Significance
Glioblastoma (GBM) is the most aggressive brain cancer and highly resistant to therapy, including immunotherapies. It is able to escape immune recognition due to its high heterogeneity, active immunometabolic suppression, and antigen escape mechanisms. Here, we describe an immunotherapy centered around human natural killer (NK) cells engineered to simultaneously target these pathways of immune resistance. These engineered NK cells are able to block adenosine signaling in GBM via CD73 while avoiding antigen escape. We also uncover the functional cooperation between these cells’ intratumoral infiltration and impaired autophagy in GBM as a powerful approach to traffick NK cells into the GBM niche. This NK cell–based immunotherapy provides opportunities to broaden the breadth and versatility of current therapeutic regimens for GBM.
Keywords: natural killer cells, glioblastoma, autophagy, CD73, immunotherapy
Abstract
Tumor antigen heterogeneity, a severely immunosuppressive tumor microenvironment (TME) and lymphopenia resulting in inadequate immune intratumoral trafficking, have rendered glioblastoma (GBM) highly resistant to therapy. To address these obstacles, here we describe a unique, sophisticated combinatorial platform for GBM: a cooperative multifunctional immunotherapy based on genetically engineered human natural killer (NK) cells bearing multiple antitumor functions including local tumor responsiveness that addresses key drivers of GBM resistance to therapy: antigen escape, immunometabolic reprogramming of immune responses, and poor immune cell homing. We engineered dual-specific chimeric antigen receptor (CAR) NK cells to bear a third functional moiety that is activated in the GBM TME and addresses immunometabolic suppression of NK cell function: a tumor-specific, locally released antibody fragment which can inhibit the activity of CD73 independently of CAR signaling and decrease the local concentration of adenosine. The multifunctional human NK cells targeted patient-derived GBM xenografts, demonstrated local tumor site–specific activity in the tissue, and potently suppressed adenosine production. We also unveil a complex reorganization of the immunological profile of GBM induced by inhibiting autophagy. Pharmacologic impairment of the autophagic process not only sensitized GBM to antigenic targeting by NK cells but promoted a chemotactic profile favorable to NK infiltration. Taken together, our study demonstrates a promising NK cell–based combinatorial strategy that can target multiple clinically recognized mechanisms of GBM progression simultaneously.
Glioblastoma (GBM) is the most common and deadliest malignant type of primary brain tumor (1). GBM patients are poorly responsive to traditional treatments, resulting in a grim prognosis that has only modestly improved over the past several decades, motivating the hunt for new treatment approaches (2). So far, chimeric antigen receptor (CAR)-engineered natural killer (NK) cells targeting single GBM antigens—EGFR, EGFRvIII, or ErbB2/HER2—have been limited to the use of NK cell lines, and the overall response rates have been disappointingly low and inconsistent (3–5). These responses appear to mirror the clinical hurdles of single antigen-targeted CAR-T therapies for GBM (6–9). CAR-T cells, administered to target single GBM antigens via intracavitary, intraventricular, or intravenous routes, have so far resulted in inconclusive durable responses (8).
Preclinical and patient data have pointed to the heterogeneity of the GBM tumor microenvironment (TME) as a uniquely complex obstacle to overcome ( 10). This is reflected in immunotherapies tested so far having struggled to improve GBM patient overall survival (OS) in phase III clinical trials (11, 12). GBM induces localized lymphopenia to drive disease progression and resist treatment (13). In addition, the tumor’s heterogeneity is broad, with each of the known GBM subtypes—classical, mesenchymal, neural, and proneural—displaying diverse genetic and epigenetic signatures associated with distinct and variable cell plasticities (14). Not surprisingly, the outgrowth of antigen escape variants has been recorded clinically with most GBM-associated antigens to date, resulting in immune evasion and resistance to treatment (7, 15). And though strategies including dual antigen-targeting or programmable tumor-sensing CARs—so far primarily in the context of adoptive T cell therapy—have been evaluated preclinically to combat such evasion, GBM employs mechanisms beyond antigen escape to avoid targeting. Treatment evasion by GBM is fueled by a heavily immunosuppressive, hypoxic TME, which provides a niche unfavorable to NK cell effector function (16). A subset of GBM cells, glioma stem-like cells (GSCs), contribute to treatment resistance and are poorly recapitulated by conventional GBM model cell lines, including U87MG (17). Metabolic and functional pathways, moreover, converge to fuel the tumor’s invasiveness by driving exhaustion of immune cells (18). For instance, immunometabolic dysregulation of NK cell function in GBM is driven in part by the activity of ecto-5′-nucleotidase (CD73). CD73 is a hypoxic ectoenzyme that we and others have found to be associated with a negative prognosis and has emerged as an attractive clinical target (19, 20). In addition, we have previously shown that the CD73-driven accumulation of extracellular adenosine (ADO) leads to significant purinergic signaling–mediated impairment of NK cell activity (21, 22).
Although they are among the most abundant lymphocytes found within the GBM TME, NK cells are still present in insufficient amounts in these tumors and exhibit a highly dysfunctional phenotype (23, 24). The need to recapitulate NK cell function lost to multiple complex mechanisms not only presents a significant challenge to traditional CAR-NK therapy but requires a greater presence specifically within the tumor tissue to mount meaningful clinical responses.
Here, we describe an example of a multifunctional, engineered human NK cell–based therapy for glioblastoma developed around the programmed targeting of three clinically recognized pathways of GBM progression: antigen escape, immunometabolic suppression, and poor intratumoral NK cell presence. We achieved dual antigen recognition by modifying NK cells with multi-CARs to target disialoganglioside (GD2) and ligands to NK group 2D (NKG2D), which are widely expressed on human GBM (25, 26). Within the same NK cells, we engineered the concomitant local release of an antibody fragment that impairs immunosuppressive purinergic signaling by blocking the activity of CD73 via the cleavage of a tumor-specific linker. This cleavage is dependent on the activity of proteases that are up-regulated in the tumor microenvironment (27). Such local release is able to avoid systemic toxicities owing to its tumor-specific activation that occurs independently of CAR-based signaling.
We report the homing of such multifunctional NK cells was enhanced when administered in conjunction with autophagy inhibitors in patient-derived GBM xenografts. Disabling autophagy further revealed a sophisticated and complex reorganization of anti-GBM immunological responses which could contribute to enhanced CAR-NK effector function. The clinical efficacy of adding an autophagy inhibitor to GBM therapy has shown that such treatments are clinically safe and well tolerated (28). We reveal a nuanced and potentially important role for autophagy inhibitors in adoptive human NK therapy.
These studies aim to expand the repertoire of GBM-targeting NK cell–based immunotherapy and describe a first example of addressing, simultaneously, the challenge of tumor antigen heterogeneity, an immunosuppressive TME, and insufficient intratumoral trafficking of NK cells.
Results
NK Cells Engineered with a Multifunctional Responsive CAR-Based Construct Can Target Multiple Antigens Widely Present in Human GBM.
We previously reported that the expression of NT5E, the gene which encodes the ectoenzyme CD73, is a negative prognostic factor for patient survival in GBM (19). Our analyses also showed that NK cell infiltration into GBM is associated with a better prognosis in patients overexpressing NT5E, indicating that NK cell presence and activation are beneficial to overall GBM patient outcomes.
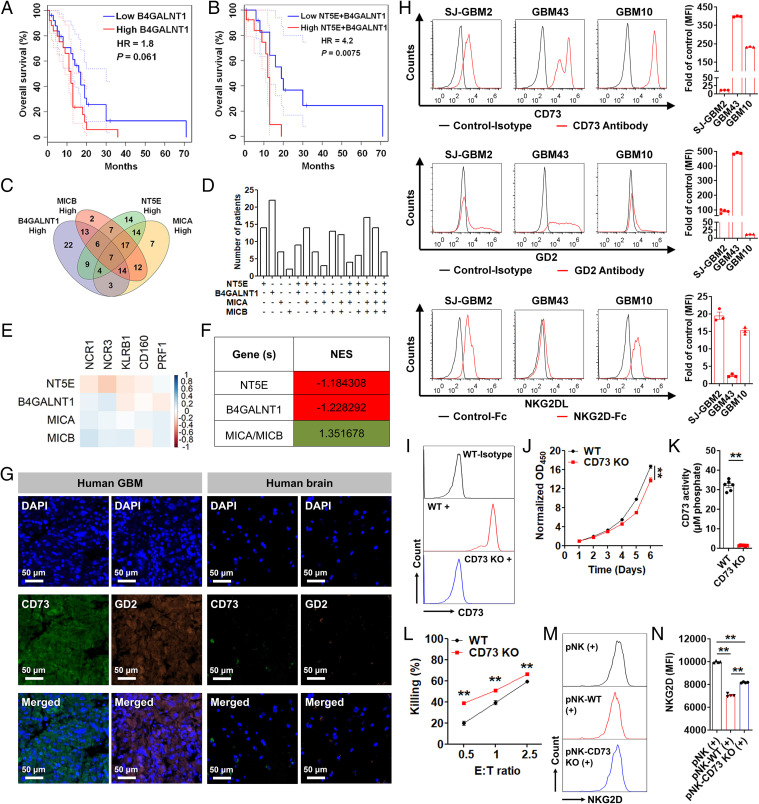
Apart from CD73, the glycolipid disialoganglioside GD2 is well known as a tumor-associated antigen and is frequently overexpressed on malignant glioma in addition to several other cancer types, and its expression in these tumors is associated with increased cell growth, mobility, and invasiveness (29). By analyzing public GBM patient data, we found that BGALNT41, the gene encoding the GD2-producing enzyme β-1,4 N-acetylgalactosaminyltransferase 1, has a negative prognostic value, as measured by a worse overall survival (OS), in GBM patients, particularly those exhibiting the mesenchymal subtype (hazard ratio, HR) = 1.8, P = 0.061; Fig. 1A). GBM patients with the mesenchymal subtype, the most aggressive form of GBM (30), are the most likely to be negatively prognostically affected due to overexpression of CD73 or GD2.
