Abstract
We previously identified a gene encoding a putative GTPase, GTPBP1, which is structurally related to elongation factor 1α, a key component of protein biosynthesis machinery. The primary structure of GTPBP1 is highly conserved between human and mouse (97% identical at the amino acid level). Expression of this gene is enhanced by gamma interferon in a monocytic cell line, THP-1. Although counterparts of this molecule in Caenorhabditis elegans and Ascaris suum have also been identified, the function of this molecule remains to be clarified. In the present study, our immunohistochemical analyses on mouse tissues revealed that GTPBP1 is expressed in some neurons and smooth muscle cells of various organs as well as macrophages. Immunofluorescence analyses revealed that GTPBP1 is localized exclusively in cytoplasm and shows a diffuse granular network forming a gradient from the nucleus to the periphery of the cells in smooth muscle cell lines and macrophages. To investigate the physiological role of GTPBP1, we used targeted gene disruption in embryonic stem cells to generate GTPBP1-deficient mice. The mutant mice were born at the expected Mendelian frequency, developed normally, and were fertile. No manifest anatomical or behavioral abnormality was observed in the mutant mice. Functions of macrophages, including chemotaxis, phagocytosis, and nitric oxide production, in mutant mice were equivalent to those seen in wild-type mice. No significant difference was observed in the immune response to protein antigen between mutant mice and wild-type mice, suggesting normal function of antigen-presenting cells of the mutant mice. The absence of an eminent phenotype in GTPBP1-deficient mice may be due to functional compensation by GTPBP2, a molecule we recently identified which is similar to GTPBP1 in structure and tissue distribution.
Gamma interferon (IFN-γ) is a key monocyte-activating cytokine (3, 17, 20, 27). With IFN-γ stimulation, the human monocytic cell line THP-1 expresses a number of genes associated with the antigen-presenting function of macrophages, including major histocompatibility complex class II (MHC-II), CD74 (MHC-II-associated invariant chain), and interleukin-1β (7, 26; our unpublished observation). To identify genes involved in the function of macrophages, we had previously carried out PCR-based cDNA subtraction and subsequent differential display on mRNA obtained from IFN-γ-treated and untreated THP-1 cells. In so doing, we found a novel gene encoding protein bearing GTP-binding motifs, the characteristic of the GTPase superfamily, and we designated this gene GP-1 (22). We later renamed the gene GTPBP1, as recommended by the Human Gene Nomenclature Committee.
GTPBP1 is highly conserved between human and mouse (97% identical over the entire protein). In Northern blot analyses on mouse tissues, transcripts of the gene are evident in various tissues, with a relatively high expression in brain, thymus, and lung. GTPBP1 is similar to the putative GTPases of nematodes, AGP-1 of Ascaris suum and CGP-1 of Caenorhabditis elegans (11). The sequence similarity in total protein between GTPBP1 of mouse and CGP-1 of C. elegans is about 45%, the amino acid sequences of four GTP-binding motifs (G1 to G4) of them being practically identical. Therefore, we proposed a novel GTPase subfamily, the GP-1 family, composed of human and mouse GTPBP1, AGP-1 of A. suum, and CGP-1 of C. elegans. The primary structure of members of GP-1 family is related to those of elongation factor 1α (eEF-1α) and elongation factor Tu (EF-Tu), key components of protein synthesis machinery in eukaryotes and prokaryotes, respectively. By a TBLASTN (protein query versus nucleotide database) search against expressed sequence tag databases, using the amino acid sequence of mouse GTPBP1 as a query, we found uncharacterized sequences highly similar to GTPBP1 in zebra fish (GenBank accession no. AI436986) and in Drosophila melanogaster (GenBank accession no. AA949198). They presumably represent members of this family in the species. On the other hand, sequences closer to those of GP-1 family than to those of EFs were not evident in yeast and prokaryotes. Therefore, the members of the GP-1 family may play some role, probably essential for and unique to multicellular organisms.
We have now done an immunohistochemical analysis on various mouse organs and immunofluorescence analyses on cultured cells to examine cellular and subcellular localizations of GTPBP1 protein. In addition, we developed mice carrying a mutation in the GTPBP1 gene by using targeted gene disruption.
MATERIALS AND METHODS
Antibodies.
Polyclonal antibodies recognizing near the amino terminus (GP1a) and carboxyl terminus (GP1b) of the GTPBP1 protein were raised by immunizing rabbits with keyhole limpet hemocyanin (KLH)-conjugated synthetic oligomer peptides CGETIYVIGQGSDGTE and CSGGRRRGGQRHKVKS, respectively. Antibodies were purified from immune sera, using peptide affinity chromatography, and specificity was verified by immunoblot analyses and indirect immunofluorescence analyses on COS-7 cells transfected with a mouse GTPBP1 expression vector. For preabsorption experiments, peptides were used in 100-fold molar excesses. Anti-influenza virus hemagglutinin (HA) monoclonal antibody (clone 12CA5) and phycoerythrin-labeled anti-mouse macrophage antibody (clone F4/80) were purchased from Boehringer Mannheim and Caltag Laboratories, respectively. Absence of cross-reaction of fluorescein isothiocyanate (FITC)-labeled goat anti-mouse immunoglobulin G (IgG) polyclonal antibody (PharMingen) to rabbit antibody and of Cy-3-labeled goat anti-rabbit IgG polyclonal antibody (Amersham Pharmacia) to mouse IgG was confirmed.
