Significance
Many current gene editing techniques rely on use of a donor template to elicit correction. While functional correction remains the strongest indicator of successful editing, many candidates are initially screened via in vitro–based outcomes. Our work here indicates that peptide nucleic acids have an intrinsic capability to aggregate with single-stranded DNA donor templates, which may lead to false positives in PCR-based readouts. To mitigate this issue, we have validated several methods including size exclusion, nuclease digestion, and purification columns, which reversed the appearance of false signals. This is critical to enable efficient development of optimal gene editing agents, which could impact the treatment of many diseases ranging from severe combined immunodeficiencies to sickle cell anemia to cystic fibrosis.
Keywords: gene editing, peptide nucleic acid, hematopoietic stem cell, DNA repair
Abstract
Triplex gene editing relies on binding a stable peptide nucleic acid (PNA) sequence to a chromosomal target, which alters the helical structure of DNA to stimulate site-specific recombination with a single-strand DNA (ssDNA) donor template and elicits gene correction. Here, we assessed whether the codelivery of PNA and donor template encapsulated in Poly Lactic-co-Glycolic Acid (PLGA)-based nanoparticles can correct sickle cell disease and x-linked severe combined immunodeficiency. However, through this process we have identified a false-positive PCR artifact due to the intrinsic capability of PNAs to aggregate with ssDNA donor templates. Here, we show that the combination of PNA and donor templates but not either agent alone results in different degrees of aggregation that result in varying but highly reproducible levels of false-positive signal. We have identified this phenomenon in vitro and confirmed that the PNA sequences producing the highest supposed correction in vitro are not active in vivo in both disease models, which highlights the importance of interrogating and eliminating carryover of ssDNA donor templates in assessing various gene editing technologies such as PNA-mediated gene editing.
Genetic manipulation of DNA holds tremendous potential to alleviate some of the most devastating disorders known to man that are caused by genetic defects. In an attempt to address these important unmet medical needs, many different gene modification techniques have been developed over the last several decades, with multiple approaches now being turned into clinical therapies. Successful gene therapy treatments approved for clinical use include Strimvelis for adenosine deaminase (ADA) deficiency severe combined immunodeficiency (SCID) and Zynteglo for transfusion-dependent beta-thalassemia, respectively (1), and there are now many additional gene therapy treatments similarly utilizing ex vivo genetic manipulation of hematopoietic stem and progenitor cells (HSPCs) advancing toward clinical approvals as well (2). However, while these treatments have been shown or are likely to show that they are curative for each of their respective diseases, they remain suboptimal and difficult to use as they require first removing HSPCs from a patient, significant ex vivo processing, followed by transplantation, which often needs to be proceeded by chemotherapy conditioning, which in-and-of itself holds significant risk to the patient. As such, various in vivo gene modification approaches are also being explored including addition of extrachromosomal DNA via adeno-associated virus (AAV) vectors or direct in vivo gene editing of host DNA. In fact, this year a patient with a genetic condition that causes blindness called Leber’s congenital amaurosis 10 became the first to receive a CRISPR–Cas9 gene therapy administered directly into their body using an AAV vector (EDIT-101) (3).
Another promising early-stage gene editing technology that could be used in vivo to alleviate the burden of ex vivo treatments is called triplex gene editing (4). This involves the use of a DNA mimetic called a peptide nucleic acid (PNA) to specifically bind to a genomic target sequence near the mutation of interest, and following strand invasion of the PNA into the DNA, a stable triplex is formed. The nucleobase spacing of PNA is similar to DNA, which allows base-specific pairing (A and T and C and G) between PNA and RNA/DNA, and the lack of a negatively charged backbone facilitates high-affinity binding. It was previously shown that pyrimidine PNAs are able to form PNA/DNA/PNA triplex structures within double-stranded DNA at complementary purine sites by strand invasion of the duplex DNA. It is thought the cell addresses the triplex through nucleotide excision repair (4, 5), and if a single-strand DNA (ssDNA) donor is also present at the repair site, then editing and correction of the mutation may occur. Codelivery of PNA and wild-type DNA template in Poly Lactic-co-Glycolic Acid (PLGA)-based nanoparticles (NP) has been reported to induce the recombination of short donor DNAs with genomic DNA, in effect enabling targeted and specific gene correction in various cell types (6–10) in utero (11) and in vivo in adult β-thalassemia (12, 13) and cystic fibrosis mouse models (7, 8, 14), although these results have not yet been independently replicated and reported by outside groups.
To date, there has been limited exploration of the structure to activity relationship of PNA designs and the efficiency of triplex gene editing for sickle cell disease (SCD) and severe combined immunodeficiency disease (SCID). Our goal here was to describe the design, synthesis, and structure–activity relationship (SAR) of a hit-to-lead series of more than 300 PNAs. Our initial set of PNA designs consisted of 49 PNAs targeting the HBB E6V allele for in vitro correction of the mutation in bone marrow cells from Townes mice. We subsequently extended the series to include 18 PNAs that targeted the IL2RG R226H mutation in SCID and conducted in vitro and in vivo testing of efficacy. Whereas there is controversy in the field as to the degree of HSPC correction needed in order to alleviate SCD, ranging from estimates of ∼10 to 30% (15–17), it is widely appreciated that SCID requires very low-level correction given the subsequent in vivo expansion of functional immune progenitors from the wild-type HSPCs. Furthermore, it was previously shown in a SCID-X1 mouse models that only ∼10% editing efficiency in HSPCs was required to fully correct the disease, with ∼1% editing resulting in some B and T cell recovery (18), thus our rationale was to assess PNA technology in this point mutation–induced disease, which should be highly sensitive to low rates of gene correction.
Here, we report that by using digital droplet PCR (ddPCR) to assess gene editing by PLGA NP treatment, we observed a PNA-dependent false-positive signal (FPS) originating from donor DNA carryover. Some PNAs tended to cause aggregates with the donor DNA template, and this aggregate was difficult to remove from the cellular substrate in sample processing prior to PCR. We hypothesize that this carried over donor DNA initially elongated in a linear fashion to which the primers could then bind, transforming it into a template for exponential PCR amplification. By highlighting the importance of this ddPCR artifact in producing false-positive gene editing signals, we hope this paper serves as a guide in the design and performance of any gene editing technologies utilizing ssDNA, including PNA-mediated triplex gene editing.
Results
SCD PNA Probe Screening.