Fig. 1.
Functional and prognostic role of CD73, GD2, and NKG2DL in GBM. (A and B) OS of GBM patients with the mesenchymal subtype based on NT5E and NT5E + B4GALNT1 expression, respectively. (C) Venn diagram showing the number of GBM patients with high expression of at least one of the four genes. (D) Bar graph showing the GBM patient distribution based on gene expression in C. (E) Correlation between normalized expression of selected genes. Pearson's correlation coefficients are shown with continuous gradient colors. (F) Correlation between normalized expression of the indicated genes and the NK signature gene set. Correlation expressed as NES. (G) A representative human GBM tumor section double stained with anti-CD73 antibody (green) and anti-GD2 antibody (orange). A normal brain tissue (cerebrum) was used as a control. (Scale bar: 50 μm; 200× magnification.) (H) Surface expression of CD73, GD2, and NKG2DL on patient-derived GBM cells. Results are reported as fold change over control (n = 3 or 4). (I) Representative histograms depicting the surface expression of CD73 on GBM43 WT and GBM43 CD73 KO cells. (J) In vitro proliferation of GBM43 WT and GBM43 CD73 KO cells (n = 6). (K) Enzymatic activity of CD73 on GBM43 WT and GBM43 CD73 KO cells (n = 6). (L) In vitro cytotoxicity of pNK cells against GBM43 WT and GBM43 CD73 KO cells at indicated E/T ratios over 4 h (n = 5). (M and N) NKG2D expression (MFI) on pNK cells after coincubation with GBM43 WT and GBM43 CD73 KO cells at an E/T ratio of 0.5/1 over 48 h (n = 4). Data represent independent samples. Data are shown as mean ± SEM, **P < 0.01. P values in J, K, and L were determined using the two-tailed Student’s t test and in N using one-way ANOVA analysis.
An analysis of public transcriptional GBM patient datasets further shows that coexpression of NT5E (CD73) and B4GALNT1 (GD2) has a significant negative prognostic value in GBM, leading to a shorter OS compared to GD2 expression alone (HR = 4.2, P = 0.0075; Fig. 1B).
Based on these observations, we classified GBM patients using The Cancer Genome Atlas (TCGA) RNA sequencing (RNA-seq) data into high/low groups based on 50% upper and lower quartiles to identify the expression spread of these genes among GBM patients. By doing this, we identified 151/156 patients overexpressing either NT5E or B4GALNT1. Because we recently described the relationship between CD73 expression and NK cell activation via down-regulated NKG2D, we also stratified patients based on genes encoding ligands to this NK cell–activating receptor, namely MICA or MICB. A Venn diagram showing the association of gene expression among patient numbers of these four genes indicates individual or overlapping patient subsets expressing one or multiple genes (Fig. 1 C and D). In addition, an analysis of patient TCGA RNA-seq data showed that neither NT5E nor BGALNT41 expression in GBM patients correlated with other clinical features, including gender, race, or prior treatments (SI Appendix, Fig. S1).
Stimulated by these observations, we next questioned whether there is any correlation between NK cell presence and expression of either inhibitory genes NT5E or B4GALNT1 or activating genes MICA and MICB in GBM. In doing so, we were interested in defining the functional association between CD73, GD2, and NK cell presence and activation in GBM. We did this by using an NK cell signature gene set (NCR1, NCR3, KLRB1, CD160, and PRF1) (31) to represent NK cells. An analysis of GBM RNA-seq patient datasets (TCGA) revealed NT5E and B4GALNT1 to have a negative correlation with individual genes representing the NK signature set in GBM (r < 0), suggesting that the antigens encoded by these genes act to suppress NK cell function and proliferation in GBM. Alternatively, NKG2D ligand (NKG2DL)-encoding genes, MICA and MICB, overall revealed a positive correlation with individual NK signature genes (r > 0) (Fig. 1E).
Further enrichment analysis against the full NK signature gene set using GBM patient data with a high expression of specific genes indicated a negative correlative expression (normalized enrichment score [NES] < 1) for genes NT5E and B4GALNT1 and a positive correlative expression (NES > 1) for genes MICA and MICB (Fig. 1F and SI Appendix, Table S1).
These results confirmed that not only are NT5E and B4GALNT1 negative prognostic factors in GBM but enhanced NK cell presence in GBM can benefit patients with poor prognosis because of an overexpression of these genes.
To verify overexpression of CD73 and GD2 at the protein level using GBM patient tumors, we performed immunohistochemical staining of human GBM tissue sections. We observed that both CD73 and GD2 are highly expressed in human GBM but not in normal human brain tissues (Fig. 1G). In addition, to validate the relevance of these targets on patient-derived GBM cells, we found that the two protumorigenic markers CD73 and GD2 are also widely and heterogeneously expressed on different types of patient-derived GBM cells, including primary pediatric (SJ-GBM2), adult primary (GBM43), and adult recurrent (GBM10) brain tumor cells (Fig. 1H).
We had previously shown that CD73-produced adenosine is inhibitory to metabolism and cytotoxicity of NK cells (32). To functionally study the effect of CD73 on GBM, we generated and used CRISPR CD73 knockout (KO) patient-derived GBM cells. We found that GBM cells lacking CD73 (GBM43 CD73 KO; Fig. 1I) displayed a significantly delayed in vitro growth rate (Fig. 1J) and an impaired enzymatic activity of CD73 (Fig. 1K and SI Appendix, Fig. S2) when compared to GBM43 wild-type (WT) cells (GBM43 control). In addition, the primary NK (pNK) cells’ lytic activity against GBM43 CD73 KO cells was significantly higher than that against GBM43 WT cells (Fig. 1L). Furthermore, while a coculture of human NK cells with both GBM43 WT and GBM43 CD73 KO cells significantly decreased the expression of activating NK cell receptor NKG2D (MFI), the decrease was significantly more pronounced on NK cells cultured in the presence of GBM43 WT cells than GBM43 CD73 KO cells (Fig. 1 M and N), indicating a direct role for CD73 in down-regulating NK cell activation. Similarly, direct treatment with adenosine also significantly down-regulated NKG2D expression on pNK cells (SI Appendix, Fig. S3). These results indicated that, even though the cognate ligands to NKG2D (NKG2DL) are abundantly present on GBM (Fig. 1H) (25), NKG2D becomes down-regulated on NK cells in a CD73-rich environment such as that found in the GBM TME (Fig. 1 M and N and SI Appendix, Fig. S3) (33).
Based on these data, we hypothesized that cotargeting CD73 and GD2 can restore NK cell function by, among others, rescuing NKG2D-based NK cell activation, enhancing the intratumoral presence of NK cells, lowering adenosine-mediated immunometabolic suppression, and directly impairing GBM progression, thus improving anti-GBM responses.
To achieve combinatorial targeting of these GBM antigens (CD73, GD2, and NKG2DL), we first engineered a bicistronic vector to express two individual CARs simultaneously, yielding NK cells expressing both a GD2.CD28.CD3ζ-CAR and NKG2D.DAP10.CD3ζ-CAR. The GD2.CD28.CD3ζ-CAR was then further engineered in tandem, following a cleavable, tumor-sensitive linker, with an anti-CD73 single-chain variable fragment (scFv) to generate CD73 scFv-GD2.CD28.CD3ζ-CAR (SI Appendix, Fig. S4). This resulted in the localized release of a CD73-blocking antibody fragment in the GBM TME independent of CAR activation. We surmised that a localized release of anti-CD73 scFv can achieve a blockade of CD73 activity more broadly in the TME compared to a variant attached as part of a cell-expressed construct while responsiveness to the TME provides a mechanism to control its release.
