Abstract
New Approach Methodologies (NAMs) that employ artificial intelligence (AI) for predicting adverse effects of chemicals have generated optimistic expectations as alternatives to animal testing. However, the major underappreciated challenge in developing robust and predictive AI models is the impact of the quality of the input data on the model accuracy. Indeed, poor data reproducibility and quality have been frequently cited as factors contributing to the crisis in biomedical research, as well as similar shortcomings in the fields of toxicology and chemistry. In this article, we review the most recent efforts to improve confidence in the robustness of toxicological data and investigate the impact that data curation has on the confidence in model predictions. We also present two case studies demonstrating the effect of data curation on the performance of AI models for predicting skin sensitisation and skin irritation. We show that, whereas models generated with uncurated data had a 7–24% higher correct classification rate (CCR), the perceived performance was, in fact, inflated owing to the high number of duplicates in the training set. We assert that data curation is a critical step in building computational models, to help ensure that reliable predictions of chemical toxicity are achieved through use of the models.
Keywords: artificial intelligence, data curation, data quality, data reproducibility, QSAR
Introduction
Efforts to reduce, refine and replace animal tests (according to the Three Rs principles) have accelerated in the last two decades.1,2 The Strategic Roadmap, published by the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) in 2018,3 called for the development of alternative ‘New Approach Methodologies’ (NAMs) to help reduce the animal testing of chemical and medical agents. More recently, the United States Environmental Protection Agency (US EPA) declared a commitment to “eliminate all mammal study requests and funding by 2035.”4
The prediction of chemical toxicity by using studies and methods that could represent alternatives to animal testing has been an active research area at the intersection of toxicology, chemistry, molecular modelling, and regulatory science.5,6 Many modern in silico toxicity prediction tools, from read-across7 to quantitative structure–activity relationship (QSAR) modelling,8 rely upon knowledge on a range of adverse health effects that has been derived from experimental data on chemicals tested under different study protocols. The read-across relies on extrapolations from data-rich to data-poor compounds, based on perceived chemical ‘sameness’ or the presence of so-called ‘toxicity alerts’.9–11 QSAR models employ various statistical and artificial intelligence (AI) approaches toward forecasting the putative adverse effects of new compounds. Both approaches are pursued actively to facilitate the replacement and reduction of animal testing in toxicology and risk assessment; however, these tools have limitations and their application has been challenged.12
Toxicologists and chemists alike are well aware that similar compounds, including those obtained by minor modifications of a parent molecule through metabolism, may have very different properties regarding toxicity, efficacy, or inter-individual variability.13 The presence of such pairs or groups of compounds with similar structures but different activity (often called ‘activity cliffs’)14 represents a significant challenge to both read-across and QSAR modelling. The difficulty of addressing this challenge explains why the results derived from QSAR and other in silico models are often met with caution.15 Nevertheless, computational predictions are appealing, as they are considerably less resource-intensive when compared to in vivo experimental testing. Also, as they are non-invasive and involve no animal use, they are usually not subject to any associated ethical approval requirements. As a result of this conflict between attractiveness and concerns over accuracy, computational predictions are usually used in combination with other evidence, or only for the initial screening/ranking of compounds for further testing.16 Thus, increasing the reliability of predictions is a critical challenge to overcome, to help ensure the increased use of computational models of chemical toxicity as NAMs.
The recent emergence of AI methods has generated optimistic expectations in chemical toxicology, as researchers in this field have striven to reduce or replace animal testing with NAMs.17,18 Despite this excitement and current progress, the main limitation of AI models remains the quality of the toxicology and chemistry data used to train the models. Indeed, a key consideration that can be universally applied to any computational modelling approach is the need for careful data curation before initiating any model development.19 While data preparation has been repeatedly emphasised as a critical element to ensure rigour and reproducibility of experimental and computational research,15,20 many publications seem to overlook this critical step.21–25 In this article, we highlight several challenges associated with experimental data reproducibility, in order to reinforce the need for high-quality data curation as an essential initial step when developing predictive and robust AI models that could eventually reduce or replace animal testing.
Concordance in the outcomes of in vivo toxicology studies
Chemical toxicity mechanisms are complex and involve many interconnected molecular pathways, multiple cell types and different organ systems. The interpretation of experimental toxicology studies is further compounded by differences among studies in terms of assay protocols, exposure conditions and duration, chemical purity, strain, sex, and dose selection. Other sources of biological and stochastic variability may also weigh in on the analysis of concordance of toxicology outcomes from studies performed in the various animal test systems. The challenges associated with comparing results from across studies are well documented. For instance, evaluation of the sources of variability in ‘no effect levels’ derived from systemic toxicity studies in rodents has concluded that about one-third of the total variance evident cannot be accounted for solely by considering the obvious differences in study characteristics.26 One possible explanation for the challenges in replicating the ‘no effect level’ across studies stems from choices in dose selection; indeed, reproducible dose–response data sets tend to have higher numbers of dose groups with fewer animals in each dose group. Another study found that, for the chemicals that were tested independently in the same Draize test on rabbit eyes, the reproducibility of the toxicity classification ranged from 73 to %94.27 The discordance was most pronounced for the compounds with weak or reversible effects — which is a consistent challenge in experimental toxicology. In a recent study, Rooney et al.28 found that 40% of chemicals classified initially as moderate irritants were classified as mild or non-irritants in the second test.
