Significance
Spatial organization of mitochondria is vital for cellular function. In many specialized cell types, mitochondria are immobilized at specific subcellular loci through interactions with the cytoskeleton. One of the most striking mitochondrial configurations occurs in mammalian sperm, where mitochondria wrap around the flagellum. Malformation of the mitochondrial sheath causes infertility, but the molecular structures underlying this intricate arrangement are unknown. Here, we analyzed the mitochondrial sheath in sperm from three mammalian species. We find that although mitochondrial dimensions and cristae architecture vary across species, molecular assemblies mediating intermitochondria and mitochondria–cytoskeleton interactions are conserved. These findings yield important insight into sperm physiology and evolution and are relevant for other polarized cell types, such as muscles, neurons, photoreceptors, and hair cells.
Keywords: mitochondria–cytoskeleton contacts, cryoelectron tomography, cryo-FIB milling, cross-linking mass spectrometry, subtomogram averaging
Abstract
Mitochondria–cytoskeleton interactions modulate cellular physiology by regulating mitochondrial transport, positioning, and immobilization. However, there is very little structural information defining mitochondria–cytoskeleton interfaces in any cell type. Here, we use cryofocused ion beam milling-enabled cryoelectron tomography to image mammalian sperm, where mitochondria wrap around the flagellar cytoskeleton. We find that mitochondria are tethered to their neighbors through intermitochondrial linkers and are anchored to the cytoskeleton through ordered arrays on the outer mitochondrial membrane. We use subtomogram averaging to resolve in-cell structures of these arrays from three mammalian species, revealing they are conserved across species despite variations in mitochondrial dimensions and cristae organization. We find that the arrays consist of boat-shaped particles anchored on a network of membrane pores whose arrangement and dimensions are consistent with voltage-dependent anion channels. Proteomics and in-cell cross-linking mass spectrometry suggest that the conserved arrays are composed of glycerol kinase-like proteins. Ordered supramolecular assemblies may serve to stabilize similar contact sites in other cell types in which mitochondria need to be immobilized in specific subcellular environments, such as in muscles and neurons.
In many cell types, mitochondria collectively form a dynamic network whose members divide, fuse, and communicate with one another (1–3). Through interactions with the cytoskeleton, mitochondria are transported—sometimes across large distances—and positioned in response to dynamic stimuli (4, 5). Interactions with the cytoskeleton can also restrain mitochondria to specific subcellular locations. In neurons, axonal mitochondria can be immobilized by interactions with the microtubule or actin cytoskeletons (6–8). In cardiac and skeletal muscle, mitochondrial distribution is regulated by interactions with myofibrils and intermediate filaments (9, 10). However, despite the prevalence of intermitochondria and mitochondria–cytoskeleton interactions and their integral roles in cellular function, there is very little information on the molecular architectures of these interaction sites in any cell type.
One of the most striking mitochondrial configurations occurs in amniote sperm, in which mitochondria are arranged in a spiral around the axoneme, defining a region called the midpiece (11, 12). Mitochondria are among the few organelles retained in sperm throughout their maturation, during which they otherwise lose most of their cytoplasm and organelles en route to becoming highly streamlined cells specialized for finding and fusing with the egg. The extensive mitochondrial sheath in amniote sperm may be an adaptation needed to power the large, long flagellum in these lineages. Variations in midpiece morphometry affect sperm motility and competitiveness (13, 14), and different species rely on energy from mitochondrial respiration to different extents (15, 16), warranting comparative studies of mitochondrial structure across species.
The core of the midpiece is the flagellar cytoskeleton, composed of the microtubule-based axoneme and accessory elements called outer dense fibers (ODFs). A poorly characterized network of cytoskeletal filaments called the submitochondrial reticulum lies between the ODFs and the mitochondria. The submitochondrial reticulum copurifies with the outer mitochondrial membrane (OMM), suggesting that they are intimately associated (17, 18). Mitochondria wrap around the cytoskeleton and are in turn ensheathed by the plasma membrane. As a consequence of this arrangement, each mitochondrion has three distinct surfaces (19): one facing the axoneme, one facing the plasma membrane, and one facing neighboring mitochondria. Thin-section electron microscopy (EM) (19) and freeze-fracture EM (20, 21) revealed that each surface is characterized by a unique membrane protein profile. In particular, these studies uncovered an ordered array of particles on the axoneme-facing surface of sperm mitochondria. Notwithstanding the insight gained from these methods, such techniques require harsh sample preparation steps that can distort fine cellular structure and limit achievable resolution (22). As such, the molecular landscape of intermitochondrial and mitochondrial–cytoskeleton contacts in the sperm midpiece remains largely unexplored.
Assembly of the mitochondrial sheath occurs late in spermiogenesis and involves an intricately choreographed series of events (23, 24). Initially, spherical mitochondria are broadly distributed in the cytoplasm. Mitochondria are then recruited to the flagellum, forming ordered rows along the flagellar axis. Finally, mitochondria elongate and twist around the axoneme. While our understanding of the molecular details of these processes is cursory at best, studies on gene-disrupted mice have implicated several proteins in mitochondrial sheath morphogenesis. For instance, mice expressing mutant forms of kinesin light chain 3 (KLC3) have malformed midpieces, hinting at a role for microtubule-based transport (25). Other examples are the voltage-dependent anion channels (VDACs), which are highly abundant mitochondrial proteins that mediate transport of metabolites, ions, and nucleotides like ATP across the OMM (26). Male mice lacking VDAC3 are infertile, and their sperm cells have disorganized mitochondrial sheaths (27), so VDACs may also have unappreciated roles in mitochondrial trafficking; indeed, KLC3 binds mitochondria through VDAC2 (25). Similarly, disrupting sperm-specific isoforms of glycerol kinase (GK) leads to gaps in the mitochondrial sheath despite proper initial alignment of spherical mitochondria (28, 29). Mice lacking spermatogenesis-associated protein 19 (SPATA19) (30) and glutathione peroxidase 4 (GPX4) (31, 32) also have structurally abnormal mitochondria.
Here, we use cryofocused ion beam (cryo-FIB), milling-enabled cryoelectron tomography (cryo-ET) to image the mitochondrial sheath in mature sperm from three mammalian species. We take advantage of the uniquely multiscale capabilities of cryo-ET to unveil aspects both of the overall organization of the mitochondrial sheath and of the molecular structures important for its assembly. We find that mitochondria are tethered to their neighbors through intermitochondrial linkers and to the underlying cytoskeleton through conserved protein arrays on the OMM. These arrays were first described by deep-etch, freeze-fracture EM in guinea pig sperm (20). Here, we resolve the three-dimensional structures of the OMM arrays in a near-native state and at molecular resolution, revealing how they anchor onto the mitochondrial membrane and how they interact with the flagellar cytoskeleton. Subtomogram averaging reveals that the arrays consist of twofold-symmetric, boat-shaped particles anchored on a lattice of OMM pores whose arrangement and dimensions are consistent with VDACs. Proteomics and in-cell, cross-linking mass spectrometry (XL-MS) suggest that the arrays consist of GK-like proteins. Our data thus show that although mitochondrial dimensions and cristae architecture vary across species, the architecture of the mitochondria–cytoskeleton interface is conserved at the molecular level.
Results
Mitochondrial Dimensions and Cristae Organization Vary across Species.
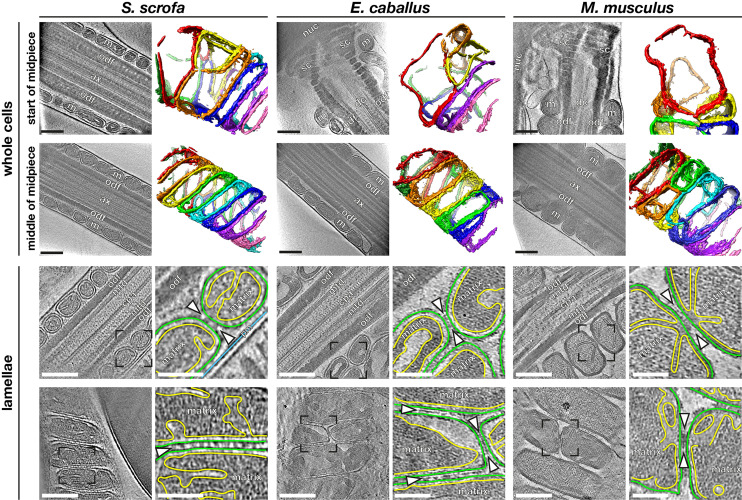
We imaged the mitochondrial sheath in mature sperm from three mammalian species, namely the pig (Sus scrofa), the horse (Equus caballus), and the mouse (Mus musculus) (Fig. 1). These species differ in terms of sperm size, motility patterns, and metabolism. To visualize the overall organization of the mitochondrial sheath, we imaged whole sperm with a Volta phase plate (VPP) (33, 34). Neural network–based segmentation (35) of the mitochondrial membrane allowed us to visualize mitochondrial organization in three dimensions (Fig. 1 A–F).