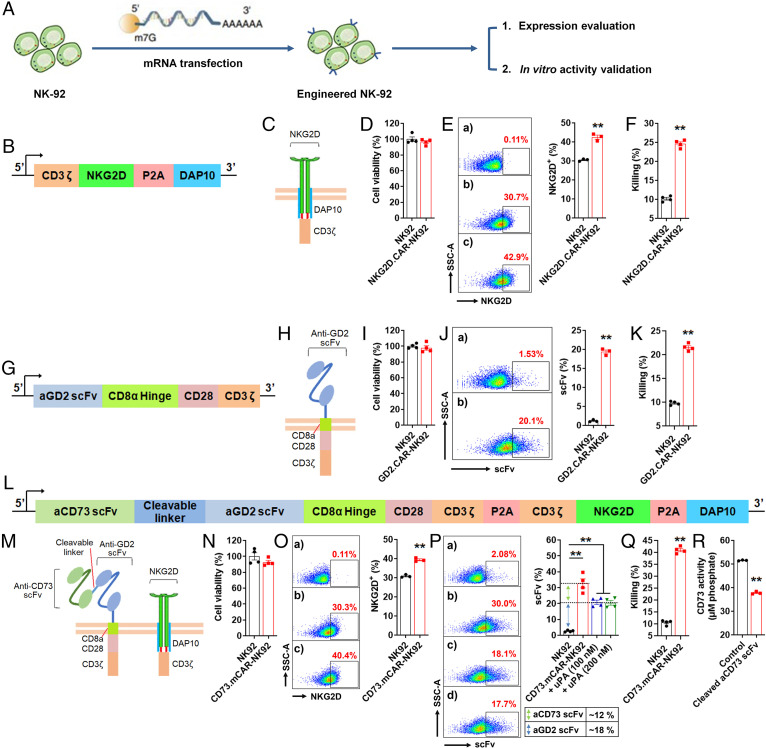
To validate the functionality of individual genetic components of the multifunctional construct, we first engineered NK cells to generate gene-modified cells that express separate elements of the construct. In this step only, we engineered the NK cell line (NK-92) and used messenger RNA (mRNA) transfection to do so (Fig. 2A) in order to initially validate the activity of the multitargeting construct. We thus engineered NK-92 cells expressing, separately, an NKG2D.DAP10.CD3ζ-CAR (Fig. 2 B and C), a GD2.CD28.CD3ζ-CAR (Fig. 2 G and H), and the multifunctional CAR bearing the full construct (Fig. 2 L and M). In all cases (Fig. 2 D, I, and N), the CAR-expressing NK cells retained high viability (>95%). At the same time (Fig. 2E), robust NKG2D.DAP10.CD3ζ-CAR expression on NK-92 cells was achieved. NKG2D.DAP10.CD3ζ-CAR-NK92 cells also showed higher cytotoxic activity against GBM43 cells compared to nontransfected cells (Fig. 2F). Expression, on NK cells, of the GD2.CD28.CD3ζ-CAR (Fig. 2J) also resulted in a correspondingly higher cytotoxicity against GBM43 target cells (Fig. 2K). This confirmed that the GD2.CD28.CD3ζ-CAR construct was capable of effectively recognizing and eliminating GD2-expressing GBM cells. When engineered to express the complete multifunctional sequence, NK-92 cells showed a significant increase in NKG2D expression (Fig. 2O), anti-GD2 scFv expression, and anti-CD73 scFv expression (Fig. 2P). Furthermore, compared to control NK-92 cells, the engineered NK-92 cells that expressed the entire construct (CD73.mCAR-NK92) showed significantly improved killing of GBM43 cells after 8 h coincubation at an effector/target (E/T) ratio of 5 (Fig. 2Q). Additionally, the cleaved anti-CD73 scFv from CD73.mCAR-NK92 cells resulted in significantly decreased enzymatic activity of CD73 in GBM43 cells (Fig. 2R and SI Appendix, Fig. S2), indicating that the anti-CD73 scFv is functional and specific for CD73 and can abrogate adenosine accumulation. Collectively, these results validate the use of CD73, GD2, and NKG2DL as combinatorial GBM targets and provide a rationale for investigating their use for NK cell–based GBM immunotherapy.
Fig. 2.
Functional validation of genetic construct components. (A) A schematic illustration showing the process of engineering NK-92 cells and evaluation of their activity. (B) Transgene structure encoding NKG2D.DAP10.CD3ζ-CAR. (C) Schematic illustration of NKG2D.DAP10.CD3ζ-CAR structure. (D) Cell viability (%) of NK-92 cells after 48 h transfection with NKGD2.CAR gene (n = 4). (E, Left) NKG2D expression on NKG2D.CAR-NK92 cells (n = 3). a-b represent NK-92 cells stained with isotype and APC-anti-NKG2D antibodies. c represents NKG2D.CAR-NK92 cells stained with APC-anti-NKG2D antibody. (Right) NKG2D expression (%) on control NK-92 and NKG2D.CAR-NK92 cells. (F) In vitro cytotoxicity of NKG2D.CAR-NK92 and control NK-92 cells against GBM43 cells at an E/T ratio of 5 over 8 h (n = 4). (G) Transgene structure encoding GD2.CD28.CD3ζ-CAR. (H) Schematic illustration of GD2.CD28.CD3ζ-CAR structure. (I) Cell viability (%) of NK-92 cells after 48 h transfection with GD2.CAR gene (n = 4). (J, Left) Anti-GD2 scFv expression determined by flow cytometry on GD2.CAR-NK92 cells (n = 3). a represents NK-92 cells stained with Biotin-protein L + APC-streptavidin antibodies. b represents GD2.CAR-NK92 cells stained with Biotin-protein L + APC-streptavidin antibodies. (Right) GD2.CAR expression (%), on control NK-92 cells and GD2.CAR-NK92 cells. (K) In vitro cytotoxicity of GD2.CAR-NK92 and control NK-92 cells against GBM43 cells at an E/T ratio of 5 over 8 h (n = 4). (L) Transgene structure of the full multifunctional construct: tumor-responsive anti-CD73 scFv-secreting dual-specific CAR targeting NKG2DL and GD2. (M) Schematic illustration of the full construct. (N) The cell viability (%) of NK-92 cells after 48 h transfection with the full construct gene (n = 4). (O) NKG2D expression on CD73.mCAR-NK92 cells. a–b represent NK-92 cells stained with isotype and APC-anti-NKG2D antibodies; c represents CD73.mCAR-NK92 cells stained with APC-anti-NKG2D. (P, Left) Expression of anti-CD73 scFv and anti-GD2 scFv on CD73.mCAR-NK92 cells determined by flow cytometry. a represents NK-92 cells stained with Biotin-protein L + APC-streptavidin; b–d represent non-treated, 100 nM uPA-treated, and 200 nM uPA-treated CD73.mCAR-NK92 cells stained with Biotin-protein L + APC-streptavidin, respectively. (Right) Calculation of the anti-CD73 scFv and anti-GD2 scFv expression (%) on CD73.mCAR-NK92 cells. (Q) In vitro cytotoxicity of NK-92 and CD73.mCAR-NK92 cells against GBM43 cells at an E/T ratio of 5 over 8 h (n = 4). (R) Enzymatic activity of CD73 on GBM43 cells after incubation with cleaved aCD73 scFv following release from uPA-treated CD73.mCAR-NK92 cells (n = 3). Data represent independent samples. Data are shown as mean ± SEM, **P < 0.01. P values in E, F, J, K, O, Q, and R were determined using the two-tailed Student’s t test and in P using one-way ANOVA analysis.
Multifunctional Engineered Human NK Cells Can Target Patient-Derived GBM Cells While Sparing Normal Cells.
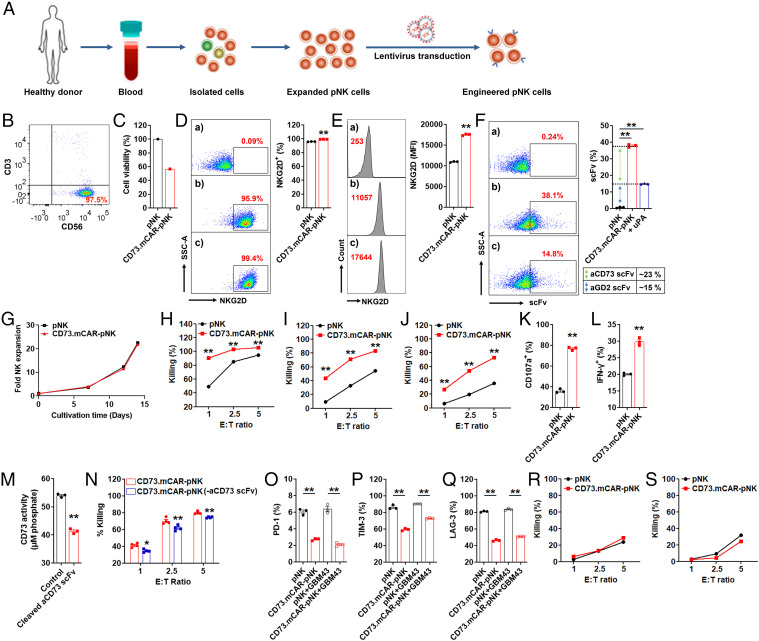
We next tested the ability of human peripheral blood NK cells to express the multifunctional CAR-based construct following viral transduction. To generate virus-transduced primary human NK (pNK) cells, we isolated and expanded pNK cells from healthy adult donors and then engineered them by lentiviral transduction in the presence of DEAE-dextran to express the full multifunctional construct (referred to as CD73.mCAR-pNK) (Fig. 3 A and B and SI Appendix, Fig. S5 A, F, and K), respectively. After two rounds of transduction, an expected, a donor-specific decrease in the viability of the NK cells immediately after the transduction procedure, mainly due to DEAE-dextran (34) (Fig. 3C and SI Appendix, Fig. S6 B, G, and L), was accompanied by significantly increased expression of NKG2D, anti-GD2 scFv, and anti-CD73 scFv fragments (Fig. 3 D–F and SI Appendix, Fig. S5 C, H, and M). However, engineered pNK cells were able to reach high viability. Specifically, CD73.mCAR-pNK cells were able to expand substantially over 2 wk in commercial NK-MACS medium without the use of feeders (Fig. 3G and SI Appendix, Fig. S5 D, I, and N) while retaining high cell viability (∼ 90%) and stable gene expression (SI Appendix, Fig. S6).
