Significance
Past studies have depicted Trichodesmium as a diazotrophic genus. This genus contributes significantly to nitrogen fixation in ocean surface waters under the form of large filaments and colonies. As a result of this unusual lifestyle, Trichodesmium is not abundant in the bacterial cellular size fraction most microbial ecologists have focused on with metagenomics thus far, and not a single environmental genome was recovered. Using large cellular size fractions of Tara Oceans, we have recovered environmental genomes of Trichodesmium and revealed a fundamental aspect of this genus: the existence of species that lost the ability to fix nitrogen (along with other critical functional traits associated with nitrogen fixation) yet are abundant and widespread on the surface of the oceans.
Keywords: Trichodesmium, metagenomics, Tara Oceans, nitrogen fixation, ecology and evolution
Abstract
Filamentous and colony-forming cells within the cyanobacterial genus Trichodesmium might account for nearly half of nitrogen fixation in the sunlit ocean, a critical mechanism that sustains plankton’s primary productivity. Trichodesmium has long been portrayed as a diazotrophic genus. By means of genome-resolved metagenomics, here we reveal that nondiazotrophic Trichodesmium species not only exist but also are abundant and widespread in the open ocean, benefiting from a previously overlooked functional lifestyle to expand the biogeography of this prominent marine genus. Near-complete environmental genomes for those closely related candidate species reproducibly shared functional features including a lack of genes related to nitrogen fixation, hydrogen recycling, and hopanoid lipid production concomitant with the enrichment of nitrogen assimilation genes. Our results elucidate fieldwork observations of Trichodesmium cells fixing carbon but not nitrogen. The Black Queen hypothesis and burden of low-oxygen concentration requirements provide a rationale to explain gene loss linked to nitrogen fixation among Trichodesmium species. Disconnecting taxonomic signal for this genus from a microbial community’s ability to fix nitrogen will help refine our understanding of the marine nitrogen balance. Finally, we are reminded that established links between taxonomic lineages and functional traits do not always hold true.
Plankton in the sunlit ocean includes a wide range of microbial lineages with different functional capabilities that influence global biogeochemical cycles and climate (1–6). The primary productivity of plankton is constrained by the amount of bioavailable nitrogen (7, 8), a critical element for cellular growth and division. Few bacterial and archaeal populations can code for the catalytic (nifHDK) and biosynthetic (nifENB) proteins required for biological nitrogen fixation, transferring a valuable source of nitrogen from the atmosphere to the plankton (9–11). These populations are called diazotrophs and represent key marine players that sustain plankton primary productivity in large oceanic regions (9).
Cyanobacterial species within the genus Trichodesmium first described in 1830 (12) are among the most prominent marine nitrogen fixers (13), possibly accounting for half the biological nitrogen fixation in the sunlit ocean (14–16). While other makers provide different trends (e.g., in ref. 17), phylogeny of the 16S ribosomal RNA (rRNA) genes points to three distinct Trichodesmium clades covering Trichodesmium thiebautii and Trichodesmium hildebrandtii (clade I), Trichodesmium tenue and Trichodesmium contortum (clade II), and Trichodesmium erythraeum, and Trichodesmium havanum (clade III) (18). For now, only cultures of T. erythraeum and T. thiebautti have been characterized with genomics. Insights from culture representatives and oceanic expeditions have provided a wealth of information regarding their biogeography, functional lifestyles, and nitrogen fixation regulation mechanisms (14, 19). Most notably, Trichodesmium cells are capable of forming large blooms of filaments and colonies in the sunlit ocean (13, 20–22), regulate nitrogen fixation rates on a daily basis using a dedicated circadian rhythm (23, 24) as well as an associated microbiome (25–27), and possess genes linked to the recycling of hydrogen, a by-product of nitrogen fixation (28–30). Decades of scientific insights have shaped a dogma depicting the numerous Trichodesmium cells as carbon and nitrogen fixers with a highly beneficial role for plankton productivity.
Recently, large-scale, single-cell, and genome-resolved metagenomic surveys have dramatically expanded the genomic characterization of free-living marine microbes (31–34) and lead to new insights into primary processes in the surface of the open ocean, including nitrogen fixation (34). However, a major focus on small planktonic size fractions typically excluded organisms such as Trichodesmium that form filaments and colonies and occur in larger size fractions (21). This gap is partially filled by a recent metagenomic survey that reconstructed and manually curated nearly 2,000 bacterial genomes abundant in large size fractions of the Tara Oceans expeditions (35), which included five near-complete environmental Trichodesmium genomes. They correspond to Trichodesmium genomes recovered without the need for cultivation, providing a venue to study the ecology and evolution of populations within this genus and occurring in the open ocean. While three genomes resolve to T. erythraeum and T. thiebautti, the remaining ones reveal the long-overlooked existence of nondiazotrophic Trichodesmium species that are not only abundant in the open ocean but also expand the biogeography of this prominent marine genus.
Results
A Set of Trichodesmium Environmental Genomes from the Sunlit Ocean.
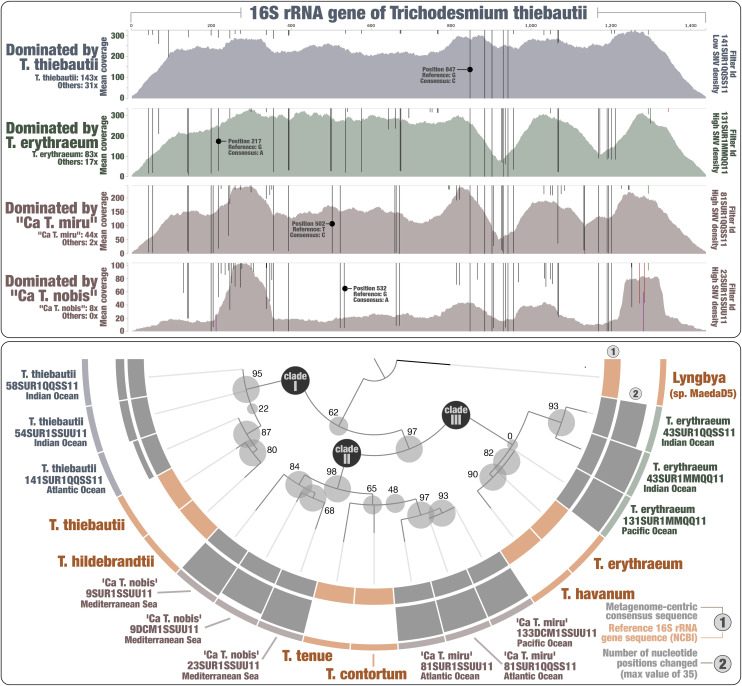
A genome-resolved metagenomic survey was recently performed to target planktonic populations abundant in polar, temperate, and tropical sunlit oceans using nearly 1,000 metagenomes (total of 280 billion reads) derived from the Tara Oceans expeditions (36) and encompassing eight plankton size fractions ranging from 0.8 μm to 2 mm (37) (Dataset S1). Notably, bacterial metagenome-assembled genomes (MAGs) characterized from this dataset covered five distinct Trichodesmium populations (35) (Fig. 1 and Dataset S2).
Fig. 1.
Hybrid phylogeny of the 16S rRNA gene of Trichodesmium species using references and metagenomes. Top panel displays read recruitments for the T. thiebautii 16S rRNA gene across four metagenomes, each dominated by a single Trichodesmium species (mapping stringency of >95% identity over >95% of the read length). Single-nucleotide variants identified by anvi’o (when using default parameters) were visualized with an amplitude reaching 100% when all reads contained the same nucleotide type. The bottom panel displays a hybrid phylogenetic analysis using both reference sequences and 12 metagenome-centric consensus sequences. The number of changed nucleotide positions compared to the reference (T. thiebautii 16S rRNA gene) is presented for each consensus sequence. The phylogenetic tree was decorated with associated data and visualized using anvi’o.