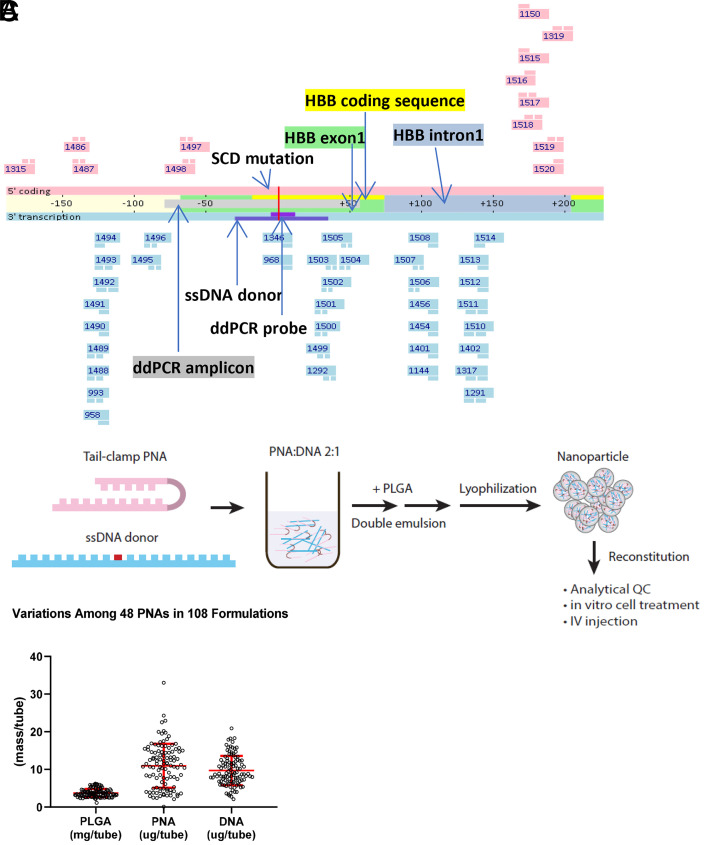
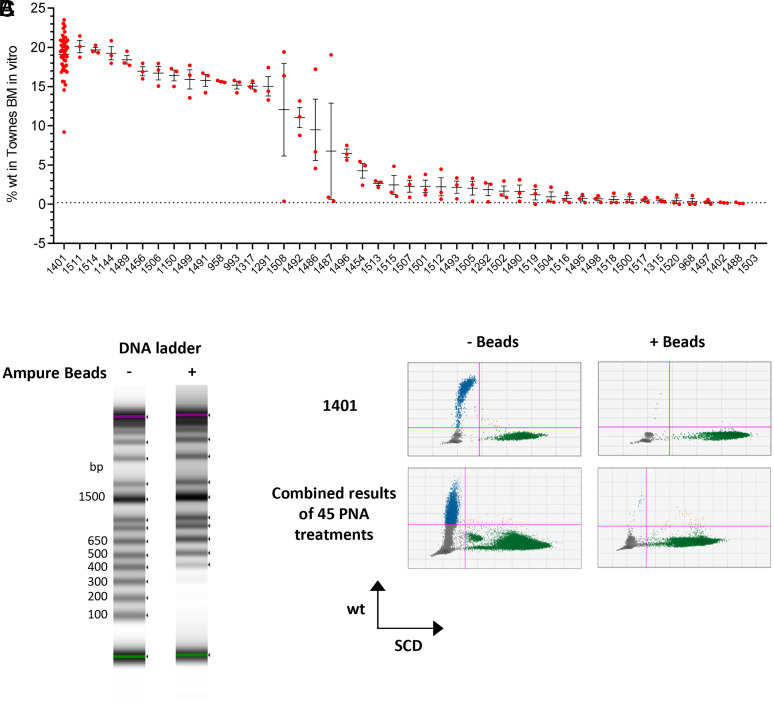
As a proof-of-concept study, we tested if tail-clamp PNAs binding to human HBB gene surrounding the SCD mutation site were able to correct the mutation. We designed and synthesized 49 PNAs across the region (Fig. 1A) and randomly picked 10 PNAs for an in vitro dsDNA invasion assay. A total of 4 out of 10 PNAs were able to invade dsDNA amplicon at physiological conditions (SI Appendix, Fig. 1). We further generated individual NP for all the PNAs with a 65-nucleotide donor DNA that carries the wild-type sequence spanning the target site by PLGA double emulsion (DE) (Fig. 1B). The NP on average (n = 108) contain 10.9 µg PNA, 9.7 µg DNA, and 3.7 mg PLGA per tube, or 295 pmol PNA/mg PLGA and 131 pmol DNA/mg PLGA (Fig. 1C), consistent with published PLGA NP data (10). We treated bone marrow cells isolated from Townes mice carrying a homozygous human HBB SCD mutation (HBBS/S) for 3 d with each PLGA NP and then ran ddPCR to measure the frequency of wild-type sequence. The ddPCR assay was evaluated by wild-type and HBBS/S DNA mixture samples for accuracy in measuring wild-type sequence frequency (SI Appendix, Fig. 2). We detected wild-type sequences above 2% in 33/45 PNA treatments (Fig. 2A). Among them, treatments from PNA 1401 NP consistently generated high level of wild-type sequence in the range from 15 to 24% (Fig. 2A). We also measured wild-type sequence frequency by targeted next generation sequencing on the same DNA samples. We observed a significant correlation between ddPCR and NGS results on the ranking of PNAs based on wild-type sequence frequency. However, the frequencies were lower in NGS results compared to ddPCR (SI Appendix, Fig. 3).
Fig. 1.
Design and formulation of PNAs for targeting HBB SCD mutation. (A) PNA binding sites across HBB E6V (SCD mutation site as position 0). PNAs colored in pink contain the coding sequence and bind to the antisense strand. PNAs colored in blue contain the antisense sequence and bind to the coding strand. The donor DNA and ddPCR detection probes contain sequences from the antisense strand that is transcribed. (B) PLGA DE to generate NP that contain PNA and donor DNA. (C) PLGA NP composition of 45 individual PNAs in 108 formulations described in terms of PLGA (mg/tube), PNA, and DNA (µg/tube).
Fig. 2.
False-positive editing signals from PNA NP–treated bone marrow cells. (A) Bone marrow cells from Townes HBBS/S mice were treated by PLGA NPs for 3 d. The genomic DNA were extracted for ddPCR. Each PLGA NP treatment is in triplicate. (B) Size exclusion (Ampure XP) beads mixed with samples at 0.65:1 efficiently remove DNA fragments smaller than 300 bp. The 1-kb DNA ladder was incubated with Ampure XP beads at v:v 1:0.65. The samples were analyzed on Agilent TapeStation D1000 ScreenTape. (C) The DNA samples from A were combined and treated by Ampure XP beads. The ddPCR results of PNA1401 and all PNA combined were shown. Error bar indicates SD; n = 3.
False-Positive Editing Results Due to Donor DNA Carryover.
