Significance
Medicinal plants with extensive ethnobotanical histories, particularly those from Asia, have contributed to the approval of natural compounds as pharmaceutical drugs. In contrast, Samoan traditional medicine is relatively understudied. Working with traditional healers via an ethical, data sovereignty–driven collaboration led by indigenous Samoan researchers, we elucidate the chemical biology of the poorly understood but often-used Samoan traditional medicine “matalafi,” the homogenate of Psychotria insularum leaves commonly used to treat inflammation-associated illnesses. Our approach unifies genomics, metabolomics, analytical biochemistry, immunology, and traditional knowledge to delineate the mode of action of the traditional medicine rather than by the more common reductionist approach of first purifying the bioactive principles, which can be used to better understand the ethnobotany of traditional medicine.
Keywords: traditional medicine, chemical biology, genomics, metabolomics, iron homeostasis
Abstract
The leaf homogenate of Psychotria insularum is widely used in Samoan traditional medicine to treat inflammation associated with fever, body aches, swellings, wounds, elephantiasis, incontinence, skin infections, vomiting, respiratory infections, and abdominal distress. However, the bioactive components and underlying mechanisms of action are unknown. We used chemical genomic analyses in the model organism Saccharomyces cerevisiae (baker’s yeast) to identify and characterize an iron homeostasis mechanism of action in the traditional medicine as an unfractionated entity to emulate its traditional use. Bioactivity-guided fractionation of the homogenate identified two flavonol glycosides, rutin and nicotiflorin, each binding iron in an ion-dependent molecular networking metabolomics analysis. Translating results to mammalian immune cells and traditional application, the iron chelator activity of the P. insularum homogenate or rutin decreased proinflammatory and enhanced anti-inflammatory cytokine responses in immune cells. Together, the synergistic power of combining traditional knowledge with chemical genomics, metabolomics, and bioassay-guided fractionation provided molecular insight into a relatively understudied Samoan traditional medicine and developed methodology to advance ethnobotany.
Compounds from natural resources are reliable lead templates of new pharmaceuticals, having persisted through evolutionary selection to control fundamental molecular pathways. Of the 1,562 newly approved drugs from 1981 to 2019, 64% were either natural products, derived from natural products, or based upon natural product scaffolds, biological macromolecules, or botanical drugs (1). In 2008 alone, 50% of the 255 drugs in various stages of development originated from plant natural products, demonstrating the large and yet-untapped potential of plant natural products (2).
Medicinal plants in effect have been trialed for activity through centuries of ethnobotanical use, making traditional medicines an attractive yet challenging source for further investigation (3). For example, the anti-malarial drug artemisinin was isolated from Artemisia annua L., a well-documented herb in traditional Chinese medicine used for the treatment of fever and malaria (4). Samoan medicinal plants also comprise such a resource, as evident by the phorbol ester prostratin, a compound sourced from the medicinal plant Homalanthus nutans (G.Forst) Guill., which is in development as a latency reversing agent of HIV (5–7).
Another medicinal plant commonly used in Samoa is Psychotria insularum A.Gray (Rubiaceae), a small tree approximately 2 m in height with small white flowers and glossy red berries. Known locally as “matalafi,” the homogenate of P. insularum leaves (fresh leaf juice) is used to treat inflammation associated with fever, body aches, swelling, wounds, incontinence, skin infections, elephantiasis, vomiting, respiratory infections, and abdominal distress (8–12). Despite these diverse uses and wide distribution across the Pacific and in South America, the medicinal use of P. insularum has only been documented in the Samoan islands. Although model organisms have been used to validate a subset of traditional uses (10–13), the molecular mechanisms of P. insularum remain enigmatic. Thus far, a class of bioactive compounds common in Psychotria species (cyclotides) have been investigated, but these compounds have not been found in P. insularum (14, 15); hence the bioactive compounds mediating the biological activities of P. insularum remain to be identified. Here, we identified iron chelation as a mechanism of action of the P. insularum homogenate using chemical genomic analysis of a yeast deletion mutant library. We carried out bioassay-guided fractionation that led to the isolation and purification of rutin and nicotiflorin as bioactive constituents of the P. insularum homogenate, then translated our results from yeast to mammalian immune cells and validated the traditional use of the homogenate (and rutin) as an anti-inflammatory agent.
Results
Chemical Genomic Analysis Shows Metal Ion Homeostasis Is Affected by the P. insularum Homogenate.
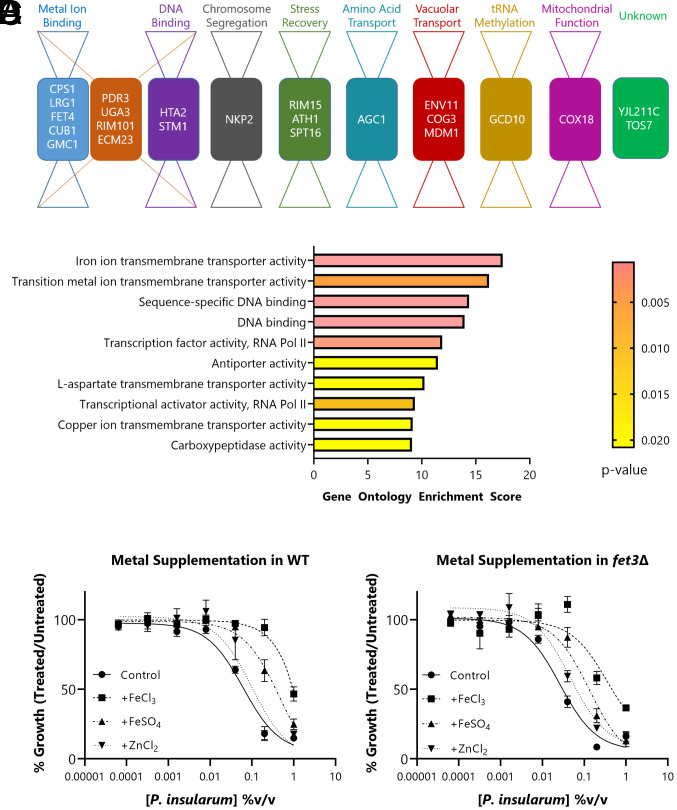
To understand how the P. insularum homogenate mediates its bioactivity, we prepared P. insularum leaf homogenate with a traditional healer and conducted competitive haploinsufficiency profiling genomic analysis screens for bioactivity using a pooled heterozygous deletion library of Saccharomyces cerevisiae. This analysis is an established method to determine mechanism of action of thousands of compounds and extracts wherein loss of mechanistically important genes results in hypersensitivity (i.e., reduced viability) (16–18). Cells were grown either in the presence or absence of 0.005% volume/volume (v/v) P. insularum homogenate, a concentration that reduced the growth of the pooled library by 20% compared to untreated cells. Using a false discovery rate (FDR) cutoff value of 0.025 against the chemical genomic profile of the P. insularum homogenate (SI Appendix, Fig. S1), we identified 23 gene deletions annotated to eight broad functions (metal ion binding, DNA binding, chromosome segregation, stress recovery, amino acid transport, vacuolar transport, transfer RNA (tRNA) methylation, and mitochondrial function) (Fig. 1A and SI Appendix, Table S1). To statistically evaluate over-representation within these 23 genes for specific molecular functions, gene ontology (GO) enrichment analysis was conducted using YeastEnrichr (19). There was significant enrichment for a multitude of processes including iron transport, DNA binding, transcription, transport of the amino acid L-aspartate, and carboxypeptidase activity (Fig. 1B and SI Appendix, Table S2). Notably, the top-ranked enrichment scores were iron ion transmembrane transporter activity (GO: 0005381) and transitional metal ion transmembrane transporter activity (GO: 0046915).
Fig. 1.
Chemical genomic analysis of the P. insularum homogenate. (A) Bow-tie representation of molecular functions of the 23 genes buffering the cellular response to the P. insularum homogenate. The 23 genes were identified from the barcode-sequencing haploinsufficiency analysis of 6,000 heterozygous deletion strains using a statistical cutoff to detect significant differences in growth in the presence of the homogenate compared to the control (SI Appendix, Fig. S1). (B) GO enrichment analysis using YeastEnrichr distinguishes iron transport as being significantly over-represented within the 23 genes. The enrichment score is a combined score that integrates P and z-score of expected and observed classification of a gene set to molecular functions. Percent growth of (C) WT or (D) fet3Δ in increasing concentrations of P. insularum homogenate with the absence or addition of 100 µM FeCl3, FeSO4, or ZnCl2 compared to growth in media without P. insularum homogenate. Data shown are average and SD of three independent experiments, each with three technical replicates.