Several additional examples come from the efforts to define a ‘reference set’ of chemicals with conclusive and human-relevant adverse effects in rodent studies.29 For the rat uterotrophic assay, 70 compounds were identified in more than one study; among these, 75% concordance was observed between studies with most discordant results attributable to the differences in study design (e.g. injection versus oral dosing).30 For the Hershberger Bioassay (a short-term test to evaluate androgen disruption in rats),31 authors found that, of 25 chemicals tested in more than one study, 28% had discordant results between studies. In addition, of the 65 chemicals tested in Hershberger studies and other in vivo studies with androgen-responsive endpoints, 43% indicated disagreements.32 Differences in study designs or physiology of the animal models were cited by the authors as potential reasons for discordant outcomes.
In addition, a recent analysis of skin sensitisation data showed that the Local Lymph Node Assay (LLNA), which is the most commonly used animal test, has low specificity (high rate of false positives) compared to human data.33 Additionally, in a previous analysis, we found the LLNA to be less reproducible than validated non-animal methods, such as the in chemico Direct Peptide Reactivity Assay (DPRA) and the in vitro KeratinoSens and human Cell Line Activation Test (h-CLAT).34
The impact of data curation on QSAR modelling
Researchers in the field of chemical toxicology are striving to improve the quality and reproducibility of both the data and the models. Rigorous data preparation and curation are essential, in order to support the development of robust and reproducible QSAR models.8 It has been shown repeatedly in the field of QSAR modelling that data curation strongly affects the predictive accuracy of the models.15,35–37 This experience suggests that data curation and rigorous external model validation should be made mandatory when employing computational models in regulatory assessment, to account for data inconsistency or relevance and avoid overly optimistic evaluation of a computational model’s power. Chemical and biological data curation is not the only factor that affects the accuracy and utility of in silico approaches as alternatives to animal testing. For instance, improper statistical analysis has been heavily discussed as a factor as well,38–40 but inaccurate chemical and biological data is still the central issue in modelling chemogenomics data.8
Lack of concordance between the outcomes of animal studies for the same chemical is a known challenge in regulatory science, but computational toxicologists do not widely recognise it. However, this challenge should be acknowledged and identified when building QSAR models, mostly because prediction error cannot be significantly smaller than experimental measurement error.15 When analysing duplicates, the assay reproducibility can be estimated. Divergent data points should be further investigated for the availability of additional data. In the case of none being identified, the respective compounds can be set aside and predicted by consensus QSAR models, i.e. multiple QSAR models using different sets of descriptors and/or AI algorithms.41
As was recently reported,42 some of the data contained in the European Chemicals Agency (ECHA) Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) database are not from the guideline studies, but were predicted by using QSAR models or read-across. Therefore, these data points should be used advisedly for building new models, since models that predict compound categories that have been predicted by similar models are likely to suffer from inflated accuracy. Also, much of the data in this database has been marked as ‘not reliable’ by ECHA. As part of a more robust data ecosystem, experimental and predicted data employed by regulatory agencies need to be better structured and properly annotated in order to be easily integrated within informatics systems. It also needs not to be mined from free text present in reports. Another critical issue is the variability or errors in reported units of measurement, especially the use of concentrations or doses in weight rather than in molar units, since the biological effect of a chemical is due to the number of molecules present and not their weight (even though the weight can affect pharmacokinetic properties or dispersion in the environment).15 To provide an example of data harmonisation, the ChEMBL43 database has standardised all its data to nanomolar units.
In vivo test reproducibility is not the only factor that affects the accuracy and utility of in silico approaches as alternatives to animal testing. Recently,42 we have reported the collection, curation and integration of the most extensive publicly available data sets for acute toxicity tests collectively known as the ‘six-pack’ (acute oral toxicity, acute dermal toxicity, acute inhalation toxicity, skin irritation and corrosion, eye irritation and corrosion, and skin sensitisation). The data used in this study were collected from multiple sources, including scientific publications, the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods on behalf of ICCVAM, and the REACH study results database (https://iuclid6.echa.europa.eu/reach-study-results). As a result of curation, the sizes of the data sets were considerably diminished before QSAR model development. Upon inspection, we found many data points initially processed as experimental records to be, in fact, predictions made by either QSAR models, read-across, or expert-based systems. In addition, many experimental data points have been flagged as ‘not reliable’. The ‘shrinkage’ was substantial for most of the data sets that were examined:
for the skin sensitisation endpoint, we reduced the data set from 10,861 records to 1000;
for skin irritation/corrosion, the data set was reduced from 5274 to 1012;
for eye irritation and corrosion, the data set was reduced from 7322 to 3547;
for acute dermal toxicity, from 29,824 to 2622;
for acute inhalation, from 8176 to 681; and
lastly, because acute oral toxicity data was already extensively curated, the data set was reduced from 8994 records to 8495.
More details about the data curation and how many compounds were reduced at each step can be found elsewhere.42 The models were implemented on a freely available web application termed Systemic and Topical Toxicity (STopTox; https://stoptox.mml.unc.edu/).
