Abstract
The recent increase in extensively drug resistant bacterial pathogens and the associated increase of morbidity and mortality demonstrate the immediate need for new antibiotic backbones with novel mechanisms of action. Here, we report the development of the PepSAVI-MS pipeline for bioactive peptide discovery. This highly versatile platform employs mass spectrometry and statistics to identify bioactive peptide targets from complex biological samples. We validate the use of this platform through the successful identification of known bioactive peptides from a botanical species, Viola odorata. Using this pipeline, we have widened the known antimicrobial spectrum for V. odorata cyclotides, including antibacterial activity of cycloviolacin O2 against A. baumannii. We further demonstrate the broad applicability of the platform through the identification of novel anticancer activities for cycloviolacins by their cytotoxicity against ovarian, breast and prostate cancer cell lines.
Graphical Abstract
INTRODUCTION
Difficult-to-treat nosocomial and community acquired fungal and extensively drug resistant bacterial infections are increasingly commonplace, new viral diseases are emerging and spreading rapidly, and cancer remains a leading cause of death worldwide. In the US, there are almost two million hospital-acquired bacterial infections each year, resulting in ~100,000 deaths1. The emergence of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) pathogens has led to a significant increase in multidrug resistant (MDR) infections in the clinic with associated increases in morbidity and mortality2. Additionally, the recent emergence, resurgence, and spread of viruses, including Zika, SARS, West Nile, Ebola, and MERS3, for which there are limited or no direct-acting antivirals, highlight the susceptibility of the human population to future potentially untreatable pandemics. Furthermore, despite continued progress in anticancer therapeutics, limitations to current lead compounds of nonspecific toxicity, poor drug penetration, and multidrug resistance have emphasized the need for discovery of anticancer therapeutics with novel mechanisms of action4. With many bacteria now unresponsive to multiple classes of antimicrobial compounds and cancer the leading cause of death worldwide, there is an undeniable and desperate need to identify novel pharmacological chemistries and accelerate their development through new methods and innovative technologies.
Natural products have long been sources of virtually all traditional medicinal preparations and have been the single most successful source of lead compounds for drug discovery5. Specifically, plants have played a significant role in the treatment of human ailments since prehistoric times. The teas and tinctures of times past are one source to drug discovery, allowing the ethnobotanically-guided isolation and characterization of pharmacologically active compounds for the treatment of bacterial and fungal infections, cancers, and other ailments. Despite historical relevance and past success, challenges associated with natural product discovery have slowed progress in drug discovery efforts. Additionally, natural product discovery efforts have largely focused on small molecule constituents. However, recent discoveries have revealed ribosomally-synthesized, post-translationally-modified peptide natural products (RiPPs) with substantial structural diversity and bioactivity potential, including novel mechanisms of action6,7. While traditionally studied for antifungal and antibacterial properties, recent studies have piqued interest in these peptides as potential anticancer therapeutics8. Peptides offer several advantages over other small and large molecule drug candidates – including greater efficacy, selectivity and specificity relative to small organic molecules, better tissue penetration, and reduced immunogenicity and manufacturing costs relative to proteins/antibodies. Advances in peptide modification, formulation, and delivery methods can address known limitations of peptidic drug candidates9, including modification of peptide length/content to increase selectivity10, stapling and/or peptidomimetic conversion techniques to improve pharmacokinetic properties11, as well as encapsulation methods to protect from proteolytic degradation10. However, RiPPs fall outside the scope of standard therapeutic screening approaches and elude detection via standard -omic workflows due to their large size, structural diversity and high level of post-translational modification; therefore, systematic approaches for discovery and characterization are in nascent development12. Developing natural product screens for antimicrobially active RiPPs (AMPs) has the potential for discovery of new bioactive compounds with novel mechanisms of action able to address the growing problem of antimicrobial resistance.
Current methods for RiPP discovery often rely on bioassay guided fractionation13 or genomic mapping12,14,15 (eg. Pep2Path, RiPPquest) to facilitate downstream analysis. Relying on iterative rounds of chromatographic separations, bioassay guided fractionation is extremely time consuming and provides no structural information until late in the discovery process. Additionally, this approach often leads to replication of previously known compounds as a bias towards highly abundant and highly active compounds is evident. While alternative genomic mapping approaches can provide structural information from the beginning, prior genomic sequencing and knowledge of peptide biosynthetic pathways is required. Furthermore, this approach is unable to provide direct bioactivity information on the target peptide thus necessitating downstream isolation and activity screening to determine function and biological activity. Recent approaches to address some of these limitations have been proposed16 (eg. Compound Activity Mapping); however, these platforms are specific to small molecule identification and do not translate efficiently towards advancing untargeted proteomics approaches.
To this end, we have developed the PepSAVI-MS (Statistically-guided bioactive peptides prioritized via mass spectrometry) pipeline for the screening and identification of cationic bioactive peptides from natural product sources. For streamlined identification of bioactive peptides, our platform employs only one round of crude fractionation and relies on the power of mass spectrometry and statistics to assign bioactivity to individual components. Furthermore, we implement physiologically relevant whole cell bioassays to obtain activity information early in the discovery process, thus focusing on only active bioactive peptide targets. Our PepSAVI-MS pipeline is adaptable to any natural product source of peptides and can test against multiple physiological targets of various cell lines or organisms, including bacteria, fungi, viruses, protozoans, and cancer cells. Herein, we demonstrate the PepSAVI-MS pipeline using plant material and focusing on peptides with antimicrobial and/or anticancer properties. Proof-of-principle studies for a known AMP from the plant Viola odorata are presented to validate this workflow. Additionally, we use this pipeline to explore novel antibacterial and anticancer activities of the Viola odorata cyclotide, cycloviolacin O2.
EXPERIMENTAL
Source materials and growth conditions.
Viola odorata seedlings purchased from Strictly Medicinal Seeds (formerly Horizon Herbs; Williams, OR) were grown to mature rosette stage (~6 weeks) in standard greenhouse conditions. Seedlings were planted in nutrient-rich soil under controlled temperature (63.5 - 68.5°F) and light cycle (14-hr light) conditions. Aerial tissue was harvested with immediate flash freezing under liquid nitrogen, and stored at −80°C until subsequent extraction.