Fig. 3.
Generation of CD73.mCAR-pNK cells and in vitro evaluation of their activity. (A) Process for generating CD73.mCAR-pNK cells. (B) Flow cytometry dot plot showing the purity of isolated peripheral blood-derived NK (pNK) cells (CD56+CD3−) after expansion. (C) Cell viability (%) of pNK cells immediately after two rounds of transduction. We set control pNK as 100%. (D and E) NKG2D expression on CD73.mCAR-pNK cells. D indicates % level. E indicates MFI level. a–b represent pNK cells stained with isotype and APC-anti-NKG2D antibodies. c represents CD73.mCAR-pNK cells stained with APC-anti-NKG2D antibody. (F) Expression of anti-CD73 scFv and anti-GD2 scFv on CD73.mCAR-pNK cells. uPA (100 nM) was used to release the anti-CD73 scFv fragment. (Left) Representative flow cytometric dot plot. a represents pNK cells stained with Biotin-Protein L + APC-streptavidin, b–c represent non-treated and uPA-treated CD73.mCAR-pNK cells, respectively stained with Biotin-Protein L + APC-streptavidin. (Right) Calculation of anti-CD73 scFv and anti-GD2 scFv expression (%). (G) Fold expansion of pNK and CD73.mCAR-pNK cells in NK MACS medium. (H–J) In vitro cytotoxicity of control pNK and CD73.mCAR-pNK cells against SJ-GBM2, GBM43, and GBM10 cells at indicated E/T ratios over 4 h, respectively (n = 3 or 5). (K) Degranulation of pNK and CD73.mCAR-pNK cells (% CD107) after 4 h coculture with GBM43 cells (E/T ratio, 5:1) (n = 3). (L) IFN-γ production by pNK and CD73.mCAR-pNK cells (% IFN-γ) after 4 h coculture with GBM43 cells (E/T ratio, 5:1) (n = 3). (M) CD73 activity on GBM43 cells after incubation with cleaved aCD73 scFv following cleavage from uPA-treated CD73.mCAR-NK cells (n = 3). (N) In vitro cytotoxicity of pNK and CD73.mCAR-pNK (following aCD73 scFv cleavage) cells against GBM43 cells at indicated E/T ratios over 4 h (n = 4). (O–Q) Expression (%) of PD-1, TIM-3, and LAG-3 on both pNK and CD73.mCAR-pNK cells after 12 h coculture with or without GBM43 cells (E/T ratio, 5:1) (n = 3). (R and S) In vitro cytotoxicity of pNK and CD73.mCAR-pNK cells against nonmalignant neural cells hCMEC/D3 and HCN-2 at indicated E/T ratios over 4 h (n = 4). Data shown here stand for one representative from four donors. “n” means independent repeats. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01. P values in D, E, H, I, J, K, L, M, and N were determined using the two-tailed Student’s t test and in F, O, P, and Q using one-way ANOVA analysis.
We further evaluated the cytotoxicity of CD73.mCAR-pNK cells against patient-derived GBM targets, showing efficient killing of SJ-GBM2 (pediatric), GBM43 (adult), and GBM10 (recurrent) cells to significantly higher levels than those of control nonengineered NK (referred to as pNK) cells (Fig. 3 H, I, and J and SI Appendix, Fig. S5 E, J, and O). Live imaging of the killing of GBM targets demonstrates the dynamic nature of this process and a higher killing specificity of GBM cells by CD73.mCAR-pNK cells (SI Appendix, Fig. S7 and Movie S1) compared to that by pNK cells (SI Appendix, Fig. S7 and Movie S2). Stimulation by GBM cells contributed to a significantly increased NK cell degranulation and intracellular production of IFN-γ by CD73.mCAR-pNK cells (Fig. 3 K and L). After incubation with anti-CD73 scFv cleaved from CD73.mCAR-pNK cells through urokinase-type plasminogen activator (uPA) treatment, GBM43 cells showed a significantly reduced ability to produce adenosine because of the loss of active CD73 (Fig. 3M and SI Appendix, Fig. S2). In addition, CD73.mCAR-pNK cells lacking the anti-CD73 scFv displayed significantly decreased cytotoxic ability against target GBM43 cells after coculture at various E/T ratios for 4 h (Fig. 3N). When compared with pNK cells, the CD73.mCAR-pNK cells showed significantly lower expression of PD-1, TIM-3, and LAG-3, respectively, upon contact with or without GBM43 target cells, suggesting that they may have improved resistance to functional exhaustion (Fig. 3 O–Q) (35).
In order to address potential off-target effects because of the multispecific targeting of any potential expression of these antigens on nontumor tissues, we evaluated the ability of CD73.mCAR-pNK cells to target healthy cells. CD73.mCAR-pNK cells did not preferentially kill normal cells, specifically those belonging to neural lineages, including hCMEC/D3 and HCN-2 cells. CD73.mCAR-pNK cells exhibited effector activity comparable to that of pNK cells against healthy brain cells (Fig. 3 R and S). Although multiple mechanisms may be involved in the targeting of such cells, including a distinct pattern of inhibitory ligand expression, the low cytotoxicity rates induced by CD73.mCAR-pNK cells could be due to the relatively lower expression profiles for the targeted ligands on these cells compared with GBM cells (SI Appendix, Fig. S8).
Also, the pNK cells underwent the same transduction procedure to individually express either the NKG2D.CAR or the GD2.CAR constructs. In each case, CAR expression yielded significantly improved in vitro cytotoxic activity against GBM43 target cells (SI Appendix, Fig. S9), thus further validating the functionality of each individual component of the multifunctional CAR.
Multifunctional Engineered Human NK Cells Efficiently Target Patient-Derived GBM Tumors In Vivo.
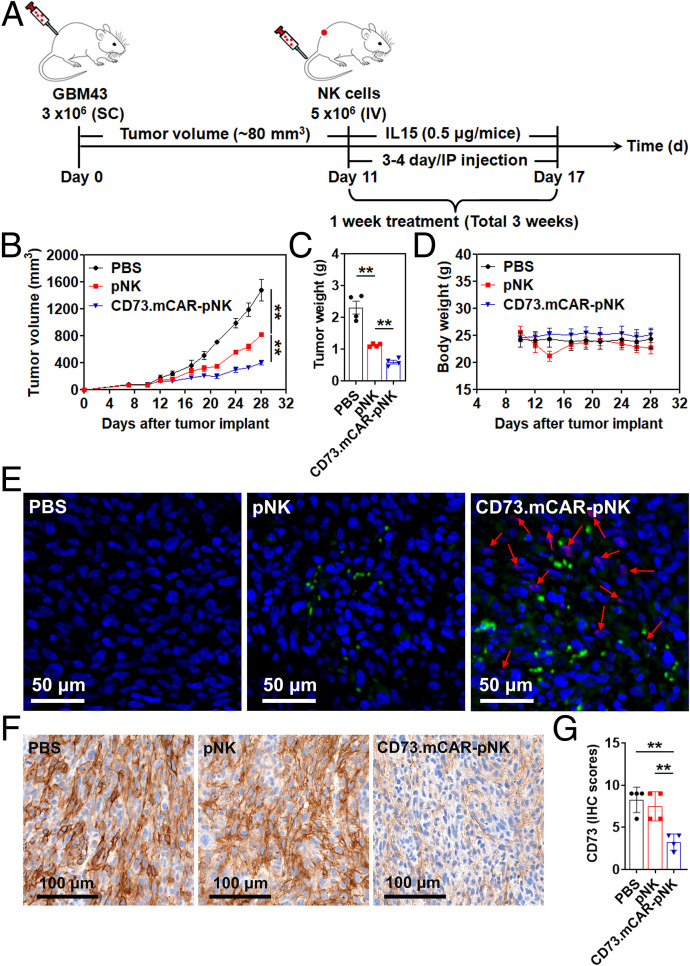
To validate the in vivo antitumor activity of multifunctional engineered CD73.mCAR-pNK cells, we established a subcutaneous xenograft model by engrafting patient-derived GBM43 cells into nonobese diabetic (NOD)-Rag (-)-gamma chain (-) (NRG) mice. The treatment schedule is summarized in Fig. 4A. Treatment with pNK cells significantly delayed tumor growth compared with mice in the phosphate-buffered saline (PBS) control group (mean tumor volume: 815.75 mm3 versus 1,475 mm3, **P < 0.01; mean tumor weight: 1.13 g versus 2.31 g, **P < 0.01; Fig. 4 B and C). Compared to pNK cell–treated mice, CD73.mCAR-pNK cells mediated more potent antitumor responses and significantly reduced the tumor growth rate (mean tumor volume: 401.5 mm3 versus 815.75 mm3, **P < 0.01; mean tumor weight: 0.59 g versus 1.13 g, **P < 0.01; Fig. 4 B and C). There was no significant decrease in body weight of the mice in all groups throughout the entire treatment period (Fig. 4D).
Fig. 4.