Construction of the expression vector.
A mouse GTPBP1 cDNA clone isolated from a mouse brain cDNA library (30) was inserted into the mammalian expression vector pBJ1, downstream of the SRα promoter (25). Subsequently, double-stranded oligomer DNA coding for the HA epitope (MYPYDVPDYA) was added just downstream of the initiation codon to obtain the HA-tagged GTPBP1 expression construct pSRHAGP1. COS-7 cells were transfected with the construct by lipofection using Transfectam reagent (Promega). Twenty-four hours after the transfection, the cells were harvested and reseeded on coverslips.
Immunofluorescence analysis.
Cells cultured overnight on coverslips coated with fibronectin were washed twice with phosphate-buffered saline (PBS) and treated with fixative solution (PBS containing 4% paraformaldehyde) for 15 min at room temperature. After being washed three times with PBS, cells were treated with ethanol for 2 min, with permeabilizing buffer (PBS containing 0.1% Triton X-100 and 2% bovine serum albumin) for 2 min, and with blocking buffer (PBS containing 2% bovine serum albumin) for 30 min. The cells were stained with primary antibodies for 1 h, washed 3 times with PBS, stained with FITC- or Cy3-labeled secondary antibodies for 1 h, and washed three times with PBS. A fluorescence microscope (Axioplan 2; Carl Zeiss) was used for microscopy.
Immunoblot analysis.
Tissues and cultured cells were homogenized in lysis buffer (1% sodium dodecyl sulfate [SDS], 60 mM Tris-HCl [pH 6.8], 10% glycerol), and then lysates were subjected to electrophoresis on SDS-polyacrylamide gels, under reducing conditions, and transferred onto nitrocellulose membranes (Trans-Blot; Bio-Rad). Membranes were probed with anti-GTPBP1 antibody and subsequently with peroxidase-conjugated goat anti-rabbit IgG. Signals were visualized by chemiluminescence, using ECL reagent (Amersham Pharmacia). In some analyses, filters were reprobed with antibodies after being treated with stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.7) for 30 min at 60°C and extensively washed with Tris-buffered saline containing 0.2% Tween.
Immunohistochemical analysis.
Anesthetized female C57BL/6 mice underwent intracardiac perfusion with fixative solution. The tissues were removed, immersed in fixative solution at 4°C for 16 h, dehydrated, and embedded in paraffin. Sections (6 μm) were deparaffinated, treated with methanol-H2O2 for inactivation of endogenous peroxidase activity, and stained with anti-GTPBP1 antibody, GP1a. As a negative control, nonimmunized rabbit serum was also used in the same experiments. For immunohistochemical detection, we used the avidin-biotin-peroxidase technique (Vectastain ABC kit; Vector Laboratories, Burlingame, Calif.) and peroxidase was developed using diaminobenzidene. Immunostained sections were counterstained with methyl green.
Construction of the targeting vector.
A mouse GTPBP1 cDNA clone was used as a probe to isolate genomic DNA clones corresponding to the GTPBP1 loci from a mouse genomic DNA library. In the targeting vector, pKO-GP1, the genomic region encoding amino acids 58 to 193 (in the previously published amino acid sequence [22]) of GTPBP1 was replaced with a phosphoglycerate kinase (PGK)-neomycin phosphotransferase (neoR)-poly(A) cassette. The vector contained homologous genomic DNA fragments 6.2 and 1.25 kb on either side of the PGK-neoR cassette. The PGK-thymidine kinase (tk)-poly(A) cassette and pMC1-diphtheria toxin A (DT)-poly(A) cassette (28) were ligated on the 3′ and 5′ ends, respectively.
Generation of GTPBP1-mutant mice.
Culture and transfection of TT2 (29) embryonic stem (ES) cells were done as previously described (14, 16, 31). Transfected ES cells were selected in medium containing G418 (200 μg/ml) and ganciclovir (2 μM). Drug-resistant ES cell clones were screened by PCR with a flanking primer (5′-CTTGTCCTGGCATTCCCCTACACT-3′) and a PGK-promoter-specific primer (5′-TGCTAAAGCCCATGCTCCAGACTG-3′), which yields a 1.25-kb product in the case of proper homologous recombination. PCR-positive clones were analyzed by Southern blot analyses to verify homologous recombination. Homologous recombinant ES cell clones were injected into ICR eight-cell stage embryos, and the embryos were implanted into the uteri of pseudopregnant ICR mice to generate chimeric mice. Male chimeras were mated with C57BL/6 female mice. F1 offspring were genotyped to select heterozygous mutant mice. Heterozygous mutant mice were intercrossed, and homozygous mutant mice were obtained. Genotypes of the mice were routinely determined with tail DNA, using PCR. A mutant allele-specific primer (5′-GCCTACCCGCTTCCATTGCTCAG-3′), a wild-type-specific primer (5′-GCTAGTTCCTGAGGAGATACTCGA-3′), and a common primer (5′-ATCCTCTTACGAGAACGTCAAGAAG-3′) were mixed in each reaction mixture, and PCR products about 350 and 500 bp in length appeared from the mutated and wild-type alleles, respectively. The results of PCR were occasionally confirmed by Southern blot analyses. Mice at ages of 8 to 16 weeks were used for the functional analyses.
