Abstract
The winged-helix (WH) BF-1 gene, which encodes brain factor 1 (BF-1) (also known as foxg1), is essential for the proliferation of the progenitor cells of the cerebral cortex. Here we show that BF-1-deficient telencephalic progenitor cells are more apt to leave the cell cycle in response to transforming growth factor β (TGF-β) and activin. We found that ectopic expression of BF-1 in vitro inhibits TGF-β mediated growth inhibition and transcriptional activation. Surprisingly, we found that the ability of BF-1 to function as a TGF-β antagonist does not require its DNA binding activity. Therefore, we investigated whether BF-1 can inhibit Smad-dependent transcriptional responses by interacting with Smads or Smad binding partners. We found that BF-1 does not interact with Smads. Because the identities of the Smad partners mediating growth inhibition by TGF-β are not clearly established, we examined a model reporter system which is known to be activated by activin and TGF-β through Smads and the WH factor FAST-2. We demonstrate that BF-1 associates with FAST-2. This interaction is dependent on the same region of protein which mediates its ability to interfere with the antiproliferative activity of TGF-β and with TGF-β-dependent transcriptional activation. Furthermore, the interaction of FAST-2 with BF-1 is mediated by the same domain which is required for FAST-2 to interact with Smad2. We propose a model in which BF-1 interferes with transcriptional responses to TGF-β by interacting with FAST-2 or with other DNA binding proteins which function as Smad2 partners and which have a common mode of interaction with Smad2.
The neocortex develops from the progenitor cells of the rostral neural plate, the telencephalic neuroepithelial cells. Following a period of uniform proliferation, cerebral cortical progenitors generate neurons asynchronously beginning at E11 in the mouse. The progenitors undergo asymmetric cell divisions in which one daughter cell leaves the cell cycle to differentiate while the other continues to divide. The fraction of cells with asymmetric divisions increases steadily over a period of several days. Toward the end of the neurogenetic period (E17), both daughter cells differentiate, resulting in depletion of the progenitor pool (2, 32). The duration of the neurogenetic period is an important determinant of the number of neurons generated within the cerebral cortex. Thus, the mechanisms which regulate the proliferation of progenitor cells and the timing of their withdrawal from the cell cycle are of central importance in the development of the brain.
The progenitor cells of the telencephalon are identifiable as early as the eight-somite stage (E8.5) by the expression of the Winged-Helix (WH) protein brain factor 1 (BF-1) (also known as foxg1) (11, 29). WH proteins are a family of putative transcriptional regulators with diverse roles in development, characterized by a highly conserved DNA binding structure, the WH domain (14, 15). We have previously shown that the BF-1 gene plays a critical role in the development of the cerebral hemispheres of the brain. Targeted disruption of the BF-1 gene in mice leads to severe defects in the development of telencephalic structures, e.g., the cerebral cortex and basal ganglia. The loss of BF-1 results in an accelerated rate of neuronal differentiation and the shortening of the neurogenetic period in the embryonic cerebral cortex (37). Although BF-1 is expressed by E8.5 in telencephalic progenitors, the disruption of the BF-1 gene has no apparent effect on the behavior of cerebral cortical progenitor cells until about E11.5, after neuronal differentiation has begun. These observations suggested that BF-1 may regulate the response of cerebral cortical progenitors to environmental cues which act at this stage in development to control their withdrawal from the cell cycle.
Transforming growth factor β (TGF-β) and related peptides inhibit the proliferation of many types of epithelial cells in the embryo and are present in the developing brain during the period of neurogenesis (6, 7, 26). TGF-β ligands signal through cell surface receptor kinases, which phosphorylate cytoplasmic Smad proteins. Receptor-specific Smad proteins (24) associate with Smad4, translocate to the nucleus, and direct transcriptional activation by interacting with a DNA binding partner. For the activin-responsive genes Mix.2 and goosecoid, these partners have been identified as the WH proteins FAST-1 (foxh1) and FAST-2 (foxh2), respectively (3, 18, 21). While a number of other DNA binding partners of Smad proteins have been discovered, the identities of the partners which mediate most TGF-β responses remain unknown.
We find that isolated telencephalic progenitor cells from BF-1−/− mutant embryos have an increased sensitivity to growth inhibition by TGF-β and activin compared with cells from their normal littermates. BF-1−/− mutant embryos yield a limited number of neuroepithelial cells, making biochemical studies impractical. To investigate the mechanisms by which BF-1 regulates cellular responses to TGF-β, we developed cell lines with inducible expression of BF-1 and used an in vitro transcriptional reporter system in these cells. The mink lung epithelial cell line Mv1Lu was selected because the TGF-β signal transduction pathway is well characterized in this line. Using this model system, we found that BF-1 antagonizes the antiproliferative activity of TGF-β and inhibits TGF-β-dependent transcriptional activation. Unexpectedly, we discovered that the DNA binding activity of BF-1 is not required for these functions, raising the possibility that BF-1 might act by interacting with components of the TGF-β signaling pathway. We provide evidence that BF-1 can form a complex in the cell with the Smad partner FAST-2. Studies to characterize the functional regions of the BF-1 protein revealed a common domain which is required to antagonize the antiproliferative activity of TGF-β, to inhibit TGF-β-dependent transcriptional activation, and to interact with FAST-2. These observations, together with the identification of the domain in the FAST-2 protein which mediates its interaction with BF-1, lead to a model in which BF-1 interferes with multiple TGF-β responses by associating with DNA binding proteins which function as Smad partners.
MATERIALS AND METHODS
Constructs.
Site-directed mutagenesis (pALTER; Promega) of the mouse BF-1 cDNA (33) was used to create a PvuII restriction site at the beginning of the translated sequence, permitting the insertion of BF-1 into the expression vectors pFlag-CMV2 (Eastman Kodak) and Myc-tagged CS2 vector, beginning with the second amino acid. The BF-1(NH-AA) mutant (see Results) was generated by site-directed mutagenesis, creating a novel StuI site. Mutated plasmids were completely sequenced. Myc-tagged FAST-2 constructs were prepared by inserting FAST-2 cDNA clones 1.2 (encoding amino acids 9 to 401) and 12.1 (encoding amino acids 52 to 401) into the CS2 vector downstream of Myc epitopes (21). A3-luc reporter constructs were kindly provided by M. Whitman (3, 4). All other constructs have been described previously (12, 22, 35).
Isolation and culture of primary neuroepithelial cells.
The genetic background of the mice used in these studies (C57BL6) differs from that of the mice used in earlier studies (mixed 129 and C57BL6). While no significant differences are noted in the brain phenotype, the gestation period of the mice is 1 day longer, with mice being born at E19.5 instead of E18.5. This is associated with slower development of the embryos. Embryos at E13.5 are comparable in size and developmental stage to those previously obtained at E12.5, while embryos at E10.75 are equivalent to those previously obtained at E10.
Telencephalic neuroepithelial cells were isolated from E10.75 mouse embryos by a method modified from that of Kilpatrick and Bartlett (16). Embryos from BF-1+/− × BF-1+/− heterozygote matings were dissected in ice-cold phosphate-buffered saline. The epidermis of the head was removed, and the head was treated with pancreatin-trypsin on ice for 30 to 45 min. The mesenchyme was separated from the neural tube, and the neuroepithelium of the telencephalon was dissected. The isolated neuroepithelium was treated with 0.025% trypsin and 0.001% DNase I for 7 min to dissociate the cells. The cells were washed with Dulbecco's modified Eagle's medium (DMEM)–10% fetal bovine serum (FBS) followed by DMEM-F12 (1:1) supplemented with 2 mM l-glutamine, 6 mg of glucose per ml, N2 supplements, and 1% FBS. They were then plated onto poly-l-lysine- and laminin-coated 48-well plates at 10,000 to 40,000 cells/well in DMEM-F12 supplemented with 20 ng of fibroblast growth factor 2 per ml of FGF-2 and TGF-β or bone morphogenetic protein 4 (BMP4) at 100 pM as indicated in duplicate wells.