Pertinent to investigating the mechanism of P. insularum as a traditional medicine used by Samoan people, 9 of the 23 genes were involved in iron/zinc metabolism and are also conserved from yeast to humans (20, 21). We therefore tested whether metal supplementation would rescue the yeast growth defect in the presence of P. insularum by growing wild-type (WT) yeast in increasing concentrations of the homogenate alone, or with supplementation of iron (FeCl3, FeSO4) or zinc (ZnCl2). While iron supplementation rescued the growth defect of WT in the presence of P. insularum, zinc supplementation did not rescue the growth defect (Fig. 1C). We also assessed if metal supplementation would rescue the P. insularum–induced growth defect of the fet3Δ strain, a well-characterized mutant defective in high-affinity iron uptake (22, 23). As in WT, the iron supplementation rescued the growth defect in fet3Δ (Fig. 1D), likely through low-affinity iron uptake, while zinc supplementation did not rescue the growth defect. The iron-specific rescue was not a general cytoprotective effect because supplementation with exogenous iron did not rescue generalized growth inhibition induced by atorvastatin (cholesterol synthesis inhibitor) or cycloheximide (protein synthesis inhibitor) (SI Appendix, Fig. S2). These findings directed us to an iron homeostasis mechanism of action of the P. insularum homogenate. Since several iron transporter deletion strains (fet3Δ, ftr1Δ, fet4Δ, arn1Δ, arn3Δ, arn4Δ, fet5Δ, and fth1Δ) involved in different aspects of iron homeostasis exhibited the hypersensitive phenotype to the P. insularum homogenate (SI Appendix, Fig. S3), it was unlikely that a single protein (24), in this case iron transporter was the physical target of the homogenate.
P. insularum Increases the Expression of Proteins Involved in Iron Transport Systems.
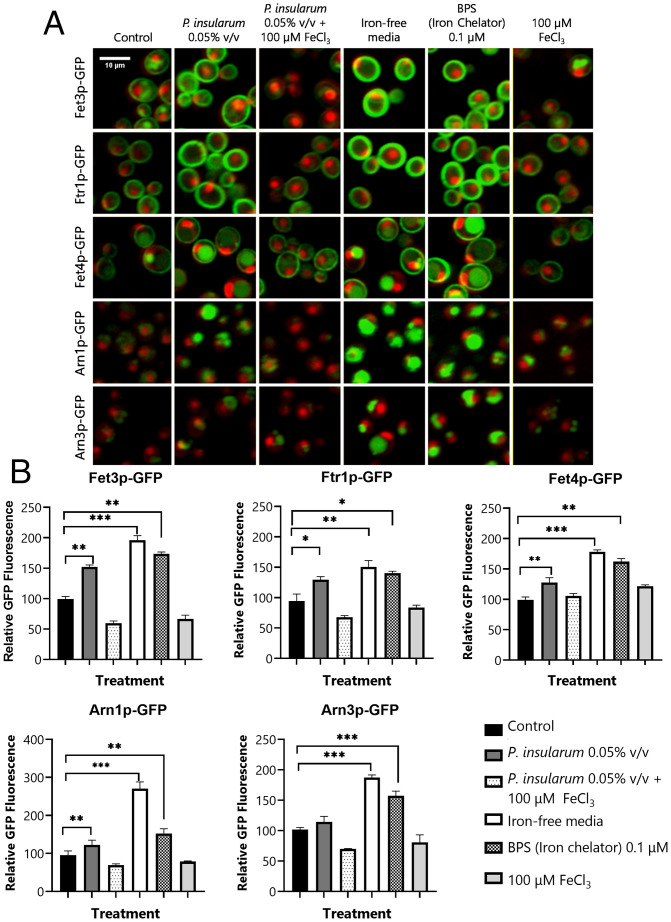
Given that iron supplementation reduced the growth inhibitory effects of the P. insularum homogenate, we hypothesized that P. insularum homogenate was reducing intracellular iron availability, possibly via an extracellular iron-chelating mechanism. A hallmark phenotype of low intracellular iron is increased expression of iron transporter proteins (20, 21, 25). We thus monitored the expression levels of 84 proteins involved in iron transport and metabolism in the presence of the P. insularum homogenate or an extracellular iron chelator bathophenanthroline disulfonic acid (BPS) relative to untreated cells. Each protein was green fluorescent protein (GFP) tagged, imaged, and quantified using confocal fluorescence microscopy relative to the constant red fluorescent protein (RFP) markers for the nucleus and cytoplasm (RedStar and mCherry, respectively) (26). Overall, the effects of the P. insularum homogenate were markedly similar to BPS, with identical changes in 13 proteins, of which 5 were noteworthy (Fig. 2 A and B). Relative to the untreated control, the P. insularum homogenate or BPS significantly increased the expression levels of the high-affinity iron transporters Fet3p-GFP and Ftr1p-GFP. The low-affinity iron transporter Fet4p-GFP also increased after treatment with the P. insularum homogenate or BPS, albeit less dramatically than the high-affinity iron transporters. Similarly, Arn1p-GFP (involved in the uptake of siderophore-bound iron) exhibited significantly increased abundance in the presence of the P. insularum homogenate when compared with the control. These transporters, together with Fet3p, Ftr1p, and Fet4p, are known to be up-regulated under conditions of iron deprivation (20, 21, 25), further suggesting the P. insularum homogenate induces an iron-deficient condition. This prediction was further supported where these significant increases in protein abundance with the P. insularum homogenate were mirrored with growth in iron-free media, and moreover, these increased levels were restored to control levels with iron supplementation (Fig. 2 A and B).
Fig. 2.
Protein expression levels of iron transporters are highly responsive to the P. insularum homogenate. (A) Protein abundance of iron transporters under control (ddH2O) and treatment conditions (0.05% v/v P. insularum, 0.05% v/v P. insularum homogenate with 100 µM FeCl3 supplementation, iron-free media, 0.1 µM BPS iron chelator, or media supplemented with 100 µM FeCl3) was monitored using confocal fluorescent microscopy analysis of GFP-tagged iron transporters relative to nuclear and cytoplasm markers tagged with high RedStar2 intensity and low-intensity mCherry RFP, respectively. (B) Quantification of GFP fluorescence for each of the five iron transporters across the control and the five treatments. GFP quantification of 300 to 400 cells was achieved using ACAPELLA software version 2.0 (PerkinElmer) that identified cells based on the mCherry and RedStar signals and prescribed the quantification of GFP as previously described (25). Three independent experiments with three technical replicates each were conducted. *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test comparison to control media.
P. insularum Reduces Intracellular Iron Content and Heme Synthesis via Iron Chelation.
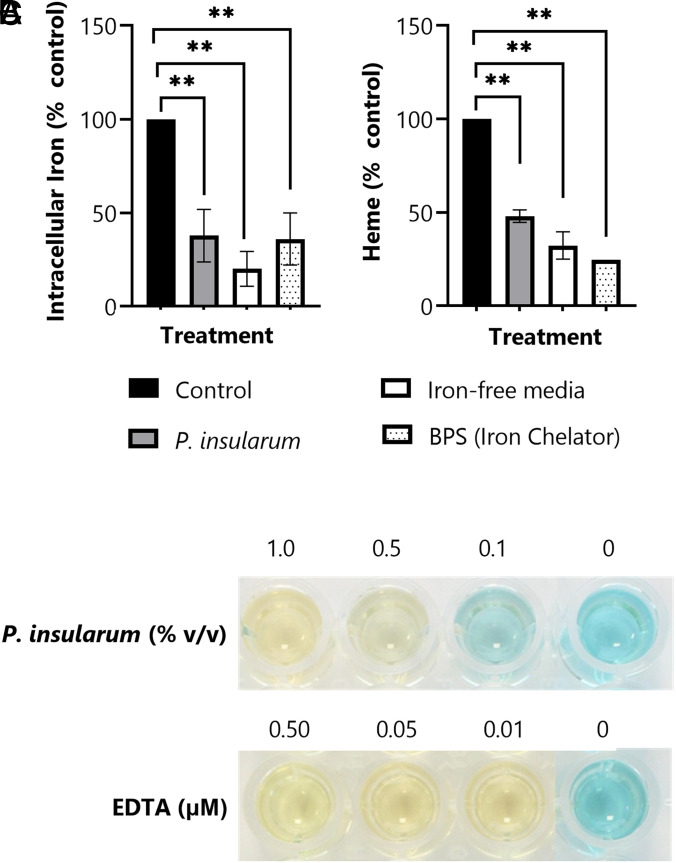
Our results thus far predict that the bioactive principle of the P. insularum homogenate reduces iron availability potentially via iron chelation. To further test this model, we measured intracellular iron levels using inductively coupled plasma mass spectrometry (ICP-MS) (27). Iron levels in lysates from untreated WT cells, or those treated with the P. insularum homogenate, grown in iron-free media or in the presence of BPS, were analyzed to quantify intracellular iron content. Consistent with our prediction, the P. insularum homogenate elicited a 60% decrease in total intracellular iron in cells compared to untreated cells (P = 0.008) (Fig. 3A). Significant reductions in intracellular iron were also detected in cells grown in the BPS iron chelator control (P = 0.008) (60%) or cells grown in iron-free media (80%) compared to untreated cells (P = 0.002).