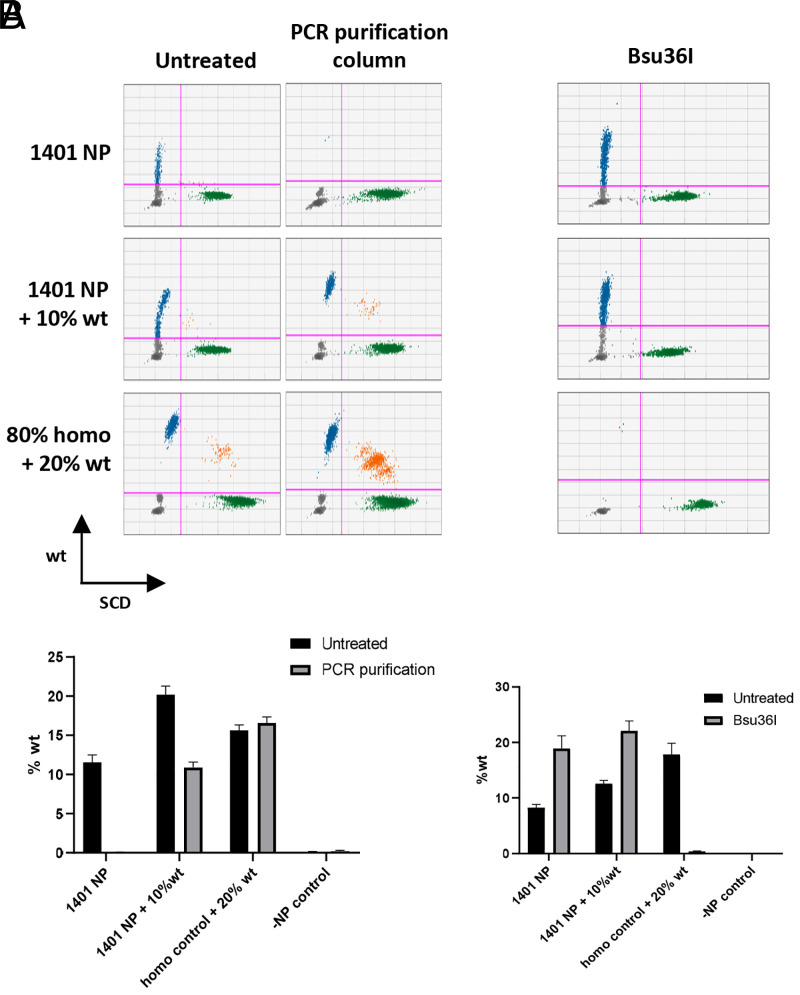
Next, we examined if ddPCR results reflect authentic gene editing in cells. We speculated that PNA or donor DNA carryover in the final genomic DNA preparation may interfere with the PCR and generate skewed results. We therefore treated the genomic DNA samples with size exclusion beads to remove small fragments (Fig. 2B). After bead treatment, the percentage of wild-type sequences dropped down to baseline levels (Fig. 2C), suggesting that detected wild-type sequences were not from genomic DNA. We also used an orthogonal approach to remove small fragments from genomic DNA using PCR purification columns with high-cutoff binding buffer and observed the same results (Fig. 3A). We further questioned whether positive signal was from genomic DNA by subjecting the sample to Bsu36I treatment. The HBB+/+ sequence contains a Bsu36I digestion site that is disrupted by the presence of a SCD mutation. If PLGA NP treatment is able to correct the SCD mutation, the corrected genomic DNA will be digested by Bsu36I, which will result in loss of wild-type droplets in ddPCR. However, we failed to detect any decrease of wild-type sequence frequencies of Bsu36I-treated genomic DNA from PLGA NP–treated cells, in contrast with the spike-in control (Fig. 3B). It is important to note that any single-strand donor DNA present in the sample would not be digested by Bsu36I because the enzyme is double-strand specific. These results indicate that the wild-type sequence detected by ddPCR is not from genomic DNA and therefore is not a result of gene editing.
Fig. 3.
False-positive results do not originate from genomic DNA. DNA samples from PNA1401 NP–treated cells were purified by (A) PCR purification column; (B) Bsu36I. Wild-type genomic DNA from Townes HBB+/+ mice were spiked into NP-treated or untreated HBBS/S DNA. The wild-type sequences were not detected by ddPCR after purification but persist post–Bsu36I digestion, which eliminates amplification of wild-type sequence from spiked-in wild-type DNA. PCR purification or Bu36I digestion is in triplicate. Error bar indicates SD; n = 3.
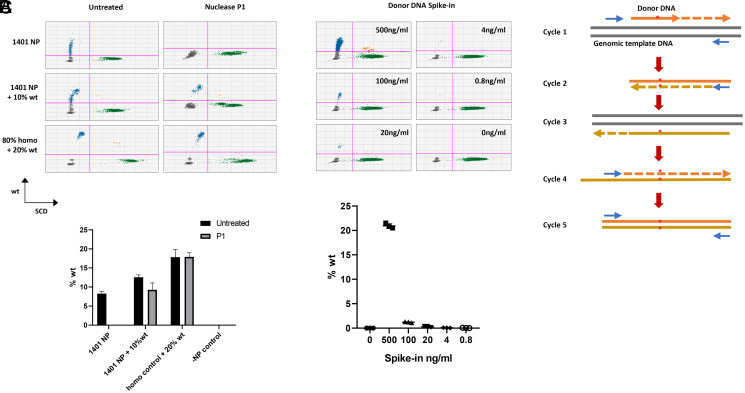
Next, we suspected that the false detection of wild-type sequence originated from donor DNA contamination, as the donor template is the only reagent which contains wild-type sequence in the system. We treated the genomic DNA samples with nuclease P1 that specifically digests ssDNA to remove hypothesized carryover donor DNAs. After nuclease treatment, we failed to detect wild-type sequence above background levels (Fig. 4A). Furthermore, we were able to mimic the NP-treated positive results by simply spiking donor DNA into Townes HBBS/S genomic DNA (Fig. 4B). These results demonstrate that carryover donor DNA is the source of the FPS, and the level of FPS correlates with the amount of donor DNA present in the sample.
Fig. 4.
Donor DNA contamination leads to false-positive results. (A) DNA samples from PNA1401 NP–treated cells were digested by nuclease P1. ddPCR failed to detect wild-type sequences from PNA1401 NP–treated samples, in contrast to the spiked-in wild-type genomic DNA control. (B) Serial amounts of donor DNA were spiked into untreated Townes HBBS/S genomic DNA at 5 ng/μL. After spike-in, 8 μL DNA was used for ddPCR. Error bar indicates SD; n = 3. (C) Proposed model for donor DNA–originated false editing.