Luciferase and β-gal assays.
Mv1Lu cells were transfected with Lipofectamine (GIBCO BRL) and treated with 100 pM TGF-β1 (R&D Systems) for 18 to 24 h. Luciferase activity was measured with a luciferase assay kit (Promega), and β-galactosidase (β-gal) activity was measured with a chemoluminescence detection kit from Tropix. In some cases, cell lysates were also analyzed by Western blotting to check the expression of transfected vectors under different conditions.
RT-PCR.
Total RNA from neuroepithelium or cell lines was prepared with Tri-reagents and reverse transcribed with random hexamers and Superscript II reverse transcriptase (GIBCO-BRL). TGF-β receptor mRNAs were amplified with the primer pairs 5′-GTC CGC AGC TCC TCA TCG TGT TG-3′ and 5′-GGT GGT GCC CTC TGA AAT GAA AG-3′ for TGFβRI and 5′-CCC GGG GCA TCG CTC ATC TC-3′ and 5′-AAT TTC TGG GCG CCC TCG GTC TCT-3′ for TGFβRII. Glyceraldehyde-3-phosphate dehydrogenase was amplified using the primers 5′-GTG GCA AAG TGG AGA TTG TTG CC-3′ and 5′-GAT GAT GAC CCG TTT GGC TCC-3′, and activin receptor type IIB was amplified using 5′-TCC CTA CGG CCA TGT GGA CAT CCA-3′ and 5′-ATG CAG GTA TGA GAG GCC TCG TGA-3′. Amplification was performed for 30 cycles.
Generation of BF-1-inducible Mv1Lu cell lines.
To construct a vector in which the BF-1 coding sequence is under the control of the tetracycline operator, the SstI-StuI fragment of BF-1 cDNA clone mN3 (33), which encodes the full-length BF-1 protein, was inserted to the XbaI site of pUHD-10-3-hygromycin plasmid (28). The pUHD-10.3-hygromycin–BF-1 plasmid was transfected with Lipofectamine into Mv1Lu (14tTA) cells, in which the tTA expression vector is stably integrated. The transfected cells were cultured in selection medium containing 1 mg of G418 (GIBCO BRL) per ml and 0.3 mg of hygromycin (Boehringer Mannheim) per ml. The medium was changed every 2 days for 2 weeks. The colonies were ring-cloned, and each colony was analyzed by Western blotting with polyclonal antibody against the BF-1 N terminus (BNF1; 1:1,000). Of 32 clones we analyzed, 8 had high levels of BF-1 expression, 10 had moderate levels of BF-1, and 14 had undetectable BF-1 expression. Clone 8 is one of the high-level expressers. A mutant form of BF-1, BF-1(NH-AA), and several Flag-tagged BF-1 constructs were also transfected into 14tTA cells to yield various stable cell lines. Lines with comparable protein expression as monitored by Western blotting with anti-Flag antibody were selected for further studies.
Mv1Lu cells were maintained in minimal essential medium supplemented with 2 μg of tetracycline per ml and 10% FBS along with antibiotics and l-glutamine. COS1 cells were cultured in high-glucose DMEM (DMEM HG) supplemented with 10% FBS, l-glutamine, and antibiotics (excluding tetracycline).
[3H]thymidine incorporation assay. (i) Mv1Lu cells.
Cells in 24-well plates were labeled with [3H]thymidine (2 μCi/well) (Amersham) in serum-free medium for 2 h. At the end of the labeling step, the cells were washed with phosphate-buffered saline and lysed in 0.5 ml of 0.5% sodium dodecyl sulfate. Cell lysates were mixed with an equal volume of 20% cold trichloroacetic acid and left on ice for at least 30 min. The mixture was filtered through a fiberglass filter and washed sequentially with 10% trichloroacetic acid and 95% ethanol. Filters were then dried and counted in 5 ml of EconoFluor 2 (DuPont) scintillation fluid. Counts from triplicate wells were averaged and plotted.
(ii) Primary neuroepithelial cells.
Cells in a 48-well plate were cultured for 18 h and then labeled with [3H]thymidine (2 μCi/well in 200 μl) in growth medium for 6 h. The cells were washed and lysed as described above. Counts from duplicate wells were averaged and plotted.
Contact release assay.
Mv1Lu cells were cultured to confluence and then maintained at confluence for another 5 days to achieve quiescence in the presence of 2 μg of tetracycline per ml. The cells were then replated at a 1:5 ratio to release them from contact inhibition. [3H]thymidine incorporation was measured 15 h after replating. When added, TGF-β1 at a final concentration of 100 pM was supplied at the time of replating. Induction of BF-1 was achieved by withdrawal of tetracycline 48 h prior to replating.
T2 RNase protection assay.
Total RNA was prepared from BF-1-induced and uninduced Mv1Lu cells by using Tri-reagent as specified by the manufacturer (Molecular Research Center, Inc.). Total RNA (10 μg) in 100 μl of 70% ethanol was mixed with 32P-labeled p15ink4b riboprobe (109 cpm/μg) and 3,000 cpm of low-specific-activity (107 cpm/μg) mouse glyceraldehyde-3-phosphate dehydrogenase (Ambion) internal control riboprobe. The mixture was precipitated with 70% ethanol and redissolved in 25 μl of 80% formamide 1× hybridization buffer [40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4), 400 mM NaCl, 1 mM EDTA]. Hybridization was performed by first denaturing the mixture at 80°C for 10 min and then hybridizing it overnight at 60°C. At the end of the hybridization, 300 μl of T2 RNA endonuclease (GIBCO BRL) digestion buffer (50 mM sodium acetate [pH 4.6], 100 mM NaCl, 2 mM EDTA) was added to the hybrid mixture, which was then incubated for 2 h at 30°C. The digested products were ethanol precipitated, denatured, and resolved in a 7 M urea–6% polyacrylamide gel.
Gel mobility shift assay.
Proteins comprising the BF-1 binding domain (BD) and BF-1 BD mutation (NH-AA) were made by in vitro translation using reticulocyte lysates (GIBCO BRL). The DNA binding assay was performed with 1 ng of radiolabeled S2 probe essentially as described previously (33).
Immunoprecipitation and immunoblotting.
COS1 cells were cotransfected with various Flag- or Myc-tagged expression vectors by the DEAE-dextran method. Cells receiving TGF-β treatment were also transfected with a constitutively active TGF-β receptor, TβR-I (T204D) (35). At 40 to 48 h after transfection, the cells were treated with low-serum medium (DMEM-HG plus 0.2% FBS), plus or minus 0.5 nM TGF-β, for 1 h and then lysed in 1 ml of TNE buffer (10 mM Tris [pH 8.0], 0.15 M NaCl, 1 mM EDTA, 1% NP-40) plus protease inhibitors. Cell lysates were precleared with protein A- and G-coupled agarose beads and incubated with Myc (9E10; Santa Cruz Biotechnology) or M2 Flag (Eastman Kodak) monoclonal antibodies for 3 h. Immunoprecipitates and aliquots of cell lysates before immunoprecipitation were separated on sodium dodecyl sulfate–7 or 12% polyacrylamide gel and transferred to an Immobilon-P membrane. The membrane was then probed with Flag (0.8 μg/ml) or Myc (50 ng/ml) antibody and incubated with horseradish peroxidase-conjugated goat anti-mouse antibody and detected by chemiluminescence (Pierce).