The Trichodesmium MAGs were affiliated to T. erythraeum (one population), T. thiebautii (two closely related populations), and two candidate species we tentatively named “Candidatus Trichodesmium miru” (one population) and “Candidatus Trichodesmium nobis” (one population) that form a distinct evolutionary clade (average nucleotide identity of 94%) distantly related from both T. thiebautii and T. erythraeum (average nucleotide identity <91%) (SI Appendix, Fig. S1 and Dataset S2). The five MAGs have a length ranging between 5.4 Mbp and 6.8 Mbp, with an estimated completion >90% (average of 96%). They were mostly detected in the Indian Ocean, with the exception of Ca. T. miru that occurred mostly in the Atlantic Ocean (Dataset S3). Furthermore, their niche partitioning between the Pacific Ocean, Red Sea, and Mediterranean Sea revealed different distribution patterns between the four species. For instance, Ca. T. nobis was markedly more detected in the Mediterranean Sea. As expected, signal across size fractions indicates that all five populations mostly occur under the form of filaments and colonies in the sunlit ocean. We note that Ca. T. miru was substantially more detected in the largest size fraction (180 to 2,000 μm), suggesting it forms larger aggregates in the Atlantic Ocean compared to the other Trichodesmium populations mostly detected in the Indian Ocean. In terms of overall signal across metagenomes regardless of oceanic regions or size fractions, the two candidate species were abundant but substantially less so compared to T. thiebautii and T. erythraeum populations (Dataset S3). On the other hand, Ca. T. miru was detected in 29 Tara Ocean stations, being markedly more widespread compared to MAGs from the other species, which were detected in a number of stations ranging from 19 to 23 (Datasets S2 and S3).
Marker Genes Point to the Discovery of Trichodesmium Lineages.
The 16S rRNA gene sequences are missing in most MAGs (including within Trichodesmium) due to their high evolutionary stability and occurrence in multicopy (e.g., ref. 34). While in uncharted territory, it is, in theory, possible to retrieve the missing gene of a bacterial population using a closely related reference to recruit metagenomic reads, provided this population dominates the signal within the range of the affiliated genus in the sequence space. Since each of the four Trichodesmium species dominated the metagenomic signal at times within the scope of this genus (e.g., in parts of the Atlantic Ocean for Ca. T. miru), we took this opportunity to recover their 16S rRNA genes. We used the 16S rRNA gene of T. thiebautii as bait and performed a stringent read recruitment (to minimize nonspecific mapping) for metagenomic triplicates targeting each species (Dataset S4). For each metagenome, gene positions that differed from the reference among recruited reads were changed accordingly to the most prevalent nucleotide type. Using this approach, we could create 12 metagenome-centric consensus 16S rRNA gene sequences for Trichodesmium. As expected, metagenomes dominated by T. thiebautii were highly coherent with the reference sequence, requiring only five to eight changes that denote slight differences between the culture and dominant population in one hypervariable region (Fig. 1). In contrast, metagenomes dominated by other species required between 25 and 35 changes covering multiple regions of the gene (Dataset S4), denoting this time greater distances between Trichodesmium species.
We then performed a phylogenetic analysis using the 12 consensus sequences along with reference 16S rRNA genes retrieved from the National Center for Biotechnology Information (NCBI), recapitulating the three Trichodesmium clades while nesting the candidate species into clade II. NCBI blast confirmed that the candidate species contain a 16S rRNA gene signal most closely related to handpicked colonies tentatively affiliated to T. tenue and T. contortum (18) among the metagenomes considered (percent identity >99% for Ca. T. miru and >98.5% for Ca. T. nobis). Yet we detected nucleotide differences in multiple regions of the 16S rRNA gene when using T. tenue and T. contortum as baits for mapping (SI Appendix, Fig. S2). In addition, phylogenetic analysis of the gene marker hetR revealed a cluster corresponding to Ca. T. miru (SI Appendix, Fig. S3) distant from T. tenue and T. contortum. This marker was previously organized into four distinct Trichodesmium clusters (38). Altogether, the gene markers indicate that Ca. T. miru and Ca. T. nobis correspond to previously uncharacterized Trichodesmium species.
The Case for Trichodesmium Species Lacking Nitrogen Fixation Gene Apparatus.
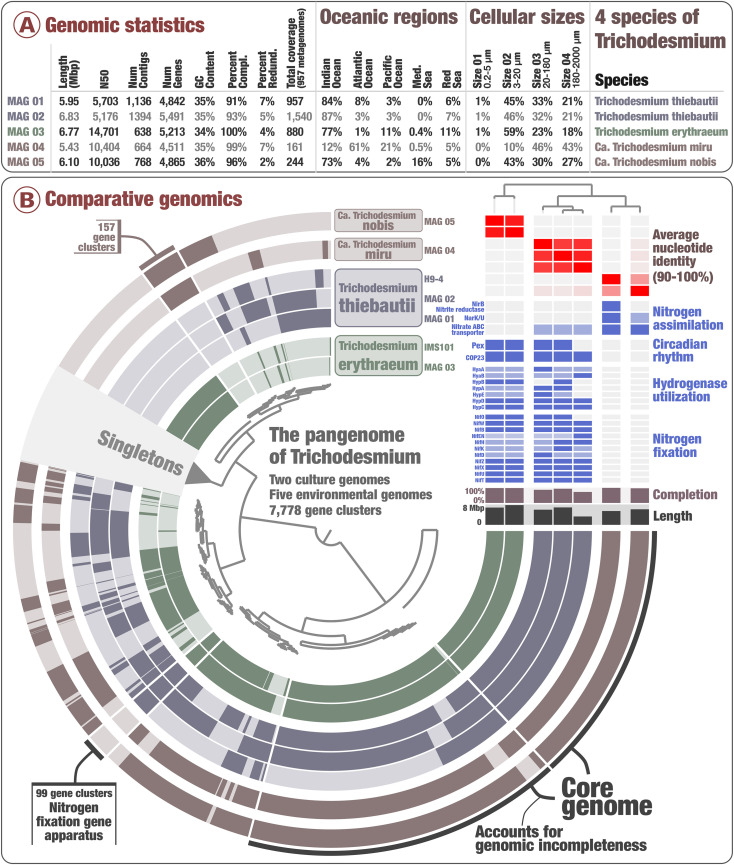
We performed a comparative genomic survey of the genus Trichodesmium by considering the five newly identified MAGs plus two reference genomes from cultivation and accessed from the NCBI: T. erythraeum IMS101 (closed genome) and T. thiebautii H9-4 (fragmented and only 72% complete). Our pangenomic analysis of these seven genomes with a total of 33,249 genes resulted in 7,778 gene clusters (Fig. 1 and Dataset S5). We collapsed singletons (2,542 gene clusters only detected in a single genome) and grouped some of the remaining gene clusters into bins based on their occurrence across genomes: 1) a core-genome (2,183 gene clusters), 2) gene clusters characteristic of T. erythraeum and T. thiebautii (“erythraeum/thiebautii” bin; n = 99), and 3) gene clusters characteristic of the two candidate species (“miru/nobis” bin; n = 157). The overall pangenomic trends for these four Trichodesmium species revealed a relatively large pan-genome but also denoted species-specific gene clusters that might be linked to different lifestyles for Trichodesmium clades I, II, and III.
In order to provide a global view of functional capabilities across four Trichodesmium species, we accessed functions in their gene content using Pfam (39) within the anvi’o pangenomic workflow (40) (Dataset S5), COG20 functions, categories and pathways (41), KOfam (42), and Kyoto Encyclopedia of Genes and Genomes (KEGG) modules and classes (43) within the anvi’o genomic workflow (44) (Dataset S6), and the Rapid Annotation using Subsystem Technology (RAST) annotation (45) (Dataset S7). The two most prominent functional capabilities of Trichodesmium are photosynthesis and nitrogen fixation. As expected, a large set of photosynthetic genes occurred in the seven genomes. The same was true for gas vesicle genes, for instance. On the other hand, multiple lines of evidence emerging from the inspection of all functional annotations and cross-validated by complementary metagenomic investigations point to Ca. T. nobis and Ca. T. miru lacking the ability to fix nitrogen from the atmosphere, providing a case for the occurrence, in plain sight, of nondiazotrophic marine Trichodesmium species.