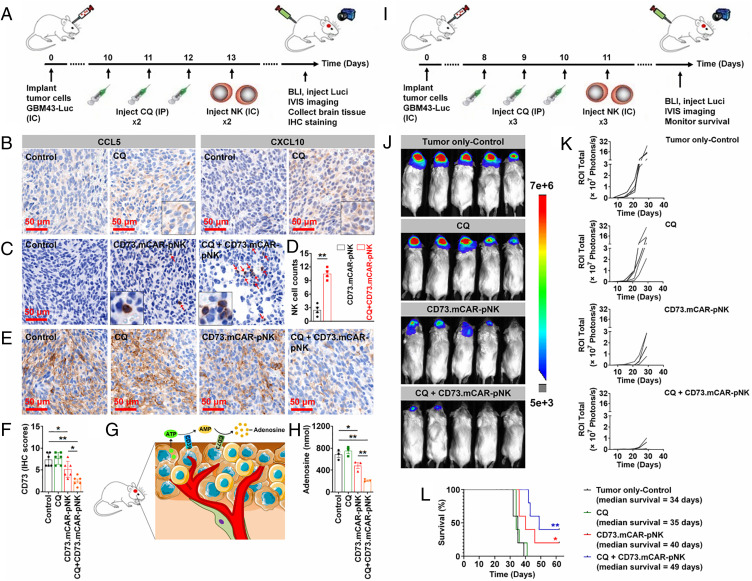
In vivo activity of CD73.mCAR-pNK cells in a patient-derived GBM xenograft model. (A) Schematic diagram illustrating the in vivo treatment program. (B) Tumor growth of individual groups. (C) Tumor weight of the mice after necropsy on day 28 post-tumor implantation. (D) Body weight of the mice during the treatment period. (E) Immunofluorescence staining of NK cells (green) and cleaved anti-CD73 scFv (arrows; red) performed on representative tumor sections using anti-NKp46 antibody and Protein L. (Scale bar: 50 μm; 200× magnification.) (F) IHC staining of CD73 performed on representative tumor sections using anti-CD73 antibody. (Scale bar: 100 μm; 200× magnification.) (G) IHC scores of CD73 expression performed on representative tumor sections in each group. Data were measured from independent samples. Data are shown as mean ± SEM, **P < 0.01. P values in B, C, and G were determined using one-way ANOVA analysis.
NK cell infiltration into GBM xenograft tumors was next investigated by immunofluorescence staining. As shown in Fig. 4E, some adoptively transferred NK cells (green dots) were observed in tumor tissues from pNK cell–treated mice, while substantially higher NK cell numbers were detected in tumors from CD73.mCAR-pNK cell–treated mice. In addition, the locally cleaved anti-CD73 scFv (arrows; red dots) that are able to bind CD73+ tumor cells in vivo were detected in CD73.mCAR-pNK cell–treated mice in the vicinity of tumor cells (Fig. 4E). In vitro data reported here and in previous in vivo studies (36) demonstrated the need for intratumoral protease activity to trigger ligand cleavage. For that reason, we do not expect its activation in blood and, as a result, any detectable presence of scFv in the circulation. The presence of anti-CD73 scFv was observed separately from CD73.mCAR-pNK staining, indicating the anti-CD73 scFv was successfully cleaved. Moreover, we observed a significant decrease in expression of CD73 on tumors in the CD73.mCAR-pNK treatment group compared to other treatment groups (Fig. 4 F and G), indicating an effective reduction of CD73 on cancer cells following treatment with engineered NK cells.
Functional Targeting of Autophagy in GBM Sensitizes the Tumor to Enhanced NK Cell Homing and Activity.
Autophagy is considered a critical cell survival mechanism driving GBM tumorigenesis and leading to its resistance to therapy (37). It has been reported that targeting autophagy is able to sensitize GBM to temozolomide treatment (38). Recently, Mgrditchian et al. reported that blocking autophagy through genetic inhibition of BECN1 augments infiltration of NK cells into melanoma, thereby suppressing tumor growth (39). Stimulated by these observations, we investigated the effects of blocking autophagy in GBM, either genetically by targeting the BECN1 gene through the generation and use of knockdown patient-derived GBM cells (BECN1− GBM43) or pharmacologically via treatment with CQ, a common, FDA-approved autophagy inhibitor (SI Appendix, Fig. S10).
The in vitro viability and proliferation of BECN1− GBM43 cells were unaffected by BECN1 knockdown (Fig. 5A). The impact of BECN1 knockdown on GBM tumor growth and NK cell infiltration was then further evaluated in vivo in Rag1 KO mice, which bear no mature B and T lymphocytes but possess a mature NK cell compartment (Fig. 5B). Tumors from the left flanks of mice engrafted with BECN1− GBM43 cells exhibited significantly slower growth and a smaller size than those arising from control GBM43 cells (Fig. 5C). A significantly higher expression of NKp46 was detected in tumors lacking BECN1, demonstrating a deeper infiltration of NK cells into GBM lacking the ability to perform autophagy compared with control tumors (Fig. 5 D and E). In addition, NK cells in BECN1− tumors showed a higher distribution both at the tumor periphery and in intratumoral areas (SI Appendix, Fig. S11).
Fig. 5.
Effects of targeting autophagy in GBM on NK cell function and homing. (A) In vitro proliferation of GBM43 WT and BECN1− GBM43 cells (n = 3 or 5). (B) Schematic of the experimental design. (C) Tumor growth was monitored and recorded on the indicated days. IHC staining for (D and E) NK cells (Top), (F and G) CCL5 (Middle), and (H and I) CXCL10 (Bottom) on WT and BECN1- tumors. (Scale bar: 50 μm; 200× magnification.) (J) Viability of GBM43 cells after treatment with various concentrations of CQ for 24 h (n = 4). (K) IC50 value of CQ against GBM43 cells. (L and M) Expression of CCL5 and CXCL10 mRNA in CQ-treated GBM43 cells (n = 4). (N and O) Quantification of CCL5 and CXCL10 in the supernatant of GBM43 cells following treatment with CQ for 24 h in the presence or absence of various inhibitors (n = 3). (P) Schematic diagram illustrating the in vivo CQ treatment program. (Q) Tumor growth of individual mice in each group (n = 4). (R) IHC staining for CCL5 (Left) and CXCL10 (Right) performed on representative tumor sections from each group. (Scale bar: 50 μm; 200× magnification.) (S) IHC scores of CCL5 and CXCL10 in control and CQ-treated tumors. (T) Schematic of in vitro experimental direct contact bilayer blood–brain barrier (BBB) model setup. (U) Transmigration of pNK cells through experimental BBB (n = 3). (V) Correlation between normalized expression of selected genes. Pearson's correlation coefficients are shown with continuous gradient colors. (W) Correlation between normalized expression of the entire NK gene set and individual genes. Correlation expressed as NES. Data shown represent independent samples. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01. P values in N, O, and U were determined using one-way ANOVA analysis and in E, G, I, L, M, Q, and S using the two-tailed Student’s t test.
To evaluate whether this increased NK cell infiltration contributed to inhibition in tumor growth, BECN1− GBM43 and control GBM43 cells were implanted subcutaneously into the left/right flanks of NRG mice, which bear no functional B and T lymphocytes as well as no functional NK cells (SI Appendix, Fig. S12A), respectively, and tumor formation and growth were monitored. Tumors from the left flanks of mice engrafted with BECN1− GBM43 cells showed a significantly higher retardation in growth than those from control GBM43 cells (SI Appendix, Fig. S12B). However, the growth rates of tumors originating from BECN1− GBM43 cells in NRG mice were visibly higher than those from the same cells in Rag1 KO mice (SI Appendix, Fig. S12B and Fig. 5C), further indicating that the increase in tumor-infiltrating NK cells driven in response to autophagy blockade contributes to an enhanced antitumor activity (SI Appendix, Fig. S13).
Because inhibiting autophagy resulted in higher NK cell presence in GBM in vivo, we hypothesized that this intervention engages a mechanism that drives NK cells into the GBM milieu. Here, we surmised that specific chemokines might be involved in the autophagy-mediated promotion of NK cell infiltration in GBM. Because of the paucity of insight into the chemokine-mediated trafficking of NK cells to GBM, we used IHC staining to explore the expression levels of chemokines known to be associated with NK cell trafficking to tumors, including CCL5 and CXCL10. As shown in Fig. 5 F–I, the expression of both CCL5 and CXCL10 was up-regulated from relatively low baseline levels in control tumors to significantly higher levels in BECN1− tumors, suggesting that driving NK cells to the tumor bed partially relied on the ability of autophagy-defective tumors to secrete the chemokines CCL5 and CXCL10.
We next studied the effects of CQ-mediated inhibition of autophagy on GBM and found, firstly, that CQ impaired in vitro viability and proliferation of GBM43 cells in a dose-dependent manner after 24 h treatment (half-maximal inhibitory concentration, IC50 = 67.10 μM; Fig. 5 J and K). In addition, CQ-treated GBM43 cells showed a substantial up-regulation of both CCL5 and CXCL10 at the transcriptional level (Fig. 5 L and M). Meanwhile, the chemokines CCL2 and CXCL12 showed decreased mRNA levels in GBM43 cells upon inhibition of autophagy (SI Appendix, Fig. S14). To further evaluate changes in the in vitro protein levels of these chemokines (CCL5 and CXCL10) and gain more insight into the underlying molecular mechanisms, we quantified their concentrations in the conditioned media of CQ-treated GBM43 cells by enzyme-linked immunosorbent assay in the absence and presence of pharmacological inhibitors driving signaling pathways for these chemokines. This included the PI3K inhibitor LY294002, NF-κB inhibitor BAY11-7082, and JNK inhibitor SP600125. CQ-treated GBM43 cells secreted significantly higher amounts of both CCL5 and CXCL10 compared with untreated control cells (Fig. 5 N and O). The elevated production of these chemokines was significantly inhibited by LY294002 or BAY11-7082 but not by SP600125. This suggests a mechanistic link between the enhanced expression of CCL5 and CXCL10 upon inhibition of autophagy on GBM and activation of the PI3K/NF-κB pathway.
