Significance
Many drugs have off-target effects that involve membrane proteins. We explore a general mechanism for such effects based on the notion that first, membrane proteins are energetically coupled to their host bilayer, and second, many drugs are amphiphiles that partition into and alter lipid bilayer properties. Using the prototypical potassium channel KcsA, we show that structurally diverse drugs regulate membrane protein function through changes in bilayer properties and nonspecific drug binding. These nonspecific mechanisms likely underlie pleiotropic, off-target drug effects observed with structurally and functionally diverse membrane proteins when the concentrations at which a drug alters the function of a desired target protein overlaps with the concentration at which it alters bilayer properties.
Keywords: lipid bilayer properties, lipids, membrane protein regulation, drugs, ion channels
Abstract
The hydrophobic coupling between membrane proteins and their host lipid bilayer provides a mechanism by which bilayer-modifying drugs may alter protein function. Drug regulation of membrane protein function thus may be mediated by both direct interactions with the protein and drug-induced alterations of bilayer properties, in which the latter will alter the energetics of protein conformational changes. To tease apart these mechanisms, we examine how the prototypical, proton-gated bacterial potassium channel KcsA is regulated by bilayer-modifying drugs using a fluorescence-based approach to quantify changes in both KcsA function and lipid bilayer properties (using gramicidin channels as probes). All tested drugs inhibited KcsA activity, and the changes in the different gating steps varied with bilayer thickness, suggesting a coupling to the bilayer. Examining the correlations between changes in KcsA gating steps and bilayer properties reveals that drug-induced regulation of membrane protein function indeed involves bilayer-mediated mechanisms. Both direct, either specific or nonspecific, binding and bilayer-mediated mechanisms therefore are likely to be important whenever there is overlap between the concentration ranges at which a drug alters membrane protein function and bilayer properties. Because changes in bilayer properties will impact many diverse membrane proteins, they may cause indiscriminate changes in protein function.
Cell membranes regulate membrane protein function by providing the necessary lipids and the appropriate bulk bilayer environment for function (1–5). The bilayer-mediated regulation arises because membrane proteins are hydrophobically coupled to their host lipid bilayer. As membrane proteins undergo conformational transitions (change shape), from a state I to a state II, the hydrophobic coupling gives rise to changes in the organization of adjacent lipids, which has an energetic cost (), where and are the bilayer deformation energies incurred in each state. The total energetic cost of a transition between two protein conformations (I and II) thus is the sum of contributions from conformational rearrangements within the protein () and rearrangements within the bilayer to minimize the exposure of hydrophobic amino acids to the aqueous environment (). When the constraints on lipid packing around a protein do not allow for perfect hydrophobic matching, resulting in residual exposure of hydrophobic amino acids, the associated residual exposure energy will also contribute to (6). Thus, the equilibrium constant between I and II is:
where and will vary depending on protein shape and conformational transitions.
varies with changes in bilayer physical properties (3), meaning that changes in bilayer properties will alter the protein’s conformational equilibrium and function. This bilayer-mediated regulation of protein function is nonspecific; any membrane protein is subject to this regulation, which becomes important when , where kB is Boltzmann’s constant and T temperature in Kelvin. The bilayer’s role in regulating membrane protein function by amphiphiles (including many drugs) is important because amphiphiles reversibly partition into the bilayer/water interface and thereby alter bilayer properties (7). Amphiphilic drugs thus may act through both direct binding to the desired target(s) and bilayer-mediated mechanisms. Distinguishing between direct and indirect bilayer-mediated regulation will help understand the mechanism(s) underlying the pleiotropic effects observed with many biologically active molecules.
To develop a framework for understanding the mechanisms of drug-induced effects on membrane proteins, we used KcsA as a prototypical, proton (H+)-gated (8) ion channel and examined how changes in bulk bilayer properties and diverse drugs with varying bilayer (7, 9, 10) and clinical off-target effects (11–13) alter its function. We used gramicidin channels, whose bilayer-mediated regulation is well understood (7), to determine the drugs’ bilayer-modifying potency. To calibrate the drugs’ bilayer-modifying effect to its concentration in the bilayer, we estimated the mole fraction of each drug using membrane or octanol partition coefficient. We determined the drugs’ affinity for the purified KcsA to compare to its effective and bilayer-modifying concentrations.
We first show that changes in bulk bilayer properties, specifically thickness, regulate KcsA gating and that thin bilayers promote the activated state. A more complex picture emerges when we explore the effects of various (amphiphilic) drugs, in which a drug’s effect(s) varied with bilayer thickness. Yet, the drug mole fraction in the bilayer and changes in bilayer properties predicted the drugs’ effects on KcsA gating, which enabled us to tease apart direct and bilayer-mediated effects. Our results provide a strategy for determining whether a drug’s pleiotropic effects result from direct, nonspecific, and bilayer-mediated mechanisms.
Results
Bilayer Thickness and KcsA Gating Kinetics.
KcsA function was quantified using a fluorescence quench assay, in which KcsA is reconstituted into large unilamellar vesicles (LUVs) loaded with the aqueous fluorophore 8-aminonaphthalene-1,3,6-trisulfonate (ANTS). KcsA is H+ gated (8), and lowering the pH increases its permeability to Tl+ (14), an ANTS fluorescence quencher, meaning that the rate of fluorescence quench (Tl+ influx) reflects the number of open KcsA channels.
We have previously shown, using a mixture of phospholipids with different chain lengths, KcsA activity is promoted in thin, as compared to thick, bilayers (15); similar results were obtained by (16). To rule out the possibility of lateral lipid redistribution (17), here we use LUVs composed of DC22:1PC/DC22:1PG 3:1 (C22:1) or DC18:1PC/POPG 3:1 (C18:1) homogenous acyl chain lengths for each thickness, resulting in thick (34 Å) or thin bilayers (27 Å), respectively (18).
To establish whether there are differences in KcsA reconstitution yield in C22:1 and C18:1, we determined the amount of KcsA per vesicle in each bilayer type (SI Appendix, Fig. S1). The numbers of KcsA per LUV in C22:1 (16.4 ± 1.2, mean ± SD, and n = 4) and C18:1 (22.8 ± 6.9 and n = 4) did not differ significantly (two-sample Student’s t test assuming equal variance P = 0.14) (SI Appendix, Fig. S1). We did not assess the channels’ orientation, but it can be inferred from Heginbotham et al. (19) that channel incorporation is highly asymmetric, with the majority of channels oriented with their cytosolic (proton sensitive) domain facing the intravesicular solution.
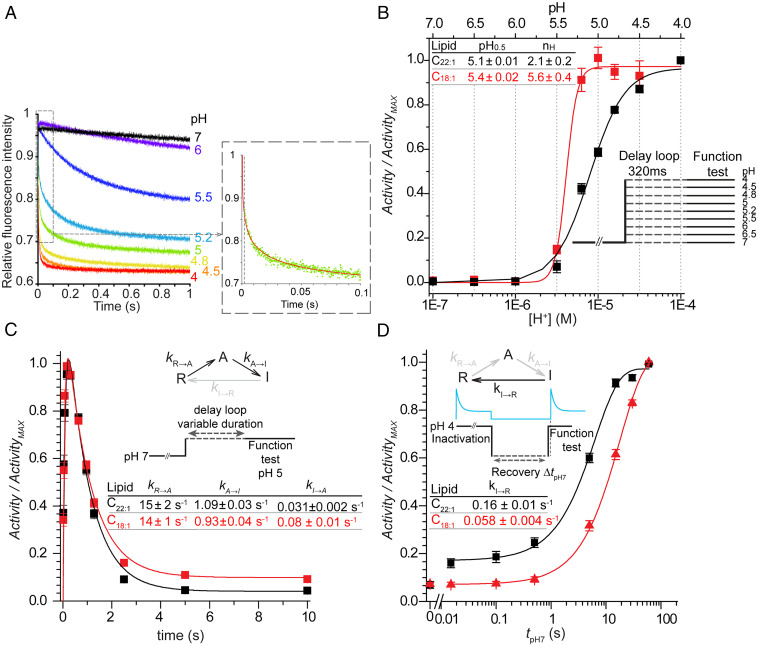
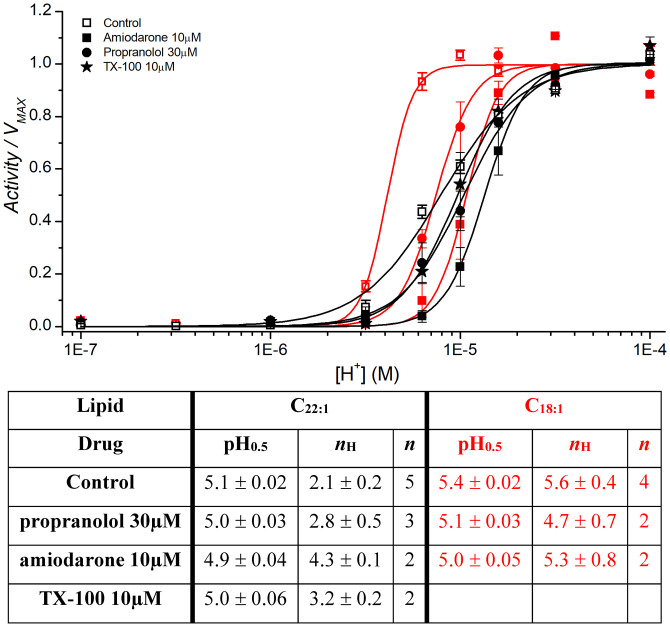
We first compared the effects of bilayer thickness on H+ activation by determining H+ dose–response curves in C22:1 or C18:1. The reconstituted channels were incubated at pHs between 7 and 4 (in the delay loop) for 320 ms, close to the time for maximal activity for each pH, and then tested for open channels using the initial slope of the fluorescence quench time course as a measure of activity (Fig. 1A; protocol in SI Appendix, Fig. S2). The initial slope (Activity) is plotted as function of pH and fit with a standard Hill function yielding the pH at the midpoint of activation (pH0.5) and the Hill coefficient (nH) (Materials and Methods) (Fig. 1B and SI Appendix, Fig. S2). In C18:1, pH0.5 was ∼0.3 pH units higher ([H+] twofold lower) than in C22:1, suggesting that KcsA activation occurs more readily in thin the bilayers; the cooperativity, as quantified by nH, was ∼2.5-fold greater in C18:1 than in C22:1.