Fig. 3.
P. insularum treatment impacts iron homeostasis. (A) Intracellular iron levels in WT cells grown in control media or treatment media containing 0.05% v/v P. insularum homogenate, iron-free media, or 0.1 µM BPS iron chelator. Cells were grown to midlog, lysed, and intracellular iron was quantified using ICP-MS. Results shown are average and SD from three independent experiments with three technical replicates each. **P < 0.01; Student’s t test comparison to control media. (B) Heme levels in WT cells grown in the same conditions as panel A. Protein was extracted and heme levels were measured using the triton methanol method via a standard curve with known concentrations of hemin and normalization to control levels. Results presented are the average and SD calculated from three independent experiments with three technical replicates each. **P < 0.01; Student’s t test comparison to control media. (C) Iron chelation activity of the P. insularum homogenate compared to EDTA iron chelation was measured in a cell-free CAS assay. A color change from blue to yellow is indicative of iron chelation.
To further investigate the effect of the P. insularum homogenate on fundamental iron-dependent processes such as heme synthesis (28), we quantified intracellular heme using a colorimetric assay (29). The P. insularum homogenate conferred a significant 50% reduction (P = 0.002) in heme levels relative to untreated cells (Fig. 3B). Cells grown in iron-free media and in the BPS iron chelator control also exhibited significantly reduced heme levels by 70% (P = 0.006) and 75% (P = 0.001), respectively, relative to untreated cells. These results show that the P. insularum homogenate reduced heme synthesis via an iron-dependent mechanism.
The results thus far suggest iron chelation, so we next directly measured iron chelation using the cell-free chrome azurol S (CAS) assay. In this assay, iron chelation is indicated with a blue-to-yellow color transformation (30). Because the pink color of iron-chelated BPS obscures the blue-to-yellow color change, we used EDTA as the positive control. Indistinguishable from the EDTA control, the P. insularum homogenate chelated iron at 0.5% (v/v) and 1% (v/v) (Fig. 3C), which were the same concentrations that significantly inhibited yeast growth (Fig. 1 C and D), thereby demonstrating that the P. insularum homogenate mediates bioactivity via an iron chelation mechanism.
Bioassay-Guided Fractionation Identifies Two Flavonol Glycosides from the P. insularum Homogenate.
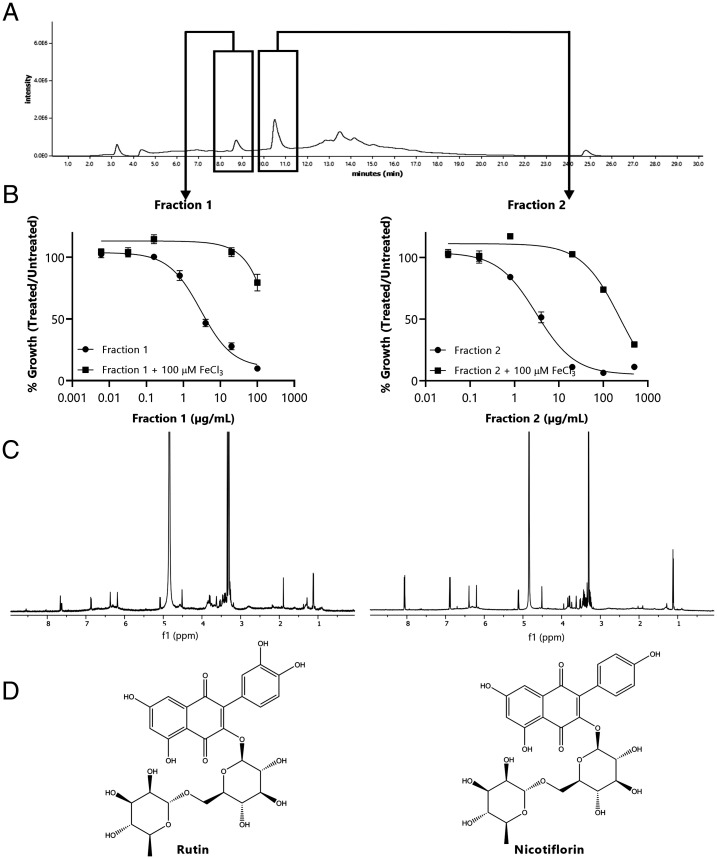
A major challenge in the field of traditional medicine is to identify the bioactive component from crude extracts (31–33). To identify the bioactive component in the P. insularum homogenate used in Samoan traditional medicine, we conducted bioassay-guided fractionation of the homogenate by monitoring subfractions for the rescue of yeast cell growth with iron supplementation in tandem with nuclear magnetic resonance (NMR) spectroscopic analysis of the purified fractions. Initial fractionation of the homogenate used reversed-phase chromatography on a polymeric (polystyrene-divinyl benzene copolymer; HP-20) resin, followed by HP-20ss purification. Further purification was achieved using size-exclusion chromatography (LH20) and then a final purification using reversed-phase octadecyl-derivatized silica (C18) high performance liquid chromatography (HPLC) to yield low milligram quantities (0.4 to 3.3 mg) of the flavonol glycosides rutin and nicotiflorin, the structures of which were determined by NMR analysis (Fig. 4 A–D and SI Appendix, Fig. S4). In addition, a third bioactive fraction was isolated, although our attempts to characterize this fraction were unsuccessful because of significantly broad, unresolved, aromatic, and aliphatic NMR signals that are characteristic of polymeric flavonoids such as condensed polymeric tannins. To verify our results, NMR spectra of commercial samples of rutin and nicotiflorin were identical with those isolated from P. insularum (SI Appendix, Fig. S5). Also consistent with the compounds isolated from P. insularum, each commercially obtained compound-chelated iron in the CAS assay (SI Appendix, Fig. S6). Interestingly, commercially obtained rutin reduced yeast growth at concentrations as low as 63 µM, and this growth defect as well that at higher concentrations was suppressed with iron supplementation, while commercially obtained nicotiflorin did not inhibit yeast growth at the highest 1.7-mM concentration tested (SI Appendix, Fig. S7). The lack of growth inhibition bioactivity for authentic nicotiflorin could be explained by the absence of a catechol (ortho-dihydroxybenzene), known to reduce metal-chelating ability. Indeed, the iron-chelating activity of nicotiflorin was less than that of rutin (SI Appendix, Fig. S6), suggesting a higher concentration of nicotiflorin is required to elicit growth inhibition in yeast. Nonetheless, nicotiflorin purified from the P. insularum homogenate inhibited yeast growth more than the authentic commercial compound, perhaps explained by the presence of minor unresolved compounds seen in the spectra of the purified samples (SI Appendix, Fig. S5). In P. insularum–derived rutin and nicotiflorin, broad and unresolved resonances around 6.5 ppm within the 1H NMR spectrum are consistent with the presence of condensed tannins, which could explain the distinct bioactivity of P. insularum–derived nicotiflorin. Based upon 1H NMR integration values, we estimate the impurity to be between 5 and 10% of each sample, but given the broadness of the signal, it is difficult to determine with certainty.
Fig. 4.
Bioactivity-guided HPLC purification and NMR analyses led to the isolation and identification of rutin and nicotiflorin. The purification of P. insularum homogenate was tracked using a bioassay-guided approach in tandem with NMR analyses. (A) The HPLC chromatogram, monitored at 254 nm, showing the two peaks (boxed) that exhibited bioactivity. (B) Percent growth of WT yeast in increasing concentrations of HPLC fraction 1 or 2 with the absence or addition of 100 µM FeCl3 compared to growth in media without each fraction. (C) The 1H NMR (600 MHz, CD3OD) spectrum of each fraction was subsequently identified as (D) rutin and nicotiflorin.
The P. insularum Homogenate Reduces Production of Inflammation-Associated Cytokines by Activated Immune Cells.