First and foremost, MAGs corresponding to the two candidate species lacked the entire nitrogen fixation gene apparatus, with the corresponding gene clusters occurring in the erythraeum/thiebautii pangenomic bin (Fig. 2). This lack of signal was recapitulated across all functional annotations (Dataset S8) after removing few false positives easily identified by NCBI blast (Dataset S6). For instance, we found that genes with COG20 function incorrectly annotated as “Nitrogenase ATPase subunit NifH/coenzyme F430 biosynthesis subunit CfbC” correspond, in reality, to “ferredoxin: protochlorophyllide reductase.” Just two nifU-related genes were detected (with NCBI blast confirmation) in those MAGs; however, their occurrence in nondiazotrophic lineages indicates they are not reliable markers for nitrogen fixation. More relevant nitrogen fixation gene markers (nifHDK and nifENB) were successfully detected in a wide range of newly identified marine diazotrophic MAGs spanning multiple phyla (34, 35), suggesting our functional workflow could detect those markers in newly identified Trichodesmium species. It did not.
Fig. 2.
The pangenome of Trichodesmium. A displays genomic statistics for the five environmental Trichodesmium MAGs along with their environmental signal among 937 Tara Oceans metagenomes. B displays the Trichodesmium pangenome that covers 33,249 genes and 7,776 gene clusters from seven genomes of Trichodesmium corresponding to four distinct species. The top-right corner of B holds average nucleotide identity between genomes, and a selection of RAST functional features decorate this pangenome, which was visualized with the anvi’o interactive interface (44). Finally, genomes were organized based on the average nucleotide identity metric.
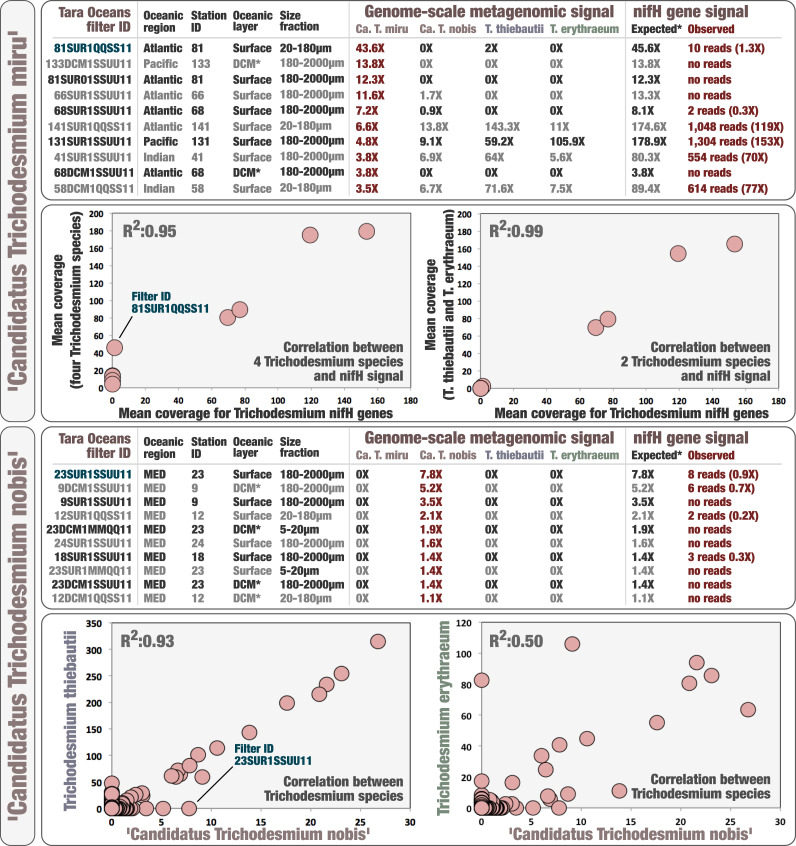
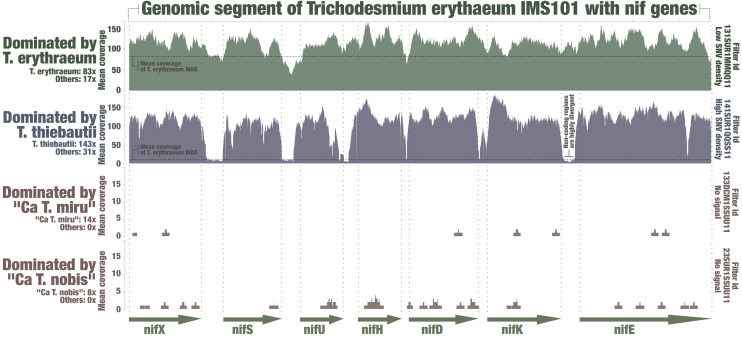
Perhaps more problematically, reconstructing genomes from metagenomes often suffers from quality and completion issues, which could explain the lack of nif genes in some Trichodesmium MAGs. As a first effort to address this, we investigated the occurrence of a comprehensive marine nifH gene database (see Materials and Methods) across the Tara Oceans metagenomes, which includes nifH genes for T. erythraeum and T. thiebautii with >95% sequence identity. We used a sufficiently low mapping stringency (>80% sequence identity over 80% of the read length) to capture this genus in the nucleotidic sequence space. The nifH gene sequence has long been documented to display very low genetic diversity among Trichodesmium species (18, 46, 47). Yet we found that Tara Oceans metagenomes with a high signal for Ca T. miru did not contain the expected signal for Trichodesmium nifH genes (Fig. 3 and Dataset S3). For instance, at Station 133 in the Pacific Ocean (deep chlorophyll maximum layer, 180 to 2,000 μm size fraction), where only Ca T. miru was detected with a mean coverage of 13.8× (i.e., its genome was sequenced nearly 14 times in this metagenome), not a single metagenomic read matched to the >750-nucleotides-long nifH genes from T. erythraeum and T. thiebautii or any of the other nifH genes in the database. Overall, the correlation between the coverages of Trichodesmium genomes and their nifH genes better correlated in those samples when excluding the two candidate species (R2:0.99). Ca T. nobis displayed a higher biogeographic overlap with T. erythraeum and T. thiebautii; nevertheless, we could detect a clear discrepancy between its genomic occurrence and signal for Trichodesmium nifH genes in metagenomes of the Mediterranean Sea (Fig. 3 and Dataset S3). To expand this search for the missing signal beyond the nifH gene, we then mapped metagenomic reads against the entire closed genome of T. erythraeum using an even lower mapping stringency (70% identity over 70% of the read length), revealing this time a lack of signal for the entire nif operon in samples dominated by the candidate species (Fig. 4). In contrast, in samples that were dominated by T. thiebautii, the entire nif operon was perfectly recovered, except for the divergent intergenic regions. Thus, metagenomic signals for nifH and related genes were coherent with the comparative genomic insights, both pointing to a lack of the nitrogen fixation gene apparatus in the two closely related candidate species.
Fig. 3.