Moreover, we formulated a different panel of PNAs (n = 24) targeting the CFTR G542X mutation in cystic fibrosis in PLGA NPs with the SCD corrective donor DNA as described (Formulation/Characterization, Materials and Methods) and tested in the in vitro Townes whole bone marrow (WBM) model for HBB gene correction. We postulated that these PNAs would serve as nonbinding PNAs to determine if FPS was independent of PNA affinity to the cognate target site. We observed FPS in 14 out of 24 PNAs, indicating that the FPS is not dependent on the PNA cognate target (SI Appendix, Fig. 4), which supports donor DNA as the source of FPS.
How does donor DNA contamination lead to false-positive editing results? We proposed that donor DNA can serve as primer in the initial round of PCR to generate wild-type template for sequential amplification. The wild-type sequence can be amplified exponentially in five cycles after the initial step (Fig. 4C). This could occur in any PCR-based methods used to measure gene editing. It is striking, however, that different PNAs lead to different extent of correction DNA carryover at very different yet highly reproducible percentage, indicating that a “scramble PNA” is an inappropriate control that is historically common in the field.
PNAs Aggregate with Single-Strand Donor DNA.
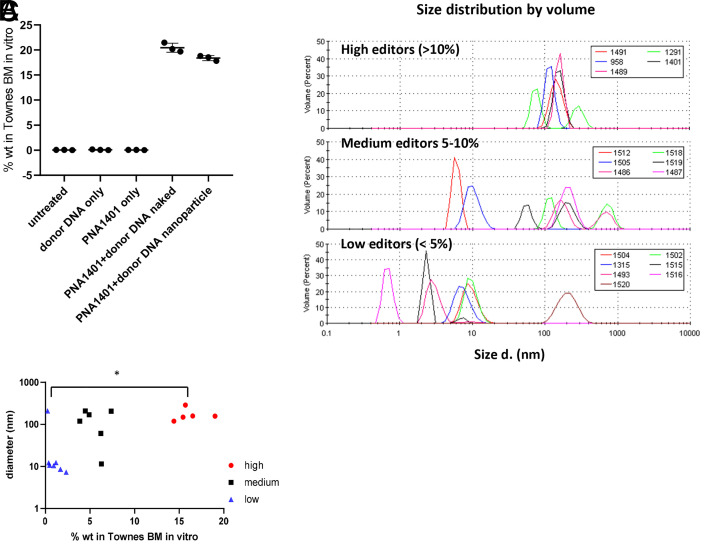
Both PNA and donor DNA are required for false-positive results. We failed to detect wild-type sequences above background if bone marrow cells were treated with PNA or donor DNA alone (Fig. 5A). Furthermore, we found that naked PNA and donor DNA mixture is sufficient to generate false editing results, independent of PLGA DE formulation (Fig. 5A). We also noticed that FPS is PNA dependent (Fig. 2A) and hypothesized that different PNAs may have different degrees of aggregation with the donor DNA, which leads to different levels of FPS. We observed PNA/DNA aggregation during PLGA formulation when PNA and DNA were mixed at high concentration (>10 mg/mL total nucleic acid). To determine if there were differences in the aggregation potential of different PNAs and DNA, we took a subset of the SCD PNA panel and mixed the sample with DNA in phosphate buffered saline (20 μg/mL) and analyzed the mixture by dynamic light scattering (Fig. 5B). This concentration was chosen as it reflects the PNA/DNA concentration within cell culture experiments and is dilute enough to be suitable for light scattering measurements. In some cases, aggregates of 100 to 200 nm in diameter formed, and those PNAs were found to more likely to give a strong FPS (10 to 20% editing frequencies), whereas PNAs that did not aggregate or formed small aggregates (∼10 nm) led to weak FPS (<5%) (Fig. 5C). We observed a significant correlation between PNA/donor DNA aggregate size and FPS.
Fig. 5.
PNA and donor DNA form aggregates. (A) ddPCR signals from in vitro Townes BM when treated with donor DNA only, 1401 PNA only, a mixture of PNA and DNA, and PLGA NP formulated DNA/PNA with an untreated negative control for 3 d. (B) Plots of PNA/DNA aggregate size as measured by volume-weighted dynamic light scattering. The plots are separated for clarity into PNAs with average editing efficiency > 10% (Top), 5 to 10% (Middle), and <5% (Lower). (C) Dot plot of gene editing efficiency in BM as a function of mean PNA/DNA aggregate size. Horizontal bar indicates statistical comparison with equal variance by Student’s t test; *P < 0.05.
Utilizing SCID-X1 as a Secondary Model to Test PNA Triplex Gene Editing.
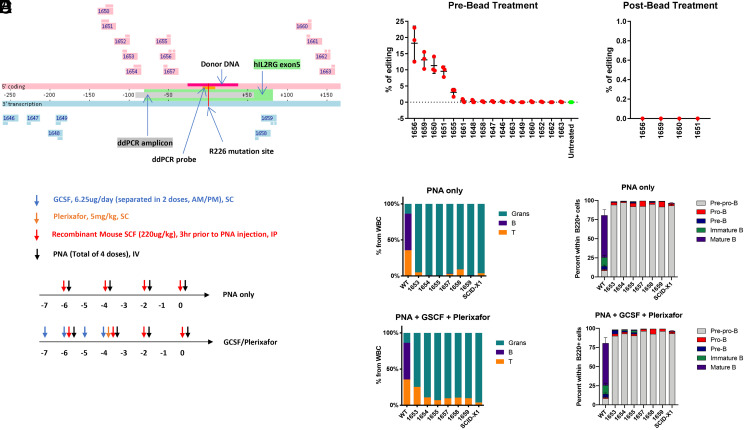
To validate whether the lack of editing efficacy was disease specific, we next designed and assessed PNA editing in X-linked SCID (SCID-X1). Like SCD, we chose a SCID-X1 animal model that harbors a human IL2RG R226H point mutation and would only require a single base edit for disease reversion. 16 PNAs targeting the human IL2RG gene were formulated by PLGA DE (Fig. 6A). To identify favorable candidates for in vivo testing, we initially performed an in vitro screen on CD117-enriched BM cells harvested from SCID-X1 mice. Cells were harvested 72 h post PLGA NP incubation and subjected to ddPCR analyses for correction of IL2RG. We observed four positive candidates ranging from 10 to 25% editing efficiency (Fig. 6B); however after size exclusion bead treatment, all candidates reverted to baseline, demonstrating FPS. Since we were unable to identify optimally favorable candidates from the in vitro screen, our strategy was to choose six PNA candidates binding closest to the mutation site for in vivo testing.
Fig. 6.
