Significance
For plants grown in a crowded environment, PHYTOCHROME INTERACTING FACTOR 7 (PIF7) plays a critical role by initiating a series of adaptive growth responses. Here, we demonstrate that, in addition to transcription activity and subcellular localization, the PIF7 protein level, which is stringently regulated, is also important for shade avoidance responses. We identified two ubiquitin-specific proteases, UBP12 and UBP13, which positively regulate rapid plant growth in response to shade light. These two ubiquitin proteases directly interact with PIF7 and protect the latter from destruction by 26S proteasomes. The dynamic changes of PIF7 abundance regulated by UBP12 and UBP13 provide insight into the roles of posttranslational modifications of PIF7 in integrating environmental changes with endogenous responses.
Keywords: shade avoidance response, cell elongation, PIF7, UBP12, UBP13
Abstract
Changes in light quality caused by the presence of neighbor proximity regulate many growth and development processes of plants. PHYTOCHROME INTERACTING FACTOR 7 (PIF7), whose subcellular localization, DNA-binding properties, and protein abundance are regulated in a photoreversible manner, plays a central role in linking shade light perception and growth responses. How PIF7 activity is regulated during shade avoidance responses has been well studied, and many factors involved in this process have been identified. However, the detailed molecular mechanism by which shade light regulates the PIF7 protein level is still largely unknown. Here, we show that the PIF7 protein level regulation is important for shade-induced growth. Two ubiquitin-specific proteases, UBP12 and UBP13, were identified as positive regulators in shade avoidance responses by increasing the PIF7 protein level. The ubp12-2w/13–3 double mutant displayed significantly impaired sensitivity to shade-induced cell elongation and reproduction acceleration. Our genetic and biochemical analysis showed that UBP12 and UBP13 act downstream of phyB and directly interact with PIF7 to maintain PIF7 stability and abundance through deubiquitination.
Light is a critical environmental factor to all life on earth. Besides providing energy to photosynthesis, sunlight also acts as a signal to regulate plants growth and development (1). When grown under a crowded environment, the presence of neighboring vegetation modifies the light quality experienced by plants by reducing the red/far-red light (R/FR) ratio (2). To reduce the degree of existing or future shade caused by neighbors, a number of adaptive responses in plant architecture and function are triggered, including rapid elongation of stems and petioles, upward bending of leaves, reduced leaf lamina expansion, and early flowering. Collectively, these morphological adaptations are referred to as shade avoidance responses (SARs) (2, 3).
Plants use photoreceptors to perceive changes of ambient light environment (4–6). The reduced R/FR ratio of light is mainly sensed by the photoreceptor phytochrome B (phyB) (4, 5). After perception by phyB, shade light (SL) converts phyB from the far-red light–absorbing active form (Pfr) to the red light–absorbing inactive form (Pr) (7–9), allowing dephosphorylation and accumulation of a subset of basic helix–loop–helix transcription factors, PHYTOCHROME INTERACTING FACTORS (PIFs) (10, 11). Activated PIFs bind to promoters of shade response genes to promote their expression. Of the PIFs reported, PIF4, PIF5, and PIF7 promote SARs redundantly with PIF7 playing a major role (11–14). The abundance and phosphorylation of PIF7 are regulated by light quality; however, detailed molecular mechanisms regarding to how the posttranslational modifications of PIF7 are regulated are still largely unknown (11, 15).
Protein ubiquitination is a critical posttranslational modification mediating a large number of eukaryotic cellular processes and signaling pathways. Proteins polyubiquitinated by specific ubiquitin E3 ligases are subsequently sent to 26S proteasomes for degradation, which is essential for protein turnover and resetting of signaling pathways (16, 17). Like phosphorylation, protein ubiquitination is a dynamic and reversible process. A ubiquitinated protein can be deubiquitinated by deubiquitinating enzymes (DUBs), which rescue the targeted proteins from destruction (18). The Arabidopsis genome encodes more than 1,000 E3 ligases but not more than 100 DUBs (19, 20). This suggests that DUBs likely have multiple targets. UBP12 and UBP13, two closely homologous proteins from the Ubiquitin-Specific Proteases (UBPs/USPs) subfamily (19, 21, 22), regulate many aspects of plant growth and development including pathogen immunity, leaf senescence, photoperiodic flowering, circadian clock, cell size, root meristem maintenance, and JA signaling pathway (22–29). To gain a better understanding of the ubiquitination dynamics in plant growth and development, additional functions and substrates of UBP12 and UBP13 would need to be identified and characterized. Using genetic and molecular approaches, we demonstrate here that UBP12 and UBP13 play critical roles in accelerating SARs. These UBPs promote shade-mediated changes of plant architecture by direct binding to PIF7 and preventing its degradation.
Results
UBP12 and UBP13 Positively Regulate SARs.