Metagenomic signal for nifH gene in the context of Ca T. miru and Ca T. nobis. The top panel summarizes metagenomic read recruitment results for five Trichodesmium MAGs (genome-scale with mapping stringency of >90% identity over >80% of the read length) and three known Trichodesmium nifH genes (gene-centric with mapping stringency of >80% identity over >80% of the read length) across 10 Tara Oceans metagenomes with highest mean coverage for Ca T. miru. It also shows the correlation between mapped metagenomic reads for Trichodesmium nifH genes and either the cumulative mean coverage of all four Trichodesmium species (five MAGs) or the cumulative mean coverage of just T. thiebautii and T. erythraeum (three MAGs) across the same set of 10 metagenomes. The bottom panel displays the correlation between Ca. T. nobis and two Trichodesmium species across all Tara Oceans metagenomes. It also summarizes metagenomic read recruitment results for five Trichodesmium MAGs (genome scale) and three known Trichodesmium nifH genes (gene centric) across 10 Tara Oceans metagenomes with highest mean coverage for Ca T. miru and no detection of the other MAGs (MED: Mediterranean Sea). *DCM: deep chlorophyll maximum.
Fig. 4.
Metagenomic signal for the nif operon of T. erythraeum. The figure displays read recruitment for the T. erythraeum IMS101 nif operon across four metagenomes, each dominated by a single Trichodesmium species (low mapping stringency of >70% identity over >70% of the read length; anvi’o visualization).
Hydrogen is a by-product of nitrogen fixation that cyanobacterial diazotrophs reutilize to gain energy (28–30). We found that functions related to the recycling of hydrogen (hyaABD and hypABCDE) were only missing in MAGs corresponding to the two candidate species (Fig. 2 B, Top Right and Dataset S8), which was also supported by a lack of metagenomic read recruitments for the hyaABD and hypABCDE genes of T. erythraeum in relevant metagenomes with low mapping stringency, echoing results for the nif operon (SI Appendix, Fig. S4). Furthermore, it has recently been suggested on the basis of comparative genomics that nonheterocyst-forming cyanobacterial diazotrophs (including Trichodesmium) produce hopanoid lipids as a mechanism to control intracellular oxygen concentrations to the benefit of the nitrogenase enzyme activity (48). We found that functions related to the production of hopanoid lipids (squalene synthase, squalene-hopene cyclase, and hpnABGH) were missing in MAGs corresponding to the two candidate species (Datasets S7 and S8). Nondiazotrophs in the surface ocean may have no reasons to contain genes for the recycling of hydrogen or production of hopanoid lipids; however, they do need to assimilate biologically available nitrogen molecules. We found the nitrite/nitrate transporter gene nark in single copy in T. erythraeum and T. thiebautii (COG20 functions), yet it occurred in two copies in Ca T. miru and in three copies in Ca T. nobis. This function provides a means to catalyze the uptake of nitrogen-enriched small molecules (49). RAST annotation also detected an enrichment of this functional annotation in the candidate species. Furthermore, COG20 functions and RAST annotation identified the gene nirB allowing dissimilatory nitrate reduction (anaerobic respiration and nitrogen metabolism) in Ca T. miru. NCBI best blast hits linked this gene to the distantly related phyla Lentisphaera and Verrucomicrobia [including Rubritalea marina isolated from a sponge (50)] rather than to more closely related lineages within Cyanobacteria. In addition, the gene appears next to a cyanobacterial transposase. Thus, we observed a lack of hydrogen recycling and hopanoid lipid production genes concomitant with the enrichment of genes related to nitrogen assimilation and metabolism in the candidate species, coherent with a nondiazotrophic lifestyle within Cyanobacteria.
Some cyanobacterial diazotrophs possess circadian rhythms that regulate nitrogen fixation and photosynthesis activities temporally and spatially (51–54). While all four Trichodesmium species contained genes coding for the core circadian clock proteins (kaiABC) linked to photosynthesis regulation (54), the pangenomic analysis revealed two gene clusters corresponding to the circadian oscillating protein COP23 and only missing in MAGs corresponding to the candidate species (Dataset S8). Furthermore, RAST annotation only identified one gene for COP23 and one gene for Pex (another function linked to circadian rhythm), which were only detected in T. erythraeum and T. thiebautii (Fig. 2 B, Top Right). The cyanobacterial nitrogen fixation rhythm can be disconnected from light variations in laboratory experiments (23, 55); however, genes related to this mechanism have yet to be fully understood. Our results echo previous work on the COP23 and Pex genes (56–59), suggesting that at least some of these genes play a central role in the circadian rhythm of nitrogen fixation in T. erythraeum and T. thiebautii.
Finally, we cross-validated these insights by performing six additional Tara Oceans genome-resolved metagenomic surveys guided by the known distribution of four Trichodesmium species and designed to only target either Ca T. miru or Ca T. nobis by means of single assemblies or small coassemblies (Dataset S9). First, we used Hidden Markov Models (HMMs) for the nifHDK and nifENB genes designed to cover the entire bacterial spectrum and found no trace of these gene markers in the six raw metagenomic assemblies, which contain contigs as short as 1,000 nt (for perspective, the nifB gene of T. erythraeum is 1,470 nt long). Then, we characterized and manually curated the Trichodesmium MAG of the targeted candidate species in each metagenomic assembly. Their completion ranged between 90.1% and 98.6%, with good assembly metrics (Dataset S9). Their genetic content agreed with the previous lines of evidence, as they were enriched in genes for nitrate/nitrite transporter nark, contained, this time, the gene nirB in both species (systematically linking this gene to Lentisphaera and Verrucomicrobia), and, most importantly, lacked genes related to the nitrogen fixation apparatus (except for the nifU-related genes), hydrogen recycling (hyaABD and hypABCDE), and hopanoid lipid production (Dataset S10). Thus, our lines of evidence for nondiazotrophic Trichodesmium species were reproducible using various metagenomic combinations and are supported by a lack of signal for nifHDK and nifENB genes in the raw metagenomic assemblies.
Discussion
The association between Trichodesmium and nitrogen fixation has been deeply rooted in our minds due to considerable culture and fieldwork legacies, and as a result, Trichodesmium is routinely referred to as a diazotrophic genus (e.g., ref. 14). Here, we provide multiple lines of evidence indicating that the identified species Ca. T. miru and Ca. T. nobis, characterized by means of genome-resolved metagenomics and found abundant in multiple regions of the open ocean, do not have the ability to fix nitrogen from the atmosphere. These species form a sister genomic clade with distinct gene markers compared to previously characterized Trichodesmium lineages. Near-complete environmental genomes for those candidate species denoted a lack of the entire nitrogen fixation gene apparatus, hydrogen recycling, and hopanoid lipid production genes concomitant with the enrichment of genes related to nitrogen assimilation. These comparative genomic insights were supported by the absence of a metagenomic signal for Trichodesmium nifH genes in relevant stations, targeted metagenomic assemblies, and binning efforts, contrasting with the current paradigm that Trichodesmium species are necessarily capable of nitrogen fixation.