Creation of peptide libraries: Extraction.
Frozen tissue (100 g) was ground under liquid nitrogen using a mortar and pestle and aqueous extracted in 300 mL of 10% acetic acid with protease inhibitors (Roche, 1 tablet/50 mL) and stirring for 4 hr at 4°C. Insoluble material was pelleted by centrifugation at 13,000 rcf for 45 min, and 0.45 μm stericup filtration (Millipore) was used to remove remaining particulates. Protein concentration filters (MWCO 30kDa; Millipore) were used to remove high molecular weight species, and subsequent dialysis (0.1 – 1 kDa cutoff; SpectrumLabs) into 5 mM ammonium formate pH 2.7 was performed to eliminate small molecule contaminants. The sample was concentrated via vacuum centrifugation to 4 mL to generate the final crude extract. HPLC fractionation. Extracts were subjected to a 47-min SCX method using a PolySulfoethylA column (100 x 4.6 mm, 3 μm particles, PolyLC). A salt gradient (Figure S-1) was employed using a linear ramp from 5 mM ammonium formate, 20% acetonitrile, pH 2.7 to 500 mM ammonium formate, 20% acetonitrile, pH 3.0. Fractions were collected across the 47-min run in one-min intervals and desalted with three washes of 1.3 mL water using a vacuum concentrator. Peptide libraries were stored in water (400 μL/fraction) at 4°C until bioactivity assay.
Bioactivity screening: Bacterial.
Escherichia coli 25922 was obtained through ATCC and the ESKAPE pathogen strains are clinical isolates that belong to a collection acquired by the Shaw Lab [Enterococcus faecium (1449), Staphylococcus aureus (635), Klebsiella pneumoniae (1433), Acinetobacter baumannii (1403), Pseudomonas aeruginosa (1423), Enterobacter cloacae (1454)]17. Bacterial cultures were synchronized to mid-log phase by inoculating 1 mL of overnight culture into 100 mL of TSB and incubated for 3 hr, shaking at 37°C. All assays were performed in 96-well microtiter plates by adding 10 μL 2x MH broth, 20 μL of 1x MH broth, 10 μL peptide fraction, and 10 μL 0.5 OD600 bacteria culture. The addition of 2x broth ensured sufficient nutrients were available since the peptide fractions, in water, accounted for 1/5 of the final volume. TSB was used to test S. aureus due to its decreased growth rate in MHB. Ampicillin (100 μg/mL) or Tetracycline (100 μg/mL) was added respectively to E. coli or ESKAPE pathogen plates in place of the plant fraction to serve as the positive control, and water as the negative control. The prepared plates were incubated at 37°C, 275 rpm shaking for 1.5 - 4 hr depending on the growth characteristic of the organism before 5 μL of the cell viability indicator dye, resazurin (1.19 mM), was added to each well. After one additional hr of shaking and incubation, a fluorescence read of 544 nm (ex) and 590 nm (em) was collected to measure relative fluorescence units for each sample. Percent activity of each well was calculated using the formula: .
Cancer cell lines: Cell viability assay.
MDA-MB-231 breast cancer cells, OVCAR ovarian cancer cells, PC3 prostate cancer cells, or human dermal fibroblasts in the appropriate growth medium were plated into 96-well flat-bottom microtiter plates at 2 × 104 cells/well and cultured for 24 hr in a 37°C humidified incubator in order to form adherent monolayers. Medium was then removed and replaced with fresh serum-free medium with or without individual plant fractions to be assayed for cytotoxic activity. Cells were then cultured for an additional 24 hr. Two hr before the end of culture, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution was added to each well at a final concentration of 0.5 mg/mL to measure mitochondrial succinate dehydrogenase activity. Culture supernatants were discarded and formazan crystals were solubilized in 0.1 mL of dimethyl sulfoxide. Absorbance was measured at 570 nm on an ASYS Expert 96 plate-reader (Montreal Biotech Inc., Kirkland, QC). Absorbance in each treatment group was compared to the control to determine percent reduction in viability, as reflected by the change in mitochondrial succinate dehydrogenase activity.
LC-MS/MS.
Peptide libraries were analyzed via a nano-LC-ESI-MS/MS platform: nanoAcquity (Waters, Milford, MA) coupled to a TripleTOF5600 (AB Sciex, Framingham, MA). Peptide fractions were diluted to the appropriate loading level, acidified with formic acid and transferred to low-volume 96-well plates covered with adhesive plate seals. Each sample was injected onto a trap column (NanoAcquity UPLC 2G-W/M Trap 5 μm Symmetry C18, 180 μm x 20 mm: Waters) before subsequent passing onto the analytical C18 column (10k PSI, 100 Å, 1.8μm, 75μm x 250 mm: Waters). Peptide separation was carried out at a flow rate of 0.3 μL/min using a linear ramp of 5 – 50 % B (mobile phase A, 1% formic acid; mobile phase B, 1% formic acid in acetonitrile) over 30 min. The MS was operated in positive ion, high sensitivity mode with the MS survey spectrum using a mass range of 350-1600 Da in 250 ms and information dependent acquisition (IDA) of MS/MS data. The first 20 features above 150 counts threshold and having a charge state of +2 to +5 were fragmented using rolling collision energy ±5%. Auto calibration was performed every eight samples (8 hr). The data have been deposited to the ProteomeXchange Consortium via the PRIDE18 partner repository with the dataset identifier PXD004780 (Username: reviewer05751@ebi.ac.uk, Password: gbg2SK43). De-isotoped peak lists for each fraction were generated using Progenesis QI for Proteomics software (Nonlinear Dynamics, v.2.0). To align runs, a reference run was chosen from a subset of bioactive fractions (15 - 30 for V. odorata). Automatic processing settings with a retention time filter of 14 - 45 min were used to align and peak pick ions across all runs. Identified features were quantified using AUC integration of survey scan data based on the summed intensity of each de-isotoped feature. Data was exported as “peptide ion data” with the default parameters from Progenesis at the “Identify Peptides” stage in the software. Our analysis yielded 6,258 unique MS features for V. odorata.
Data reduction and statistical modeling.
