SUMMARY
Human respiratory syncytial virus (RSV) is a common cause of lower respiratory tract infections in the pediatric, elderly, and immunocompromised individuals. RSV non-structural protein NS1 is a known cytosolic immune antagonist, but how NS1 modulates host responses remains poorly defined. Here, we observe NS1 partitioning into the nucleus of RSV-infected cells, including the human airway epithelium. Nuclear NS1 coimmunoprecipitates with Mediator complex and is chromatin associated. Chromatin-immunoprecipitation demonstrates enrichment of NS1 that overlaps Mediator and transcription factor binding within the promoters and enhancers of differentially expressed genes during RSV infection. Mutation of the NS1 C-terminal helix reduces NS1 impact on host gene expression. These data suggest that nuclear NS1 alters host responses to RSV infection by binding at regulatory elements of immune response genes and modulating host gene transcription. Our study identifies another layer of regulation by virally encoded proteins that shapes host response and impacts immunity to RSV.
In brief
Pei et al. show that NS1 is present in the nucleus in primary airway epithelial, MDDCs, and A549 cells upon RSV infection. This study finds that NS1 functions as an immune antagonist by binding chromatin at the promoters and enhancers of immune response genes and modulating host gene transcription.
Graphical Abstract
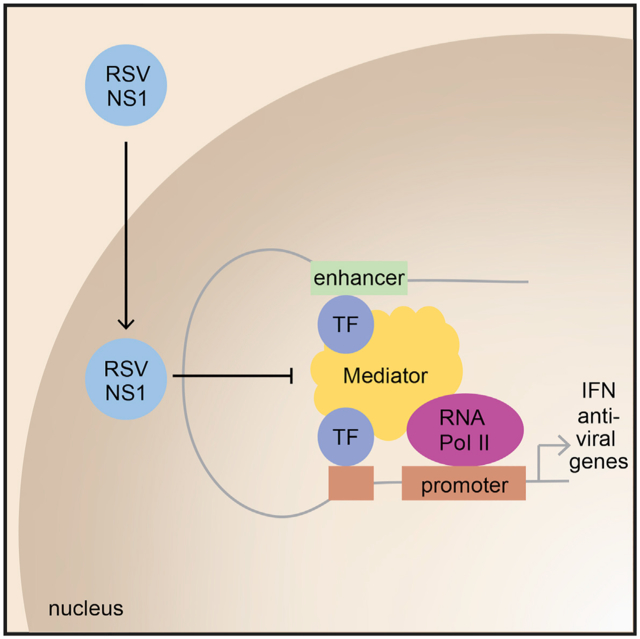
INTRODUCTION
Human respiratory syncytial virus (RSV) is a common cause of lower respiratory tract infections. Global estimates show that RSV causes high hospitalization rates and mortality in children under the age of 5 and in adults over 65 years of age (Shi et al., 2017, 2020). Furthermore, severe RSV infections contribute to the onset of childhood asthma and bronchiolitis. The disease burden in children and elderly worldwide is significant, especially with limited, effective prophylactic or treatment options available. Our lack of understanding of the host responses as well as the host-viral interactions that occur during RSV infection consequently restricts our ability to identify additional targets for therapeutic development that can be used to treat RSV infections.
RSV is a member of the Pneumoviridae family, along with the related human metapneumovirus (Rima et al., 2017). Unlike other enveloped, non-segmented, negative strand RNA viruses (NNSVs), the RSV genome contains two additional ORFs at the 3′ end encoding for the multifunctional non-structural proteins NS1 and NS2. Both NS1 and NS2 have been implicated as interferon (IFN) antagonists, but their described functions have been largely confined to the cytosol during early stages of infection. Consistent with a significant role in shaping host antiviral responses, recombinant NS1 or NS2 deletion viruses are attenuated and result in increased IFN-β mRNA levels compared to wild-type (WT)-infected cells (Jin et al., 2003; Le Nouën et al., 2014; Meng et al., 2014a; Teng et al., 2000; Whitehead et al., 1999). Of the two non-structural proteins, NS1 appears to function as a more potent suppressor of IFN signaling, with NS2 enhancing the effect of NS1 (Ling et al., 2009; Schlender et al., 2000; Spann et al., 2004). NS1 inhibits RIG-I activity by interfering with its activation or interaction with the MAVS adaptor protein (Ban et al., 2018; Boyapalle et al., 2012). NS1 also suppresses the gene expression of transcription factors IRF3/7 and may enhance the degradation of STAT2 through a proteasomal-mediated process (Lo et al., 2005; Ren et al., 2011; Spann et al., 2005). In addition, NS1 functions as an inhibitor of DC maturation (Chatterjee et al., 2017; González et al., 2008; Munir et al., 2008; Spann et al., 2004; Zhang et al., 2005) and may facilitate the proliferation and activation of Th2 cells (Munir et al., 2011), thereby promoting RSV pathogenesis. While these studies show an impact of NS1 on transcription, how the non-structural proteins impact transcription in the nucleus during RSV infection is poorly defined.
Like other NNSVs, RSV replicates in the cytoplasm. Until recently, nuclear translocation of viral proteins and their contributions to modulating host responses were not well described, with the exception of a few reports (Atreya et al., 1998; Chatterjee et al., 2017; Spann et al., 2005; Tan et al., 2013). The RSV matrix protein (M) was the first RSV virally encoded protein to be identified in the nucleus, but, to date, the function of M in the nucleus is not completely characterized (Ghildyal et al., 2003, 2005, 2009). We recently reported that RSV NS1 adopts a structural fold similar to the N-terminal domain of RSV matrix protein and other NNSV matrix proteins (Chatterjee et al., 2017). Coupled with this is the observation that RSV infection stimulates increased expression of many genes (Chatterjee et al., 2017; Dave et al., 2014; Martínez et al., 2007). RSV-induced gene-expression differences include chemokine and cytoskeletal-related genes at earlier time points of infection and IFN and inflammatory response genes later in infection. Infection with recombinant RSVs containing mutations in the C-terminal helix of NS1 that is absent in RSV matrix results in differential gene-expression patterns more similar to mock-infected cells than to WT RSV-infected cells (Chatterjee et al., 2017). Altogether, these observations suggest that NS1 may play a previously underappreciated role in modulating host responses in the nucleus through changes in gene expression and contribute to RSV viral replication and pathogenesis.
To assess additional functions of NS1, we generated a new RSV NS1-specific monoclonal antibody. These efforts revealed that RSV NS1 partially partitions into the nucleus upon RSV infection in primary human lung epithelial cells. This finding demonstrates essential functions of NS1 nuclear localization during infections. Similar functions were identified in immune cells, including human monocyte derived dendritic cells (hMDDCs), and in NS1 transfections in adenocarcinomic human alveolar basal epithelial (A549) cells. Importantly, chromatin immunoprecipitation sequencing (ChIP-seq) for NS1 revealed enrichment of NS1 peaks in transcriptional regulatory elements, including promoters and enhancers of differentially expressed genes (DEGs) during RSV infection. This pattern is consistent with a role in disrupting transcription of genes involved in host immune response. These results highlight an unexpected nuclear function for RSV NS1 that reshapes the host response to infection and presents previously unrecognized opportunities to use RSV NS1 to develop new therapeutic approaches.
RESULTS
NS1 partitions into the nucleus and interacts with host nuclear components
RSV infects a wide range of cell types; however, the first site of contact is the upper airway, and the most common disease is respiratory bronchiolitis due to infection of the epithelia of the small airways (Aherne et al., 1970; Buckingham et al., 2000; DeVincenzo et al., 2005; Hall et al., 1981; Hall et al., 2009; Liesman et al., 2014; Pickles and DeVincenzo, 2015). Among the airway cell types, human in vivo and in vitro data indicate that RSV preferentially infects the ciliated epithelial cells (Mata et al., 2012; Smith et al., 2014; Villenave et al., 2013; Villenave et al., 2012; Zhang et al., 2002, 2011). To further characterize the role of NS1 during infection, we first developed a monoclonal antibody against recombinant NS1, which we verified for specificity by western blot analysis (Figure S1A) and immunofluorescence. We next used isolated primary cells from the tracheobronchial epithelium of donated human lungs (hTECs) for infection with RSV A2 at a multiplicity of infection (MOI) of 1 and analyzed the resulting infected cells by immunostaining with the NS1 antibody. We used the RSV A2 strain since this is the prototypic virus of the A strain viruses that alternates dominance with B strain viruses (Meng et al., 2014b; Pandya et al., 2019). Sequence alignment of NS1 from different virus strains show >95% identity among the group A viruses and >85% identity among group B viruses when compared to group A2 (Figure S1B). After 72 h post-infection (hpi), we found that NS1 is predominantly localized to the cytoplasm as expected (Figures 1A, 1B, and S1C). Interestingly, we also observed that RSV infection induces partitioning of NS1 into the nucleus. Mock-infected controls showed no staining for NS1 (Figures 1A and 1B). By comparison, immunostaining for RSV nucleoprotein revealed that nucleoprotein remains completely cytoplasmic, likely in inclusion bodies, which is consistent with prior observations (Blanchard et al., 2020; García et al., 1993; Lifland et al., 2012). Previous studies also showed that human immune cells, including hMDDCs, can be infected by RSV (Chatterjee et al., 2017; Munir et al., 2011; Munir et al., 2008). To test whether NS1 also partitions into the nucleus of immune cells, we generated and infected hMDDCs with RSV. At 24 hpi, we find that NS1 is also present in the nucleus of RSV-infected hMDDCs compared to mock infected (Figures S2A and S2B), whereas nucleoprotein is restricted to the cytoplasm (Figure S2C). Pierson correlation coefficient (PCC) analysis of intensity profile data measured across the cells demonstrates a significant correlation of DAPI and RSV NS1 staining compared to mock NS1 or RSV nucleoprotein (Figure S2D). Together, these data demonstrate that NS1 partitioning into the nucleus is a phenomenon consistently observed during RSV infection.
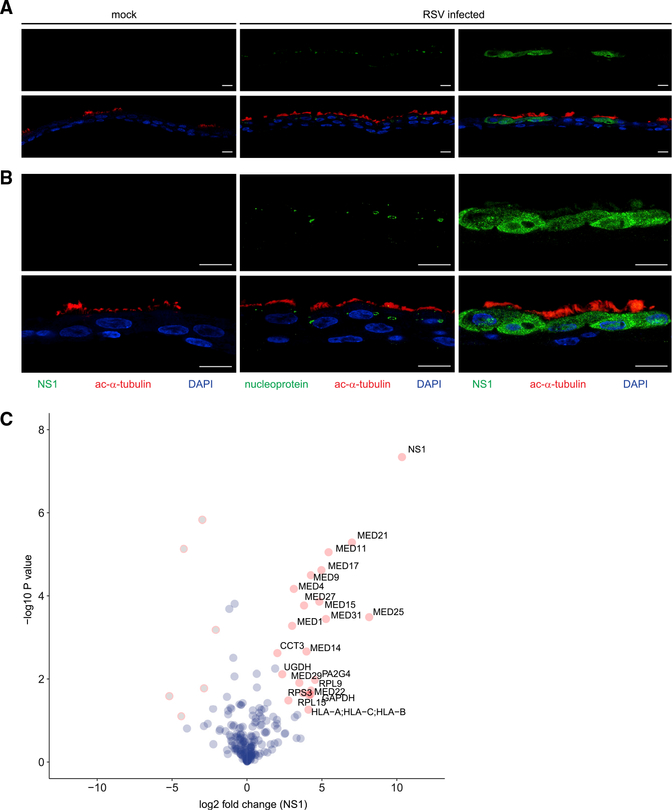
Figure 1. NS1 partitions into the nucleus in RSV-infected airway epithelial cells and associates with host nuclear proteins.
hTECs differentiated on supported membranes using air-liquid interface culture were treated with PBS (mock) or RSV at MOI of 1 on the apical surface for 72 h. Fixed cells on membranes were processed through paraffin embedding and then immunostained using antibodies to RSV NS1 (green; left and right panels) or nucleoprotein (green; middle panel) and co-stained with cilia marker anti-acetylated tubulin (ac-α-tubulin, red).