Case studies
As illustrative examples of the impact of data curation on QSAR modelling accuracy, we describe here two study cases: a direct comparison between modelling the uncurated and curated data for: (i) skin sensitisation endpoint from the REACH database (https://iuclid6.echa.europa.eu/); and (ii) rabbit Draize Skin Irritation/Corrosion data available in the ICE database (https://ice.ntp.niehs.nih.gov/).44 The curated and uncurated data sets for each endpoint were modelled using the same protocol: ECFP4-like circular fingerprints with 2048 bits and an atom radius of 2 (Morgan2) calculated in RDKit (http://www.rdkit.org) along with the Random Forest45 algorithm implemented in KNIME.46 Both data sets were balanced by using undersampling before modelling, in order to equalise the number of toxic and non-toxic compounds. We followed a five-fold external cross-validation procedure to estimate the predictive power of the models.47 The full set of compounds with known experimental activities was divided into five subsets of similar size (external folds), although Random Forest parameters were not extensively tuned for these comparisons. The curated and uncurated data sets and the KNIME workflow for model development are available on GitHub (https://github.com/alvesvm/atla_curation).
The original skin sensitisation data set comprised 10,588 records for 9801 unique chemicals, composed of many types of assays and study categories. Data from different OECD (Organisation for Economic Co-operation and Development) Test Guideline (TG) skin sensitisation assays (TG 406, 411, 429 and 442B)48–50 were available. Only 3309 data points had defined chemical structures, which composed the ‘uncurated set’, containing 818 sensitisers and 2491 non-sensitisers. Next, we curated the data following the best practices widely accepted by the cheminformatics community.20 In the ‘curated set’, in vitro, ‘weight-of-evidence’ categories as well as in vivo studies labelled as ‘disregarded study’ were discarded; only the data corresponding to the Local Lymph Node Assay (LLNA; TG 42950 and 442B51) were retained, resulting in 1275 data points. Upon chemical standardisation, removal of mixtures, inorganics, and the neutralisation and removal of counterions, 532 compounds (187 sensitisers and 345 non-sensitisers) were retained. With these data, we developed binary QSAR models to predict LLNA skin sensitisation outcomes according to the best practices of QSAR modelling.36 The statistical characteristics of binary QSAR models are summarised in Table 1. Although Random Forest parameters were not highly tuned for this comparison, we can observe that models generated with uncurated data showed a 7% higher correct classification rate (CCR), 16% higher specificity, 10% higher positive predictive value (PPV) and 3% higher negative predictive value (NPV). The sensitivity was 2% lower than that for the ‘curated’ data set models. These results show artificially higher model performance when developed from the uncurated set; however, this model performance is deceptive because of the high number of duplicates of the training set compounds found in the external folds of the models.
Table 1.
Statistical characteristics of QSAR models for REACH skin sensitisation LLNA data and rabbit skin irritation data at the 24-hour timepoint, built with uncurated and curated data.
| Skin Sensitisation Model | |||||
| CCR | Sensitivity | PPV | Specificity | NPV | |
|
| |||||
| Uncurated data set | 0.75 | 0.72 | 0.76 | 0.77 | 0.74 |
| Curated data set | 0.68 | 0.74 | 0.66 | 0.61 | 0.71 |
|
Skin Irritation Model | |||||
| CCR | Sensitivity | PPV | Specificity | NPV | |
|
| |||||
| Uncurated data set | 0.87 | 0.94 | 0.92 | 0.79 | 0.84 |
| Curated data set | 0.63 | 0.54 | 0.66 | 0.72 | 0.61 |
CCR = correct classification rate; PPV = positive predictive value; NPV = negative predictive value.
The original rabbit skin irritation/corrosion data set comprised 13,844 records collected from the REACH database. Rooney et al.28 established a curated data set by flagging studies with methodological deviations and curated the data set based on US EPA and the Global Harmonisation System protocols. The final data set comprised 2624 test records, representing 990 chemicals. The data set contains records for several timepoints. For this analysis, we kept the data only for the 24-hour timepoint—thus, 4160 chemicals with defined chemical structure constituted the ‘uncurated set’, comprising 2993 irritants and 1167 nonirritants. The complete protocol for the curation of this data set can be found elsewhere.28 Here, chemicals were standardised, mixtures and inorganics removed, and for the records containing counterions, the molecules were neutralised and counterions were removed. The ‘curated set’ contains 159 compounds (79 irritants and 80 non-irritants). Binary QSAR models to predict rabbit skin irritation were developed following the same protocol for skin sensitisation and the results are also summarised in Table 1. We can observe that models generated with uncurated data showed a 24% higher correct classification rate (CCR), 40% higher sensitivity, 26% higher PPV, 7% higher specificity and 23% higher NPV. The difference between the statistics of models developed with curated and uncurated sets was more pronounced than in the case of skin sensitisation. The models generated with uncurated rabbit skin irritation data also showed an artificially higher performance than the models developed with curated data, which were not predictive. Obviously, the statistics of the models generated with the uncurated set are deceiving, and the use of these models could lead to making less scientifically sound decisions.