Areas of interest in the bioactivity profile were selected for subsequent data reduction and modeling. The bioactivity region for each V. odorata data set was defined for each pathogen based on the observed bioactivity profile as follows: ovarian cancer (18-22), breast cancer (18-22), prostate cancer (18-22), A. baumannii (17-21), E. faecium (17-21), P. aeruginosa (18-21), E. coli (18-25), F. graminearum (19-24). Using the PepSAVI-MS software, background ions were eliminated through retention time, mass, and charge state filters to reduce the data to potential compounds of interest. Peak-picked data believed to belong to the same MS feature were binned together if their m/z values were within a 0.05 Da window and their charge states were identical. Next, workflow-informed filtering criteria were applied using the following rules: 1) m/z intensity maximum must fall inside the range of the bioactivity “area of interest”, 2) the m/z intensity of species meeting the first criteria must be <1% of its respective maximum peak intensity in the areas bordering said “area of interest”, 3) there must be non-zero abundance in the fraction following the maximum intensity fraction, 4) the maximum intensity must be > 1,000 in active window, 5) all charge states > +10 are excluded. All m/z species meeting these filtering criteria were modeled using the elastic net estimator with a quadratic penalty parameter specification of 0.001 to determine each species’ contribution to the observed overall bioactivity profile. The resulting list contains candidate compounds ranked in order of when they entered the model, such that the highest ranked compounds have the greatest likelihood to be contributing to the bioactivity.
Isolation of cyO2.
Dried aerial plant material from Viola odorata (Alfred Galke GmbH, Bad Grund, DE) was used to isolate cyO2. Cyclotide isolation was carried out according to Herrmann et al. with minor modifications19. Briefly, ground plant material was subjected to extraction with 60% methanol in water, followed by filtration and liquid–liquid extraction with dichloromethane. The polar phase was diluted and acidified before subjected to a strong cation exchange chromatography to capture cyclotides of net positive charge. Individual cyclotides, i.e. cyO2, were then isolated using RP-HPLC. Final cyO2 was of >95% purity assessed by RP-HPLC (214 nm).
Validation of cyO2 activity.
Minimum inhibitory concentration determination of cyO2 against E. coli and A. baumannii was performed in Mueller-Hinton media using a broth microdilution method. Bacteria were grown overnight and added to the assay at 105 CFU/mL and turbidity/clearing was observed after 20 hr incubation. Assay was performed in triplicate starting with 10 or 25 μM cyO2 for E. coli or A. baumannii, respectively, and using 2x serial dilution. Mean MICs of 5 and 15 μM were obtained for each species, respectively. The mean IC50 value of 6.6 μg/mL for cyO2 against the PC3 prostate cancer cell line was determined using duplicate measurements of the MTT cell viability assay as described previously.
RESULTS AND DISCUSSION
Overview.
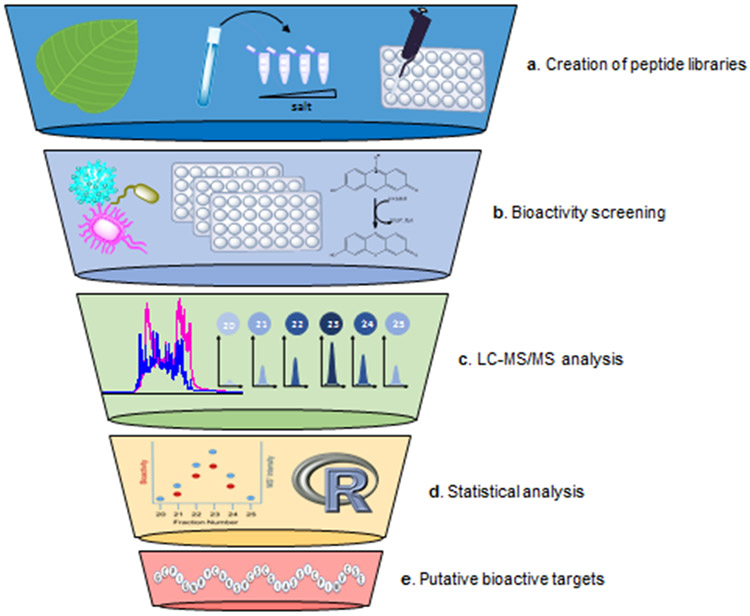
Relying on a multi-pronged approach, the PepSAVI-MS pipeline utilizes selective extraction and fractionation of peptide source material, bioactivity screening, and statistics-guided mass spectrometry-based peptidomics for the targeted identification and characterization of only putative bioactive compounds (Figure 1). We demonstrate use of the PepSAVI-MS pipeline focusing bioactive peptides from the plant kingdom and their activity against a panel of microbial pathogens and cancer cell lines.
Figure 1:
Overall workflow for the PRISMS screening platform including (a) creation of peptide libraries through extraction and fraction of crude extracts (b) whole-cell bioactivity screening of peptide libraries against pathogen targets of interest (c) LC-MS/MS analysis of active peptide libraries and (d) statistical modeling of LC-MS/MS datasets vs. active bioactivity regions for determination of (e) putative bioactive peptide targets.
Plant selection, cultivation and harvesting.
Plant species are selected based on several criteria including known bioactivity of the plant, traditional use, and availability. While many plants are commonly used for their perceived health-related benefits, the bioactive peptides from these species have not been comprehensively evaluated. Seeds/seedlings of selected species are obtained through commercial sources and grown in a laboratory greenhouse. Tissue is harvested with immediate flash freezing in liquid nitrogen for future extraction.
Peptide library creation (Figure 1a).