(A and B) Shown are representative photomicrographs from three experiments with (A) low-power observation and (B) high-power magnification. Scale bar: 10 μm. Merged images in the bottom rows of (A) and (B) demonstrate that NS1 localizes in cytoplasm and nucleus of cells, while nucleoprotein is present in aggregates within the cytoplasm of ciliated cells during RSV infection.
(C) AP-LC-MS/MS reveals cytoplasmic and nuclear interactors of RSV NS1. Volcano plot shows p values and fold enrichment over unrelated control (n = 3). Significant interactors are depicted in red, other proteins in blue (non-significant) or gray (depleted).
To define the molecular basis for NS1 regulation of gene expression, we sought to identify NS1 interactors. To this end, we performed affinity purification of streptavidin II (SII) and hemagglutinin (HA) tagged NS1 (SII-HA NS1) in HEK293T cells followed by tandem mass spectrometry (AP-LC-MS/MS). In total, we identified 21 proteins in NS1 purifications that were significantly enriched relative to purification of an unrelated control protein, FluAV NS1 (Figure 1C). Of these 21 identified proteins, 13 proteins identified with high probability were subunits of the Mediator complex, including subunits MED1, MED 14, and MED25. Mediator proteins comprise a nuclear-localized, multi-subunit protein complex that is part of the preinitiation complex required for RNA polymerase II transcription and is a known regulator of many innate immune response genes (Carlsten et al., 2013; Soutourina, 2018). These results align with an earlier proteomics study performed in A549 cells transfected with NS1 that also identified Mediator complex as an interactor of NS1 but with different subunits (Wu et al., 2012).
Nucleocytoplasmic trafficking of NS1
The presence of NS1 in the nucleus and interactions between NS1 and host nuclear components across different cell lines prompted us to further investigate how NS1 partitions into the nucleus, given that RSV replication occurs in the cytoplasm (Rima et al., 2017). We find that RSV infection of A549 cells induces partitioning of NS1 into the nucleus relative to a mock-infected control that showed no staining for NS1 (Figure 2A; Figure S3), as observed in the hTECs and hMDDCs. Intensity profile measurements reveal that NS1 was distributed throughout the nucleus, with relatively less staining in the nucleoli. The nuclear distribution of NS1 was similar to the localization of Mediator as shown by immunostaining of the MED1 subunit, a component of the middle module of the Mediator complex.
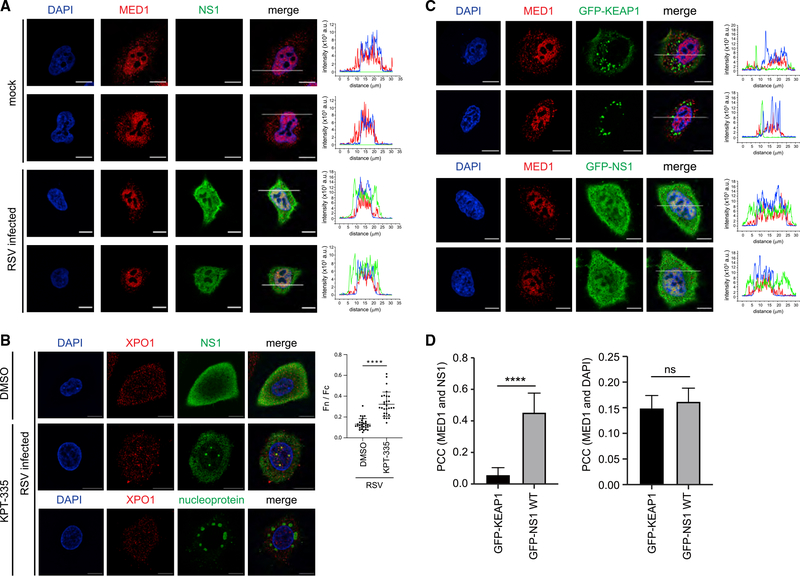
Figure 2. Nucleocytoplasmic partitioning of NS1 is facilitated through interactions with host transporters.
(A) Representative confocal micrographs of A549 cells that were mock infected (top rows) or infected with RSV A2 at a MOI of 1 for 24 h (bottom rows). Cells were fixed and stained with anti-NS1 antibody (green), anti-Mediator 1 (MED1) (red), or DAPI (blue). Scale bar: 10 μm. Right panels: intensity profile analysis was performed along the white line using ZEN 2.6 software to display the distribution of the three fluorophore signals.
(B) Representative confocal images of A549 cells that were infected with RSV A2 at a MOI of 1 for 24 h and incubated with DMSO (control, left panel) or KPT-335 (1 μM, middle panel). Cells were fixed and stained with anti-exportin 1 (XPO1) (red), anti-NS1 antibody (green), or DAPI (blue). Scale bar: 10 μm. Cells were analyzed using ImageJ to determine the NS1 fluorophore signal of nucleus relative to cytoplasm (Fn / Fc, right part). Data represent mean ± SD. Statistical analysis was performed using Student’s t test (****p < 0.0001).
(C) Representative confocal micrographs of A549 cells that were transfected either with GFP-KEAP1 (top rows) or GFP-NS1 (bottom rows) for 24 h. Cells were fixed and stained with anti-MED1 antibody (red) or DAPI (blue) and visualized for GFP fluorescence (green). Scale bar: 10 μm. Right panels: intensity profile analysis was performed along the white line using ZEN 2.6 software to display the distribution of the three fluorophore signals.
(D) PCC was determined between MED1 and NS1 or MED1 and KEAP1 channels for GFP-NS1 and GFP-KEAP1 transfected cells as shown in (C). Representative of 3 experiments. (****p < 0.0001; ns, not significant)
We next treated A549 cells with the inhibitor KPT-335 prior to RSV infection. KPT-335 binds to the nuclear transporter exportin 1 (XPO1; also known as chromosomal maintenance 1 or CRM1) in the groove where the nuclear export signal of cargoes bind, sequestering XPO1 and XPO1 binding proteins in the nucleus (Etchin et al., 2013; Jorquera et al., 2019; Perwitasari et al., 2014). Here, we find that KPT-335 treatment increased accumulation of NS1 in the nucleus compared to untreated cells, confirming its presence in the nucleus (Figure 2B; Figure S4). We also observe colocalization of NS1 with XPO1 in the nucleus, which is consistent with previous observations (Wu et al., 2012). Interestingly, NS1 and XPO1 appeared to concentrate in foci within the nucleus (Figure 2B), the significance of which is not clear at present. Furthermore, this pattern of colocalization with XPO1 is absent when KPT-335 treated cells are stained for RSV nucleoprotein. To further examine whether NS1 associates with XPO1, we performed coimmunoprecipitation (coIP) assays in A549 cells transfected with FLAG-tagged NS1. Results show that FLAG-NS1 immunoprecipitates endogenous XPO1 (Figure S4D), suggesting that XPO1 may play a role in regulating nucleocytoplasmic trafficking of NS1.
Because NS1 is a small protein of approximately 15 kDa and lacks a canonical nuclear localization signal or nuclear export signal, passive diffusion of NS1 across the nuclear pore complex and into the nucleus may account for this observation. To assess whether NS1 employs a specific mechanism to localize in the nucleus, we tagged NS1 with a GFP molecule, which increases the overall molecular weight to >42 kDa. We find that GFP-NS1 is localized in the same regions of the nucleus as MED1 and DAPI (Figures 2C and 2D), similar to our observations with untagged NS1 produced during RSV infection (Figure 1; Figures 2A and 2B). In contrast, transfection of a control molecule GFP-tagged KEAP1, an adaptor protein for a Cullin3 ubiquitin E3 ligase, remained cytoplasmic despite its overexpression. Altogether, these results demonstrate that NS1 is actively transported into the nucleus and suggest that NS1 may perform roles beyond IFN antagonism in the cytoplasm.
NS1 binds to regulatory elements on chromatin
Having demonstrated that NS1 produced by RSV infection or NS1 ectopically expressed through transfection localizes in the nucleus, we next evaluated sub-nuclear localization to uncover NS1 nuclear function. NS1 colocalization with Mediator and DAPI and exclusion from nucleoli suggested a potential role in transcriptional regulation. We separated FLAG-NS1-transfected A549 cells into cytosolic, soluble nuclear, and chromatin fractions and performed FLAG coIP assays with antibodies against Mediator subunits (MED1, MED14, and MED25) identified by AP-MS (Figure 1C). NS1 was present in all fractions, whereas all three MED components were primarily localized to the soluble nuclear fraction (Figure 3A). CoIP of FLAG-NS1 from the soluble nuclear fraction revealed interactions with MED1, MED14, and MED25 (Figure 3A). Cell fractionation also showed NS1 in the soluble nuclear and chromatin fractions; the quality of fractionation is shown by detection of Mediator and the chromatin binding factor CTCF in these fractions as expected (Figure 3B).
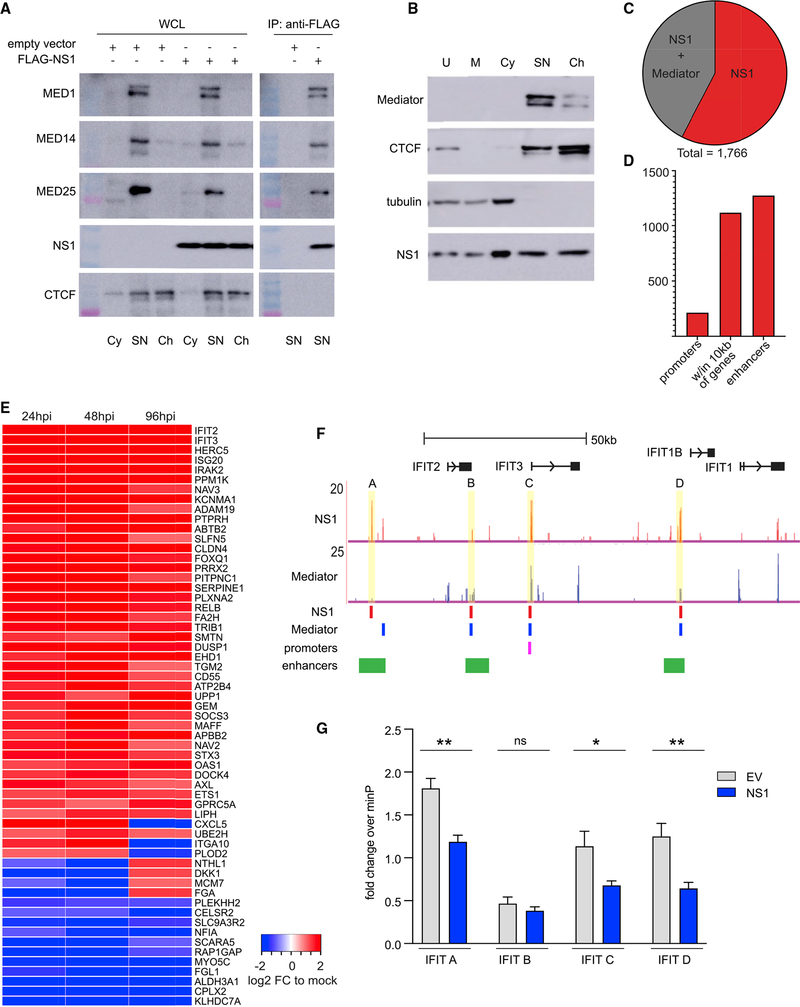
Figure 3. NS1 binds at transcriptional regulatory elements of genes differentially expressed during RSV infection.
(A) Western blots show co-IP of FLAG-NS1 with MED1, MED14, and MED25. WCL, whole-cell lysate; Cy, cytoplasmic fraction; SN, soluble nuclear fraction; Ch, chromatin fraction.
(B) Western blots of subcellular fractionated A549 cells expressing HA-NS1. U, unfractionated; M, membrane; Cy, cytosol.
(C) Overlap of NS1 and Mediator ChIP-seq peaks in A549 cells.
(D) Bar chart shows number of NS1 ChIP-seq peaks in functional genomic regions.
(E) Heatmap shows differentially expressed genes with 10 kb of NS1 peaks at 24, 48, and 96 hpi with RSV (RNA-seq).