Data curation challenges in the era of Big Data
The rapid growth of publicly available toxicology data52 does not obviate the need for careful data curation before model development. Addressing and ensuring data quality and assay reproducibility is the first step in curating chemogenomics data. Several studies have pointed out errors in chemical structures deposited in public databases.35,53 Even a relatively small number of erroneous data points can significantly affect modelling outcomes, especially when assessing the external model accuracy.54 Many investigations have shown that the model’s predictivity is affected by the accuracy of the chemical structure, choice of descriptors, modelling algorithms and data curation.55–57 To help address this issue, Mansouri et al.58 have developed a KNIME workflow to curate and correct errors in the structure and harmonise chemical identity (such as name, CAS number, and any other respective identifiers) by using environmental fate data sets. Gadaleta et al.59 developed a workflow for retrieving chemical structures from several web-based databases. The workflow automatically compares these data and performs structural cleaning. The developers of many publicly accessible databases have made substantial efforts to ensure that chemical structures in their databases are correct. For example, ChemSpider has promoted a crowdsourced collaboration to curate chemical structures derived from public compound databases.60 More recently, the US EPA CompTox Chemistry Dashboard52 performed curation of the substances linked to chemical structures and integrated with other diverse types of publicly available data from multiple sources.
The recent efforts to provide curated chemical databases do facilitate the curation process for modelling purposes. However, it is essential to emphasise that different curation levels may be needed for data storage/management compared to data modelling. Many data points are related to substances that are not, in principle, suitable for regular QSAR modelling, such as mixtures, macromolecules, some organometallics, inorganics and counterions. Although mixtures can be modelled,61 they require unique modelling and model validation techniques. Also, leaving many duplicate compounds in the modelling data sets can lead to artificially high reported accuracies.25 By eliminating duplicates, imputed data, or data evaluated as ‘not reliable’ before embarking on QSAR model development, more reliable predictions with a higher impact on both experimental toxicological studies or regulatory decision support can be achieved.
Considerable advances have been made in implementing the best practices to support both the wider acceptance and understanding of QSAR model-based predictions and their limitations. A decade ago, Fourches et al.54 highlighted major steps for chemical curation, followed by biological data curation steps for QSAR modelling.41 However, chemogenomics data sets have grown, and some of the steps proposed in the original study, such as manual data curation, have become impractical. To this day, many essential aspects of data preparation and modelling are not available in a single standalone program. Data curation pipelines have been implemented in KNIME58,62 and RDKit/Python.63 The recent RDKit pipeline63 helps with curating chemical structures, but it does not address the 3 biological curation essential for QSAR modelling. The KNIME workflow proposed by Neves et al.62 is the first attempt to combine all critical steps of both chemical and biological data curation within a single tool. This workflow helps automate data curation for large data sets, flagging mixtures and conflicting replicates. However, flagged data 7 points still need manual curation, a challenging step to overcome. As data sets grow, losing a few data points might not be critical to the success of QSAR modelling. Still, when working with a small data set, especially when associated with a ‘neglected’ disease (e.g. schistosomiasis64 or leishmaniasis65), every data point counts.
In the papers by Fourches et al.,41,54 the authors describe the first step of chemical curation as the “removal of mixtures and inorganics”. Although this is still valid as the first step, it is worth noting that the current chemical standardisation packages usually blindly remove the smaller fragments. If the smaller components are small organic solvents or counterions, this automatic step is acceptable. However, in the case of mixtures, the user might be keeping the mixture’s datapoint as its largest component. Therefore, we highlight the importance of identifying the mixtures before chemical standardisation. The open-source Indigo’s Component Separator node66 available for KNIME46 can be used to separate chemical components.
When identifying duplicates, SMILES strings should not be used.41,54 InChIKey can be used, but only after the structures are standardised (i.e. all specific chemotypes such as aromatic rings and nitro groups are represented in the same way).67 Two-dimensional chemical descriptors, such as the commonly used Morgan fingerprints68 or MACCS keys,69 do not differentiate enantiomers by default. Different chemicals (stereoisomers) will not be identified as duplicates by SMILES or InChIKey without chemical standardisation and removing the stereocentres. However, stereoisomers will appear as duplicates when using two-dimensional descriptors. In this case, if the experimental activity associated with both chemicals is the same, the model’s predictivity will be overestimated; but if the two compounds have different activity, the model accuracy will decline. If the modeller opts to use two-dimensional descriptors, only one enantiomer should be kept if the biological response agrees. If they disagree, both enantiomers should be removed. Conversely, the user should use descriptors that differentiate enantiomers.
When collecting data from large repositories such as ChEMBL70 or PubChem,71 it is crucial to avoid combining data derived from different experimental protocols. These data should be curated independently and only employed together in a QSAR campaign when a high concordance has been established between both assays. However, data from different protocols can be utilised to develop an in silico framework that hierarchically addresses an endpoint’s prediction, usually with higher accuracy. This is particularly interesting, not only to make better use of all data available for one endpoint, but also to improve a model’s predictive power.
Integrative knowledge-driven experimental design for reducing animal testing
The challenge of reproducibility has been acknowledged in all areas of science.72–77 To employ computational models as a reliable means to reduce animal testing, we need to ensure model transparency and reproducibility. To make computational models transparent and reproducible, all the curated data employed need to be made publicly available following the FAIR (Findability, Accessibility, Interoperability and Reusability) data principles.78 In the field of molecular modelling, this means that the chemical structures and associated data should be made available in a machine-readable format. The data must be well organised and structured in databases (e.g. Integrated Chemical Environment (ICE),44 Tox21,79 ToxCast,80 ChEMBL,70 PubChem71) and made publicly available.81 In addition, all of the scripts used to process or model the data should be available. In the context of the regulatory use of QSAR models, the harmonised template for summarising and reporting key information on QSAR models, namely the QSAR Model Reporting Format (QMRF), is recommended, especially when validation studies are available.