Immunodetection of retinoblastoma protein (Rb) was performed with anti-Rb monoclonal antibody G3-245 (1 μg/ml; Pharmingen) and horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Pierce) and chemiluminescence detection (Pierce).
RESULTS
Telencephalic progenitor cells lacking BF-1 are more responsive to growth inhibition by TGF-β and activin.
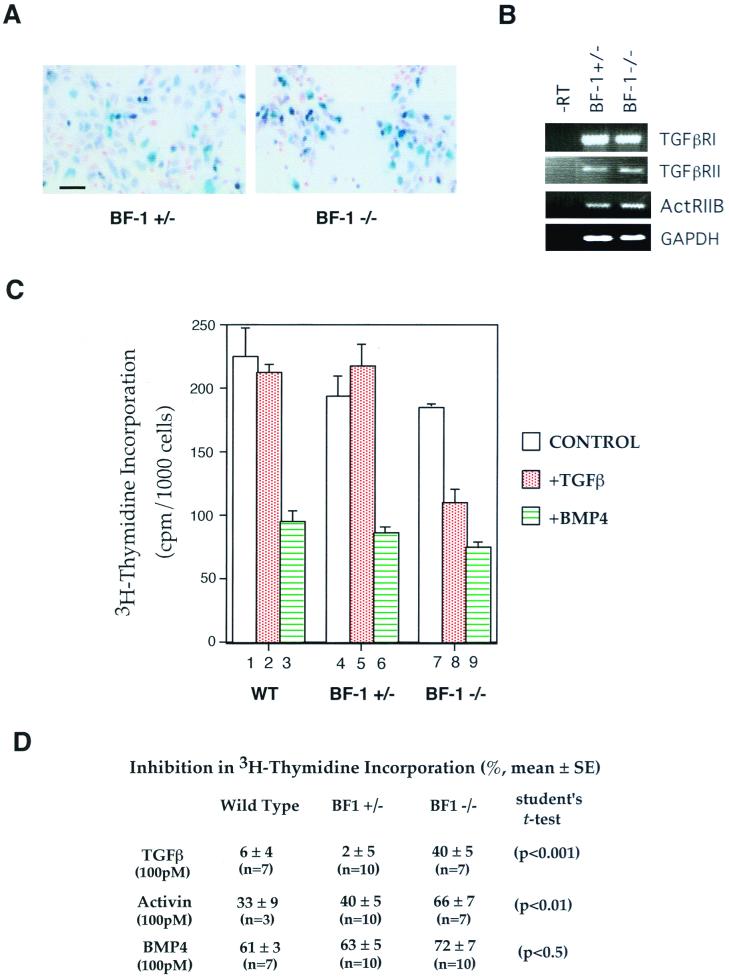
To test the possibility that BF-1 regulates the response of neural progenitors to extracellular signals, we examined [3H]thymidine incorporation in cells isolated from the dorsal telencephalon of E10.75 BF-1−/− mutant embryos and littermates. Cells from embryos at E10.75 were selected for study because at this age the BF-1−/− mutants are indistinguishable from wild-type (WT) and BF-1+/− heterozygous embryos. The dorsal telencephalic neuroepithelium was dissected, separated from the adjacent mesenchyme, dissociated into single cells, and plated. We routinely obtained populations of cells in which >80% were derived from the telencephalon, as monitored by staining for β-gal activity in the BF-1+/− heterozygote and the BF-1−/− mutant (Fig. 1A). Cells from these embryos expressed β-gal under the control of the BF-1 promoter. Therefore, the β-gal staining shows that the cells are of telencephalic origin. The variability in staining intensity between cells may reflect the gradient of BF-1 expression within the telencephalon. The higher overall staining intensity in the mutant cells can be attributed to the fact that each cell has two copies of the β-gal gene whereas the heterozygous cells have only one copy.
FIG. 1.
Telencephalic neuroepithelial cells from BF-1 homozygous mutant embryos are more sensitive to growth inhibition by TGF-β. (A) Neuroepithelial cells isolated from E10.75 BF-1+/− heterozygous and BF-1−/− mutant embryos were cultured for 24 h and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and nuclear fast red. Cells staining blue are telencephalic neuroepithelial cells with activated BF-1 promoter. Strong and weak staining indicates cells with high and low BF-1 promoter activity, respectively. Bar, 25 μm. (B) RT-PCR for TGF-β receptor type I and II and activin receptor type IIB from RNA isolated from neuroepithelial cells. (C) Neuroepithelial cells isolated from WT, BF-1+/− and BF-1−/− embryos were cultured in the presence or absence of TGF-β or BMP4 (both at 100 pM) for 24 h. Control medium included 1% FBS and 20 ng of FGF-2 per ml. [3H]thymidine was added during the last 6 h, and the amount of [3H]thymidine incorporated was determined as described in Materials and Methods. The [3H]thymidine counts from duplicate wells of a representative experiment were normalized to the cell number. (D) The inhibition of [3H]thymidine incorporation by TGF-β, activin, or BMP4 (mean ± standard error) is reported.
FGF-2 promotes the survival and proliferation of neuroepithelial cells (16). [3H]thymidine incorporation is similar in WT and in BF-1+/− heterozygotes and BF-1−/− homozygous mutants in telencephalic progenitors in the presence of both FGF-2 and 1% FBS (Fig. 1C, columns 1, 4, and 7). Very little [3H]thymidine incorporation was observed when cells were cultured in media with FGF-2 or serum alone (data not shown). These results show that at E10.75, the loss of BF-1 function does not substantially alter the ability of the cerebral cortical progenitors to proliferate in response to growth factors. Next, we investigated the response of cerebral cortical progenitors to TGF-β, activin, and BMP4. We found that telencephalic progenitor cells from WT and BF-1+/− heterozygotes were not growth inhibited by TGF-β (Fig. 1C and D) while those from BF-1−/− mutant embryos showed a 40% inhibition of [3H]thymidine incorporation in response to TGF-β. Activin also had a greater antiproliferative activity on mutant cells, reducing [3H]thymidine incorporation by 66% versus 33 to 40% in progenitor cells isolated from normal littermates (Fig. 1D). By comparison, BMP4 inhibited [3H]thymidine incorporation to a similar level in all three populations of cells (Fig. 1C and D). We showed by RT-PCR that receptors for TGF-β and activin were present in the telencephalic neuroepithelium at this stage in development in WT as well as BF-1−/− mutant embryos (Fig. 1B).
BF-1 antagonizes TGF-β-mediated growth arrest in cultured cells.
