Abstract
Filamentous fungi are highly productive cell factories, often used in industry for the production of enzymes and small bioactive compounds. Recent years have seen an increasing number of synthetic-biology-based applications in fungi, emphasizing the need for a synthetic biology toolkit for these organisms. Here we present a collection of 96 genetic parts, characterized in Penicillium or Aspergillus species, that are compatible and interchangeable with the Modular Cloning system. The toolkit contains natural and synthetic promoters (constitutive and inducible), terminators, fluorescent reporters, and selection markers. Furthermore, there are regulatory and DNA-binding domains of transcriptional regulators and components for implementing different CRISPR-based technologies. Genetic parts can be assembled into complex multipartite assemblies and delivered through genomic integration or expressed from an AMA1-sequence-based, fungal-replicating shuttle vector. With this toolkit, synthetic transcription units with established promoters, fusion proteins, or synthetic transcriptional regulation devices can be more rapidly assembled in a standardized and modular manner for novel fungal cell factories.
Keywords: synthetic biology toolkit, Modular Cloning, hybrid transcription factor, inducible promoter, transcriptional regulation, filamentous fungi
Introduction
Filamentous fungi are widely used as cell factories: organic acids, small-molecule drugs, and homologous as well as heterologous proteins expressed in fungi are applied in various industries, and fungal biotechnology is considered as an innovation driver for a circular economy.1 Not only are fungi excellent workhorses for protein production because of their natural capacity for protein secretion, but also, fungal genomes contain a large number of biosynthetic gene clusters (BGCs) encoding potentially useful natural products. The core enzymes of these natural-product-producing clusters are usually nonribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs), or terpene synthases (TPSs). Advanced bioinformatics tools predict about 30–70 BGCs per fungal species.2 It has become obvious that next to known natural products, fungal genomes hold an enormous amount of untapped biosynthetic potential in the form of transcriptionally silent, uncharacterized BGCs.2 These “cryptic” BGCs, which are usually not expressed under laboratory conditions, can potentially provide new leads for novel natural products. Single species like Aspergillus nidulans or Penicillium rubens contain over 30 NRPSs and PKSs that are responsible for natural product biosynthesis, most of which are still awaiting characterization.3,4
Synthetic biology has revolutionized metabolic engineering by providing new tools to create modular, synthetic genetic circuits for controlled activation and/or fine-tuned expression of specific genes or complete BGCs, thereby optimizing the production of endogenous or exogenous proteins and secondary metabolites.5−11 In addition to “rewiring” pathways that are already transcriptionally active, such tools can be used for the activation of transcriptionally silent BGCs and the discovery of novel natural products. Synthetic genetic circuits provide a new way of transcriptional regulation by mimicking natural regulatory mechanisms. Synthetic transcription factors (STFs) can be employed to achieve transcriptional regulation and in their simplest design are fusions between the DNA-binding domain (DBD) of a known transcription factor and a transcriptional regulator (activator or repressor). As the DBD of a TF binds to its specific upstream activating sequence (UAS) in the targeted promoter, the strength of the regulation can be increased by integrating additional UASs in a synthetic promoter. These systems are further tunable by utilizing inducible promoters to titrate the protein levels of the corresponding TFs or other genetic switches. By the use of such synthetic transcriptional regulators, gene activation or repression can be achieved in a controlled manner, or transcription can be fine-tuned for each gene individually.5−7 Synthetic expression systems have previously been demonstrated in Aspergillus species,5,6,8,10Trichoderma reesei,9P. rubens,7 and Ustilago maydis.11 For instance, the bacterial doxycycline/tetracycline-inducible system has been adopted for Aspergillus species and U. maydis, providing inducer-based transcriptional regulation.5,8,11 STF-based regulatory systems show transferability among a variety of different fungi.6,12 Next to methods that require introducing genetic parts permanently into the host organism genome, plasmid-based alternatives are also available for filamentous fungi, as well as CRISPR-based technologies for transcriptional regulation.13−15 All of these synthetic-biology-based tools provide new alternatives to further aid the exploitation of fungal workhorses.
Targeted DNA delivery and precise genome editing are often required for the construction of STF-regulated genetic circuits. Engineering of nondomesticated strains is often time-consuming, and engineering efforts show low efficiency. The targeting efficiency of the integrated donor DNA to the designated loci can be increased by using long homologous fragments of genomic DNA of the host organism. More accurate genome editing is possible with strains devoid of the fungal homologues of the ku70 or ku80 genes, as homology-directed repair (HDR) will be favored over the nonhomologous end joining (NHEJ) DNA repair pathway.16 In some (nondomesticated) fungal isolates, genome engineering can be less efficient because of the presence of the NHEJ machinery, resulting in more random integration events. In such strains, DNA delivery using nonintegrative fungal shuttle vectors can be advantageous, as this method does not rely on genomic integrations. The AMA1 sequence provides autonomous vector replication and therefore supports episomal DNA delivery in several species of filamentous fungi, and shuttle vectors containing this sequence are commonly used.17 Such vectors enable rapid genetic circuit assembly for gene expression in the fungal host. Fungal shuttle vectors are commonly used to deliver the in vivo expressed components of the CRISPR-Cas (CRISPR-associated protein) genome editing technology in filamentous fungi,18 which further allows for swift and reliable genomic engineering.
Modular toolkits allow rapid construction of genetic circuits, various STFs, and protein fusions in a combinatorial manner through recombination of already available genetic parts or incorporation of new genetic parts into the established system.19 Standardized, characterized genetic parts are key elements for rapid and modular construction of novel genetic circuits. In modular cloning systems, typically the genetic elements (as PCR products or synthetic DNA) are first inserted into entry vectors (level 0) to create genetic parts. These basic genetic parts (also called modules) are then used for the next step of the assembly into transcription units (level 1), which can be further combined into genetic circuits containing multiple transcription units (level 2).19 The Golden Gate Assembly-based Modular Cloning (MoClo) system supports the assembly of several transcription units on a single plasmid, where the number of units is limited only by the host’s tolerance for the size of plasmid DNA.19 A limitation of the Golden Gate Assembly line is the initial cloning step, which often requires the removal of type IIS recognition sites used by MoClo through PCR amplification or DNA synthesis. This initial work can be reduced by using parts made available through repositories for synthetic toolkits, which could contribute to more rapid assembly of novel synthetic circuits for various organisms. Synthetic modular vector collections (toolkits) are publicly available for bacteria,20 various yeasts,21,22 plants,23 and mammalian host24 cell lines. Although collections of Golden Gate-based vectors were recently established in Aspergillus niger(25) (GoldenMOCS) and deposited on Addgene for metabolic pathway construction25 or in Sordaria macrospora and P. rubens(26) for protein fusions and gene deletions, a substantial collection of generic tools for synthetic biology applications in filamentous fungi is not yet deposited and available in global nucleic acid repositories.