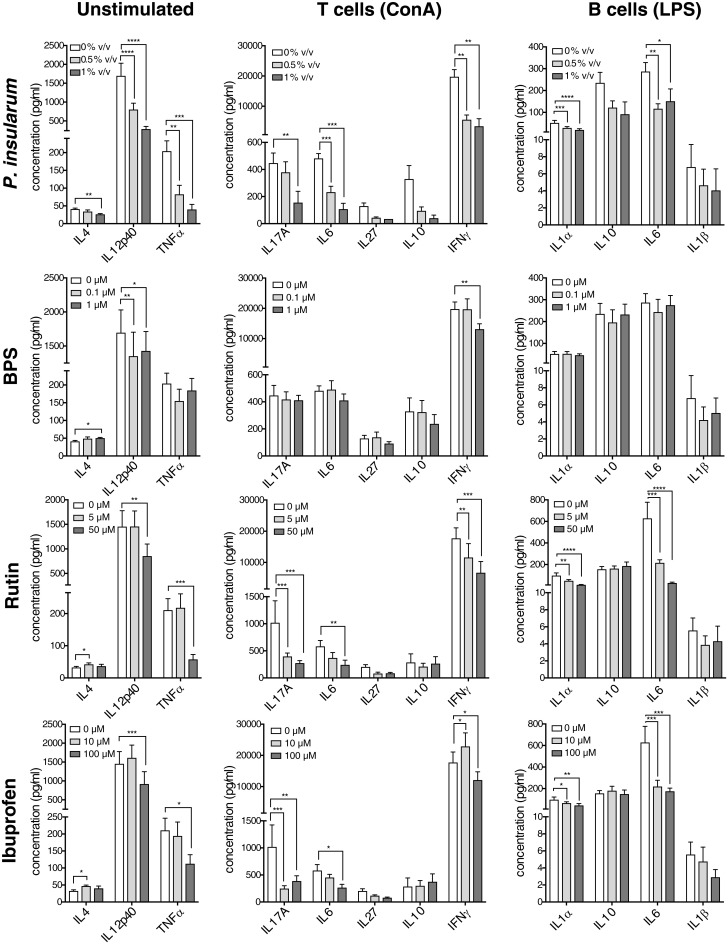
Iron chelation is involved in many bioactivities including antioxidant, antibacterial, and anti-inflammatory activities (32, 34). To determine if the P. insularum homogenate reduces inflammation, for which it is prescribed in traditional Samoan medicine, we compared the effects of varying concentrations of the P. insularum homogenate with the bioactive component rutin, the iron chelator BPS, and the anti-inflammatory drug ibuprofen on established mediators of innate and adaptive immunity. Specifically, we selected cytokines that highlighted proinflammatory and anti-inflammatory T helper (Th) subsets: Th1 (cell-mediated immunity), Th2 (wound healing), Th17 (inflammation and antifungal immunity), and regulatory T cells.
To investigate adaptive immunity, cytokines were measured in unstimulated and stimulated splenocytes in the presence of P. insularum homogenate, rutin, BPS, or ibuprofen (Fig. 5). The treatments did not reduce viability at any of the tested concentrations (SI Appendix, Fig. S8). In unstimulated splenocytes, the baseline production of the Th1-inducing IL12p40 and the fever-inducing cytokine TNFα were significantly reduced with the P. insularum homogenate, which was consistent with the activity of rutin, BPS, and ibuprofen. The cytokine IL4, which is critical for Th2-driven wound healing and tissue repair, was significantly increased with ibuprofen, BPS, and rutin treatments, while the P. insularum homogenate significantly reduced IL4 levels. In splenocytes stimulated with concanavalin A (ConA) to activate T cells, the P. insularum homogenate significantly reduced levels of the Th1 (IFNγ) and Th17 (IL6 and IL17A) proinflammatory cytokines, which aligns with the reduction in baseline IL12p40 and TNFα. A similar effect was observed with rutin and ibuprofen, whereas only the Th1 cytokines were affected by BPS. Likewise, splenocytes stimulated with lipopolysaccharide (LPS) to activate B cells exhibited reduced levels of proinflammatory, fever-inducing cytokines (IL1α, IL1β, and IL6) in the Th17 subgroup with P. insularum homogenate as well as rutin and to a lesser extent ibuprofen.
Fig. 5.
Anti-inflammatory activity of P. insularum homogenate on activated murine immune cells. Cytokine production by unstimulated splenocytes (baseline responses), ConA-stimulated splenocytes (T cell activation; 72 h) and LPS-stimulated splenocytes (B cell activation; 72 h) in the presence of P. insularum homogenate, the bioactive component rutin, the iron chelator BPS, and the anti-inflammatory drug ibuprofen. Shown are the mean and SE from three independent biological replicates; each independent replicate performed with splenocytes pooled from three animals. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; repeated-measure two-way ANOVA followed by Sidak’s multiple comparison test compared to vehicle treatment.
To investigate innate immunity, we measured the two main cytokines produced by bone marrow–derived macrophages. The P. insularum homogenate significantly increased production of the anti-inflammatory cytokine IL10, significantly reduced production of the proinflammatory Th1-inducing cytokine IL12p40, and also significantly increased viability and proliferation based on the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (SI Appendix, Fig. S9). Notably, BPS and ibuprofen produced a similar IL10 response, albeit to a lesser extent than that observed from the P. insularum homogenate. In addition, while BPS treatment significantly down-regulated IL12p40 production, ibuprofen failed to show the same effect. Together, these results indicate the P. insularum homogenate and the bioactive component rutin reduce proinflammatory and fever-inducing pathways while increasing anti-inflammatory pathways, which are both consistent with traditional use of the P. insularum homogenate to reduce inflammation, reduce fever, and enhance wound-healing.
LC-MS/MS Metabolomics Confirm Iron Chelation Activity of Rutin and Nicotiflorin.
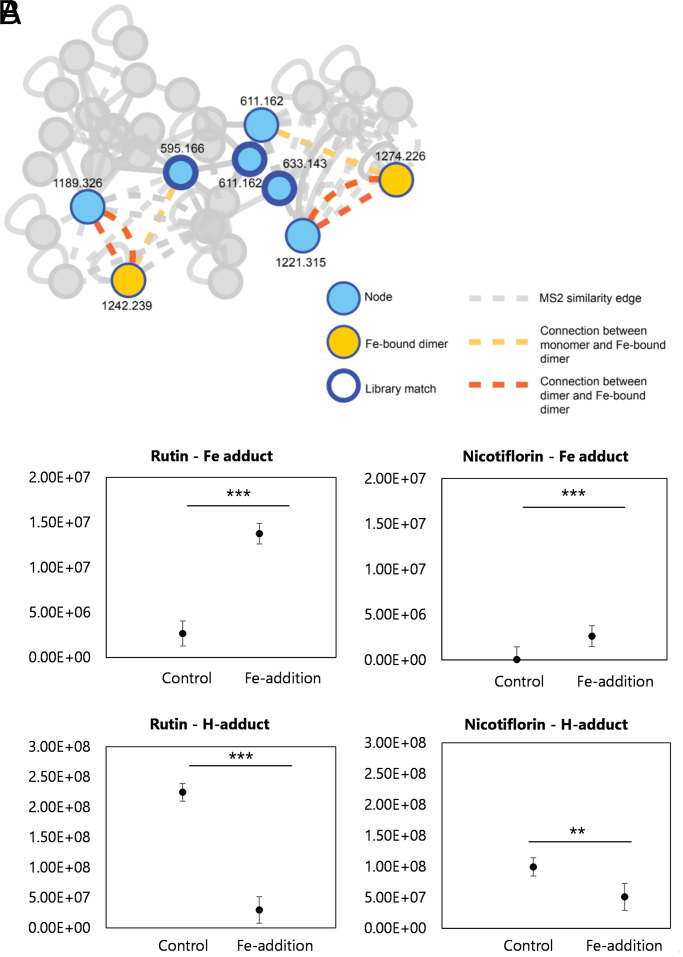
To confirm the ability of rutin and nicotiflorin to chelate iron, we used liquid chromatography with tandem mass spectrometry (LC-MS/MS) metabolomics analysis with postcolumn pH adjustment and addition of iron to the column eluent along with ion identity molecular networking within the Global Natural Product Social Molecular Networking (GNPS) platform to detect iron-chelating compounds (35, 36). This analysis identifies defined mass offsets of metal-bound molecular ions relative to their metal-free counterparts. Both pure compounds (rutin, m/z 611.162; nicotiflorin, m/z 595.166) were detected as Fe-bound dimers [2M + Fe + 2H]+ (rutin dimer + Fe, m/z 1,274.226; nicotiflorin dimer + Fe, m/z 1,242.239) when run with iron addition (Fig. 6A). These peaks were absent in control experiments without cation supplementation. The detected peak area of the rutin–iron adduct ion was roughly one order of magnitude greater than that of the nicotiflorin–iron adduct (Fig. 6B), with both analyzed at the same concentration. Given their structural similarity and therefore a likely similar ionization response factor, this implies faster binding kinetics for rutin over nicotiflorin. Overall, these findings confirm that rutin and nicotiflorin contribute to the iron chelation mechanism of the P. insularum homogenate.
Fig. 6.
Native spray metal metabolomics network identifies iron-binding activity of rutin and nicotiflorin. (A) Post-LC infusion of Fe3+ or H+ with subsequent MS/MS-based GNPS analysis highlights rutin and nicotiflorin were the only compounds in the full network to complex with iron based on ion identity molecular networking. Edges are based on MS/MS similarity (gray in the absence of binding iron). Both pure compounds (rutin, m/z 611.162; nicotiflorin, m/z 595.166) were detected to bind iron as dimers with strong connections to monomeric (yellow edges) and dimeric (orange edges) molecular ions of each compound alone, with these peaks absent in control experiments without iron supplementation. Nodes corresponding to binding of an H+ ion (blue) are shown with a thick outline representing a library match to MoNA MS/MS library in GNPS. (B) Integrated peak intensities of dimer peak (2M + H adduct) and iron-bound dimer peak (2M + Fe – 2H adduct) in the control (no infusion) and Fe-infusion samples. Shown are the results from three replicates with the average indicated with a horizontal line. Statistical significance between integrated peak areas was calculated using a Student’s t test (*P < 0.05; **P < 0.01).