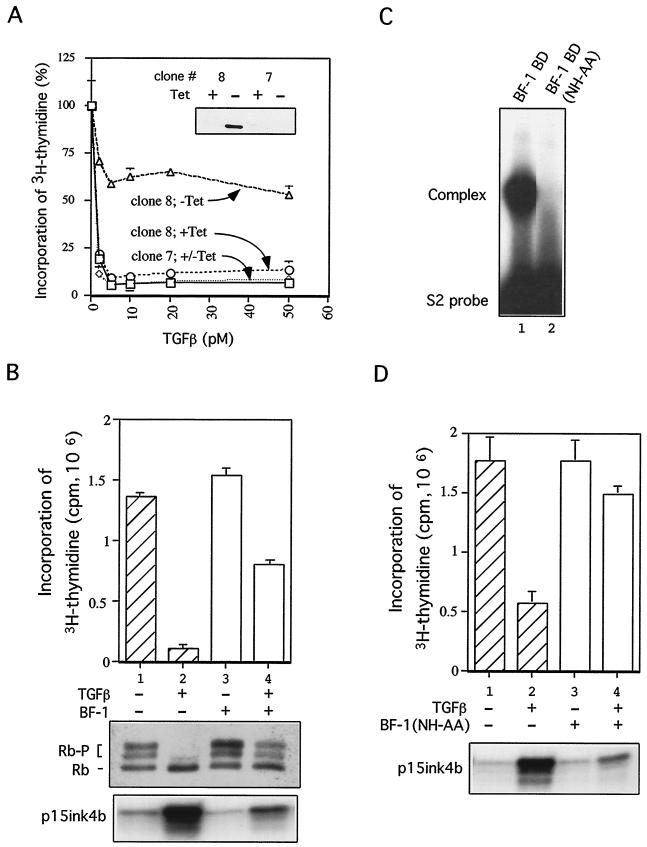
To facilitate studies of how BF-1 controls cellular responses to TGF-β, we developed a model system, a mink lung epithelial cell line (Mv1Lu) with tetracycline transactivator (9)-inducible expression of BF-1 (clone 8). Another line (clone 7), which does not express ectopic BF-1, was used as a control. Expression of BF-1 did not alter the growth rate (data not shown), indicating that ectopic BF-1 does not directly stimulate cell proliferation and is not toxic to the cells. However, BF-1 expression resulted in reduced responsiveness to growth inhibition by TGF-β (Fig. 2A). BF-1 expression also overcame the ability of TGF-β to block reentry into the cell cycle in cells released from contact inhibition (Fig. 2B, upper panel). Growth inhibition by TGF-β is associated with its ability to block Rb hyperphosphorylation (19). We observed that ectopic expression of BF-1 in Mv1Lu cells resulted in the hyperphosphorylation of Rb even when these cells were exposed to TGF-β (Fig. 2B, middle panel).
FIG. 2.
(A) BF-1 blunts the growth-inhitory activity of TGF-β. (Inset) Western blot analysis with anti-BF-1 antibody. BF-1 is expressed in clone 8 (lane 2) upon tetracycline withdrawal but not in clone 7 (lane 4). Exponentially growing cells not expressing BF-1 are growth inhibited by TGF-β. Induction of BF-1 (clone 8, −Tet) results in a reduced response to TGF-β. [3H]thymidine incorporation in cells exposed to TGF-β is expressed as a percentage of that in cells not exposed to TGF-β. (B) [3H]thymidine incorporation in cells released from contact inhibition in the presence or absence of TGF-β is shown in the top panel. Cells were cultured for 5 days at contact density to achieve quiescence. BF-1 expression was induced by withdrawal of tetracycline for 48 h prior to replating cells with or without TGF-β (100 pM) for 15 h. When BF-1 is expressed, [3H]thymidine incorporation is increased 10-fold (compare lanes 2 and 4). Hyperphosphorylation of Rb in BF-1-expressing cells treated with TGF-β is shown in the middle panel. Western blot analysis with anti-Rb antibody was performed to determine the amount of hyperphosphorylated (Rb-P) and hypophosphorylated (Rb) forms of Rb in these cells. BF-1 overcomes inhibition of Rb phosphorylation by TGF-β (lanes 2 and 4); an RNase protection assay for p15ink4b is shown in the bottom panel. Induction of p15ink4b by TGF-β is reduced in the presence of BF-1 (lane 4). (C) A 2-amino-acid substitution in the WH domain of BF-1 disrupts DNA binding activity. The DNA binding domain (WH domain) of BF-1 or the WH domain with the NH-AA mutation was expressed by translation in reticulocyte lysates. A gel mobility shift assay demonstrates high-affinity binding of the WT BF-1 protein to the S2 double-stranded oligonucleotide (lane 1). This binding activity is abolished by the NH-AA mutation (lane 2). (D) BF-1(NH-AA) antagonizes TGF-β-mediated cell cycle arrest (top panel). Cells cultured with tetracycline to repress BF-1(NH-AA) are inhibited from reentering S phase by TGF-β upon replating at low density (lanes 1 and 2). When BF-1(NH-AA) expression is induced, TGF-β is unable to inhibit [3H]thymidine incorporation. BF-1(NH-AA) inhibits the induction of p15ink4b expression by TGF-β (bottom panel). An RNase protection assay for p15ink4b expression is shown. TGF-β induction of p15ink4b expression is reduced in the presence of BF-1(NH-AA) (compare lanes 2 and 4).
A mutation of BF-1 which abolished DNA binding does not alter its ability to antagonize TGF-β.
Because BF-1 has previously been shown to function as a transcriptional repressor (20), we tested whether DNA binding activity was essential for inhibiting the activity of TGF-β. Based on the structure of HNF-3γ complexed with DNA (5), we designed a mutation in the WH domain of BF-1 of two residues, N165 and H169, predicted to be involved in critical contacts with DNA. Mutation of these two amino acids to alanine (NH-AA mutant) abolished DNA binding activity in reticulocyte lysates expressing the mutant binding domain (Fig. 2C).
To examine the role of DNA binding in BF-1 function, we generated an Mv1Lu cell line in which BF-1 containing the NH-AA mutation, BF-1(NH-AA), was expressed under the control of the tetracycline transactivator. The mutant BF-1 protein was expressed at levels comparable to the expression of WT BF-1 in clone 8 (described above) but lacked DNA binding activity (data not shown). We found that the mutant protein is active in antagonizing TGF-β activity. Expression of BF-1(NH-AA) promoted entry into S phase in the presence of TGF-β (Fig. 2D, upper panel, lane 4), suggesting that BF-1 interferes with TGF-β activity through a mechanism independent of DNA binding.
BF-1 inhibits TGF-β-dependent gene expression.
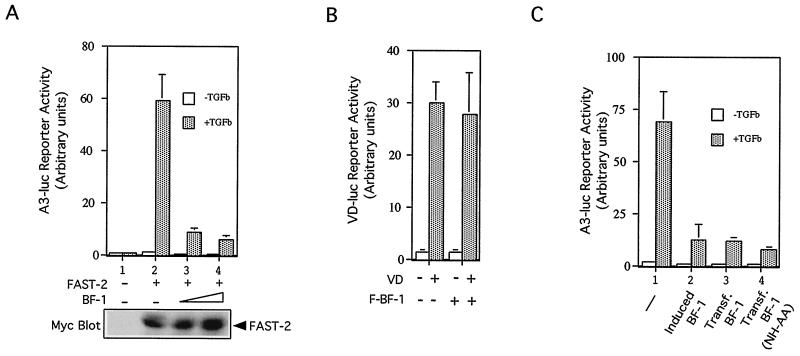
To determine whether BF-1 interferes with TGF-β-dependent gene expression, we examined the effect of ectopic BF-1 on the induction of the cyclin-dependent kinase (CDK) inhibitor p15. Increased expression of p15 in response to TGF-β results in the inhibition of CDK activity and blockade of Rb phosphorylation by G1 cyclin-dependent kinases (10, 28). In Mv1Lu cells expressing BF-1 (Fig. 2B, lower panel) and BF-1(NH-AA) (Fig. 2D, lower panel), induction of p15 mRNA levels by TGF-β was inhibited. The regulation of the p15 promoter is not well understood. Therefore we examined a well-characterized TGF-β-responsive promoter, the A3-luc reporter, as a model to gain further insight into how BF-1 may be interfering with TGF-β activity. A3-luc has previously been shown to be activated in Mv1Lu cells by TGF-β or activin in a Smad2-Smad4- and FAST-2-dependent manner (4, 22). We found that BF-1 inhibited TGF-β-induced transcriptional activation of the A3-luc reporter gene by about 75 to 85% without inhibiting the expression of FAST-2 (Fig. 3A). The BF-1(NH-AA) mutant also blocked the activation of the A3-luc reporter (Fig. 3C).