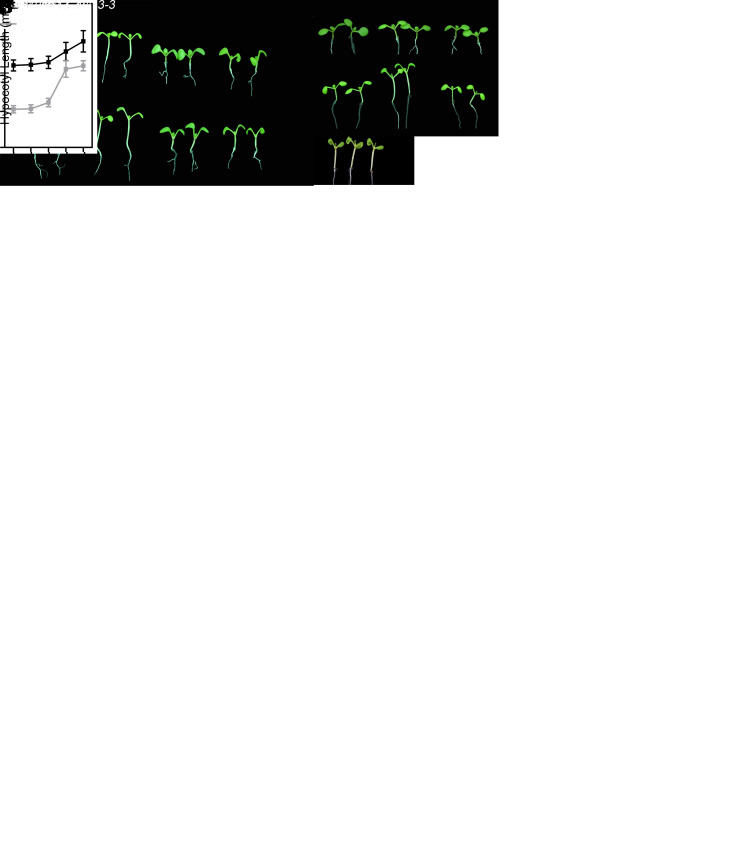
UBP12 and UBP13 loss-of-function plants display short petioles and dwarfism phenotype (22, 23), suggesting these two UBPs are involved in regulating cell elongation during plant development. Light is the most important environmental factor regulating cell elongation. Changes in light quality dramatically alter the stem growth and hyponasty of plants. To examine whether UBP12 and UBP13 are involved in regulating growth and development mediated by changes in light quality, we analyzed the responses of various UBP12/UBP13 mutants and overexpressors to SL with a reduced ratio of R/FR (low R/FR). The hypocotyl elongation of the ubp13-3 single mutant did not show any difference to that of wild type (WT) under both normal white light (WL) and SL conditions. However, ubp12-2w, a weak double mutant of UBP12 and UBP13 (22), showed reduced sensitivity to SL-induced hypocotyl growth. Moreover, ubp12-2w/13–3, a stronger double mutant, displayed a more severe phenotype than ubp12-2w with near complete insensitivity to SL (Fig. 1 A and B). The results from the scanning electron microscopy showed that the short hypocotyl phenotype of ubp12-2w/13–3 double mutant under shade condition was mainly caused by an inhibition of cell elongation but not a reduction in cell number (SI Appendix, Fig. S1). In contrast to ubp12-2w and ubp12-2w/13–3, transgenic plants overexpressing UBP12 and UBP13 showed slightly longer hypocotyls than WT when grown under SL (SI Appendix, Fig. S2). We also investigated the petiole length and flowering time of WT and ubp12-2w/13–3 under both WL and SL conditions. Consistent with our observations with seedlings, petiole elongation and flowering acceleration induced by SL were greatly impaired in ubp12-2w/13–3 compared to WT (Fig. 1 C–F). Taken together, our physiological results indicate that UBP12 and UBP13 act as positive regulators of SARs.
Fig. 1.
UBP12 and UBP13 are positive regulators of SAR. (A and B) Hypocotyl phenotypes (A) and hypocotyl lengths (B) of 8-d-old seedlings grown under WL or SL conditions. (Scale bar: 1 cm.) The values shown are mean ± SD (n ≥ 20). *P < 0.05 and **P < 0.01; based on Student’s t test. (C and D) Phenotypes of 3-wk-old WT (Col-0) and ubp12-2w/13–3 grown under WL or SL conditions. Plants grown under WL for 10 d were shifted to SL or kept under WL for an additional 10 d. (Scale bar: 1 cm.) (E and F) Petiole length of the third and fourth leaf (E) and leaf number (F) from plants analyzed in C and D. The values shown are mean ± SD (n ≥ 10). *P < 0.05 and **P < 0.01; based on Student’s t test.
To further determine whether the deubiquitination catalytic activity of UBP12 is required for shade responses, we complemented the deficiency in ubp12-2w/13–3 with UBP12 (WT) and UBP12 (C208S), which encodes a catalytically inactive form of UBP12 (22, 24). Transgenic plants with equivalent UBP12 expression levels were used to examine their responses to SL (SI Appendix, Fig. S3E). UBP12 (WT), but not UBP12 (C208S), can partially but significantly rescue the hypocotyl and petiole responses of ubp12-2w/13–3 to SL (SI Appendix, Fig. S3 A–D). These results indicate that the DUB activity of UBP12 is critical for its function in SARs.
We also investigated the transcript and protein levels of UBP12 and UBP13 under WL and SL. The transcription levels of UBP12 and UBP13 were slightly reduced after the transfer from WL to SL for 8 h (SI Appendix, Fig. S4 A and B); however, UBP12 and UBP13 protein abundance was not significantly changed by SL treatment (SI Appendix, Fig. S4 C and D). These results suggest SL may regulate the functions of UBP12 and UBP13 through increasing their catalytic activities or their association with substrates but not their expression levels.
UBP12 and UBP13 Promote SARs Downstream from phyB, and Their Action Is Dependent on PIF7.