Fig. 1.
Mitochondrial dimensions and cristae organization vary across species. (A–F) Slices through VPP cryotomograms (Left) and corresponding three-dimensional segmentations (Right) of mitochondria from the start (A–C) or middle (D–F) of the midpiece from pig (A and D), horse (B and E), and mouse (C and F) sperm. (G–L) Slices through cryotomograms of FIB-milled pig (G and H), horse (I and J), and mouse (K and L) sperm midpieces. Right panels show digital zooms of the regions boxed out in the Left panels. The OMM is traced in green, the inner mitochondrial membrane in yellow, and the plasma membrane in blue. Arrowheads indicate intermitochondrial linker complexes. Labels: nuc, nucleus; sc, segmented columns; m, mitochondria; odf, outer dense fibers; dc, distal centriole; ax, axoneme; mtd, microtubule doublets; cpa, central pair apparatus; and pm, plasma membrane. (Scale bars: [A–L] Left panels, 250 nm, and [G–L] Right panels, 100 nm.)
To investigate variations in mitochondrial width along the midpiece, we first measured the width of each mitochondrion at multiple points along its length. We then divided mitochondria into groups based on their positions along the midpiece, as measured by their distance from the head (SI Appendix, Fig. S1). The midpiece is ∼10 µm long in both pig and horse sperm but ∼20 µm long in mouse sperm, so each group represents ∼2 µm in the pig and the horse and ∼4 µm in the mouse. We found that mouse sperm mitochondria are ∼1.5 times wider than pig and horse sperm mitochondria overall (SI Appendix, Fig. S1A). In all three species studied, most mitochondria in the middle (∼60%) of the midpiece are crescent-shaped tubes (Fig. 1 D–F) with consistent widths along their lengths (SI Appendix, Fig. S1B). Mitochondria at the proximal end of the midpiece are larger than their more distal counterparts (Fig. 1 A–C and SI Appendix, Fig. S1A). Moreover, proximal mitochondria have more variable shapes, evidenced by greater variation in their widths at different point along their lengths (SI Appendix, Fig. S1B). Because mitochondria wrap around the axoneme, variations in mitochondrial dimensions both across species and along the proximodistal axis of the flagellum affect the overall diameter and rigidity of the midpiece, likely fine-tuning the hydrodynamics of sperm motility.
To visualize the internal organization of sperm mitochondria in a near-native state, we imaged sperm thinned by cryo-FIB milling (Fig. 1 G–L). This revealed unexpected diversity in the internal ultrastructure of mitochondria across mammalian species, especially in terms of cristae morphology. Horse sperm mitochondria have an expanded intermembrane space and a condensed matrix (Fig. 1 I and J). Mouse sperm mitochondria have an expanded matrix, with a narrow intermembrane space and thin cristae (Fig. 1 K and L). Pig sperm mitochondrial morphology is intermediate (Fig. 1 G and H), and although the mitochondrial matrix was dense, we could identify individual complexes that resembled ATP synthase on cristae of FIB-milled mitochondria (SI Appendix, Fig. S2 A and B), which was confirmed by subtomogram averaging (SI Appendix, Fig. S2B).
Interspecies differences in cristae morphology correlate with measurements of matrix volume relative to mitochondrial volume (SI Appendix, Fig. S2D). In this regard, horse sperm mitochondria resemble “condensed” mitochondria, which correlate with higher rates of oxidative activity in a number of different cell types, including developing germ cells, neurons, and liver (36–38). Indeed, horse sperm are dependent on oxidative phosphorylation (39), whereas pig (16) and mouse sperm (40, 41) are thought to rely largely on glycolytic mechanisms.
Intermitochondrial Junctions Are Associated with Linker Complexes.
Mitochondria are closely packed within the mitochondrial sheath, but it is unclear whether or how individual organelles communicate with their neighbors. To address this, we imaged intermitochondrial junctions captured in FIB-milled sperm lamellae. We observed transmitochondrial cristae alignment in mouse sperm (Fig. 1 K and L) (44/91 junctions across 14 tomograms) but not in pig or in horse sperm (Fig. 1 G–J) (0/67 junctions across 11 tomograms and 0/41 junctions across eight tomograms, respectively). Transmitochondrial cristae alignment has also been observed in muscle tissue of various organisms and is proposed to mediate electrochemical coupling between adjacent mitochondria (42). It is particularly curious, however, that transmitochondrial cristae alignment in sperm appears to be species specific.
We found that intermitochondrial junctions are characterized by intermitochondrial linker complexes in all three species (arrowheads in Fig. 1 G–L and SI Appendix, Fig. S2C). These intermitochondrial linkers span the ∼8-nm distance between the outer membranes of neighboring mitochondria. In mouse sperm, these linkers are specifically associated with sites of transmitochondrial cristae alignment (Fig. 1 K and L); in the pig and in the horse, they are positioned at regularly spaced intervals along intermitochondrial junctions (Fig. 1 H and J). Electron-dense intermitochondrial junctions were also seen in cardiomyocytes by classical EM (42–44). Thus, it is plausible that the as-yet-unidentified linker complexes that we visualize here represent a more general structural mechanism for orchestrating intermitochondrial communication in various cell types.
Ordered Protein Arrays at the Mitochondria–Cytoskeleton Interface Are Conserved across Species.
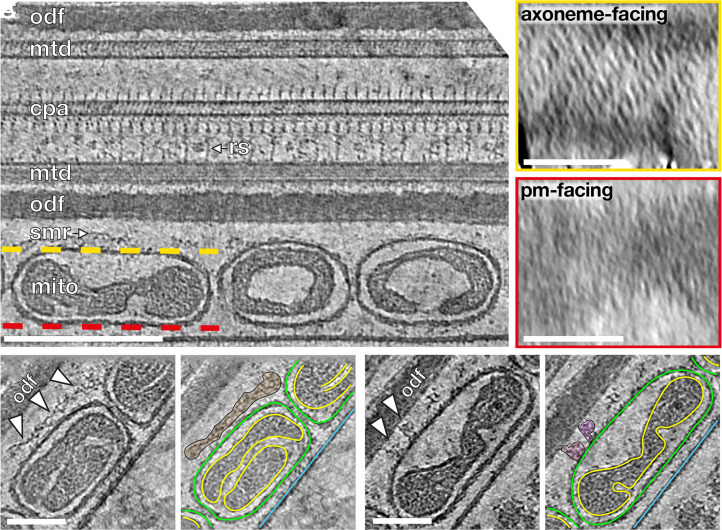
To determine how mitochondria interact with the flagellar cytoskeleton, we imaged the mitochondria–cytoskeleton interface in cryo-FIB–milled lamellae (Fig. 2). Previous studies using freeze-fracture EM on guinea pig sperm (20) and mouse sperm (21) and thin-section EM on golden hamster sperm (19) uncovered an ordered array of particles arranged in a ladder-like pattern on the axoneme-facing surface of sperm mitochondria. We found these arrays on the axoneme-facing surface of the OMM in our tomograms as well (Fig. 2A, yellow box) across all three species and along the entire midpiece (SI Appendix, Fig. S3 A–F). We observed direct interactions between the arrays and either the ODFs or the cytoskeletal filaments surrounding the ODFs (Fig. 2 B and C and SI Appendix, Fig. S4 A–L), indicating that these arrays tether mitochondria to the midpiece cytoskeleton. The highly ordered arrays are absent from the plasma membrane surface (Fig. 2A, red box), which is consistent with freeze-fracture EM studies that detect only very short rows scattered randomly across this surface (20).
Fig. 2.