FIG. 3.
Transcriptional activation by TGF-β is inhibited by BF-1. (A) The A3-luc reporter construct, Rous sarcoma virus (RSV)–β-gal, and Myc-FAST-2 were cotransfected into Mv1Lu cells with or without BF-1 expression vector. The cells were treated with 100 pM TGF-β for 24 h before being harvested for luciferase and β-gal assays. Luciferase activity was normalized to cotransfected RSV-β-gal expression. The mean and standard error from duplicate wells is plotted. Very little luciferase activity is detected in the absence of FAST-2 (lane 1). FAST-2 causes a 40-fold transcriptional activation by TGF-β (lane 2). Increasing amounts of cotransfected BF-1 (50 and 100 ng) inhibit FAST-2-mediated A3 luc reporter expression (lanes 3 and 4). In the lower panel, cell lysates from the same experiment were subjected to Western blotting with anti-Myc antibody to monitor the expression levels of FAST-2. (B) BF-1 does not inhibit VD-luc reporter expression. VD-luc reporter, RSV-β-gal, and VD receptor were cotransfected into Mv1Lu cells in the presence or absence of BF-1. The cells were treated with VD (10 nM) or left untreated for 24 h before being harvested. Data were analyzed and plotted in the same way as described for panel A. (C) Inhibition of TGF-β- and FAST-2-dependent activation of A3-luc by levels of BF-1 which do not interfere with cell proliferation (see also Fig. 2B). Clone 8 (Fig. 2A) Mv1Lu cells were cultured with tetracycline (lanes 1, 3, and 4) or without tetracycline to induce BF-1 expression (lane 2). Inhibition by induced BF-1 is comparable to that achieved by cotransfection with an expression plasmid for WT BF-1 (lane 3) or BF-1(NH-AA) (lane 4).
In our studies with transfected cells, we limited the levels of expressed BF-1 so that transcription was not globally repressed. Under these conditions, BF-1 did not inhibit either the basal or vitamin D (VD)-dependent transcriptional activation of a VD receptor-responsive promoter (Fig. 3B). In contrast, BF-1 was observed to reduce expression from the A3-luc reporter by 25 to 40% in the absence of TGF-β. This effect was dependent on the presence of the FAST-2 response elements (Fig. 3A and C). We also found that the induced BF-1 levels achieved by tetracycline withdrawal in clone 8 Mv1Lu cells are sufficient to inhibit A3-luc activation by TGF-β (Fig. 3C, lanes 2 and 3). These levels of BF-1 did not interfere with normal cell growth, indicating that transcription is not globally inhibited in these stably transfected cells. Taken together, these results suggest that inhibition of TGF-β-dependent transcriptional activation by BF-1 is mediated through a specific mechanism.
BF-1 associates with FAST-2.
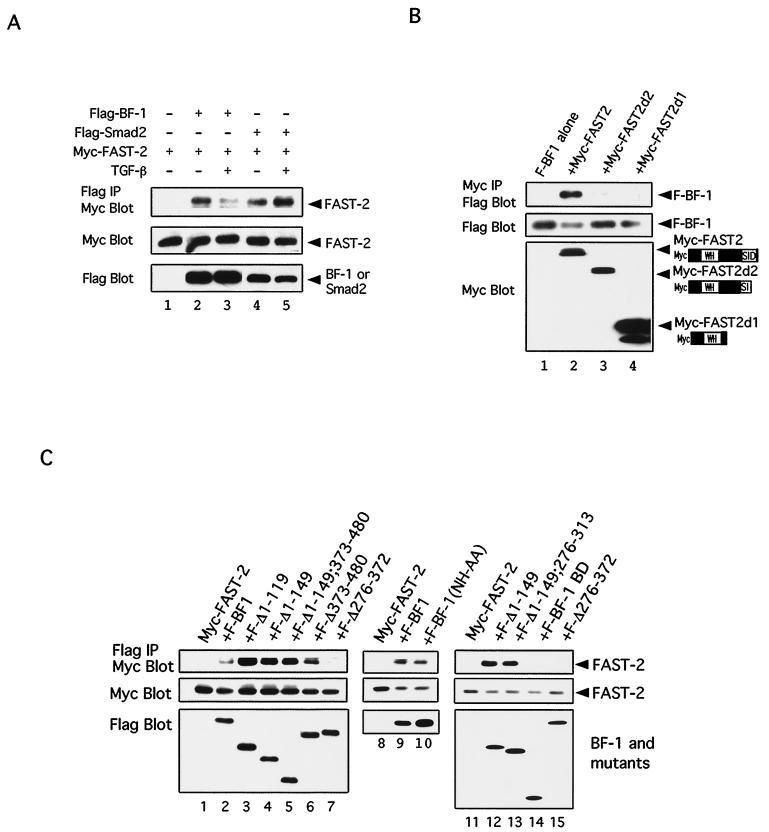
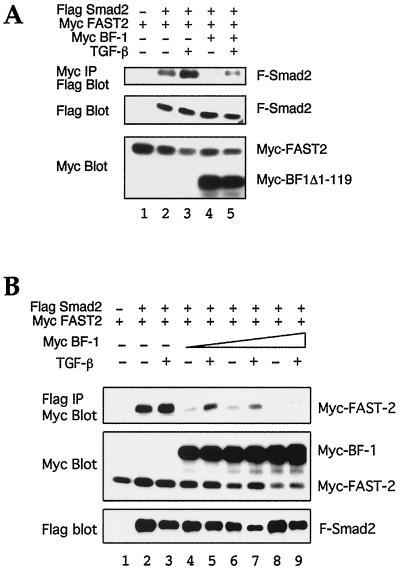
To investigate DNA binding-independent mechanisms of BF-1 action, we looked for interactions between BF-1 and components of TGF-β signal transduction pathways. Because BF-1 is a nuclear protein, we focused our attention on proteins which can act in the nucleus. We detected no interaction between BF-1 and Smad1, Smad2, Smad3, or Smad4 (data not shown), and so we investigated whether BF-1 might interact with the DNA binding partners of Smad proteins. Because we could demonstrate an effect of BF-1 on FAST-2 and TGF-β-dependent transcriptional activation of the A3-luc reporter, we looked for an interaction between BF-1 and FAST-2. Flag-tagged BF-1 and Myc-tagged FAST-2 were found to coimmunoprecipitate when expressed together in COS cells with either anti-Flag (Fig. 4A) or anti-Myc (Fig. 4B) antibody. The expression levels of each of the constructs were monitored by Western analysis. Controls with other epitope-tagged proteins and alternately tagged BF-1 and FAST-2 demonstrated that the interaction was not mediated by the epitope tags (data not shown). The efficiency of coimmunoprecipitation for BF-1 and FAST-2 was similar to that observed for Smad2 and FAST-2 (Fig. 4A, lanes 2 and 5). The interaction between BF-1 and FAST-2 was reduced upon exposure of the cells to TGF-β (lanes 2 and 3), while the Smad2–FAST-2 interaction was enhanced (lanes 4 and 5). To delineate the region of the FAST-2 protein which is required for association with BF-1, we examined the ability of BF-1 to coimmunoprecipitate a series of truncated FAST-2 proteins (Fig. 4B). Deletions which disrupted the C-terminal Smad interaction domain abolished the association with BF-1.