Among the five phytochrome photoreceptors, phyB plays a major role in light quality detection (2, 4, 5). phyB loss-of-function mutants show elongated hypocotyls and petioles and early flowering even under high R/FR light condition. To determine the genetic relationship between phyB and UBP12/UBP13, we generated a triple mutant, phyB/ubp12-2w/13–3, and compared its sensitivity to SL with phyB. We found that ubp12-2w/13–3 greatly rescued the constitutive shade response of phyB (Fig. 2 A and B and SI Appendix, Fig. S5), indicating that UBP12 and UBP13 act downstream of phyB in SARs.
Fig. 2.
The promotion of shade responses by UBP12 and UBP13 depends on the function of PIF7. (A) Phenotypes of WT (Col-0), phyB, ubp12-2w/13–3, and phyB/ubp12-2w/13–3 under WL and SL conditions. (Scale bar: 1 cm.) (B) Hypocotyl lengths of plants shown in A. The values shown are mean ± SD (n ≥ 20). *P < 0.05 and **P < 0.01; based on Student’s t test. (C) The hypocotyl response of WT (Col-0) and ubp12-2w/13–3 grown under WL and SL to PIC treatment. The seedlings were grown on the medium with different concentrations (0, 0.01, 0.1, 1.0, and 2.0 µM) of PIC and kept under WL for 4 d before being transferred to shade or remaining under WL for an additional 4 d. (Scale bar: 1 cm.) (D and E) Hypocotyl lengths of seedlings indicated in C under WL (D) and SL (E). The values shown are mean ± SD (n ≥ 20). *P < 0.05 and **P < 0.01; based on Student’s t test. (F–I) The expression of shade-induced genes in WT (Col-0) and ubp12-2w/13–3 under WL and SL conditions. The values shown are mean ± SD. *P < 0.05 and **P < 0.01; based on Student’s t test. (J and K) Genetic interaction between PIF7 and UBP12/UBP13 in SAR. Hypocotyl phenotypes and lengths are shown in J and K, respectively. (Scale bar: 1 cm.) The values shown are mean ± SD (n ≥ 20). *P < 0.05 and **P < 0.01; ns, not significant (P ≥ 0.05); based on Student’s t test. (L and M) The expression of shade response genes in various genotypes shown in J and K. Plants were grown under WL for 7 d, and then half of them were treated under SL for 4 h. The values shown are mean ± SD. *P < 0.05 and **P < 0.01; ns, not significant (P ≥ 0.05); based on Student’s t test.
Previous studies demonstrated after perception by phyB, SL increases auxin accumulation greatly and rapidly by activation of auxin biosynthesis genes (11, 30, 31). To investigate whether UBP12/UBP13-mediated SARs are dependent on auxin biosynthesis, we analyzed the responses of WT and ubp12-2w/13–3 to different concentrations of picrolam (PIC), an analog of auxin. Fig. 2 C–E shows that hypocotyls of ubp12-2w/13–3 were more sensitive to PIC than those of WT under both WL and SL conditions. Exogenous PIC application significantly rescued the short hypocotyl phenotype of ubp12-2w/13–3 under SL (Fig. 2 C and E). Consistent with the physiological results, transcript levels of auxin biosynthesis genes YUC8 and YUC9, and auxin response genes IAA19 and IAA29 in WT were induced after transfer to SL for 4 h. The induction, however, was largely impaired in ubp12-2w/13–3 (Fig. 2 F–I). Our results demonstrate a critical role of UBP12 and UBP13 in linking SL perception and auxin biosynthesis.
The accumulation of newly synthesized auxin, which is required for the rapid growth in response to SL, is regulated by a group of PIFs, with PIF7 playing a prominent role (11). SL relieves the repression from phyB and increases the activity and abundance of PIF7, which directly binds to promoter regions of many auxin biosynthesis genes to activate their expression (11, 15). To determine the epistatic relationship between PIF7 and UBP12/UBP13, we crossed pif7 with ubp12-2w/13–3, UBP12-OE (UBQ10::UBP12-HA), and UBP13-OE (UBQ10::UBP13-HA), and homozygous plants were used for shade response analyses. Consistent with previous reports, pif7 single mutant showed greatly decreased sensitivity to shade-induced growth, although it could still respond to SL (Fig. 2 J and K), which is likely caused by the functional redundancy of PIFs. The responses of ubp12-2w/13–3 hypocotyls to SL were even more severe than those of pif7, losing almost all of their shade responsiveness (Fig. 2 J and K), suggesting that, in addition to PIF7, UBP12 and UBP13 may also regulate the functions of other PIFs. The hypocotyl phenotype of ubp12-2w/13–3 under both WL and SL were not significantly altered by pif7 (Fig. 2 J and K), suggesting that PIF7 and UBP12/UBP13 act in the same pathway in regulating shade-induced growth. Moreover, UBP12-OE and UBP13-OE were not able to rescue the insensitivity phenotype of pif7 to SL (Fig. 2 J and K). Consistent with the genetic results, shade-induced expression of YUC8 and IAA19 in pif7/UBP12-OE and pif7/UBP13-OE were significantly impaired compared to UBP12-OE and UBP13-OE but similar to those in pif7 (Fig. 2 L and M). Taken together, all these data support the notion that regulation of SARs mediated by UBP12/UBP13 is dependent on PIF7 function.
UBP12 and UBP13 Directly Interact with PIF7.