Discussion
Natural products for human diseases have always been important, and traditional remedies such as Samoan medicinal plants have significantly added to this armamentarium (5–13). Here, we have added to this knowledge base by ascertaining the mechanism of action of a traditional Samoan medicinal plant (P. insularum); we identified and correlated mechanism of action with the traditional use of the medicine and ultimately determined bioactive principles. First, we used haploinsufficiency genomic analysis in yeast to determine that iron availability was integral to the mechanism of action of the P. insularum homogenate. We described a bioassay-guided fractionation scheme that led to the discovery of bioactive iron chelators, rutin and nicotiflorin, in the P. insularum homogenate. The anti-inflammatory effects of the P. insularum homogenate and rutin on unstimulated splenocytes, ConA-activated T cells, and LPS-activated B cells were as potent as the anti-inflammatory drug ibuprofen. The similarity in the anti-inflammatory activity of the P. insularum homogenate with the extensively used ibuprofen would be consistent with the diverse uses of the P. insularum homogenate in Samoan traditional medicine. Overall, using a crude traditional homogenate with methodology integrating genomics, metabolomics, analytical chemistry and immunology, we identified 1) a mechanism of action that underpinned the traditional anti-inflammatory remedy and 2) potential applications of the traditional medicine not previously informed by traditional knowledge.
Our results fit and extend an established system using yeast as a model system for elucidating mechanisms of iron metabolism in humans (20–23). For example, the natural product and first-line treatment for type 2 diabetes prescribed for 150 million persons worldwide, metformin, was shown in yeast to induce an iron-deficiency state (37, 38). Indeed, iron chelators, such as deferasirox, deferiprone, and deferoxamine, initially approved for iron overload disorders associated with transfusions impacting 1.62 billion people (39), are also now described as prospective agents against common diseases such as cancer (40), neurodegenerative diseases (41), cardiovascular disease (42), and diabetes (43). As inflammation and iron chelation are fundamental to these diseases (44–46), this raises the possibility for applications of P. insularum beyond traditional use.
We identified rutin and nicotiflorin as bioactive components of P. insularum homogenate in treating inflammation and associated symptoms in Samoan traditional medicine. More broadly, our findings fit with anti-inflammatory and neuroprotective effects of rutin and nicotiflorin in treating neurodegenerative diseases such as Parkinson’s and Alzheimer’s (47–49). These results reiterate that rutin and nicotiflorin are bioactive compounds of current interest and moreover suggest that there may be uses for the P. insularum homogenate not informed by traditional use in Samoa but informed here with unbiased chemical genomic analyses. While there is no record of P. insularum in a traditional use against hyperlipidemia-associated diseases, our chemical genomic analysis highlighted the sensitivity of the RIM101 gene deletion to the P. insularum homogenate. RIM101 in yeast is a major regulator of lipotoxicity, the process of lipid-dependent cell death that is associated with obesity, diabetes, and cardiovascular disease (50). This observation may be of direct relevance to Samoa given the genetic predisposition to obesity in Samoa (51). Also of current interest, molecular docking studies of 33 molecules predicted rutin as the strongest contender in binding the active site of the SARS-CoV-2 main 3C-L protease for inhibiting viral replication and potentially treating the COVID-19 pandemic (52). Antilipotoxicity and antiviral activities by rutin are not unprecedented (53–56), albeit not used to date to inform new uses of traditional medicine. Our results thus broaden the therapeutic potential and applications of P. insularum in Samoan traditional medicine.
It is also plausible that the iron chelation activity of the P. insularum homogenate (by rutin, nicotiflorin, and other condensed tannins) inhibits ferroptosis, an iron-dependent programmed cell death triggered by intracellular phospholipid peroxidation. Pathological cell death associated with several diseases including inflammation, Alzheimer’s, Parkinson’s, and Huntington’s diseases, periventricular leukomalacia, renal insufficiency, carcinogenesis, and kidney degeneration have shown hallmarks of ferroptosis (57). Regulating ferroptosis through iron chelation becomes a potential management tool for these diseases. Ferroptosis is a fairly new categorization and not yet well-studied when it comes to traditional medicine, with only a few studies in traditional Chinese medicine (58). Given flavonoids are prominent components of many plant extracts used in traditional medicine, it would be interesting to determine if antioxidant and iron chelation mechanisms mediating antiferroptosis activity are also mechanisms amenable to traditional medicine.
To further develop the P. insularum homogenate, additional analyses of natural variation in the chemical biology of P. insularum must be investigated. Spatial and temporal variation can lead to differences in chemical composition, but also cultivar differences have been shown to alter the levels and types of flavonoids present within plants (59). As the P. insularum in our study was collected from P. insularum cultivated by a traditional healer, future studies should include P. insularum collected from cultivated as well as wild sources in Samoa and additionally wild P. insularum from other South Pacific islands, where interestingly, P. insularum is not reportedly used in traditional medicine. Likewise, a pharmacodynamic understanding of P. insularum is necessary. Since iron chelators alter the microbiome and immune cells respond to microbiota (60), it is plausible that the P. insularum homogenate consumed orally alters the intestinal microbiome, which could also mediate additional effects of the homogenate.
This study incorporated sophisticated yeast chemical genomic analyses to identify the biological mechanism of action of a traditional medicine as a whole entity as prepared by traditional healers, and this mechanism was then monitored in bioactivity-guided fractionation to identify the bioactive components. This combinatorial approach provides molecular insight into traditional knowledge and provides insight into potential new uses of traditional medicine. As recently deemed to be increasingly important for advancing ethnobiology and natural product drug discovery, albeit rarely accomplished (61–63), our study integrated indigenous researcher–community engagement with interdisciplinary methodologies to discover and develop the great pharmaceutical potential in traditional knowledge regarding natural products as used in traditional medicine.
Materials and Methods
Plant Collection.
Leaves were collected in 2013 and 2018 from an individual P. insularum plant (GPS coordinates: Latitude S 13°48’53”, Longitude W 171°53’6”) via engagement with Seti Fa’aifo, the landowner of the property and distant relative to S.M.S. Both Seti Fa’aifo and botanist S.T.F. positively identified P. insularum. Permit for plant collection was provided by the Ministry of Natural Resources and Environment (MNRE) in Samoa. Voucher specimen (SROS_2021_407) has been deposited at Scientific Research Organisation of Samoa (SROS).
Leaf Homogenate Preparation.
Within an hour after collection, leaves were washed with sterile water, processed through a juicer (Breville BJE200), and the resulting leaf juice was transported on ice to Wellington, New Zealand, for analysis under the auspices of the MNRE permit, the memorandum of understanding between SROS and Victoria University of Wellington, and the import permit from the Ministry of Primary Industries New Zealand. The juice was centrifuged, and the supernatant was treated as the homogenate after a final filtration through a 0.2-µm filter with a 30-mm polyethersulfone membrane (Biofil).
Strains, Media, and Chemicals.
All yeast strains used in this study were in the BY4741 background. Haploid yeast deletion strains (MATa; Open Biosystems) carrying the geneticin-selectable marker were maintained on yeast peptone dextrose (YPD) medium containing 200 µg/mL geneticin (Life Technologies). GFP/RFP-tagged yeast strains were maintained on synthetic complete media buffered to pH 7 with 25 mM HEPES (SCH) without histidine (selection for GFP) and containing hygromycin and nourseothricin antibiotics (selection for RFP) (26). HEPES was purchased from Formedium. BPS, rutin, cycloheximide, atorvastatin and ibuprofen were purchased from Sigma-Aldrich. Nicotiflorin was supplied by AvaChem Scientific. EDTA was purchased from AppliChem GmbH.
Chemical Genomic Analysis.
An aliquot of a pooled heterozygous deletion library of yeast (Open Biosystems) was treated with 0.005% v/v of the P. insularum homogenate for 20 generations at 30 °C. Genomic DNA from treated cells was isolated using YeaStar Genomic DNA Kit (ZymoResearch). Then the unique 20-bp barcodes flanking each gene deletion (UPTAG upstream and DNTAG downstream of each gene deletion) were amplified, sequenced, demultiplexed, and quantified to record logFC, FDR, and logCPM for every gene deletion of the heterozygous library as previously described (64).
Functional Enrichment Analysis.
Over-representation of genes sensitive to the P. insularum homogenate for specific GO terms in the molecular function category was investigated using YeastEnrichr (19). Significant enrichment was defined via an enrichment score that integrates the log of the P less than 0.05 from the Fisher exact test for random gene sets and the z-score of the deviation of the observed rank for each GO term in the P. insularum homogenate-sensitive gene set from the expected rank for each term.