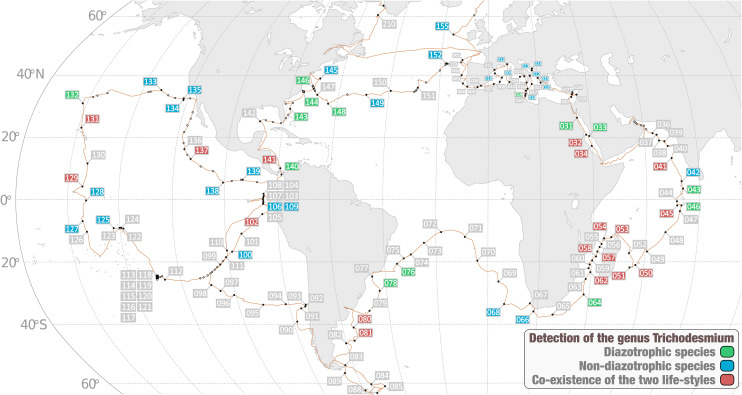
The discovery of nondiazotrophic Trichodesmium species elucidates nanoscale secondary ion mass spectrometry (NanoSIMS) field work observations of cells fixing carbon but not nitrogen (60, 61) and impacts our understanding of the ecology and evolution of this prominent genus. First, Ca. T. miru and Ca. T. nobis were mostly detected in large planktonic size fractions, echoing trends for known diazotrophic Trichodesmium species and suggesting filaments and colonies are the norm for this genus regardless of nitrogen fixation. Second, a critical consequence of this gene loss is that Trichodesmium apparently has a much broader marine biogeographic distribution compared to its nitrogen fixation capability. Indeed, T. erythraeum and T. thiebautii remained undetected in 27 stations with clear signal for nondiazotrophic Trichodesmium species (based on genome-wide metagenomic read recruitment; see Fig. 5 and SI Appendix, Fig. S5 and Dataset S3). These stations cover the Indian, Atlantic, and Pacific Oceans as well as the Mediterranean Sea. Dedicated environmental surveys and cultures are needed to better delineate optimal growth parameters and the niche partitioning of the different Trichodesmium species at large scale. A preliminary insight from the metadata associated with 20 Tara Oceans samples that show largest difference in coverage between diazotrophic and nondiazotrophic Trichodesmium MAGs suggests that the latter occur more in colder waters containing more nitrate and phosphate but less iron as compared to waters enriched in T. erythraeum and T. thiebautii (Dataset S3). In our view, these biogeographic distributions stress the need to disconnect the taxonomic signal for Trichodesmium from the ability of a microbial community to fix nitrogen. Far from contesting the paramount importance of T. erythraeum and T. thiebautii for nitrogen fixation in the open ocean (in the context of dozens of other abundant cyanobacterial and heterotopic bacterial populations), we merely suggest that differentiating diazotrophic from nondiazotrophic Trichodesmium populations might be needed to refine our understanding of the nitrogen balance in the oceans and seas. Counting Trichodesmium colonies to survey the biomass of marine diazotrophs, as routinely performed for decades (e.g., ref. 62), might lead to erroneous nitrogen fixation rate estimations. Finally, the joint study of diazotrophic and nondiazotrophic Trichodesmium populations under culture conditions could bolster our understanding of the underlying genetic mechanisms for nitrogen fixation in Trichodesmium and beyond. As a humble step in this direction, it could be hypothesized that some circadian rhythm genes missing in Ca. T. miru and Ca. T. nobis but present in T. erythraeum and T. thiebautii are specifically linked to the regulation of nitrogen fixation, rather than photosynthesis, among Trichodesmium diazotrophs.
Fig. 5.
Detection of Trichodesmium species across the Tara Oceans stations. The world map describes stations in which we detected 1) only T. erythraeum and/or T. thiebautii (diazotrophic species), 2) only Ca. T. miru and/or Ca. T. nobis (putative nondiazotrophic species), or 3) species from both groups. This survey covers all size fractions >0.2 μm.
Nondiazotrophic Trichodesmium populations might have diverged from a diazotrophic lineage in order to fill distinct ecological niches at the surface of the open ocean (gene loss hypothesis), echoing within a genus results observed at the level of Cyanobacteria that revealed its complex evolutionary history (14, 63, 64) and suggested repeated losses of the nif genes within this phylum (65). The nirB gene linked to dissimilatory nitrate reduction in low-oxygen environments might provide a clue to understanding mechanisms behind this ecologically important evolutionary process, which, based on our lines of evidence, occurred in some Trichodesmium species but not in others. First, this gene was only detected in the two candidate species and is most closely related to Lentisphaera- and Verrucomicrobia-related genes. Second, nirB genes have been observed in the genomic content of noncyanobacterial epibionts living at the surface of Trichodesmium colonies (66, 67), suggesting a possible complementary functional role contributing to their interactions. The temporality of nirB gene acquisition and loss of nitrogen fixation, hydrogen recycling, and hopanoid lipid production genes in the candidate species is currently unknown. Nevertheless, one could wonder whether lateral gene transfers between Trichodesmium colonies and their epibionts have provided a nondiazotrophic evolutionary path for the common ancestor of Ca T. miru and Ca T. nobis, opening exciting prospects regarding the ecology and evolution of this model planktonic lineage in the context of gene flows.
While one may intuitively see nitrogen fixation as a great functional asset within plankton, nondiazotrophic Trichodesmium species remind us of the cost of maintaining a lifestyle centered in part on nitrogen fixation. For instance, hopanoid lipid production may be one of several critical strategies Trichodesmium diazotrophs exploit to the benefit of the nitrogenase. Here, we suggest that loss of nitrogen fixation capabilities can be explained by the burden of low oxygen concentration requirements on the ecology and evolution of marine diazotrophs. Nondiazotrophic Trichodesmium species were found to be more abundant compared to their diazotrophic counterparts in various oceanic regions, possibly the best evidence we have thus far to support the hypothesis of a significant marine nitrogen fixation metabolic burden. In addition, Ca. Trichodesmium nobis strongly correlated with T. thiebautii (R2 = 0.93), the most abundant of the four species within the scope of Tara Oceans metagenomes. The considerable overlap between these two species in the Indian Ocean especially, with one prevalent diazotroph and a less abundant one (ratio of about 1/15) lacking the ability to fix nitrogen fits well with the principles behind the Black Queen hypothesis (68). This hypothesis states that gene loss can provide a selective advantage as long as the function is dispensable. Ca. T. nobis might have lost its ability to fix nitrogen in part because it could benefit from the nitrogen fixation of T. thiebautii, the “leaky helper” mentioned by Morris et al. (68). Thus, both nitrogen fixation constraints and nitrogen availability can provide a rational for the existence of abundant nondiazotrophic Trichdoesmium species.
Conclusion
The cyanobacterial genus Trichodesmium includes some of the most prominent marine nitrogen fixing species, which have been extensively studied in culture and the field for decades. Here, we explored the genome-resolved metagenomic content of two previously uncharacterized Trichodesmium species relatively abundant in the surface of oceans and seas. Critically, multiple lines of evidence point to the existence of nondiazotrophic Trichodesmium species with distinct biogeographic distributions. Establishing that newly identified microbial populations lack critical functional traits from the sole perspective of environmental genomics could be perceived as a precarious endeavor. This is especially true when observations are not in line with those resulting from decades of cultivation. Yet this approach comes with its own strengths, including reference-free de novo metagenomic assemblies on one hand and metagenomic read recruitments using reference genomes and gene markers as bait on the other. In the case of Ca. T. miru and Ca. T. nobis, genome-resolved metagenomics and read recruitments applied to the considerable metagenomic legacy of Tara Oceans provided strong evidence that two closely related Trichodesmium species have lost the ability to fix nitrogen from the atmosphere. Culture representatives for these candidate species are needed to move beyond those metagenomic insights, but already, we are reminded that long-established links between taxonomic lineages and functional traits supporting our understanding of the ocean microbiome do not always hold true.
Materials and Methods
Tara Oceans Metagenomes.
We analyzed a total of 937 Tara Oceans metagenomes available at the European Bioinformatics Institute under project PRJEB402 (https://www.ebi.ac.uk/ena/browser/view/PRJEB402). Dataset S1 reports general information (including the number of reads and environmental metadata) for each metagenome.
Biogeography of MAGs.
We performed a mapping of all metagenomes to calculate the mean coverage and detection of MAGs. Briefly, we used the Burrows–Wheeler Aligner (BWA) version 0.7.15 (minimum identity of >90% over >80% of the read length) and a FASTA file containing the 1,888 nonredundant MAGs from Delmont et al. (35) to recruit short reads from all 937 metagenomes. We considered MAGs were detected in a given filter when >25% of their length was covered by reads to minimize nonspecific read recruitments (34). The number of recruited reads below this cutoff was set to zero before determining vertical coverage and percent of recruited reads.
Metagenome-Centric Consensus 16S rRNA Gene Sequences.