The genetic relationship between PIF7 and UBP12/UBP13 suggested the possibility that their encoded proteins may have a direct biochemical relationship. To explore this hypothesis, we checked for possible interaction between PIF7 and UBP12 or UBP13. Yeast two-hybrid assays showed PIF7 interacts with UBP12 and UBP13 in yeast cells (Fig. 3A). The interaction was confirmed by in vitro assays using GST-UBP12 or GST-UBP13 to pull down purified MBP-PIF proteins. PIF7 and PIF3 associated strongly with UBP12 and UBP13, whereas PIF4 and PIF5 had a weak interaction with UBP13 but not UBP12 (Fig. 3B). Therefore, besides PIF7, UBP12/UBP13 may also regulate the functions of several other PIFs. Together, these results supported the notion that PIF7 can directly interact with UBP12/UBP13. In addition to in vitro interactions, strong fluorescence signals were observed in the nuclei of cells cotransformed with PIF7 combined with UBP12 or UBP13 in a bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana (Fig. 3C). Moreover, coimmunoprecipitation (co-IP) assays in Arabidopsis further confirmed the in vivo association between UBP12 and PIF7 under both WL and SL conditions (Fig. 3D).
Fig. 3.
PIF7 interacts with UBP12 and UBP13 in vitro and in vivo. (A) Interaction between PIF7 and UBP12 or UBP13 in yeast two-hybrid assays. (B) In vitro pull-down assays of UBP12 and UBP13 with PIF transcription factors. * indicates the main band of each MBP-PIF protein. (C) BIFC assay showing PIF7 interacts with UBP12 and UBP13 in Nicotiana benthamiana. (D) co-IP assay of extracts derived from WT (Col-0) and 35S::PIF7-FLASH plants expressing PIF7 with a FLASH tag (3×FLAG, 6×MYC, and 6×HIS). An anti-FLAG M2 affinity agarose gel was used for immunoprecipitation. The input and the immunoprecipitation products were detected by an anti-MYC and an anti-UBP12 antibody, respectively. P-PIF7, phosphorylated form of PIF7.
Regulation of PIF7 Protein Abundance Is Important for SARs.
Previous studies demonstrated posttranslational modifications are critical for PIF7 to regulate shade avoidance (11, 15). SL treatment results in rapid dephosphorylation of PIF7 (15), which promotes PIF7 to translocate from the cytoplasm to the nucleus. In addition to phosphorylation, ubiquitination is another important modification of PIF transcription factors, which regulates the stability of PIFs (10, 32–34). A previous study showed SL increases the amount of PIF7, which could be reversed by WL (11). To clarify whether PIF7 abundance is important for its function, we overexpressed PIF7 tagged with a MYC tag in WT (35S::PIF7-MYC) and analyzed the phenotype and PIF7 protein levels of four independent transgenic lines. The transgenic lines #12 and #13, with lower PIF7 protein levels, showed a similar phenotype as WT, whereas lines #14 and #16, with much higher PIF7 levels than #12 and #13, showed significantly elongated hypocotyls even under WL (SI Appendix, Fig. S6), indicating a close relationship between PIF7 abundance and its function in promoting cell elongation. To determine if the PIF7 protein level is dynamically regulated in response to SL, we used 35S::PIF7-FLASH (35S::PIF7-9×Myc-6×His-3×FLAG) transgenic plants to investigate PIF7 stability under WL and SL. In the presence of cycloheximide (CHX), which blocks new protein synthesis, PIF7 protein levels decreased under both WL and SL, but a faster degradation rate was observed under WL (SI Appendix, Fig. S7). PIF7 degradation was arrested by MG132, indicating the involvement of 26S proteasomes (SI Appendix, Fig. S7). Our results demonstrated PIF7 is an unstable protein, but SL can protect it from degradation by 26S proteasomes. Previous studies showed that phosphorylation is necessary for subsequent ubiquitination and degradation of many transcription factors (32–34). PIF7 can be phosphorylated as well, which is critical for its subcellular localization and functions (15). To examine the possible relationship between the phosphorylation and ubiquitination of PIF7 in SARs, we generated 35S::PIF7(2A)-MYC transgenic plants in which S139 and S141 were mutated to alanine to mimic the unphosphorylated state of these residues (15). We found that compared to 35S::PIF7-MYC#12, whose phenotype is similar to WT (Col-0), 35::PIF7(2A)-MYC showed significantly elongated hypocotyls under both WL and SL (SI Appendix, Fig. S8 A and B). Moreover, 35::PIF7(2A)-MYC plants expressed much higher PIF7-MYC protein levels than 35S::PIF7-MYC#12 (SI Appendix, Fig. S8C). Note that the PIF7 transcript level in 35::PIF7(2A)-MYC was lower than that in 35S::PIF7-MYC#12 (SI Appendix, Fig. S8D). Our evidence indicated that phosphorylation of PIF7 is critical for the regulation of its stability. Shade light promotes the dephosphorylation of PIF7, which enhances its stability, resulting in a greater PIF7 abundance to promote shade-induced rapid growth.
UBP12 and UBP13 Increase the Stability of PIF7.