Ordered protein arrays on the OMM interact with the cytoskeleton. (A) Slice through a cryotomogram of an FIB-milled, horse sperm midpiece showing mitochondria (mito), the submitochondrial reticulum (smr) ODFs (odf), microtubule doublets (mtd), and the central pair apparatus (cpa). Note how individual complexes (like the radial spoke, rs) are visible in the raw tomogram. The ordered protein array is only found on the axoneme-facing surface (yellow) of midpiece mitochondria and not on the plasma membrane–facing surface (red). (B and C) Slices through a cryotomogram of an FIB-milled, horse sperm midpiece showing how the array directly interacts with the submitochondrial reticulum to anchor mitochondria to the flagellar cytoskeleton (arrowheads). In Right panels, the OMM is traced in green, the inner mitochondrial membrane in yellow, and the plasma membrane in blue. (Scale bars: [A] Left, 250 nm, Insets, 100 nm, and [B and C] 100 nm.)
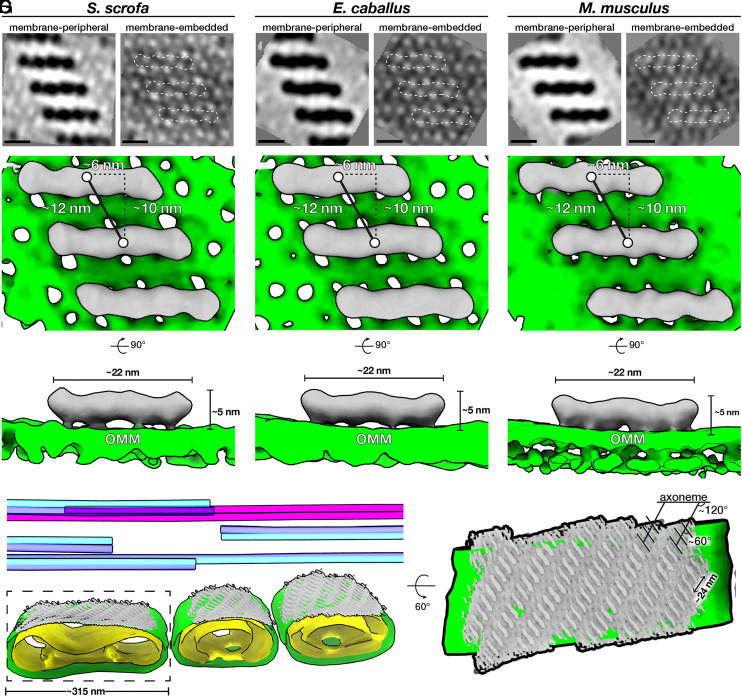
We then aligned and averaged subvolumes containing the protein arrays and the underlying OMM from the axoneme-facing surface (Fig. 3 and SI Appendix, Table S1). Our averages revealed ∼22-nm-long, twofold-symmetric, boat-shaped structures connected via four densities to a porous membrane (Fig. 3 and SI Appendix, Fig. S3 G–I). Each boat-shaped particle rises ∼5 nm above the membrane and consists of two tilde-shaped densities arranged end-to-end. The boat-shaped structures form rows in which each particle is related to its closest neighbors by a ∼10-nm translation perpendicular to the particle long axis and a ∼6-nm shift along this axis, yielding a center-to-center spacing of ∼12 nm (Fig. 3 D–F). Each row is oriented ∼120° to the long axis of the flagellum, and adjacent rows are spaced ∼12 nm apart, forming extensive arrays on the axoneme-facing surface of the OMM (Fig. 3G). Remarkably, the averages we obtained from the three species were highly similar, both in terms of individual particle dimensions and in terms of their supramolecular arrangement (Fig. 3 and SI Appendix, Fig. S3). This conservation suggests that these arrays are a crucial structural element of the mitochondrial sheath.
Fig. 3.
Ordered protein arrays at the mitochondria–cytoskeleton interface are conserved across species. (A–C) Subtomogram averages of the protein arrays and underlying OMM after applying twofold symmetry (note that density is black). (D–F) Isosurface renderings of the subtomogram averages in A–C with boat-shaped particles in gray and the OMM in green. (G, Left) Segmentation of the tomogram shown in Fig. 2A, with the OMM in green, the IMM in yellow, microtubule doublets in blue, and the cpa in pink. Subtomogram averages of boat-shaped particles are colored gray and plotted back into their positions and orientations in the tomogram. (Right) Rotated and zoomed-in view of the axoneme-facing surface of a mitochondrion. The axoneme is oriented horizontally, so the ladder-like arrays are oriented ∼120° to the flagellar long axis, and individual particles within the array are oriented ∼60° to this axis. (Scale bars, 10 nm.)
Our averages revealed that the OMM underlying the protein arrays is studded with ∼3 to 4-nm pores arranged in a pseudolattice with a center-to-center spacing of ∼5 nm (Fig. 3 A–C and SI Appendix, Fig. S3 G–I). These pore sizes are consistent with the diameters of the VDACs, which are known to form ordered arrays in the OMM (45–48). VDACs are known to localize to the sperm midpiece (49, 50); indeed, our label-free quantitative proteomics experiments show that VDAC2 and VDAC3 are among the most abundant OMM proteins in pig sperm (SI Appendix, Table S2). Furthermore, the lattice dimensions in our averages closely match those of VDAC in purified Neurospora OMM (45, 51). The lattice can be modeled by fitting multiple copies of the VDAC2 crystal structure (52) (SI Appendix, Fig. S5A). We oriented VDAC2 in the membrane plane based on its known topology (53, 54); however, at the current resolution, we cannot determine the orientation around the pore axis. Thus, in our model, each boat-shaped particle stretches across eight VDAC molecules (Fig. 3).
GK-Like Proteins Are Probable Constituents of the Conserved Arrays at the Mitochondria–Cytoskeleton Interface.
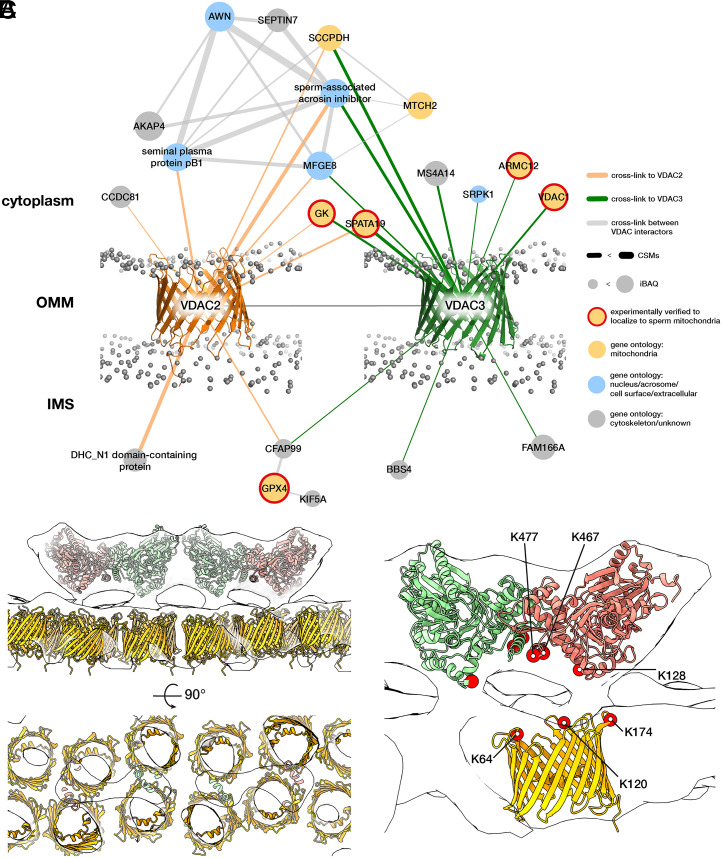
To search for possible constituents of the protein arrays on the VDAC lattice, we used in-cell XL-MS (55, 56) to find potential VDAC2/VDAC3 interaction partners on the OMM (Fig. 4). We treated pig sperm cells with the cross-linker disuccinimidyl sulfoxide (DSSO), which covalently links free lysines that are within ∼3 nm (Cα-Cα) of each other. To increase confidence, we screened for protein interactions supported with at least two cross-link spectral matches (CSMs) (Materials and Methods).
Fig. 4.