FIG. 4.
BF-1 associates with FAST-2. (A) Comparison of the association between FAST-2 and BF-1 with the association of FAST-2 and Smad-2. Flag-tagged BF-1, Flag-tagged Smad2, and Myc-tagged FAST-2 were transfected into COS cells as indicated. BF-1 and Smad2 coimmunoprecipitate with FAST-2 with similar efficiencies in the absence of TGF-β. TGF-β treatment reduces the association of FAST-2 with BF-1 and enhances its association with Smad2. (Top panel) Cell lysates were immunoprecipitated with anti-Flag antibody and blotted with anti-Myc antibody. (Middle panel) Western blot of cell lysates with anti-Myc antibody. (Bottom panel) Western blot of cell lysates with anti-Flag antibody. (B) Flag-tagged BF-1 was transfected into COS1 cells alone or with several different Myc-tagged FAST-2 constructs. (Top panel) Western blot of cell lysates immunoprecipitated with anti-Myc antibody and probed with anti-FLAG antibody. (Middle panel) Western blot of cell lysates with anti-Flag antibody. (Bottom panel) Western blot of cell lysates with anti-Myc antibody. (C) Requirement of amino acids 314 to 372 in BF-1 for antagonism of TGF-β activity and association with FAST-2. Myc-tagged FAST-2 alone or with different Flag-tagged BF-1 constructs was transfected into COS1 cells. (Top panel) Western blot of cell lysate immunoprecipitated with anti-Flag antibody and probed with anti-Myc antibody. (Middle panel) Western blot of cell lysates with anti-Myc antibody. (Bottom panel) Western blot of cell lysates with anti-Flag antibody.
The same region of the BF-1 protein is required to antagonize TGF-β activity and to associate with FAST-2.
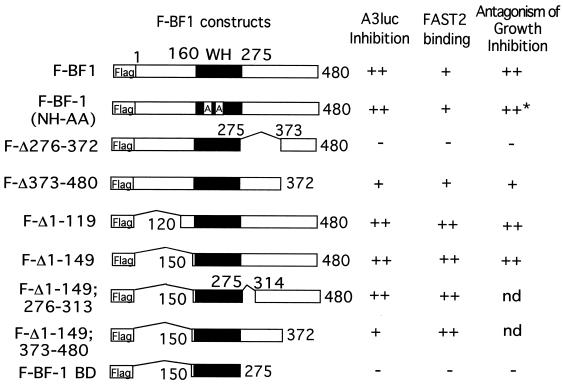
To evaluate whether the activities of BF-1 (i) to antagonize the antiproliferative activity of TGF-β, (ii) to inhibit transcriptional activation by TGF-β, and (iii) to associate with FAST-2 are related, we compared the structural requirements of each of these functions. Examination of a series of stable cell lines in Mv1Lu, expressing mutants of the BF-1 protein, revealed that a region of BF-1 adjacent to the WH domain (amino acids 276 to 372) was required to antagonize growth inhibition by TGF-β (Fig. 5B and 6). This region of BF-1 was also required for inhibition of TGF-β and FAST-2-dependent transcriptional activation from the A3-luc reporter (Fig. 5C and 6). Each of the BF-1 polypeptides was Flag tagged to permit quantitation of their expression levels (Fig. 5A). The Flag epitope did not alter the activity of the full-length BF-1 protein (compare Fig. 3C and Fig. 5C). We then determined which region of BF-1 was required for association with FAST-2. The DNA binding activity of BF-1 was not required for its interaction with FAST-2 (Fig. 4C, lane 10). However, mutations of BF-1 which abolish its ability to antagonize TGF-β activities, e.g., growth inhibition and stimulation of A3-luc transcription, also destroy its ability to associate with FAST-2 (Fig. 4C, lanes 12 to 15; summarized in Fig. 6). Further studies showed that deletion of amino acids 276 to 313 in BF-1 did not affect its ability to antagonize TGF-β-mediated transcriptional activation or to associate with FAST-2 (Fig. 6). These results narrow the critical domain in BF-1 required for interference with TGF-β signaling to amino acids 314 to 372. Our data do not exclude the possibility that BF-1 and FAST-2 interact through an intermediary protein.
FIG. 5.
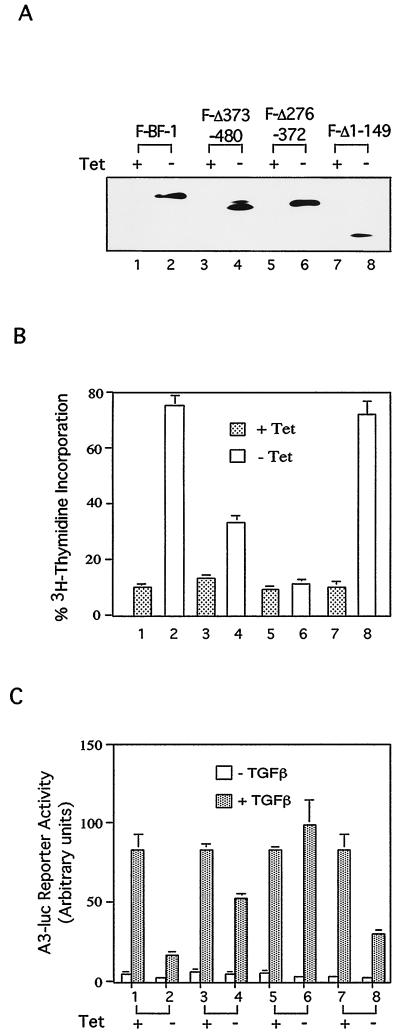
Four Mv1Lu cell lines with inducible expression of different Flag-tagged BF-1 constructs (A) were used in growth inhibition (B) and A3-luc reporter (C) assays. (A) Expression of Flag-tagged BF-1 (F-BF-1) (lanes 1 and 2) and three deletion mutants of BF-1 (F-Δ373–480 [lanes 3 and 4], F-Δ276–372 [lanes 5 and 6], and F-Δ1–149 [lanes 7 and 8]) was induced by tetracycline withdrawal and monitored by Western blotting. (B) Inhibition of [3H]thymidine incorporation by TGF-β is antagonized by Flag-tagged BF-1 and F-Δ1–149 (lanes 2 and 8) and partially antagonized by F-Δ373–480 (lane 4). F-Δ276–372 is inactive in this assay (lane 6). (C) Activation of the A3-luc reporter by TGF-β in Mv1Lu cells cotransfected with FAST-2 is inhibited by Flag-tagged BF-1 and F-Δ1–149 (lanes 2 and 8) and partially inhibited by F-Δ373–480 (lane 4). F-Δ276–372 is inactive in this assay (lane 6).
FIG. 6.
Schematic diagram of BF-1 constructs used in the experiments in Fig. 4 and 5B and C. The relative efficiency of each construct in coimmunoprecipitating FAST-2 is compared with their ability to inhibit TGF-β-stimulated A3 luc reporter expression and to antagonize the growth-inhibitory activity of TGF-β. ++, strong activity; +, low to moderate activity; −, no activity; nd, not determined. The asterisk indicates that the BF-1(NH-AA) protein in this stable cell line is not Flag tagged.