Considering UBP12 and UBP13 are DUBs which directly interact with PIF7, it is likely that these two UBPs regulate SARs by removing ubiquitin from polyubiquitinated PIF7 to rescue it from destruction. To check this possibility, we generated UBP12-OE/35S::PIF7-FLASH plants by crossing 35S::PIF7-FLASH with pUBQ10::UBP12-HA. PIF7 transcript levels were not significantly changed in different genetic backgrounds (SI Appendix, Fig. S9A). We then compared FLASH-tagged PIF7 protein abundance in WT and UBP12-OE under WL and SL. Consistent with previous studies (11, 15), SL promoted the conversion of PIF7 from the phosphorylated to the dephosphorylated form. PIF7 protein levels in WT were significantly increased by SL treatment, and MG132 can block PIF7 degradation in WL (Fig. 4A). By contrast, in UBP12-OE background, PIF7 protein levels were greatly increased compared to those in WT, and MG132 treatment had less effect on PIF7 protein levels (Fig. 4A). These results suggest that PIF7 in UBP12-OE is more stable than that in WT. To compare PIF7 protein levels in WT and ubp12-2w/13–3, we also generated ubp12-2w/13–3/35S::PIF7-FLASH by crossing ubp12-2w/13–3 with 35S::PIF7-FLASH, but the FLASH-tagged PIF7 levels in the double-mutant background were too low to be detected. To circumvent this issue, we crossed ubp12-2w/13–3 with another transgenic plant, 35S::PIF7-MYC#14, with a much higher PIF7 expression level (SI Appendix, Fig. S6). We found that PIF7-MYC abundance was significantly decreased in ubp12-2w/13–3 background compared to WT; however, PIF7-MYC abundance in the double mutant can be elevated by MG132, indicating its instability (Fig. 4B). Note that PIF7 transcript levels were not significantly altered by the MG132 treatment (SI Appendix, Fig. S9B). To confirm the important role of UBP12 in protecting PIF7 from destruction, we investigated the time course of PIF7 protein degradation in WT and UBP12-OE background under WL and SL by adding CHX to inhibit new protein biosynthesis. The results of immunoblots showed that PIF7 degradation in UBP12-OE was significantly blocked compared to that in WT under both WL and SL conditions (SI Appendix, Fig. S10). To determine whether the deubiquitinating catalytic activity of UBP12 was required for its role in regulating PIF7 stability, we compared the PIF7-FLASH protein level in WT, UBP12-OE, and UBP12(C208S)-OE plants harboring the 35S::PIF7-FLASH transgene. PIF7 transcript levels were comparable in plants of these three genotypes (SI Appendix, Fig. S11A). The protein level of FLASH-tagged PIF7 was greatly increased in UBP12-OE compared to WT (Fig. 4C). However, in UBP12(C208S)-OE, which expressed an equal UBP12-HA protein level to UBP12-OE, PIF7-FLASH abundance was not significantly changed compared to WT (Fig. 4C). We further checked the ubiquitination status of PIF7 in WT, UBP12-OE, and UBP12(C208S)-OE backgrounds. To preserve polyubiquitinated PIF7, we added MG132 to block 26S proteasome activity. We found that the PIF7-FLASH protein in the WT background was highly polyubiquitinated under WL compared to SL. Polyubiquitination of PIF7 was significantly reduced upon expression of UBP12 but not UBP12 (C208S) mutant lacking in UBP catalytic activity (Fig. 4D). Moreover, overexpressing UBP12 but not the UBP12 (C208S) mutant increased hypocotyl elongation of 35S::PIF7-FLASH under both WL and SL (SI Appendix, Fig. S11 B and C), which is consistent with the increased PIF7 protein level in UBP12-OE/35S::PIF7-FLASH plants (Fig. 4C). Taken together, our results confirmed the notion that UBP12 and UBP13 are required to deubiquitinate PIF7 and maintain its abundance in SARs.
Fig. 4.
UBP12 and UBP13 increase PIF7 stability. (A) PIF7 protein levels in WT (Col-0) and UBP12-OE background under WL and SL conditions. At 7 d old, 35S::PIF7-FLASH and UBP12-OE/35S::PIF7-FLASH seedlings grown under WL were moved to shade or remained under WL with or without 50 µM MG132 for 4 h, and then samples were harvested for protein extraction. P-PIF7, phosphorylated form of PIF7. (B) PIF7 protein levels in WT (Col-0) and ubp12-2w/13–3 background under WL and SL conditions. 35S::PIF7-MYC and ubp12-2w/13–3/35S::PIF7-MYC plants were treated as indicated in A. (C) PIF7 protein levels in WT (Col-0), 35S::PIF7-FLASH, UBP12-OE/35S::PIF7-FLASH, and UBP12(C208S)-OE/35S::PIF7-FLASH under WL and SL. At 7 d old, WL-grown seedlings were transferred to SL or left under WL for 4 h before being collected for protein extraction. (D) The polyubiquitination status of PIF7 in various genotypes. Seedlings were grown under WL for 10 d and then were treated with 50 µM MG132 under SL or WL for 4 h. Protein extracts were immunoprecipitated (IP) by using Ni-NTA nickel beads (Qiagen). IP products were analyzed by Western blotting using anti-Ubiquitin and anti-MYC antibodies. (Bottom) Shorter exposure of Middle. Molecular weight standards are indicated. (E and F) Hypocotyl phenotypes (E) and measurements of hypocotyl lengths (F) of WT (Col-0), ubp12-2W/13–3, 35S::PIF7-MYC#14, and ubp12-2w/13–3/35S::PIF7-MYC#14 grown under WL and SL. (Scale bar: 1 cm.) The values shown are mean ± SD (n ≥ 20). *P < 0.05 and **P < 0.01; based on Student’s t test. (G and H) The expression of shade-induced genes under WL and SL in plants indicated in E and F. The values shown are mean ± SD. *P < 0.05 and **P < 0.01; based on Student’s t test.