Fig. 1.
Effects of bilayer thickness on KcsA gating. KcsA was reconstituted into either C22:1 (black squares, lines) or C18:1 (red squares, lines). (A) Changes in the time courses of fluorescence quench for KcsA reconstituted in C18:1 bilayers at the indicated pHs. (Inset) A single repeat of a quench at pH5 (green dots) with a stretched exponential fit (red line). Dashed line at 2 ms denotes the instrumental dead time. (B) Normalized Activity-[H+] curves in C22:1 (black) or C18:1 (red). The Activity was determined from the initial slopes of the quench curves (A) at each pH. The Activity-[H+] curve for each experiment (n = 5 for C22:1; n = 4 for C18:1) was fit with a Hill function to determine ActivityMAX, pH0.5, and nH. The pH0.5 (pH0.5 = −log([H+]0.5) and nH values from multiple experiments were averaged. The Activity-[H+] curves were constructed from the average Activity at each pH, normalized to ActivityMAX (squares), which were fit with Hill function (solid lines). (C) Time courses of KcsA activation and inactivation at pH 5 in C22:1 or C18:1. Activity/ActivityMAX is plotted as function of incubation time. The Inset shows a three-state KcsA gating model (20), which was used to determine the rate constants using the QuB software suite (22) in C22:1 (mean ± SEM, n = 32) and C18:1 (n = 24). (D) Time course of recovery from inactivation in C22:1 or C18:1. KcsA was incubated at pH 4 for 10 min, to reach full inactivation, and then allowed to recover at pH 7 for varying durations. kI→R was determined by fitting a single exponential function to Activity as function of recovery time: (mean ± SEM, n = 21 or n = 10 in C22:1 or C18:1, respectively). The difference in kI→R in C18:1 compared to that in C22:1 is statistically significant (two-sample Student’s t test assuming equal variance P = 3·10−7).
To determine how activation and inactivation gating are altered by changes in bilayer thickness, we analyzed the time course of activation using a gating model with transitions between three states (20, 21): resting (R); activated (A); and inactivated (I), obtaining the underlying rate constants (kR→A, kA→R and kA→I, kI→A) in C22:1 and C18:1 bilayers (Fig. 1C). Time course was determined at pH 5, near the pH for half-maximal activity in both bilayers, which provides for an optimal dynamic range (Fig. 1B). The LUV-reconstituted KcsA was incubated at pH 5 between 15 ms (minimum delay loop setting) and 10,000 ms before measuring the Activity by mixing with Tl+, and the Activity was plotted as function of incubation time (protocol in SI Appendix, Fig. S4). The resulting biphasic time course was fit with a three-state model (21) (Fig. 1 C, Inset) using QuB (22). In C22:1, kR→A= 15 ± 2 s−1 and kA→I = 1.10 ± 0.03 s−1 (n = 34); in C18:1, kR→A = 15 ± 1 s−1 and kA→I = 0.94 ± 0.04 s−1 (n = 24). The rate constants for return from the inactivated to the activated state, kI→A, were 0.03 ± 0.002 s−1 (n = 34) and 0.08 ± 0.01 s−1 (n = 24) in C22:1 and C18:1, respectively. Using this simple three-state model, the rate constants for deactivation (from the activated to the resting state), kA→R, were < 10−4 s−1 in either system.
The rate of recovery from inactivation (kI→R) was determined by incubating LUV-reconstituted KcsA for 10 min at pH 4 to reach steady-state inactivation followed by incubating at pH 7 for varying durations and testing for channel activity 2 ms after exposure to pH 4 (Fig. 1D; protocol in SI Appendix, Fig. S4) (at pH 4, activation is nearly complete by 2 ms [SI Appendix, Fig. S6], and the changes in Activity reflect the population of channels available for activation). Plotting the results as function of incubation time at pH 7 and fitting with a single exponential, the rate of recovery from inactivation (kI→R) in C18:1 (0.06 ± 0.004 s−1 n = 10), was ∼2.5-fold slower than in C22:1 (0.16 ± 0.01 s−1 n = 21).
Inactivation is pH independent (20, 23) (SI Appendix, Fig. S6), and the differences between the steady-state activity after 10 s incubation at pH 5 (Fig. 1C) and the initial recovery levels after incubation for 10 min at pH 4 (Fig. 1D) are due to differences in protocol (activating/inactivating and testing for function at pH 5, close to pH0.5, in Fig. 1C; inactivating at pH 4 and monitoring recovery at pH 7 in Fig. 1D). The fraction of active channels after 10 min incubation at pH 4 in the latter experiments were similar in C22:1 and C18:1 (the tpH7 = 0 data points in Fig. 1D), 0.069 ± 0.014 s−1 (n = 9) and 0.073 ± 0.007 s−1 (n = 5), respectively (Fig. 1C). The recovery after 15 ms at pH 7, however, differs markedly between C18:1 and C22:1, with a rapid initial recovery at pH 7 in the thick bilayer. When recovering at pH 5.2, we do not observe this rapid phase of recovery in either C22:1 or C18:1 (SI Appendix, Fig. S7); the 15-ms activity at pH 5.2 is similar and equal to the activity after 10 min in pH 4. Measuring the rate of recovery from inactivation informs us about the fraction of resting channels that are ready for activation (20, 23, 24). Comparing the results at pH 7 and 5.2, we conclude that the rapid phase of reactivation at pH 7 in C22:1 results from a rapid deprotonation that is promoted in the thick bilayer. This likely reflects the differential bilayer thickness–dependence of channel states in C18:1 compared to C22:1, with the resting state being stabilized (relative to the active and inactive states) in C22:1.
Indeed, at pH 4 at which the channels were maximally activated in both bilayers (Fig. 1B), the nonnormalized Activity in C22:1 was twofold greater than in C18:1 (SI Appendix, Fig. S5) despite a lower pH0.5 and nH than in C18:1. This is not due to differences in reconstitution yield (SI Appendix, Fig. S1). The slower recovery from inactivation and lower Activity after 15-ms recovery in C18:1, as compared to C22:1, (Fig. 1D and SI Appendix, Fig. S7) thus suggests that KcsA in C18:1 may enter the inactivated state from the resting state, making them unavailable for activation. This could reflect the relatively extended conformation of KcsA in the resting state (25), which would be stabilized in thick bilayers. In contrast, the relatively compact activated and inactivated states (26, 27) are likely to be stabilized in thin bilayers. A similar bilayer thickness–dependence in the fraction of activatable nicotinic acetylcholine receptors was observed previously (28).
Drug Regulation of KcsA Function: Role of the Bilayer-Mediated Mechanism.
Having established that changes in lipid bilayer thickness alter KcsA gating, we explored whether changes in bilayer properties induced by bilayer-modifying drugs also alter KcsA function. We chose compounds with varying physico-chemical profiles, clinical off-target effects, and bilayer-modifying potencies, as sensed by gramicidin channels (9, 10, 29). The structures are shown in SI Appendix, Table S1. Amiodarone (13), troglitazone (withdrawn) (12), and fluoxetine (30) have multiple membrane protein targets and off-target effects, whereas propranolol (11, 31) pindolol (11) and pioglitazone (12) are more specific with few off-target effects. We also tested two molecules not used therapeutically but with well-defined mechanisms of action that allowed them to serve as “markers” at either end of the spectrum of possible mechanisms: direct protein interaction–mediated mechanisms (tetrabutylammonium, TBA) and bilayer-mediated mechanisms (Triton X-100 and TX-100). TBA is an open-channel blocker that binds in the KcsA pore close to the cytoplasmic end of the selectivity filter (32, 33) and does not alter bilayer properties at the concentrations used here (SI Appendix, Fig. S8). TX-100 is a detergent that alters bilayer properties and the gating of N-type calcium, voltage-gated sodium, and GABAA channels at the concentrations at which it alters bilayer properties (34–36), indicating that TX-100’s effects are mediated through the bilayer. We similarly expected that TX-100’s modulation of KcsA function would be solely bilayer mediated.
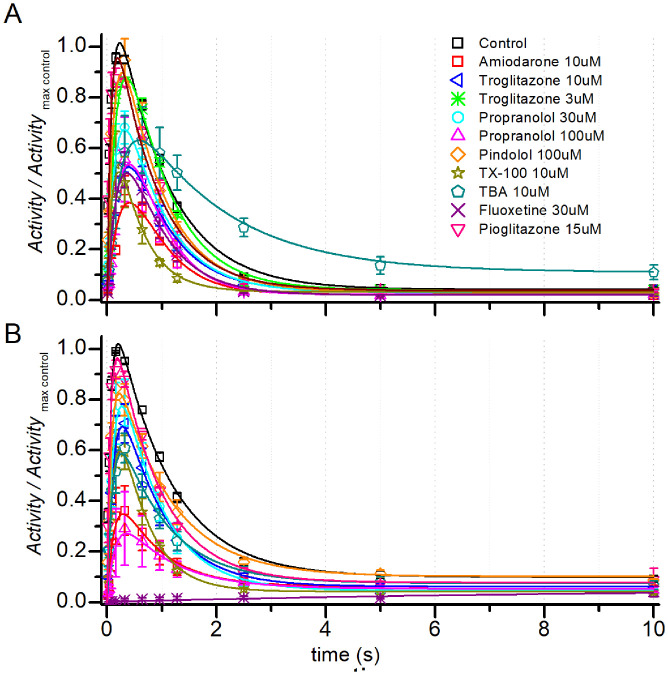
The drugs inhibited KcsA Peak activity in both C22:1 (Fig. 2A) and C18:1 (Fig. 2B) with varying potencies and altered the time course of KcsA activation and inactivation. To analyze the effects on KcsA gating, we determined kR→A, kA→I, and kI→R in the absence and presence of drugs. Though pharmacologically and structurally diverse (SI Appendix, Table S1), the drugs had similar effects on KcsA gating, In C22:1, the drugs reduced kR→A, (Fig. 3A), increased kA→I (Fig. 3B), reduced the maximal Activity (Peak) (Fig. 3C), and reduced kI→R (Fig. 3D). This commonality suggests that their effects may be mediated by common (nonspecific) mechanisms.
Fig. 2.