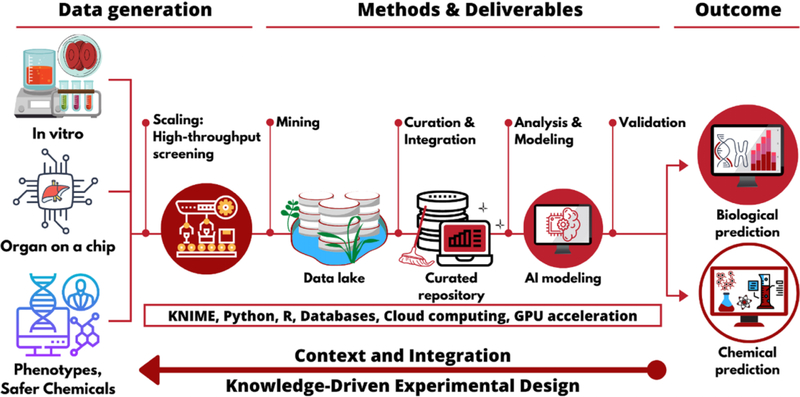
Figure 1 illustrates a knowledge-driven approach to enable highly accurate AI models for chemical toxicity prediction that can be used to reduce or replace animal testing. As more robust in vitro and organ-on-chip data become available, after validation and acceptance for regulatory use, these assays need to be scaled for highthroughput screening. All the data resulting from such screening campaigns, and all of the scripts used to process or model the data, need to be stored and managed, following the FAIR data principles,78 and curated based on the standard approaches described above. Subsequent cheminformatics analysis, as well as generation and validation of AI models, should be carried out following the best practices promoted by the OECD37 and universally accepted by the community. Only then should these models be used to predict biological responses and, potentially, be employed to design safer chemicals.
Figure 1.
Integrative knowledge-driven experimental design for reducing animal testing.
Towards a robust data ecosystem for toxicology
The main goal of this article is to discuss the importance of data curation in QSAR modelling. Data curation is the most critical step in guaranteeing predictive models. We reason that only those models that were built with data processed in strict compliance with mandatory curation protocols should be employed in regulatory toxicology. These models can then be used for the reliable toxicity assessment of compounds lacking experimental data.
In recent years, recognition of the need to develop a research data ecosystem that promotes FAIR data principles has grown, along with resources to foster the development of the ecosystem.82 Contributors, users and developers of this ecosystem must also recognise the critical need for curation and the role of curated data repositories. Data repositories must provide accurate records of deposited data sets, even if this leads to issues for modellers, such as duplicated records. Raising awareness of these issues within the users of these repositories should be emphasised. The deposition of curated data sets within repositories for further future use should also be promoted. Positive steps in this direction already exist, e.g. PubChem and the Chemical Effects in Biological Systems (CEBS) database provide submitted study data, while repositories such as ICE and ToxRefDB are focused on data sets targeted at modellers.
Forming communities of best practice among users of data repositories to discuss, store, share and adopt common curation approaches will not only enable and facilitate data curation for QSAR modelling, but — more importantly — it will also ensure that researchers and regulatory agencies will be able to link and combine different sources of data easily. Successful implementation of a standardised toxicological data ecosystem would create an unprecedented ability to analyse historical data and employ modern data analytics to derive knowledge and generate smarter, faster and safer regulatory policies.
Conclusions
It is exciting that computational alternatives to animal testing have received a high level of media attention.83–85 We support further efforts in this direction. We expect that, as high-quality data accumulates and mechanistic understanding informs new experimental approaches, non-animal — especially computational — models, will reduce or replace animal testing. To achieve this goal, we posit that any new data sets should be generated and managed following FAIR principles, regardless of their size. However, chemogenomics data curation would still be necessary before computational modelling, since — as explained in this article — not all data points are suitable for QSAR modelling. We strongly caution against overinterpreting results from models built on non-curated data sets. Following the editorial requirements implemented by the ACS Journal of Chemical Information and Modeling, we suggest that all scientific journals should include a section on data curation in any manuscript submitted for publication.86 Any computational model must be consistent with the OECD principles for the validation, for regulatory purposes, of (Quantitative) Structure–Activity Relationship models.37 We hope that this article will help establish the mandatory practice of robust data curation in computational toxicology.
Acknowledgements
VMA thanks the Lush Prize. The authors thank Dr Mary S. Wolfe and Dr Stephen S. Ferguson who kindly reviewed an earlier version of this manuscript and provided valuable suggestions and comments.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This study was supported, in part, by the National Institute of Environmental Health Sciences Grant P42 ES027704.
Footnotes
Declaration of conflicting interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship and/or publication of this article: AT and ENM are co-founders of Predictive, LLC, which develops computational methodologies and software for toxicity prediction. All the other authors declare no conflicting interests.
References
- 1.Flecknell P. Replacement, reduction and refinement. ALTEX 2002; 19: 73–78. [PubMed] [Google Scholar]
- 2.Patlewicz G and Fitzpatrick JM. Current and future perspectives on the development, evaluation, and application of in silico approaches for predicting toxicity. Chem Res Toxicol 2016; 29: 438–451. [DOI] [PubMed] [Google Scholar]
- 3.ICCVAM. A strategic roadmap for establishing new approaches to evaluate the safety of chemicals and medical products in the United States, https://ntp.niehs.nih.gov/pub-health/evalatm/natl-strategy/index.html (2018, accessed 27 January 2021).