Because UBP12 and UBP13 directly interact with PIF7 to promote its stability, we hypothesized the insensitivity of ubp12-2w/13–3 to SL treatment was caused by a reduced PIF7 protein level. To confirm this hypothesis, we crossed ubp12-2w/13–3 with 35S::PIF7-MYC#14, which expressed PIF7 protein at a high level (SI Appendix, Fig. S6) and displayed a constitutive shade response. We found 35S::PIF7-MYC#14 could significantly rescue the phenotype of ubp12-2w/13–3 (Fig. 4 E and F). Consistent with the genetic results, the expression of shade-induced genes YUC8 and IAA19 was also increased in ubp12-2w/13–3/PIF7-OE#14 under SL compared to that in ubp12-2w/13–3 (Fig. 4 G and H). Taken together, our data demonstrate that the regulation of PIF7 protein stability mediated by UBP12 and UBP13 is critical for shade-induced adaptive growth.
Discussion
Protein instability is a hallmark of transcription factors because the fine-tuning of their levels enables rapid responses and adaptations to changing cellular conditions. Upon transfer of plants from WL to SL, there is no significant change in the PIF7 transcript levels, suggesting that PIF7 regulation in response to shade is largely a posttranslational event (11). Here, we demonstrated that the dynamic regulation of PIF7 protein abundance is critical for plants to respond rapidly in a shade environment. PIF7, which is unstable in WL, becomes stabilized upon transfer to SL. The decline of PIF7 levels can be inhibited by MG132 (SI Appendix, Fig. S7 and Fig. 4 A and B). Previous studies demonstrated changes in light quality induce rapidly reversible phosphorylation of PIF7, which is critical for its subcellular localization (15). In this study, we showed that phosphorylation of PIF7 plays an important role in regulating PIF7 stability as well (SI Appendix, Fig. S8). SL induces the conversion of PIF7 from the phosphorylated form to the dephosphorylated stable form, resulting in increased PIF7 levels.
The dynamic of protein abundance is determined by the balance between ubiquitination and deubiquitination, which are usually mediated by E3 ligases and UBPs, respectively. In Arabidopsis, members of the UBP subfamily are involved in different cellular processes and signaling pathways. A large number of studies demonstrated that UBPs play important roles in embryo development (35), pollen development and transmission (36), chromatin modification (37), pathogen defense (27), leaf development and senescence (21, 23), and peptide and hormone signal transduction (26, 29). Despite the multifunctions of UBPs during the life cycle of plants, the roles of UBPs in integrating environmental changes to plant endogenous responses are still largely unknown. Here, we have identified UBP12 and UBP13 as the DUBs for PIF7 in SARs. The ubp12-2w/13–3 mutant is blocked in shade-induced cell elongation and flowering acceleration (Fig. 1). Our in-depth genetic analyses indicated UBP12 and UBP13 act as positive regulators of the phyB-PIF7-auxin cascade in SARs, whose functions are dependent on PIF7 (Fig. 2 and SI Appendix, Fig. S5). These two UBPs form a complex with PIF7 in the nucleus (Fig. 3) and deubiquitinate PIF7 to rescue it from degradation (Fig. 4).
Our study uncovers an important role of PIF7 stability regulation in SARs and demonstrates that UBP12 and UBP13 play critical roles in regulating PIF7 abundance in responding to changes in the light environment. Under normal light conditions, phyB is photoconverted by red light from the inactive Pr to the active Pfr, which interacts with PIF7, resulting in the rapid phosphorylation of PIF7. This posttranslational modification of PIF7 promotes its polyubiquitination, resulting in the degradation of most PIF7 protein through 26S proteasomes. UBP12 and UBP13 are required to maintain basal PIF7 levels and expression of auxin biosynthetic genes in this process. Under shade light, with reduced R/FR ratio, phyB is deactivated by far-red light, and the PIF7 protein is converted from the phosphorylated form to the dephosphorylated form. UBP12/UBP13-mediated PIF7 deubiquitination was enhanced, resulting in an increased accumulation of PIF7 protein in the nucleus, which activates the expression of auxin biosynthetic genes (SI Appendix, Fig. S12).
When plants are grown in a crowded environment, the photoreceptors sense the reduction of R/FR ratio in ambient light, and these plants rapidly undergo adaptive growth to avoid shade-induced stress. However, if plants remain under shade light for a prolonged period of time, they become susceptible to pathogen attacks. Moreover, flowering time is accelerated, resulting in reduced fertilization, thus compromising crop yield. Our results and those reported previously demonstrate UBP12 and UBP13 are positive regulators of SARs but negative regulators of plant defense (27), suggesting these two UBPs play an important role in maintaining the balance between plant defense and SAR-induced rapid growth. We hope they will also inspire the formulation of new strategies to design crop plants with shade tolerance and enhanced immunity.
Methods
Plant Materials and Growth Conditions.
