., Sedat S. e., Phan Quynh S., Variation in the horticultural characteristics of native Fragaria virginiana and F. chiloensis from North and South America. J. Am. Soc. Hortic. Sci. 128, 201–208 (2003). [Google Scholar]
- 55.Liu B., et al., Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant. Biol. 35, 62–67 (2013). [Google Scholar]
- 56.Walker B. J., et al., Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanke M., Waack S., Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (suppl. 2), ii215–ii225 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Majoros W. H., Pertea M., Salzberg S. L., TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Bromberg Y., Rost B., SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35, 3823–3835 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blanco E., Parra G., Guigó R., Using geneid to identify genes. Curr. Protoc. Bioinformatics Chapter 4, Unit 4.3 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Burge C., Karlin S., Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Tang S., Lomsadze A., Borodovsky M., Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 43, e78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campbell M. A., Haas B. J., Hamilton J. P., Mount S. M., Buell C. R., Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7, 327 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haas B. J., et al., Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quevillon E., et al., InterProScan: Protein domains identifier. Nucleic Acids Res. 33, W116–W120 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarailograovac M., Chen N., Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics 25, 4.10.11–14.10.14 (2009). [Google Scholar]
- 67.Price A. L., Jones N. C., Pevzner P. A., De novo identification of repeat families in large genomes. Bioinformatics 21 (suppl. 1), i351–i358 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Edgar R. C., Myers E. W., PILER: Identification and classification of genomic repeats. Bioinformatics 21 (suppl. 1), i152–i158 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Xu Z., Wang H., LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bao W., Kojima K. K., Kohany O., Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nussbaumer T., et al., MIPS PlantsDB: A database framework for comparative plant genome research. Nucleic Acids Res. 41, D1144–D1151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y., et al., MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z., et al., KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259–263 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han M. V., Thomas G. W. C., Lugo-Martinez J., Hahn M. W., Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987–1997 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Li Y., Pi M., Gao Q., Liu Z., Kang C., Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 6, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H., et al., 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKenna A., et al., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang C. C., et al., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang J., Lee S. H., Goddard M. E., Visscher P. M., GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang C., Dong S. S., Xu J. Y., He W. M., Yang T. L., PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019). [DOI] [PubMed] [Google Scholar]
- 88.Liu X., Fu Y. X., Exploring population size changes using SNP frequency spectra. Nat. Genet. 47, 555–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw genomic reads generated in this study have been deposited in the NCBI Sequence Read Archive (BioProject nos. PRJNA743176 and PRJNA757203). The genome assembly and annotation files are available at the Genome Database for Rosaceae (F. daltoniana: https://www.rosaceae.org/Analysis/11885161; F. pentaphylla: https://www.rosaceae.org/Analysis/12137892; F. mandschurica: https://www.rosaceae.org/Analysis/12137893; F. nilgerrensis: https://www.rosaceae.org/Analysis/12137894; F. viridis: https://www.rosaceae.org/Analysis/12137895).