- 4.US Environmental Protection Agency. EPA directive to prioritize efforts to reduce animal testing, https://www.epa.gov/sites/production/files/2019-09/documents/image2019-09-09-231249.pdf (2019, accessed 15 June 2021).
- 5.Thomas RS, Bahadori T, Buckley TJ, et al. The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency. Toxicol Sci 2019; 169: 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs J and McKeague M. Green toxicology: connecting green chemistry and modern toxicology. Chem Res Toxicol 2020; 33: 2919–2931. [DOI] [PubMed] [Google Scholar]
- 7.Schultz TW, Amcoff P, Berggren E, et al. A strategy for structuring and reporting a read-across prediction of toxicity. Regul Toxicol Pharmacol 2015; 72: 586–601. [DOI] [PubMed] [Google Scholar]
- 8.Muratov EN, Bajorath J, Sheridan RP, et al. QSAR without borders. Chem Soc Rev 2020; 49: 3525–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barratt MD. Prediction of toxicity from chemical structure. Cell Biol Toxicol 2000; 16: 1–13. [DOI] [PubMed] [Google Scholar]
- 10.Myatt GJ, Ahlberg E, Akahori Y, et al. In silico toxicology protocols. Regul Toxicol Pharmacol 2018; 96: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madden JC, Enoch SJ, Paini A, et al. A review of in silico tools as alternatives to animal testing: principles, resources and applications. Altern Lab Anim 2020; 48: 146–172. [DOI] [PubMed] [Google Scholar]
- 12.Greene N and Pennie W. Computational toxicology, friend or foe? Toxicol Res 2015; 4: 1159–1172. [Google Scholar]
- 13.Guha R. The ups and downs of structure–activity landscapes. Methods Mol Biol 2011; 672: 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maggiora GM. On outliers and activity cliffs — why QSAR often disappoints. J Chem Inf Model 2006; 46: 1535. [DOI] [PubMed] [Google Scholar]
- 15.Dearden JC, Cronin MTD and Kaiser KLE. How not to develop a quantitative structure–activity or structure–property relationship (QSAR/QSPR). SAR QSAR Environ Res 2009; 20: 241–266. [DOI] [PubMed] [Google Scholar]
- 16.Hansson SO and Rudén C. Priority setting in the REACH system. Toxicol Sci 2006; 90: 304–308. [DOI] [PubMed] [Google Scholar]
- 17.Ciallella HL and Zhu H. Advancing computational toxicology in the big data era by artificial intelligence: data-driven and mechanism-driven modeling for chemical toxicity. Chem Res Toxicol 2019; 32: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luechtefeld T, Rowlands C and Hartung T. Big-data and machine learning to revamp computational toxicology and its use in risk assessment. Toxicol Res (Camb) 2018; 7: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu X, Ilyas IF, Krishnan S, et al. Data cleaning: overview and emerging challenges. In: Proceedings of the 2016 international conference on management of data, 26 June 2016–1 July 2016, San Francisco, CA, USA. New York, NY: ACM, pp. 2201–2206. [Google Scholar]
- 20.Fourches D, Muratov E and Tropsha A. Curation of chemogenomics data. Nat Chem Biol 2015; 11: 535–535. [DOI] [PubMed] [Google Scholar]
- 21.Pogodin PV, Lagunin AA, Rudik AV, et al. AntiBac-Pred: a web application for predicting antibacterial activity of chemical compounds. J Chem Inf Model 2019; 59: 4513–4518. [DOI] [PubMed] [Google Scholar]
- 22.Gao H, Struble TJ, Coley CW, et al. Using machine learning to predict suitable conditions for organic reactions. ACS Cent Sci 2018; 4: 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luechtefeld T, Marsh D, Rowlands C, et al. Machine learning of toxicological big data enables read-across structure activity relationships (RASAR) outperforming animal test reproducibility. Toxicol Sci 2018; 165: 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaka Y and Uesawa Y. Optimization of a deep-learning method based on the classification of images generated by parameterized deep snap a novel molecular-image-input technique for quantitative structure–activity relationship (QSAR) analysis. Front Bioeng Biotechnol 2019; 7: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alves VM, Borba J, Capuzzi SJ, et al. Oy vey! A comment on “machine learning of toxicological big data enables read-across structure activity relationships outperforming animal test reproducibility.” Toxicol Sci 2019; 167: 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham LL, Watford SM, Pradeep P, et al. Variability in in vivo studies: defining the upper limit of performance for predictions of systemic effect levels. Comput Toxicol 2020; 15: 100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luechtefeld T. Analysis of Draize eye irritation testing and its prediction by mining publicly available 2008–2014 REACH data. ALTEX 2016; 33: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooney JP, Choksi NY, Ceger P, et al. Analysis of variability in the rabbit skin irritation assay. Regul Toxicol Pharmacol 2021; 122: 104920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wignall JA, Shapiro AJ, Wright FA, et al. Standardizing benchmark dose calculations to improve science-based decisions in human health assessments. Environ Health Perspect 2014; 122: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinstreuer NC, Ceger PC, Allen DG, et al. A curated database of rodent uterotrophic bioactivity. Environ Health Perspect 2016; 124: 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.OECD. Hershberger Bioassay in rats (H assay) (OECD TG 441) (including OECD GD 115 on the weanling Hershberger Bioassay). In: Revised Guidance Document 150 on standardised test guidelines for evaluating chemicals for endocrine disruption. Paris: Organisation for Economic Co-operation and Development, 2018, pp. 463–476. [Google Scholar]