Metal Rescue Assay.
Yeast cultures at 5 × 105 cells/mL were grown in SCH and supplemented with or without FeCl3, FeSO4, or ZnCl2, together with increasing concentrations of P. insularum homogenate. Cells were subsequently incubated at 30 °C until control cells reached midlog (optical density [OD] = 0.4 to 0.6) and then quantified via absorbance at 590 nm using a PerkinElmer Envision plate reader.
Intracellular Iron Quantification.
Yeast cultures at 5 × 105 cells/mL were grown in SCH at 30 °C until control cells reached midlog (OD = 0.4 to 0.6), and intracellular iron was measured in yeast cells using ICP-MS as previously described (27). Briefly, cells were digested in 3% nitric acid at 96 °C for 16 h, supernatant was collected, and iron content was quantified using Element 2 High Resolution ICP-MS (Thermo Scientific).
Confocal Fluorescence Microscopy.
Protein expression was quantified by fluorescence of a GFP-tagged protein of interest relative to the RFP fluorescence of internal markers of the cytoplasm and nucleus (mCherry and RedStar, respectively). Strains were grown in liquid SCH with and without P. insularum homogenate at 30 °C for 15 h, diluted to ∼1 × 108 cells/mL, transferred to 384 clear bottom microtitre plates (PerkinElmer), and visualized using a high-throughput spinning disk confocal microscope (General Electric IN Cell Analyzer 6500HS with 60× (numerical aperture 0.9) air objective for screening 84 GFP-tagged strains or PerkinElmer Opera with 60× (numerical aperture 1.2) water objective for select GFP-tagged strains). GFP excitation was at 488 nm and detected with a 520/35 filter. mCherry and RedStar excitation was at 561 nm and detected with a 600/40 filter. An exposure of 400 ms with five Z-stacks 0.5 µm apart was used for all images. GFP quantification of 300 to 400 cells was achieved using ACAPELLA software version 2.0 (PerkinElmer) that identified cells based on the mCherry and RedStar signals and prescribed the quantification of GFP as previously described (26).
Heme Quantification.
Yeast cultures at 5 × 105 cells/mL were grown in SCH at 30 °C until control cells reached midlog (OD = 0.4 to 0.6), and protein was extracted as previously described (65). Heme was then quantified by the triton-methanol protocol as previously described (29). In brief, equal amounts of clarified cell lysate was added to 5% triton in methanol, absorbance was read at 405 nm using a PerkinElmer Envision plate reader, and heme was quantified relative to a hemin (Sigma-Aldrich) standard curve (10 to 500 nM).
Colorimetric Iron Chelation Assay.
Iron chelation was directly measured using a modified CAS assay as previously described (30). Briefly, a solution comprising CAS, FeCl3, and hexadecyltrimethylammonium bromide was prepared in the presence and absence of various dilutions of P. insularum homogenate or EDTA. A color change from blue to yellow was indicative of iron chelation activity.
Mouse Husbandry.
Male and female C57BL/6 mice (6- to 12-wk old) were obtained from the Malaghan Institute of Medical Research and housed in the School of Biological Sciences Animal Facility at Victoria University. All experiments with animals were carried out in this animal facility and were approved by the Victoria University of Wellington Animal Ethics Committee (protocols 2014-R23 and 25295).
Immune Response Assays.
To assess T cell responses, splenocytes were isolated from C57BL/6 mice as previously described (66) and cultured (1 × 106 cells/well) for 72 h in the presence or absence of ConA (a polyclonal T cell mitogen; 1 μg/mL; Sigma). For each experiment, splenocytes were pooled from three animals, and three independent experiments were performed for each assay. To assess B cell responses, splenocytes were isolated and stimulated for 72 h with LPS (a polyclonal B cell mitogen; 200 ng/mL). Cytokines were measured in the culture supernatants by enzyme-linked immunosorbent assay (BD Biosciences) for IL12p40, IL4, and IL10 according to manufacturer instructions, and plates were read at 450 nm using a multiwell Enspire Multilabel plate reader (PerkinElmer). Additionally, TNFα, IL6, IL27, IL17A, IFNγ, IL1α, and IL1β were measured in culture supernatants from unstimulated, ConA-stimulated, or LPS-stimulated splenocytes using a LEGENDplex Mouse Inflammation Panel 13-plex (Biolegend) by flow cytometry according to manufacturer instructions and acquired on a FACS Canto II flow cytometer (BD Biosciences). Data analysis was conducted using LEGENDplex software (Biolegend). Primary bone marrow–derived macrophage cultures were prepared from C57BL/6 mice as described (67) using IL-3 and GM-CSF (both at 5 ng/mL; Peprotech) for 7 to 10 d. Macrophages (1 × 105 cells/well) were primed overnight with IFNγ (20 U/mL; Peprotech) before stimulation with LPS (200 ng/mL; Sigma) for 24 h. Cell viability was assessed via MTT assay; briefly, cells were incubated for 2 h with the MTT solution (5 mg/mL; Sigma) before adding the solubilizer (10% sodium dodecyl sulfate, 45% dimethylformamide) overnight, and MTT reduction was measured by absorbance at 550 nm in a multiwell Enspire Multilabel plate reader (PerkinElmer). Statistical significance between multiple groups assessed using repeated-measure two-way ANOVA followed by Sidak’s multiple comparison test.
NMR, MS, HPLC, and TLC Analyses for Compound Isolation.
NMR analyses were carried out using a Varian Direct Drive 600-MHz NMR spectrometer operating at 600 MHz for 1H and 150 MHz for 13C nuclei, respectively. Samples were dissolved in deuterated solvent (Cambridge Isotope Labs), and the residual protonated solvent peak was used as an internal reference (CD3OD: δH 3.31, δC 49.0; CDCl3: δH 7.21, δC 77.0). High-resolution electrospray ionization mass spectrometry (HRESIMS) data were acquired using an Agilent 6530 Accurate-Mass Q-TOF LC-MS equipped with a 1260 Infinity binary pump. Data-dependent electrospray LC-MS/MS analysis was performed with a Q-Exactive orbitrap mass spectrometer (Thermo Scientific) operating in both positive and negative ion modes. HPLC purification was carried out using an Agilent Technologies 1260 Infinity instrument equipped with a quaternary pump and dual detection capability using the diode array and evaporative light scattering detector modules. Thin-layer chromatography (TLC) analyses were performed on Macherey-Nagel Polygram Sil G/UV254 plates and developed by first visualizing under ultraviolet light (λ = 254 nm), then dipping in 5% H2SO4/CH3OH, followed by 0.1% vanillin/ CH3CH2OH before heating.
Bioactivity-Guided Fractionation.
Filtered P. insularum juice (200 mL) was passed through Diaion HP-20 poly(styrene-divinyl benzene) copolymer resin (Supelco) (100 mL) that was pre-equilibrated with three column volumes each of acetone, methanol, and dH2O. The column was then eluted with three column volumes each of dH2O, 30, 75, and 100% acetone in dH2O (v/v), to which bioactivity was predominantly confined to the 75% acetone in dH2O fraction. The 75% aqueous acetone fraction was then loaded onto Diaion HP-20ss (Supelco) by evaporating the fraction onto HP-20ss (5 mL) and transferring as a slurry in dH2O onto a larger bed of the stationary phase (15 mL). The column was washed with dH2O and eluted with three bed volumes each of 30, 40, 50, 60, 75, and 100% acetone in dH2O. Fractions (10 mL) were collected in test tubes and evaporated in vacuo. Bioactive fractions found within the 50% aqueous acetone fraction were further subjected to size-exclusion chromatography using Sephadex LH20 (GE Healthcare Life Sciences) (2.5 × 90 cm) eluting with 50% methanol in dH2O (v/v, 750 mL) at 17 mL/h collected in bulk, followed by 70% acetone in dH2O (v/v, 400 mL) at a flow rate of 25 mL/h collecting 5-mL fractions. Fractions were dried in vacuo, combined on the basis of TLC analysis (butanol:acetic acid:dH2O, 4:1:2, v/v), and their iron chelation bioactivity determined. Subsequent purification utilizing reversed-phase HPLC with octadecyl-derivatized silica (C18) (Phenomenex Prodigy: 4.6 × 250 mm, 5 µm) and a binary gradient solvent system consisting of increasing volumes of acetonitrile in dH2O (0 to 10 min, 20 to 30% acetonitrile in dH2O; 10 to 20 min, held at 30% acetonitrile in dH2O; 20 to 23 min, 30 to 90% acetonitrile in dH2O; 23 to 30 min, held at 90% acetonitrile in dH2O) resulted in the isolation of rutin (0.4 mg; Rt 8.9 min) and nicotiflorin (3.3 mg; Rt 10.2 min). HRESIMS provided the deprotonated molecular ion for rutin m/z 609.1458 [M-H]− (calcd C27H29O16 m/z 609.1459, Δ = −0.3 ppm) and nicotiflorin m/z 593.1509 [M-H]− (calcd C27H29O15 m/z 593.1512, Δ = −0.44 ppm), respectively. Full NMR characterization of purified compounds was performed and validated by comparison of NMR data with that of authentic commercial standards.