Preparation of crude extracts from plant material includes aqueous extraction and size exclusion to selectively target AMP-like molecules, i.e. water-soluble compounds that are smaller than 10 kDa20,21. Extracts are crudely fractionated using strong cation exchange (SCX) chromatography such that individual peptides elute over multiple sequential fractions. The distribution of bioactive peptide abundance across sequential fractions is reflected in the distribution of activity seen in bioactivity data for the same fractions. As peptides are eluted using volatile salts during SCX, peptide libraries are simply desalted via vacuum concentration for compatibility with downstream bioassays. Peptide libraries can be created from constitutive expression, as well as from abiotic and biotic stresses known to induce production of defense compounds22-25. Bioactivity screening. Peptide libraries are assayed for growth inhibition against pathogens of interest using physiologically relevant whole-cell bioactivity screens (Figure 1b). Library fractions are incubated with a microbial or cancer cell culture and the presence of bioactive peptides in a given fraction will result in inhibition of culture growth during the incubation period. For bacterial assays, the remaining viable cells are quantified indirectly by spectrophotometric measurement of the irreversible intracellular reduction of resazurin26. For anticancer bioactivity, cytotoxicity assays are performed using MTT-based assays to measure mitochondrial succinate dehydrogenase activity with absorbance measurements at 570 nm. Values for each fraction are compared to positive and negative controls containing a known therapeutic or water, respectively, to determine a percent activity of each library fraction, where a small value of remaining viable cells indicates high activity. Species with bioactivity profiles of interest are prioritized for LC-MS/MS and downstream statistical analysis.
MS profiling,data reduction and statistical modeling to identify putative bioactive peptides.
Peptide libraries are analyzed via nanoLC-ESI-MS/MS to obtain accurate intact mass and relative intensity information for peptide constituents contained within each library fraction (Figure 1c). The resulting data array is processed using the developed PepSAVI-MS software package. Using this function, unwanted compounds can be eliminated using mass, charge, and retention time filters. Remaining compounds are binned to condense all of the hits for a given m/z ratio into a single feature. Binned datasets are then filtered using the following workflow-informed criteria to narrow the library to those peptides most likely contributing to the bioactivity profile: 1) m/z intensity maximum must fall inside the range of the bioactivity “area of interest”, 2) the m/z intensity of species meeting the first criteria must be <1% of its respective maximum peak intensity in the areas bordering said “area of interest”, 3) there must be non-zero abundance in fraction following the maximum intensity fraction, 4) the maximum abundance must be > 1,000 in active window, 5) all charge states > +10 are excluded. This filtering results in a reduction of each data set to those peptides with their highest abundance in the bioactive region. Subsequently, sparse penalized linear regression modeling (elastic net27,28, including lasso29) is applied to correlate the relative abundance of post-filtered peptides and percent activity across library fractions in order to identify the specific peptide(s) causing bioactivity (Figure 1d). Because multiple peptides could jointly contribute to the bioactivity across a group of active fractions, a simple marginal correlation analysis strategy may produce inaccurate or spurious results for any individually contributing peptide. However, sparse penalized linear regression is able to account for this co-contribution to activity and results in more accurate identification of putative bioactive peptide species under the theoretical model. Elastic net regularization is an appropriate penalization specification as it elicits a sparse set of peptides contributing to bioactivity levels, helping to identify peptides most likely responsible for the corresponding bioactivities without strict limitation to a set number of peptides being selected. Identification of such a set facilitates prioritization of peptides for subsequent characterization and validation (Figure 1e). The elastic net path for fixed choice of quadratic penalty parameter can be obtained through a transformation of the predictor data and a subsequent application of the LARS algorithm30. Thus, in order to obtain a list of candidate compounds most likely to be contributing to the bioactivity, the order in which compounds' coefficients first obtain a nonzero value is tracked along the course of the algorithm, with coefficients that become nonzero earlier presumed to be better candidates for further investigation.
Peptide characterization.
Prioritized peptides are characterized via appropriate strategies depending on the target(s) of interest. While a top-down approach31 is used for detection of peptides of interest and may be sufficient for characterization, a hybrid approach of chemical or enzymatic reaction followed by LC-MS/MS analysis may be necessary to facilitate sequencing and identification of post-translational modifications (PTMs). The accurate intact mass and MS/MS spectrum of the peptide species is obtained from the initial LC-MS/MS data collection. If necessary, further characterization is carried out utilizing the library fraction containing the most abundant amount of the peptide species of interest. Reduction and alkylation of cysteine residues is used to determine the number of disulfide bonds. Proteolysis is used to assess disulfide bond connectivity and possible cyclizations (e.g. cyclotides containing a single glutamic acid residue can be linearized through cleavage with endoproteinase GluC32). Paired Lys-C and Lys-N digestions can facilitate determination of termini by allowing identification with high confidence of b- and y- ions from the isobaric precursor ions33. Complementary tandem MS fragmentation via CID and ECD/ETD is used in combination to maximize peptide primary sequence determination and post-translational modification localization31,34,35. De novo peptide sequencing will be facilitated using PEAKS software (Bioinformatics Solutions Inc.)36, supplemented with composition-based sequencing37,38 and homology-based database searches39 to address plant species with limited sequencing information available. Structural mapping to delineate disulfide bond connectivity is carried out in combination with multi-enzyme digestions35, as appropriate, to dissect these key features that often confer bioactivity.
Isolation and in vitro/in vivo validation of prioritized bioactive peptides.
Bioactive peptides that have been characterized are validated and their requisite bioactivity characterized. In vivo confirmation/validation of a bioactive peptide requires isolation to a high degree of purity to allow testing without interference from contaminating co-eluting compounds. Split-flow UPLC-MS or de novo synthesis of peptides will permit necessary biological and pharmacological testing to determine if a peptide is a satisfactory lead compound that should be further evaluated.
Platform validation.