In vivo effects of CQ-mediated autophagy inhibition on GBM tumor growth and chemokine secretion were further tested in a subcutaneous xenograft model by engrafting patient-derived GBM43 cells into NRG mice. The treatment schedule is summarized in Fig. 5P. As shown in Fig. 5Q, CQ treatment showed obvious inhibition of tumor growth compared with the PBS group. Additionally, tumor tissues from CQ-treated mice also revealed a significantly higher presence of the chemokines CCL5 and CXCL10 (Fig. 5 R and S).
To determine the involvement of CCL5 and CXCL10 in the migration of NK cells, we tracked the migration of activated pNK cells toward CCL5 and CXCL10 gradients using an in vitro blood–brain barrier model setup by a direct contact coculture of primary human astrocytes and hCMEC/D3 human cerebral microvessel endothelial cells (Fig. 5T) (40). As shown in Fig. 5U, activated pNK cells had a significantly increased ability to migrate along CCL5 (100 ng/mL) as well as CXCL10 (100 ng/mL) ligand gradients.
In addition to the increase in NK cell intratumoral infiltration, we found that CQ-mediated inhibition of autophagy had an effect on the in vitro expression of NKG2DL and GD2 on GBM43 cells. Specifically, NKG2DL expression (both percentage, % and mean fluorescence intensity, MFI) on GBM43 cells increased significantly after treatment with CQ at various concentrations for 24 h (SI Appendix, Fig. S15A). Expression of GD2 (MFI) on GBM43 cells decreased after treatment with CQ at concentrations of 50 μM or higher. Conversely, the percentage expression (%) of GD2 decreased in response to up to 100 μM CQ, after which it remained constant (∼46%) compared to that of control cells (∼58%) (SI Appendix, Fig. S15B). To further investigate the impact of the inhibition of autophagy on the in vitro cytotoxic capacity of NK cells, we incubated CQ-treated GBM43 cells with pNK cells for 4 h at an E/T ratio of 5. We found that treatment with CQ sensitized GBM43 cells to superior killing by pNK cells (SI Appendix, Fig. S15C).
Because of the strong link we observed between NK cell migration and CCL5 and CXCL10 expression in GBM, we were interested in finding out whether a correlation between CXCL5 and CCL10 expression and NK cell presence is evident in patient GBM data. To do so, we analyzed the correlation between the expression profiles of genes encoding these two eponymous chemokines (CCL5 and CXCL10) and the NK cell signature gene set in GBM using an RNA-seq patient dataset (TCGA) in order to infer the relationship between the expression of these chemokine genes and NK cell presence and function. Gene expression analysis revealed a positive correlation between CCL5 and CXCL10 and individual NK signature genes (r > 0; Fig. 5V). Additionally, enrichment analysis against the entire NK signature gene set using the same GBM patient data with high expression of specific genes indicated a positive correlative expression (NES > 1) for genes CCL5 and CXCL10 (Fig. 5W and SI Appendix, Table S1).
Together, these data demonstrate that targeting autophagy improves the infiltration of NK cells into the GBM tumor bed aided by higher levels of the chemokines CCL5 and CXCL10. In addition, disabling autophagy can also potentiate NK cell–mediated cytotoxicity against GBM cells by reprogramming their phenotypic signatures in favor of enhanced NK cell functionality, such as increased NKG2DL expression.
Activity of the Combination of Multifunctional Genetically Engineered Human NK Cells with CQ against Orthotopic Patient-Derived GBM Xenografts.
To assess the anti-tumor efficacy achieved by the combination of CQ with CD73.mCAR-pNK cells, we established an orthotopic GBM xenograft model in NRG mice by intracranial injection of firefly luciferase–expressing GBM43 (GBM43-Luc) cells (SI Appendix, Fig. S16).
Firstly, we focused on the evaluation of the mechanisms involved in the combination therapy through a 2-wk treatment program (Fig. 6A). Overall (SI Appendix, Fig. S17 A and B), mice treated with CQ alone showed no significant effect on tumor growth compared with those in the control group. Conversely, mice that were treated with either CD73.mCAR-pNK cells or CQ + CD73.mCAR-pNK cells showed an obvious reduction in tumor growth as determined by bioluminescence imaging. In particular, tumors on half of the mice treated with CQ + CD73.mCAR-pNK cells showed significant growth arrest during the treatment period. Also, no significant change in body weight of the mice in all groups was observed (SI Appendix, Fig. S18C). Similar to subcutaneous xenograft studies, tumor tissues of mice in CQ-treated groups displayed robustly up-regulated CCL5 and CXCL10 chemokine expression (Fig. 6B). As shown in Fig. 6 C and D, a significantly higher NK cell infiltration was found in GBM tumors of CQ + CD73.mCAR-pNK cell–treated mice compared to mice which received CD73.mCAR-pNK cells alone. Compared to control or CQ-treated groups, the tumors of mice in both CD73.mCAR-pNK cell– and CQ + CD73.mCAR-pNK cells–treated groups displayed decreased CD73 expression, with the latter group showing the most substantial loss of CD73 expression (Fig. 6 E and F). This was associated with a significantly decreased level of extracellular adenosine detected in the tumors of treated mice, most prominently in mice treated with CQ + CD73.mCAR-pNK cells (Fig. 6 G and H and SI Appendix, Fig. S18).
Fig. 6.
In vivo activity of CD73.mCAR-pNK cells and the combination immunotherapy with the pharmacological inhibition of autophagy in an orthotopic patient-derived GBM xenograft model. (A–H) Validation of the mechanisms involved in the combination therapy (four mice/group). (A) Schematic diagram showing the treatment program. (B) IHC staining of CCL5 (Left) and CXCL10 (Right) performed on indicated tumor sections in each group. (Scale bar: 50 μm; 200× magnification.) (C) IHC staining for NK cells performed on tumor sections from indicated groups. (Scale bar: 50 μm; 200× magnification.) (D) Quantification of NK cell infiltration into tumors. Here, cell counts were recorded in four consecutive high-power fields. (E) IHC staining for CD73 performed on tumor sections from indicated groups. (Scale bar: 50 μm; 200× magnification.) (F) IHC scores of CD73 expression performed on tumor sections. (G) Schematic diagram showing extracellular adenosine production in the GBM tumor microenvironment. (H) Adenosine concentration in local brain tissues of mice in each group (n = 3). (I–L) Evaluation of the efficacy of the combination therapy in GBM (five mice/group). (I) Schematic diagram showing the treatment program. (J) Bioluminescence imaging of mice on day 29 after tumor implantation (3-wk treatment program). (K) Tumor growth for individual mice over time monitored using bioluminescence imaging. (L) Kaplan–Meier plot showing mouse survival. Data were measured from independent samples. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01. P values in D were determined using the two-tailed Student’s t test and in F and H using one-way ANOVA analysis. P values in L were determined using log-rank test.
We further evaluated the effects of the combination treatment on the survival benefit of treated GBM-bearing mice using the same orthotopic GBM xenograft model (Fig. 6I). As shown in Fig. 6 J and K, both CD73.mCAR-pNK cell– and CQ + CD73.mCAR-pNK cell–treated groups showed a clear inhibitory effect on tumor growth, with the CQ + engineered NK cell–treated group showing the greatest benefit. In addition, the combination of CQ and CD73.mCAR-pNK cells also prolonged mice survival with a median survival of 49 d, superior to other treatment groups (Fig. 6L).
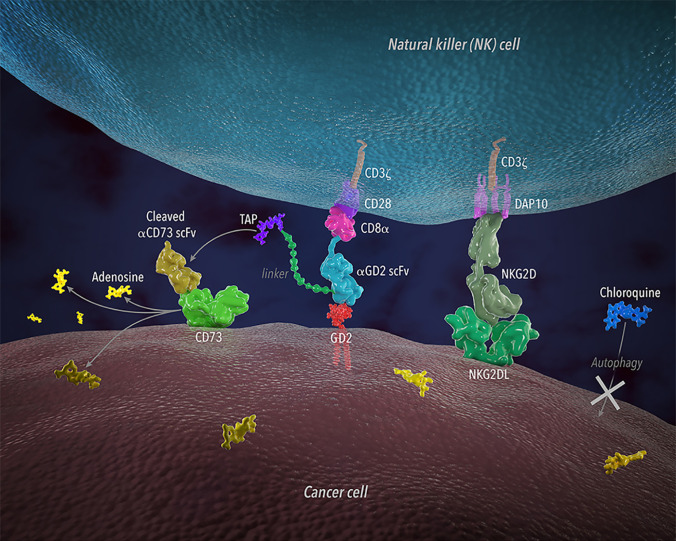
Taken together, all the above studies demonstrated the potent and specific activity of CD73.mCAR-pNK cells against patient-derived adult GBM. These responses were associated with the induction of significant changes in chemokine secretion on GBM following cotherapy with CQ, indicating a reorganization of the GBM TME induced upon disabling autophagy (Fig. 7).