Negative editing results in SCID mouse model. (A) PNA binding sites across IL2RG R226H (R226H mutation site as position 0). PNAs colored in pink contain the coding sequence and bind to the antisense strand. PNAs colored in blue contain the antisense sequence and bind to the coding strand. The donor DNA and ddPCR detection probes contain sequences from the coding strand. (B) Bone marrow cells from SCID-X1 mice were treated by indicated PLGA NP for 3 d. The genomic DNA were extracted for ddPCR. The DNA samples that showed positive results were treated with Ampure XP beads, and ddPCR was run again. The editing signals were eliminated by Ampure XP beads. Error bar indicates SD; n = 3. (C) Dosing regimen of PNA candidates and mobilizing agents (G-CSF and plerixafor) within a 7-d treatment window for in vivo screening. (D) Composition of myeloid and lymphoid lineages of peripheral blood 16 wk post PNA treatment. Percentage of granulocytes, B, and T cells are quantified from white blood cell population, normalized to total Ter119− cells set as 100%. (E) Percentage of B cell progenitors in bone marrow within B220+ cell population at 16 wk post PNA treatment.
In Vivo Correction of SCID-X1.
As SCID-X1 is a disease that lacks the normal production of T, B, and natural killer cells, we first sought to observe the production of these cell populations as an indication of gene correction. Given previous reports of administration of recombinant mouse SCF prior to PLGA NP treatment greatly improved editing efficacy (13), we similarly introduced this to our dosing regimen (Fig. 6C) and also compared this with the addition of HSPC-mobilizing agents. Both plerixafor and granulocyte colony stimulating factor (G-CSF) stimulate the bone marrow to release stem cells into peripheral blood, and we reasoned that increasing the pool of cycling hematopoietic stem cells (HSCs) may enhance their exposure to PNA-mediated editing.
Compared to wild-type mice, we observed that SCID-X1 mice proportionally overproduce granulocytes given the lack of other subpopulations. However, no change was indicated in peripheral blood profiles of mice treated with PLGA NPs (Fig. 6D), with or without mobilizing, which indicates no functional correction of SCID-X1. We next sought to specifically focus on analyzing changes in the bone marrow, consisting of B cell progenitors and HSCs. Since the most robust readout of phenotypic SCID-X1 correction is the generation of B cells, which was not observed in the peripheral blood, we interrogated whether B cell progenitors had developed post treatment. As shown in Fig. 6E, we observe a conserved B cell progenitor profile between untreated and NP-treated SCID-X1 mice, again signaling that the lack of B cells was not due to a bottleneck effect.
Discussion
Our initial goal in this study was to describe the design, synthesis, formulation, and SAR of a hit-to-lead series of SCD and SCID PNAs and evaluate triplex gene editing ex vivo and in vivo on two target sites: HBB E6V and IL2RG R226H, respectively. To make this PNA SAR evaluation, we varied the PNA designs while using a well-documented means of delivery and substrate for biological readout (13, 14). Our PLGA NP were made as previously described (7, 10, 13, 19) that had similar size and composition as the published data (10, 14). However, during the study we identified an FPS using both ddPCR and NGS, and after investigation, we attributed FPS to donor DNA template contamination. A similar concern that contaminating DNA can lead to FPS with CRISPR/Cas9 editing was also recently reported (20), which compelled us to examine the effect of removing potential DNA contaminants from our system.
In this work, removal of low–molecular weight DNA was sufficient to abolish the editing signal, leading us to the conclusion that the ddPCR- and NGS-positive editing data were due to FPS. Within our SCD SAR studies, we had several interesting FPS observations. First, the extent of the FPS observed was very much PNA-sequence dependent and highly reproducible (Figs. 2A and 6B). The FPS was not detected by ddPCR when cells were treated with donor DNA only (Fig. 5A). Furthermore, we did not observe false-positive results from 13/45 SCD PNAs and 11/16 IL2RG PNAs but consistently detected 15 to 25% corrected sequence from PNAs 1401. The results suggest that PNA is required for FPS, and different PNAs lead to different levels of signal. It was previously shown that treating cells with donor DNA alone did not cause amplification of the corrected sequence (10) and that a scrambled PNA control, when formulated with corrective template, displayed background levels of gene editing in a beta thalassemic model (13). The high prevalence of negative results from our screening indicates that a scramble or random PNA is not sufficient to serve as negative control for triplex gene editing. From our SCD screen, we determined that roughly one-third of PNAs across the structurally diversified panel elicited an FPS.
A series of PNA/DNA aggregation studies showed that some PNAs tended to aggregate with the donor DNA template more than others and that the PNAs which aggregated were more likely to give rise to FPS. We hypothesize that during cell culture studies, these PNAs bind donor DNA, aggregate, and then are retained on the BM cells, making it difficult to remove donor DNA prior to ddPCR or NGS. The gamma miniPEG side-chain was developed to improve known issues of PNA water solubility, self-aggregation and precipitation, as well as prevent nonspecific binding and aggregation of PNA with dsDNA. Additionally, helical preorganization imparted by the gamma substitution increases both affinity and specificity of binding to complementary DNA and RNA. These findings are based on study of a limited number of fully and partially substituted gamma miniPEG PNA compounds that are relatively small compared to larger PNA tail clamps developed for gene editing (2 to 3 versus 10 kDa). Our results demonstrate that miniPEG substitution is not universally beneficial at preventing tail-clamp aggregation with dsDNA because some of the most highly substituted miniPEG tail clamps severely aggregated with noncomplementary dsDNA. In our experience, it is not possible to generalize the impact of a PNA backbone substitution based on study of one or a few compounds. Rather, we believe that large number of PNA constructs of a particular type must be studied to draw meaningful conclusions. From our findings, it appears that nucleobase composition and other determinants inherent to individual compounds are significant factors that dictate the favorable or unfavorable properties of a particular tail-clamp.
We tested the ability of our IL2RG-targeted PNA formulations to induce corrective gene repair in SCID-X1 mice and observed the same outcome. Upon treatment with size exclusion beads, candidates with positive editing signals identified from an in vitro screen were eliminated (Fig. 6B). In agreement, functional assessment of PNA-mediated in vivo correction was incapable of restoring B or T cells (Fig. 6 D and E), a phenotypic hallmark of SCID-X1 disease correction. This further suggests with a secondary model that previous studies performed solely in vitro may have included overrepresented false editing signals. However, other variables may have also contributed to the negative results obtained in vivo, which cannot be excluded.