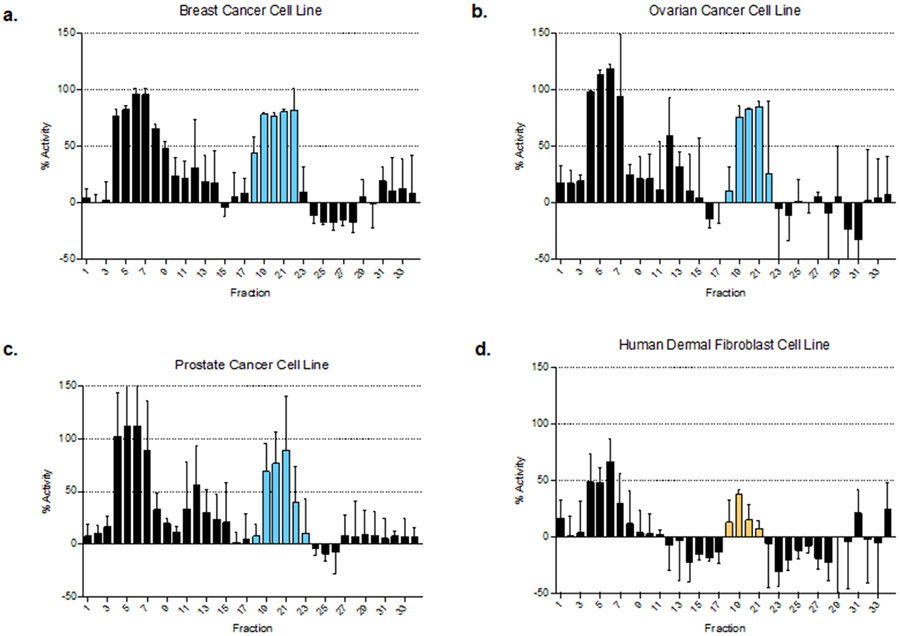
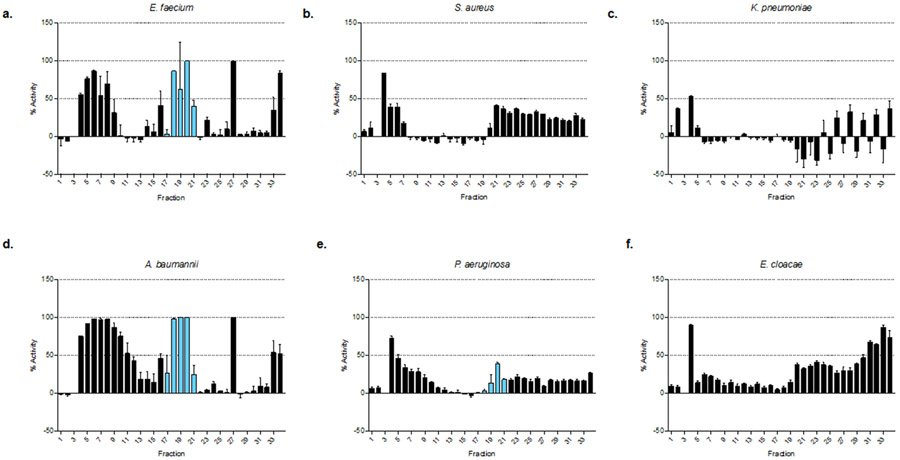
To validate the PepSAVI-MS pipeline, we demonstrate successful detection and identification of a known AMP from the botanical species Viola odorata. Viola odorata, commonly known as sweet violet, contains many cyclotides40 - including cycloviolacin O2 (cyO2). CyO2 is a small, cysteine rich cyclotide comprised of 30 amino acids (MWmonoisotopic: 3138.37 Da), which has been shown to have diverse activity against many Gram-negative bacteria (E. coli, K. pneumoniae, and P. aeruginosa)41, as well as several cancer cell lines42,43. Following the PepSAVI-MS pipeline as described above, V. odorata plants were grown, harvested, extracted, and crudely fractionated for creation of its peptide library. Using the known activities of cyO2 as a guide for assay selection, antibacterial bioassays were performed against E. coli (Figure 2a) and a panel of clinical strains representing the ESKAPE pathogens17 (Figures 4 and S-2). Each bioassay yielded robust bioactivity profiles unique to each microbe tested. Fractions containing the mass corresponding to cyO2 demonstrated growth inhibition for A. baumannii, P. aeruginosa and E. coli, but not for E. faecium, S. aureus, K. pneumoniae, or E. cloacae. As demonstrated previously44, the crude nature of SCX separation often results in group isolation of cyclotides (Figure S-3). Hence, these fractions also contained additional cyclotides that have the potential to be contributing to the aforementioned activity. The majority of the remaining fractions did not show growth inhibition, indicating the source of activity was due to constituents within the fractions rather than the extract itself. To further demonstrate the broad applicability of the platform, V. odorata fractions were screened for activity against human breast (MDA-MB-231), prostate (PC3), and ovarian (OVCAR) carcinoma cell lines and yielded robust activity profiles with increased cytotoxicity for these neoplastic cells in comparison to a non-cancerous human dermal fibroblast cell line (Figure 3). Viola odorata peptide library was also screened for growth inhibition against the filamentous fungus, Fusarium graminearum (PH-1), which similarly yielded a robust activity profile across the cyclotide-containing fractions (Figure S-4). To determine which of the detected cyclotides could be contributing to the aforementioned bioactivity, V. odorata peptide library was subjected to LC-MS/MS and statistical analysis with the developed PepSAVI-MS software package. Retention time alignment and peak-picking of detected ions identified 6,258 unique features across V. odorata fractions 11 – 43. To validate our automated workflow for data processing, extracted ion chromatograms of cyO2 peptide were plotted for manual versus automated data extraction (Figure 2b). Extracted ion chromatograms show that cyO2 was detected at varying abundances across fractions 18 – 22 and validate the use of automation for data array generation. Following the known mass and charge properties of bioactive peptides, exported peptide ion data was filtered to include masses between 2 and 15 kDa having charge states between +2 and +9 and eluting between 14 and 45 minutes. Remaining data was then binning using a 0.05 Da mass window with identical charge state requirements. Using the workflow informed criteria (described above) for data filtering and reduction with the bioactivity region specified as fractions 17 – 25, the number of possible candidates contributing to bioactivity was reduced to 225. This resulting list of candidate peptides was then subjected to statistical modeling of all bioactivity data sets, using the sparse penalized regression method, elastic net28. CyO2 from V. odorata was within the top 20 candidates for 2 of the 7 active species when modeled against the averaged bioactivity data. Furthermore, modeling individual bioactivity replicates with the requirement that any true component must be pulled out in at least two of the three replicates can further reduce the number of considered compounds for each data set. CyO2 was characterized via a multi-step mass spectrometry-based approach (Figure S-5) and these results are in agreement with previously published findings, thus confirming the identity of cyO240,45. MS2 sequence coverage across the peptide allowed for distinction from another cyclotide, cyO9, present in sweet violet with the same intact mass as cyO2 and differing only by three residues. Only one other cyclotide corresponding to the intact mass of cyO17 appeared in the top 20 candidates after data reduction and statistical modeling of the E. coli bioactivity data set. No other known cyclotides appeared in the top 20 candidates for any of the other bioactivity data sets. While this does not eliminate the possibility that other cyclotides could be contributing to the activity, our experiments suggest that cyO2 was the main contributor. To allow for greater access to the scientific community, the statistical analysis methods established for this platform have been developed into an R package, PepSAVIms, that is publically available through CRAN (https://cran.r-project.org/package=PepSAVIms). Included with this package are two vignettes providing an in depth description of the functionality of the PepSAVI-MS pipeline package, as well as the specific bioinformatics implementation performed in this manuscript.