Fig. 7.
Illustration showing the multifunctional, tumor-responsive engineered NK cells and their working mechanisms against GBM.
Discussion
GBM is a particularly challenging tumor to treat due to its intratumor heterogeneity, whereby cells with varying molecular, genetic, and epigenetic characteristics contribute to its ability to evade standard treatments. This complexity results in the need for varying therapeutic strategies which can act in concert to mount meaningful therapeutic responses. The limitations of single antigen-based therapies have been recognized in clinical studies through tumor outgrowth variants. The stringent requirements for target selection place constraints on the targeting strategies that are able to result in effective and sustained responses (41). While combinatorial antigenic targeting with multitargeted CAR-engineered cells has demonstrated an ability to address antigen escape in GBM, poor immune cell infiltration and immunometabolic reprogramming caused by the heavily hypoxic GBM microenvironment have stymied durable responses. The underwhelming responses have resulted in a lack of improvement in the OS of GBM patients treated with CAR-T therapies in phase III trials (42). In the current study, we describe an approach wherein we simultaneously addressed three critical hurdles associated with resistance to immunotherapy for GBM: antigen escape, hypoxic metabolic reprogramming, and poor immune cell infiltration. We developed an approach that strategically combines bifunctional CARs targeting GD2 along with NKG2DL and the tumor-responsive, locally triggered release of CD73-blocking antibody fragments into a single gene-modified NK cell product.
Modulating the strength of antigen recognition to target antigens present in low amounts can enhance activation signals in settings of poor antigenic density, but it also potentiates off-target toxicities for those antigens that are present on normal cells. Targeting a combination of antigens that couples direct antigen recognition (via CAR-based activation) with the sustained, spatially controlled release of soluble antibody fragments subdues potential off-target effects and provides targeting combinations in patients for which these antigens show altered expression over time or in individual tumors. Accordingly, we armed NK cells to express a trifunctional construct which can target heterogeneous combinations of antigens responsively to their expression levels locally and while sparing healthy cells. Part of this is aided by the low expression of at least one of the targeted markers on these cells. The triggerable release of CD73 antibody fragments by GBM tumor-associated proteases (TAPs) results in low concentrations of CD73 in the local TME, a sustained response that depends on CAR expression but is independent of its activation, providing two related but independent mechanisms of tumor recognition.
Several studies have demonstrated that the phenotype and function of GBM tumor-resident NK cells are markedly altered and characterized by significantly lower levels of many activating receptors (24). Mechanistically, GBM patients utilize their own immunosuppressive TME to impair infiltrated NK cell function in favor of GBM escape from immune surveillance and NK-mediated cytotoxicity. These immune escape mechanisms are rooted in GBM’s heterogeneity and represent a complex network of pathways that promote GBM progression (43). NKG2D–NKG2DL interactions play a vital role in activating the anticancer immune response (44). However, NKG2D expression on the surface of NK cells is significantly decreased in response to the high extracellular concentrations of adenosine produced by the ectoenzyme CD73 in the GBM TME. This study, we induced up-regulation of NKG2D by engineering NK cells to express an NKG2D-based CAR, which we previously showed can enhance antitumor responses in combination with the CD73 blockade. The immunosuppression of NK cells via the adenosinergic axis encompasses NK cell metabolism alongside various effector functions (21, 45). Because of the wide expression of CD73, localizing the targeted blockade of this enzyme may be preferential to the systemic administration of anti-CD73 therapies, which we achieved through a tumor-responsive, locally released anti-CD73 scFv. The sustained, low concentration release of anti-CD73 scFv is likely to sustain the metabolic function of NK cells that would otherwise be lost in a setting of unencumbered CD73 enzymatic activity. In that respect, CAR-mediated signaling in combination with the local CD73 blockade is likely to result in antitumor responses that are not only measurable by cytotoxicity against cancer cells but also by their impaired metabolic activity, in turn sustaining the longer-term retention of NK cells in the tumor.
A temporal modulation of antigens also occurs in response to treatment (46). NKG2DL expression was previously found to inversely correlate with GBM cell maturity, with stem-like GBM cells expressing a more substantial, though heterogeneous, pattern of expression of these ligands (46). The increase in NKG2DL we observed in response to CQ might sensitize GBM to effector function via the NKG2DL-R axis but also promote the indirect loss of NKG2D on effector cells through a phenomenon that can be lessened by the induction of NKG2D expression via CARs. The reorganization of the GBM TME induced by CQ included a decrease in GD2 expression, potentially lessening the apoptotic burden imposed on tumor-infiltrating cells induced by gangliosides on GBM (47). To recapitulate some of the stem cell–like properties associated with resistance to therapy owing to the presence of GSCs in patient GBM tumors, we utilized patient-derived xenografts that were generated with cells sourced from patients with primary GBM.
Antigenic targeting may not be sufficient to induce sustained anti-GBM responses. Clinical data have indicated that intratumoral NK cell presence associates positively with patient outcome and survival (48, 49). However, NK cells are present in low amounts in GBM. Among regulating a variety of pathophysiological functions in GBM, chemokine receptor/ligand interactions can drive NK cell trafficking alongside their signaling gradient to result in improved homing (50). However, scant evidence of chemokine-dependent trafficking has been shown in intracranial GBM. Mechanistically, we found that inhibition of autophagy in GBM cells triggered the up-regulation of chemokines, particularly CCL5 and CXCL10. However, we cannot rule out contributions from others to the intratumoral trafficking of NK cells. Conversely, expression of CCL2 and CXCL12, both recognized for their contributing roles to the recruitment of immunosuppressive cells such as tumor-associated macrophages, was significantly decreased in response to autophagy blockade (51, 52). Our preclinical data further demonstrated that inhibition of autophagy potentiates anti-GBM activity of NK cells via mechanisms that may involve a sensitization to their killing by modulating antigen level expression and the activation of proapoptotic pathways on GBM. These data point to novel actionable responses induced by the inhibition of autophagy and CAR-based antigenic targeting. Our data also highlight the sophisticated reorganization of the GBM TME induced by CQ. As an adjuvant, CQ was shown to enhance clinical responses to anti-GBM therapy in double-blinded phase III studies (53). CQ was shown to be able to cross the blood–brain barrier in vivo (54) while preclinical studies have demonstrated that concentrations of 50 mg/kg are suitable for administration regimens against orthotopic glioblastoma xenografts (55). The multiple effects of CQ on the phenotypic signatures and chemokine profiles of GBM in vivo, alongside a limited ability to control tumor burden alone, however, point to a nuanced role in combination regimens which can nonetheless uniquely enhance CAR-based adoptive transfer immunotherapy. Any potential toxicity of CQ, however, needs to be factored into the therapy administration regimens (56,57). The use of the less toxic metabolite hydroxychloroquine is one strategy to mitigate toxicity associated with CQ (58). In our study, we observed no CQ-dependent toxicity to NK cells in vivo.
Overall, this study demonstrated that the development of multifunctional genetically engineered human NK (CD73.mCAR-pNK) cells can result in effective anti-GBM activity supported by a concerted approach of overcoming tumor heterogeneity and multiple immunosuppressive features of the GBM TME. We also unveiled that targeting autophagy functions as an immuno-modulator to promote the homing of effector CAR-NK cells into GBM tumor sites while reprogramming the GBM TME toward sensitization to CAR-based targeting. As a demonstration of the use of human CAR-NK cells in immunotherapy of glioblastoma, we show that while human CAR-NK therapy is a viable option, optimal responses rely on administration regiment, dosage, and frequency of adoptive transfer. It is our belief that the findings from this study will pave the way to fully exploit NK cell–based immunotherapy in clinical settings to treat GBM.
Materials and Methods
In this study, we built a cooperative NK cell–based therapeutic platform for GBM, which combines multispecific, tumor microenvironment-responsive engineered human NK cells with autophagy inhibitors. We performed multiple in vitro and in vivo experiments to evaluate the therapeutic efficacy.
All the cells used were cultured and handled according to required protocols. Peripheral blood samples were collected from human volunteers by the Purdue University Student Health Service Center under the Institutional Review Board (IRB, protocol no. 1804020540) with written informed consent. Written informed consent was obtained from all subjects involved in the study. All procedures performed in studies involving human participants were approved by Purdue University's IRB. All animal-related experiments were performed in full compliance with animal protocols approved by the Purdue University Institutional Animal Care and Use Committee. For each experiment, the number of mice, statistical analyses, and numbers of experimental replicates are described in the figure legends. Investigators were not blinded during follow-up of mice and evaluation of the in vivo experiments.
A detailed description of materials and methods is provided in SI Appendix, Materials and Methods.
Acknowledgments
This work was supported by V Foundation for Cancer Research (Grant No. D2019-039) and the Walther Cancer Foundation (Embedding Tier I/II Grant No. 0186.01). We also gratefully acknowledge the support of the Collaborative Core for Cancer Bioinformatics, the Biological Evaluation Shared Resource, and the Flow Cytometry Shared Resource with support from the Purdue Center for Cancer Research, NIH Grant P30 CA023168, the Indiana University Simon Cancer Center NIH Grant P30 CA082709, and the Walther Cancer Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107507118/-/DCSupplemental.