We performed a mapping of metagenomes against a reference 16S rRNA gene in order to retrieve a corresponding signal for each of the four Trichodesmium species. Briefly, we used BWA version 0.7.15 (minimum identity of >95% over >95% of the read length) and a FASTA file containing the 16S rRNA gene sequence T. thiebautii (AF013027) to recruit short reads from 12 metagenomes. We then used the anvi’o version 7 metagenomic workflow (44, 69) to create a CONTIG database (the reference 16S rRNA gene sequence) and PROFILE databases for each metagenome, with the flag “–report-variability-full” in order to report every single nucleotide variation (full mode), or without it (default mode). After merging the PROFILE databases, we used the anvi’o interactive interface with inspection mode to visualize the coverage of this gene across metagenomes in the context of single nucleotide variants (default mode). We then used the program “anvi-gen-gene-consensus-sequences” with the flag “–contigs-mode” to generate metagenome-centric consensus 16S rRNA gene sequences (full mode).
Phylogenetic Inferences Using 16S rRNA Gene Sequences.
We performed a phylogenetic analysis of the metagenome-centric consensus 16S rRNA gene sequences (see Metagenome-Centric Consensus 16S rRNA Gene Sequences) and reference 16S rRNA genes retrieved from NCBI. Briefly, we used the online platform GenomeNet (https://www.genome.jp/) to generate a phylogenetic tree at the nucleotide level and using as parameters the function “build” of ETE3 version 3.1.1 (70) and MAFFT version 6.861b with the linsi options (71) for alignment. Columns with more than 10% of gaps were removed from the alignment using trimAl version 1.4.rev6 (72). Finally, maximum likelihood tree was inferred using PhyML version 20160115 run with the Generalized Time Reversible model and parameters “-o tlr –alpha e –bootstrap 100 –pinv e –nclasses 4 -f m” (73). Branch supports were computed out of 100 bootstrapped trees. We used anvi’o to visualize the phylogenomic tree in the context of additional information.
Phylogenomic Analysis of Cyanobacterial Genomes.
We used PhyloSift (74) version 1.0.1 with default parameters to infer associations between genomes in a phylogenomic context. Briefly, PhyloSift 1) identifies a set of 37 marker gene families in each genome, 2) concatenates the alignment of each marker gene family across genomes, and 3) computes a phylogenomic tree from the concatenated alignment using FastTree (75) version 2.1. We used anvi’o to visualize the phylogenomic tree in the context of additional information.
Phylogenetic Analysis of hetR Genes.
The hetR gene sequences were aligned with the L-INS-i algorithm of MAFFT version 7.475 (71), and sites with >80% gaps were trimmed using Goalign (76). IQ-TREE version 2.0.6 (77) was used for the phylogenetic reconstruction, with the ModelFinder Plus option: the TPM3+F+G4 model was estimated to be the best-fit model and chosen accordingly. Branch supports were computed through nonparametric bootstraps on 100 replicates. The tree was rooted and visualized with the anvi’o version 7 phylogenetic workflow (44) (manual mode).
Pangenomic Analysis of MAGs.
We used the anvi’o pangenomic workflow (40) to compute and visualize the pangenome of Trichodesmium. Briefly, the workflow consisted of three main steps: 1) we generated an anvi’o genome database to store DNA and amino acid sequences as well as functional annotations of each gene in the seven Trichodesmium genomes under consideration, 2) we computed the Trichodesmium pangenome from a genome database by identifying “gene clusters,” and 3) we displayed the pangenome to visualize the distribution of gene clusters across genomes. The gene clusters represent sequences of one or more predicted open reading frames grouped together based on their homology at the translated DNA sequence level. To compute the Trichodesmium pangenome, we used the program “anvi-pan-genome” with the flag “–use-ncbi-blast” and default parameters. This program 1) calculates similarities of each amino acid sequence in every genome against every other amino acid sequence using blastp (78), 2) removes weak hits using the “minbit heuristic,” which was originally described in the Integrated Toolkit for Exploration of microbial Pan-genomes (79), 3) uses the Markov Cluster Algorithm (MCL) (80) to identify gene clusters in the remaining blastp search results, 4) computes the occurrence of gene clusters across genomes and the total number of genes they contain, 5) performs hierarchical clustering analyses for gene clusters (based on their distribution across genomes) and for genomes (based on gene clusters they share) using Euclidean distance and Ward clustering by default, and, finally, 6) generates an anvi’o pan database that stores all results for downstream analyses and was used to visualize the Trichodesmium pangenome in the interactive interface.
Functional Inferences of MAGs.
We inferred functions among the Trichodesmium genes using 1) Pfam (39) from within the anvi’o pangenomic workflow, 2) COG20 functions, categories and pathways (41), KOfam (42), and KEGG modules and classes (43) within the anvi’o genomic workflow (44), and 3) the RAST online platform (45). Regarding the KEGG modules, we calculated their level of completeness in each genomic database using the anvi’o program “anvi-estimate-metabolism” with default parameters. Ref. 81 describes this program in more detail. Lastly, we used online NCBI blasts to identify false positives regarding specific nitrogen fixation gene markers. False positives correspond to functional annotations for which NCBI blast identified a different function with lower e-values and bit scores.
Metagenomic Signal for the Extended nifH Gene Database.
We performed a mapping of metagenomes to calculate the mapped reads and mean coverage of sequences in an extended nifH gene database (see ref. 35 for more details). Briefly, we used BWA version 0.7.15 (minimum identity of 80%) and a FASTA file containing the sequences to recruit short reads from 937 Tara Oceans metagenomes.
Metagenomic Signal for the Reference T. erythraeum IMS101.
We performed a mapping of metagenomes against the reference genome T. erythraeum IMS101 in order to retrieve a corresponding signal for each of the four Trichodesmium species. Briefly, we used BWA version 0.7.15 (minimum identity of >70% over >70% of the read length) and a FASTA file containing the entire genome T. erythraeum IMS101 to recruit short reads from 12 metagenomes. We then used the anvi’o metagenomic workflow to create a CONTIG database (the genome) and PROFILE databases for each metagenome. After merging the PROFILE databases, we used the anvi’o interactive interface with inspection mode to visualize the coverage of regions of interest (e.g., the nif operon) across metagenomes.
Reproducing the Recovery of MAGs for the Candidate Species.
As a supplement of the initial large-scale, genome-resolved metagenomic survey (35, 37), we also performed additional genome-resolved metagenomic surveys by taking advantage of our knowledge of their distribution across the Tara Oceans metagenomes. Briefly, we used metagenomic reads as inputs for six metagenomic single assemblies or coassemblies using MEGAHIT (82) version 1.1.1 and simplified the scaffold header names in the resulting assembly outputs using anvi’o (44). We then completed the anvi’o manual binning workflow to extract Trichodesmium MAGs from these assemblies. For each targeted species, we used a distinct set of eight relevant metagenomes for mapping in order to effectively use differential coverage for binning. Finally, we studied the functional repertoire of recovered Trichodesmium MAGs using anvi’o, as described in Functional Interferences of MAGS.
Search for nifHDK and nifENB Genes in Raw Assemblies.
We used HMM models designed to cover the entire bacterial spectrum of nifHDK and nifENB genes to search for these gene markers in raw metagenomic assemblies performed to extract the genomic content of the candidate species. The e-value cutoff was set to e-100.
Supplemental Information.
We provided a supplemental information document describing the anvi’o workflows used in the study. Each section of the document includes the list of anvi’o programs and a brief explanation of the workflow.