Figure 2.
V. odorata fractions (a) bioactivity vs. Escherichia coli with the growth-inhibition defined bioactivity region in blue where % activity indicates the decrease in bacterial aerobic metabolism quantified by resazurin reduction. (b) Aligned cyO2 elution profile with comparison of manual (yellow) and automated (gray) extraction of the detected cyO2 charge states.
Figure 4:
Viola odorata peptide library bioactivity profile against (a) breast (MDA-MB-231), (b) ovarian (OVCAR), and (c) prostate (PC3) cancer cell lines and (d) non-cancerous human dermal fibroblasts. The cytotoxicity-defined bioactivity region of cyO2 against the given cell line is defined with blue bars for cancer cell lines and yellow bars for non-cancerous cell lines.
Figure 3:
Bioactivity data of Viola odorata fractions vs. the ESKAPE pathogens: (a) Enterococcus faecium, (b) Staphylococcus aureus, (c) Klebsiella pneumoniae, (d) Acinetobacter baumannii, (e) Pseudomonas aeruginosa, (f) Enterobacter cloacae. The growth inhibition-defined bioactivity region of cyO2 is depicted using blue bars for the species deemed to demonstrate activity.
To confirm that these bioassays are capable of detecting AMP activity at concentrations relevant for the observed activity, the minimum inhibitory concentration (MIC) of isolated cyO2 against E. coli was determined. Using a broth microdilution assay in 96-well plate format, the MIC was determined to be 5 μM, which lies in between the previously reported MIC of 2.2 μM41 and MIC50 of 6.8 μM13 for the same peptide on E. coli using different protocols. The concentration of cyO2 in the most abundant fraction (21) that killed or inhibited these strains was determined to be ~300 μM via area-under-the-curve integration of RPLC-MS peaks, which is in excess of the MIC needed for growth inhibition.
Novel findings.
In addition to platform validation, unknown activities of cyO2 from V. odorata have been revealed in this study. Previously, it was demonstrated that cyO2 was active against a subset of the ESKAPE pathogens, including K. pneumoniae, and P. aeruginosa, and inactive against the tested Gram-positive bacteria, including S. aureus and E. faecium41. Results from our study are in agreement with the activity of cyO2 in the cyclotide fractions against E. coli and P. aeruginosa, and inactivity against E. faecium and S. aureus. However, we could not detect activity in these fractions against K. pneumoniae, which is likely due to high strain variability among this bacterial species46,47. Additionally, our assay detects strong activity of the cyclotide fractions against A. baumannii, of which there are no known reported studies of activity (Figure 4). The MIC for the ESKAPE pathogen representing novel activity, A. baumannii, was determined to be 15 μM using isolated cyO2 and thus supports that cyO2 is the main contributor to the activity seen in these fractions. Throughout this study we have noticed variation in the sensitivity of peptidyl activity due to differences in media conditions and salt concentrations (Figure S-6). We suspect that small differences even within different brands of the same medium could affect activity48,49 necessitating validation of new lots of media through testing of established fraction libraries to confirm consistency with previously collected bioactivity data. While our study shows some examples of changes in activity due to strain variability, the majority of findings remain consistent across different strains of the same species.
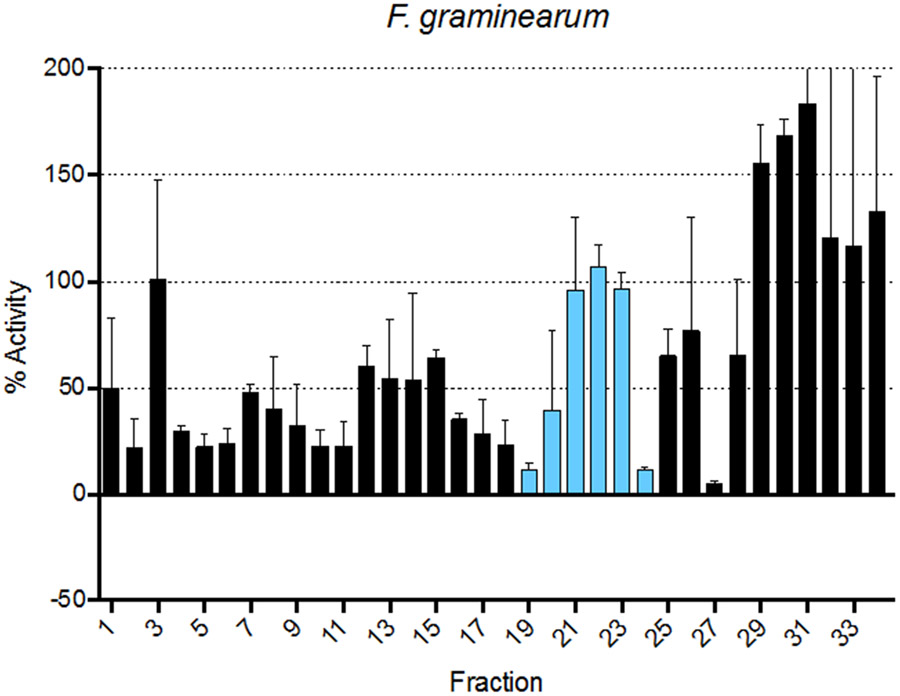
Screening of V. odorata fractions against a panel of human cancer cell lines similarly demonstrates proof-of-principle, as well as an additional novel finding. In previous studies, cyO2 has been shown to have anticancer activity against multiple breast and ovarian cancer cell lines42,43. Although our screen used different cell lines, strong activity was observed against the MDA-MB-231 breast cancer cell line, as well as the OVCAR ovarian cancer cell line across fractions in which cyO2 was eluted. Additionally, this is the first demonstration of the ability of purified cyO2 to kill PC3 prostate cancer cells, with a determined IC50 of 6.6 μg/mL (2.1 μM). Furthermore, we demonstrate that the cyclotide fractions can kill the filamentous fungus F. graminearium and suspect the presence of an additional peptide(s) from V. odorata possessing antifungal capabilities (fractions 28 – 36). These studies not only highlight the wide applicability of this platform but also the capabilities of antimicrobial peptides as broad-spectrum therapeutics.