Modeling the OMM array as GK proteins anchored on VDACs. (A) The VDAC2/VDAC3 interactome derived from in-cell XL-MS of pig sperm. Protein nodes are colored according to their known subcellular localizations (yellow with red border, experimentally verified to localize to sperm mitochondria; yellow, gene ontology: mitochondria; blue, gene ontology: nucleus/acrosome/cell surface; and gray, gene ontology: cytoskeleton/unknown). Gray spheres indicate the phosphate groups of a simulated lipid bilayer which was structurally aligned based on the simulation for monomeric mouse VDAC1 (PDB 4C69) obtained from the MemProtMD server (97). (B) Modeling the OMM array as GK-like proteins anchored on VDACs. A GK-like dimer-of-dimers homology model (red and green) and VDAC2 homology models (yellow) were fitted into the pig subtomogram average map (white). (C) The positions of cross-linked Lys residues (red circles) are consistent with GK and VDAC orientation assignments in our model. Note that the cross-links are shown for VDAC2, but we detect an additional cross-link between K140 on GK and VDAC3 (SI Appendix, Fig. S6).
We first screened candidate proteins based on their known subcellular localizations (Fig. 4A). VDAC2/VDAC3 cross-linked to mitochondria-associated proteins as well as to sperm head–associated proteins. This is consistent with immunofluorescence studies localizing VDAC2/VDAC3 both to the midpiece and to the acrosome, a large vesicle capping the anterior sperm nucleus (49, 50, 57). Of the proteins in the mitochondria-associated interaction hub, four proteins are particularly noteworthy because they are known to localize to the OMM and because their disruption results in dysplasia of the mitochondrial sheath: armadillo repeat-containing protein 12 (ARMC12) (58), SPATA19 (30), GPX4 (31, 32), and GK (28, 29).
To distinguish among these candidates, we compared the location of the cross-links with the known topology of VDAC in the OMM (53, 54). GPX4 would interact on the side facing the intermembrane space, whereas SPATA19, ARMC12, and GK would interact on the cytoplasmic face. Although we detect ARMC12 in mature pig sperm, in the mouse, it is only present in developing spermatids and disappears from mature sperm (58), which is inconsistent with the fact that the OMM arrays are present in cauda epididymal mouse sperm. SPATA19 and GK are both highly abundant (SI Appendix, Table S2), as would be expected for proteins forming extensive arrays. Assuming an average protein density of ∼1.43 g/cm3 (59), which corresponds to ∼0.861 Da/Å3, we estimate that each boat-shaped particle in the array has a molecular weight of ∼250 kDa. SPATA19 is a small protein with an estimated molecular weight of ∼18 kDa. To fit into our EM densities, it must either be present in multiple copies or form a complex with other proteins. In contrast, GK has an estimated molecular weight of ∼60 kDa and is known to form S-shaped dimers (∼120 kDa) that are conserved from bacteria (60, 61) to eukaryotes (62, 63).
To build a GK–VDAC model based on our subtomogram average, we used rigid-body fitting to place two GK dimers end-to-end into a boat-shaped density (SI Appendix, Fig. S5B and Fig. 4B). These fits defined a clear orientation for GK, with the N termini pointing upwards and the C-terminal helices facing the OMM (Fig. 4B). To validate our fits, we mapped the cross-linked lysines onto the resulting model (Fig. 4C). All cross-links were between the cytosolic face of VDAC2 and the OMM-facing surface of GK, which is consistent with the orientation expected from our fits. These fits show that the molecular dimensions and relative positions of candidate proteins are consistent with our subtomogram average maps, but we cannot define specific interaction sites at the current resolution. We also attempted to analyze interactions between the putative GK-like proteins and the underlying cytoskeleton, and although we detect cross-links on the putative cytoskeleton-facing side of GK (SI Appendix, Fig. S6), interpretation is complicated by the fact that the precise protein compositions of the submitochondrial reticulum and the ODFs are unclear.
Assigning GK-like proteins as constituents of the ordered OMM arrays at the mitochondria–cytoskeleton interface is also supported by recent genetic studies. Sperm from mice lacking sperm-specific GK isoforms have disorganized mitochondrial sheaths (28, 29). In these mice, spherical mitochondria properly align along the flagellum but fail to properly elongate and coil around it, leaving gaps along the midpiece where the ODFs are exposed (29). This phenotype is consistent with our data showing contacts between GK protein arrays and the underlying cytoskeleton (Fig. 2 B and C and SI Appendix, Fig. S4 A–L) and by experiments showing that the submitochondrial reticulum remains attached to the OMM in detergent-extracted sperm (18). Although sperm-specific GK isoforms do not show GK activity in vitro (64), we cannot completely exclude that they function in mitochondrial metabolism in situ, especially since knockout mice have higher ATP levels than wild-type (28). If the arrays that we observe indeed consist of GK-like proteins and if these proteins are in fact metabolically active, then their preferential orientation toward the flagellar cytoskeleton could enhance metabolite shuttling between mitochondria and the axoneme.
Discussion
In this study, we used cryo-FIB milling–enabled cryo-ET to image the sperm mitochondrial sheath in three mammalian species. Our data reveal that overall, mitochondrial dimensions are remarkably consistent in sperm from the same species (Fig. 1 and SI Appendix, Fig. S1). This contrasts with other mitochondria-rich tissues such as muscle, in which there are massive variations in mitochondrial size and morphology within individual cells (65). In addition, we did not observe mitochondrial nanotunnels in any of the species we examined, in contrast to their relative abundance in muscle tissue (3, 65). However, we do observe discrete linker proteins that tether neighboring mitochondria in all three species. It will be important to investigate the precise molecular identity of these linkers and to define whether they serve purely structural roles or also act as conducting channels that coordinate metabolite shuttling, energy production, and calcium signaling across the entire midpiece. Hinting at possible interorganelle coordination, we observed transmitochondrial cristae alignment at specialized junctions between neighboring mitochondria in mouse sperm (Fig. 1). Thus, the concept of an interconnected mitochondrial reticulum that occurs in muscle (2) may also be applicable to the mammalian sperm midpiece and warrants further consideration.
In this study, we imaged either ejaculated or mature cauda epididymal sperm. However, mammalian sperm must undergo a plethora of biochemical and morphological changes in the female reproductive tract before they become fertilization competent. These processes are collectively known as capacitation and cause extensive metabolic changes in sperm (66). To cope with increased energy demands, rates of both glycolysis and oxidative phosphorylation increase during capacitation in mouse sperm (67). Future work could use cryo-ET to establish whether these metabolic changes relate to any structural changes in midpiece mitochondria, for instance at the level of cristae architecture or intermitochondrial junctions.
Our data also show that mitochondrial dimensions and cristae architecture vary across species (Fig. 1), providing possible structural bases for interspecific differences in mitochondrial energetics. Horse sperm mitochondria appear to take on a more condensed appearance than their counterparts in pig and mouse, which may correlate with increased reliance on oxidative phosphorylation in horse sperm. Although studies on horse sperm physiology are limited (68), condensed mitochondrial profiles are suggested to be a sign of calcium overload in other species (69, 70), so cross-species studies exploring the link between mitochondrial morphology and possible differences in mitochondrial calcium homeostasis are clearly necessary. In general, the natural diversity of sperm enables comparative studies of how mitochondrial structure varies with sperm metabolism, which will undoubtedly contribute to our broader understanding of how mitochondrial form relates to function.
Our data show that despite this diversity, the molecular underpinnings of mitochondrial sheath architecture are conserved, at least in mammals. Specifically, we identified intermitochondrial linkers that tether adjacent mitochondria (Fig. 1 and SI Appendix, Fig. S2). We also describe ordered protein arrays on the axoneme-facing surface of sperm mitochondria, which were first identified by freeze-fracture EM. Here, we characterize the three-dimensional architecture of these arrays in a near-native state and at molecular resolution, revealing how they are anchored on the OMM and how they interact with the flagellar cytoskeleton. In-cell subtomogram averaging reveals that these arrays consist of boat-shaped particles anchored on a lattice of OMM pores (Figs. 2 and 3). Proteomics and in-cell XL-MS suggest that these arrays consist of GK-like proteins anchored on VDAC lattices in the OMM (Fig. 4). Given that VDACs are ubiquitous OMM proteins, our findings motivate efforts to explore whether they also regulate mitochondria–cytoskeleton interactions in other cell types.
The OMM arrays may function to regulate the precise elongation and coiling of mitochondria, contributing to the striking consistency within the mitochondrial sheath. In mature sperm, these arrays may help maintain the integrity of mitochondria-cytoskeleton contacts, stabilizing them against shear stresses during sperm motility and hyperactivation. However, it is unclear what determines the organization of these arrays in the first place. Our averages do not hint at direct interactions between boat-shaped particles. Instead, their spacing may be defined by the organization of the underlying VDAC lattice. Another intriguing possibility is that the arrays are organized by their cytoskeletal binding partners; the periodicity of relevant motifs on the submitochondrial reticulum could dictate the spacing of the OMM arrays.