Modular assemblies provide high flexibility with regard to assembly compared with systems that leave an “assembly scar” after cloning. As the genetic parts in such systems are flanked with Type IIs restriction enzyme cut sites because the restriction happens outside their recognition sequence, the created cohesive sequences can be used for one-pot “scarless” cloning approaches. These cohesive linker sequences mark the predetermined location for the genetic element in an assembled transcription unit and are used for the assembly of multiple transcription units as well. For example, in the standard MoClo language,19 a transcription unit for cytosolic proteins consists of promoters (P), untranslated regions (U), coding sequences (CDS) and terminators (T), and four-base-pair linker sequences are used to connect them to each other and to the receiving backbone (e.g., GGAG-(P)-TACT-(U)-AATG-(CDS)-GCTT-(T)-CGCT). This hierarchical structure provides a platform for rapid and easily automatable assembly of multigene constructs but on the other hand creates limitations for interchanging building blocks from other modular systems. Numerous modular assemblies have aimed to improve the standard MoClo assembly,20,21,24 but by changing the linker sequences for transcription unit assembly and failing to consider backward compatibility, this creates incompatibility among the different modular assembly systems.
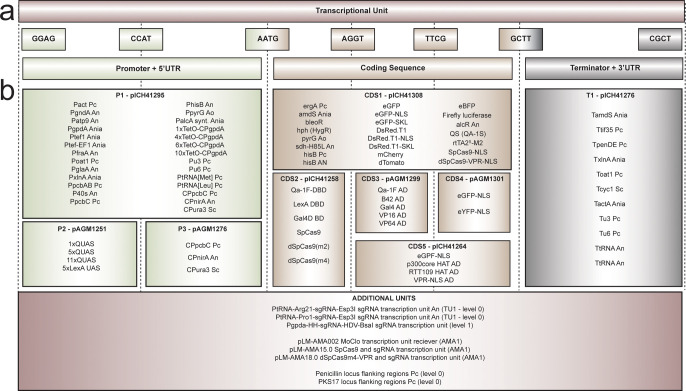
This Fungal Modular Cloning Toolkit consists of 96 genetic parts as MoClo-compatible entry vectors, including synthetic and native fungal promoters, terminators, selection markers, various CDSs for transcriptional activation and DNA-binding domains, fluorescent reporters, and the AMA1 sequence for fungal autonomous replication as well as CRISPR components such as Cas9, dCas9 sequences, and single guide RNA (sgRNA) transcription units for filamentous fungi (Figure 1). This generic modular toolkit, which provides the building blocks for rapid construction of complex genetic circuits, should be of great use to the field of fungal synthetic biology and accelerate the discovery of bioactive compounds as well as optimization of their production.
Figure 1.
List of vectors in the Fungal Modular Cloning Toolkit. (a) Location of genetic parts in a transcription unit with their corresponding linker sequences. (b) List of parts of the toolkit, containing promoters (P1), UASs (P2), UAS-compatible core promoters (P3), coding sequences with various fusion possibilities (CDS1–5), terminators (T1), complete transcription units (TUs), and additional vectors (sgRNA transcription units, flanking sequences, and AMA1 vectors). Abbreviations (Pc, An, Ania, Ao, Sc) indicate the origin of the template (P. rubens, A. niger, A. nidulans, A. oryzae, S. cerevisiae, respectively).
Results and Discussion
In this work, we describe a modular synthetic biology toolkit for use in filamentous fungi. Most of the genetic parts in this toolkit originate from Aspergillus or Penicillium species or from other established synthetic fungal systems for gene regulation, heterologous expression, and genetic engineering.5−7,27,28 It is a common observation that promoters and other genomic elements of filamentous fungi are interchangeable among fungal species and are therefore widely used in heterologous filamentous fungal systems.6,12 The parts of this MoClo toolkit were analyzed in P. rubens unless the genetic part was already established or characterized in previous studies as listed in Table 1. All of the vectors were constructed using the standardized MoClo system, which was discussed in detail by Weber et al..19 This collection of basic genetic parts provides a tool for rapid assembly of various combinations of parts into multigene genetic circuits, which can be delivered to the host organism through genomic integration or using episomal AMA1 vectors.
Table 1. Genetic Modules and Other Vectors in the Fungal Toolkit for Modular Cloning (FTK)a.