Time courses of KcsA activation and inactivation at pH 5 in the absence (Control) or presence of drugs in (A) C22:1 or (B) C18:1 Activity/ActivityMAX Control, in which each point was normalized to the maximum activity in the absence of drug for that experiment. Symbols represent mean ± SEM, and n = 34 and 24 for control in C22:1 and C18:1 respectively, in the presence of drugs n varied between 3 and 10; in case of n = 2 (pindolol), Mean ± range/2 is used. The three-state KcsA gating model (20) was used to fit the (curves) using QuB (22).
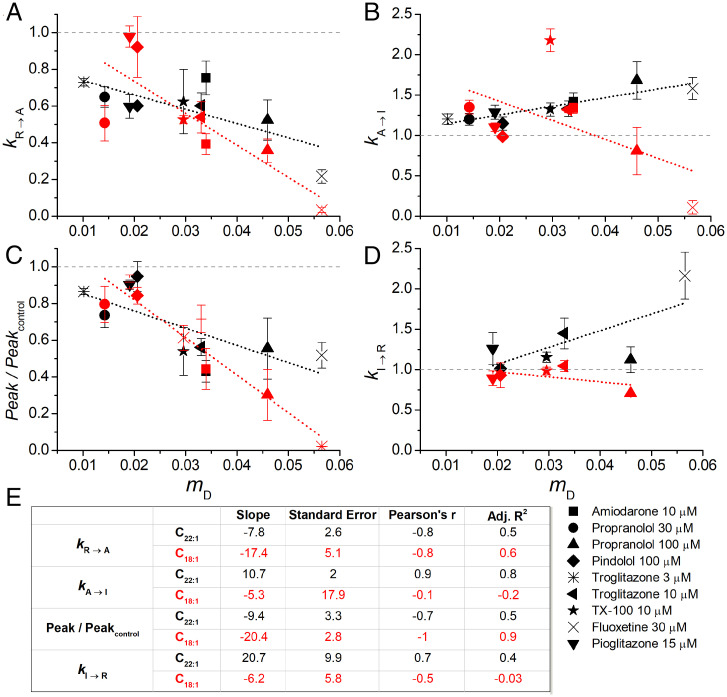
Fig. 3.
Drug-induced changes in KcsA gating in C22:1 or C18:1 versus the drugs’ mole fraction in the bilayer (mD). (A) kR→A, (B) kA→I, (C) Peak activity, and (D) kI→R. The points denote mean ± SEM, n = 3 to 10, and mean ± range/2 (n = 2); the dashed lines denote linear fits to the data. (E) Slopes and SEs of fit, Pearson’s ρ, and the adjusted R2 (Adj. R2) for each of the fits. Fluoxetine was excluded from analysis of kA→I, because there was no observable inactivation over the time scale tested.
The drugs altered KcsA gating in both C22:1 and C18:1 but differed quantitatively with greater effects on kA→I and kI→R in C22:1.
We therefore examined the relative contribution of the bilayer-mediated regulation by 1) comparing the relationship between mole fractions of the drugs in the bilayer (mD) and the magnitude of their effects on KcsA rate constants; 2) comparing the drugs’ effects in bilayers of different thickness; and 3) determining the drugs’ affinity to purified KcsA.
Drug partitioning and KcsA function.
We determined the drugs’ mole fraction in the membrane by measuring their bilayer partition coefficient (KP) using isothermal titration calorimetry. LUVs composed of C22:1PC were titrated into a drug solution, and the associated partitioning heats were measured (SI Appendix, Fig. S9A). To establish that heats of dilution of either drug or LUVs do not interfere with the signal, we also titrated LUV suspensions into buffer and buffer into a drug solution (SI Appendix, Fig. S9A). Buffer compositions were matched in all experiments. The partition coefficient KP, enthalpy (ΔH), Gibbs free energy (ΔG), and entropy (ΔS) were calculated by fitting injection heats plotted as a function of injection number with a function that assumes transition between two immiscible phases (lipid and aqueous) (Eq. 1) (SI Appendix, Fig. S9B) (9, 37). KP for amiodarone, propranolol, and troglitazone are listed in SI Appendix, Fig. S9C. KP for TX-100 (38) and fluoxetine (39) are from previous studies. Pioglitazone (9), pindolol, and TBA had too-low heats of partitioning to determine KP, and we used ALogP for pioglitazone and pindolol as a proxy for KP. The drugs’ concentration in the membrane then was determined from KP using Eq. 2, and the mole fraction was calculated using Eq. 3. SI Appendix, Table S1 summarizes the information for all the tested drugs.
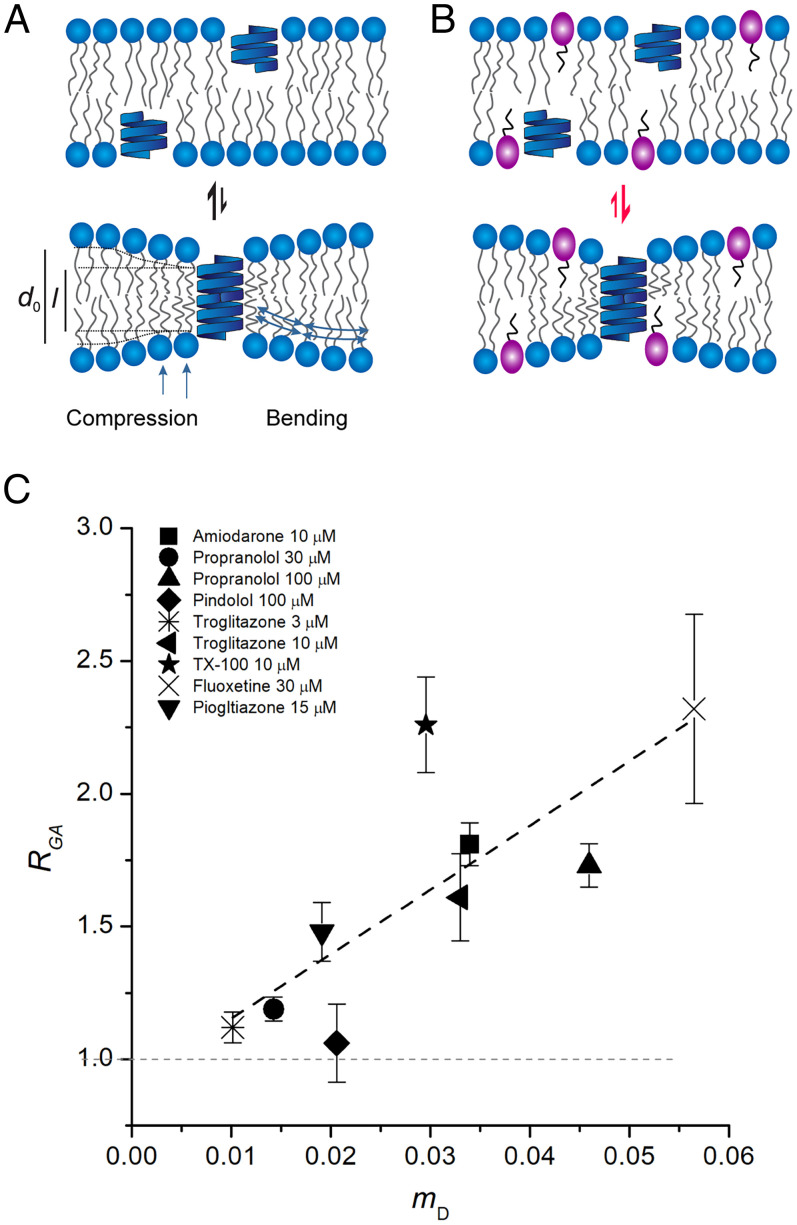
To determine the drugs’ bilayer-modifying potencies, as sensed by a bilayer-spanning channel, we used the drug-induced changes in gramicidin channel activity, measured using a fluorescence quench assay similar to that used to quantify KcsA function (9, 10, 29, 35). The quench rates obtained with gramicidin (RGA) increase linearly with the drug’s mole fraction (mD) (9, 38–41) (SI Appendix, Table S1 and Fig. 4). These results reveal that the bilayer-modifying effect per molecule in the bilayer was similar for all the tested molecules regardless of their individual structure, suggesting that these molecules increase bilayer elasticity (42–44).
Fig. 4.
Drug-induced changes in bilayer properties as a function of the drug’s mole fraction in the bilayer (C22:1 bilayers). (A) Gramicidin forms channels by monomer dimerization involving a symmetrical compression and bending of the bilayer. The resulting bilayer deformation incurs an energetic cost, ΔGbilayer, which dictates the equilibrium distribution between nonconducting monomers and conducting dimers and thereby gramicidin channel activity. (B) Bilayer-modifying drugs partition into the bilayer and alter the ΔGbilayer resulting in changes in monomer–dimer equilibrium thereby altering gramicidin activity. If bilayer-modifying drugs increase bilayer elasticity (softening), thereby reducing ΔGbilayer, the equilibrium is shifted toward dimer formation thereby increasing gramicidin activity. (C) Changes in gramicidin function (monomer↔dimer equilibrium) monitored as the normalized change in ANTS quench rate (quench rate in the presence of drug normalized to the rate in the absence of drug), RGA. Mean ± SEM of RGA plotted as a function of the corresponding drug mD, (n = 3 to 5), mean ± range/2 for n = 2. The data were fit by a straight line (black, dashed line) with slope 24.2 ± 7.1 (mean ± SEM); Pearson’s R2 correlation coefficient (ρ) and the adjusted R2 were 0.8 and 0.6, respectively.
The bilayer deformations that result from changes in KcsA conformation will differ (and be more complex) than those associated with gramicidin channel formation (Fig. 4A) (45–47), and the changes in KcsA function may include contributions from drug binding to the protein. Thus, the relationship between KcsA gating and mD would be expected to differ from that for gramicidin channels. Using the correlation between drug-mediated effects on gramicidin activity and mD (Fig. 4) as a benchmark for (simple) bilayer-mediated regulation we can use the correlation between drug-induced changes in KcsA function and mD (Fig. 3) to assess the bilayer contributions to changes in KcsA function. The closer the correlation between changes in KcsA gating and mD (Fig. 3) is to that between RGA and mD (Fig. 4), the greater the relative contribution of the bilayer-mediated regulation to the changes in KcsA function. We therefore plotted the magnitude of the drug-induced changes in KcsA gating, as reflected in changes in kR→A (Fig. 3A), kA→I (Fig. 3B), Peak activity (Fig. 3C) and kI→R (Fig. 3D), as function of mD. Based on the slopes and the Adjusted R2 and Pearson’s correlation coefficient, which reflect the strength and direction of the correlation, all the gating changes correlate with mD (Fig. 3), though the strength of the correlations and direction of the changes varied for the different gating steps.