BF-1 can interfere with the association between FAST-2 and Smad2.
Because the interaction between FAST-2 and BF-1 is dependent on the Smad interaction domain of FAST-2, we examined whether BF-1 could affect the ability of FAST-2 to associate with Smad2. When Myc-tagged BF-1Δ1–119 was cotransfected with Myc-tagged FAST-2 and Flag-tagged Smad2 in COS cells, a reduction in the amount of Flag-tagged Smad protein which is coimmunoprecipitated with Myc-tagged FAST-2 was observed (Fig. 7A). The full-length BF-1 protein was also capable of interfering with the formation of the FAST-2–Smad2 complex. When Myc BF-1 was cotransfected with Myc-tagged FAST-2- and Flag-tagged Smad2, the amount of Myc-tagged FAST-2 which coimmunoprecipitated with Flag-tagged Smad2 was reduced (Fig. 7B).
FIG. 7.
BF-1 interferes with the formation of the FAST-2–Smad2 complex. (A) Interference by BF-1Δ1–119. (Top panel) Flag blot of proteins immunoprecipitated with Myc antibody. Flag-tagged Smad2 (F-Smad2) coimmunoprecipitates with Myc-lagged FAST-2 when cotransfected into COS cells. Formation of this complex is enhanced by TGF-β. Coexpression of Myc-tagged BF-1Δ1–119 reduces the amount of Flag-tagged Smad2 which is coimmunoprecipitated with Myc-tagged FAST-2. (Middle and bottom panels) Western blots of cell lysates indicate the relative amounts of Flag-tagged Smad2 (middle) and Myc-tagged FAST-2 or Myc-tagged BF-1Δ1–119 (bottom). (B) Interference by BF-1. (Top panel) Myc blot of proteins immunoprecipitated with Flag antibody. Coexpression of Myc-tagged BF-1 reduces the amount of Flag-tagged Smad2 which is coimmunoprecipitated with Myc-tagged FAST-2. (Middle and bottom panels) Western blots of cell lysates indicate the relative amounts of Myc-tagged FAST-2 or Myc-tagged BF-1 (middle) and Flag-tagged Smad2 (bottom).
DISCUSSION
BF-1 antagonizes TGF-β activity through a DNA binding-independent mechanism.
We demonstrate that the WH transcription factor BF-1 functions as an antagonist of TGF-β. This conclusion is based on results of both (i) loss-of-function studies in primary neuroepithelial cell cultures and (ii) gain-of-function studies in a cell line with inducible expression of BF-1. Cerebral cortical progenitor cells isolated from BF-1−/− mutant embryos are more responsive to growth inhibition by TGF-β and activin than are cells isolated from their normal littermates. These differences are observed in progenitor cells isolated from embryos at E10.75. At this stage, no differences are observed in the morphology of the telencephalon between BF-1−/− mutant embryos and their normal littermates. Bromodeoxyuridine labeling reveals no differences in the rate of proliferation in the cerebral cortical progenitor population (37). Isolated BF-1−/− mutant cells have a similar proliferative response to mitogens in vitro as do cells obtained from WT and BF-1+/− heterozygotes. Thus, the altered response to the antiproliferative activity of TGF-β and activin is the earliest phenotype we detected in the BF-1−/− mutant cerebral cortical progenitor.
We also examined the activity of BF-1 in Mv1Lu cell lines generated to express BF-1 and mutant forms of BF-1. We find that ectopic expression of BF-1 in mink lung epithelial cells does not significantly alter their rate of proliferation. However, BF-1 expression results in a reduction in the response of these cells to the antiproliferative activity of TGF-β. BF-1 inhibits the ability of TGF-β to block the hyperphosphorylation of the Rb protein and to stimulate the expression of the CDK inhibitor p15. BF-1 can also antagonize transcriptional activation by TGF-β of a FAST-2-dependent reporter gene, A3-luc.
A 2-amino-acid mutation (NH-AA) within the third α-helix of the WH domain abolishes the ability of BF-1 to bind to a high-affinity site on double-stranded DNA. However, this mutation does not alter the ability of BF-1 to inhibit TGF-β-mediated growth arrest and TGF-β-dependent transcriptional activation. This result suggested that BF-1 may have functions which do not require DNA binding. Other transcription factors also have important functions which are independent of their ability to bind to DNA. Truncated forms of eve and msx-1, which disrupt their DNA binding domains, can function as transcriptional repressors (1, 34). In addition, mice with a DNA binding-defective glucocorticoid receptor are viable whereas glucocorticoid receptor-deficient mice die shortly after birth (27). We cannot exclude the possibility that the BF-1(NH-AA) mutant can bind to DNA sequences other than known BF-1 sites. However, because we have targeted the mutations to critical residues in the binding helix of BF-1, any DNA binding activity of the BF-1(NH-AA) mutant is likely to utilize an atypical mode of interaction with DNA. We favor the interpretation that BF-1 may be antagonizing TGF-β function through a DNA binding-independent mechanism.
Association of BF-1 with a Smad partner.
The activity of a DNA binding-defective form of BF-1 raised the possibility that BF-1 could be interfering with TGF-β function by interacting with components of the TGF-β signaling pathway. Several mechanisms have previously been described in which TGF-β signal transduction is negatively regulated by interference with Smad transcriptional complexes. Smad6 associates with Smad1, thereby blocking signaling through the activating Smads (13, 25). The homeodomain protein evi has been suggested to antagonize TGF-β signals by undergoing a direct interaction with Smad3 (17). The homeodomain protein TGIF and the oncoproteins Ski and SnoN act as transcriptional corepressors (23, 30, 31, 36). In these examples, negative regulation is achieved through interactions with Smad proteins. We did not obtain any evidence for interactions between BF-1 and Smad proteins.
We find that BF-1 forms a complex with FAST-2 in cells. We provide evidence that the region of the BF-1 protein which is required for its interaction with FAST-2 is also essential for its ability to inhibit TGF-β-stimulated A3-luc expression and to antagonize the antiproliferative activity of TGF-β. These results suggest a common mechanism for these three activities of BF-1. However, FAST-2 is not known to mediate the antiproliferative activity of TGF-β. BF-1 antagonizes this activity of TGF-β in Mv1Lu cells which do not express FAST-2. Furthermore, the low expression levels of FAST-2 in the telencephalic neuroepithelium (C. Dou et al., unpublished results) suggest that other DNA binding partners of Smad proteins mediate TGF-β family signals in the developing brain. Thus, it is likely that BF-1 interferes with TGF-β responses which are not mediated by FAST-2.