| vector name | Addgene ID | part type | unit description | recipient MoClo backbone | template | source and references |
|---|---|---|---|---|---|---|
| pFTK001 | 171273 | P1 | Pact Pc20g11630 promoter | pICH41295 | P. rubens DS54468 | (27) |
| pFTK002 | 171274 | P1 | PgndA An11g02040 promoter | pICH41295 | A. niger N402 | (7), 27 |
| pFTK003 | 171275 | P1 | Patp9 An04g08190 promoter | pICH41295 | A. niger N402 | (27) |
| pFTK004 | 171276 | P1 | PgpdA ANIA_08041 promoter | pICH41295 | pDONR221-AMDS | (27) |
| pFTK005 | 171277 | P1 | Ptef1 ANIA_04218 promoter | pICH41295 | pFC334 (Addgene ID 87846) | (32) |
| pFTK006 | 171278 | P1 | Ptef EF1-subunit ANIA_02063 promoter | pICH41295 | A nidulans FGSC A4 | (33) |
| pFTK007 | 171279 | P1 | PfraA An16g04690 promoter | pICH41295 | A. niger N402 | (5) |
| pFTK008 | 171280 | P1 | Poat1 Pc18g03600 promoter | pICH41295 | P. rubens Wisconsin 54–1255 | (34) |
| pFTK009 | 171281 | P1 | PglaA An03g06550 promoter | pICH41295 | pEBA520 | (5) |
| pFTK010 | 171282 | P1 | PxlnA ANIA_03613 promoter | pICH41295 | A nidulans FGSC A4 | (35) |
| pFTK011 | 171283 | P1 | PpcbAB Pc21g21390 promoter | pICH41295 | P. rubens DS54468 | (27) |
| pFTK012 | 171284 | P1 | P40s An0465 promoter | pICH41295 | pDSM-JAK-108 | (7), 36 |
| pFTK013 | 171285 | P1 | PpcbC Pc21g21380 promoter | pICH41295 | P. rubens DS54468 | (27) |
| pFTK014 | 171286 | P1 | PAnHisB AN6536.2 promoter | pICH41295 | A. niger N402 | (37) |
| pFTK015 | 171287 | P1 | PpyrG AO090011000868 promoter | pICH41295 | pMF21.1 | (38) |
| pFTK016 | 171288 | P1 | 1x TetO UAS + CPgpdA (fused) | pICH41295 | pVG2.2 | (5) |
| pFTK017 | 171289 | P1 | 4x TetO UAS+ CPgpdA (fused) | pICH41295 | pVG2.2 | (5) |
| pFTK018 | 171290 | P1 | 6x TetO UAS + CPgpdA (fused) | pICH41295 | pVG2.2 | (5) |
| pFTK019 | 171291 | P1 | 10x TetO UAS + CPgpdA (fused) | pICH41295 | pVG2.2 | (5) |
| pFTK020 | 171292 | P1 | PalcA synt_NoCrea (ANIA_08979) promoter | pICH41295 | A nidulans FGSC A4 | (39) |
| pFTK021 | 171293 | P1 | Pu3 hom., Putp25, P. rubens Pol-III promoter | pICH41295 | P. rubens DS54468 | (28) |
| pFTK022 | 171294 | P1 | Pu6 hom., P. rubens Pol-III promoter | pICH41295 | P. rubens DS54468 | (28) |
| pFTK023 | 171295 | P1 | PtRNA[Met] P. rubens Pol-III promoter | pICH41295 | P. rubens DS54468 | (28) |
| pFTK024 | 171296 | P1 | PtRNA[Leu] P. rubens Pol-III promoter | pICH41295 | P. rubens DS54468 | (28) |
| pFTK025 | 171297 | P1 | CPpcbC Pc21g21380 (no UAS) core promoter | pICH41295 | P. rubens Wisconsin 54–1255 | (7) |
| pFTK026 | 171298 | P1 | CPnirA AN0098 (no UAS) core promoter | pICH41295 | A nidulans FGSC A4 | (7) |
| pFTK027 | 171299 | P1 | CPura3 YEL021W (no UAS) core promoter | pICH41295 | S. cerevisiae CEN.PK113–7D | (7) |
| pFTK028 | 171300 | P2 | 1xQUAS UAS (for fusion) | pAGM1251 | synthetic DNA | (7), 40 |
| pFTK029 | 171301 | P2 | 5xQUAS UAS (for fusion) | pAGM1251 | synthetic DNA | (7), 40 |
| pFTK030 | 171302 | P2 | 11xQUAS UAS (for fusion) | pAGM1251 | synthetic DNA | (7), 40 |
| pFTK031 | 171303 | P2 | 5xLexA_UAS UAS (for fusion) | pAGM1251 | synthetic DNA | (41) |
| pFTK032 | 171304 | P3 | CPpcbC Pc21g21380 core promoter (for fusion) | pAGM1276 | A nidulans FGSC A4 | (7) |
| pFTK033 | 171305 | P3 | CPnirA AN0098 core promoter (for fusion) | pAGM1276 | S. cerevisiae CEN.PK113-7D | (7) |
| pFTK034 | 171306 | P3 | CPura3 YEL021W core promoter (for fusion) | pAGM1276 | A. niger N402 | (7) |
| pFTK035 | 171307 | CDS1 | ergA Pc22g15550 terbinafine, selection marker | pICH41308 | P. rubens DS54468 | (42) |
| pFTK036 | 171308 | CDS1 | amdS ANIA_08777 acetamidase, selection marker | pICH41308 | pDONR221-AMDS | (29) |
| pFTK037 | 171309 | CDS1 | bleoR phleomycin, selection marker | pICH41308 | pDSM-JAK-109 | (29), 36 |
| pFTK038 | 171310 | CDS1 | hph hygromycin selection marker (hygR) | pICH41308 | pAN7.1 | (43) |
| pFTK039 | 171311 | CDS1 | pyrG AO090011000868 orotidine 5′-phosphate decarboxylase, selection marker | pICH41308 | pMF21.1 | (38) |
| pFTK040 | 171312 | CDS1 | sdh-H85L An14g04400 succinate dehydrogenase, selection marker | pICH41308 | A. niger N402 | (44) |
| pFTK041 | 171313 | CDS1 | hisB Pc20g11690 histidine, selection marker | pICH41308 | P. rubens DS54468 | (37) |
| pFTK042 | 171314 | CDS1 | hisB AN6536.2 histidine, selection marker | pICH41308 | pSE1.6 | (37) |
| pFTK043 | 171315 | CDS1 | eGFP fluorescent reporter | pICH41308 | pLM2_30 (Addgene ID 154222) | (7) |
| pFTK044 | 171316 | CDS1 | eGFP-NLS fluorescent reporter | pICH41308 | pLM2_30 (Addgene ID 154222) | (7) |
| pFTK045 | 171317 | CDS1 | eGFP-SKL fluorescent reporter | pICH41308 | pLM2_30 (Addgene ID 154222) | (7) |
| pFTK046 | 171318 | CDS1 | DsRed.T1 fluorescent reporter | pICH41308 | pDSM-JAK-109 | (7), 27, 36 |
| pFTK047 | 171319 | CDS1 | DsRed-NLS fluorescent reporter | pICH41308 | pDSM-JAK-109 | (7), 27, 36 |
| pFTK048 | 171320 | CDS1 | DsRed.T1-SKL fluorescent reporter | pICH41308 | pDSM-JAK-109 | (27), 36, 45 |
| pFTK049 | 171321 | CDS1 | mCherry fluorescent reporter | pICH41308 | pURA3_1147651 cP_mCherry | (6) |
| pFTK050 | 171322 | CDS1 | dTomato fluorescent reporter | pICH41308 | pMF30.1 | (46) |
| pFTK051 | 171323 | CDS1 | eBFP fluorescent reporter | pICH41308 | pLM2_30 (Addgene ID 154222) with Y66H/Y145F mutations | (7) |
| pFTK052 | 171324 | CDS1 | firefly luciferase reporter | pICH41308 | pVG4.1 | (5) |
| pFTK053 | 171325 | CDS1 | alcR ANIA_08978 transcriptional activator | pICH41308 | A nidulans FGSC A4 | (39) |
| pFTK054 | 171326 | CDS1 | QS (QA-1S) codon optimized, quinic acid repressor | pICH41308 | pAC-Qsco, (Addgene ID 46106) | (40) |