All plants used in this study are of the Col-0 accession. pif7-1 (CS68809) and phyB-9 (CS6217) were obtained from ABRC (Arabidopsis Biological Resource Center). ubp12-2w (GABI_244E11), ubp13-3 (SALK_132368), and ubp12-2w/13–3 double mutant were obtained from Xiaofeng Cao’s laboratory, and their properties were previously described (22). The 35S::PIF7-FLASH was kindly provided by Lin Li, Fudan University, China (11). UBQ10::UBP12-HA and UBQ10::UBP13-HA were previously described (23, 26). For constitutive overexpression, DNA fragments including full-length open reading frame (ORF) of PIF7, PIF7(2A) mutant, and the UBP12(C208S) mutant form with an HA tag were generated by PCR using the indicated primers (SI Appendix, Table S1) and cloned into pBA-35S::GWR-MYC or UBQ10::GWR vectors, respectively, by using a gateway cloning approach (Invitrogen). These constructs were introduced into the GV3101 strain of Agrobacterium and transformed into WT (Col-0) or ubp12-2w/13–3 plants using the floral dipping method (38). ubp12-2w/13–3/UBP12-OE and ubp12-2w/13–3/UBP12(C208S)-OE plants were obtained by transforming Agrobacterium harboring UBQ10::UBP12-HA and UBQ10::UBP12(C208S)-HA into ubp12-2w/13–3. pif7/ubp12-2w/13–3, pif7/UBP12-OE, and pif7/UBP13-OE were obtained by crossing pif7-1 with ubp12-2w/13–3, UBQ10::UBP12-HA, and UBQ10::UBP13-HA, respectively. The phyB/ubp2-2w/13–3 triple mutant was generated by crossing phyB-9 with ubp12-2w/13–3. UBP12-OE/35S::PIF7-FLASH and UBP12(C208S)-OE/35S::PIF7-FLASH plants were obtained by crossing 35S::PIF7-FLASH with UBQ10::UBP12-HA and UBQ10::UBP12(C208S)-HA, respectively. ubp12-2w/13–3/PIF7-MYC#14 was generated by crossing ubp12-2w/13–3 with 35S::PIF7-MYC#14 transgenic plant. All plants were grown at 22 °C under long day condition (16 h light/8 h dark) for general growth and seeds harvesting. For phenotype and molecular analyses, we used constant WL (50 μE ⋅ m−2 ⋅ s−1) or simulated shade as previously described (11, 30).
Hypocotyl Measurements.
Surface-sterilized seeds were planted on 1/2 Murashige and Skoog (MS) medium containing 1% sucrose and 0.8% agar. After 2 d of vernalization, the plates were placed under WL for 4 d and then were transferred to SL or remained under WL for another 4 d before measurements. PIC (Sigma) treatments were previously described (31). Briefly, seeds plated on 1/2 MS medium containing various concentrations of PIC were incubated under WL for 4 d before being transferred to SL or WL conditions. ImageJ software was used to quantify hypocotyl lengths. The experiments were performed with three biological replicates with at least 20 seedlings measured for each independent experiment.
RNA Extraction and Real-Time PCR.
Seedlings grown under WL for 7 d were transferred to SL or remained at WL for 4 h. Total RNAs were extracted using a Plant Total RNA Extraction Kit (Qiagen). A total of 1 μg total RNA was used for complementary DNA (cDNA) synthesis using a Bio-Rad Reverse Transcriptase Kit. Real-time PCR was performed by using SYBR Premix Ex Taq II (Bio-Rad) on a Bio-Rad Real-time PCR system according to the manufacturer’s instructions. Three biological replicates were done for each experimental set. Similar results were obtained, and one set of representative results was shown after normalized against ACTIN2.
Immunoblotting.
Seedlings grown under WL conditions for 7 d were transferred to SL or still remained under WL for an additional 4 h. About 20 seedlings were harvested for protein extraction. Plant tissues were ground to fine powder in liquid nitrogen. Total proteins were extracted with protein extraction buffer (100 mM Tris HCl pH 7.8, 4 M urea, 5% sodium dodecyl sulfate [SDS], 15% glycerol, 1 mM DTT, 1 mM phenylmethanesulfonyl fluoride [PMSF], and 1 mM protease inhibitor mixture). For CHX treatment, 7-d-old seedlings grown under WL were shifted to SL for 4 h to accumulate PIF7 protein to a high level. Seedlings were then transferred to WL or remained under SL condition for the indicated time in the presence of 200 µM CHX combined with or without 50 µM MG132. Protein levels were detected with the indicated antibodies. TUBULIN probed with an anti–α-tubulin antibody (Sigma) was used as an internal loading control. The experiments were repeated three times with independent biological samples, and similar results were obtained. One set of representative results was shown.
Yeast Two-Hybrid Assay.
Full-length UBP12 and UBP13 cDNA were cloned into a pGBKT7 vector, and full-length PIF7 cDNA was cloned into the pGADT7 vector by using a fusion method (Clontech). Yeast two-hybrid assays were performed as described (39).
BiFC.
Full-length PIF7 cDNA was cloned into pEarley Gate201-nYFP, and full-length cDNA of UBP12 and UBP13 were cloned into pEarley Gate202-cYFP using the Gateway method. BiFC assays were performed as described (39).
Pull-Down Assay.