Isolation of peritoneal macrophages.
Mice were intraperitoneally injected with 2 ml of 4% Brewer thioglycollate solution. Four days later, exudating cells were obtained by lavage of the peritoneal cavity with 6 ml of PBS. The number of cells obtained from each mouse was determined using a Neubauer hemacytometer. Aliquots of harvested cells were stained with phycoerythrin-labeled anti-mouse macrophage antibody (F4/80) and analyzed on a flow cytometer (FACScan; Becton Dickinson).
Adhesion assay.
Adhesion assays were done as previously reported (24), with some modification. In brief, thioglycollate-elicited peritoneal cells suspended in RPMI 1640 medium supplemented with 10% fetal calf serum were plated at a density of 2 × 105 cells per well in 96-well flat-bottom plates and incubated for 50 min at 37°C. Cells in plates were fixed in methanol and stained with 10% Giemsa's solution. Plates were washed five times with water and dried. Retained dye was solubilized in methanol and measured as the absorbance at 450 nm on a microplate reader (model 550; Bio-Rad).
Phagocytosis assay.
Cell suspensions in RPMI 1640 medium supplemented with 10% fetal calf serum were plated at a density of 106 cells per well in 24-well tissue culture plates and allowed to adhere to the plates overnight at 37°C. Nonadherent cells were then removed by washing the plates with RPMI 1640 medium. BODIPY FL-labeled zymosan particles (Molecular Probes) were opsonized with IgG and added to the plates (107/well). Plates were incubated for 15 min at 37°C, treated with lyticase (1,000 U/ml) for 5 min, and washed with PBS. Subsequently, the cells were fixed with fixative solution for 10 min, harvested, and analyzed using a flow cytometer.
Analysis of immune response to KLH.
Wild-type and homozygous mutant mice were genotyped by PCR to select mice bearing the I-Ak/k genotype. Selected mice were immunized at the base of the tail with 50 μg of KLH protein emulsified in complete Freund's adjuvant. After 8 days, these mice were killed and bilateral inguinal and para-aortic lymph nodes were isolated. Single-cell suspensions were made from lymph nodes in RPMI 1640 medium supplemented with 10% horse serum. Cells were seeded in 96-well flat-bottom culture plates (4 × 105/well) in the presence or absence of KLH (10 μg/ml) and cultured for 5 days. [3H]thymidine (1 μCi/well, 6.7 Ci/mmol) was added to the culture in the last 18 h, and the incorporation of [3H]thymidine was measured by scintillation counting.
RESULTS
Subcellular localization of GTPBP1.
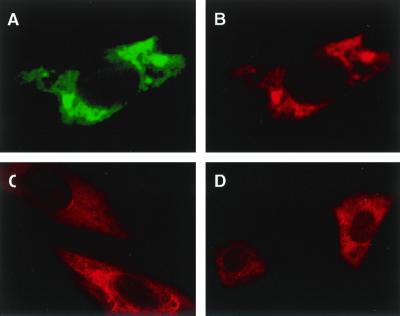
To determine the subcellular localization of GTPBP1, we carried out indirect immunofluorescence analyses on several types of cultured cells. COS-7 cells were transiently transfected with an HA-tagged GTPBP1 expression vector, pSRHAGP1, and grown on fibronectin-coated coverslips. The cells were double stained with anti-HA monoclonal antibody (12CA5) and anti-GTPBP1 polyclonal antibody (GP1a), and signals were detected using FITC-labeled anti-mouse IgG antibody and Cy3-labeled anti-rabbit IgG antibody, respectively (Fig. 1A and B). As expected, fluorescence signals of the two antibodies showed the same distribution. No fluorescence signal was observed when GP1a was preabsorbed by excess amounts of the peptide used to raise the antibody or when mock-transfected COS-7 cells were stained (not shown). Forced expression of GTPBP1 led to no change in the morphology of the cells and structures of actin filaments and microtubules in COS-7 cells (not shown). The rat aortic smooth muscle cell line, A-10 (13), and mouse peritoneal macrophages elicited by stimulation with thioglycollate solution, both of which express abundant intrinsic GTPBP1 protein, were also stained with GP1a (Fig. 1C and D). In all of these cells, GTPBP1 is present exclusively in the cytoplasm, and punctate or irregular distribution with much of the signal in the juxtanuclear region was observed. The distribution in A-10 cells and in macrophages appeared as finer granules than those seen in COS cell transfectants. The forced expression of the molecule or addition of the HA tag sequence at the amino terminus of the molecule may affect intracellular distribution in the transfectants. The expression of GTPBP1 was observed in another rat aortic cell line, A7r5 (13), and mouse L cells, and staining patterns were similar to those observed in A-10 cells and macrophages (not shown). Absence of specific fluorescence in the nucleus was also confirmed by confocal microscopic analyses (not shown). In some cells, GTPBP1 was also enriched in a region adjacent to the plasma membrane in podosomes and in cell edges.
FIG. 1.