| pFTK055 | 171327 | CDS1 | rtTA2S-M2 (TetR-3xVP16) transcriptional activator | pICH41308 | pVG2.2 | (5) |
| pFTK056 | 171328 | CDS1 | SpCas9-NLS | pICH41308 | pYTK036 (Addgene ID 65143) | (14) |
| pFTK057 | 171329 | CDS1 | dSpCas9(m4)-VPR-NLS | pICH41308 | pYTK036 (Addgene ID 65143), pAG414GPD (Addgene ID 63801) | (14), 47 |
| pFTK058 | 171330 | CDS2 | QF DBD from QA-1F (for fusion) | pICH41258 | pAC-7-QFBDAD (Addgene ID 46096) | (40) |
| pFTK059 | 171331 | CDS2 | LexA DBD (for fusion) | pICH41258 | FRP718_PACT1(-1-520)-LexA-ER-haB42-TCYC1 (Addgene ID 58431) | (48) |
| pFTK060 | 171332 | CDS2 | Gal4D BD (for fusion) | pICH41258 | S. cerevisiae CEN.PK113-7D | (49) |
| pFTK061 | 171333 | CDS2 | SpCas9 (for fusion) | pICH41258 | pYTK036 (Addgene ID 65143) | (14), 21 |
| pFTK062 | 171334 | CDS2 | dSpCas9(m2) (for fusion) | pICH41258 | pYTK036 (Addgene ID 65143) | (14), 21 |
| pFTK063 | 171335 | CDS2 | dSpCas9(m4) (for fusion) | pICH41258 | pYTK036 (Addgene ID 65143) | (14), 21 |
| pFTK064 | 171336 | CDS3 | QF AD from QA-1F (for fusion) | pAGM1299 | pAC-7-QFBDAD (Addgene ID 46096) | (7), 40 |
| pFTK065 | 171337 | CDS3 | B42 AD (for fusion) | pAGM1299 | FRP718_PACT1(-1-520)-LexA-ER-haB42-TCYC1 (Addgene ID 58431) | (48) |
| pFTK066 | 171338 | CDS3 | Gal4 AD (for fusion) | pAGM1299 | S. cerevisiae CEN.PK113-7D | (49) |
| pFTK067 | 171339 | CDS3 | VP16 AD (for fusion) | pAGM1299 | pVG2.2 | (5), 49 |
| pFTK068 | 171340 | CDS3 | VP64 AD (for fusion) | pAGM1299 | pcDNA-dCas9-VP64 (Addgene ID 47107) | (47) |
| pFTK069 | 171341 | CDS4 | eGFP-NLS fluorescent reporter (for fusion) | pAGM1301 | pLM2_30 (Addgene ID 154222) | (7) |
| pFTK070 | 171342 | CDS4 | eYFP-NLS fluorescent reporter (for fusion) | pAGM1301 | pLM2_30 (Addgene ID 154222) with S65G/V68L/S72A/T203Y mutations | (7) |
| pFTK071 | 171343 | CDS5 | eGPF-NLS fluorescent reporter (for fusion) | pICH41264 | PX458 (Addgene ID 48138) | (7) |
| pFTK072 | 171344 | CDS5 | p300core HAT AD, Homo sapiens E1A binding protein p300 (for fusion) | pICH41264 | pcDNA-dCas9-p300 (Addgene ID 61357) | (15), 47, 50 |
| pFTK073 | 171345 | CDS5 | RTT109 HAT AD (for fusion) | pICH41264 | S. cerevisiae CEN.PK113-7 | (51) |
| pFTK074 | 171346 | CDS5 | VPR-NLS AD (for fusion) | pICH41264 | pAG414GPD (Addgene ID 63801) | (47) |
| pFTK075 | 171347 | T1 | TamdS ANIA_08777 terminator | pICH41276 | pDONR221-AMDS | (29) |
| pFTK076 | 171348 | T1 | Ttif35 Pc22g19890 terminator | pICH41276 | pDSM-JAK-108 | (7), 36 |
| pFTK077 | 171349 | T1 | TpenDE Pc21g21370 terminator | pICH41276 | P. rubens Wisconsin 54–1255 | (36) |
| pFTK078 | 171350 | T1 | TxlnA ANIA_03613 terminator | pICH41276 | A nidulans FGSC A4 | (35) |
| pFTK079 | 171351 | T1 | Toat1 Pc18g03600 terminator | pICH41276 | P. rubens Wisconsin 54–1255 | (34) |
| pFTK080 | 171352 | T1 | Tcyc1 YJR048W terminator | pICH41276 | pDSM-JAK-109 | (36) |
| pFTK081 | 171353 | T1 | TactA (Tact1) ANIA_06542 P. rubens terminator | pICH41276 | pDSM-JAK-108 | (7), 36 |
| pFTK082 | 171354 | T1 | Tu3 hom., Tutp25, P. rubens Pol-III terminator | pICH41276 | P. rubens DS54468 | (28) |
| pFTK083 | 171355 | T1 | Tu6 hom., P. rubens Pol-III terminator | pICH41276 | P. rubens DS54468 | (28) |
| pFTK084 | 171356 | T1 | TtRNA[Met] A. niger Pol-III terminator | pICH41276 | A. niger N402 | (28) |
| pFTK085 | 171357 | T1 | TtRNA[Met] A. niger Pol-III terminator | pICH41276 | A. niger N402 | (28) |
| pFTK086 | 171358 | TU | P-ANtRNA[Arg21]-sgRNA-dummy-Esp3I, Pol-III sgRNA transcription unit | pICH41331 | A. niger N402 | (31) |
| pFTK087 | 171359 | TU | P-ANtRNA[Pro1]-sgRNA-dummy-Esp3I, Pol-III sgRNA transcription unit | pICH41331 | A. niger N402 | (31) |
| pFTK088 | 171360 | TU | AMA1 sequence (short), entry vector providing fungal replication | pICH41331 | pDSM-JAK-109 | (25), 36 |
| pFTK089 | 171361 | TU | penicillin gene cluster P. rubens 5′ flanking region | pICH41331 | P. rubens DS54468 | (7) |
| pFTK090 | 171362 | TU | penicillin gene cluster P. rubens 3′ flanking region | pICH41331 | P. rubens DS54468 | (7) |
| pFTK091 | 171363 | TU (level 1) | pks17 Pc21g16000 (conidial pigment biosynthesis) P. rubens 5′ flanking region | pICH47732 (lvl1) | P. rubens DS54468 | (28) |
| pFTK092 | 171364 | TU (level 1) | pks17 Pc21g16000 (conidial pigment biosynthesis) P. rubens 3′ flanking region | pICH47772 (lvl1) | P. rubens DS54468 | (28) |
| pFTK093 | 171365 | TU (level 1) | sgRNA transcription unit (MoClo lvl1 unit), P-gpdA-HH-sgRNA-HDV-Ttrpc | pICH47761 (lvl1) | pFC334 (Addgene ID 87846), pLM-AMA18.0 dCas9-VPR (Addgene ID 138945) | (14) |
| pFTK094-LM-AMA002.0 | 171366 | AMA1 | pLM-AMA002, fungal shuttle vector with bleoR marker for MoClo entry vectors | n/a | pDSM-JAK-109 | (36) |
| pFTK095-LM-AMA015.0 | 171367 | AMA1 | pLM-AMA15.0, CRISPR/Cas9 genome editing with HH-sgRNA-HDV transcription unit, ergA and bleoR fungal markers | n/a | pDSM-JAK-109, pYTK036 (Addgene ID 65143), pLM-AMA15.0 Cas9 (Addgene ID 138944) | (14) |
| pFTK096-LM-AMA018.0 | 171368 | AMA1 | pLM-AMA18.0, CRISPRa/dSpCas9-VPR transcriptional activator with HH-sgRNA-HDV transcription unit, ergA and bleoR fungal markers | n/a | pDSM-JAK-109, pYTK036 (Addgene ID 65143), pAG414GPD (Addgene ID 63801), pLM-AMA18.0 dCas9-VPR (Addgene ID 138945) | (14) |
Units in the toolkit are described using a vector name, an Addgene ID, a part type specifying the function of the part (P, promoter; CDS, coding sequence; T, terminator; TU, transcription unit; AMA1, AMA1-sequence-based fungal replicating vector), a short description of the vector, its recipient Modular Cloning destination vector, the source of the genetic element, and its applications(s).