Drug effects on KcsA vary with bilayer thickness.
In the case of gramicidin channels, drug effects vary with bilayer thickness. This thickness-dependence arises, because the bilayer deformation and therefore ΔGbilayer increases with increasing hydrophobic mismatch (7, 48). We similarly find that drug effects on KcsA varied with bilayer thickness, indicating that bilayer-mediated mechanism plays a role in drug effects on KcsA.
The drugs reduced pH0.5 and increased nH in both C22:1 and C18:1, with more pronounced changes in C18:1 (Fig. 5). In C18:1, the changes in kR→A (Fig. 3A) and Peak activity (Fig. 3C) were better correlated to mD than in C22:1, with greater slope, smaller error, and greater ρ and adjusted R2. In C18:1, kA→I (Fig. 3B) showed a different trend from C22:1 and the drug-induced changes in kI→R are less than in C22:1 (Fig. 3D). Thus, in C18:1, the distribution between the inactivated and resting states appears to be less sensitive to changes in bilayer properties than in C22:1, whereas the distribution between the resting and activated states is more sensitive. Similar stabilization of the inactivated state and slowed recovery from inactivation has been demonstrated previously for voltage-dependent sodium channels (9, 35, 49).
Fig. 5.
Effects of bilayer-modifying drugs on KcsA activation in C22:1 (black) or C18:1 (red) bilayers. KcsA was incubated with the drugs for 10 min at the indicated concentrations for 10 min after which, Activity at the indicated pH was determined (Materials and Methods and SI Appendix, Fig. S2). The results for each experiment were fitted with a Hill equation , and the resulting averages are plotted as function of [H+] (n = 3 to 5; mean ± SEM; n = 2, and mean ± range/2). Comparing the means of pH0.5 and nH from control experiments in C22:1 to those in C18:1 using two-sample Student’s t test, the difference is statistically significant (assuming equal variance P = 0.0005 and 0.001 for pH0.5 and nH respectively).
TBA does not alter bilayer properties at the concentrations used here (SI Appendix, Fig. S8). Because we interrogate only channels with their cytosolic domain (which encompasses the pH sensor) (50) exposed to the extravesicular milieu, TBA’s effects on KcsA gating (kR→A, kA→I, Peak activity and kI→R) will be due to binding at the cytosolic end of KcsA. TBA reduced kR→A, kA→I, and Peak activity in C22:1, with less effect in C18:1 (SI Appendix, Fig. S10). The reduction of Peak activity is consistent with a pore-block; the reduction in kA→I, which contrasts with the other drugs, may arise because KcsA cannot inactivate when TBA is bound in the permeation path, the so-called “foot-in-the-door” mechanism (51, 52).
The bilayer thickness–dependent effects of TBAs therefore may reflect that TBA has different affinities to different channel states (with different ΔGdef).
We also determined the relationship between drug-induced changes in KcsA function and bilayer properties, as reported by gA, by plotting the changes in kR→A, kA→I, Peak activity, and kI→R as a function of RGA (SI Appendix, Fig. S11). The trends observed with mD (Fig. 3) were conserved with RGA: kR→A and Peak decreased in both C22:1PC and C18:1PC, and kA→I and kI→R increased in C22:1PC and deceased C18:1PC. The Adjusted R2 and Pearson’s correlation coefficient indicate stronger correlation with mD (Fig. 3E) than with RGA (SI Appendix, Fig. S11E). These differences likely reflect differences in the mechanisms involved: simple bilayer-mediated in the case of gramicidin channels (7, 10, 39, 53); and multiple mechanisms in the case of KcsA. In mechanistic terms: drug effects on KcsA function depend on drug partitioning into the bilayer, but, unlike for gramicidin, changes in bulk bilayer properties alone do not account for the aggregate effects that also may involve changes in lipid–KcsA interface and direct drug–KcsA interactions. Thus, the weaker correlation between KcsA function and RGA suggests that drug-induced changes in KcsA function are due to a combination of mechanisms that include ΔGbilayer, ΔGres, and ΔGprotein.
Drug binding to KcsA.
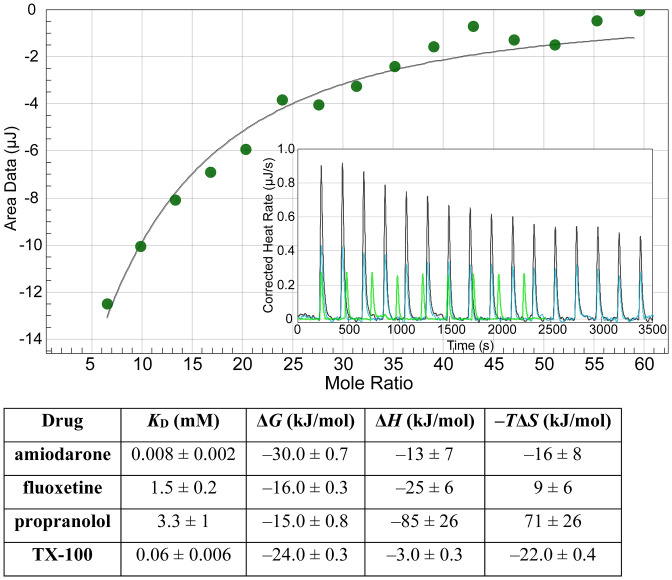
Drug-induced changes in KcsA function are likely due to a combination of indirect (bilayer-mediated) and direct (binding) drug–channel interactions. We therefore determined the drug’s affinity for KcsA using isothermal titration calorimetry. Binding isotherms for KcsA was determined in a 5 mM n-Decyl-β-D-Maltopyranoside (DM) solution (Fig. 6), and the affinity (KD), Gibbs free energy (ΔG), enthalpy (ΔH), and entropy (ΔS) were determined by fitting the plot of binding heats as function of drug/KcsA mole fraction with an independent binding model (NanoAnalyze software), which assumes single or multiple independent binding sites. Amiodarone, fluoxetine, propranolol, and TX-100 all bound to purified KcsA. Amiodarone had the highest affinity, KD = 8 µM, which overlaps with the concentrations that alter KcsA function. Fluoxetine, propranolol, troglitazone, and TX-100 had orders of magnitude lower affinities (KD > 60 µM), and there was no overlap with the nominal concentrations needed to alter KcsA function (Fig. 3). The effects of fluoxetine, troglitazone, propranolol, TX-100, and troglitazone therefore are primarily bilayer mediated. Amiodarone, however, may alter KcsA function by both direct and bilayer-mediated mechanisms, though direct binding may not lead to measurable changes in protein function. Thus, even though amiodarone binds to KcsA at concentrations at which we observe the changes in function, these changes may be due to drug-induced changes in bilayer properties [we did not observe heats of injection above control with 100 µM troglitazone, though troglitazone partitioning into lipid bilayers can be determined using isothermal titration calorimetry (9); the lack of an signal signifies no binding].
Fig. 6.
Drug binding to purified KcsA was determined using isothermal titration calorimetry. Fluoxetine binding to purified KcsA. (Inset) Trace plotting the rate of heat change as a function of time as fluoxetine is titrated into the sample cell containing KcsA (black), fluoxetine titrated into buffer solution in the absence of KcsA (blue), and buffer solution titrated into 14 μM KcsA (green) (the injection duration and KcsA concentration in the cell was different in the latter control, as compared to the experiment in which fluoxetine was injected; the heats for this control did not change with injection number). Buffer solution composition is the same in the sample cell and syringe and contained 5 mM detergent and DMSO if it was required for drug solubilization. The sample cell contained 28 µM KcsA, and the injection syringe contained 30 mM fluoxetine, both dissolved in DM-containing KcsA elution Buffer B (Materials and Methods). Heats of injection, corrected for the heats of dilution, are plotted as a function of the Fluoxetine/KcsA mole fraction (green dots) and analyzed using an independent binding model which assumes a single or multiple independent binding sites (solid black line). The parameters were determined using an independent binding model fit (NanoAnalyze) to the heats of injection (mean ± SEM, n = 3 to 7).
Discussion
Many drugs are amphiphiles, meaning they will accumulate at the bilayer/solution interface and thereby alter lipid bilayer properties like thickness, curvature, and elasticity (54). This, in turn, means that drugs may alter membrane protein function by, at least, two mechanisms: binding to the target protein and, if the drug-induced changes in bilayer properties are large enough, bilayer-mediated regulation. We show this to be the case by demonstrating the following: that KcsA gating varies with changes in bilayer thickness; that structurally diverse drugs inhibit KcsA function with potencies that vary with the drugs’ mole fraction in the membrane; and that the magnitude of the drug-induced changes in KcsA function is bilayer thickness dependent (Figs. 2, 3, and 5).
We first discuss the effects of changes in bilayer thickness. We then turn to drug-induced changes in channel function and how they depend on bilayer thickness. We finally consider strategies for distinguishing between direct and bilayer-mediated changes in membrane protein function and the implications for drug development.
Bilayer Thickness Modulates Channel Function.