Immunofluorescence analysis of cultured cells on the intracellular distribution of GTPBP1. COS-7 cells were transfected with an HA-tagged mouse GTPBP-1 expression vector, pSRHAGP1, and cultured on coverslips. Forty-eight hours after the transfection, cells were double stained with anti-HA monoclonal antibody (12CA5) and anti-GTPBP1 polyclonal antibody (GP1a). Staining signals with 12CA5 were visualized by FITC-labeled anti-mouse IgG (A), and the GP1a staining signal was visualized by Cy3-labeled anti-rabbit IgG (B). A rat aortic smooth muscle cell line, A10 (C), and mouse peritoneal macrophages (D) were also cultured on coverslips and stained with GP1a. Magnifications, ×850 (A, B, and C) and ×340 (D).
Tissue distribution of GTPBP1 protein in various mouse tissues.
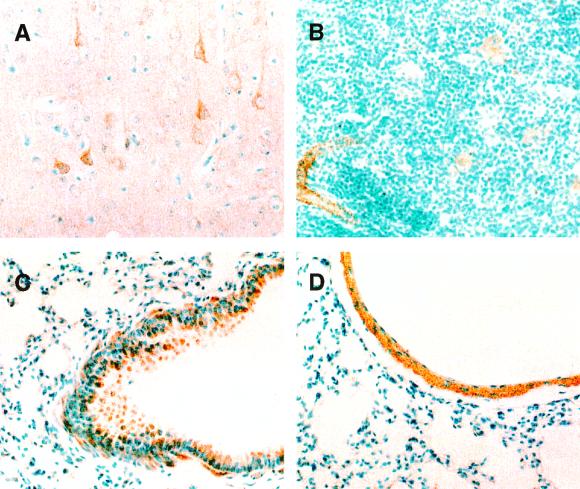
To determine the distribution of GTPBP1 molecule in tissues, we analyzed various organs of C57BL/6 mice by immunohistochemistry. Paraffin-embedded tissue sections were stained with the anti-GTPBP1 polyclonal antibody, GP1a. Figure 2A shows a section of the cerebral cortex. GTPBP1 is expressed in some neurons but not in neuroglia cells. In the thymus, GTPBP1 is found in small arteries as well as in macrophages and dendritic cells in the medulla, but not in thymocytes (Fig. 2B). In the lung, GTPBP1 is expressed in bronchi (Fig. 2C) and pulmonary arteries (Fig. 2D). GTPBP1 is expressed in bronchial epithelial cells and submucosal smooth muscle cells. In pulmonary arteries, smooth muscle cells of tunica media are stained. Table 1 summarizes the results of immunohistochemical analyses on various mouse organs. In addition to neurons and macrophages, GTPBP1 is expressed in smooth muscle cells in a broad range of organs, for example, blood vessels, respiratory and digestive tracts, uterus, and urinary bladder.
FIG. 2.
Immunohistochemical localization of GTPBP1 in several mouse organs. Expression of GTPBP1 is seen in some neurons in the cerebral cortex (A), arterial smooth muscle cells and macrophages in thymus (B), bronchial epithelial cells and peribronchial smooth muscle cells (C), and smooth muscle cells of the pulmonary artery (D). Magnifications, ×200.
TABLE 1.
GTPBP1 immunoreactivity in mouse tissues
| Organ or tissue | Structure or cell type | GTPBP1 intensity |
|---|---|---|
| Brain (cerebral cortex) | Neurons | ++/−a |
| Neuroglia cells | − | |
| Thymus | Thymocytes | − |
| Epithelial cells | − | |
| Macrophages/dendritic cells | + | |
| Spleen | Lymphocytes | − |
| Macrophages | + | |
| Lung | Pneumocytes | − |
| Bronchial epithelium | ++ | |
| Peribronchial smooth muscle | +++ | |
| Bronchial cartilage | − | |
| Heart | Myocardium | + |
| Endocardium | − | |
| Arteries and veins | Endothelium | − |
| Tunica media (smooth muscle) | +++ | |
| Tunica adventitia | − | |
| Esophagus | Epithelium | + |
| Muscularis mucosae | + | |
| Stomach | Gastric mucosa | − |
| Muscularis mucosae | + | |
| Muscularis propria | ++ | |
| Small intestine | Epithelium | − |
| Muscularis mucosae | + | |
| Muscularis propria | ++ | |
| Colon | Epithelium | − |
| Muscularis mucosae | + | |
| Muscularis propria | ++ | |
| Liver | Hepatocytes | − |
| Sinusoidal endothelium | − | |
| Bile duct epithelium | − | |
| Kidney | Glomeruli | − |
| Renal tubules | − | |
| Urinary bladder | Epithelium | − |
| Smooth muscle | ++ | |
| Uterus | Endometrium | − |
| Myometrium | ++ |
Some neurons do not express GTPBP1.
Generation of GTPBP1-mutant mice.