A collection of functional native or synthetic promoters and terminators are essential for a synthetic biology toolkit. The Fungal Modular Cloning Toolkit provides 20 promoters, three core promoters, and 11 terminators (Table 1). These genomic elements were previously used in synthetic genetic circuits in Aspergillus or Penicillium with varying strain background, media, and cultivation methods (Table 1).5−7,28 Others were benchmarked previously in P. rubens using fluorescent reporters in a BioLector microbioreactor.27
Constitutive Promoters
Constitutive promoters deliver stable expression across different growth environments and growth phases. Strong constitutive promoters like the commonly used promoter of gpdA (ANIA_08041)29 from the glycolytic pathway are often used to drive gene expression in Aspergillus or Penicillium. The gpdA promoter is used to constitutively express various genes as well as fungal selection markers, ribozyme self-cleaved sgRNA, or expression of STFs.5,18 The promoter of the TEF1 (translation-elongation factor 1a) gene is another common strong and constitutive fungal promoter that has been used for polygalacturonase production and the expression of the SpCas9 encoding gene.18 The constitutive promoter of the 40S ribosomal protein S8 (An0465, 40S, RPS8) has been shown to provide stable expression of fluorescent reporters, STFs for scalable transcriptional activation,7 and expression of dSpCas9-VPR from Streptococcus pyogenes for CRISPR-based transcription activation (CRISPRa).14 The promoter of gndA (An11g02040, 6-phosphogluconate dehydrogenase) was shown to give an intermediate strength of transcription27 and proven to be weaker than the constitutive An0465 promoter in P. rubens.7 The well-studied promoters of the bidirectional penicillin biosynthesis genes pcbAB (Pc21g21390) and pcbC (Pc21g21380) are commonly used as strong promoters. Although pcbAB and pcbC are under the control of regulation by both nutritional and developmental factors, they provide a strong transcription rate in lactose-based cultivations.27 Our toolkit also includes the constitutive promoter of oliC31 (An04g08190, mitochondrial ATP synthase subunit 9), which was shown to provide expression comparable to the promoter of pcbAB in Penicillium(27) as well as the constitutive promoter of the housekeeping γ-actin (Pc20g11630) from P. rubens. Besides reliable and constitutive promoters, stimulus-responsive feedback loops may require expression of the regulators at certain time points of the cultivation. Therefore, a set of inducible promoters (PXlnA by xylose, POAT1 by amino acids, PglaA by maltose, PTet by tetracycline, and PalcA by aldehydes) are incorporated.
Synthetic Promoters
An increasing number of promoter libraries have been designed for yeast and filamentous fungi by the creation of synthetic promoters for STFs through the combination of various upstream activating sequence (UAS) elements and different core (or minimal) promoters (CPs).6,7 Transcription-factor-based specific activation/repression mechanisms interact with the designated UAS elements, but a CP sequence is required to recruit general transcription factors and the RNA polymerase II for transcription initiation.30 As part of this toolkit, a collection of CPs are included (CPpcbC from P. rubens, CPNirA from A. nidulans, and CPURA3 from S. cerevisiae), which in combination with UASs compatible with a DBD of an STF (1x, 5x, or 11x QUAS for QA-1f-DBD, 5x LexA UAS for LexA-DBD) can create synthetic promoters with expression levels ranging from hardly detectable to similar to that of highest expressed native genes.7 Moreover, entry vectors are provided for the construction of bacterial-originated tetracycline-inducible (Tet-On) synthetic genetic circuits, including the rtTA2S-M2 (modified TetR-3xVP16) STF and its synthetic promoters using 1, 4, 6, or 10 repeats of TetO UASs.5
Synthetic Transcription Factors
Various STFs (transcriptional activators or repressors) can be constructed using transcription factor domain fusions, where a selected regulator domain can be recruited to a promoter region of the gene of interest.5−7 These STFs often consist of direct fusion of a DBD and an activation domain (AD). On the basis of the ability of the DBD of a transcription factor to bind to its UAS, these STF fusion proteins can be used to design synthetic transcriptional regulators or genetic control circuits. Viral ADs are widely used to create potent STFs, most commonly VP16 or its tandem repeats (VP64, VP160) from herpes simplex virus. Numerous DBDs of transcription factors have been shown to be functional in filamentous fungi, like the bacterial TetR-based STF from the Tet expression system in A. niger and A. fumigatus,5 the qa-1F-based STF (qa-1F-DBD-VP16, QF) from Neurospora crassa in P. rubens,7 the bacterial Bm3R1-based STF (Bm3R1-VP16) in A. niger, T. reesei, and several yeasts,6 and the Gal4 and LexA DBDs, which are frequently used in synthetic expression systems. In Aspergillus species, the often-utilized Tet-On/Tet-Off system provides precise, reversible, and efficiently controlled gene expression using rtTA and rTA STFs, respectively. With the Tet-On system, induced gene activation can be achieved in a titratable manner by addition of the tetracycline derivative doxycycline, whereas induced repression can be achieved using the tetracycline-controlled transactivator (tTA) component to quantitatively reduce gene expression using the Tet-Off system.5 The Fungal Modular Cloning Toolkit contains a collection of DBDs (from the qa-1F, Gal4, LexA, and TetR transcription factors) and transcriptional activation domains (from the qa-1F, Gal4, and B42 transcription factors), VP16 and its four tandem repeats VP64, the tripartite activator VPR (VP64-p65-Rta), and histone acetyltransferases (p300core and Rtt109).