(F) NS1 ChIP-seq peaks coincide with an enhancer (A), promoter (C), and Mediator peaks (B–D). Tracks: ChIP-seq as reads per million (RPM). Red and blue bars: called peaks (HOMER); GeneHancer track shows overlapping promoter (pink) and enhancers (green) (UCSC browser).
(G) Bar graph shows fold change + standard deviation of luciferase activity of reporters with NS1-bound regions in the IFIT locus (A–D as in (F) relative to minimal promoter reporter (minP) in A549 cells transfected with empty vector (EV) or NS1 and treated with LMW polyI:C for 24 h. Unpaired t test, *p < 0.05, **p < 0.005, ns, not significant. All are representative of ≥2 experiments.
Nuclear proteins can impact gene transcription by directly or indirectly modulating chromatin-associated proteins. Given the evidence that NS1 localizes in the nucleus and is chromatin associated, we next mapped the epigenome-wide chromatin binding of NS1. We performed ChIP-seq for NS1 in A549 cells expressing HA-NS1 or empty vector-HA (EV-HA). For comparison, we performed ChIP-seq for Mediator in RSV-infected, HA-NS1 or EV-HA-transfected A549 cells. NS1 or Mediator ChIP-seq identified 1,756 and 21,020 peaks, respectively, with 43% of NS1 peaks coinciding with Mediator peaks (Figure 3C). Mediator peaks in RSV-infected, HA-NS1 and EV-HA-transfected cells were >90% overlapping, while anti-HA ChIP in EV-HA cells yielded only 81 weak peaks that did not overlap with any of the NS1 or Mediator peaks. To determine whether NS1 peaks are located near functional genomic elements, we performed a genome annotation analysis (Heinz et al., 2010; Yu et al., 2015). We found that NS1 peaks, like Mediator peaks, were highly enriched (log2 fold change >2) in promoters and 5′ untranslated regions (UTR) regions compared to the non-coding regions that comprise the vast majority of the genome (Figure S5A). The majority (63.5%) of NS1 peaks are located within 10 kb of genes, 12% coincide with promoters, and 72.6% overlap enhancers defined in the EnhancerAtlas (Gao et al., 2016) (Figure 3D). Enhancers are genomic elements that regulate the timing and level of gene transcription in response to developmental and environmental cues and are highly tissue specific (Andersson et al., 2014). Further analysis of our ChIP-seq results show that nearly all of the enhancer NS1 peaks (~95%) overlap with enhancers found in lung tissue, suggesting that NS1 may selectively bind to regulatory elements that control genes essential for respiratory tissue functions, including response to respiratory infection. By comparison, a smaller percentage of Mediator peaks are associated with all enhancers or lung tissue enhancers (39%, 82%, Figure S5B). Forty percent of genes with NS1-bound promoters were differentially expressed during RSV infection (log2 fold change > abs(1) and false discovery rate [FDR] < 0.05, DESeq2, Figure S5B). The greater coincidence of NS1 peaks with lung tissue enhancer regions and with promoters of DEGs in RSV infection suggests that NS1 may play a modulatory role in controlling host transcriptional immune response to RSV in the nucleus.
To determine whether NS1 preferentially binds near DEGs during RSV infection, we identified NS1 peaks located within 10 kb of genes. Although genes may be regulated by more distant enhancers, we limited the distance to 10 kb because confidently assigning enhancer-gene pairs becomes more difficult with increasing distance (Chatterjee et al., 2017; Fulco et al., 2016, 2019; Gasperini et al., 2019). In almost half of the cases, NS1 peaks are located within 10 kb of DEGs during RSV infection (47.5%, 464/977; log2 fold change > abs(1) and FDR < 0.05, DESeq2). Of these, the most differentially expressed across three time points post-infection are shown in a heatmap in Figure 3E, including viral immune response genes IFIT2, IFIT3, IRAK2, OAS1, RELB, and CXCL5. There are several NS1 peaks within 10 kb of genes in the IFIT locus, including one in the IFIT3 promoter, and three in enhancers near IFIT2 and IFIT1B (Figure 3F). Three of the NS1 peaks also coincide with Mediator peaks. To directly test whether NS1 can suppress transcription driven by these regulatory elements, we cloned these NS1-binding regions into luciferase reporter constructs and co-transfected them into cells with NS1 or empty vector and treated with poly(I:C). NS1 significantly decreased the transcriptional activity of all but one of the NS1 binding regions (Figure 3G). Together, these results demonstrate that RSV NS1 binds preferentially in the regulatory elements of genes with altered expression during RSV infection.
The C-terminal helix of NS1 is important for modulating gene transcription
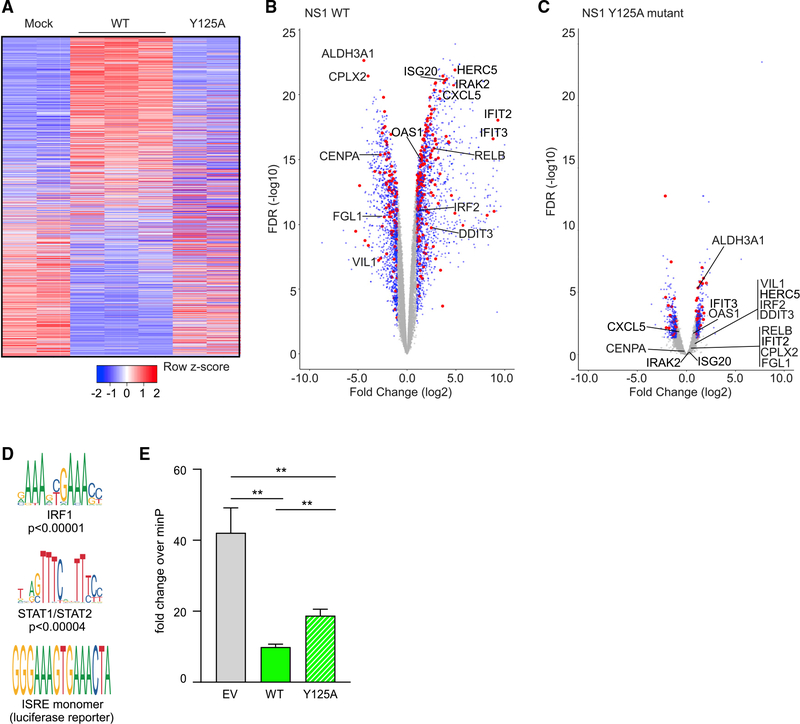
We previously established that NS1 contains a C-terminal alpha helix 3 and mutation of this helix results in a recombinant RSV that is attenuated and induces lower expression of host immune response genes; however, the mechanism by which the C terminus controls this process was not known (Atreya et al., 1998; Chatterjee et al., 2017; Zhang et al., 2016). To determine the impact of the mutation on host gene expression, we compared RNA sequencing (RNA-seq) from cells 96 hpi with NS1 WT or a NS1 C-terminal helix mutant, Y125A (Figure S6A). Figure 4A shows differential expression of genes within 10 kb of NS1 peaks in WT NS1 RSV infection compared to mock (log2 fold change > abs(1) and FDR <0.05, DESeq2). In contrast, expression of these same genes in RSV NS1 Y125A infection is quite similar to mock infection. Volcano plots show significant fold change in the expression of genes within 10 kb of NS1 peaks in RSV NS1 WT infection (red), while in RSV NS1 Y125A mutant infection, the fold changes are much lower and often non-significant, including IFIT2, IFIT3, CXCL5, IRAK2, IRF2, ISG20, OAS1, and RELB (Figures 4B and 4C). Given these dramatic differences, we predicted that the NS1 Y125A mutation may impact the cellular distribution of NS1. However, we find that the Y125A mutation does not alter the localization of NS1 as FLAG-NS1 Y125A localizes to the cytoplasm and nucleus similar to FLAG-NS1 WT (Figure S6B). Together, these results suggest that RSV NS1 can act in the nucleus to modulate the expression of genes involved in the host response to infection, and that the NS1 Y125 residue may be important for this function.
Figure 4. The C-terminal helix of NS1 is important for modulating gene transcription.
(A) Heatmap shows DEG within 10 kb of NS1 peaks at 96 hpi with mock, NS1 WT, or Y125A RSV (RNA-seq).
(B and C) Volcano plots show (B) WT NS1 or (C) NS1 Y125A versus mock infection (as in A). Red: DEG <10 kb from an NS1 peak; blue: DEG >10 kb from an NS1 peak; gray: non-DEG. Selected genes are labeled.
(D) Two transcription factor motifs enriched in NS1 peaks. The ISRE motif, which is highly similar to IRF1 and STAT1/STAT2 motifs, drives transcription of the luciferase reporter in (E).
(A)–(D) are representative of 2 experiments.
(E) Bar graph shows fold change + standard deviation of ISRE reporter luciferase activity relative to minimal promoter reporter (minP) in A549 cells transfected with empty vector (EV), WT, or Y125A NS1 and treated with LMW polyI:C for 24 h. Unpaired t test; **p < 0.005. Representative of >3 experiments.
To determine whether chromatin binding is impacted by the Y125A mutation, we performed ChIP for HA-tagged NS1 Y125A, NS1 WT, or for endogenous Mediator in transfected A549 cells. NS1 WT and NS1 Y125A proteins expressed at near equal levels (Figure S7A). ChIP-qPCR for NS1 WT peak regions nearest to differentially expressed IFIT genes (Figure 3F, yellow highlights) demonstrates that NS1 Y125A binding to chromatin at these sites is not significantly different from NS1 WT (Figure S7B). Mediator binding at the IFIT loci also was not different in the NS1 WT or NS1 Y125A-transfected cells. Mediator binding was higher than NS1 as expected near a known Mediator site in the GAPDH promoter; there is no NS1 peak here (Figure S7C). These results suggest that the Y125 residue of NS1 may not be important for its interaction with chromatin but may impact NS1 host transcriptional regulation of immune response genes via other interactions.
We hypothesized that nuclear NS1 may function by binding to and disrupting the activity of TFs and other chromatin-associated factors that regulate the transcription of immune response genes. Motif enrichment analysis of NS1 peaks and comparison to publicly available factor ChIP-seq peaks (ENCODE) (Gerstein et al., 2012) revealed significantly enriched TF motifs and TF peaks, including IFN downstream effectors IRF1 and STAT1/2 at 50%–75% of NS1 peaks (Figure 4D). To directly test whether the Y125A mutation impacts NS1 suppression of transcription driven by IFN-responsive elements, we expressed a luciferase reporter containing the IFN-stimulated response element (ISRE), which is highly similar to the IRF1 and STAT1/2 motifs (Figure 4D) and exhibits robust response downstream of IFN activation (Veals et al., 1992). We expressed the reporter in A549 cells with NS1 WT, NS1 Y125A, or EV and stimulated with the double-stranded RNA (dsRNA) mimic poly(I:C). NS1 WT significantly decreased the transcriptional activity of the ISRE reporter and this suppression was significantly abrogated by the NS1 Y125A mutant (Figure 4E). These data suggest that NS1 may interact with or displace one or more TF and transcriptional regulatory factors to modulate host immune response genes.
DISCUSSION
Host responses to viral infections determine disease outcome, and, therefore, it is important to understand interactions such as those characterized here for RSV NS1. The impact of RSV NS1 in the cytosolic control of early IFN responses has been previously described (Ban et al., 2018; Boyapalle et al., 2012; Ling et al., 2009; Lo et al., 2005; Schlender et al., 2000; Spann et al., 2004; Spann et al., 2005; Swedan et al., 2011). Our findings here reveal that NS1 partitions to the nucleus upon RSV infection, including primary human lung epithelial cells, and impacts transcription, suggesting that NS1 regulates host gene expression.
NS1 localization in the nucleus has been previously shown when NS1 is expressed upon transient transfection (Chatterjee et al., 2017; Munday et al., 2010; Spann et al., 2004; Swedan et al., 2011; Swedan et al., 2009; Tan et al., 2013; Wu et al., 2012). However, it was not clear whether NS1 also partitions to the nucleus during natural infection. Using a new mouse monoclonal antibody that we generated against recombinant RSV NS1 protein, we detected NS1 in the nucleus by immunofluorescence. This unique reagent allowed us to visualize in RSV-infected primary human airway epithelial cells, hMDDCs, and A549s that NS1 is present both in the cytoplasm and in the nucleus. This finding is striking because it demonstrates that NS1 protein may not be retained entirely in the cytoplasm by another viral factor, such as NS2 as has been reported (Swedan et al., 2011). Our observations here of the nuclear and cytoplasmic partitioning is consistent with the multifunctional nature of NS1.