CONCLUSIONS
As demonstrated herein, the developed PepSAVI-MS pipeline is broad-spectrum, high-throughput, and has the potential to expedite the search for new bioactive peptides from plants. Our platform overcomes many traditional limitations of drug discovery efforts through achieving increased efficiency by directly targeting and characterizing only those species contributing to the bioactivity. This may eliminate many laborious rounds of bioassay-guided fractionation or the need for characterization of all molecular species present, regardless of demonstrated activity. Furthermore, the developed platform is highly versatile as it is adaptable to any natural product source of peptides and can test against diverse physiological targets, including bacteria, fungi, viruses, protozoans, and cancer cells for which there is a developed bioassay. With this platform there is little bias towards high abundance compounds as the trend of activity values is more important than the raw abundance values. While the PepSAVI-MS pipeline has been developed to target cationic AMPs, the platform is adaptable to target other types of compounds by changing the type of chromatography performed. The PepSAVI-MS pipeline opens the door for investigating purpose-guided natural product extracts with a new lens, and has the potential to lead to the discovery of novel chemistries at the forefront of modern drug discovery.
Supplementary Material
Figure 5:
Viola odorata peptide library bioactivity profile against the filamentous fungus Fusarium graminearum with the modeled bioactivity region depicted in blue.
ACKNOWLEDGEMENTS
We acknowledge the Pakistan-U.S. Science and Technology Cooperation Program (Phase 4 funding to L.M.H. and A.J.) and Awais Altaf, Brian Gau, Nazia Kanwal, Brian Kelly, Sumaira Kousar, Alicia Malone, and William Slade for financial and technical assistance, respectively, in the preliminary development of the PepSAVI-MS pipeline.
Footnotes
ASSOCIATED CONTENT
Supporting information consists of additional figures depicting SCX fractionation of V. odorata (S-1), bioactivity screening of V. odorata library vs. the remaining ESKAPE pathogens (S-2) and F. graminearum (S-4), SCX elution profiles of other detected cyclotides (S-3), mass spectral characterization of cyO2 (S-5) and different media effects on the observed bioactivity profiles (S-6).
REFERENCES
- (1).Klevens RM; Edwards JR; Richards CL Jr.; Horan TC; Gaynes RP; Pollock DA; Cardo DM Public Health Rep. 2007, 122, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Boucher HW; Talbot GH; Bradley JS; Edwards JE; Gilbert D; Rice LB; Scheld M; Spellberg B; Bartlett J Clin. Infect. Dis 2009, 48, 1–12. [DOI] [PubMed] [Google Scholar]
- (3).Howard CR; Fletcher NF Emerg Microbes Infect 2012, 1, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Guzman-Rodriguez JJ; Ochoa-Zarzosa A; Lopez-Gomez R; Lopez-Meza JE Biomed Res Int 2015, 2015, 735087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Harvey AL; Edrada-Ebel R; Quinn RJ Nat Rev Drug Discov 2015, 14, 111–129. [DOI] [PubMed] [Google Scholar]
- (6).Arnison PG; Bibb MJ; Bierbaum G; Bowers AA; Bugni TS; Bulaj G; Camarero JA; Campopiano DJ; Challis GL; Clardy J; Cotter PD; Craik DJ; Dawson M; Dittmann E; Donadio S; Dorrestein PC; Entian KD; Fischbach MA; Garavelli JS; Göransson U; Gruber CW; Haft DH; Hemscheidt TK; Hertweck C; Hill C; Horswill AR; Jaspars M; Kelly WL; Klinman JP; Kuipers OP; Link AJ; Liu W; Marahiel MA; Mitchell DA; Moll GN; Moore BS; Muller R; Nair SK; Nes IF; Norris GE; Olivera BM; Onaka H; Patchett ML; Piel J; Reaney MJ; Rebuffat S; Ross RP; Sahl HG; Schmidt EW; Selsted ME; Severinov K; Shen B; Sivonen K; Smith L; Stein T; Sussmuth RD; Tagg JR; Tang GL; Truman AW; Vederas JC; Walsh CT; Walton JD; Wenzel SC; Willey JM; van der Donk WA Nat. Prod. Rep 2013, 30, 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Essig A; Hofmann D; Munch D; Gayathri S; Kunzler M; Kallio PT; Sahl HG; Wider G; Schneider T; Aebi MJ Biol. Chem 2014, 289, 34953–34964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Schweizer F Eur. J. Pharmacol 2009, 625, 190–194. [DOI] [PubMed] [Google Scholar]
- (9).Vlieghe P; Lisowski V; Martinez J; Khrestchatisky M Drug Discov. Today 2010, 15, 40–56. [DOI] [PubMed] [Google Scholar]