CRISPR Elements
Next to STFs, catalytically inactive CRISPR-Cas proteins can provide new alternatives for the delivery of transcriptional regulators to the target. The CRISPR/Cas9-based systems require the expression of both the Cas protein and a locus-specific sgRNA in the host organism. The toolkit provides entry vectors for both catalytically active (spCas9) and dead (dSpCas9) Cas9 versions from S. pyogenes, which is the most widely applied Cas protein in filamentous fungi. Catalytically active Cas9 provides opportunities for genome editing, whereas dCas9 can be applied to deliver transcriptional regulators to a desired genomic locus through protein fusion of regulator domains. CRISPRa (activation) and CRISPRi (interference) can provide a genome-editing-free alternative for transcriptional activation and repression, respectively. In comparison with the use of STFs, CRISPRa/i tools can provide genome-editing-free transcriptional regulation in filamentous fungi, guiding the regulator to the desired genomic locus, resulting in transcriptional activation (dCas9-VP64 and dCas9-VP64-p65-Rta “VPR”)13,14 or epigenome editing (dCas9-p300).15 The toolkit provides various options for CRISPR sgRNA delivery. A sgRNA “plug-and-play” transcription unit carrying (level 1) vector is included, in which the transcript is under control of the gpdA RNA polymerase II (Pol II) promoter, resulting in a transcript that is self-cleaved using the hammerhead and hepatitis delta virus ribozymes flanking the sgRNA (HH-sgRNA-HDV).14 Ribozyme-based sgRNA delivery is widely used in filamentous fungi,18 as it relies only on an established promoter in the host and ribozyme sequences that work across multiple species. Although the delivery of the ribosome-self-cleaved sgRNAs has been shown to work in numerous fungal applications, in some cases RNA polymerase III (Pol III)-transcribed sgRNA delivery could be advantageous, as the created transcript does not need further processing.18,31 Therefore, the toolkit provides entry vectors containing a collection of Pol III promoters and corresponding terminators (tRNA-Met, tRNA-Leu, U6, and U3) established in P. rubens(28) as well as sgRNA transcription units using tRNA promoters (tRNA-Arg and tRNA-Pro) established in A. niger(31) (Table 1). To assemble a functional transcription unit, the latter utilizes the Esp3I restriction enzyme for insertion of the sgRNA target sequence into the sgRNA transcription unit, whereas the former ones are provided as entry vectors (Figure S1). Two previously established AMA1-based fungal CRISPR vectors with terbinafine and phleomycin markers are also part of this toolkit: pLM-AMA-18.0 for CRISPR-based transcriptional activation and pLM-AMA-15.0 for CRISPR-based genome editing in P. rubens, both with a blue/white selection-aided user-friendly sgRNA “plug-and-play” module to aid rapid library construction.14 The toolkit provides a collection of commonly used transcriptional activation domains (VP16, VP64, and VPR), histone acetyltransferases (p300core and Rtt109), and fluorescent reporters for possible fusion variations.
Fluorescent Reporters
Fluorescent reporters are often used to validate genetic circuits, protein expression, and localization through fusions. This toolkit provides a collection of CDSs of fluorescent and bioluminescent reporters (GFP, DsRed, dTomato, mCherry, YFP, BFP, firefly luciferase) with a nuclear localization sequence (NLS) or serine-lysine-leucine peroxisomal localization (SKL) or without any localization tags, established in Aspergillus and Penicillium species (Table 1). Reporters can be used to demonstrate functionality of genetic circuits or as fusion proteins to validate the expression of the gene of interest.
Selection Markers
The toolkit contains a collection of the most commonly used fungal selection markers (ergA, amdS, pyrG, ble, hph, sdh2, and hisB) as entry vectors. Table 1 shows DNA sources of the markers and their established applications. Overexpression of the native squalene epoxidase (ergA) gene has been shown to provide resistance against terbinafine in a broad range of fungi as well as in Penicillium. In Aspergillus, Trichoderma, and Penicillium species lacking acetamidase activity, overexpression of the acetamidase (amdS) gene provides selection on media containing acetamide as a sole nitrogen source that can be counterselected using fluoroacetamide. The orotidine 5′-phosphate decarboxylase (pyrG) gene from A. oryzae is widely applied in Aspergillus, with examples in Penicillium and Neurospora, as a strong, recyclable, auxotrophic selection marker that can be counterselected using 5-fluoroorotic acid or fully supplemented using uracil or uridine. Overexpression of the bacterial resistance genes as phleomycin (ble) or hygromycin B phosphotransferase (hph) provides selection in numerous Aspergillus and Penicillium strains as well as in N. crassa for phleomycin (glycopeptide antibiotic of the bleomycin family) or hygromycin (aminoglycosidic antibiotic), respectively. The succinate dehydrogenase (sdh2) gene from A. niger is also included, with a single histidine-to-leucine point mutation in the third cysteine-rich cluster (H269L), which has been shown to play a role in conferring resistance to the fungicide carboxin in A. flavus. After generation of a histidine-auxotrophic strain, delivery of the key gene of histidine biosynthesis can provide selection. For the creation of such strains, the toolkit provides entry vectors on the native hisB genes from A. niger and P. rubens.