Given the size of NS1, NS1 may diffuse through nuclear pore. However, KPT-335, an inhibitor that binds to the nuclear export transporter XPO1 and inhibits its exit from the nucleus, increased the accumulation of NS1 in the nucleus and its colocalization with XPO1. This observation suggests that XPO1 may play a role in regulating NS1 nucleocytoplasmic trafficking. Furthermore, when we tagged NS1 with a large fluorophore, we also detected GFP-tagged NS1 in the nucleus. In contrast, a GFP-tagged control protein remained completely cytosolic. These results suggest that NS1 may associate with factors that facilitate nucleocytoplasmic transport of host proteins, such as those identified in a previous proteomic study that also identified Mediator subunits associated with NS1 (Wu et al., 2012).
Viral structural proteins from other mononegaviruses have been shown to inhibit IFN signaling by binding to importins and preventing trafficking of cargoes into the nucleus that stimulate antiviral responses. This mechanism of IFN inhibition has been described for measles virus nucleoprotein (Takayama et al., 2012) and Ebola virus VP24 protein (Xu et al., 2014). Other viral matrix proteins, including RSV and Nipah virus matrix, traffic through the nucleus as a prerequisite for virion assembly and budding; however, the biological significance of the nuclear shuttling of matrix remains unclear (Coleman and Peeples, 1993; Günther et al., 2020; Pentecost et al., 2015; Ringel et al., 2020; Wang et al., 2010). Notably, nucleocytoplasmic trafficking of viral structural proteins is facilitated by nuclear localization signals (NLSs) and/or sequences enriched with lysine or arginine residues that can mimic NLSs (Tessier et al., 2019; Yarbrough et al., 2014). While RSV NS1 is a non-structural protein that lacks a canonical NLS, its structure is strikingly similar to the N-terminal domain of RSV matrix protein despite little sequence similarity (Chatterjee et al., 2017). Additional studies are warranted to further define the mechanism of NS1 transport into the nucleus and whether related matrix proteins encode the ability to modulate host gene expression during viral infection. Importantly, insights from such studies are likely to define host targets that may limit RNA infections and/or NS1-dependent immune suppression.
Here, we show that nuclear RSV NS1 binds to chromatin, in particular, to regulatory elements near genes that are differentially expressed during RSV infection. In addition, we demonstrate that NS1 binds these regulatory elements and suppresses gene transcription. Many of these NS1-bound regions also coincide with regions that bind Mediator, a key transcriptional regulatory complex recruited by transcription factors that subsequently contributes to assembly of the RNA polymerase II preinitiation complex (PIC) on core gene promoters (Carlsten et al., 2013; Soutourina, 2018). Mediator functions as a physical bridge between enhancers and promoters, forming chromatin loops that are essential for transcriptional regulation (Park et al., 2005; Siersbæk et al., 2017; Soutourina, 2018), including many immune response genes. To date, there are limited descriptions of viral proteins interacting with Mediator, but these report transcriptional regulation of viral genes (Léonard et al., 2006; Lester and DeLuca, 2011; Milbradt et al., 2011; Mittler et al., 2003; Verbruggen et al., 2011; Vojnic et al., 2011). Some viruses, including HIV, HSV, and IAV, suppress host gene transcription via interference with CDK9/P-TEFb and activation of RNA Pol II (Mbonye et al., 2013; Rivas et al., 2016). Our findings here demonstrate a viral protein that interacts with Mediator and gene regulatory elements to target host gene transcription and counteract host antiviral mechanisms during RSV infection. Our collective results from AP-MS and ChIP-seq experiments support direct association of RSV NS1. Moreover, we show that this interaction targets regulatory elements that are associated with immune response genes, including those near the IFIT genes, and inhibits their transcription.
We previously established that NS1 contains a C-terminal alpha helix 3 that is critical for mediating IFN antagonism. This helix is unique in comparison to RSV M and mutation of this helix results in a recombinant RSV that is attenuated and induces lower expression of host immune response genes, suggesting that NS1 may have a greater role in host immune evasion than previously appreciated (Atreya et al., 1998; Chatterjee et al., 2017; Zhang et al., 2016). The studies here not only support our observations but also show that this structural element may not directly impact NS1 nuclear localization or chromatin binding. Instead, residues within this helix may reduce host transcription of immune response genes. The suppression of host transcription could be due to altered affinity of mutant NS1 Y125A for the same transcriptional regulatory factors as WT NS1 and/or interaction with other transcriptional regulatory proteins. Either could result in greater interference with host gene transcription, contributing to the suppression of immune response genes we observe with RSV NS1 Y125A infection (Chatterjee et al., 2017). These studies point to a multifunctional role for viral proteins, like RSV NS1, in perturbing the transcriptional control mechanisms that shape host immune response to infection. While similar mechanisms have been previously described for DNA viruses, this is not a common mode of action for RNA viral proteins, which are largely cytosolic and typically cause acute infections with little evidence of dormancy and reactivation. Because transcriptional control mechanisms alter the chromatin landscape, which is inheritable, these findings suggest a new mechanism by which RSV could prevent the formation of long-term immunological memory, particularly in the lung airway epithelium, where RSV and other pulmonary pathogens cause primary damage.
Limitations of the study
Our study is based on our findings using a new monoclonal antibody that we developed against recombinant RSV NS1 protein to detect NS1 in the nucleus of primary hTECs, hMDDCs, and A549 cell line upon RSV infection. NS1 is an abundant, multifunctional protein, and we do not exclude that NS1 is also localized at other sites in the cell, including in the cytoplasm and at the mitochondria. While it is possible that NS1 can also diffuse into the nucleus, we used the inhibitor of XPO1, KPT-335, as a method to increase visualization of untagged NS1 in the nucleus and used a GFP-tagged NS1 to increase the molecular weight beyond the nuclear diffusion limit in these studies. Further studies are warranted to identify the host factor(s) that facilitates the direct import and export of NS1 to and from the nucleus and the potential impact of other RSV proteins on the nucleocytoplasmic trafficking of NS1.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daisy W. Leung (dwleung@wustl.edu).
Materials availability
All unique/stable reagents generated in this study will be made available upon request, but we may require a complete Materials Transfer Agreement if there is potential for commercial application.
Data and code availability
ChIP-seq and RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-RSV NS1 | This paper | N/A |
| Mouse monoclonal anti-RSV nucleoprotein | Abcam | Cat#ab94806; RRID: AB_10674696 |
| Rabbit polyclonal anti-MED1 | Bethyl Laboratories | Cat#A300-793A; RRID: AB_577241 |
| Rabbit polyclonal anti-MED14 | Bethyl Laboratories | Cat#A301-044A; RRID: AB_2266461 |
| Mouse monoclonal anti-MED25 | Santa Cruz Biotechnology | Cat#sc-393759 |
| Rabbit monoclonal anti-XPO1 | Abcam | Cat#ab180144; RRID: AB_2847862 |
| Mouse monoclonal anti FLAG | Sigma-Aldrich | Cat#F3165; RRID: AB_259529 |
| Rabbit monoclonal anti-Acetyl-alpha-Tubulin | Sigma-Aldrich | Cat#SAB5600134 |
| Alexa Fluor 488-conjugated goat anti-mouse IgG | Thermo Fisher Scientific | Cat#A-11029; RRID: AB_2534088 |
| Alexa Fluor 594-conjugated goat anti-rabbit IgG | Thermo Fisher Scientific | Cat#A-11037; RRID: AB_2534095 |
| Alexa Fluor 488-conjugated donkey anti-mouse IgG | Thermo Fisher Scientific | Cat#A-21202; RRID: AB_141607 |
| Alexa Fluor 555-conjugated donkey anti-rabbit IgG | Thermo Fisher Scientific | Cat#A-31572; RRID: AB_162543 |
| Peroxidase affiniPure goat anti mouse IgG | Jackson ImmunoResearch | Cat#115-035-003; RRID: AB_10015289 |
| Peroxidase affiniPure goat anti rabbit IgG | Jackson ImmunoResearch | Cat#111-035-003; RRID: AB_2313567 |
| Goat polyclonal anti-RSV | Fisher Scientific | Cat#AB1128MI |
| HRP-conjugated donkey anti-goat IgG | Fisher Scientific | Cat#AP180PMI |
| Rabbit polyclonal anti-GAPDH | Abcam | ab9485; RRID: AB_307275 |
| Rabbit polyclonal anti-CTCF | Cell Signaling Technology | 2899; RRID: AB_2086794 |
| Mouse monoclonal anti-tubulin | Sigma-Aldrich | T9026-100Ul; RRID: AB_477593 |
| Rabbit polyclonal anti-HA | Abcam | Ab9110; RRID: AB_307019 |
| Normal rabbit IgG | Millipore | 12-370; RRID: AB_145841 |
| Rat monoclonal anti-CD209-PerCP-Cyanine5.5 | Fisher Scientific | Cat#50-158-32; RRID AB_1106983 |
| Mouse monoclonal anti-CD141-APC | Fisher Scientific | Cat#17141942; RRID: AB_2662437 |
| Mouse monoclonal anti-CD83-FITC | Fisher Scientific | Cat#50-968-3 |
| Mouse monoclonal anti-CD1c-PE | ThermoFisher Scientific | Cat#25-0015-42; RRID: AB_2573324 |
| Mouse monoclonal anti-HLA-DR-Alexa Fluor 700 | Fisher Scientific | Cat#5016938 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Human respiratory syncytial virus (RSV, A2 strain) | ATCC | Cat#VR-1540 |
| Human respiratory syncytial virus NS1 Y125A | Chatterjee et al., 2017 | N/A |
| Escherichia coli BL21(DE3) | Novagen | Cat#69450 |
|
| ||
| Biological samples | ||
|
| ||
| Fetal Bovine serum | Sigma-Aldrich | Cat#F4135 |
| Donkey serum | Sigma-Aldrich | Cat#D9663; RRID: AB_2810235 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat#BP9704-100 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| KPT-335 | RayBiotech | Cat#331-21369-1 |
| NS1 protein | Chatterjee et al., 2017 | N/A |
| Lipofectamine 2000 transfection reagent | Thermo Fisher Scientific | Cat#11668019 |
| Dulbecco’s modified Eagle’s medium (DMEM) | Thermo Fisher Scientific | Cat#11965 |
| OptiMEM medium | Thermo Fisher Scientific | Cat#31985070 |
| RPMI 1640 media | GIBCO | Cat#224000-071 |
| Trypsin-EDTA (0.05%) | Thermo Fisher Scientific | Cat#25300054 |
| Penicillin-streptomycin | Gen Clone | Cat#25-512 |
| F-12K medium | GIBCO | Cat#11330-032 |
| Poly(I:C) | Invivogen | Cat#TLVL-PICW |
| EGS (ethylene glycol-bis(succinic acid N-hydroxysuccinimide ester | Thermo | Cat#21565 |
| Formaldehyde | Sigma-Aldrich | Cat#252549-25ml |
| LysC | Wako Chemicals | Cat#121-05063 |
| Trypsin | Promega | Cat#V5280 |
| Glycine | Fisher Scientific | Cat#BP3811 |
| PBS | Sigma-Aldrich | Cat#D8537 |
| GM-CSF | R&D Systems | Cat#415ML010 |
| IL-4 | R&D Systems | Cat#35050-061 |
| GlutaMAX | GIBCO | Cat#35050-061 |
| 2-mercapthoethanol | Sigma-Aldrich | Cat#M3148 |
| DMSO | ThermoFisher Scientific | Cat#BP231100 |
| Lymphoprep | StemCell Technologies, Inc. | Cat#7861 |
| Dynabeads Prot A ref 10002D | Thermo Fisher Scientific | Dynabeads Prot A ref 10002D |
| DNA extraction kit (MinElute PCR) | QIAGEN | Cat. No. / ID: 28004 |
| RNA extraction kit (RNAeasy Micro) | QIAGEN | Cat. No. / ID: 74004 |
| Power SYBR Green PCR Master Mix | Fisher Scientific | Cat # 4367659 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Subcellular Protein Fractionation Kit | Thermo Fisher Scientific | Cat#78840 |
| Nano-Glo system | Promega | Cat#N1110 |