Our initial effort testing PNA 968 or 1401 (Fig. 1A and SI Appendix, Table S1) PLGA NP in SC-1 cells that carry HBB E6V mutation failed to generate any reproducible editing signals above background by ddPCR or NGS. We also tried to edit wild-type HBB gene in CD34+ HSPC from normal donors, HEK293, or K562 cells into SCD mutant and failed to detect mutant sequences by ddPCR. The only cells where we were able to detect robust levels of corrected sequences were bone marrow cells from mice. Our FPS therefore stems from promiscuous PNA/DNA aggregate formation followed by carryover of donor DNA and the susceptibility of mouse bone marrow cells to retain these aggregates during sample processing. In addition, when premixed naked 1401 PNA and DNA was added to the cells, we observed a similar FPS as when 1401 PNA was encapsulated, indicating that the NP played no role in the FPS observed (Fig. 5A).
Taken together, our findings reveal the importance of removing donor DNA to enable PCR-based gene editing readouts, especially for PNA-induced triplex gene editing.
Materials and Methods
PNA Design.
For a given target site (e.g., HBB E6V or IL2RG R226H), the genomic region on coding strand 250 bp up and downstream of the mutation site was scanned for purine-enriched sequences. Sequences containing more than 7 continuous purine bases or more than 10 bases with only 1 pyrimidine base were considered as candidate target regions for binding tail-clamp PNAs. The upstream and downstream 10 bp next to the clamp region were analyzed together with the clamp region for intra- and intermolecular self-complementation. The formation of secondary structure was minimized by adjusting the length of the tail and clamp regions, or by introducing bases into the self-complementary regions to replace regular bases. The bases are N7-deazaguanine (dg) for guanine, 2,6-diaminopurine (d) for adenine, and 2-thiouracil (s1) for thymine. Cytosine bases in the clamp arm of PNA were replaced by pseudoisocyosine (j) to improve Hoogsteen paring at neutral pH. If the clamp region contains pyrimidine, double glycine residues were introduced at the corresponding position at the clamp arm of PNA as a skip. In some PNAs, γ-miniPEG was introduced to up to 50% of the residues (13).
The PNA probes of HBB EV6 (SCD), IL2RG R226H (SCID), and CFTR G542X (CF) are listed in SI Appendix, Tables S1–S3.
The donor DNA sequence for HBB SCD is as follows:
mut->wt.
T*T*G*CCCCACAGGGCAGTAACGGCAGACTTCTCCTCAGGAGTCAGGTGCACCATGGTGTCTGT*T*T*G.
wt->mut.
T*T*G*CCCCACAGGGCAGTAACGGCAGACTTCTCCACAGGAGTCAGGTGCACCATGGTGTCTGT*T*T*G.
The donor DNA sequence for IL2RG R226H (SCID) is as follows:
A*A*C*GCTACACGTTTCGTGTTCGGAGCCGCTTTAACCCACTCTGTGGAAGTGCTCAGCATTG*G*A*G.
*, phosphorothioated internucleotide linkages.
PNA Synthesis.
All PNAs were synthesized on an Intavis automated peptide synthesizer using Fmoc solid-phase peptide synthesis protocol on rink-amide Tentagel resin, Rapp Polymere. Multistep synthesis was performed such that each monomer addition cycle was composed of three major subcycles, which were 1) deprotection of Fmoc group, 2) addition of a monomer or amino acid, and 3) capping of any unreacted amine groups. Between each subcycle, the resin was washed five times with dimethyl formamide (DMF). Specifically, 45 mg resin (6umol) was placed in the reaction column and swollen with dichloromethane (DCM) for 15 min. The resin was then treated twice with 1,200 μL 20% piperidine/DMF for 5 min each to deprotect the Fmoc group. After five washes with DMF, 42 μmol 0.2-M monomer or amino acid solution was added, 84 μmol 0.3-M diisopropylethylamine and 39 μmol 0.2-M 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate in anhydrous DMF were then added, and the reaction mixture was shaken for 30 min. The column was then drained, and the resin was washed five times with DMF, and then to that was added 1,200 μL capping solution, which was 5% acetic anhydride and 6% lutidine in DMF, with shaking for 5 min. Monomer addition cycles were repeated till the end of the sequence was reached. To serve as a “purification handle” during purification by reversed-phase chromatography, the ultimate Fmoc group was not removed following the final cycle. To cleave PNA from the resin, the resin was washed five times with DCM and then dried. Each crude PNA was obtained by treating the dried resin with Trifluoroacetic acid/m-Cresol, 95:5, V/V for 2 h; the resin was then removed by filtration and PNA subsequently precipitated by adding cold diethyl ether to the filtrate. The precipitate was pelleted by centrifugation, and the pellet washed twice with diethyl ether. PNA was dissolved in a 6-mL 1/1, V/V, water/acetonitrile mixture, and the solution was lyophilized to dryness. After lyophilization, crude PNA was dissolved in 2 mL 5% by volume of acetonitrile in water and purified using a Thermo Preparative high performance liquid chromatography system, a C18 reversed-phase column with monitoring at 260 nm. Running buffers were buffer A (0.1% TFA in water) and buffer B (0.1% TFA in acetonitrile), and the elution gradient was 5 to 60% B over 30 min at a column at room temp. Fractions containing N terminally Fmoc-protected PNA were analyzed by analytical HPLC and subjected to liquid chromatography-mass spectrometry on a Waters quadrupole time-of-flight platform to confirm identity of each PNA and to choose product fractions for subsequent pooling. Pooled fractions were lyophilized, and each purified Fmoc-PNA was then dissolved in 1 mL dry dimethyl sulfoxide and transferred to a tube containing a piperazine resin (250 mg). This mixture was shaken for 24 h and monitored by HPLC until complete Fmoc removal was apparent. Resin was then removed by filtration, and the filtrate was subjected once again to purification by reversed-phase HPLC. As before, product fractions were analyzed by analytical HPLC and LCMS, and pure fractions were pooled and subsequently lyophilized to provide final pure PNAs as dried powders.
Formulation/Characterization.