We find that mitochondria–cytoskeleton contact sites in the sperm midpiece can be quite variable (SI Appendix, Fig. S4). Occasionally, OMM arrays make direct contact with the ODFs (SI Appendix, Fig. S4 A–C, E, F, and I); sometimes, there are considerable gaps between them that are bridged either by the submitochondrial reticulum (SI Appendix, Fig. S4 D, G, H, and J) or by unidentified linkers (SI Appendix, Fig. S4 K and L). This may be explained by variability in both the sizes and shapes of the ODFs themselves (SI Appendix, Fig. S4 M–R). Defining the precise mechanisms by which the OMM arrays tether to the underlying cytoskeleton is an important avenue for future research, in part because it has important implications for understanding sperm motility. In mammalian sperm, forces from axoneme bending are transmitted to the connecting piece through the ODFs, which are physically coupled to the microtubule doublets along most of the principal piece but not in the midpiece (71, 72). The observation that mitochondria are anchored to the ODFs through extensive ordered arrays adds another level of complexity to the multilayered sliding mechanics of the mammalian sperm flagellum, affecting, for example, the extent to which the ODFs can slide relative to each other or to the axoneme. Indeed, the infertility of mice lacking sperm-specific GK isoforms appears to be caused by motility defects arising from the fragmented mitochondrial sheath (28, 29). Intermitochondrial linkers that physically tether neighboring mitochondria may also affect the extent to which the midpiece can bend during movement. On a larger scale, interspecies variations in mitochondrial dimensions and packing coupled with known differences in overall midpiece length could result in subtle changes in sperm motility. Furthermore, the stiffness of the midpiece changes during sperm maturation, becoming less rigid as sperm transit through the epididymis (73, 74). Tracing how the molecular structures that we describe here form and change during spermiogenesis and epididymal maturation are likely to provide further insight in this respect. Ultimately, combining structural information from cryo-ET with motility analysis and mathematical modeling could help illuminate how midpiece morphometry affects swimming behavior.
The methods we use here can also be used to study other specialized organelle configurations in the highly-streamlined sperm cell. For example, it is unclear how exactly the sperm head anchors to the tail or how the acrosomal vesicle is laminated to the nucleus. Beyond sperm, such methods could be used to gain molecular insight into the organization of organelles in other terminally differentiated and polarized cell types like neurons, photoreceptors, or hair cells. For example, exploring whether similar arrays are present at the mitochondria–cytoskeleton interface in other differentiated cell types—and whether they use a similar pool of protein components—is an area ripe for study. In striated muscle, proper mitochondrial positioning is critical for muscle function and depends on direct associations between mitochondria and intermediate filaments (9, 75). Similarly, in skin cells, mitochondrial organization depends on keratin (76). The structural bases for these associations are unknown, but cryo-ET and in-cell XL-MS may prove useful in these contexts as well.
Materials and Methods
Sperm Collection and Preparation.
Pig sperm samples were obtained from Varkens KI Nederland and prepared for imaging within a few hours of delivery. Sperm were layered onto a discontinuous gradient consisting of 4 mL of 35% Percoll (GE Healthcare) underlaid with 2 mL of 70% Percoll both in Hepes-buffered saline (HBS: 20 mM Hepes, 137 mM NaCl, 10 mM glucose, 2.5 mM KCl, 0.1% kanamycin, and pH 7.6) and centrifuged at 750 × g for 15 min at room temperature (RT). Pelleted cells were washed once in phosphate-buffered saline (PBS: 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, and pH 7.4) and resuspended in PBS for further processing. All solutions were iso-osmotic at RT (290 to 300 mOsm/kg).
Horse semen was collected from mature Warmblood stallions using a Hanover artificial vagina in the presence of a teaser mare. Semen was filtered through gauze and kept at RT until further processing. Semen was diluted in INRA96 (IMV Technologies) to a sperm concentration of 30 × 106 cells/mL. Sperm were then centrifuged through a Percoll gradient (as described above for the pig) for 10 min at 300 × g followed by 10 min at 750 × g (77). The pellet was resuspended in 1 mL of PBS, centrifuged for 5 min at 750 × g, and finally resuspended in PBS.
Mouse sperm were obtained from the cauda epididymis of adult male C57BL/6 mice as described in ref. 78. Male mice were culled as described in ref. 79, and the cauda epididymides were dissected with the vas deferens attached. Tissues were placed in a 500-µL droplet of modified Biggers, Whitten, and Whittingham media (BWW: 20 mM Hepes, 91.5 mM NaCl, 4.6 mM KCl, 1.7 mM D-glucose, 0.27 mM sodium pyruvate, 44 mM sodium lactate, 5 U/mL penicillin, and 5 µg/mL streptomycin, adjusted to pH 7.4 and an osmolarity of 300 mOsm/kg). Sperm were gently pushed out from the vas deferens, after which two incisions were made with a razor blade in the cauda. Spermatozoa were allowed to swim out of the cauda into the BWW over a period of 15 min at 37 °C, after which the tissue was removed and sperm were loaded onto a 27% Percoll density gradient and washed by centrifugation at 400 × g for 15 min. The sperm pellet was resuspended in BWW and centrifuged at 400 × g for 2 min to remove excess Percoll, then finally resuspended in BWW.
Cryo-EM Grid Preparation.
Typically, 3 µL of sperm suspension was pipetted onto a glow-discharged Quantifoil R 2/1 200-mesh holey carbon grid. Sperm cell density was ∼2 to 3 × 106 cells/mL (for whole-cell tomography) or 20 to 30 × 106 cells/mL (for cryo-FIB milling). Approximately 1 µL of bovine serum albumin–conjugated gold (Aurion) was added and the grids blotted manually opposite the side of cell deposition for ∼3 s (for whole-cell tomography) or for ∼5 to 6 s (for cryo-FIB milling) using a manual plunge-freezer (Max Planck Institute Martinsreid). Grids were immediately plunged into a liquid ethane–propane mix (37% ethane) (80) cooled to liquid nitrogen temperature. Grids were stored under liquid N2 until imaging.
Cryo-FIB Milling.
Grids were mounted into modified Autogrids (Thermo Fisher Scientific) for mechanical support. Clipped grids were loaded into an Aquilos (Thermo Fisher Scientific) dual-beam, cryo-FIB/scanning electron microscope (SEM). SEM imaging was performed at 2 kV and 13 pA, whereas FIB imaging for targeting was performed at 30 kV and 10 pA. Milling was performed with stage tilts between 15° and 18°, so lamellae were inclined 8 to 11° relative to the grid. Each lamella was milled in four stages: an initial rough mill at 1-nA beam current, an intermediate mill at 300 pA, a fine mill at 100 pA, and a polishing step at 30 pA. Lamellae were milled with the wedge premilling technique described in ref. 81 and with expansion segments as described in ref. 82.
Tilt Series Acquisition.
Tilt series were acquired on either a Talos Arctica (Thermo Fisher Scientific) operating at 200 kV (all lamellae datasets and the whole-cell VPP horse dataset) or a Titan Krios (Thermo Fisher Scientific) operating at 300 kV (whole-cell VPP pig and mouse datasets). Both microscopes were equipped with a postcolumn energy filter (Gatan) operated in zero-loss imaging mode with a 20-eV energy-selecting slit. All images were recorded on a K2 Summit direct electron detector (Gatan) in either counting or superresolution mode with dose-fractionation. Tilt series were collected using SerialEM (83) at target defocus values between −4 and −6 µm (conventional defocus-contrast) or between −0.5 and −1.5 µm (for tilt series acquired with the VPP). Tilt series were typically recorded using either strict or grouped dose-symmetric schemes, either spanning ± 56° in 2° increments or ± 54° in 3° increments, with total dose limited to ∼100 e−/Å2.
Tomogram Reconstruction.
Frames were aligned either postacquisition using Motioncor2 1.2.1 (84) or on-the-fly using Warp (85). Frames were usually collected in counting mode; when superresolution frames were used (whole-cell VPP pig dataset), they were binned 2× during motion correction. Tomograms were reconstructed in IMOD (86) using weighted back-projection, with a simultaneous iterative reconstruction technique (SIRT)–like filter (87) to aid visualization and segmentation. Defocus-contrast tomograms were contrast transfer function corrected in IMOD using ctfphaseflip. VPP tomograms were left uncorrected.