Data Availability
All study data are included in the article or SI Appendix.
References
- 1.A. C. Tan, D. M. Ashley, G. Y. López, M. Malinzak, H. S. Friedman, M. Khasraw, Management of glioblastoma: State of the art and future directions. CA. A Cancer J. Clin. 70, 299–312 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Ray S., Bonafede M. M., Mohile N. A., Treatment patterns, survival, and healthcare costs of patients with malignant gliomas in a large US commercially insured population. Am. Health Drug Benefits 7, 140–149 (2014). [PMC free article] [PubMed] [Google Scholar]
- 3.Han J., et al., CAR-engineered NK cells targeting wild-type EGFR and EGFRvIII enhance killing of glioblastoma and patient-derived glioblastoma stem cells. Sci. Rep. 5, 11483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genßler S., et al., Dual targeting of glioblastoma with chimeric antigen receptor-engineered natural killer cells overcomes heterogeneity of target antigen expression and enhances antitumor activity and survival. OncoImmunology 5, e1119354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C., et al., ErbB2/HER2-specific NK cells for targeted therapy of glioblastoma. J. Natl. Cancer Inst. 108, djv375 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Brown C. E., et al., Bioactivity and safety of IL13Rα2-redirected chimeric antigen receptor CD8+ T cells in patients with recurrent glioblastoma. Clin. Cancer Res. 21, 4062–4072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown C. E., et al., Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N. Engl. J. Med. 375, 2561–2569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Rourke D. M., et al., A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9, eaaa0984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed N., et al., Her2-specific chimeric antigen receptor–modified virus-specific t cells for progressive glioblastoma: A phase 1 dose-escalation trial. JAMA Oncol. 3, 1094–1101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirosh I., Suvà M. L., Tackling the many facets of glioblastoma heterogeneity. Cell Stem Cell 26, 303–304 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Weller M., et al., ACT IV trial investigators, Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 18, 1373–1385 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Reardon D. A., et al., OS10.3 randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: CheckMate 143. Neuro Oncol. 19, iii21 (2017). [Google Scholar]
- 13.Kim W. J., et al., Clinical observation of lymphopenia in patients with newly diagnosed glioblastoma. J. Neurooncol. 143, 321–328 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Friedmann-Morvinski D., Glioblastoma heterogeneity and cancer cell plasticity. Crit. Rev. Oncog. 19, 327–336 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Chuntova P., Downey K. M., Hegde B., Almeida N. D., Okada H., Genetically engineered T-cells for malignant glioma: Overcoming the barriers to effective immunotherapy. Front. Immunol. 9, 3062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pombo Antunes A. R., et al., Understanding the glioblastoma immune microenvironment as basis for the development of new immunotherapeutic strategies. eLife 9, e52176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Hora C. C., Schweiger M. W., Wurdinger T., Tannous B. A., Patient-derived glioma models: From patients to dish to animals. Cells 8, 1177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woroniecka K., et al., T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin. Cancer Res. 24, 4175–4186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Matosevic S., NT5E/CD73 as correlative factor of patient survival and natural killer cell infiltration in glioblastoma. J. Clin. Med. 8, 1526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Association for Cancer Research, Targeting CD73 may improve immunotherapy efficacy in glioblastoma. Cancer Discov. 10, 175 (2020). [Google Scholar]
- 21.Wang J., Matosevic S., Adenosinergic signaling as a target for natural killer cell immunotherapy. J. Mol. Med. (Berl.) 96, 903–913 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Lupo K. B., Chambers A. M., Matosevic S., Purinergic targeting enhances immunotherapy of CD73+ solid tumors with piggyBac-engineered chimeric antigen receptor natural killer cells. J. Immunother. Cancer 6, 136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kmiecik J., Zimmer J., Chekenya M., Natural killer cells in intracranial neoplasms: Presence and therapeutic efficacy against brain tumours. J. Neurooncol. 116, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Close H. J., et al., Expression profiling of single cells and patient cohorts identifies multiple immunosuppressive pathways and an altered NK cell phenotype in glioblastoma. Clin. Exp. Immunol. 200, 33–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss T., et al., NKG2D-dependent antitumor effects of chemotherapy and radiotherapy against glioblastoma. Clin. Cancer Res. 24, 882–895 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Longee D. C., et al., Disialoganglioside GD2 in human neuroectodermal tumor cell lines and gliomas. Acta Neuropathol. 82, 45–54 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Hu J., et al., Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proc. Natl. Acad. Sci. U.S.A. 108, 15984–15989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Hernández M., et al., Targeting autophagy for cancer treatment and tumor chemosensitization. Cancers (Basel) 11, E1599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nazha B., Inal C., Owonikoko T. K., Disialoganglioside GD2 expression in solid tumors and role as a target for cancer therapy. Front. Oncol. 10, 1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedele M., Cerchia L., Pegoraro S., Sgarra R., Manfioletti G., Proneural-mesenchymal transition: Phenotypic plasticity to acquire multitherapy resistance in glioblastoma. Int. J. Mol. Sci. 20, 2746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Böttcher J. P., et al., NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers A. M., et al., Adenosinergic signaling alters natural killer cell functional responses. Front. Immunol. 9, 2533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crane C. A., et al., TGF-beta downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro-oncol. 12, 7–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong Y., et al., Rosuvastatin enhances VSV-G lentiviral transduction of NK cells via upregulation of the low-density lipoprotein receptor. Mol. Ther. Methods Clin. Dev. 17, 634–646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judge S. J., Murphy W. J., Canter R. J., Characterizing the dysfunctional NK cell: Assessing the clinical relevance of exhaustion, anergy, and senescence. Front. Cell. Infect. Microbiol. 10, 49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desnoyers L. R., et al., Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med. 5, 207ra144 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Yang K., Niu L., Bai Y., Le W., Glioblastoma: Targeting the autophagy in tumorigenesis. Brain Res. Bull. 153, 334–340 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Yan Y., et al., Targeting autophagy to sensitive glioma to temozolomide treatment. J. Exp. Clin. Cancer Res. 35, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mgrditchian T., et al., Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 114, E9271–E9279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulczar C., Lubin K. E., Lefebvre S., Miller D. W., Knipp G. T., Development of a direct contact astrocyte-human cerebral microvessel endothelial cells blood-brain barrier coculture model. J. Pharm. Pharmacol. 69, 1684–1696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sampson J. H., Maus M. V., June C. H., Immunotherapy for brain tumors. J. Clin. Oncol. 35, 2450–2456 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Akhavan D., et al., CAR T cells for brain tumors: Lessons learned and road ahead. Immunol. Rev. 290, 60–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeCordova S., et al., Molecular heterogeneity and immunosuppressive microenvironment in Glioblastoma. Front. Immunol. 11, 1402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raulet D. H., Gasser S., Gowen B. G., Deng W., Jung H., Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 31, 413–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambers A. M., Matosevic S., Immunometabolic dysfunction of natural killer cells mediated by the hypoxia-CD73 axis in solid tumors. Front. Mol. Biosci. 6, 60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flüh C., et al., NKG2D ligands in glioma stem-like cells: Expression in situ and in vitro. Histochem. Cell Biol. 149, 219–233 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Yeh S.-C., et al., Glycolipid GD3 and GD3 synthase are key drivers for glioblastoma stem cells and tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 113, 5592–5597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S., et al., Prognostic significance of tumor-infiltrating natural killer cells in solid tumors: A systematic review and meta-analysis. Front. Immunol. 11, 1242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Matosevic S., Functional and metabolic targeting of natural killer cells to solid tumors. Cell. Oncol. (Dordr.) 43, 577–600 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Müller N., et al., Engineering NK cells modified with an EGFRvIII-specific chimeric antigen receptor to overexpress CXCR4 improves immunotherapy of CXCL12/SDF-1α-secreting glioblastoma. J. Immunother. 38, 197–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Würth R., Bajetto A., Harrison J. K., Barbieri F., Florio T., CXCL12 modulation of CXCR4 and CXCR7 activity in human glioblastoma stem-like cells and regulation of the tumor microenvironment. Front. Cell. Neurosci. 8, 144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang A. L., et al., CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 76, 5671–5682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weyerhäuser P., Kantelhardt S. R., Kim E. L., Re-purposing chloroquine for glioblastoma: Potential merits and confounding variables. Front. Oncol. 8, 335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou W., et al., Chloroquine against malaria, cancers and viral diseases. Drug Discov. Today 25, 2012–2022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu S. P. C., et al., Temozolomide, sirolimus and chloroquine is a new therapeutic combination that synergizes to disrupt lysosomal function and cholesterol homeostasis in GBM cells. Oncotarget 9, 6883–6896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferrante A., Goh D. H. B., The effect of anti-malarial drugs on human natural killer cells in vitro. Parasite Immunol. 6, 571–580 (1984). [DOI] [PubMed] [Google Scholar]
- 57.Austin Taylor M., Bennett M., Kumar V., Schatzle J. D., Functional defects of NK cells treated with chloroquine mimic the lytic defects observed in perforin-deficient mice. J. Immunol. 165, 5048–5053 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Cook K. L., et al., Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin. Cancer Res. 20, 3222–3232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article or SI Appendix.