- 32.Browne P, Kleinstreuer NC, Ceger P, et al. Development of a curated Hershberger database. Reprod Toxicol 2018; 81: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alves VM, Capuzzi SJ, Muratov E, et al. QSAR models of human data can enrich or replace LLNA testing for human skin sensitization. Green Chem 2016; 18: 6501–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alves VM, Capuzzi SJ, Braga RC, et al. A perspective and a new integrated computational strategy for skin sensitization assessment. ACS Sustain Chem Eng 2018; 6: 2845–2859. [Google Scholar]
- 35.Young D, Martin T, Venkatapathy R, et al. Are the chemical structures in your QSAR correct? QSAR Comb Sci 2008; 27: 1337–1345. [Google Scholar]
- 36.Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol Inform 2010; 29: 476–488. [DOI] [PubMed] [Google Scholar]
- 37.OECD. Guidance Document on the Validation of (Quantitative) Structure–Activity Relationship [(Q)SAR] Models. OECD series on testing and assessment, No. 69. Paris: OECD Publishing, 2014, 154 pp. [Google Scholar]
- 38.Golbraikh A and Tropsha A. Beware of q2! J Mol Graph Model 2002; 20: 269–76. [DOI] [PubMed] [Google Scholar]
- 39.Todeschini R, Ballabio D and Grisoni F. Beware of unreliable Q2! A comparative study of regression metrics for predictivity assessment of QSAR models. J Chem Inf Model 2016; 56: 1905–1913. [DOI] [PubMed] [Google Scholar]
- 40.Alexander DLJ, Tropsha A and Winkler DA. Beware of R2: simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J Chem Inf Model 2015; 55: 1316–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fourches D, Muratov E and Tropsha A. Trust, but verify II: a practical guide to chemogenomics data curation. J Chem Inf Model 2016; 56: 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borba JVVB, Alves V, Braga R, et al. STopTox: an in-silico alternative to animal testing for acute systemic and TOPical TOXicity. ChemRxiv 2020; chemrxiv.13283930.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaulton A, Hersey A, Nowotka M, et al. The ChEMBL database in 2017. Nucleic Acids Res 2017; 45: D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell S, Abedini J, Ceger P, et al. An integrated chemical environment with tools for chemical safety testing. Toxicol In Vitro 2020; 67: 104916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breiman LEO. Random forests. Mach Learn 2001; 45: 5–32. [Google Scholar]
- 46.Berthold MR, Cebron N, Dill F, et al. KNIME: the Konstanz Information Miner. In: Preisach C, Burkhardt H, Schmid-Thieme L, Gaul W, Vichi M, Weihs C, et al. (eds) Studies in classification, data analysis, and knowledge organization. Berlin, Heidelberg: Springer, 2008, pp. 319–326. [Google Scholar]
- 47.Cherkasov A, Muratov EN, Fourches D, et al. QSAR modeling: where have you been? Where are you going to? J Med Chem 2014; 57: 4977–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.OECD. Test No. 406: Skin Sensitisation. OECD Guidelines for the Testing of Chemicals, Section 4. Paris: Organisation for Economic Co-operation and Development, 1992, 9 pp. [Google Scholar]
- 49.OECD. Test No. 411: Subchronic Dermal Toxicity: 90-day Study. OECD Guidelines for the Testing of Chemicals, Section 4. Paris: Organisation for Economic Co-operation and Development, 1981, 9 pp. [Google Scholar]
- 50.OECD. Test No. 429: Skin Sensitisation: Local Lymph Node Assay. OECD Guidelines for the Testing of Chemicals, Section 4. Paris: Organisation for Economic Co-operation and Development, 2010, 20 pp. [Google Scholar]
- 51.OECD. Test No. 442B: Skin Sensitization: Local Lymph Node Assay: BrdU-ELISA or –FCM. OECD Guidelines for the Testing of Chemicals, Section 4. Paris: Organisation for Economic Co-operation and Development, 2018, 15 pp. [Google Scholar]
- 52.Williams AJ, Grulke CM, Edwards J, et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform 2017; 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olah M, Mracec M, Ostopovici L, et al. WOMBAT: world of molecular bioactivity. In: Oprea TI (ed) Chemoinfomatics in drug discovery. New York, NY: Wiley-VCH, 2005, pp. 221–239. [Google Scholar]
- 54.Fourches D, Muratov E and Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminfomatics and QSAR modeling research. J Chem Inf Model 2010; 50: 1189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansouri K, Kleinstreuer N, Abdelaziz AM, et al. CoMPARA: collaborative modeling project for androgen receptor activity. Environ Health Perspect 2020; 128: 027002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mansouri K, Abdelaziz A, Rybacka A, et al. CERAPP: collaborative estrogen receptor activity prediction project. Environ Health Perspect 2016; 124: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu H, Tropsha A, Fourches D, et al. Combinatorial QSAR modeling of chemical toxicants tested against Tetrahymena pyriformis. J Chem Inf Model 2008; 48: 766–784. [DOI] [PubMed] [Google Scholar]