Tomogram Segmentation.
Segmentation was performed semiautomatically using the neural network-based workflow implemented in the TomoSeg package in Electron Micrograph Analysis (EMAN) 2.21 (35). Microtubules were traced manually in IMOD. Segmentation was then manually refined in Chimera 1.12 (88) or in ChimeraX (89). Visualization was performed in ChimeraX.
Subtomogram Averaging of ATP Synthase and Ladder-Like Arrays.
Subtomogram averaging with missing wedge compensation was performed using Particle Estimation for Electron Tomography (PEET) 1.13.0 (90, 91) on defocus-contrast tomograms of cryo-FIB–milled lamellae. Resolution was estimated using the Fourier shell correlation at a cutoff of 0.5 (90). Alignments were generally performed first on binned data, after which aligned positions and orientations were transformed to less-binned data using scripts provided by Daven Vasishtan. Details of acquisition parameters and particle numbers are summarized in SI Appendix, Table S1.
Alignment strategies for mitochondrial complexes were designed to take advantage of their defined orientations relative to the membrane plane. Particles were picked manually, and their initial orientations were defined using stalkInit. Initial references were either a randomly chosen particle (for ladder-like arrays) or an average of all particles after roughly aligning them based on their initial orientations (for ATP synthase). Independent alignments using independent initial references were performed for datasets from different species. Alignments allowed for large rotational search ranges around the particle long axis (defined as the y-axis, perpendicular to the membrane plane), with limited search ranges around the x- and z-axes (the membrane plane).
All initial alignments were performed without symmetry. After visual inspection of the maps, twofold symmetry was applied for ladder-like arrays. Symmetrization involved using the aligned positions from the unsymmetrized runs as seed points and rotating particles around the axis of symmetry to generate virtual particles. A symmetrized volume was generated by averaging all particles and virtual particles and used as a reference for a final, restricted alignment.
Plotbacks were generated in IMOD by first running createAlignedModel to generate model files reflecting updated particle positions and orientations after alignment. The relevant subtomogram average was then thresholded for visualization and saved as an isosurface model, which was then placed back into the tomograms using clonemodel.
Measurements and Quantification.
All measurements of mitochondrial width were performed in IMOD on VPP tomograms filtered with a SIRT-like filter. Mitochondrial width was measured in the nonmissing wedge direction at five points along the length of each mitochondrion. Only mitochondria that were entirely in the field of view were included in the measurements. Tomograms and corresponding measurements were then grouped based on their locations relative to the connecting piece, which were determined based on low-magnification images used for targeting during data acquisition.
Internal mitochondrial ultrastructure was quantified from tomograms from cryo-FIB–milled lamellae. The volume occupied by the matrix (Vmatrix, the volume enclosed by the inner mitochondrial membrane [IMM]) was measured relative to the volume occupied by the entire mitochondrion (Vmito, the volume enclosed by the OMM). Mesh volumes were extracted from segmentations using imodinfo. Because neural network–based segmentation often resulted in gaps, mitochondrial membranes were segmented manually in IMOD for quantification. Only slices in which both the IMM and OMM were clearly defined were used for segmentation.
Cross-Linking, Lysis, Digestion, and Peptide Fractionation.
All proteomics and XL-MS experiments were performed on Percoll-washed pig sperm prepared as described above. For cross-linking, ∼300 × 106 cells were used, each from three different animals. Briefly, pelleted sperm cells were resuspended in 540 µL of PBS supplemented with DSSO (Thermo Fisher Scientific) to a final concentration of 1 mM. The reaction mix was incubated for 30 min at 25 °C with 700 rpm shaking in a ThemoMixer C (Eppendorf) and subsequently quenched for 20 min by adding Tris·HCl (final concentration 50 mM). Cross-linked cells were spun down at 13 × 800 × g for 10 min at 4 °C, after which the supernatant was removed. Cells were then lysed according to a protocol modified from ref. 92. Cells were resuspended in 1 mL of lysis buffer [100 mM Tris·HCl pH 8.5, 7 M Urea, 1% Triton X-100, 5 mM tris(2-carboxyethyl)phosphine (TCEP), 30 mM chloroacetamide (CAA), 10 U/mL DNase I, 1 mM MgCl2, and 1% benzonase (Merck Millipore, Darmstadt, Germany), 1 mM sodium orthovanadate, phosphoSTOP phosphatases inhibitors, and cOmplete Mini ethylenediamine tetraacetic acid-free protease inhibitors]. Cells were sonicated on ice for 2 min using an ultrasonic processor (UP100H, Hielscher) at 80% amplitude. The proteins were then precipitated according to ref. 93 and the dried protein pellet resuspended in digestion buffer (100 mM Tris·HCl pH 8.5, 1% sodium deoxycholate [Sigma-Aldrich], 5 mM TCEP, and 30 mM CAA). Trypsin and Lys-C proteases were added to a 1:25 and 1:100 ratio (weight/weight), respectively, and protein digestion performed overnight at 37 °C. The final peptide mixtures were desalted with solid-phase extraction C18 columns (Sep-Pak, Waters) and fractionated with an Agilent 1200 HPLC pump system (Agilent) coupled to a strong cation exchange (SCX) separation column (Luna SCX 5 µm to 100 Å particles, 50 × 2 mm, Phenomenex), resulting in 25 fractions.
Liquid Chromatography with MS.
Approximately 1,000 ng of peptides from each biological replicate before SCX fractionation were first injected onto an Agilent 1290 Infinity ultra high-performance liquid chromatography system (Agilent) on a 50-cm analytical column packed with C18 beads (Dr. Maisch Reprosil C18, 3 µm) coupled online to an Orbitrap HF-X (Thermo Fisher Scientific). For this classical bottom-up analysis, we used the following liquid chromatography with MS/MS (LC-MS/MS) parameters: after 5 min of loading with 100% buffer A (H2O with 0.1% formic acid), peptides were eluted at 300 nL/min with a 95-min gradient from 13 to 40% of buffer B (80% acetonitrile and 20% H2O with 0.1% formic acid). For MS acquisition, we used an MS1 Orbitrap scan at 60,000 resolution, automatic gain control (AGC) target of 3 × 106 ions and maximum inject time of 20 ms from 375 to 1,600 m/z; the 15 most-intense ions were submitted to MS2 Orbitrap scan at 30,000 resolution, AGC target of 1 × 105 ions, and maximum inject time of 50 ms (isolation window of 1.4 m/z, normalized collision energy at 27%, and dynamic exclusion of 16 s). The SCX fractions were analyzed with same Agilent HPLC and the same nano-column coupled on-line to an Orbitrap Lumos mass spectrometer (Thermo Fisher Scientific). For these runs, we used a gradient from 6 to 39% buffer B over 100 min with specific MS settings for DSSO cross-links: survey MS1 Orbitrap scan at 60,000 resolution from 375 to 1,500, AGC target of 4 × 105 ions and maximum inject time of 50 ms and MS2 Orbitrap scan at 30,000 resolution, AGC target of 5 × 104 ions, and maximum inject time of 100 ms for detection of DSSO signature peaks (difference in mass of 37.972 Da). The four ions with this specific difference were analyzed with a MS3 Ion Trap scans at AGC target of 2 × 104 ions, maximum inject time of 150 ms for sequencing selected signature peaks (representing the individual peptides).
Data Processing.
The three raw files obtained with classical bottom-up approach were analyzed with MaxQuant version 1.6.17 with all the automatic settings adding Deamidation (N) as dynamic modification against the S. scrofa reference proteome (Uniprot version of 08/2020 with 49,795 entries). With this search, we were able to calculate intensity-based absolute quantification values and created a smaller FASTA file to use for analysis of cross-linking experiments. Raw files for cross-linked cells were analyzed with Proteome Discoverer software suite version 2.4.1.15 (Thermo Fisher Scientific) with the incorporated XlinkX node for analysis of cross-linked peptides as described in ref. 94. Data were searched against the smaller FASTA created in house with “MS2_MS3 acquisition strategy.” For the XlinkX search, we selected full tryptic digestion with three maximum missed cleavages, 10 ppm error for MS1, 20 ppm for MS2, and 0.5 Da for MS3 in Ion Trap. For modifications, we used static Carbamidomethyl (C) and dynamic Oxidation (M), Deamidation (N), and Met-loss (protein N-term). The crosslinked peptides were accepted with a minimum score of 40, minimum score difference of 4, and maximum false discovery rate rate set to 5%; further standard settings were used.