Acknowledgments
I thank Michael D. Lee, A. Murat Eren, Eric Pelletier, Eric A. Webb, Jed A. Fuhrman, and others who joined an international online gathering triggered by the post of the study’s preprint for technical support, editorial suggestions, and constructive discussions regarding Trichodesmium and nitrogen fixation in the sunlit ocean. I would like to specifically thank Clara Martinez-Pérez and Francisco M. Cornejo-Castillo for pointing to the NanoSIMS fieldwork observations and hopanoid lipid production genes, respectively. I also would like to thank Paul Frémont regarding world map production for the relative distribution of Trichodesmium species and Morgan Gaia regarding the phylogeny of hetr genes. In addition, this survey was especially made possible by two scientific endeavors: the sampling and sequencing efforts by the Tara Oceans consortium and the bioinformatics and visualization capabilities afforded by anvi’o. As a result, I would like to thank everyone that contributed to Tara Oceans and anvi’o over the years. In addition, Tara Oceans (which includes the Tara Oceans and Tara Oceans Polar Circle expeditions) would not exist without the leadership of the Tara Oceans Foundation and the continuous support of 23 institutes (https://oceans.taraexpeditions.org/). Finally, part of the computation was performed using the platine, titane, and curie high performance computing machine provided through Grand Equipement National de Calcul Intensif (GENCI) Grants t2011076389, t2012076389, t2013036389, t2014036389, t2015036389, and t2016036389.
Footnotes
The author declares no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112355118/-/DCSupplemental.
Data Availability
First, the genomic resource (bacterial and archaeal MAGs from the surface of the oceans) is publicly available in Genoscope at https://www.genoscope.cns.fr/tara/. The link provides access to the 11 raw metagenomic coassemblies from Delmont et al. (37) as well as the FASTA file for 1,888 MAGs, including the five corresponding to Trichodesmium. In addition, the 1) FASTA files for the initial five Trichodesmium MAGs, 2) anvi’o files corresponding to the metagenomic read recruitments used to generate 16S rRNA metagenomic consensus sequences, 3) anvi’o CONTIGS databases for reference Trichodesmium MAGs and isolate genomes (including functional annotations), 4) anvi’o files corresponding to the Trichodesmium pangenome, 5) anvi’o files corresponding to the metagenomic read recruitments for reference genome T. erythraeum IMS101 (different mapping stringencies included), 6) targeted genome-resolved metagenomic surveys for the candidate Trichodesmium species (contigs >2.5 kbp, anvi’o files and summaries), 7) anvi’o CONTIGS databases for the Trichodesmium MAGs extracted from the targeted genome-resolved metagenomic surveys (including functional annotations), 8) HMM models for six nitrogen fixation gene markers, 9) the supplemental tables and information, and, finally, 10) the raw assemblies (contigs >1 kbp) for targeted, genome-resolved metagenomic surveys are available in Figshare (DOI: 10.6084/m9.figshare.14207321). All other study data are included in the article and/or supporting information.
References
- 1.Sanders R., et al., The biological carbon pump in the North Atlantic. Prog. Oceanogr. 129, 200–218 (2014). [Google Scholar]
- 2.Boyd P. W., Toward quantifying the response of the oceans’ biological pump to climate change. Front. Mar. Sci. 2, 77 (2015). [Google Scholar]
- 3.Charlson R. J., Lovelock J. E., Andreae M. O., Warren S. G., Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326, 655–661 (1987). [Google Scholar]
- 4.Falkowski P. G., Barber R. T., Smetacek V., Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Arrigo K. R., Marine microorganisms and global nutrient cycles. Nature 437, 349–355 (2005). [DOI] [PubMed] [Google Scholar]
- 6.De Vargas C., et al., Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Moore C. M., et al., Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710 (2013). [Google Scholar]
- 8.Tyrrell T., The relative influences of nitrogen and phosohorus on oceanic primary production. Nature 400, 525–531 (1999). [Google Scholar]
- 9.Zehr J. P., Capone D. G., Changing perspectives in marine nitrogen fixation. Science 368, eaay9514 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Zehr J. P., Jenkins B. D., Short S. M., Steward G. F., Nitrogenase gene diversity and microbial community structure: A cross-system comparison. Environ. Microbiol. 5, 539–554 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Dos Santos P. C., Fang Z., Mason S. W., Setubal J. C., Dixon R., Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13, 162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenberg C. G., Neue Beobachlungen über blutartige Erscheinungen in Aegypten, Arabien und Sibirien, nebst einer Uebersicht und Kritik der früher bekannnten. Ann. Phys. (1830). [Google Scholar]
- 13.Capone D. G., Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229 (1997). [Google Scholar]
- 14.Bergman B., Sandh G., Lin S., Larsson J., Carpenter E. J., Trichodesmium—A widespread marine cyanobacterium with unusual nitrogen fixation properties. FEMS Microbiol. Rev. 37, 286–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karl D., et al., The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature 388, 533–538 (1997). [Google Scholar]
- 16.Dore J. E., Brum J. R., Tupas L. M., Karl D. M., Seasonal and interannual variability in sources of nitrogen supporting export in the oligotrophic subtropical North Pacific Ocean. Limnol. Oceanogr. 47, 1595–1607 (2002). [Google Scholar]
- 17.Orcutt K. M., et al., Characterization of Trichodesmium spp. by genetic techniques. Appl. Environ. Microbiol. 68, 2236–2245 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janson S., Bergman B., Carpenter E. J., Giovannoni S. J., Vergin K., Genetic analysis of natural populations of the marine diazotrophic cyanobacterium Trichodesmium. FEMS Microbiol. Ecol. 30, 57–65 (1999). [Google Scholar]
- 19.McKinna L. I. W., Three decades of ocean-color remote-sensing Trichodesmium spp. in the World’s oceans: A review. Prog. Oceanogr. 131, 177–199 (2015). [Google Scholar]
- 20.Dyhrman S. T., et al., Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439, 68–71 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Pierella Karlusich J. J., et al., Global distribution patterns of marine nitrogen-fixers by imaging and molecular methods. bioRxiv [Preprint] (2020). (Accessed 26 October 2021). [DOI] [PMC free article] [PubMed]
- 22.Klawonn I., et al., Distinct nitrogen cycling and steep chemical gradients in Trichodesmium colonies. ISME J. 14, 399–412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y. B., Dominic B., Mellon M. T., Zehr J. P., Circadian rhythm of nitrogenase gene expression in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. strain IMS 101. J. Bacteriol. 180, 3598–3605 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez I. B., Ho T. Y., Diel nitrogen fixation pattern of Trichodesmium: The interactive control of light and Ni. Sci. Rep. 4, 4445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frischkorn K. R., Haley S. T., Dyhrman S. T., Coordinated gene expression between Trichodesmium and its microbiome over day-night cycles in the North Pacific Subtropical Gyre. ISME J. 12, 997–1007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M. D., et al., The Trichodesmium consortium: Conserved heterotrophic co-occurrence and genomic signatures of potential interactions. ISME J. 11, 1813–1824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frischkorn K. R., Rouco M., Van Mooy B. A. S., Dyhrman S. T., The Trichodesmium microbiome can modulate host N2 fixation. Limnol. Oceanogr. Lett. 3, 401–408 (2018). [Google Scholar]
- 28.Wilson S. T., Foster R. A., Zehr J. P., Karl D. M., Hydrogen production by Trichodesmium erythraeum Cyanothece sp. and Crocosphaera watsonii. Aquat. Microb. Ecol. 59, 197–206 (2010). [Google Scholar]
- 29.Wilson S. T., Kolber Z. S., Tozzi S., Zehr J. P., Karl D. M., Nitrogen fixation, hydrogen cycling, and electron transport kinetics in Trichodesmium erythraeum (cyanobacteria) strain ims101. J. Phycol. 48, 595–606 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Eichner M., Basu S., Gledhill M., de Beer D., Shaked Y., Hydrogen dynamics in trichodesmiumcolonies and their potential role in mineral iron acquisition. Front. Microbiol. 10, 1565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks D. H., et al., Author correction: Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 3, 253 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Tully B. J., Graham E. D., Heidelberg J. F., The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5, 170203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pachiadaki M. G., et al., Charting the complexity of the marine microbiome through single-cell genomics. Cell 179, 1623–1635.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delmont T. O., et al., Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delmont T. O., et al., Heterotrophic bacterial diazotrophs are more abundant than their cyanobacterial counterparts in metagenomes covering most of the sunlit ocean. 10.1101/2021.03.24.436778(2021). [DOI] [PMC free article] [PubMed]
- 36.Sunagawa S., et al., Tara Oceans Coordinators, Tara Oceans: Towards global ocean ecosystems biology. Nat. Rev. Microbiol. 18, 428–445 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Delmont T. O., et al., Functional repertoire convergence of distantly related eukaryotic plankton lineages revealed by genome-resolved metagenomics. bioRxiv [Preprint] (2020). (Accessed 26 October 2021).