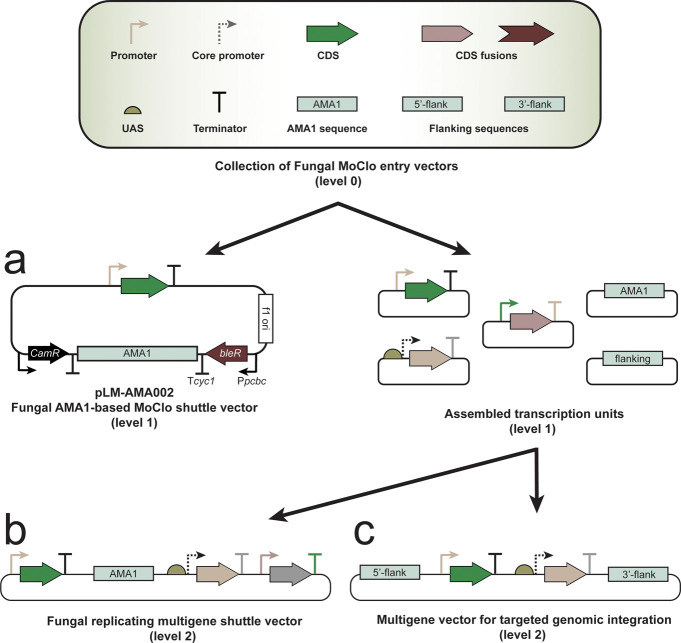
Several options exist for the introduction of assembled transcription units in fungi; if the assembled constructs include the AMA1 sequence, it can be delivered as an episomal vector (Figure 2a,b), or multigene constructs can be integrated to a genomic locus using homologous flanking sequences (Figure 2c). In the toolkit, fungal shuttle vectors with an AMA1 sequence are included. The AMA1 sequence supports autonomous plasmid replication in numerous filamentous fungi as well as flanking regions for homologous recombination-based genomic integration into P. rubens at the frequently used penicillin (Pc21g21370-Pc21g21390) and PKS17 (Pc21g16000) loci. A 50% shorter version of the AMA1 sequence is also provided on a MoClo entry vector, which can be incorporated in complex MoClo-language-based constructs. This truncated sequence can be amplified by PCR and showed transient vector propagation while maintaining selection pressure; without selection, more rapid loss of the vector was detected compared with a full-size AMA1 vector in A. niger.25 As this sequence is integrated on a MoClo entry vector, it is possible to incorporate it into a MoClo multigene construct (level 2), turning the original bacterial vector into a fungal replicating episomal vector (Figure 2b). Fungal shuttle vectors can be assembled in Escherichia coli and delivered into Aspergillus, Penicillium, potentially other fungi in the Aspergillaceae family, or any other AMA1- and selection-marker-compatible fungal host. The vector allows rapid assembly and validation of transcription units, providing alternatives for genomic integration (Figure 2c).
Figure 2.
Transcription unit construction using the MoClo system and delivery platforms. A schematic representation of the recombination and assembly of the MoClo entry vectors into transcription units is shown. Transcription units can be assembled into (a) fungal shuttle vectors or (b, c) multigene constructs that can be delivered (b) as AMA1-based episomal vectors or (c) via genomic integration by homologous recombination.
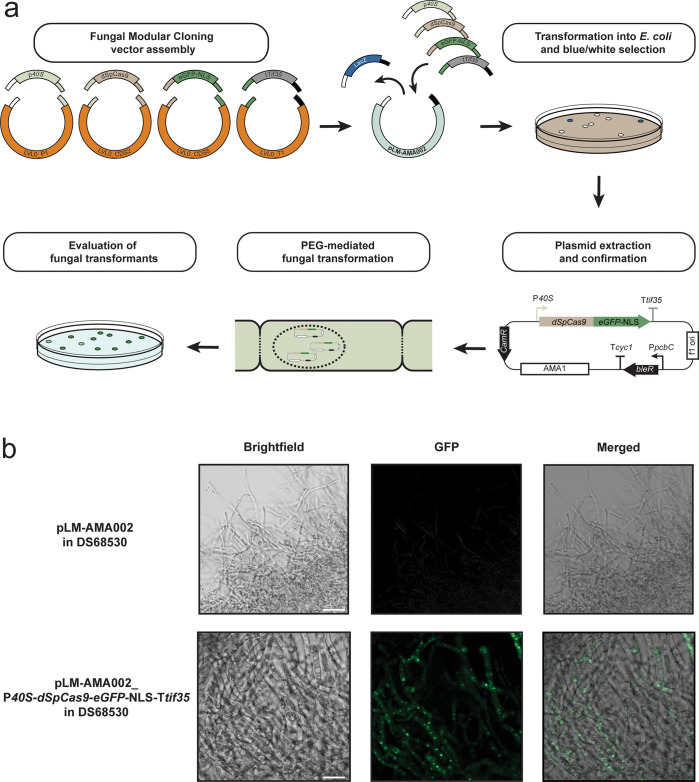
For this toolkit, a shuttle vector (pLM-AMA002) analogous to a MoClo system “level 1” backbone was built, thus providing a MoClo entry vector-compatible fungal transcription unit delivery platform (Figures 2a and 3). As the assembly follows the MoClo language,19 the vector uses BsaI restriction enzyme-generated GGAG and CGCT fusion sites to receive the compatible MoClo entry vectors. The fungal shuttle vector additionally contains a lacZα fragment, which is replaced during the assembly of the transcription unit, allowing for convenient blue/white screening of successful clones. The created transcription-unit-carrying vectors can directly be transformed into fungal hosts using phleomycin as a selection marker. To test our MoClo-adapted and AMA1-based fungal shuttle vector for expressing a gene of interest, a transcription unit was assembled that expresses a fusion protein of the catalytically dead Cas9 protein (dSpCas9) from S. pyogenes and a green fluorescent protein with SV40 nuclear localization (eGFP-NLS) reporter. The genetic parts were rapidly assembled into a transcription unit on the pLM-AMA002 fungal shuttle vector through the first two steps (level 0 construction and level 1 assembly) of MoClo assembly (Figure 3a). The restriction-ligation-based assembly resulted in an AMA1 vector expressing a direct fusion of dSpCas9 and eGFP-NLS driven by a constitutive promoter. The created vector was delivered to P. rubens, and the expression of the protein fusion was validated using fluorescence microscopy, which showed expression of nucleus-localized GFP (Figure 3b). The construction of this expression platform required the integration of the coding sequence of the gene of interest into the appropriate position-predetermined MoClo entry vector. As numerous entry vectors from the toolkit can be utilized, the assembly and validation time of a transcription unit can be significantly reduced. After successful validation of additional new entry vectors, no more sequencing is required in later assembly steps. With the high efficiency of MoClo assembly, transcription units can be rapidly assembled in a single cloning step. Meanwhile, multigene genetic circuits can be constructed in two cloning steps (carrying up to seven transcription units per assembly).19
Figure 3.