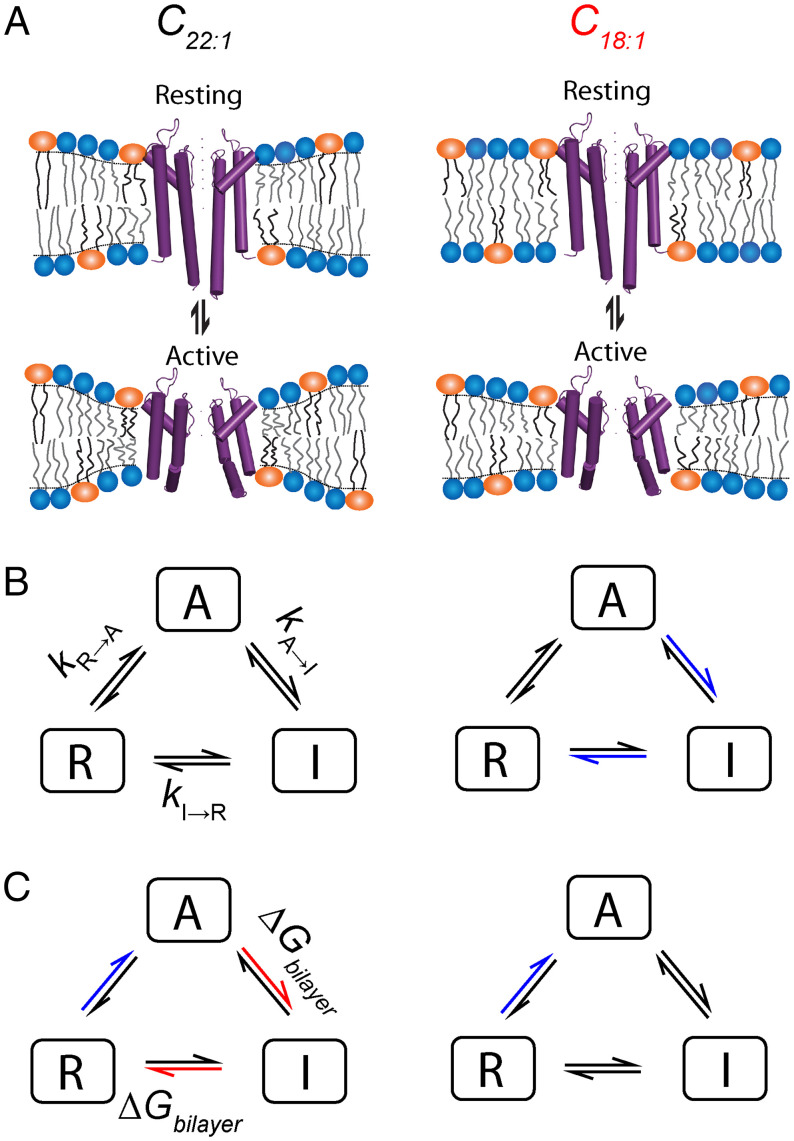
KcsA are sensitive to changes in lipid bilayer thickness, consistent with the notion of strong hydrophobic coupling between KcsA and its host bilayer (55). Comparing to C22:1, we observe a modest reduction in kA→I, an increase in kI→A, and an ∼2.5-fold decrease in kI→R in C18:1, which indicates that both the activated (A) and inactivated (I) states are stabilized, relative to the resting (R) state in the thin bilayer (Figs. 1 and 7 A and B). This conclusion is consistent with the increase in pH0.5 and greater cooperativity observed in thin bilayers and suggests that the A state is stabilized (relative to R) in C18:1, cf refs. 56 and 57. Thus, change in hydrophobic mismatch shifts KcsA conformational distribution, with little effect on each conformation, consistent with Callahan et al. (16).
The bilayer/protein interface is similar in A and I (26) and differs from that in R (25, 27), we thus expect change in bilayer thickness to have a minor effect on the distribution between A and I (Fig. 7 A and B). The bilayer contribution to the free energy difference between A/I and R is given by
where and are bilayer deformation energies associated with the activated (and inactivated) and resting states, respectively. The difference between the conformations of A (or I) and R (Fig. 7A) (27) suggests that could be significant, with R being stabilized in thick bilayers, which would account for the rapid phase of recovery at pH 7 in C22:1 (Fig. 1D) as well as the higher (nonnormalized) quench rate in C22:1 (SI Appendix, Fig. S5). It also would account for why Callahan et al. (16) found that the nonactivating E71A KcsA mutant was difficult to activate as bilayer thickness is increased.
Fig. 7.
Bilayer-mediated regulation of KcsA. (A) Conformational changes during KcsA gating deform the lipid bilayer and will contribute to conformational equilibrium. The difference in the protein/bilayer interface and coupling, as compared to gramicidin, channels will produce energy terms, ΔGres, in addition to ΔGbilayer. The magnitude of ΔGbilayer will vary with hydrophobic mismatch and therefore will be different in bilayers composed of C22:1 and C18:1. Similarly, in the presence of bilayer-modifying drug, the bilayer softening will alter ΔGbilayer and ΔGres associated with KcsA conformational change and may also alter ΔGprotein by binding KcsA. KcsA gating models comparing bilayer thickness (B) and drug (C) effects on rates in C22:1 and C18:1. (B) In thin bilayers, channel, inactivation, and recovery from inactivation are slowed compared to thick bilayers (blue arrows). (C) Drug effects also differ in bilayers of different thickness. In thin bilayers, drug-induced changes in kA→I and kI→R show negligible dependence on mD, as compared to thick bilayers where we observe rate increases (red arrows). In contrast kR→A and Peak activity are reduced in both bilayers and have similar dependence on mD.
Our conclusion that there is a bilayer thickness/hydrophobic mismatch–dependent shift in the distribution among KcsA states also provides insight into our results with TBA. TBA binding causes minimal changes in KcsA structure (58), yet TBA’s effects on channel gating depend on bilayer thickness (SI Appendix, Fig. S9), suggesting that TBA’s interactions with the channel pore are coupled to KcsA interactions with the host bilayer.
Bilayer-Mediated Regulation and Drug-Induced Changes in KcsA Function.
The drugs slowed KcsA activation in both thick and thin bilayers, and they accelerated the rate of inactivation and the recovery from inactivation in thick bilayers, with negligible effects in thin bilayers (Fig. 3). These hydrophobic mismatch–dependent drug effects are markers of bilayer-mediated regulation, because conformational transitions in membrane proteins tend to involve the proteins’ bilayer-spanning domains (7) (Fig. 7C). In the case of KcsA, the host bilayer lipids regulate both stability and function (55, 59–61). Disruption of KcsA–lipid interactions may shift the distribution among conformations and therefore functional states. The correlation between the drugs’ effects on KcsA gating and mD (Fig. 3) and bilayer-modification potency (RGA) (SI Appendix, Fig. S11) suggest that drug effects are mediated by such a disruption in KcsA/lipid interactions.
Gramicidin activity is strictly coupled to changes in bilayer properties; thus, the closer the dependence of KcsA gating on mD resembles that observed with gramicidin (Fig. 4), the greater the role of bilayer-mediated mechanism. In C22:1, the drug-induced reduction in kR→A and Peak activity showed less correlation with the bilayer-modifying potency than in C18:1, suggesting that the ΔGbilayer contribution to the R↔A equilibrium is less in C22:1 (thick) than in C18:1 (thin) bilayers (Fig. 3 A and C and Fig. 7 B and C). This likely reflects differences in the bilayer deformation associated with R and A in the two systems. The slow recovery from inactivation in thin, compared to thick bilayers, suggests that A/I (and/or the relevant transition states) are stabilized in thin bilayers. Drug-induced increases in bilayer elasticity may further stabilize I/A in thin bilayers, leading to the reduced effect on kI→R. The stronger correlations between the changes in kA→I and kI→R and mD (Fig. 3 B and D) and RGA (SI Appendix, Fig. S11 B and D) in thick, compared to thin, bilayers similarly suggest that both the A↔I and the I↔R equilibria have significant ΔGbilayer contributions in thick bilayers.
Drug-induced interference with the local lipid adaptation to KcsA (e.g., drug binding at regions with imperfect hydrophobic matching) (6) may also be involved in the drug-induced changes in channel function, decreasing kR→A and Peak activity and increasing kA→I and kI→R by weakening the energetic coupling at the protein/lipid interface. Indeed, regions with imperfect hydrophobic matching may be hotspots for nonspecific but direct pharmacological modification of membrane protein function. The orientation of M4 in ELIC depends on interactions with phosphatidylglycerol, which may be modulated by exogenous amphiphiles (62).
Bilayer-Mediated versus Direct Effects.
Promiscuous, hydrophobic drugs like amiodarone (10) and troglitazone (9) alter KcsA function at concentrations at which they alter the function of multiple membrane proteins (and gramicidin channels). Though these drugs may bind to different membrane proteins with similar affinities, the interpretation is that drugs that alter bilayer properties and KcsA function at similar concentrations, alter membrane protein function because of their bilayer-perturbing effects.
Except for amiodarone, the tested drugs’ KD for purified KcsA were markedly higher than the nominal concentrations that altered KcsA function (Fig. 6). The drug concentrations in the bilayer, of course, may be much higher than the nominal aqueous concentrations and reach millimolar (SI Appendix, Table S1). In any case, the differences between the effects in C22:1 and C18:1 demonstrate hydrophobic mismatch-dependence, meaning that these drugs act via a bilayer-mediated mechanism.
At concentrations at which these and other drugs alter bilayer properties as monitored using gramicidin channels, they promote inactivation of voltage-dependent calcium and sodium channels (NaV) (10, 35, 49), desensitize GABAA receptors (36), and promote inactivation of KcsA, indicating that these changes in the function of structurally and functionally diverse proteins are due to bilayer-mediated regulation. In contrast to their effects on KcsA, however, troglitazone, pioglitazone, and TX-100 had modest effects on NaV activation/inactivation, and TX-100 slowed recovery from inactivation. The interpretation of these divergent effects on structurally and functionally different channels is that a membrane protein’s sensitivity to changes in bilayer properties depends also on the protein–bilayer interface and changes therein, as the protein transitions between different conformations.
Implications for Drug Development.
The canonical mechanism for pharmacological regulation of membrane protein function is that the drug in question binds to (specific) sites on the proteins, thereby shifting the distribution among protein conformation, which leads to changes in function. Here, we show that structurally diverse and therapeutically unrelated drugs also can regulate ion channel function by drug-induced changes in bilayer properties (54) and protein–bilayer interfacial interactions (6, 54). This bilayer-mediated regulation has implications for drug development, because many drugs and drug-leads are hydrophobic/amphiphilic and therefore tend to partition into the bilayer/solution interface (e.g., refs. 63 and 64), which will alter lipid bilayer properties (42–44), as well as protein/bilayer interfacial interactions and nonspecific binding to hydrophobic pockets in the protein. Though distinct in terms of their impact on the energetics of conformational changes, these mechanisms are related if the drug has to partition into the bilayer to mediate its effect through either mechanism. That is, aside from the need to partition to alter bulk bilayer properties, the drug must partition to alter the interactions at the bilayer–protein interface; it also may need to partition to access a binding pocket in the transmembrane domain of the protein—though such direct drug binding only will alter function if the drug’s affinity differs among protein conformations.