Interactome Analysis, Homology Modeling, and Cross-Link Mapping.
The interaction map for VDAC proteins was generated in R (95) using the igraph package (version 1.2.4.2). Only protein interactions supported with at least two CSMs were included in the final network. Homology models of pig GK and pig VDAC2 were generated in Robetta (96) and fitted into subtomogram average maps by rigid body fitting in ChimeraX. Cross-links were mapped onto the resulting models using ChimeraX.
Acknowledgments
We thank Dr. M. Vanevic for excellent computational support; Dr. D. Vasishtan for sharing scripts for subtomogram averaging; Ingr. C. T. W. M. Schneijdenberg and J. D. Meeldijk for managing the Utrecht University EM Square facility; Stal Schep (Tull en het Waal, The Netherlands) for providing horse semen; M. W. Haaker and Dr. M. Houweling for providing mouse reproductive tracts; Prof. F. Förster and Prof. A. Akhmanova for their comments on early versions of the manuscript; and Prof. E. Y. Jones for insightful discussions. This work benefitted from access to the Netherlands Center for Electron Nanoscopy with support from operators Dr. R. S. Dillard and Dr. C. Diebolder and IT support from B. Alewijnse. R.Z.-C., J.F.H., and A.J.R.H. acknowledge support from Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) funding the Netherlands Proteomics Centre through the X-omics Road Map program (Project 184.034.019). This work was funded by NWO Start-Up Grant 740.018.007 to T.Z.-B.-M. M.R.L. is supported by a Clarendon Fund—Nuffield Department of Medicine Prize Studentship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110996118/-/DCSupplemental.
Data Availability
Subtomogram average maps have been deposited to the EM Data Bank (EMDB) with the following accession numbers: EMDB-12354–EMDB-12357. The model of putative GK-like proteins anchored on a VDAC array has been deposited to the Protein Data Bank (PDB) with the accession number PDB ID 7NIE. MS data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD025562.
References
- 1.Viana M. P., et al., Mitochondrial fission and fusion dynamics generate efficient, robust, and evenly distributed network topologies in budding yeast cells. Cell Syst. 10, 287–297.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Glancy B., et al., Mitochondrial reticulum for cellular energy distribution in muscle. Nature 523, 617–620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent A. E., Turnbull D. M., Eisner V., Hajnóczky G., Picard M., Mitochondrial nanotunnels. Trends Cell Biol. 27, 787–799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore A. S., Holzbaur E. L. F., Mitochondrial-cytoskeletal interactions: Dynamic associations that facilitate network function and remodeling. Curr. Opin. Physiol. 3, 94–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenton A. R., Jongens T. A., Holzbaur E. L. F., Mitochondrial dynamics: Shaping and remodeling an organelle network. Curr. Opin. Cell Biol. 68, 28–36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang J. S., et al., Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137–148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Sheng Z. H., Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol. 202, 351–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutnick A., Banghart M. R., West E. R., Schwarz T. L., The light-sensitive dimerizer zapalog reveals distinct modes of immobilization for axonal mitochondria. Nat. Cell Biol. 21, 768–777 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milner D. J., Mavroidis M., Weisleder N., Capetanaki Y., Desmin cytoskeleton linked to muscle mitochondrial distribution and respiratory function. J. Cell Biol. 150, 1283–1298 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone M. R., et al., Absence of keratin 19 in mice causes skeletal myopathy with mitochondrial and sarcolemmal reorganization. J. Cell Sci. 120, 3999–4008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fawcett D. W., A comparative view of sperm ultrastructure. Biol. Reprod. Suppl. 2, 90–127 (1970). [PubMed] [Google Scholar]
- 12.Fawcett D. W., The mammalian spermatozoon. Dev. Biol. 44, 394–436 (1975). [DOI] [PubMed] [Google Scholar]
- 13.R. C. Firman, L. W. Simmons, Sperm midpiece length predicts sperm swimming velocity in house mice. Biol. Lett. 6, 513–516 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. S. Fisher, E. Jacobs-Palmer, J.-M. Lassance, H. E. Hoekstra, The genetic basis and fitness consequences of sperm midpiece size in deer mice. Nat. Commun. 7, 13652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourmente M., Villar-Moya P., Rial E., Roldan E. R. S., Differences in ATP generation via glycolysis and oxidative phosphorylation and relationships with sperm motility in mouse species. J. Biol. Chem. 290, 20613–20626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marin S., et al., Metabolic strategy of boar spermatozoa revealed by a metabolomic characterization. FEBS Lett. 554, 342–346 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Olson G. E., Winfrey V. P., Identification of a cytoskeletal network adherent to the mitochondria of mammalian spermatozoa. J. Ultrastruct. Mol. Struct. Res. 94, 131–139 (1986). [DOI] [PubMed] [Google Scholar]
- 18.Olson G. E., Winfrey V. P., Mitochondria-cytoskeleton interactions in the sperm midpiece. J. Struct. Biol. 103, 13–22 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Olson G. E., Winfrey V. P., Structural organization of surface domains of sperm mitochondria. Mol. Reprod. Dev. 33, 89–98 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Friend D. S., Heuser J. E., Orderly particle arrays on the mitochondrial outer membrane in rapidly-frozen sperm. Anat. Rec. 199, 159–175 (1981). [DOI] [PubMed] [Google Scholar]
- 21.Woolley D. M., Neesen J., Vernon G. G., Further studies on knockout mice lacking a functional dynein heavy chain (MDHC7). 2. A developmental explanation for the asthenozoospermia. Cell Motil. Cytoskeleton 61, 74–82 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Al-Amoudi A., et al., Cryo-electron microscopy of vitreous sections. EMBO J. 23, 3583–3588 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otani H., Tanaka O., Kasai K., Yoshioka T., Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: Regular dispositions and synchronized changes. Anat. Rec. 222, 26–33 (1988). [DOI] [PubMed] [Google Scholar]
- 24.Ho H.-C. C., Wey S., Three dimensional rendering of the mitochondrial sheath morphogenesis during mouse spermiogenesis. Microsc. Res. Tech. 70, 719–723 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., et al., KLC3 is involved in sperm tail midpiece formation and sperm function. Dev. Biol. 366, 101–110 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Colombini M., VDAC structure, selectivity, and dynamics. Biochim. Biophys. Acta 1818, 1457–1465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson M. J., et al., Immotile sperm and infertility in mice lacking mitochondrial voltage-dependent anion channel type 3. J. Biol. Chem. 276, 39206–39212 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., et al., Glycerol kinase-like proteins cooperate with Pld6 in regulating sperm mitochondrial sheath formation and male fertility. Cell Discov. 3, 17030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada K., Kato H., Miyata H., Ikawa M., Glycerol kinase 2 is essential for proper arrangement of crescent-like mitochondria to form the mitochondrial sheath during mouse spermatogenesis. J. Reprod. Dev. 65, 155–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi Y., Shi Z., Li J., Spata19 is critical for sperm mitochondrial function and male fertility. Mol. Reprod. Dev. 82, 907–913 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Schneider M., et al., Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB J. 23, 3233–3242 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Imai H., et al., Depletion of selenoprotein GPx4 in spermatocytes causes male infertility in mice. J. Biol. Chem. 284, 32522–32532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuda Y., Laugks U., Lučić V., Baumeister W., Danev R., Electron cryotomography of vitrified cells with a Volta phase plate. J. Struct. Biol. 190, 143–154 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Danev R., Buijsse B., Khoshouei M., Plitzko J. M., Baumeister W., Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 111, 15635–15640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M., et al., Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 14, 983–985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Martino C., et al., Morphological, histochemical and biochemical studies on germ cell mitochondria of normal rats. Cell Tissue Res. 196, 1–22 (1979). [DOI] [PubMed] [Google Scholar]
- 37.Perkins G. A., Ellisman M. H., Mitochondrial configurations in peripheral nerve suggest differential ATP production. J. Struct. Biol. 173, 117–127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackenbrock C. R., Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J. Cell Biol. 37, 345–369 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davila M. P., et al., Mitochondrial ATP is required for the maintenance of membrane integrity in stallion spermatozoa, whereas motility requires both glycolysis and oxidative phosphorylation. Reproduction 152, 683–694 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Mukai C., Okuno M., Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol. Reprod. 71, 540–547 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Odet F., Gabel S., London R. E., Goldberg E., Eddy E. M., Glycolysis and mitochondrial respiration in mouse LDHC-null sperm. Biol. Reprod. 88, 95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picard M., et al., Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat. Commun. 6, 6259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X., et al., Kissing and nanotunneling mediate intermitochondrial communication in the heart. Proc. Natl. Acad. Sci. U.S.A. 110, 2846–2851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duvert M., Mazat J.-P., Barets A.-L., Intermitochondrial junctions in the heart of the frog, Rana esculenta. A thin-section and freeze-fracture study. Cell Tissue Res. 241, 129–137 (1985). [DOI] [PubMed] [Google Scholar]