LC-MS/MS Iron-Binding Metabolomics (Data Acquisition).
Binding of iron by small molecules was detected using Ion Identity Molecular Networking metabolomics as previously described (36). For microflow UHPLC-MS/MS analysis, samples were dissolved in 80% methanol (to a final concentration of 100 µg/mL) and 5 µL was injected into a Vanquish UHPLC system coupled to a Q-Exactive quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific) with an Agilent 1260 quaternary HPLC pump (Agilent) as a makeup pump. For reversed-phase chromatography, a C18 core-shell microflow column (Kinetex C18, 50 × 1 mm, 1.8-µm particle size, 100-A pore size, Phenomenex) was used. The mobile phase consisted of solvent A (H2O + 0.1% formic acid) and solvent B (acetonitrile + 0.1% formic acid). The flow rate was set to 100 µL/min. A linear gradient from 5 to 50% B between 0 to 4 min and 50 to 99% B between 4 to 6 min, followed by a 2-min washout phase at 99% B and a 4-min re-equilibration phase at 5% B. Data-dependent acquisition of MS/MS spectra was performed in positive mode. Makeup flow of ammonium acetate buffer (10 mM) + 0.2% ammonium hydroxide was set to 100 µL/min and infused postcolumn through a peak T-splitter. Iron chloride (FeCl3) was infused postcolumn and postmakeup through a second T-splitter at a flow rate of 5 µL/min and a concentration of 1 mM. Electrospray ionization (ESI) parameters were set to 40 arbitrary units (AU) sheath gas flow, auxiliary gas flow was set to 10 AU, and sweep gas flow was set to 0 AU. Auxiliary gas temperature was set to 400 °C. The spray voltage was set to 3.5 kV and the inlet capillary was heated to 320 °C. S-lens level was set to 70 V applied. MS scan range was set to 200 to 2,000 m/z with a resolution at m/z 200 (Rm/z 200) of 70,000 with one microscan. The maximum ion injection time was set to 100 ms with automatic gain control (AGC) target of 5.0E5. Up to two MS/MS spectra per duty cycle were acquired at Rm/z 200 17,000 with one microscan. The maximum ion injection time for MS/MS scans was set to 100 ms with an AGC target of 5.0E5 ions and a minimum 5% AGC. The MS/MS precursor isolation window was set to m/z 1. The normalized collision energy was stepped from 20 to 30 to 40% with z = 1 as default charge state. MS/MS scans were triggered at the apex of chromatographic peaks within 2 to 15 s from their first occurrence. Dynamic precursor exclusion was set to 5 s. Ions with unassigned charge states were excluded from MS/MS acquisition as well as isotope peaks.
LC-MS/MS Iron-Binding Metabolomics (Data Analysis).
Feature finding and ion identity networking were performed using an in-house modified version of MZmine2.37 (68), corr.17.7 available at https://github.com/robinschmid/mzmine2/releases. Feature tables, MS/MS spectra files (mgf), and ion identity networking results were exported, uploaded to the MassIVE repository, and submitted to GNPS (35) for feature-based molecular networking analysis.
MS/MS spectra were converted to .mzML files using MSconvert (ProteoWizard) (69). All raw and processed data are publicly available at ftp://massive.ucsd.edu/MSV000086287/. MS1 feature extraction and MS/MS pairing were performed with MZmine 2.37 (68) corr17.7_kai_merge2. An intensity threshold of 1E5 for MS1 spectra and of 1E3 for MS/MS spectra was used. MS1 ADAP chromatogram building was performed within a 10 ppm mass window and a minimum peak intensity of 3E5 was set. Extracted Ion Chromatograms (XICs) were deconvoluted using baseline cutoff with m/z range for MS2 pairing of 0.01 and RT range for MS2 scan pairing of 0.2. After chromatographic deconvolution, MS1 features linked to MS/MS spectra within 0.01 m/z mass and 0.2 min retention time windows. Isotope peaks were grouped and features from different samples were aligned with 10 ppm mass tolerance and 0.2 min retention time tolerance. MS1 peak lists were joined using an m/z tolerance of 10 ppm and retention time tolerance of 0.1 min; alignment was performed by placing a weight of 75 on m/z and 25 on retention time. Correlation of coeluting features was performed with the metaCorrelate module; retention time tolerance of 0.1, minimum height of 1E5 and noise level of 1E5 were used. A correlation of 0.85 was set as the threshold for the min feature shape corr., and feature height correlation was not used. The following adducts were searched: [M + H+]+, [M + Na+]+, [M + K+]+, [M + NH]2+, [M + Fe3+ - 2H+]+, with an m/z tolerance of 10 ppm, a maximum charge of 2, and maximum molecules/cluster of 2.
Peak areas and feature correlation pairs were exported as .csv files and the corresponding consensus MS/MS spectra were exported as an .mgf file. For spectral networking and spectrum library matching, all files were uploaded to the feature-based molecular networking workflow on GNPS (35) (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). For spectrum library matching and spectral networking, the minimum cosine score to define spectral similarity was set to 0.7. The Precursor and Fragment Ion Mass Tolerances were set to 0.01 Da and Minimum Matched Fragment Ions to 4, Minimum Cluster Size to 1 (MS Cluster off). When analog search was performed, the maximum mass difference was set to 100 Da. The GNPS job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=76115ecaad9c46e9aaa30413d0d435eb. Molecular networks were visualized with Cytoscape 3.7.1 (70).
The XCalibur Quant Browser (Thermo) was used to obtain integrated peak areas of both apo- and iron-bound adducts. The XCalibur processing method integrates exact mass +/− 10 ppm, utilizing ICIS Peak Detection with 7 smoothing points, a baseline window of 40, area noise factor of 5, and peak noise factor of 6. Statistical significance between integrated peak areas was calculated using an unpaired t test.
Acknowledgments
Funding from a Victoria University of Wellington (VUW) Doctoral Scholarship (S.M.-S. and V.P.) is greatly appreciated, and this research was supported in part by a contract from the Health Research Council of New Zealand (V.H.W.), for which we are also very grateful. Permission from the MNRE (Samoa) and Ministry of Primary Industries (New Zealand) to collect and import plant material are gratefully acknowledged. We thank the SROS for supporting this research and assisting with the fieldwork and Mr. Seti Fa’aifo for permitting the sampling from his property. We also acknowledge the overarching VUW/SROS memorandum of understanding that facilitated collaboration integral to this study.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.A.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100880118/-/DCSupplemental.
Data Availability
Feature finding and ion identity networking were performed using an in-house modified version of MZmine2.37 (68), corr.17.7 available at GitHub, https://github.com/robinschmid/mzmine2/releases. Feature tables, MS/MS spectra files (mgf), and ion identity networking results were exported, uploaded to the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/dataset_files.jsp?task=16c65622121c488d9123dc42c556dbff#%7B%22table_sort_history%22%3A%22main.collection_asc%22%2C%22main.collection_input%22%3A%22metadata%7C%7CEXACT%22%7D), and submitted to GNPS (35) (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=76115ecaad9c46e9aaa30413d0d435eb) for feature-based molecular networking analysis. All raw and processed data are publicly available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=16c65622121c488d9123dc42c556dbff. For spectral networking and spectrum library matching, all files were uploaded to the feature-based molecular networking workflow on GNPS (35) (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). The GNPS job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=76115ecaad9c46e9aaa30413d0d435eb. All other study data are included in the article and/or SI Appendix.