When a drug’s clinical and bilayer-modifying concentration ranges overlap, one should a priori consider whether the changes in membrane protein function may result from bilayer-mediated regulation. When the changes in function can be predicted from the drug’s bilayer-modifying potency (9, 49), the contribution of the bilayer to the overall therapeutic profile is likely to be significant. It is in this context important to note that the relevant concentrations are likely to be the drug concentrations in bilayer, which can be approximated as [Drug]m ∼ mD·[Lipid]m, where [Lipid]m ∼ 1.1 M. An mD = 0.01 thus corresponds to a [Drug]m ∼ 11 mM.
Drug-induced changes in bilayer properties, of course, may alter the function of numerous proteins, which may result in desired polypharmacology, as may be the case for the antiarrhythmic amiodarone (10, 65, 66) and antidepressant fluoxetine (30, 39), or undesired off-target effects and toxicity, as may be the case for the PPARγ agonist troglitazone (9, 67). It thus becomes important to be able to distinguish between direct yet nonspecific drug–protein interactions and bilayer-mediated interactions, which can be done using the strategies presented here. Knowing that drugs and drug candidates with desired actions are bilayer modifiers and therefore likely to be promiscuous may guide the development of molecules that retain the desired activity with less bilayer-modifying effect.
Materials and Methods
Materials.
Phospholipids DC22:1PC (1,2-dierucoyl-sn-glycero-3-phosphocholine), DC22:1PG (1,2-dierucoyl-sn-glycero-3-phospho-(1'-rac-glycerol)), DC18:1PC (1,2-dioleoyl-sn-glycero-3-phosphocholine), and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol)) were purchased from Avanti Polar Lipids (Alabaster). The naturally occurring mixture of linear gramicidin proteins from Bacillus brevis (68), denoted gramicidin D (gD), was obtained from Sigma-Aldrich (St. Louis, MO). ANTS was from Thermo Fisher Scientific (Waltham); DM was from Anatrace (Maumee); and BioBeads SM-2 from BioRad (Hercules). Phenylmethylsulfonyl fluoride (PMSF), Leupeptin, Pepstatin, IPTG (Isopropyl-β-D-thiogalactoside), and Thallium (I) Nitrate (Tl+) were from Sigma-Aldrich.
KcsA Mutagenesis, Expression, and Purification.
KcsA was expressed and purified as described previously (15) with minor modifications. Briefly, KcsA was expressed from the pQE60 vector in BL21 (DE3) T1-R cells (Sigma-Aldrich). Cells were grown with aeration at 37 °C until an OD600 ~ 1.0, and expression was induced by addition of 0.5 mM IPTG. After 3 h induction at 37 °C, the cells were spun at 5,000 rpm for 10 min at 4 °C. Cell pellets were resuspended in buffer containing 50 mM Tris·HCl, 100 mM KCl, and protease inhibitor mixture: PMSF, Leupeptin, and Pepstatin, pH 7.6, and lysed by probe sonication. Membranes were solubilized by incubation with 25 mM DM for 2 h at room temperature on a rotator followed by centrifugation at 17,500 rpm at 4 °C for 45 min. The following affinity and sizing purification steps then were carried out on AKTA Pure (GE Healthcare Life Sciences). The supernatant was applied to a Hi-Trap column (GE Healthcare Life Sciences) charged with Co2+ using 50 mL Superloop (GE Healthcare Life Sciences) and equilibrated with Buffer B (20 mM Tris·HCl, 100 mM KCl, pH 7.6, and 5 mM DM). The column then was washed with Buffer B supplemented with 30 mM imidazole. The protein was eluted with Buffer B supplemented with 300 mM imidazole. Peak fractions were combined and concentrated on Amicon molecular weight cut-off (MWCO) 50 Filter units (EMD Millipore). The concentrated samples were applied to a Superdex 200 Increase 10/300 GL column (GE) equilibrated with Buffer B, and fractions containing KcsA were pooled and assessed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentration was calculated from absorption measurement at 280 nm using an extinction coefficient of 38,500 M−1/cm−1 (69) using GENESYS 180 UV-Vis Spectrometer (Thermo Fisher Scientific).
KcsA Reconstitution.
KcsA reconstitution was done as described previously (15). Briefly, DC22:1PC and DC22:1PG or DC18:1PC and POPG in chloroform were mixed 3:1 in a 50-mL round-bottom flask for C22:1 or C18:1, respectively. The mixture was dried under N2, left overnight in a desiccator to remove the remnants of chloroform, and resuspended in a solution of ANTS 25 mM, KCl 140 mM, Hepes 10 mM, and succinic acid 10 mM, pH 7, followed by vortexing and stepwise addition of 35 mg 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulphonate (CHAPS) (final concentration 34 mM) for solubilization and sonication. Purified KcsA was added at an 11 μg:mg protein:lipid ratio and incubated at room temperature for 1 h. LUVs then were formed by incubating the lipid/CHAPS/protein mixtures with 1 g of BioBeads SM 2 at room temperature on a rotator for 2.5 h to remove the CHAPS. After separating the BioBeads from the LUVs, the LUVs were briefly sonicated and extruded through a 100-nm filter using Avanti Mini Extruder (Avanti Polar Lipids).
Stopped-Flow Assay.
The rate of Tl+ influx into ANTS-loaded LUV is proportional to the number of activated KcsA/LUV and was determined from the rate of ANTS quench by Tl+ using sequential mixing stopped-flow spectrofluorometry (15), which allows for timed and separate control of two pairs of syringes (SI Appendix, Figs. S2–S4) in an SX-20 spectrofluorometer (Applied Photophysics, Leatherhead, United Kingdom). Activating Ram 1 fills the delay loop with the contents of syringes 1 (containing LUVs with reconstituted KcsA) and 2 (containing buffer) as 1:1 mixture that then is incubated for the desired duration: 15 ms (the minimum delay loop setting) to 60 s. The pH in syringe 2 is varied to achieve the desired pH (4.0 to 7.0) in the delay loop. For example, if KcsA LUVs loaded with the fluorophore ANTS are to be incubated at pH 5 and the KcsA–LUV solution in syringe 1 is pH 7, then to achieve pH 5 in the delay loop, syringe 2 was filled with a pH 3.9 solution. After the desired incubation, the second pair of syringes are triggered; syringe 3 flushes the delay loop, and syringe 4 contains the Tl+ quencher at the desired pH, which is mixed with the content of the delay loop in the optical cell at a 1:1 ratio. The quench time course acquired following mixing in the optical cell has the dead time of 2 ms, meaning that 2 ms is required for all the contents to mix fully and the data output can be analyzed reliably after this point, which allows for quantification of Tl+ influx through open KcsA.
KcsA dose–response (H+-activation) curves were determined by varying the pH in the delay loop over the desired pH range followed by incubation for 320 ms to achieve maximal population of open channels at the pH and testing for function in the second mixing reaction (SI Appendix, Fig. S2). The time to Peak activation depends on pH, but the Activity at 320 ms at any pH was not statistically distinguishable from the Peak activity at that pH (SI Appendix, Fig. S6).
The time course of activation and inactivation was measured with a pH of 5 in the delay loop, and the KcsA-containing LUVs were incubated between 15 ms and 10 s followed by testing for function in the second mixing reaction (SI Appendix, Fig. S3).
The time course of recovery from inactivation was determined by incubating KcsA at pH 4 for 10 min to achieve maximal inactivation. Inactivated KcsA at pH 4 then was combined with a buffer in syringe 2 at pH 11.8 to obtain a final pH 7.0 in the delay loop and incubated for between 15 ms and 60 s before testing for function in the second mixing reaction by mixing 1:1 the content of the delay loop with a Tl+ solution at pH 2.3 to give a pH of 4.0 in the optical cell (SI Appendix, Fig. S4). In experiments in which we measured the recovery at pH 5.2, the pHs in the syringes 2 and 4 were 7.25 and 3, respectively.
Several repeats were recorded for each condition (pH or duration in the delay loop). The resulting quench time courses were fit with a stretched exponential function (70)
where F(t) is the fluorescence intensity as a function of time, τ0 a parameter with units of time, and β (0<β≤1) is a measure of sample dispersity and the distribution of conducting channels. β = 1.0 for a homogenous population of vesicles. The observed distribution of β varied between 1.0 and near-zero, with a tendency for β to increase as the slope decreased (71). The ANTS quench rate then was determined as at t = 2 ms (instrument dead time) from the above fit. The fitting of quench traces was done using MATLAB (MathWorks) using previously developed code (53). Slopes from the repeats recorded in each condition were imported into Origin (OriginLab) software, averaged, and the averages were plotted as a function of pH, for the dose–response experiments, or duration in the delay loop, for the activation and inactivation or recovery from inactivation experiments. All experiments were done at 25 °C.
The drug effects on KcsA function were tested by preincubating LUV-reconstituted KcsA with or without (control) drugs for 10 min at 25 °C.
Gramicidin-Based Fluorescence Assay.
We used gramicidin channels as probes for changes in bilayer properties (10, 53). Briefly, the naturally occurring mixture of linear gramicidin analogs, gD, is reconstituted into LUVs composed of C22:1PC and loaded with the fluorophore ANTS. The ANTS-loaded, gramicidin-doped LUVs were mixed with the Tl+-containing quench buffer using SX-20 stopped-flow spectrometer, which allows for measuring the quench of the ANTS fluorescence due to Tl+ entry through conducting gramicidin channels. The time course of fluorescence quench was recorded, fit with a stretched exponential function, and the quench rate was determined at t = 2 ms (Stopped-Flow Assay). Changes in quench rate reflect changes in the gramicidin monomer↔dimer equilibrium. Drug-induced changes in the equilibrium (quench rate) were determined after the gramicidin-doped LUVs were incubated for 10 min at 25 °C in the absence (control) or presence of drug, after which the quench rates were determined. The quench rate in the presence of drug was normalized to the control rate from the same experiment. Mean ± SEM of normalized rates from multiple experiments are evaluated (RGA) and plotted as a function of drug mole fraction in the membrane (mD) (Fig. 3).
Data Analysis.