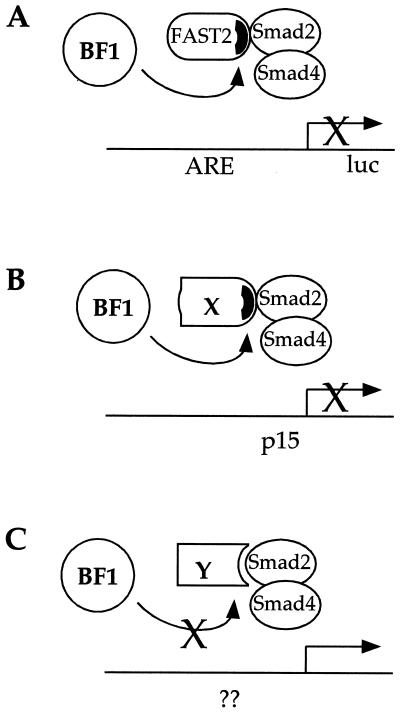
The finding that BF-1 associates with the same region of FAST-2 (the Smad interaction domain) which mediates its ability to interact with Smad proteins suggests a mechanism by which BF-1 can interfere with both FAST-2-dependent and FAST-2-independent TGF-β responses. We propose that BF-1 interacts with a subset of DNA binding proteins which are characterized by sharing with FAST-2 a mode of interaction with Smad proteins (Fig. 8A and B). Thus, BF-1 interferes with TGF-β-stimulated transcriptional activation of the A3-luc reporter through its association with FAST-2. However, the antiproliferative responses of Mv1Lu cells and neuroepithelial cells to TGF-β and/or activin are likely to be mediated through other DNA binding proteins. While transcriptional activation of the CDK inhibitor p15 has been identified as an important component of the antiproliferative response in Mv1Lu cells (28), the corresponding transcriptional targets in many other cells remain unknown and the DNA binding proteins which recruit Smad complexes to these promoters have not yet been identified. We suggest that some of these DNA binding proteins (X in Fig. 8B) will associate with Smad2 through a structure which resembles that found in FAST-2. This model also predicts that BF-1 will not interfere with all TGF-β responses, e.g., transcriptional regulation mediated through DNA binding partners which associate with Smad proteins through a distinct structural motif (Y in Fig. 8C). Our competition model further predicts that BF-1 will reduce the amount of transcriptionally active Smad complex for any level of TGF-β signal. BF-1 can have an inhibitory activity in the absence of TGF-β, for a nonzero basal level of Smad partner in the nucleus. We suggest that this is a plausible explanation for the modest repression of the A3-luc reporter by BF-1 observed in FAST-2-transfected cells in the absence of added TGF-β.
FIG. 8.
BF-1 interferes with multiple TGF-β responses by interference with FAST-2 and other Smad2 partners. (A) In this model, we propose that BF-1 associates with FAST-2 through a specific motif within the Smad interaction domain of the FAST-2 protein to inhibit TGF-β- and FAST-2-dependent transcriptional activation from the A3-luc reporter gene. ARE, activin response element. (B) BF-1 also interferes with other TGF-β responses such as the transcriptional activation of the p15 gene. The specific Smad2 partner for this and other responses remains to be established. We postulate that BF-1 will interact with a subset of these DNA binding partners, i.e., those that share with FAST-2 a mode of interaction with Smad2. (C) This model also predicts that other DNA binding partners which interact with Smad2 through a distinct mechanism will not be susceptible to interference by BF-1.
The homeodomain proteins Mixer and Milk were recently found to mediate activin- and TGF-β-induced transcription of the Xenopus goosecoid promoter (8). These proteins associate with Smad2 through a common motif, called the Smad interaction motif. This motif is also found within the Smad interaction domain of FAST-2. The expression pattern of Mixer and Milk suggests that they are not likely to function in the developing brain. However, their identification as Smad partners supports the concept that multiple DNA binding proteins can recruit Smad2 to distinct promoter elements through a common mechanism. Thus, the potential interaction targets of BF-1 may include not only WH factors related to FAST-2 but also members of other families of transcriptional regulators.
ACKNOWLEDGMENTS
We thank Abeel Mangi and Gabriela Balas for valuable assistance with these studies, M. Whitman and L. Freedman for providing reporter constructs, and R. Benezra and S. Li for critical review of the manuscript.
This work was supported by grants from the NIH to E.L. (HD29584), C.D. (F32NS10313), and MSKCC (Cancer Center Support Grant).
REFERENCES
- 1.Catron K, Zhang H, Marshall S, Inostroza J, Wilson J, Abate C. Transcriptional repression by msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caviness V J, Takahashi T, Nowakowski R. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Rubock M, Whitman M. A transcriptional partner for MAD proteins in TGF-B signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 5.Clark K, Halay E, Lai E, Burley S. Cocrystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 6.Feijen A, Goumans M, van den Eijnden-van Raaij A. Expression of activin subunits, activin receptors and follistatin in postimplantation mouse embryos suggests specific developmental functions for different activins. Development. 1994;120:3621–3637. doi: 10.1242/dev.120.12.3621. [DOI] [PubMed] [Google Scholar]
- 7.Furuta Y, Piston D W, Hogan B L M. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development. 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 8.Germain S, Howell M, Esslemont G, Hill C. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- 9.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannon G, Beach D. p15INK4B is a potential effector of TGF-B-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 11.Hatini V, Ye X, Balas G, Lai E. Dynamics of placodal lineage development revealed by targeted transgene expression. Dev Dyn. 1999;215:332–343. doi: 10.1002/(SICI)1097-0177(199908)215:4<332::AID-AJA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Hoodless P, Haerry T, Abdollah S, Stapleton M, O'Connor M, Attisano L, Wrana J. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 13.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 14.Kaestner K, Knöchel W, Martínez D. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 15.Kaufman E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 16.Kilpatrick T, Bartlett P. Cloning and growth of multipotential neural precursors: requirements for proliferation and differentiation. Neuron. 1993;10:255–265. doi: 10.1016/0896-6273(93)90316-j. [DOI] [PubMed] [Google Scholar]
- 17.Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. The oncoprotein Evi-1 represses TGF-β signalling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 18.Labbe E, Silvestri C, Hoodless P, Wrana J, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 19.Laiho M, DeCaprio J, Ludlow J, Livingston D, Massagué J. Growth inhibition by TGF-β1 linked to suppression of retinoblastoma protein phosphorylation. Cell. 1990;62:175–185. doi: 10.1016/0092-8674(90)90251-9. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Chang H, Lai E, Parker E, Vogt P. The oncogene qin codes for a transcriptional repressor. Cancer Res. 1995;55:5540–5544. [PubMed] [Google Scholar]
- 21.Liu B, Dou C, Prabhu L, Lai E. FAST-2 is a mammalian winged helix protein which mediates TGFβ signals. Mol Cell Biol. 1999;19:424–430. doi: 10.1128/mcb.19.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Pouponot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo K, Stroschein S, Wang W, Chen D, Martens E, Zhou S, Zhou Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massagué J, Chen Y. Controlling TGF-β signalling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 25.Nakao A, Afrakhte M, Moren A, et al. Identification of Smad7, a TGF-beta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 26.Pelton R, Saxena B, Jones M, Moses H, Gold L. Immunohistochemical localization of TGFβ1, TGFβ2, and TGFβ3 in the mouse embryo: expression patterns suggest multiple roles during embryonic development. J Cell Biol. 1991;115:1091–1105. doi: 10.1083/jcb.115.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichardt H, Kaestner K, Tuckermann J, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 28.Reynisdottir I, Kornelia P, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-B. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 29.Shimamura K, Hartigan D, Martinez S, Puelles L, Rubenstein J. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- 30.Stroschein S, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-β signaling by the SnoN ocoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 31.Sun Y, Liu X, Ng-Eaton E, Lodish H, Weinberg R. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc Natl Acad Sci USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi T, Nowakowski R, Caviness V J. The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- 34.Um M, Li C, Manley J. The transcriptional repressor even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wieser R, Wrana J, Massague J. GS domain mutations that constitutively activate TβR-I, downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wotton D, Lo R, Lee S, Massagué J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 37.Xuan S, Baptista C, Balas G, Tao W, Soares V, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]