| Classical monocyte isolation kit | Miltenyi Biotec | Cat#130-117-337 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-seq data | https://www.ncbi.nlm.nih.gov/geo/ GSE99298 | |
| ChIP-seq and RNA-seq data | https://www.ncbi.nlm.nih.gov/geo/ GSE155152 | |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Adenocarcinomic human alveolar basal epithelial (A549) | ATCC | Cat#CCL-185; RRID: CVCL |
| human embryonic kidney epithelial (293T) | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| human monocyte-derived dendritic cells (MDDCs) | This paper | N/A |
| human tracheobronchial epithelial cells (HTEC) | This paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| IFIT B primers forward: GGTATGCCGACCTTGAGAGAG | This paper | N/A |
| IFIT B primers reverse: TTCCCACTAAGGGTCCTGTTC | This paper | N/A |
| IFIT C primers forward: TGATGCGTGCCCTACTCTC | This paper | N/A |
| IFIT C primers reverse: CTGTGTCTCTGCTGTTCCGA | This paper | N/A |
| IFIT D primers forward: GGCTGTTTCCTTATTGTTGCTCT | This paper | N/A |
| IFIT D primers reverse: AGCAGTCCTGGTTCTGTGAG | This paper | N/A |
| GAPDH primers forward: CGCAGAGCCTCGAGGAGAAG | This paper | N/A |
| GAPDH primers reverse: ACAGGAGGACTTTGGGAACGAC | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCAGGS-3x HA-NS1 | This paper | N/A |
| pCAGGS-3x HA-NS1 (Y125A) | This paper | N/A |
| pCAGGS-FLAG-NS1 | This paper | N/A |
| pCAGGS-FLAG-NS1 (Y125A) | This paper | N/A |
| pCAGGS-FLAG-Keap1 | This paper | N/A |
| pCAGGS-GFP-NS1 | This paper | N/A |
| pCAGGS-GFP-Keap1 | This paper | N/A |
| pCAGGS-FLAG (empty) | This paper | N/A |
| pCDNA 3.1-NS1 | This paper | N/A |
| StrepII-HA NS1 | This paper | N/A |
| Flu-AV NS1 | This paper | N/A |
| MBP-His6-NS1 | Chatterjee et al., 2017 | N/A |
| ISRE luciferase reporter (pGL4.45) | Promega | Cat#E4141 |
| minP reporter (pGL4.23) | Promega | Cat#E8411 |
| Control luciferase reporter (pNL1.1.TK) | Promega | Cat#N1501 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| ZEN imaging software | ZEISS | https://www.zeiss.com/microscopy/int/products/microscope-software.html |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| MaxQuant version 1.3.0.5 | MaxQuant | https://www.maxquant.org/ |
| The R-project for statistical computing | R | www.R-project.org |
| Bowtie2 v2.2.5 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| HOMER v4.10.1 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| bamCoverage utility | https://deeptools.readthedocs.io/en/develop/content/tools/bamCoverage.html | |
| ChIPQC v1.14.0 | Carroll et al., 2014 | https://bioconductor.org/packages/release/bioc/html/ChIPQC.html |
| ChIPSeeker R package | Yu et al., 2012, 2015 | https://bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| RTA v1.9 | Illumina | https://support.illumina.com/downloads.html |
| STAR v2.5.1a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Subread:featureCount v1.4.6-p5 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| Salmon v0.8.2 | Patro et al., 2017 | https://github.com/COMBINE-lab/salmon |
| RSeQC v2.6.2 | Wang et al., 2012 | http://rseqc.sourceforge.net/ |
| EdgeR R/Bioconductor package | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Limma | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| voomWithQualityWeights | Luo et al., 2009 | https://rdrr.io/bioc/limma/man/voomWithQualityWeights.html |
| Heatmap2 R package | R | http://r.documentation.org/ |
| ggplot2 R package | R | http://r.documentation.org |
| Stats v3.4.1 R package | R | http://r.documentation.org |
| Genotify v1.2.1 | Andrews et al., 2018 | https://github.com/j-andrews7/Genotify |
|
| ||
| Other | ||
|
| ||
| BALB/c mice | The Jackson Laboratory | https://www.jax.org |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
In vivo animal studies
To generate a mouse monoclonal antibody to RSV NS1 (clone: 20E9.G12.F7.A11, IgG2a/kappa), female 6–8 week-old BALB/c mice housed 5/cage in a select pathogen-free facility were immunized using a combination of hydrodynamic injections with pCDNA 3.1 RSV NS1 (five 100 μg plasmid injections at 3–4 week intervals) and two injections of recombinantly expressed and purified NS1 protein (Chatterjee et al., 2017) in Freund’s adjuvant (CFA then IFA containing 40 ug each, one month apart). Mice were rested and given an ip boost of protein in PBS three days prior to euthanasia by CO2 inhalation and spleens were harvested 3 days later for fusion with P3X63Ag8.653 myelomas. Hybridoma supernatants were screened by ELISA, dot blot, and western blot.
All experiments were performed in accordance with procedures approved by the AAALAC-accredited Animal Studies Committee of Washington University in St. Louis and were in compliance with all relevant ethical regulations.
Cell lines
Adenocarcinomic human alveolar basal epithelial (A549) and human embryonic kidney epithelial (293T) cell lines (ATCC) were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM; ThemoFisher) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich). Cells were maintained at 37°C with 5% CO2.
Primary cell isolation and culture
Studies using deidentified primary human cells were obtained from remnant clinical specimens and healthy donors and were approved by the Institutional Review Board of Washington University School of Medicine.
Derivation of monocyte-derived dendritic cells
Peripheral blood mononuclear cells (PBMC) were isolated from deidentified healthy donors by density centrifugation with Lymphoprep (STEMCELL Technologies). From PBMC, CD14+CD16− monocytes were isolated by negative selection using the Classical Monocyte Isolation Kit (Miltenyi Biotec). Cells were resuspended at 0.5×106 cells/mL in RPMI 1640 media containing 10% FBS, 2 mM L-glutamine, 100 U pen/strep, 50 μM β-mercaptoethanol, 20 ng/mL GM-CSF and 20 ng/mL IL-4. After six days with growth media changes every 48 h, MDDC phenotype was confirmed with flow cytometry (HLA−DR+CD209+CD141+CD83−CD1c−).
Airway epithelial cell culture
Cells were isolated from surgical excess of tracheobronchial segments of lungs donated for transplantation as previously described (Horani et al., 2013). Basal epithelial cells were expanded on plastic plates prior to culture on supported membranes (Transwell, Corning Inc., Corning, NY). Cells were then differentiated to ciliated and secretory cell types using air-liquid interface conditions as described (Horani et al., 2013; You et al., 2002).
METHOD DETAILS
Plasmids
RSV NS1 was synthesized and subcloned into a pCAGGS vector containing either an N-terminal FLAG- or GFP- tag. KEAP1 (Johnson et al., 2016) was subcloned similarly. An N-terminal streptavidinII-HA tag NS1 vector (StreptII-HA NS1) was generated by Gateway cloning using the pDONR221 vector (Invitrogen).
Antibodies
All commercial antibodies are listed in the Key Resources table.
A mouse monoclonal antibody to RSV NS1 (clone: 20E9.G12.F7.A11) was generated from BALB/c mice (see in vivo animal studies). Splenic B cells were harvested and fused with P3X63Ag8.653 mouse myeloma cells. Hybridoma supernatants were screened for antibodies that bound NS1 by ELISA and western blot before expansion and subcloning by limiting dilution. Final selected monoclonal antibodies were isotyped, expanded, and purified using protein A Sepharose affinity chromatography.
Viral infection
RSV strain A2 used in the study was obtained from ATCC (VR-1540; Manassas, VA, USA). To determine the virus titers, cells cultivated in 24-well plates were inoculated with 10-fold serial dilutions of virus and incubated in 10% DMEM with methylcellulose at 37°C for 7 d. Cells were fixed with cold methanol at −80°C for 1 h. Methanol was removed and cells were incubated with 5% milk blocking buffer at 37°C for 1 h. Followed by incubation of goat anti-RSV (Fisher) and HRP-labeled donkey anti-goat (Fisher) antibodies. Cells were incubated with 0.03% 4-Chloronapthol and 1% hydrogen peroxide at 25°C. After 20 min incubation, the plate was dried upside down and plaques counted. The multiplicity of infection (MOI) was confirmed according to the virus titer from the plaque assay. RNaseq data from infections with RSV Y125A mutant NS1 was generated previously (Chatterjee et al., 2017) and re-analyzed here (GSE99298).
For infection, A549 cells and MDDCs were grown to approximately 80% confluence in cell culture plates and were infected with RSV at a MOI of 1. For hTECs, cells were differentiated on supported membranes in air-liquid interface cultures followed by RSV infection at MOI of 1. Mock infection was performed with phosphate buffered saline (PBS). After 1 hpi, the inoculum was removed by aspiration. Cells were washed twice with PBS and incubated in complete medium at 37°C for different time points until harvesting. For experiments with KPT-335 (verdinexor; RayBiotech), A549 cells were infected with RSV at a MOI of 1 or mock-infected with PBS. The inoculum was removed 1 hpi and cells were washed twice with PBS, followed by replacement of complete medium supplemented with KPT-335 (1 μM) or dimethyl sulfoxide (DMSO) at 37°C for 24 h incubation.
Transfection
For confocal microscopy
A549 cells grown to 80% confluence in 24-well cell culture plates were transfected with targeted plasmids by using Lipofectamine 2000 transfection reagent (Thermo Fisher). Briefly, 0.5 μg of plasmid and 1 μL of Lipofectamine 2000 were diluted in 50 μL of serum-free OptiMEM medium (Thermo Fisher), respectively. After 5 min-incubation, Lipofectamine 2000 OptiMEM was directly pipetted into the medium containing the diluted plasmid and incubated at room temperature for 25 min. Then the DNA-lipid complex was added into the cell culture medium, and the cells were further incubated for 24 h at 37°C. Then followed by confocal detection.
For ChIP
A549 cells were maintained in F-12K medium supplemented with 10% fetal bovine serum and 5 mM penicillin–streptomycin at 37°C with 5% carbon dioxide. 293T (human embryonic kidney cell line) were maintained in DMEM medium supplemented with 10% fetal bovine serum and 5 mM penicillin–streptomycin at 37°C with 5% carbon dioxide. A549 cells (106) in 15 cm plates were transfected with 20 μg of pCAGGS vector containing a 3x HA-tagged NS1 (HA-NS1) or 3x HA-Y125A NS1 (HA-Y125A) or no insert (empty vector) using Lipofectamine transfection reagent (Invitrogen).
Affinity purification-mass spectrometry and bioinformatics analysis
StreptII-HA NS1 was transiently transfected into 293T cells. Affinity purification of StreptII-HA NS1 or NS1 of Flu-AV (control) was performed as described previously (Holze et al., 2018). Briefly, cell lysates were incubated with streptavidin affinity resin in TAP lysis buffer followed by four washes using TAP wash buffer. Four independent affinity purifications followed by LC-MS/MS were performed. Samples were sequentially digested with LysC (Wako Chemicals USA) and trypsin (Promega), acidified with 0.1% TFA, desalted with C18 stage tips and analyzed by liquid chromatography coupled to mass spectrometry on an Orbitrap XL platform (Thermo Fisher).
Confocal immunofluorescence microscopy
For A549 and MDDC cells
After infection or transfection, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at 25°C. Cell monolayers were permeabilized with 0.2% Triton X-100 for 15 min. Cells were blocked with PBS containing 5% bovine serum albumin (BSA) for 30 min at room temperature. Next, cells were incubated for 1 h in the presence of a rabbit anti-Mediator1 antibody (anti-MED1) (1:200) and a mouse monoclonal antibody to RSV NS1 (anti-NS1) (1:100) in PBS buffer at 37°C followed by a 1 h incubation in PBS containing goat anti-mouse and anti-rabbit secondary antibodies conjugated to Alexa Fluor 488 and 594 at a dilution of 1:1000.