All quench trace plotting, fitting, and analysis, except for activation and inactivation model fitting, was performed in Origin. KcsA [H+] dose–response curves in the absence and presence of drugs were determined by plotting the quench trace slopes at varying pH as a function of pH and fitting with a Hill function in Origin. The resulting pH0.5 and nH values from each experiment were averaged and reported as mean ± SEM.
The activation and inactivation rates were determined from the initial slopes of the quench traces after gradually increasing incubation at pH 5 from 15 to 10,000 ms. The results were plotted as function of duration in the delay loop at pH 5 (SI Appendix, Fig. S2). The resulting time course was interpolated using linear interpolation (Origin software) over the time range of 0 to 10,000 ms, which yielded a 1,000-point dataset. The interpolated dataset was imported into QuB (22), a hidden Markov model software suite for fitting with a three-state model (Fig. 2, Inset), in which KcsA traverses resting–activated–inactivated state upon application of the ligand (H+). The rates we report are the following: the forward rate for Resting (R) → Active (A) transition (kR→A); the forward rate for A → Inactivated (I) transition (kA→I). The rates for A→R transitions (kA→R) were ∼104-fold smaller than kR→A when estimated using the R ↔ A ↔ I model, and we did not consider them further (if we neglected inactivation and limited the analysis to the first 320 ms, kA→R was, as expected, comparable to kR→A when estimated using an equilibrium binding, R ↔ A, model). Goodness of fit was determined in QuB using the Log Likelihood function. The very different estimates for kA→R when estimated using the R ↔ A ↔ I model and the R ↔ A model implies that our estimates for pH0.5 of the KcsA dose–response curves do not provide information about H+ binding to KcsA.
The rate of recovery from inactivation, kI→R, was determined by plotting the slopes of the quench traces as function of incubation time at pH 7 and fit with single exponential in Origin software.
Correlation of KcsA Function with RGA and mD.
To compare changes in KcsA function in the presence of drugs to effects on gramicidin channel activity (RGA) and the mole fraction (mD) each parameter, kR→A, kA→I, Peak activity (highest slope obtained at pH 5 during determination of the time course of activation/inactivation), and kI→R, determined in the presence of a drug were normalized to that value in control conditions (no drug) for each experiment. This normalization resulted in changes relative (to control) that allowed for averaging parameters among multiple experiments. The resulting averages of normalized changes for multiple experiments were plotted as function of changes in gramicidin activity in the presence of the same drugs and concentrations. The plots were fit with linear fits and Pearson’s correlation coefficient, and Adjusted R2 were used to compare correlations of different parameters. Linear fits were determined in the Origin software.
Fluorescence Size-Exclusion Chromatography Assays.
We used fluorescence size-exclusion chromatography to determine the amount of channel protein using a high-sensitivity fluorescence detector, which allows us to detect analytical quantities of protein (<5 ng) using tryptophan fluorescence, with samples injected automatically in a sequential fashion. To establish a calibration curve, we used purified KcsA solubilized in n-Dodecyl β-D-maltoside (DDM) and injecting samples with amounts of protein ranging from 1 to 100 µg. To measure the protein content in the LUVs composed of C22:1 or C18:1 with reconstituted KcsA, the LUVs were solubilized by addition of DDM (Anatrace) to a final concentration of 20 mM and incubated at room temperature for 10 min to solubilize the KcsA. For each mixture, 100-μL sample was placed into a sample cooling tray inside a high-performance liquid chromatography (HPLC) system (Shimadzu), and the samples were injected one at a time onto a Superose 6 Increase 10/300 GL column (GE Healthcare) equilibrated with buffer containing 40 mM KCl, 10 mM Hepes, 10 mM Succinic acid (pH 7.0), and 1 mM DDM. Elution of KcsA was monitored by recording the tryptophan fluorescence with a postcolumn fluorescence detector (Shimadzu) with excitation wavelength 280 nm and recording the emission above 335 nm. For calibration, defined amounts of purified KcsA solubilized in DDM were injected, which allowed for calculating the amount of protein in the proteo-liposome solubilization experiments.
Lipid Determination by Phosphate Assay.
The amount of lipid was determined by digestion followed by spectrophotometric measurement of inorganic phosphate (72); refer also to Avanti Polar Lipids https://avantilipids.com/tech-support/analytical-procedures/determination-of-total-phosphorus for protocol. Briefly, lipid samples and phosphate standard (Sigma-Aldrich) with known concentration were digested by heating in sulfuric acid and hydrogen peroxide. The inorganic phosphate was then complexed with ammonium molybdate (VI) tetrahydrate and ascorbic acid for colorimetric analysis. Optical density at 820 nm was recorded for each sample and for the standard solutions of varying concentrations. The amount of phosphate in each C22:1 or C18:1 batch was determined by comparing the OD at 820 nm to the calibration curve derived from the phosphate standards.
Drug Binding to KcsA.
Drug binding to KcsA was determined by isothermal titration calorimetry using a TA Instruments NanoITC (TA Instruments, Waters Corporation, New Castle) at 25 °C. To minimize the effects of heats of dilution, KcsA and drugs were diluted in the same buffer: 20 mM Tris·HCl, 100 mM KCl, pH 7.6, 5 mM DM, and, if the drug was dissolved in dimethyl sulfoxide (DMSO), the same final amount of DMSO was added to the KcsA–LUV solution. Final DMSO concentration did not exceed 0.5%. KcsA was loaded into the cell (volume 170 µL), and drug solution was loaded into the titration syringe with the total volume of 50 µL. KcsA and drug concentrations were adjusted to obtain optimal signal and ranged between 14 to 56 µM and 100 µM to 10 mM for KcsA and drug, respectively. Heats of binding were determined from the binding isotherms and analyzed using NanoAnalyze software (TA Instruments). Control experiments were done to determine heats of dilution for each drug by injecting drug solutions into the DM-containing buffer in the absence of KcsA, and the heats of binding (in the presence of both drug and KcsA) were corrected for the heats of dilution of the drug. To determine drug affinity (KD), the corrected heats of binding were plotted as a function of the drug/KcsA mole fraction and fitted by an independent binding model (NanoAnalyze), which assumes, single or multiple, independent binding sites.
Drug Partitioning into LUVs.
Drug partitioning into lipid bilayers was also measured by isothermal titration calorimetry. LUV and drug were each diluted in the same buffer (140 mM NaNO3 and 10 mM Hepes, pH 7) plus DMSO to match the drug’s solubility requirements (≤ 0.25%). The drug and lipid concentrations were adjusted to optimize the heats of injection and allow for saturation at the end of the titration curve. The titration syringe was loaded with the LUV suspension, and 1.5 to 3 μL aliquots were injected into the sample cell containing the drug solution. The resulting heats of injection were corrected for baseline heats and integrated to obtain injection enthalpies in NanoAnalyze software (TA Instruments). Injection enthalpies were exported into Origin software and fit with
| [1] |
where i is the injection number, δhA the injection enthalpy, δV the injection volume, CL is lipid concentration, vL the molar lipid volume, the total amount of drug in moles, VW(0) the initial volume in the sample cell, and KP the drug partition coefficient into the bilayer. Knowing KP, we then could estimate the drug’s concentration in the bilayer as (9, 37):
| [2] |
The drugs’ mole fractions in the bilayer (mD) were estimated as:
| [3] |
where [Drug]m and [Lipid]m denote the drug and lipid concentrations in the membrane phase, [Drug]nom the nominal drug concentration in the aqueous phase (moles/system volume), and Vaq and Vlip the volumes of the aqueous and lipid phase, respectively (9, 37).
Supplementary Material
Acknowledgments
We thank Dr. Olga Boudker and Krishna Reddy for advice and helpful discussions on isothermal titration calorimetry use and data analysis and Drs. Crina Nimigean, Dorothy M. Kim, and David J. Posson for generously providing us with KcsA in pQE60 vector and consultation on purification techniques. This work was supported by a grant from the Kellen Foundation to R.R. and NIH grant GM021342 to O.S.A.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113229118/-/DCSupplemental.
Data Availability
Experimental data have been deposited in the publicly accessible Open Science Framework repository https://osf.io/u49jw/?view_only=80b59dfd74b142459e0cd7ba37e3d7ed (74).
Change History
December 13, 2021: The SI Appendix has been updated.