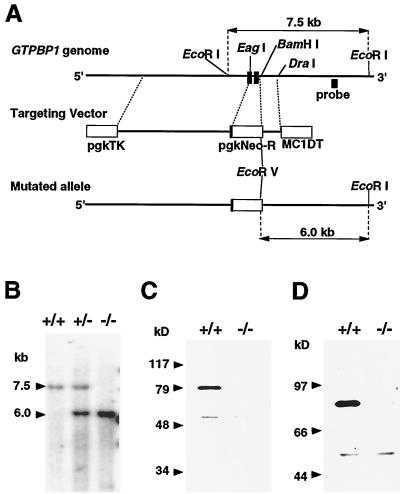
To assess the physiological role of GTPBP1, we developed mutant mice devoid of normal GTPBP1 protein. The targeting vector, pKO-GP1, was made so as to yield, by homologous recombination, a mutant allele in which the genomic region coding for most of the putative GTP-binding domain (22) of GTPBP1 was replaced by a neomycin resistance gene cassette (Fig. 3A). The targeting vector was transfected into TT2 (29) ES cells, and cells were then cultured in selection medium containing G418 and ganciclovir. Two homologous recombinant ES cell clones were obtained out of 540 double-resistant ES clones analyzed. Cells from the two ES clones were injected into eight-cell stage embryos of ICR mice. One ES cell clone gave rise to a germ line-transmitted mutant mouse strain.
FIG. 3.
Generation of GTPBP1-mutant mice. (A) Schematic depiction of a part of the mouse GTPBP1 gene, the targeting vector, and the mutant allele. Closed boxes indicate two exons coding for amino acids 17 to 77 and 78 to 193 (according to a previously published amino acid sequence [22]) of the GTPBP1 protein, respectively. The targeting vector contains a neomycin resistance gene (pgkNeo-R) for positive selection and the diphtheria toxin A gene (MC1DT) and herpesvirus thymidine kinase gene (pgkTK) for negative selection. (B) Southern blot analysis showing homologous recombination. Genomic DNA samples from a wild-type mouse, a hemizygous mutant mouse, and a homozygous mutant mouse were digested with EcoRI and EcoRV. The location of the probe, a 0.6-kb HindIII-HindIII genomic DNA fragment used for Southern blot analyses, is shown in panel A. The 7.5-kb band indicates the wild type, and the 6.0-kb band indicates the mutant allele shown in panel A. (C and D) Immunoblot analyses showing the absence of an intact GTPBP1 molecule in a homozygous mutant mouse. Lysates of brain were made from a wild type and a homozygous mutant mouse, electrophoresed in an SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane, and probed with polyclonal antibodies GP1a (C) and GP1b (D), which recognize amino- and carboxyl-terminal portions of the GTPBP1 protein, respectively.
The hemizygous mutant female and male mice were crossed to examine the phenotype caused by GTPBP1 deficiency. Among the 304 offspring analyzed, 75 were homozygous mutant mice (35 males and 40 females), as expected from Mendelian transmission. Figure 3B shows an example of a Southern blot analysis of the GTPBP1 locus of a wild-type, hemizygous mutant and homozygous mutant mice. To confirm that the homozygous mutant mice lacked intact GTPBP1 protein, we examined lysates made from brain of wild-type and homozygous mutant mice by immunoblot analysis, using polyclonal antibodies GP1a and GP1b recognizing amino- and carboxyl-terminal portions of the protein, respectively (Fig. 3C).
The homozygous mutant mice were apparently normal; the external appearances of the whole body and each organ were indistinguishable from those of wild-type mice. Mutant mice had no overt neurological and behavioral deficits; they grew up normally and were healthy at least up to the age that we have studied them (12 months). Both male and female homozygous mutant mice yielded offspring normally. In addition, no abnormality was found in histological analyses of organs in which GTPBP1 is expressed, i.e., brain, thymus, lung, spleen, and kidney.
Function of GTPBP1-deficient macrophages.
The expression of GTPBP1 is enhanced by IFN-γ in THP-1 cells, accompanying the expression of genes related to functions of macrophages, and this molecule is abundantly expressed in peritoneal macrophages elicited by stimulation with thioglycollate. Therefore, we assumed that GTPBP1 may be involved in some functions of macrophages. Although the homozygous mutant mice appeared healthy under specific-pathogen-free conditions in our mouse facility, we reasoned that homozygous GTPBP1-deficient mice may bear some deficits in the immune system.
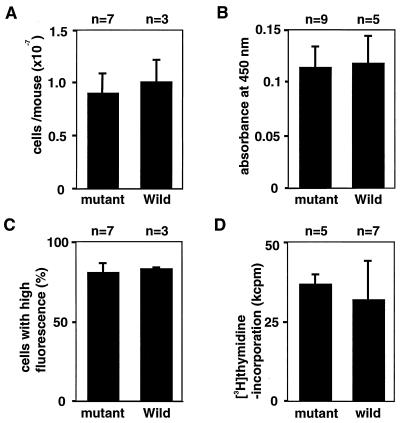
We analyzed several macrophage functions, comparing thioglycollate-elicited peritoneal macrophages obtained from wild-type and mutant mice. We assessed the chemotactic activity of macrophages by counting the number of peritoneally exudating macrophages defined by staining with monoclonal antibody F4/80 (Fig. 4A). Four days after the intraperitoneal injection of thioglycollate solution, almost the same numbers of macrophages were recovered by lavage of the peritoneal cavity of the wild-type and homozygous mutant mice. We analyzed the adhesional activity of the peritoneal macrophages, using the Giemsa assay. As shown in Fig. 4B, peritoneally exudating cells of wild-type and mutant mice adhered to the surface of culture plates at almost the same rate. Morphologies of the adhering cells were indistinguishable between macrophages obtained from wild-type and mutant mice. To investigate the possibility that GTPBP1 is involved in phagocytic activity, we analyzed Fc-receptor-mediated phagocytosis of IgG-opsonized zymozan particles. Peritoneal macrophages were incubated with fluorescent zymozan particles coated with mouse IgG, and numbers of macrophages which ingested zymozan particles were examined by flow cytometric analysis (Fig. 4C). The phagocytic activity of macrophages from wild-type and mutant mice were also equivalent in this analysis. Other than these analyses, we examined macrophages regarding expression of MHC-II molecules and Fcγ-receptor (CD16/32), phagocytosis of polystyrene beads, and nitric oxide production under the stimulation of IFN-γ and lipopolysaccharides. However, we found no differences between wild-type and mutant mice.