- 45.Guo X. W., Mannella C. A., Conformational change in the mitochondrial channel, VDAC, detected by electron cryo-microscopy. Biophys. J. 64, 545–549 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoogenboom B. W., Suda K., Engel A., Fotiadis D., The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J. Mol. Biol. 370, 246–255 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Gonçalves R. P., Buzhynskyy N., Prima V., Sturgis J. N., Scheuring S., Supramolecular assembly of VDAC in native mitochondrial outer membranes. J. Mol. Biol. 369, 413–418 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Mannella C. A., Structure of the outer mitochondrial membrane: Ordered arrays of porelike subunits in outer-membrane fractions from Neurospora crassa mitochondria. J. Cell Biol. 94, 680–687 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon W. S., Park Y. J., Mohamed S. A., Pang M. G., Voltage-dependent anion channels are a key factor of male fertility. Fertil. Steril. 99, 354–361 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Arcelay E., Salicioni A. M., Wertheimer E., Visconti P. E., Identification of proteins undergoing tyrosine phosphorylation during mouse sperm capacitation. Int. J. Dev. Biol. 52, 463–472 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Mannella C. A., Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications. J. Struct. Biol. 121, 207–218 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Schredelseker J., et al., High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J. Biol. Chem. 289, 12566–12577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomasello M. F., Guarino F., Reina S., Messina A., De Pinto V., The voltage-dependent anion selective channel 1 (VDAC1) topography in the mitochondrial outer membrane as detected in intact cell. PLoS One 8, e81522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayrhuber M., et al., Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 105, 15370–15375 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu F., Lössl P., Rabbitts B. M., Balaban R. S., Heck A. J. R., The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell. Proteomics 17, 216–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fasci D., van Ingen H., Scheltema R. A., Heck A. J. R., Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell. Proteomics 17, 2018–2033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hinsch K. D., et al., Voltage-dependent anion-selective channels VDAC2 and VDAC3 are abundant proteins in bovine outer dense fibers, a cytoskeletal component of the sperm flagellum. J. Biol. Chem. 279, 15281–15288 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Shimada K., et al., ARMC12 regulates spatiotemporal mitochondrial dynamics during spermiogenesis and is required for male fertility. Proc. Natl. Acad. Sci. U.S.A. 118, e2018355118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quillin M. L., Matthews B. W., Accurate calculation of the density of proteins. Acta Crystallogr. D Biol. Crystallogr. 56, 791–794 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Fukuda Y., et al., Epistasis effects of multiple ancestral-consensus amino acid substitutions on the thermal stability of glycerol kinase from Cellulomonas sp. NT3060. J. Biosci. Bioeng. 121, 497–502 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Bystrom C. E., Pettigrew D. W., Branchaud B. P., O’Brien P., Remington S. J., Crystal structures of Escherichia coli glycerol kinase variant S58-->W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion. Biochemistry 38, 3508–3518 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Balogun E. O., et al., Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase of Trypanosoma brucei brucei. FASEB J. 33, 13002–13013 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Schnick C., et al., Structure and non-essential function of glycerol kinase in Plasmodium falciparum blood stages. Mol. Microbiol. 71, 533–545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan Y., Decker W. K., Huq A. H. H. M., Craigen W. J., Retrotransposition of glycerol kinase-related genes from the X chromosome to autosomes: Functional and evolutionary aspects. Genomics 59, 282–290 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Vincent A. E., et al., Quantitative 3D mapping of the human skeletal muscle mitochondrial network. Cell Rep. 26, 996–1009.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mounib M. S., Chang M. C., Effect of in utero incubation on the metabolism of rabbit spermatozoa. Nature 201, 943–944 (1964). [DOI] [PubMed] [Google Scholar]
- 67.Balbach M., et al., Metabolic changes in mouse sperm during capacitation. Biol. Reprod. 103, 791–801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meyers S., Bulkeley E., Foutouhi A., Sperm mitochondrial regulation in motility and fertility in horses. Reprod. Domest. Anim. 54 (suppl. 3), 22–28 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Okunade G. W., et al., Targeted ablation of plasma membrane Ca2+-ATPase (PMCA) 1 and 4 indicates a major housekeeping function for PMCA1 and a critical role in hyperactivated sperm motility and male fertility for PMCA4. J. Biol. Chem. 279, 33742–33750 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Jamil K., White I. G., Induction of acrosomal reaction in sperm with ionophore A23187 and calcium. Arch. Androl. 7, 283–292 (1981). [DOI] [PubMed] [Google Scholar]
- 71.Leung M. R., et al., The multi-scale architecture of mammalian sperm flagella and implications for ciliary motility. EMBO J. 40, e107410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lesich K. A., de Pinho T. G., Dang L., Lindemann C. B., Ultrastructural evidence that motility changes caused by variations in ATP, Mg2+, and ADP correlate to conformational changes in reactivated bull sperm axonemes. Cytoskeleton (Hoboken) 71, 649–661 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Jeulin C., Lewin L. M., Chevrier C., Schoevaert-Brossault D., Changes in flagellar movement of rat spermatozoa along the length of the epididymis: Manual and computer-aided image analysis. Cell Motil. Cytoskeleton 35, 147–161 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Miyata H., et al., Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science 350, 442–445 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Konieczny P., et al., Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J. Cell Biol. 181, 667–681 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steen K., et al., A role for keratins in supporting mitochondrial organization and function in skin keratinocytes. Mol. Biol. Cell 31, 1103–1111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harrison R. A. P., Mairet B., Miller N. G. A., Flow cytometric studies of bicarbonate-mediated Ca2+ influx in boar sperm populations. Mol. Reprod. Dev. 35, 197–208 (1993). [DOI] [PubMed] [Google Scholar]
- 78.Hutcheon K., et al., Analysis of the small non-protein-coding RNA profile of mouse spermatozoa reveals specific enrichment of piRNAs within mature spermatozoa. RNA Biol. 14, 1776–1790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mederacke I., Dapito D. H., Affò S., Uchinami H., Schwabe R. F., High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10, 305–315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tivol W. F., Briegel A., Jensen G. J., An improved cryogen for plunge freezing. Microsc. Microanal. 14, 375–379 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schaffer M., et al., Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197, 73–82 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Wolff G., et al., Mind the gap: Micro-expansion joints drastically decrease the bending of FIB-milled cryo-lamellae. J. Struct. Biol. 208, 107389 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 87.Zeng G. L., A filtered backprojection algorithm with characteristics of the iterative landweber algorithm. Med. Phys. 39, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pettersen E. F., et al., UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Goddard T. D., et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nicastro D., et al., The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 (2006). [DOI] [PubMed] [Google Scholar]
- 91.Heumann J. M., Hoenger A., Mastronarde D. N., Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 175, 288–299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Potel C. M., Lin M.-H., Heck A. J. R., Lemeer S., Defeating major contaminants in Fe3+- immobilized metal ion affinity chromatography (IMAC) phosphopeptide enrichment. Mol. Cell. Proteomics 17, 1028–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wessel D., Flügge U. I., A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 94.Klykov O., et al., Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 13, 2964–2990 (2018). [DOI] [PubMed] [Google Scholar]
- 95.Grant B. J., Rodrigues A. P. C., ElSawy K. M., McCammon J. A., Caves L. S. D., Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 22, 2695–2696 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Kim D. E., Chivian D., Baker D., Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Newport T. D., Sansom M. S. P., Stansfeld P. J., The MemProtMD database: A resource for membrane-embedded protein structures and their lipid interactions. Nucleic Acids Res. 47, D390–D397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Subtomogram average maps have been deposited to the EM Data Bank (EMDB) with the following accession numbers: EMDB-12354–EMDB-12357. The model of putative GK-like proteins anchored on a VDAC array has been deposited to the Protein Data Bank (PDB) with the accession number PDB ID 7NIE. MS data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD025562.