References
- 1.Bienvenüe A., Marie J. S., Modulation of protein function by lipids. Curr. Top. Membr. 40, 319–354 (1994). [Google Scholar]
- 2.Lee A. G., How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Andersen O. S., Koeppe R. E. II, Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 36, 107–130 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Marsh D., Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 1778, 1545–1575 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Nji E., Chatzikyriakidou Y., Landreh M., Drew D., An engineered thermal-shift screen reveals specific lipid preferences of eukaryotic and prokaryotic membrane proteins. Nat. Commun. 9, 4253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mondal S., Khelashvili G., Shi L., Weinstein H., The cost of living in the membrane: A case study of hydrophobic mismatch for the multi-segment protein LeuT. Chem. Phys. Lipids 169, 27–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundbaek J. A., Collingwood S. A., Ingólfsson H. I., Kapoor R., Andersen O. S., Lipid bilayer regulation of membrane protein function: Gramicidin channels as molecular force probes. J. R. Soc. Interface 7, 373–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuello L. G., Romero J. G., Cortes D. M., Perozo E., pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry 37, 3229–3236 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Rusinova R., et al. , Thiazolidinedione insulin sensitizers alter lipid bilayer properties and voltage-dependent sodium channel function: Implications for drug discovery. J. Gen. Physiol. 138, 249–270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusinova R., Koeppe R. E. II, Andersen O. S., A general mechanism for drug promiscuity: Studies with amiodarone and other antiarrhythmics. J. Gen. Physiol. 146, 463–475 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh B. N., Current antiarrhythmic drugs: An overview of mechanisms of action and potential clinical utility. J. Cardiovasc. Electrophysiol. 10, 283–301 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara T., Horikoshi H., Troglitazone and related compounds: Therapeutic potential beyond diabetes. Life Sci. 67, 2405–2416 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Heijman J., Dobrev D., Pleiotropic actions of amiodarone: Still puzzling after half a century. Naunyn Schmiedebergs Arch. Pharmacol. 386, 571–574 (2013). [DOI] [PubMed] [Google Scholar]
- 14.LeMasurier M., Heginbotham L., Miller C., KcsA: It’s a potassium channel. J. Gen. Physiol. 118, 303–314 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusinova R., Kim D. M., Nimigean C. M., Andersen O. S., Regulation of ion channel function by the host lipid bilayer examined by a stopped-flow spectrofluorometric assay. Biophys. J. 106, 1070–1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callahan K. M., Mondou B., Sasseville L., Schwartz J. L., D’Avanzo N., The influence of membrane bilayer thickness on KcsA channel activity. Channels (Austin) 13, 424–439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaven A. H., et al. , Gramicidin A channel formation induces local lipid redistribution I: Experiment and simulation. Biophys. J. 112, 1185–1197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis B. A., Engelman D. M., Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J. Mol. Biol. 166, 203–210 (1983). [DOI] [PubMed] [Google Scholar]
- 19.Heginbotham L., LeMasurier M., Kolmakova-Partensky L., Miller C., Single streptomyces lividans K(+) channels: Functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrapani S., Cordero-Morales J. F., Perozo E., A quantitative description of KcsA gating I: Macroscopic currents. J. Gen. Physiol. 130, 465–478 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ader C., et al. , Coupling of activation and inactivation gate in a K+-channel: Potassium and ligand sensitivity. EMBO J. 28, 2825–2834 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolai C., Sachs F., Solving ion channel kinetics with the QuB software. Biophys. Rev. Lett. 08, 191–211 (2013). [Google Scholar]
- 23.Chakrapani S., Cordero-Morales J. F., Perozo E., A quantitative description of KcsA gating II: Single-channel currents. J. Gen. Physiol. 130, 479–496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matulef K., Annen A. W., Nix J. C., Valiyaveetil F. I., Individual ion binding sites in the K(+) channel play distinct roles in C-type inactivation and in recovery from inactivation. Structure 24, 750–761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Morais-Cabral J. H., Kaufman A., MacKinnon R., Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414, 43–48 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Cuello L. G., Jogini V., Cortes D. M., Perozo E., Structural mechanism of C-type inactivation in K(+) channels. Nature 466, 203–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumino A., Sumikama T., Iwamoto M., Dewa T., Oiki S., The open gate structure of the membrane-embedded KcsA potassium channel viewed from the cytoplasmic side. Sci. Rep. 3, 1063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.daCosta C. J., Dey L., Therien J. P., Baenziger J. E., A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nat. Chem. Biol. 9, 701–707 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Andersen O. S., R. E. Koeppe, II, Roux B., “Gramicidin channels. Versatile tools” in Biological Membrane Ion Channels: Dynamics, Structure, and Applications, Chung S.-H., Andersen O. S., Krishnamurthy V., Eds. (Springer Verlag, New York, 2007), pp. 33–80. [Google Scholar]
- 30.Bianchi M. T., Non-serotonin anti-depressant actions: direct ion channel modulation by SSRIs and the concept of single agent poly-pharmacy. Med. Hypotheses 70, 951–956 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Wang D. W., et al. , Propranolol blocks cardiac and neuronal voltage-gated sodium channels. Front. Pharmacol. 1, 144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong C. M., Interaction of tetraethylammonium ion derivatives with the potassium channels of giant axons. J. Gen. Physiol. 58, 413–437 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M., Morais-Cabral J. H., Mann S., MacKinnon R., Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature 411, 657–661 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Lundbaek J. A., Birn P., Girshman J., Hansen A. J., Andersen O. S., Membrane stiffness and channel function. Biochemistry 35, 3825–3830 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Lundbaek J. A., et al. , Regulation of sodium channel function by bilayer elasticity: The importance of hydrophobic coupling. Effects of micelle-forming amphiphiles and cholesterol. J. Gen. Physiol. 123, 599–621 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Søgaard R., et al. , GABA(A) receptor function is regulated by lipid bilayer elasticity. Biochemistry 45, 13118–13129 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Wenk M. R., Alt T., Seelig A., Seelig J., Octyl-beta-D-glucopyranoside partitioning into lipid bilayers: thermodynamics of binding and structural changes of the bilayer. Biophys. J. 72, 1719–1731 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kragh-Hansen U., le Maire M., Møller J. V., The mechanism of detergent solubilization of liposomes and protein-containing membranes. Biophys. J. 75, 2932–2946 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapoor R., Peyear T. A., Koeppe R. E. II, Andersen O. S., Antidepressants are modifiers of lipid bilayer properties. J. Gen. Physiol. 151, 342–356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seydel J. K., Octanol-water partitioning versus partitioning into membranes. Methods Princ Med Chem 15, 35–50 (2002). [Google Scholar]
- 41.Cruickshank J. M., The clinical importance of cardioselectivity and lipophilicity in beta blockers. Am. Heart J. 100, 160–178 (1980). [DOI] [PubMed] [Google Scholar]
- 42.Evans E., Rawicz W., Hofmann A. F., “Lipid bilayer expansion and mechanical disruption in solutions of water-soluble bile acid” in Bile Acids in Gastroenterology: Basic and Clinical Advances, Hofmann A. F., Paumgartner G., Stiehl A., Eds. (Kluwer Academic Publishers, Dordrecht, 1995), pp. 59–68. [Google Scholar]
- 43.Zhelev D. V., Material property characteristics for lipid bilayers containing lysolipid. Biophys. J. 75, 321–330 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruno M. J., Rusinova R., Gleason N. J., Koeppe R. E. II, Andersen O. S., Interactions of drugs and amphiphiles with membranes: Modulation of lipid bilayer elastic properties by changes in acyl chain unsaturation and protonation. Faraday Discuss. 161, 461–480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim T., et al. , Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers. Biophys. J. 102, 1551–1560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodt A. J., Beaven A. H., Andersen O. S., Im W., Pastor R. W., Gramicidin a channel formation induces local lipid redistribution II: A 3D continuum elastic model. Biophys. J. 112, 1198–1213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun D., et al. , Molecular mechanism for gramicidin dimerization and dissociation in bilayers of different thickness. Biophys. J. 117, 1831–1844 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundbaek J. A., R. E. Koeppe, 2nd, Andersen O. S., Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl. Acad. Sci. U.S.A. 107, 15427–15430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundbaek J. A., et al. , Capsaicin regulates voltage-dependent sodium channels by altering lipid bilayer elasticity. Mol. Pharmacol. 68, 680–689 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Thompson A. N., Posson D. J., Parsa P. V., Nimigean C. M., Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. USA 105, 6900–6905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swenson R. P. Jr., Armstrong C. M., K+ channels close more slowly in the presence of external K+ and Rb+. Nature 291, 427–429 (1981). [DOI] [PubMed] [Google Scholar]
- 52.Choi K. L., Aldrich R. W., Yellen G., Tetraethylammonium blockade distinguishes two inactivation mechanisms in voltage-activated K+ channels. Proc. Natl. Acad. Sci. U.S.A. 88, 5092–5095 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ingólfsson H. I., Andersen O. S., Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev. Technol. 8, 427–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen O. S., Perspectives on how to drug an ion channel. J. Gen. Physiol. 131, 395–397 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williamson I. M., Alvis S. J., East J. M., Lee A. G., Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 83, 2026–2038 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zagotta W. N., Hoshi T., Aldrich R. W., Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J. Gen. Physiol. 103, 321–362 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yifrach O., Hill coefficient for estimating the magnitude of cooperativity in gating transitions of voltage-dependent ion channels. Biophys. J. 87, 822–830 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faraldo-Gómez J. D., et al. , Mechanism of intracellular block of the KcsA K+ channel by tetrabutylammonium: Insights from X-ray crystallography, electrophysiology and replica-exchange molecular dynamics simulations. J. Mol. Biol. 365, 649–662 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Valiyaveetil F. I., Zhou Y., MacKinnon R., Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 41, 10771–10777 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Marius P., et al. , Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys. J. 94, 1689–1698 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Cruijsen E. A., Prokofyev A. V., Pongs O., Baldus M., Probing conformational changes during the gating cycle of a potassium channel in lipid bilayers. Biophys. J. 112, 99–108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hénault C. M., et al. , A lipid site shapes the agonist response of a pentameric ligand-gated ion channel. Nat. Chem. Biol. 15, 1156–1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arcario M. J., Mayne C. G., Tajkhorshid E., Atomistic models of general anesthetics for use in in silico biological studies. J. Phys. Chem. B 118, 12075–12086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ingólfsson H. I., et al. , Phytochemicals perturb membranes and promiscuously alter protein function. ACS Chem. Biol. 9, 1788–1798 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimetbaum P., Antiarrhythmic drug therapy for atrial fibrillation. Circulation 125, 381–389 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Heijman J., Voigt N., Dobrev D., New directions in antiarrhythmic drug therapy for atrial fibrillation. Future Cardiol. 9, 71–88 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Sarafidis P. A., Thiazolidinedione derivatives in diabetes and cardiovascular disease: An update. Fundam. Clin. Pharmacol. 22, 247–264 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Hotchkiss R. D., Dubos R. J., The isolation of bactericidal substances from cultures of Bacillus brevis. J. Biol. Chem. 141, 155–162 (1941). [Google Scholar]
- 69.Heginbotham L., Kolmakova-Partensky L., Miller C., Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berberan-Santos M., Bodunov E., Valeur B., Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential). Chem. Phys. 315, 171–182 (2005). [Google Scholar]
- 71.Alejo J. L., Blanchard S. C., Andersen O. S., Small-molecule photostabilizing agents are modifiers of lipid bilayer properties. Biophys. J. 104, 2410–2418 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen P. S., Toribara T. Y., Warner H., Microdetermination of Phosphorus. Anal. Chem. 28, 1756–1758 (1956). [Google Scholar]
- 73.Giaginis C., Theocharis S., Tsantili-Kakoulidou A., Contribution to the standardization of the chromatographic conditions for the lipophilicity assessment of neutral and basic drugs. Anal. Chim. Acta 573-574, 311–318 (2006). [DOI] [PubMed] [Google Scholar]
- 74.R. Rusinova, Mechanisms underlying drug-mediated regulation of membrane protein function. Open Science Framework. https://osf.io/u49jw/?view_only=80b59dfd74b142459e0cd7ba37e3d7ed. Deposited 8 October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Experimental data have been deposited in the publicly accessible Open Science Framework repository https://osf.io/u49jw/?view_only=80b59dfd74b142459e0cd7ba37e3d7ed (74).