FIG. 4.
Macrophage function and immune response of GTPBP1 mutant mice. (A) Mice were intraperitoneally injected with Brewer thioglycollate solution. Four days later, the number of macrophages in the peritoneal cavity of each mouse was counted. Mean values + SD for three wild-type and seven mutant mice are shown. (B) Adhesional activity of peritoneal macrophages to tissue culture-treated plastic surfaces. Cells were plated into 96-well culture plates, incubated for 50 min at 37°C, stained with 10% Giemsa's solution, and extensively washed. Retained dye was measured as the absorbance at 450 nm on a microplate reader. Mean values + SD for five wild-type and nine mutant mice are shown. (C) Phagocytic activity of peritoneal macrophages. Peritoneal macrophages were incubated with BODIPY FL-labeled zymosan particles coated with IgG in culture plates for 15 min, fixed with formaldehyde, harvested, and analyzed on a flow cytometer. Percentages of macrophages with high fluorescence were shown. Values are means + SD for three wild-type and seven mutant mice. (D) Immune response to KLH. Mice were subcutaneously immunized at the base of the tail with 50 μg of KLH emulsified in complete Freund's adjuvant. Eight days after the immunization, single-cell suspensions were made from draining lymph nodes. Lymph node cells were cultured for 5 days with KLH (10 μg/ml) in 96-well plates. The proliferative response in the last 18 h of the culture were measured by [3H]thymidine uptake and mean values + SD for seven wild-type and five mutant mice are shown. For both wild-type and mutant mice, counts were less than 2,000 cpm in the case of lymph node cells that were cultured in the absence of the antigen.
Immune response to protein antigen of GTPBP1-mutant mice.
It has been established that dendritic cells are the most potent antigen-presenting cells responsible for stimulating T lymphocytes in peripheral lymphoid organs. It was reported that monocyte-derived dendritic cells play a major role in transportation of antigens from peripheral tissues to draining lymph nodes in vivo (19). We found by reverse transcription-PCR analyses that GTPBP1 is expressed also in myeloid-lineage dendritic cells differentiated in vitro (data not shown). Therefore, it may be that the absence of normal functions of GTPBP1 in dendritic cells would elicit deficient T-cell responses to antigenic challenge. We analyzed the immune response to protein antigen of GTPBP1-mutant mice. Because immune response to exogenous protein antigen is affected by allotype in the MHC-II locus, mice of the I-Ak/k genotype were selected from wild-type and homozygous mutant mice and used for this experiment. Eight days after immunization with KLH to the base of the tail, almost the same numbers of the cells were obtained from inguinal and para-aortic lymph nodes of wild-type and mutant mice ([4.5 ± 1.4] × 107 and [3.3 ± 1.7] × 107 [mean ± standard deviation {SD}] per mouse, respectively). To analyze the reactivity of antigen-specific T cells and function of antigen-presenting cells, isolated lymph node cells were cultured in vitro in the absence or presence of KLH and the proliferative response was determined according to [3H]thymidine incorporation. As shown in Fig. 4D, lymph node cells from wild-type and mutant mice showed almost the same proliferative response when cultured in vitro with KLH, thereby indicating that functions of antigen-presenting cells were not affected by GTPBP1 deficiency.
DISCUSSION
Among the GTPase superfamily, the primary structure of the GP-1 family is closest to that of G proteins involved in protein translational machinery, such as initiation factors, EF-1α, EF-Tu, and GST1/RF3 (10). These G proteins play essential roles in initiation, elongation, and termination of protein translation in both prokaryotes and eukaryotes. The indirect immunofluorescence analyses in the present study revealed that the GTPBP1 molecule is localized exclusively in the cytoplasm as a diffuse granular network pattern with highly immunofluorescent signals in a perinuclear region. The localization pattern is similar to the reported localization of components of protein synthesis machinery (21), suggesting that GTPBP1 colocalizes with these molecules and functions in related protein synthesis machinery. To elucidate this issue, double immunostaining analyses with antibodies specific to ribosomal protein would be necessary.
Because of the evolutionary conservation of GTPBP1, we presumed that the homozygous mutant mice would show some abnormality. However, we found no gross morphological and behavioral abnormalities of the mutant mice. Our analysis showed that GTPBP1 is not essential for general functions of macrophages, such as chemotaxis, plastic-surface adhesion, phagocytosis, and production of nitric oxide. The development of T lymphocytes, which is potentially influenced by cells of macrophage lineage, of the mutant mice appeared normal in the flow cytometric analyses (data not shown). Proliferative response of T lymphocytes to antigenic challenge with protein antigen was also normal, indicating that antigen processing and presentation by macrophages and dendritic cells were not affected by the mutation of GTPBP1.