For hTECs
Airway epithelial cells on Transwell membranes were immunostained as described (Pan et al., 2007; You et al., 2002) using antibodies listed above. Cells on membranes were fixed with 4% paraformaldehyde for 15 min at 25°C, then washed with phosphate buffer saline (PBS) three times for 5 min at room temperature (RT). The membranes were cut from their plastic supports and divided into pieces. Membrane pieces were incubated in blocking buffer (5% donkey serum, 2% Triton X-100, in PBS) for 30 min at RT then incubated with primary antibody diluted in blocking buffer overnight at 4°C. Membranes were washed with PBS, followed by incubation with the secondary donkey antibody. Tissue sections of Transwell membranes were generated by immersing fixed cells on membrane pieces in low-melt agarose for embedding in paraffin, then sectioning. Cell sections were heated in antigen unmasking solution pH 6.0 (Vector Laboratories) in a pressure cooker (Biocare Medical) for 10 min prior to immunostaining using a rabbit anti-Acetyl-alpha-Tubulin (ac-α-tubulin) antibody (1:500) and a mouse anti-RSV NS1 (1:100) or anti-RSV Nucleoprotein antibody (1:100), followed by a 1 h incubation in PBS containing donkey anti-mouse and anti-rabbit secondary antibodies conjugated to Alexa Fluor 488 and 555 at a dilution of 1:1000. Following immunostaining, membranes were place on slides or tissue sections underwent nuclei staining using 4’, 6-diamidino-2-phenylindole (DAPI) in mounting medium (Fluoroshield; Sigma-Aldrich). Images were acquired using a Zeiss LSM 880 II FAST confocal microscope with Airyscan and adjusted globally for brightness and contrast using Adobe Photoshop.
Chromatin immunoprecipitation- (ChIP) seq and ChIP-qPCR
A549 cells were cross-linked 24 h post-transfection or 48 hours post-infection with hRSV. Cross-linking (room temperature): Cells in 15 cm plates were washed two times with PBS, incubated for 15 min with 1.25 mM EGS (ethylene glycol-bis(succinic acid N-hydroxysuccinimide ester); Sigma) in PBS, washed with 3 times PBS, incubated for 10 min with 1% formaldehyde (Sigma) in PBS. Cross-linking was quenched with the addition of 125 mM glycine (Sigma) for 5 min. Cells were washed once with PBS. Cells were scraped in ice-cold PBS, aliquoted to 106 cells per tube, snap-frozen on dry ice and stored at −80°C until lysis. Cells were lysed in an SDS buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0, dH2O) with protease inhibitors (Roche) for 20 min on ice, then chromatin was fragmented using a Bioruptor (Diagenode; Denville) to an average size of less than 300 bp. Ten percent of each sonicated sample was set aside for input; the remaining sonicated lysates were subjected to immunoprecipitation overnight at 4°C with antibody conjugated to Protein-A Dynabeads. Chromatin-IP-bead mixtures were then washed three times with buffers: low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8.0], 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8.0, 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris pH 8.0), and 1X Tris-EDTA buffer, and eluted with an SDS buffer (1% SDS, 0.1 M NaHCO3). Chromatin cross-links were reversed by overnight incubation with 5 M NaCl at 65°C and DNA was isolated using PCR MinElute spin purification columns (QIAGEN) per manufacturer instructions. At least 10 ng of ChIP or input DNA was submitted for indexed library preparation to the Washington University Genome Technology Access Center (WU GTAC). Samples were indexed and pooled (9–11 per lane) and subjected to 50 bp single-end sequencing per manufacturer protocol (Illumina HiSeq3000). ChIP-qPCR was performed with equal volumes of eluted DNA isolated as for ChIPseq with the following primers: IFITB: GGTATGCCGACCTTGAGAGAG and TTCCCACTAAGGGTCCTGTTC; IFITC: TGATGCGTGCCCTACTCTC and CTGTGTCTCTGCTGTTCCGA; IFITD: GGCTGTTTCCTTATTGTTGCTCT and AGCAGTCCTGGTTCTGTGAG; GAPDH: CGCAGAGCCTCGAGGAGAAG and ACAGGAGGACTTTGGGAACGAC.
RNA-seq
Infected cells were harvested and RNA extracted using the QIAGEN RNA Easy kit and 500 ng submitted to the WU GTAC. Samples were indexed and pooled (3 per lane) and subjected to 100 bp paired-end sequencing per manufacturer protocol (Illumina HiSeq3000).
Luciferase reporter assays
Luciferase reporter assays were performed with 2.5×104 293T cells per well of a 96-well plate transfected with pNL1.1.TK (control NanoLuc vector, Promega, Madison, WI, USA), plus WT or Y125A mutant NS1 or empty vector (pCAGGS), with or without 4 μg/mL poly(I:C) (Sigma), plus luciferase reporter plasmids: minimal promoter pGL4.23 (Promega) or ISRE reporter pGL4.45 (Promega) or minimal promoter pGL4.23 (Promega) containing one of the NS1 binding regions. Luciferase activity was measured 24 h post-transfection (triplicate wells per condition, 96-well plate) using the Promega Nano-Glo system per manufacturer protocol in a BioTek Cytation5 plate reader. All experiments were performed at least twice.
QUANTIFICATION AND STATISTICAL ANALYSIS
Colocalization analysis
The fluorescence intensity profiles and distribution of each signal were measured on the indicated line in the merge panel using ZEN 2.6 software. Pearson’s correlation coefficient (PCC) was measured in two-color images in the ImageJ analysis software package. The nuclear region was defined as a “region of interest” (ROI) over the image and the PCC value was calculated according to the formula:
where Ri and Gi refer to the intensity values of the red and green channels, respectively, and and refer to the mean intensities of the red and green channels, respectively, across the entire nuclear region in individual cells. PCC values near 1 reflect distributions of fluorescence signals that are correlated with one another. Values near zero reflect distributions of probes that are uncorrelated with one another.
ChIP- and RNA-sequencing analysis
Reads were aligned to hg19 with bowtie2 (v2.2.5) with default settings (Langmead and Salzberg, 2012). Peaks were called with HOMER (version v4.10.1) with the settings tbp = 1 and findPeaks cmd = findPeaks ns1_ha_align_tbp1/ −style factor −o auto −i ns1_input_align_tbp1/ (Heinz et al., 2010). RPKM normalized genome browser tracks were created with deepTools (v3.1.0) bamCoverage utility with settings–binSize10–extendReads150—normalize using RPKM and visualized on the UCSC genome browser 29106570. ChIPQC (v1.14.0) was used for quality control (Carroll et al., 2014). The ChIPSeeker R package (v1.16.1) and HOMER were used to annotate peaks (Heinz et al., 2010; Yu et al., 2012; Yu et al., 2015).
For new RNA-seq data acquired for this paper (0, 24, 48 hpi), reads were aligned to the Ensembl release 76 primary assembly with STAR version 2.5.1a (Dobin et al., 2013). Gene counts were imported into the R/Bioconductor package EdgeR (Robinson et al., 2010); TMM normalization size factors were calculated to adjust for differences in library size. Ribosomal genes and genes not expressed in the smallest group size minus one sample greater than one count-per-million were excluded from further analysis. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package Limma (Ritchie et al., 2015). Weighted likelihoods based on the observed mean-variance relationship of every gene and sample were then calculated for all samples with the voomWithQualityWeights (Luo et al., 2009). The performance of all genes was assessed with plots of the residual standard deviation of every gene to their average log-count with a robustly fitted trend line of the residuals. Differential expression analysis was performed using R/Bioconductor package Limma (Ritchie et al., 2015) and the results were filtered for Benjamini-Hochberg false-discovery rate adjusted p values ≤ 0.05. Heatmaps and volcano plots were generated with R packages heatmap2 and ggplot2. Genotify (v1.2.1) was used for manual gene curation (Andrews et al., 2018). Graphing and statistical analyses were performed using the R ggplot2 (3.2.1) and stats (3.4.1) packages and with Graphpad Prism (v8.2.1).
ChIP-qPCR analysis
Triplicate PCR wells were averaged and input wells were also scaled to 100% (10% of total input chromatin had been set aside from each sample prior to ChIP): input Ct value −3.322. Percent input for each ChIP sample was calculated as % input = 100 * 2^([scaled input]-[sample]). Log2 fold change was plotted in figure by 2ˆ(% input-ChIP sample / % input IgG).
Luciferase reporter assay analysis
The average ratios of the firefly to NanoLuc luciferase replicate values for each condition were compared to the average ratio for the minimal promoter reporter to determine relative luciferase activity. Statistical analyses were performed using Graphpad Prism 8. A p value < 0.05 was considered statistically significant. For all graphs, mean values ± standard deviation (SD) are shown.
Interaction proteomics data analysis
Mass spectrometry raw files were processed with MaxQuant version 1.3.0.5 or 1.4.0.654 using the built-in Andromeda engine to search against human proteome (UniprotKB, release 2012_06) containing forward and reverse sequences. In MaxQuant, the label-free quantitation (LFQ) algorithm and Match Between Runs option were used. Only proteins identified on the basis of at least two peptides and a minimum of three quantitation events in at least one experimental group were considered. LFQ protein intensity values were log-transformed, and missing values were filled by imputation. Specific enrichment was determined by multiple equal variance t tests with permutation-based false-discovery rate (FDR) statistics, performing 250 permutations. FDR thresholds and S0 parameters were empirically set to separate background from specifically enriched proteins. Data was analyzed using Perseus version 1.5.2.11. Results were plotted using R (www.R-project.org). Sequence logo was generated using WebLogo56.
Statistical analysis
Unless otherwise indicated, all data are representative of the mean and standard deviation of at least two replicates. Statistical analyses were performed using Graphpad Prism 8. A p value < 0.05 was considered statistically significant and are indicated in the figures.
Supplementary Material
Highlights.
NS1 partitions into the nucleus upon RSV infection
NS1 coimmunoprecipitates with chromatin-associated proteins
NS1 binds regulatory elements of differentially expressed genes in RSV infection
NS1 modulates host gene transcription
ACKNOWLEDGMENTS
We thank members of the D.W.L. and J.E.P. labs for assistance and support. We also acknowledge the support of NIH (R01AI107056, R01AI40758 to D.W.L.; P01AI120943 to G.K.A. and D.W.L.; R01HL146601 to S.L.B.) ERC-CoG (ProDAP 817798 to A.P.), Children’s Discovery Institute (PDII2018702 to D.W.L.), Foundation for Barnes-Jewish Hospital (to S.L.B.), and Siteman Cancer Center (P30CA091842 to N.R.B.). Sequencing was provided by the WU GTAC, which is partially supported by P30CA91842 and ICTS/CTSA UL1TR000448.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109803.