- 38.Hynes A. M., Webb E. A., Doney S. C., Waterbury J. B., Comparison of cultured Trichodesmium (Cyanophyceae) with species characterized from the field. J. Phycol. 48, 196–210 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Bateman A., et al., The Pfam protein families database. Nucleic Acids Res. 28, 263–266 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delmont T. O., Eren A. M., Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ 6, e4320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galperin M. Y., et al., COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49, D274–D281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aramaki T., et al., KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eren A. M., et al., Anvi’o: An advanced analysis and visualization platform for 'omics data. PeerJ 3, e1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aziz R. K., et al., The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben‐Porath J., Carpenter E. J., Zehr J. P., Genotypic relationships in Trichodesmium (Cyanophyceae) based on nifH sequence comparisons. J. Phycol. 29, 806–810 (1993). [Google Scholar]
- 47.Zehr J. P., Limberger R. J., Ohki K., Fujita Y., Antiserum to nitrogenase generated from an amplified DNA fragment from natural populations of Trichodesmium spp. Appl. Environ. Microbiol. 56, 3527–3531 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornejo-Castillo F. M., Zehr J. P., Hopanoid lipids may facilitate aerobic nitrogen fixation in the ocean. Proc. Natl. Acad. Sci. U.S.A. 116, 18269–18271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolesnikow T., Schröder I., Gunsalus R. P., Regulation of narK gene expression in Escherichia coli in response to anaerobiosis, nitrate, iron, and molybdenum. J. Bacteriol. 174, 7104–7111 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheuermayer M., Gulder T. A. M., Bringmann G., Hentschel U., Rubritalea marina gen. nov., sp. nov., a marine representative of the phylum ‘Verrucomicrobia’, isolated from a sponge (Porifera). Int. J. Syst. Evol. Microbiol. 56, 2119–2124 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Cohen S. E., Golden S. S., Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 79, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell-Pedersen D., et al., Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimers A. M., Knoop H., Bockmayr A., Steuer R., Cellular trade-offs and optimal resource allocation during cyanobacterial diurnal growth. Proc. Natl. Acad. Sci. U.S.A. 114, E6457–E6465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondo T., Ishiura M., The circadian clock of cyanobacteria. BioEssays 22, 10–15 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Gaudana S. B., et al., Rhythmic and sustained oscillations in metabolism and gene expression of Cyanothece sp. ATCC 51142 under constant light. Front. Microbiol. 4, 374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muñoz-Marín M. D. C., et al., The transcriptional cycle is suited to daytime N2 fixation in the unicellular cyanobacterium “Candidatus atelocyanobacterium thalassa” (UCYN-A). mBio 10, e02495-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takai N., Ikeuchi S., Manabe K., Kutsuna S., Expression of the circadian clock-related gene pex in cyanobacteria increases in darkness and is required to delay the clock. J. Biol. Rhythms 21, 235–244 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Kutsuna S., Kondo T., Aoki S., Ishiura M., A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180, 2167–2174 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen H. M., Chien C. Y., Huang T. C., Regulation and molecular structure of a circadian oscillating protein located in the cell membrane of the prokaryote Synechococcus RF-1. Planta 199, 520–527 (1996). [DOI] [PubMed] [Google Scholar]
- 60.Eichner M. J., et al., Chemical microenvironments and single-cell carbon and nitrogen uptake in field-collected colonies of Trichodesmium under different pCO2. ISME J. 11, 1305–1317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Pérez C., et al., The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat. Microbiol. 1, 16163 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Luo Y. W., et al., Database of diazotrophs in global ocean: Abundance, biomass and nitrogen fixation rates. Earth Syst. Sci. Data 4, 47–73 (2012). [Google Scholar]
- 63.Tomitani A., Knoll A. H., Cavanaugh C. M., Ohno T., The evolutionary diversification of cyanobacteria: Molecular-phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. U.S.A. 103, 5442–5447 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bothe H., Schmitz O., Yates M. G., Newton W. E., Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol. Mol. Biol. Rev. 74, 529–551 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Latysheva N., Junker V. L., Palmer W. J., Codd G. A., Barker D., The evolution of nitrogen fixation in cyanobacteria. Bioinformatics 28, 603–606 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Frischkorn K. R., Rouco M., Van Mooy B. A. S., Dyhrman S. T., Epibionts dominate metabolic functional potential of Trichodesmium colonies from the oligotrophic ocean. ISME J. 11, 2090–2101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee M. D., et al., Transcriptional activities of the microbial consortium living with the marine nitrogenfixing cyanobacterium Trichodesmium reveal potential roles in community-level nitrogen cycling. Appl. Environ. Microbiol. 84, e02026-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris J. J., Lenski R. E., Zinser E. R., The Black Queen hypothesis: Evolution of dependencies through adaptive gene loss. mBio 3, e00036-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eren A. M., et al., Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 6, 3–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guindon S., et al., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Darling A. E., et al., PhyloSift: Phylogenetic analysis of genomes and metagenomes. PeerJ 2, e243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Price M. N., Dehal P. S., Arkin A. P., FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Institut Pasteur, Goalign. https://github.com/evolbioinfo/goalign. Accessed 21 October 2021. [Google Scholar]
- 77.Minh B. Q., et al., IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 79.Benedict M. N., Henriksen J. R., Metcalf W. W., Whitaker R. J., Price N. D., ITEP: An integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 15, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Dongen S., Abreu-Goodger C., Using MCL to extract clusters from networks. Methods Mol. Biol. 804, 281–295 (2012). [DOI] [PubMed] [Google Scholar]
- 81.Meren Lab, anvi-estimate-metabolism [program]. https://merenlab.org/software/anvio/help/main/programs/anvi-estimate-metabolism/. Accessed 21 October 2021. [Google Scholar]
- 82.Li D., Liu C. M., Luo R., Sadakane K., Lam T. W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
First, the genomic resource (bacterial and archaeal MAGs from the surface of the oceans) is publicly available in Genoscope at https://www.genoscope.cns.fr/tara/. The link provides access to the 11 raw metagenomic coassemblies from Delmont et al. (37) as well as the FASTA file for 1,888 MAGs, including the five corresponding to Trichodesmium. In addition, the 1) FASTA files for the initial five Trichodesmium MAGs, 2) anvi’o files corresponding to the metagenomic read recruitments used to generate 16S rRNA metagenomic consensus sequences, 3) anvi’o CONTIGS databases for reference Trichodesmium MAGs and isolate genomes (including functional annotations), 4) anvi’o files corresponding to the Trichodesmium pangenome, 5) anvi’o files corresponding to the metagenomic read recruitments for reference genome T. erythraeum IMS101 (different mapping stringencies included), 6) targeted genome-resolved metagenomic surveys for the candidate Trichodesmium species (contigs >2.5 kbp, anvi’o files and summaries), 7) anvi’o CONTIGS databases for the Trichodesmium MAGs extracted from the targeted genome-resolved metagenomic surveys (including functional annotations), 8) HMM models for six nitrogen fixation gene markers, 9) the supplemental tables and information, and, finally, 10) the raw assemblies (contigs >1 kbp) for targeted, genome-resolved metagenomic surveys are available in Figshare (DOI: 10.6084/m9.figshare.14207321). All other study data are included in the article and/or supporting information.