- (10).Mandal SM; Roy A; Ghosh AK; Hazra TK; Basak A; Franco OL Front. Pharmacol 2014, 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Walensky LD; Bird GH J. Med. Chem 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mohimani H; Kersten RD; Liu WT; Wang M; Purvine SO; Wu S; Brewer HM; Pasa-Tolic L; Bandeira N; Moore BS; Pevzner PA; Dorrestein PC ACS Chem. Biol 2014, 9, 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Henriques ST; Huang YH; Castanho MA; Bagatolli LA; Sonza S; Tachedjian G; Daly NL; Craik DJ J. Biol. Chem 2012, 287, 33629–33643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Medema MH; Paalvast Y; Nguyen DD; Melnik A; Dorrestein PC; Takano E; Breitling R PLoS Comput. Biol 2014, 10, e1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Skinnider MA; Johnston CW; Edgar RE; Dejong CA; Merwin NJ; Rees PN; Magarvey NA Proc. Natl. Acad. Sci. U. S. A 2016, 113, E6343–e6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kurita KL; Glassey E; Linington RG Proc. Natl. Acad. Sci. U. S. A 2015, 112, 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fleeman R; LaVoi TM; Santos RG; Morales A; Nefzi A; Welmaker GS; Medina-Franco JL; Giulianotti MA; Houghten RA; Shaw LN J. Med. Chem 2015, 58, 3340–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vizcaino JA; Csordas A; del-Toro N; Dianes JA; Griss J; Lavidas I; Mayer G; Perez-Riverol Y; Reisinger F; Ternent T; Xu QW; Wang R; Hermjakob H Nucleic Acids Res. 2016, 44, D447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Herrmann A; Burman R; Mylne JS; Karlsson G; Gullbo J; Craik DJ; Clark RJ; Göransson U Phytochemistry 2008, 69, 939–952. [DOI] [PubMed] [Google Scholar]
- (20).Hammami R; Ben Hamida J; Vergoten G; Fliss I Nucleic Acids Res. 2009, 37, D963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Adochite RC; Moshnikova A; Carlin SD; Guerrieri RA; Andreev OA; Lewis JS; Reshetnyak YK Mol. Pharm 2014, 11, 2896–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Garcia-Olmedo F; Molina A; Alamillo JM; Rodriguez-Palenzuela P Biopolymers 1998, 47, 479–491. [DOI] [PubMed] [Google Scholar]
- (23).Epple P; Apel K; Bohlmann H Plant Physiol. 1995, 109, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lee SC; Hwang BK Plant Mol. Biol 2006, 61, 95–109. [DOI] [PubMed] [Google Scholar]
- (25).van Loon LC; Rep M; Pieterse CM Annu. Rev. Phytopathol 2006, 44, 135–162. [DOI] [PubMed] [Google Scholar]
- (26).Sarker SD; Nahar L; Kumarasamy Y Methods 2007, 42, 321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zou H; Hastie TJ Roy. Stat. Soc. Ser. B. (Stat. Method.) 2005, 67, 301–320. [Google Scholar]
- (28).Zou H; Hastie T elasticnet: Elastic-Net for Sparse Estimation and Sparse PCA (http://CRAN.R-project.org/package=elasticnet). 2012. [Google Scholar]
- (29).Tibshirani R J R Stat Soc Series B Stat Methodol. 1996, 58, 267–288. [Google Scholar]
- (30).Efron B; Hastie T; Johnstone I; Tibshirani R Ann. Statist 2004, 32, 407–499. [Google Scholar]
- (31).Guerrero A; Lerno L; Barile D; Lebrilla CB J. Am. Soc. Mass. Spectrom 2015, 26, 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Poth AG; Colgrave ML; Philip R; Kerenga B; Daly NL; Anderson MA; Craik DJ ACS Chem. Biol 2011, 6, 345–355. [DOI] [PubMed] [Google Scholar]
- (33).Brownstein NC; Guan X; Mao Y; Zhang Q; DiMaggio PA; Xia Q; Zhang L; Marshall AG; Young NL Rapid Commun. Mass Spectrom 2015, 29, 659–666. [DOI] [PubMed] [Google Scholar]
- (34).Hayakawa E; Menschaert G; De Bock P-J; Luyten W; Gevaert K; Baggerman G; Schoofs LJ Proteome Res. 2013, 12, 5410–5421. [DOI] [PubMed] [Google Scholar]
- (35).Ni W; Lin M; Salinas P; Savickas P; Wu SL; Karger BL J. Am. Soc. Mass. Spectrom 2013, 24, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ma B; Zhang K; Hendrie C; Liang C; Li M; Doherty-Kirby A; Lajoie G Rapid Commun. Mass Spectrom 2003, 17, 2337–2342. [DOI] [PubMed] [Google Scholar]
- (37).Spengler B J Am Soc Mass Spectrom 2004, 15, 703–714. [DOI] [PubMed] [Google Scholar]
- (38).Langsdorf M; Ghassempour A; Rompp A; Spengler B Rapid Commun Mass Spectrom 2010, 24, 2885–2899. [DOI] [PubMed] [Google Scholar]
- (39).Ma B; Johnson R Molecular & cellular proteomics : MCP 2012, 11, O111 014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ireland DC; Colgrave ML; Craik DJ Biochem. J 2006, 400, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Pränting M; Lööv C; Burman R; Göransson U; Andersson DI J. Antimicrob. Chemother 2010, 65, 1964–1971. [DOI] [PubMed] [Google Scholar]
- (42).Gerlach SL; Rathinakumar R; Chakravarty G; Göransson U; Wimley WC; Darwin SP; Mondal D Biopolymers 2010, 94, 617–625. [DOI] [PubMed] [Google Scholar]
- (43).Lindholm P; Göransson U; Johansson S; Claeson P; Gullbo J; Larsson R; Bohlin L; Backlund A Mol. Cancer Ther 2002, 1, 365–369. [PubMed] [Google Scholar]
- (44).Göransson U; Sjogren M; Svangard E; Claeson P; Bohlin LJ Nat. Prod 2004, 67, 1287–1290. [DOI] [PubMed] [Google Scholar]
- (45).Göransson U; Herrmann A; Burman R; Haugaard-Jönsson LM; Rosengren KJ ChemBioChem 2009, 10, 2354–2360. [DOI] [PubMed] [Google Scholar]
- (46).Lai YC; Yang SL; Peng HL; Chang HY Infect. Immun 2000, 68, 7149–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wu KM; Li LH; Yan JJ; Tsao N; Liao TL; Tsai HC; Fung CP; Chen HJ; Liu YM; Wang JT; Fang CT; Chang SC; Shu HY; Liu TT; Chen YT; Shiau YR; Lauderdale TL; Su IJ; Kirby R; Tsai SF J. Bacteriol 2009, 191, 4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Park IY; Cho JH; Kim KS; Kim YB; Kim MS; Kim SC J. Biol. Chem 2004, 279, 13896–13901. [DOI] [PubMed] [Google Scholar]
- (49).Schwab U; Gilligan P; Jaynes J; Henke D Antimicrob. Agents Chemother 1999, 43, 1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.