References
- 1.Newman D. J., Cragg G. M., Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Harvey A. L., Natural products in drug discovery. Drug Discov. Today 13, 894–901 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Dias D. A., Urban S., Roessner U., A historical overview of natural products in drug discovery. Metabolites 2, 303–336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu Y., The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 17, 1217–1220 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Gustafson K. R., et al. , A nonpromoting phorbol from the samoan medicinal plant Homalanthus nutans inhibits cell killing by HIV-1. J. Med. Chem. 35, 1978–1986 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Beans E. J., et al. , Highly potent, synthetically accessible prostratin analogs induce latent HIV expression in vitro and ex vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 11698–11703 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desimio M. G., Giuliani E., Ferraro A. S., Adorno G., Doria M., In vitro exposure to prostratin but not bryostatin-1 improves natural killer cell functions including killing of CD4+ T cells harbouring reactivated human immunodeficiency virus. Front. Immunol. 9, 1514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whistler A. W., Samoan Herbal Medicine (Isle Botanica, Honolulu, HI, 1996). [Google Scholar]
- 9.Castro L., Tsuda K., Samoan Medicinal Plants and Their Usage (Agricultural Development in the American Pacific Project, University of Hawai’i at Manoa, 2001) [Google Scholar]
- 10.Cox P. A., et al. , Pharmacological activity of the Samoan ethnopharmacopoeia. Econ. Bot. 43, 487–497 (1989). [Google Scholar]
- 11.Norton T. R., et al. , Pharmacological evaluation of medicinal plants from Western Samoa. J. Pharm. Sci. 62, 1077–1082 (1973). [DOI] [PubMed] [Google Scholar]
- 12.Dunstan C. A., et al. , Evaluation of some Samoan and Peruvian medicinal plants by prostaglandin biosynthesis and rat ear oedema assays. J. Ethnopharmacol. 57, 35–56 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Frankova A., et al. , In vitro antibacterial activity of extracts from Samoan medicinal plants and their effect on proliferation and migration of human fibroblasts. J. Ethnopharmacol. 264, 113220 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Gerlach S. L., Burman R., Bohlin L., Mondal D., Göransson U., Isolation, characterization, and bioactivity of cyclotides from the Micronesian plant Psychotria leptothyrsa. J. Nat. Prod. 73, 1207–1213 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Gerlach S. L., et al. , A systematic approach to document cyclotide distribution in plant species from genomic, transcriptomic, and peptidomic analysis. Biopolymers 100, 433–437 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Hillenmeyer M. E., et al. , The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science 320, 362–365 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee A. Y., et al. , Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344, 208–211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F., et al. , A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 370, 974–978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuleshov M. V., et al. , Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eide D. J., The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr. 18, 441–469 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Bleackley M. R., Macgillivray R. T. A., Transition metal homeostasis: From yeast to human disease. Biometals 24, 785–809 (2011). [DOI] [PubMed] [Google Scholar]
- 22.De Silva D. M., Askwith C. C., Eide D., Kaplan J., The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270, 1098–1101 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., Dancis A., A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271, 1552–1557 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Smith A. M., Ammar R., Nislow C., Giaever G., A survey of yeast genomic assays for drug and target discovery. Pharmacol. Ther. 127, 156–164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott C. C., Protchenko O., Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7, 20–27 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bircham P. W., et al. , Secretory pathway genes assessed by high-throughput microscopy and synthetic genetic array analysis. Mol. Biosyst. 7, 2589–2598 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Tamarit J., Irazusta V., Moreno-Cermeño A., Ros J., Colorimetric assay for the quantitation of iron in yeast. Anal. Biochem. 351, 149–151 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Lange H., Kispal G., Lill R., Mechanism of iron transport to the site of heme synthesis inside yeast mitochondria. J. Biol. Chem. 274, 18989–18996 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Pandey A. V., Joshi S. K., Tekwani B. L., Chauhan V. S., A colorimetric assay for heme in biological samples using 96-well plates. Anal. Biochem. 268, 159–161 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Alexander D. B., Zuberer D. A., Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45 (1991). [Google Scholar]
- 31.Harvey A., Strategies for discovering drugs from previously unexplored natural products. Drug Discov. Today 5, 294–300 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Phillipson J. D., Phytochemistry and medicinal plants. Phytochemistry 56, 237–243 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Wolfender J. L., Marti G., Thomas A., Bertrand S., Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 1382, 136–164 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Crielaard B. J., Lammers T., Rivella S., Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov. 16, 400–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M., et al. , Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid R., et al. , Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 12, 3832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L., Meng S., Germain-Lee E. L., Radovick S., Wondisford F. E., Potential biomarker of metformin action. J. Endocrinol. 221, 363–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stynen B., et al. , Changes of cell biochemical states are revealed in protein homomeric complex dynamics. Cell 175, 1418–1429.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming R. E., Ponka P., Iron overload in human disease. N. Engl. J. Med. 366, 348–359 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Torti S. V., Torti F. M., Iron and cancer: More ore to be mined. Nat. Rev. Cancer 13, 342–355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward R. J., Zucca F. A., Duyn J. H., Crichton R. R., Zecca L., The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13, 1045–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Haehling S., Jankowska E. A., van Veldhuisen D. J., Ponikowski P., Anker S. D., Iron deficiency and cardiovascular disease. Nat. Rev. Cardiol. 12, 659–669 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Simcox J. A., McClain D. A., Iron and diabetes risk. Cell Metab. 17, 329–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coussens L. M., Werb Z., Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruparelia N., Chai J. T., Fisher E. A., Choudhury R. P., Inflammatory processes in cardiovascular disease: A route to targeted therapies. Nat. Rev. Cardiol. 14, 133–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furman D., et al. , Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toker G., Küpeli E., Memisoğlu M., Yesilada E., Flavonoids with antinociceptive and anti-inflammatory activities from the leaves of Tilia argentea (silver linden). J. Ethnopharmacol. 95, 393–397 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Spagnuolo C., Moccia S., Russo G. L., Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur. J. Med. Chem. 153, 105–115 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Pan R. Y., et al. , Sodium rutin ameliorates Alzheimer’s disease-like pathology by enhancing microglial amyloid-β clearance. Sci. Adv. 5, eaau6328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockenfeller P., et al. , Diacylglycerol triggers Rim101 pathway-dependent necrosis in yeast: A model for lipotoxicity. Cell Death Differ. 25, 767–783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minster R. L., et al. , A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat. Genet. 48, 1049–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das S., et al. , An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 13, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manzoni A. G., et al. , Hyperlipidemia-induced lipotoxicity and immune activation in rats are prevented by curcumin and rutin. Int. Immunopharmacol. 81, 106217 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Khlifi R., et al. , Erica multiflora extract rich in quercetin-3-O-glucoside and kaempferol-3-O-glucoside alleviates high fat and fructose diet-induced fatty liver disease by modulating metabolic and inflammatory pathways in Wistar rats. J. Nutr. Biochem. 86, 108490 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Zandi K., et al. , Antiviral activity of fisetin, rutin and naringenin against Dengue virus type-2. J. Med. Plants Res. 5, 5534–5539 (2011). [Google Scholar]
- 56.Yarmolinsky L., Huleihel M., Zaccai M., Ben-Shabat S., Potent antiviral flavone glycosides from Ficus benjamina leaves. Fitoterapia 83, 362–367 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Dixon S. J., et al. , Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu W. H., Li C. H., Jiang T. L., Ferroptosis pathway and its intervention regulated by Chinese materia medica. Zhongguo Zhongyao Zazhi 43, 4019–4026 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Johnson H. E., Banack S. A., Cox P. A., Variability in content of the anti-AIDS drug candidate prostratin in Samoan populations of Homalanthus nutans. J. Nat. Prod. 71, 2041–2044 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chieppa M., Giannelli G., Immune cells and microbiota response to iron starvation. Front. Med. (Lausanne) 5, 109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox P. A., Saving the ethnopharmacological heritage of Samoa. J. Ethnopharmacol. 38, 181–188 (1993). [DOI] [PubMed] [Google Scholar]
- 62.Leal M. C., et al. , Fifty years of capacity building in the search for new marine natural products. Proc. Natl. Acad. Sci. U.S.A. 117, 24165–24172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vandebroek I., et al. , Reshaping the future of ethnobiology research after the COVID-19 pandemic. Nat. Plants 6, 723–730 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Robinson D. G., et al. , Design and analysis of Bar-seq experiments. G3: Genes/Genomes/Genet. 4, 11–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang T., et al. , An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28, 795–798 (2011). [DOI] [PubMed] [Google Scholar]
- 66.O’Sullivan D., et al. , Treatment with the antipsychotic agent, risperidone, reduces disease severity in experimental autoimmune encephalomyelitis. PLoS One 9, e104430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X., Goncalves R., Mosser D. M., The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 83, 14.1.1–14.1.14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pluskal T., Castillo S., Villar-Briones A., Oresic M., MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chambers M. C., et al. , A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Feature finding and ion identity networking were performed using an in-house modified version of MZmine2.37 (68), corr.17.7 available at GitHub, https://github.com/robinschmid/mzmine2/releases. Feature tables, MS/MS spectra files (mgf), and ion identity networking results were exported, uploaded to the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/dataset_files.jsp?task=16c65622121c488d9123dc42c556dbff#%7B%22table_sort_history%22%3A%22main.collection_asc%22%2C%22main.collection_input%22%3A%22metadata%7C%7CEXACT%22%7D), and submitted to GNPS (35) (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=76115ecaad9c46e9aaa30413d0d435eb) for feature-based molecular networking analysis. All raw and processed data are publicly available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=16c65622121c488d9123dc42c556dbff. For spectral networking and spectrum library matching, all files were uploaded to the feature-based molecular networking workflow on GNPS (35) (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). The GNPS job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=76115ecaad9c46e9aaa30413d0d435eb. All other study data are included in the article and/or SI Appendix.