Full-length coding sequences of PIF3, PIF4, PIF5, and PIF7 were fused in frame to sequences encoding the MBP tag by cloning into a pMAL-GWR -MYC vector. To make GST-UBP12 and GST-UBP13 fusion proteins, UBP12 and UBP13 were cloned into the pGEX-GWR-HA vector. MBP-tagged PIF3, PIF4, PIF5, and PIF7 were expressed in Escherichia coli BL21 and purified using an amylose resin (NEB). Purified GST–UBP12 and GST–UBP13 proteins retained on the glutathione agarose beads were incubated with equal amounts of MBP, MBP-PIF3, MBP-PIF4, MBP-PIF5, or MBP-PIF7 for 4 h under 4 °C. After four times of washing with wash buffer, 50 µL 2× SDS loading buffer was added to each sample and boiled at 95 °C for 10 min. After centrifugation, the pull-down products were detected by immunoblotting using an anti-MBP or an anti-GST antibody.
co-IP.
co-IP was carried out as described previously (39). WT (Col-0) and 35S::PIF7-FLASH seedlings were used to detect possible interaction between PIF7 and UBP12. Seedlings that were 10 d old and grown under WL conditions were transferred to SL or kept under WL for 4 h before being analyzed. Tissues were grounded to fine powder in liquid nitrogen and homogenized in IP buffer (50 mM Tris HCl pH 7.5, 1 mM ethylenediaminetetraacetic acid [EDTA], 75 mM NaCl, 0.5% Triton X-100, 5% Glycerol, 2 mM DTT, and 1 mM protease inhibitor mixture). After centrifugation at 13,000 rpm for 15 min under 4 °C, the supernatant was mixed with 40 μL anti-FLAG M2 Affinity agarose gel (Sigma) and incubated at 4 °C for 4 h. The beads were washed five times with washing buffer (50 mM Tris HCl pH 8.0, 150 mM NaCl, and 0.1% Triton X-100). Bound proteins were eluted from the affinity beads with a 2× SDS loading buffer boiled at 95 °C for 10 min. Immunoprecipitated products were analyzed by immunoblot using an anti-MYC or an anti-UBP12 antibody.
Acknowledgments
We are grateful to Dr. Xiaofeng Cao of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China, for providing ubp12-2w (GABI_244E11), ubp13-3 (SALK_132368), and ubp12-2w/13-3 double mutants and to Dr. Lin Li from Fudan University, China, for the 35S::PIF7-FLASH transgenic seeds. This research was supported by the National Research Foundation (NRF), Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) program. The Disruptive and Sustainable Technologies for Agricultural Precision is an interdisciplinary research group of the Singapore-MIT Alliance for Research and Technology Centre supported by the NRF, Prime Minister’s Office, Singapore under its CREATE program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103633118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lau O. S., Deng X. W., Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Casal J. J., Shade avoidance. Arabidopsis Book 10, e0157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerdán P. D., Chory J., Regulation of flowering time by light quality. Nature 423, 881–885 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Franklin K. A., Whitelam G. C., Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96, 169–175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith H., Phytochromes and light signal perception by plants—An emerging synthesis. Nature 407, 585–591 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Li J., Li G., Wang H., Wang Deng X., Phytochrome signaling mechanisms. Arabidopsis Book 9, e0148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halliday K. J., Koornneef M., Whitelam G. C., Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 104, 1311–1315 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schepens I., Duek P., Fankhauser C., Phytochrome-mediated light signalling in Arabidopsis. Curr. Opin. Plant Biol. 7, 564–569 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Keller M. M., et al. , Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67, 195–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C., Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Li L., et al. , Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvāo V. C., et al. , PIF transcription factors link a neighbor threat cue to accelerated reproduction in Arabidopsis. Nat. Commun. 10, 4005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Y., et al. , Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8, 348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y., et al. , The ELF3-PIF7 interaction mediates the circadian gating of the shade response in Arabidopsis. iScience 22, 288–298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X., et al. , Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7, e31636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swatek K. N., Komander D., Ubiquitin modifications. Cell Res. 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finley D., Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijman S. M., et al. , A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Yan N., Doelling J. H., Falbel T. G., Durski A. M., Vierstra R. D., The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124, 1828–1843 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon J., Parry G., Estelle M., The ubiquitin-proteasome pathway and plant development. Plant Cell 16, 3181–3195 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., et al. , Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 55, 844–856 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Cui X., et al. , Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162, 897–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S. H., Jeong J. S., Seo J. S., Park B. S., Chua N. H., Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 223, 1447–1460 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Lee C. M., et al. , GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 10, 3750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhaeren H., et al. , UBP12 and UBP13 negatively regulate the activity of the ubiquitin-dependent peptidases DA1, DAR1 and DAR2. eLife 9, e52276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong J. S., Jung C., Seo J. S., Kim J. K., Chua N. H., The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29, 1406–1424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewan R., et al. , Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 191, 92–106 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Derkacheva M., et al. , H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat. Plants 2, 16126 (2016). [DOI] [PubMed] [Google Scholar]
- 29.An Z., et al. , Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc. Natl. Acad. Sci. U.S.A. 115, 1123–1128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao Y., et al. , Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y., et al. , TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol. 176, 1850–1861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y., Khanna R., Carle C. M., Quail P. H., Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen H., et al. , Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20, 1586–1602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P. H., Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Doelling J. H., Yan N., Kurepa J., Walker J., Vierstra R. D., The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 27, 393–405 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Doelling J. H., et al. , The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145, 801–813 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sridhar V. V., et al. , Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447, 735–738 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H., Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y., et al. , TCP transcription factors associate with PHYTOCHROME INTERACTING FACTOR 4 and CRYPTOCHROME 1 to regulate thermomorphogenesis in Arabidopsis thaliana. iScience 15, 600–610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.