REFERENCES
- Aherne W, Bird T, Court SD, Gardner PS, and McQuillin J (1970). Pathological changes in virus infections of the lower respiratory tract in children. J. Clin. Pathol. 23, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JM, El-Alawi M, and Payton JE (2018). Genotify: Fast, lightweight gene lookup and summarization. J. Open Source Softw. 3, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya PL, Peeples ME, and Collins PL (1998). The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 72, 1452–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban J, Lee NR, Lee NJ, Lee JK, Quan FS, and Inn KS (2018). Human Respiratory Syncytial Virus NS 1 Targets TRIM25 to Suppress RIG-I Ubiquitination and Subsequent RIG-I-Mediated Antiviral Signaling. Viruses 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard EL, Braun MR, Lifland AW, Ludeke B, Noton SL, Vanover D, Zurla C, Fearns R, and Santangelo PJ (2020). Polymerase-tagged respiratory syncytial virus reveals a dynamic rearrangement of the ribonucleocapsid complex during infection. PLoS Pathog. 16, e1008987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyapalle S, Wong T, Garay J, Teng M, San Juan-Vergara H, Mohapatra S, and Mohapatra S (2012). Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection. PLoS ONE 7, e29386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham SC, Bush AJ, and Devincenzo JP (2000). Nasal quantity of respiratory syncytical virus correlates with disease severity in hospitalized infants. Pediatr. Infect. Dis. J. 19, 113–117. [DOI] [PubMed] [Google Scholar]
- Carlsten JO, Zhu X, and Gustafsson CM (2013). The multitalented Mediator complex. Trends Biochem. Sci. 38, 531–537. [DOI] [PubMed] [Google Scholar]
- Carroll TS, Liang Z, Salama R, Stark R, and de Santiago I (2014). Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet. 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Luthra P, Esaulova E, Agapov E, Yen BC, Borek DM, Edwards MR, Mittal A, Jordan DS, Ramanan P, et al. (2017). Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses. Nat. Microbiol. 2, 17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman NA, and Peeples ME (1993). The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology 195, 596–607. [DOI] [PubMed] [Google Scholar]
- Dave KA, Norris EL, Bukreyev AA, Headlam MJ, Buchholz UJ, Singh T, Collins PL, and Gorman JJ (2014). A comprehensive proteomic view of responses of A549 type II alveolar epithelial cells to human respiratory syncytial virus infection. Mol. Cell. Proteomics 13, 3250–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVincenzo JP, El Saleeby CM, and Bush AJ (2005). Respiratory syncytial virus load predicts disease severity in previously healthy infants. J. Infect. Dis. 191, 1861–1868. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchin J, Sun Q, Kentsis A, Farmer A, Zhang ZC, Sanda T, Mansour MR, Barcelo C, McCauley D, Kauffman M, et al. (2013). Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia 27, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, and Engreitz JM (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, et al. (2019). Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, He B, Liu S, Zhu H, Tan K, and Qian J (2016). EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics 32, 3543–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García J, García-Barreno B, Vivo A, and Melero JA (1993). Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195, 243–247. [DOI] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, et al. (2019). A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176, 377–390.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. (2012). Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildyal R, Baulch-Brown C, Mills J, and Meanger J (2003). The matrix protein of Human respiratory syncytial virus localises to the nucleus of infected cells and inhibits transcription. Arch. Virol. 148, 1419–1429. [DOI] [PubMed] [Google Scholar]
- Ghildyal R, Ho A, Wagstaff KM, Dias MM, Barton CL, Jans P, Bardin P, and Jans DA (2005). Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin beta1 independent of importin alpha. Biochemistry 44, 12887–12895. [DOI] [PubMed] [Google Scholar]
- Ghildyal R, Ho A, Dias M, Soegiyono L, Bardin PG, Tran KC, Teng MN, and Jans DA (2009). The respiratory syncytial virus matrix protein possesses a Crm1-mediated nuclear export mechanism. J. Virol. 83, 5353–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González PA, Prado CE, Leiva ED, Carreño LJ, Bueno SM, Riedel CA, and Kalergis AM (2008). Respiratory syncytial virus impairs T cell activation by preventing synapse assembly with dendritic cells. Proc. Natl. Acad. Sci. USA 105, 14999–15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther M, Bauer A, Müller M, Zaeck L, and Finke S (2020). Interaction of host cellular factor ANP32B with matrix proteins of different paramyxoviruses. J. Gen. Virol. 101, 44–58. [DOI] [PubMed] [Google Scholar]
- Hall CB, Douglas RG Jr., Schnabel KC, and Geiman JM (1981). Infectivity of respiratory syncytial virus by various routes of inoculation. Infect. Immun. 33, 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, et al. (2009). The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 360, 588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holze C, Michaudel C, Mackowiak C, Haas DA, Benda C, Hubel P, Pennemann FL, Schnepf D, Wettmarshausen J, Braun M, et al. (2018). Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 19, 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Nath A, Wasserman MG, Huang T, and Brody SL (2013). Rho-associated protein kinase inhibition enhances airway epithelial Basal-cell proliferation and lentivirus transduction. Am. J. Respir. Cell Mol. Biol. 49, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cheng X, Traina-Dorge VL, Park HJ, Zhou H, Soike K, and Kemble G (2003). Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine 21, 3647–3652. [DOI] [PubMed] [Google Scholar]
- Johnson B, Li J, Adhikari J, Edwards MR, Zhang H, Schwarz T, Leung DW, Basler CF, Gross ML, and Amarasinghe GK (2016). Dimerization Controls Marburg Virus VP24-dependent Modulation of Host Antioxidative Stress Responses. J. Mol. Biol. 428, 3483–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorquera PA, Mathew C, Pickens J, Williams C, Luczo JM, Tamir S, Ghildyal R, and Tripp RA (2019). Verdinexor (KPT-335), a Selective Inhibitor of Nuclear Export, Reduces Respiratory Syncytial Virus Replication In Vitro. J. Virol. 93, e01684–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Nouën C, Brock LG, Luongo C, McCarty T, Yang L, Mehedi M, Wimmer E, Mueller S, Collins PL, Buchholz UJ, and DiNapoli JM (2014). Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc. Natl. Acad. Sci. USA 111, 13169–13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léonard VH, Kohl A, Hart TJ, and Elliott RM (2006). Interaction of Bunyamwera Orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J. Virol. 80, 9667–9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester JT, and DeLuca NA (2011). Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediator in virus-infected cells. J. Virol. 85, 5733–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liesman RM, Buchholz UJ, Luongo CL, Yang L, Proia AD, DeVincenzo JP, Collins PL, and Pickles RJ (2014). RSV-encoded NS2 promotes epithelial cell shedding and distal airway obstruction. J. Clin. Invest. 124, 2219–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE Jr., and Santangelo PJ (2012). Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 86, 8245–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z, Tran KC, and Teng MN (2009). Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 83, 3734–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MS, Brazas RM, and Holtzman MJ (2005). Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79, 9315–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, and Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Lombardía L, García-Barreno B, Domínguez O, and Melero JA (2007). Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells. J. Gen. Virol. 88, 570–581. [DOI] [PubMed] [Google Scholar]
- Mata M, Sarrion I, Armengot M, Carda C, Martinez I, Melero JA, and Cortijo J (2012). Respiratory syncytial virus inhibits ciliagenesis in differentiated normal human bronchial epithelial cells: effectiveness of N-acetylcysteine. PLoS ONE 7, e48037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye UR, Gokulrangan G, Datt M, Dobrowolski C, Cooper M, Chance MR, and Karn J (2013). Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLoS Pathog. 9, e1003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Lee S, Hotard AL, and Moore ML (2014a). Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. MBio 5, e01704–e01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Stobart CC, Hotard AL, and Moore ML (2014b). An overview of respiratory syncytial virus. PLoS Pathog. 10, e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbradt AG, Kulkarni M, Yi T, Takeuchi K, Sun ZY, Luna RE, Selenko P, Näär AM, and Wagner G (2011). Structure of the VP16 transactivator target in the Mediator. Nat. Struct. Mol. Biol. 18, 410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Stühler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, and Meisterernst M (2003). A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 22, 6494–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday DC, Emmott E, Surtees R, Lardeau CH, Wu W, Duprex WP, Dove BK, Barr JN, and Hiscox JA (2010). Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell. Proteomics 9, 2438–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S, Le Nouen C, Luongo C, Buchholz UJ, Collins PL, and Bukreyev A (2008). Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J. Virol. 82, 8780–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S, Hillyer P, Le Nouën C, Buchholz UJ, Rabin RL, Collins PL, and Bukreyev A (2011). Respiratory syncytial virus interferon antagonist NS1 protein suppresses and skews the human T lymphocyte response. PLoS Pathog. 7, e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, You Y, Huang T, and Brody SL (2007). RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 120, 1868–1876. [DOI] [PubMed] [Google Scholar]
- Pandya MC, Callahan SM, Savchenko KG, and Stobart CC (2019). A Contemporary View of Respiratory Syncytial Virus (RSV) Biology and Strain-Specific Differences. Pathogens 8, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, and Wei LN (2005). Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell 19, 643–653. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14 (4), 417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentecost M, Vashisht AA, Lester T, Voros T, Beaty SM, Park A, Wang YE, Yun TE, Freiberg AN, Wohlschlegel JA, and Lee B (2015). Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog. 11, e1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwitasari O, Johnson S, Yan X, Howerth E, Shacham S, Landesman Y, Baloglu E, McCauley D, Tamir S, Tompkins SM, and Tripp RA (2014). Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo. J. Virol. 88, 10228–10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles RJ, and DeVincenzo JP (2015). Respiratory syncytial virus (RSV) and its propensity for causing bronchiolitis. J. Pathol. 235, 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Liu T, Pang L, Li K, Garofalo RP, Casola A, and Bao X (2011). A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J. Gen. Virol. 92, 2153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA, Lee B, Maisner A, Rota P, and Wang L; Ictv Report Consortium (2017). ICTV Virus Taxonomy Profile: Pneumoviridae. J. Gen. Virol. 98, 2912–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel M, Behner L, Heiner A, Sauerhering L, and Maisner A (2020). Replication of a Nipah Virus Encoding a Nuclear-Retained Matrix Protein. J. Infect. Dis. 221 (Suppl 4), S389–S394. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas HG, Schmaling SK, and Gaglia MM (2016). Shutoff of Host Gene Expression in Influenza A Virus and Herpesviruses: Similar Mechanisms and Common Themes. Viruses 8, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlender J, Bossert B, Buchholz U, and Conzelmann KK (2000). Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74, 8234–8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, et al. ; RSV Global Epidemiology Network (2017). Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390, 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Denouel A, Tietjen AK, Campbell I, Moran E, Li X, Campbell H, Demont C, Nyawanda BO, Chu HY, et al. (2020). Global Disease Burden Estimates of Respiratory Syncytial Virus-Associated Acute Respiratory Infection in Older Adults in 2015: A Systematic Review and Meta-Analysis. J. Infect. Dis. 222, S577–S583. [DOI] [PubMed] [Google Scholar]
- Siersbæk R, Madsen JGS, Javierre BM, Nielsen R, Bagge EK, Cairns J, Wingett SW, Traynor S, Spivakov M, Fraser P, and Mandrup S (2017). Dynamic Rewiring of Promoter-Anchored Chromatin Loops during Adipocyte Differentiation. Mol. Cell 66, 420–435. [DOI] [PubMed] [Google Scholar]