In mammals, IFN-γ, along with IFN-α and IFN-β, plays a key role in the host defense against pathogenic microorganisms. The activation of macrophages by IFN-γ is essential for resistance to intracellular bacterial infections, such as listeriosis and tuberculosis (2, 6, 12). In addition to activating macrophages, these cytokines affect a variety of cell types and induce expression of genes essential for host defense (5, 9). The MX proteins (1, 23), GTPases induced by IFN-α and -β, are involved in resistance to several types of viruses. Although GTPBP1-deficient mice seemed healthy under pathogen-free conditions, IFN-γ-inducible expression of GTPBP1 raises the possibility that GTPBP1 is involved in host defense to a specific pathogen.
In our immunohistochemical analyses, we found that GTPBP1 is expressed in smooth muscle cells of various tissues and some neurons in the cerebral cortex as well as in macrophages (Table 1). The GTPBP1-deficient mice may have a functional abnormality in these tissues, albeit not evident in nonstressed situations. As mutant mice seemed more active and aggressive than wild-type littermates, we conducted several experiments to examine activity and aggression of these mice. However, we found no significant difference between mutant and wild type in resident intruder test (4, 8, 32) and forced swimming test (18).
A large number of mutant mice have been generated by gene targeting in ES cells, and their phenotypes are sometimes much more limited than expected. However, the absence of overt abnormality in mutant mice does not mean that the genes have no physiological function. There is the possibility that the absence of apparent defects in mutant mice may be due to genetic redundancy, i.e., other molecules with similar function and tissue distribution compensate for the defect of the function of the gene in those targeting mice. Recently, we identified cDNA of another member of the GP-1 family, GTPBP2, from a cDNA library of human macrophages and mouse brain (15). Mouse GTPBP2 protein bears the GTP-binding motif similar to those of members of the GP-1 family and shares 44% similarity with mouse GTPBP1 over the entire amino acid sequence. The tissue distribution of the GTPBP2 mostly overlaps that of GTPBP1. It is possible that GTPBP2 compensates for defects in functions of GTPBP1 in the GTPBP1-mutant mice. The mouse GTPBP1 and GTPBP2 genes are located in different chromosomal loci, and generation of GTPBP2-deficient mice is under way. To assess the possibility of functional redundancy between GTPBP1 and GTPBP2, we will develop double-knockout mice which have defects in both the GTPBP1 and GTPBP2 loci.
ACKNOWLEDGMENTS
We are grateful to S. Matsushita, A. Irie, Y. Yasunami, H. Ohkubo, M. Takeya, and H. Nishiura for valuable suggestions. M. Ohara provided helpful comments on the manuscript.
This work was supported in part by grant-in-aid 10770142 from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Aebi M, Fah J, Hurt N, Samuel C E, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1989;9:5062–5072. doi: 10.1128/mcb.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchmeier N A, Schreiber R D. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc Natl Acad Sci USA. 1985;82:7404–7408. doi: 10.1073/pnas.82.21.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cofano F, Comoglio P M, Landolfo S, Tarone G. Mouse immune interferon enhances fibronectin production of elicited macrophages. J Immunol. 1984;133:3102–3106. [PubMed] [Google Scholar]
- 4.De Felipe C, Herrero J F, O'Brien J A, Palmer J A, Doyle C A, Smith A J, Laird J M, Belmonte C, Cervero F, Hunt S P. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature. 1998;392:394–397. doi: 10.1038/32904. [DOI] [PubMed] [Google Scholar]
- 5.Farrar M A, Schreiber R D. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 6.Flesch I, Kaufmann S H. Mycobacterial growth inhibition by interferon-gamma-activated bone marrow macrophages and differential susceptibility among strains of Mycobacterium tuberculosis. J Immunol. 1987;138:4408–4413. [PubMed] [Google Scholar]
- 7.Gaffney E V, Lingenfelter S E, Koch G A, Lisi P J, Chu C W, Tsai S C. Regulation by interferon gamma of function in the acute monocytic leukemia cell line, THP-1. J Leukoc Biol. 1988;43:248–255. doi: 10.1002/jlb.43.3.248. [DOI] [PubMed] [Google Scholar]
- 8.Hilakivi-Clarke L A, Wozniak K M, Durcan M J, Linnoila M. Behavior of streptozotocin-diabetic mice in tests of exploration, locomotion, anxiety, depression and aggression. Physiol Behav. 1990;48:429–433. doi: 10.1016/0031-9384(90)90339-6. [DOI] [PubMed] [Google Scholar]