- 58.Mansouri K, Grulke CM, Richard AM, et al. An automated curation procedure for addressing chemical errors and inconsistencies in public datasets used in QSAR modelling. SAR QSAR Environ Res 2016; 27: 939–965. [DOI] [PubMed] [Google Scholar]
- 59.Gadaleta D, Lombardo A, Toma C, et al. A newsemi-automated workflow for chemical data retrieval and quality checking for modeling applications. J Cheminform 2018; 10: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams AJ. Chemspider: a platform for crowdsourced collaboration to curate data derived from public compound databases. In: Ekins S, Hupcey MAZ and Williams AJ (eds) Collaborative computational technologies for biomedical research. Hoboken, NJ: John Wiley & Sons, Inc., 2011, pp. 363–386. [Google Scholar]
- 61.Muratov EN, Varlamova EV, Artemenko AG, et al. Existing and developing approaches for QSAR analysis of mixtures. Mol Inform 2012; 31: 202–221. [DOI] [PubMed] [Google Scholar]
- 62.Neves B, Moreira-Filho J, Silva A, et al. Automated framework for developing predictive machine learning models for data-driven drug discovery. J Braz Chem Soc 2021; 32: 110–132. [Google Scholar]
- 63.Bento AP, Hersey A, Félix E, et al. An open source chemical structure curation pipeline using RDKit. J Cheminform 2020; 12: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neves BJ, Braga RC, Bezerra JCB, et al. In silico repositioning chemogenomics strategy identifies new erugs with potential activity against multiple life stages of Schistosoma mansoni. PLoS Negl Trop Dis 2015; 9: e3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomes MN, Alcântara LM, Neves BJ, et al. Computer-aided discovery of two novel chalcone-like compounds active and selective against Leishmania infantum. Bioorg Med Chem Lett 2017; 27: 2459–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.KNIME. Indigo KNIME integration: KNIME Hub, https://hub.knime.com/epam-lsop/extensions/com.epam.indigo.knime.feature/latest (accessed 16 June 2021).
- 67.Saldívar-González FI, Huerta-García CS and Medina-Franco JL. Chemoinformatics-based enumeration of chemical libraries: a tutorial. J Cheminform 2020; 12: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.RDKit. Morgan Fingerprints, http://rdkit.org/docs/GettingStartedInPython.html#morgan-fingerprints-circular-fingerprints (2020, accessed 16 June 2021).
- 69.RDKit. Module MACCSkeys, http://rdkit.org/Python_Docs/rdkit.Chem.MACCSkeys-module.html (accessed 15 June 2021).
- 70.Mendez D, Gaulton A, Bento AP, et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 2019; 47: D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Suzek T, Zhang J, et al. PubChem bioassay: 2014 update. Nucleic Acids Res 2014; 42: D1075–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anon. Editorial: The long road to reproducibility. Nat Cell Biol 2015; 17: 1513–1514. [DOI] [PubMed] [Google Scholar]
- 73.Anon. Editorial: Facilitating reproducibility. Nat Chem Biol 2013; 9: 345. [DOI] [PubMed] [Google Scholar]
- 74.Anon. Editorial: Journals unite for reproducibility. Nature 2014; 515: 7. [DOI] [PubMed] [Google Scholar]
- 75.Collins FS and Tabak LA. Policy: NIH plans to enhance reproducibility. Nature 2014; 505: 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller GW. Improving reproducibility in toxicology. Toxicol Sci 2014; 139: 1–3. [DOI] [PubMed] [Google Scholar]
- 77.Waller LA and Miller GW. More than manuscripts: reproducibility, rigor, and research productivity in the big data era. Toxicol Sci 2016; 149: 275–276. [DOI] [PubMed] [Google Scholar]
- 78.Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016; 3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richard AM, Huang R, Waidyanatha S, et al. The Tox21 10K compound library: collaborative chemistry advancing toxicology. Chem Res Toxicol 2021; 34: 189–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richard AM, Judson RS, Houck KA, et al. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 2016; 29: 1225–1251. [DOI] [PubMed] [Google Scholar]
- 81.Merz KM, Amaro R, Cournia Z, et al. Editorial: Method and data sharing and reproducibility of scientific results. J Chem Inf Model 2020; 60: 5868–5869. [DOI] [PubMed] [Google Scholar]
- 82.NIH. NIH strategic plan for data science, https://datascience.-nih.gov/nih-strategic-plan-data-science (2018, accessed 16 June 2021).
- 83.Van Noorden R. Software beats animal tests at predicting toxicity of chemicals. Nature 2018; 559: 163–163. [DOI] [PubMed] [Google Scholar]
- 84.Chakravarti D. Computerized chemical toxicity prediction beats animal testing, https://www.scientificamerican.com/-podcast/episode/computerized-chemical-toxicity-prediction-beats-animal-testing/ (2018, accessed 16 June 2021).
- 85.Hartung T. Artificial intelligence outperforms the repetitive animal tests in identifying toxic chemicals, https://phys.org/news/2018-07-artificial-intelligence-outperforms-repetitive-animal.html (2018, accessed 16 June 2021).
- 86.Merz KM, Rarey M, Tropsha A, et al. Letter from the editors. J Chem Inf Model 2015; 55: 719–720. [DOI] [PubMed] [Google Scholar]