- Smith CM, Kulkarni H, Radhakrishnan P, Rutman A, Bankart MJ, Williams G, Hirst RA, Easton AJ, Andrew PW, and O’Callaghan C (2014). Ciliary dyskinesia is an early feature of respiratory syncytial virus infection. Eur. Respir. J. 43, 485–496. [DOI] [PubMed] [Google Scholar]
- Soutourina J (2018). Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274. [DOI] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Chi B, Rabin RL, and Collins PL (2004). Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J. Virol. 78, 4363–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM, Tran KC, and Collins PL (2005). Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J. Virol. 79, 5353–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedan S, Musiyenko A, and Barik S (2009). Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J. Virol. 83, 9682–9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedan S, Andrews J, Majumdar T, Musiyenko A, and Barik S (2011). Multiple functional domains and complexes of the two nonstructural proteins of human respiratory syncytial virus contribute to interferon suppression and cellular location. J. Virol. 85, 10090–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama I, Sato H, Watanabe A, Omi-Furutani M, Sugai A, Kanki K, Yoneda M, and Kai C (2012). The nucleocapsid protein of measles virus blocks host interferon response. Virology 424, 45–55. [DOI] [PubMed] [Google Scholar]
- Tan YR, Peng D, Chen CM, and Qin XQ (2013). Nonstructural protein-1 of respiratory syncytial virus regulates HOX gene expression through interacting with histone. Mol. Biol. Rep. 40, 675–679. [DOI] [PubMed] [Google Scholar]
- Teng MN, Whitehead SS, Bermingham A, St Claire M, Elkins WR, Murphy BR, and Collins PL (2000). Recombinant respiratory syncytial virus that does not express the NS1 or M2–2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74, 9317–9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier TM, Dodge MJ, Prusinkiewicz MA, and Mymryk JS (2019). Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery. Cells 8, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE Jr., and Levy DE (1992). Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12, 3315–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen P, Ruf M, Blakqori G, Överby AK, Heidemann M, Eick D, and Weber F (2011). Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J. Biol. Chem. 286, 3681–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villenave R, Thavagnanam S, Sarlang S, Parker J, Douglas I, Skibinski G, Heaney LG, McKaigue JP, Coyle PV, Shields MD, and Power UF (2012). In vitro modeling of respiratory syncytial virus infection of pediatric bronchial epithelium, the primary target of infection in vivo. Proc. Natl. Acad. Sci. USA 109, 5040–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villenave R, Shields MD, and Power UF (2013). Respiratory syncytial virus interaction with human airway epithelium. Trends Microbiol. 21, 238–244. [DOI] [PubMed] [Google Scholar]
- Vojnic E, Mourão A, Seizl M, Simon B, Wenzeck L, Larivière L, Baumli S, Baumgart K, Meisterernst M, Sattler M, and Cramer P (2011). Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 18, 404–409. [DOI] [PubMed] [Google Scholar]
- Wang YE, Park A, Lake M, Pentecost M, Torres B, Yun TE, Wolf MC, Holbrook MR, Freiberg AN, and Lee B (2010). Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 6, e1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28 (16), 2184–2185. 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, and Murphy BR (1999). Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73, 3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Tran KC, Teng MN, Heesom KJ, Matthews DA, Barr JN, and Hiscox JA (2012). The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J. Virol. 86, 7777–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, Berry KN, Yen B, Hamilton J, Brett TJ, et al. (2014). Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 16, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough ML, Mata MA, Sakthivel R, and Fontoura BM (2014). Viral subversion of nucleocytoplasmic trafficking. Traffic 15, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, and Brody SL (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L1315–L1321. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, and He QY (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zhang L, Peeples ME, Boucher RC, Collins PL, and Pickles RJ (2002). Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76, 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G, Behera S, Singam R, Lockey RF, and Mohapatra SS (2005). Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11, 56–62. [DOI] [PubMed] [Google Scholar]
- Zhang L, Collins PL, Lamb RA, and Pickles RJ (2011). Comparison of differing cytopathic effects in human airway epithelium of parainfluenza virus 5 (W3A), parainfluenza virus type 3, and respiratory syncytial virus. Virology 421, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang L, Wang H, Zhang G, and Sun X (2016). Respiratory syncytial virus non-structural protein 1 facilitates virus replication through miR-29a-mediated inhibition of interferon-a receptor. Biochem. Biophys. Res. Commun. 478, 1436–1441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-RSV NS1 | This paper | N/A |
| Mouse monoclonal anti-RSV nucleoprotein | Abcam | Cat#ab94806; RRID: AB_10674696 |
| Rabbit polyclonal anti-MED1 | Bethyl Laboratories | Cat#A300-793A; RRID: AB_577241 |
| Rabbit polyclonal anti-MED14 | Bethyl Laboratories | Cat#A301-044A; RRID: AB_2266461 |
| Mouse monoclonal anti-MED25 | Santa Cruz Biotechnology | Cat#sc-393759 |
| Rabbit monoclonal anti-XPO1 | Abcam | Cat#ab180144; RRID: AB_2847862 |
| Mouse monoclonal anti FLAG | Sigma-Aldrich | Cat#F3165; RRID: AB_259529 |
| Rabbit monoclonal anti-Acetyl-alpha-Tubulin | Sigma-Aldrich | Cat#SAB5600134 |
| Alexa Fluor 488-conjugated goat anti-mouse IgG | Thermo Fisher Scientific | Cat#A-11029; RRID: AB_2534088 |
| Alexa Fluor 594-conjugated goat anti-rabbit IgG | Thermo Fisher Scientific | Cat#A-11037; RRID: AB_2534095 |
| Alexa Fluor 488-conjugated donkey anti-mouse IgG | Thermo Fisher Scientific | Cat#A-21202; RRID: AB_141607 |
| Alexa Fluor 555-conjugated donkey anti-rabbit IgG | Thermo Fisher Scientific | Cat#A-31572; RRID: AB_162543 |
| Peroxidase affiniPure goat anti mouse IgG | Jackson ImmunoResearch | Cat#115-035-003; RRID: AB_10015289 |
| Peroxidase affiniPure goat anti rabbit IgG | Jackson ImmunoResearch | Cat#111-035-003; RRID: AB_2313567 |
| Goat polyclonal anti-RSV | Fisher Scientific | Cat#AB1128MI |
| HRP-conjugated donkey anti-goat IgG | Fisher Scientific | Cat#AP180PMI |
| Rabbit polyclonal anti-GAPDH | Abcam | ab9485; RRID: AB_307275 |
| Rabbit polyclonal anti-CTCF | Cell Signaling Technology | 2899; RRID: AB_2086794 |
| Mouse monoclonal anti-tubulin | Sigma-Aldrich | T9026-100Ul; RRID: AB_477593 |
| Rabbit polyclonal anti-HA | Abcam | Ab9110; RRID: AB_307019 |
| Normal rabbit IgG | Millipore | 12-370; RRID: AB_145841 |
| Rat monoclonal anti-CD209-PerCP-Cyanine5.5 | Fisher Scientific | Cat#50-158-32; RRID AB_1106983 |
| Mouse monoclonal anti-CD141-APC | Fisher Scientific | Cat#17141942; RRID: AB_2662437 |
| Mouse monoclonal anti-CD83-FITC | Fisher Scientific | Cat#50-968-3 |
| Mouse monoclonal anti-CD1c-PE | ThermoFisher Scientific | Cat#25-0015-42; RRID: AB_2573324 |
| Mouse monoclonal anti-HLA-DR-Alexa Fluor 700 | Fisher Scientific | Cat#5016938 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Human respiratory syncytial virus (RSV, A2 strain) | ATCC | Cat#VR-1540 |
| Human respiratory syncytial virus NS1 Y125A | Chatterjee et al., 2017 | N/A |
| Escherichia coli BL21(DE3) | Novagen | Cat#69450 |
|
| ||
| Biological samples | ||
|
| ||
| Fetal Bovine serum | Sigma-Aldrich | Cat#F4135 |
| Donkey serum | Sigma-Aldrich | Cat#D9663; RRID: AB_2810235 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat#BP9704-100 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| KPT-335 | RayBiotech | Cat#331-21369-1 |
| NS1 protein | Chatterjee et al., 2017 | N/A |
| Lipofectamine 2000 transfection reagent | Thermo Fisher Scientific | Cat#11668019 |
| Dulbecco’s modified Eagle’s medium (DMEM) | Thermo Fisher Scientific | Cat#11965 |
| OptiMEM medium | Thermo Fisher Scientific | Cat#31985070 |
| RPMI 1640 media | GIBCO | Cat#224000-071 |
| Trypsin-EDTA (0.05%) | Thermo Fisher Scientific | Cat#25300054 |
| Penicillin-streptomycin | Gen Clone | Cat#25-512 |
| F-12K medium | GIBCO | Cat#11330-032 |
| Poly(I:C) | Invivogen | Cat#TLVL-PICW |
| EGS (ethylene glycol-bis(succinic acid N-hydroxysuccinimide ester | Thermo | Cat#21565 |
| Formaldehyde | Sigma-Aldrich | Cat#252549-25ml |
| LysC | Wako Chemicals | Cat#121-05063 |
| Trypsin | Promega | Cat#V5280 |
| Glycine | Fisher Scientific | Cat#BP3811 |
| PBS | Sigma-Aldrich | Cat#D8537 |
| GM-CSF | R&D Systems | Cat#415ML010 |
| IL-4 | R&D Systems | Cat#35050-061 |
| GlutaMAX | GIBCO | Cat#35050-061 |
| 2-mercapthoethanol | Sigma-Aldrich | Cat#M3148 |
| DMSO | ThermoFisher Scientific | Cat#BP231100 |
| Lymphoprep | StemCell Technologies, Inc. | Cat#7861 |
| Dynabeads Prot A ref 10002D | Thermo Fisher Scientific | Dynabeads Prot A ref 10002D |
| DNA extraction kit (MinElute PCR) | QIAGEN | Cat. No. / ID: 28004 |
| RNA extraction kit (RNAeasy Micro) | QIAGEN | Cat. No. / ID: 74004 |
| Power SYBR Green PCR Master Mix | Fisher Scientific | Cat # 4367659 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Subcellular Protein Fractionation Kit | Thermo Fisher Scientific | Cat#78840 |
| Nano-Glo system | Promega | Cat#N1110 |
| Classical monocyte isolation kit | Miltenyi Biotec | Cat#130-117-337 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-seq data | https://www.ncbi.nlm.nih.gov/geo/ GSE99298 | |
| ChIP-seq and RNA-seq data | https://www.ncbi.nlm.nih.gov/geo/ GSE155152 | |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Adenocarcinomic human alveolar basal epithelial (A549) | ATCC | Cat#CCL-185; RRID: CVCL |
| human embryonic kidney epithelial (293T) | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| human monocyte-derived dendritic cells (MDDCs) | This paper | N/A |
| human tracheobronchial epithelial cells (HTEC) | This paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| IFIT B primers forward: GGTATGCCGACCTTGAGAGAG | This paper | N/A |
| IFIT B primers reverse: TTCCCACTAAGGGTCCTGTTC | This paper | N/A |
| IFIT C primers forward: TGATGCGTGCCCTACTCTC | This paper | N/A |
| IFIT C primers reverse: CTGTGTCTCTGCTGTTCCGA | This paper | N/A |
| IFIT D primers forward: GGCTGTTTCCTTATTGTTGCTCT | This paper | N/A |
| IFIT D primers reverse: AGCAGTCCTGGTTCTGTGAG | This paper | N/A |
| GAPDH primers forward: CGCAGAGCCTCGAGGAGAAG | This paper | N/A |
| GAPDH primers reverse: ACAGGAGGACTTTGGGAACGAC | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCAGGS-3x HA-NS1 | This paper | N/A |
| pCAGGS-3x HA-NS1 (Y125A) | This paper | N/A |
| pCAGGS-FLAG-NS1 | This paper | N/A |
| pCAGGS-FLAG-NS1 (Y125A) | This paper | N/A |
| pCAGGS-FLAG-Keap1 | This paper | N/A |
| pCAGGS-GFP-NS1 | This paper | N/A |
| pCAGGS-GFP-Keap1 | This paper | N/A |
| pCAGGS-FLAG (empty) | This paper | N/A |
| pCDNA 3.1-NS1 | This paper | N/A |
| StrepII-HA NS1 | This paper | N/A |
| Flu-AV NS1 | This paper | N/A |
| MBP-His6-NS1 | Chatterjee et al., 2017 | N/A |
| ISRE luciferase reporter (pGL4.45) | Promega | Cat#E4141 |
| minP reporter (pGL4.23) | Promega | Cat#E8411 |
| Control luciferase reporter (pNL1.1.TK) | Promega | Cat#N1501 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| ZEN imaging software | ZEISS | https://www.zeiss.com/microscopy/int/products/microscope-software.html |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| MaxQuant version 1.3.0.5 | MaxQuant | https://www.maxquant.org/ |
| The R-project for statistical computing | R | www.R-project.org |
| Bowtie2 v2.2.5 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| HOMER v4.10.1 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| bamCoverage utility | https://deeptools.readthedocs.io/en/develop/content/tools/bamCoverage.html | |
| ChIPQC v1.14.0 | Carroll et al., 2014 | https://bioconductor.org/packages/release/bioc/html/ChIPQC.html |
| ChIPSeeker R package | Yu et al., 2012, 2015 | https://bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| RTA v1.9 | Illumina | https://support.illumina.com/downloads.html |
| STAR v2.5.1a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Subread:featureCount v1.4.6-p5 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| Salmon v0.8.2 | Patro et al., 2017 | https://github.com/COMBINE-lab/salmon |
| RSeQC v2.6.2 | Wang et al., 2012 | http://rseqc.sourceforge.net/ |
| EdgeR R/Bioconductor package | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Limma | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| voomWithQualityWeights | Luo et al., 2009 | https://rdrr.io/bioc/limma/man/voomWithQualityWeights.html |
| Heatmap2 R package | R | http://r.documentation.org/ |
| ggplot2 R package | R | http://r.documentation.org |
| Stats v3.4.1 R package | R | http://r.documentation.org |
| Genotify v1.2.1 | Andrews et al., 2018 | https://github.com/j-andrews7/Genotify |
|
| ||
| Other | ||
|
| ||
| BALB/c mice | The Jackson Laboratory | https://www.jax.org |