- 9.Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil K B, Fujiwara T, Takahashi E, Tanahashi N, Tamura T, Ichihara A, Tanaka K. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med. 1996;183:1807–1816. doi: 10.1084/jem.183.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino S, Miyazawa H, Enomoto T, Hanaoka F, Kikuchi Y, Kikuchi A, Ui M. A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 1989;8:3807–3814. doi: 10.1002/j.1460-2075.1989.tb08558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y J, Stoffel R, Tobler H, Mueller F. A newly formed telomere in Ascaris suum does not exert a telomere position effect on a nearby gene. Mol Cell Biol. 1996;16:130–134. doi: 10.1128/mcb.16.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiderlen A F, Kaufmann S H, Lohmann-Matthes M L. Protection of mice against the intracellular bacterium Listeria monocytogenes by recombinant immune interferon. Eur J Immunol. 1984;14:964–967. doi: 10.1002/eji.1830141019. [DOI] [PubMed] [Google Scholar]
- 13.Kimes B W, Brandt B L. Characterization of two putative smooth muscle cell lines from rat thoracic aorta. Exp Cell Res. 1976;98:349–366. doi: 10.1016/0014-4827(76)90446-8. [DOI] [PubMed] [Google Scholar]
- 14.Kohmura N, Yagi T, Tomooka Y, Oyanagi M, Kominami R, Takeda N, Chiba J, Ikawa Y, Aizawa S. A novel nonreceptor tyrosine kinase, Srm: cloning and targeted disruption. Mol Cell Biol. 1994;14:6915–6925. doi: 10.1128/mcb.14.10.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudo H, Senju S, Mitsuya H, Nishimura Y. Mouse and human GTPBP2, newly identified members of the GP-1 family of GTPase. Biochem Biophys Res Commun. 2000;272:456–465. doi: 10.1006/bbrc.2000.2763. [DOI] [PubMed] [Google Scholar]
- 16.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 17.Murray H W, Rubin B Y, Rothermel C D. Killing of intracellular Leishmania donovani by lymphokine-stimulated human mononuclear phagocytes. Evidence that interferon-gamma is the activating lymphokine. J Clin Investig. 1983;72:1506–1510. doi: 10.1172/JCI111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porsolt R D, Le Pichon M, Jalfre M. Depression: a new animal model sensitive to antidepressant treatments. Nature. 1977;266:730–732. doi: 10.1038/266730a0. [DOI] [PubMed] [Google Scholar]
- 19.Randolph G J, Beaulieu S, Lebecque S, Steinman R M, Muller W A. Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science. 1998;282:480–483. doi: 10.1126/science.282.5388.480. [DOI] [PubMed] [Google Scholar]
- 20.Rollag H, Degre M, Sonnenfeld G. Effects of interferon-alpha/beta and interferon-gamma preparations on phagocytosis by mouse peritoneal macrophages. Scand J Immunol. 1984;20:149–155. doi: 10.1111/j.1365-3083.1984.tb00988.x. [DOI] [PubMed] [Google Scholar]
- 21.Sanders J, Brandsma M, Janssen G M, Dijk J, Moller W. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 beta gamma delta in the endoplasmic reticulum. J Cell Sci. 1996;109:1113–1117. doi: 10.1242/jcs.109.5.1113. [DOI] [PubMed] [Google Scholar]
- 22.Senju S, Nishimura Y. Identification of human and mouse GP-1, a putative member of a novel G-protein family. Biochem Biophys Res Commun. 1997;231:360–364. doi: 10.1006/bbrc.1997.6103. [DOI] [PubMed] [Google Scholar]
- 23.Staeheli P, Haller O. Interferon-induced human protein with homology to protein Mx of influenza virus-resistant mice. Mol Cell Biol. 1985;5:2150–2153. doi: 10.1128/mcb.5.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, Takashima Y, Kawabe Y, Cynshi O, Wada Y, Honda M, Kurihara H, Aburatani H, Doi T, Matsumoto A, Azuma S, Noda T, Toyoda Y, Itakura H, Yazaki Y, Kodama T, et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 25.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virelizier J L, Perez N, Arenzana-Seisdedos F, Devos R. Pure interferon gamma enhances class II HLA antigens on human monocyte cell lines. Eur J Immunol. 1984;14:106–108. doi: 10.1002/eji.1830140120. [DOI] [PubMed] [Google Scholar]
- 27.Vogel S N, Finbloom D S, English K E, Rosenstreich D L, Langreth S G. Interferon-induced enhancement of macrophage Fc receptor expression: beta-interferon treatment of C3H/HeJ macrophages results in increased numbers and density of Fc receptors. J Immunol. 1983;130:1210–1214. [PubMed] [Google Scholar]
- 28.Yagi T, Nada S, Watanabe N, Tamemoto H, Kohmura N, Ikawa Y, Aizawa S. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal Biochem. 1993;214:77–86. doi: 10.1006/abio.1993.1459. [DOI] [PubMed] [Google Scholar]
- 29.Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- 30.Yasunami M, Suzuki K, Houtani T, Sugimoto T, Ohkubo H. Molecular characterization of cDNA encoding a novel protein related to transcriptional enhancer factor-1 from neural precursor cells. J Biol Chem. 1995;270:18649–18654. doi: 10.1074/jbc.270.31.18649. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M, Suda Y, Matsuo I, Miyamoto N, Takeda N, Kuratani S, Aizawa S. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124:101–111. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimura H, Watanabe K, Ogawa N. Psychotropic effects of ginseng saponins on agonistic behavior between resident and intruder mice. Eur J Pharmacol. 1988;146:291–297. doi: 10.1016/0014-2999(88)90305-6. [DOI] [PubMed] [Google Scholar]