PLGA (50:50 LA:GA) obtained from Durect (B6010-2P) was dissolved overnight in dichloromethane prior to use. Donor DNA template was obtained from Avecia. Solutions of PNA and DNA were made by adding nuclease-free water (Invitrogen) and the concentration of each determined by absorbance using the ε-value provided. PNA and DNA (2:1 nmol:nmol) were formulated into PLGA NP using a DE technique as described (19). A batch of PNA 1401, that had been shown repeatedly to produce a positive ddPCR “editing” signal was included in every round of formulation to act as a control for each formulation run. Particles were characterized by measuring particle diameter using a Zetasizer (Malvern). PNA and PLGA content were assessed by HPLC and DNA by a fluorescence assay. To deformulate, 0.1 mL DMSO solvent was added to a representative vial of lyophilized formulation, the vial was vortexed for 3 min, and sonicated at 60 °C for 10 min. The solubilized NP were analyzed for DNA using a fluorescence-based assay using the ssDNA binding dye Oligreen (ThermoFisher). For PNA measurement, 85 µL DMSO solution was added to 170 µL 1-M borate buffer pH 8.5 in a new tube. The tube was vortexed for 3 min, sonicated at 60 °C for 10 min, and centrifuged at 17,000 relative centrifugal force for 10 min. The sample was run on a Thermo Ultimate 3000 HPLC using an Imtakt Presto FF-C18 150- × 4.6-mm, 2-µm column at 60 °C, flow rate 0.4 mL/min, with a linear gradient of acetonitrile in 0.1% TFA in water over 25 min. The concentration of PNA was determined by quantifying the absorbance at 260 nm peak area, with extrapolation of the peak area to a standard curve prepared from known concentrations of unformulated PNA. To measure the PLGA content, 10 µL from the deformulated sample prepared in DMSO was added to 990 µL DMSO in a clean tube which was sonicated as described previously, then transferred to an HPLC vial for analysis using the same type of instrument and column as described previously, with column temperature of 22 °C, a linear gradient of acetonitrile in water, and detection using a Corona VEO detector. To calculate the PLGA concentration of the samples, the peak area was compared to the peak areas of samples of known PLGA content.
Mice.
Townes mice harboring a combination of knock-in mutations for SCD were obtained from Jackson Laboratories. Briefly, mice homozygous for Hbatm1(HBA)Tow (h-α: replaced endogenous mouse α-globin with human α-globin) and Hbbtm2(HBG1,HBB*)Tow (βS: replaced endogenous mouse major and minor β-globin with human A-γ and human sickle βS gene) have the SCD genotype and are referred to as hα/hα:βS/βS. SCD control mice, also referred to as Hbbtm3(HBG1,HBB)Tow, replaced endogenous mouse major and minor β-globin with human Aγ and wild-type βA gene. All animal experiments on SCD mice were maintained at Explora BioLabs, an Association for Assessment and Accreditation of Laboratory Animal Care-accredited commercial facility under experimental protocols as approved by the Institutional Animal Care and Use Committee oversight committee. Humanized SCID-X1 C57BL/6N mice harboring the human IL2RG R226H mutation at the mouse Il2rg locus were obtained from the Naldini Laboratory (18). All animal experiments on SCID-X1 mice were performed in compliance with the Institutional Animal Care and Use Committee and approved by Stanford University Administrative Panel on Laboratory Animal Care.
In Vitro Screening of PNA.
Bone marrow cells were harvested from 8- to 12-wk-old Townes HBBS/S or IL2RG−/− SCID-X1 mice post sacrifice, and single-cell suspensions were obtained by crushing lower-extremity bones. The WBM cells from Townes mice were treated with red blood cell lysis buffer and then frozen down. The WBM cells from IL2RG−/− SCID-X1 mice were enriched using mouse CD117 MicroBeads (Miltenyi Biotec 130-091-224) according to the manufacturer’s instructions, and enriched cell numbers were determined by counting via Muse Count and Viability Kit (Millipore MCH600103). SC-1 cell line (American Type Culture Collection CRL-8756) homozygous for the sickle cell allele at the hemoglobin beta chain locus was also evaluated.
For evaluation of the PLGA NPs, thawed SC-1 cells, HBBS/S WBM cells, or fresh CD117-enriched IL2RG−/− SCID-X1 WBM cells were seeded in 24-well plates at density of 500,000 cells/well in 300 μL media overnight. Cells were cultured in a humidified atmosphere of 5% CO2/air in Roswell Park Memorial Institute medium 1640 (Gibco 11875-093) supplemented with 20% fetal bovine serum (Sigma-Aldrich 12306C), 100 units/mL penicillin, and 100 μg/mL streptomycin (Gibco 15140-122).
Lyophilized PLGA NPs were resuspended in 650 μL same culture media and sonicated for 10 s. Then, 200 μL resuspended NP (3.5 µg PNA and 2.3 µg DNA) were added to cultured cells in triplicate and incubated for 72 h. Cells were washed three times with PBS and harvested with genomic DNA extracted using Genomic DNA Purification Kit (Promega A6780) according to the manufacturer’s instructions.
Genomic DNA Treatment.
To remove small DNA fragments, 100 μL genomic DNA was incubated with 65 μL of Ampure XP beads (Beckman Coulter A63881) for 10 min at room temperature, then washed by 80% ethanol twice, air dried, and eluted in 30 μL H2O or digested by EcoRI, then purified by PCR purification kit (PureLink, Thermo K310001) using a high-cutoff binding buffer B3.
To digest wild-type genomic DNA, 100 ng DNA was incubated with Bsu36I (NEB R0524S) overnight at 37 °C. To digest ssDNA, 100 ng genomic DNA was incubated with Nuclease P1 (NEB M0660S) in 1× NEBuffer 1.1 at 37 °C for 30 min. The digested samples were then heated at 95 °C for 10 min to inactivate the enzyme and used directly for ddPCR.
ddPCR Genotyping.
A total of 10 to 50 ng genomic DNA were used as input for ddPCR according to manufacturer’s protocol (BioRad). The primers and probes for HBB SCD and IL2RG R226H are as follows: HBB forward, CACCAACTTCATCCACGTTCAC; HBB reverse, TCTATTGCTTACATTTGCTTCTGACA; HBB wild-type probe, FAM-CAGACTTCTCCTCAGGA; HBB SCD probe, VIC-CAGACTTCTCCACAGGT; IL2RG forward, TCTCCTCAAGGAACAATCAGTG; IL2RG reverse, TGGATTGGGTGGCTCCATTC; IL2RG wild-type probe, /56-FAM/TT CGG AGC C/ZEN/G CTT TAA CC/3IABkFQ/; IL2RG mutant probe, /5HEX/TT CGG AGC C/ZEN/A CTT TAA CC/3IABkFQ/.
On-Target Amplicon Sequencing.
Sequencing libraries were prepared following a modified 16S metagenomic sequencing library preparation protocol (Illumina). Briefly, genomic DNA was amplified using Q5 High-Fidelity 2× Master Mix (NEB M0492L) by target primers with 16S adaptor overhangs. The PCR products were cleaned up by Ampure XP beads (Beckman Coulter A63881) before index PCR to add sequencing indexes from Nextera XT index kit. The final PCR products were cleaned up again by Ampure XP beads, quantified by Qubit (Thermo), and normalized to 10 nM for pooling. The pooled PCR products were diluted to 1 nM for miniSeq with 30% PhiX spike-in. The data were analyzed by DNA Amplicon app in BaseSpace (Illumina) setting Somatic Variant Frequency Threshold (Percentage) to 1. The target PCR primers for HBB E6V are as follows: HBB5: CTTACATTTGCTTCTGACAC; HBB3: TTGTAACCTTGATACCAACC.
In Vivo Screening in SCID-X1 Mice.
Male 8- to 10-wk-old SCID-X1 mice were randomly divided into three cohorts of six: untreated, PLGA NP only, and PLGA NP plus mobilizing agents (G-CSF + Plerixafor). Untreated SCID-X1 and C57BL/6 wild-type mice were used as controls to compare baseline and expected changes in phenotypically corrected mice. Six PNA candidates were chosen for in vivo testing due to their close proximity to the target site. PLGA NPs were administered according to previously published protocols (13). Briefly, SCF (220 μg/kg per mouse, recombinant mouse SCF, carrier-free, R&D #455-mc-050/CF) was injected intraperitoneally 3 h prior to PNA treatment (2.5 mg PLGA in 150 μL PBS per dose) via retro-orbital intravenous injection every 48 h for a total of four doses. Mobilizing agent G-CSF (6.25 μg/day) was administered subcutaneously daily for 4 d and once in conjunction with plerixafor (5 mg/kg) during the last day of G-CSF administration (Fig. 6C).
Mice were anesthetized with isoflurane followed by retro-orbital bleeding (∼100 μl) and blood was analyzed at 8 and 16 wk post PNA administration by flow cytometry. BM cells were harvested at the end of study (16 wk) and likewise analyzed by flow cytometry for stem and progenitor cell correction.
Flow Cytometry.
All antibodies were purchased from BioLegend accordingly.
Peripheral Blood.
Ter119 – PerCpCy5.5 (#116228), B220 – APC-Cy7 (#103224), CD11b – APC (#101212), CD3 – AF700 (#100216), CD8 – BV605 (#100744), CD19 – BV786 (#115543), CD4 – PE (#100408), Gr1 – PeCy7 (#108416), and PI (Sigma P4864).
Bone Marrow—B Cell Progenitors.
CD45 – APC (#103112), B220 – APC-Cy7 (#103224), IgM – FITC (#406506), CD24 – PeCy5 (101812), CD43 – PeCy7 (#143210), Ter119 – PerCpCy5.5 (#116228), and PI.
Bone Marrow—HSCs.
CD34 – APC (#343608), CD117 – APC-Cy7 (#105838), Lineage – AF700 (#79923), CD135 – PeCy5 (#135312), CD150 – PeCy7 (#115914), Sca1 – Pacific Blue (#108120), CD48 – PerCpCy5.5 (#103422), and PI.
Light Scattering.
Unformulated PNA and donor DNA were mixed together in PBS at 10 µg/mL each, and after incubation for 10 min at room temperature, the light scattering intensity measured using a Zetasizer (Malvern Instruments) and displayed as the volume-weighted intensity.
Acknowledgments
We thank Professor Luigi Naldini from Università Vita-Salute San Raffaele for his kind gift of the SCID-X1 mice. We would also like to thank Amelia Scheck and Cynthia Klein from Stanford University for their help with laboratory management. P.Y.H. was funded by the T32 Immunology Postdoctoral Training Grant (Grant No. 5T32AI007290).
Footnotes
Competing interest statement: Authors affiliated with Vera Therapeutics are authors on patents pertaining to peptide nucleic acid technologies and disclose their financial interests in Vera Therapeutics, in which research funding was received from private venture capital investors. A. Czechowicz discloses financial interests in the following entities working in the rare genetic disease space: Beam Therapeutics, Decibel Therapeutics, Editas Medicines, Gilead Sciences, Global Blood Therapeutics, GV, Jasper Therapeutics, Lyrik Therapeutics, Magenta Therapeutics, Prime Therapeutics, and Spotlight Therapeutics. A. Czechowicz additionally has received research funding from Jasper Therapeutics and Rocket Pharmaceuticals; however, this was not used to support the aforementioned research.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109175118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Harrison C., First gene therapy for β-thalassemia approved. Nat. Biotechnol. 37, 1102–1103 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Hirakawa M. P., Krishnakumar R., Timlin J. A., Carney J. P., Butler K. S., Gene editing and CRISPR in the clinic: Current and future perspectives. Biosci. Rep. 40, BSR20200127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.First CRISPR therapy dosed. Nat. Biotechnol. 38, 382 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Rogers, F. A., Vasquez, K. M., Egholm, M. & Glazer, P. M.. Site-directed recombination via bifunctional PNA–DNA conjugates. Proc. Natl. Acad. Sci. U.S.A. 99, 16695–16700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin J. Y., Glazer P. M., Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol. Carcinog. 48, 389–399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quijano E., Bahal R., Ricciardi A., Saltzman W. M., Glazer P. M., Therapeutic Peptide nucleic acids: Principles, limitations, and opportunities. Yale J. Biol. Med. 90, 583–598 (2017). [PMC free article] [PubMed] [Google Scholar]
- 7.McNeer N. A., et al., Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat. Commun. 6, 6952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricciardi A. S., Quijano E., Putman R., Saltzman W. M., Glazer P. M., Peptide nucleic acids as a tool for site-specific gene editing. Molecules 23, 632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleifman E. B., et al., Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chem. Biol. 18, 1189–1198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNeer N. A., et al., Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol. Ther. 19, 172–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricciardi A. S., et al., In utero nanoparticle delivery for site-specific genome editing. Nat. Commun. 9, 2481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahal R., et al., Single-stranded γPNAs for in vivo site-specific genome editing via Watson-Crick recognition. Curr. Gene Ther. 14, 331–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahal R., et al., In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nat. Commun. 7, 13304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNeer N. A., et al., Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 20, 658–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh M. M., Wu C. J., Tisdale J. F., In mixed hematopoietic chimerism, the donor red cells win. Haematologica 96, 13–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters M. C., et al., Multicenter Investigation of Bone Marrow Transplantation for Sickle Cell Disease, Stable mixed hematopoietic chimerism after bone marrow transplantation for sickle cell anemia. Biol. Blood Marrow Transplant. 7, 665–673 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Hoban M. D., et al., Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiroli G., et al., Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 9, eaan0820 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Oyaghire S. N., Quijano E., Piotrowski-Daspit A. S., Saltzman W. M., Glazer P. M., “Poly(lactic-co-glycolic acid) nanoparticle delivery of peptide nucleic acids in vivo” in Peptide Nucleic Acids: Methods and Protocols, Nielsen P. E., Ed. (Springer US, 2020), pp. 261–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rees H. A., Yeh W.-H., Liu D. R., Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat. Commun. 10, 2212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.