Transcription unit assembly from MoClo entry vectors on a pLM-AMA002 fungal shuttle vector and delivery to filamentous fungi. (a) Schematic representation of the assembly of MoClo entry vectors into a single transcription unit delivered to P. rubens on the pLM-AMA002 fungal shuttle vector. (b) Fluorescence microscopy imaging of filaments of a P. rubens strain carrying pLM-AMA002 with the dSpCas9–eGFP-NLS transcription unit, showing protein expression of the fluorescently labeled gene product. Scale bars represent 20 μm.
Taken together, this Fungal Modular Cloning Toolkit aims to accelerate synthetic biology for filamentous fungi by providing essential ready-to-use genetic parts for rapid construction of genetic circuits as well as CRISPR components for more efficient genome engineering and providing aid in biotechnological exploitation. This toolkit provides genetic parts for flexible and efficient assembly of genetic circuits for filamentous fungi in the form of 96 MoClo entry vectors and assembled transcription units. It is a collection of promoters (constitutive and inducible), terminators, activator- and DNA-binding-domains of transcription factors, fluorescent reporters, fungal selection markers, and CRISPR proteins (SpCas9 and dSpCas9) that are applicable for CRISPR-based applications. All of the vectors are built using the MoClo synthetic biology language, which allows the user to assemble numerous transcription units on a single plasmid that can later be delivered to the desired host organism by various delivery methods. To further accelerate the testing of functional transcription units, genetic parts are included that have been tested in the community and shown to be interchangeable between different fungal strains. This collection of fungal genetic parts was created using the “MoClo Toolkit”,19 and therefore, this toolkit (or an equivalent version of it) is needed for the incorporation of new genetic parts for further novel assemblies unless these parts are delivered into the assembly as vector-free DNA fragments. As most of the genetic parts of the toolkit were tested in A. nidulans, A. niger, and P. rubens strains (Table 1), this toolkit aims for compatibility with strains in the Aspergillaceae family but assumes functionality in other filamentous fungal strains. The positions of the modular entry vectors in a transcription unit assembly are represented together with location identifiers in Figure 1. Complete vector sequences are available as Genebank files in Supplementary File S1 and available on Addgene as the “Fungal Toolkit for Modular Cloning (FTK)”.
Methods
Chemicals, Reagents, Oligodeoxyribonucleotides, and Cloning
All medium components and chemicals were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands) or Merck (Darmstadt, Germany). Oligodeoxyribonucleotide primers were obtained from Merck. Enzymes were obtained from Thermo Fisher Scientific (Waltham, MA) unless otherwise stated. For the design of nucleic acid constructs, in silico restriction cloning, and inspection of Sanger sequencing results, SnapGene (GSL Biotech) was used. PCR amplifications were conducted using KAPA HiFi HotStart ReadyMix (Roche Diagnostics, Rotkreuz, Switzerland). Templates for PCR amplifications were acquired from various sources (Table 1) or ordered as synthetic DNA fragments from Thermo Fisher Scientific. All internal BpiI and BsaI cloning sites (and in some cases DraIII and Esp3I) were removed during cloning from the DNA fragments, and these sequences were manually curated for frequent codons in P. rubens. All of the vectors were constructed using the MoClo assembly system and protocol.19 The receiver backbones (established in the Modular Cloning assembly19) used for constructing the genetic parts containing entry vectors are highlighted in Figure 1b. As the linker sequences between the genetic parts in the transcription unit are based on the standard MoClo language (Figure 1a), the parts are compatible with modular systems that use this linker system.
Correctly assembled plasmids were identified with blue/white screening and confirmed by sequencing. The transcription unit expressing SpCas9–eGFP-NLS on a fungal shuttle vector (pLM-AMA002_P40s-dSpCas9-eGFP-NLS-Ttif35) was assembled using a mixture of 30 fmol of each entry vector (P40s An0465 (P1), dSpCas9(m2) (CDS2), eGFP-NLS (CDS5), and Ttif35 (T1)) and the backbone vector pLM-AMA002.
The 50% shorter AMA1 sequence25 was created by PCR and integrated into a MoClo entry vector. The autonomously replicating shuttle vector carrying the AMA1 sequence was based on the pDSM-JAK-109 backbone where the pGpda-DsRed-SKL-TpenDE transcription unit was removed using the BspTI and NotI restriction enzymes. The linear vector was treated with the Klenow Fragment of DNA polymerase I and self-ligated into a circular vector using the T4 DNA ligase according to the instructions of the manufacturer, creating a new AMA1 vector without DsRed expression. This vector was cloned with a removable LacZ gene cloning site using BspTI, based on the “level 1” receiver backbones of the MoClo system, to create pLM-AMA002.
Fungal Strains, Transformation, and Cultivation
Cultivation of fungal and bacterial strains, media composition, protoplast generation, and fungal transformation using phleomycin marker was carried out as described previously.14 A list of fungal strains created in this study with corresponding transformed donor DNA can be found in Table S1.
Fluorescence Microscopy
Transformants were further cultivated after transformation on phleomycin (50 μg/mL)-supplemented transformation solid medium for 5 days and examined using fluorescence microscopy. A small amount of hyphae was taken from the peripheral zone of the colonies and suspended in phosphate-buffered saline (58 mM Na2HPO4, 17 mM NaH2PO4, 68 mM NaCl, pH 7.3). Confocal imaging was performed on a Carl Zeiss LSM800 confocal microscope using a 20× objective and ZEN 2009 software (Carl Zeiss, Oberkochen, Germany). The GFP signal was visualized by excitation with a 488 nm argon laser (Lasos Lasertechnik, Jena, Germany), and emission was detected using a 509 nm bandpass emission filter.
Acknowledgments
The project leading to this application received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Marie Skłodowska-Curie Grant Agreement 713482 (ALERT Program). The authors thank Zsófia Büttel for building some of the entry vectors provided and Jeroen Nijland for help with depositing the library to Addgene.
Glossary
Abbreviations
- MoClo
Modular Cloning
- BGC
biosynthetic gene cluster
- NRPS
nonribosomal peptide synthetase
- PKS
polyketide synthase
- STF
synthetic transcription factor
- DBD
DNA-binding domain
- UAS
upstream activating sequence
- CRISPR
clustered regularly interspaced short palindromic repeats
- CAS
CRISPR-associated protein
- sgRNA
single guide RNA
- AD
activation domain
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.1c00260.
Author Contributions
# L.M. and C.P. contributed equally to this work. L.M. and C.P. designed and carried out all of the experiments and wrote the manuscript with critical feedback and help from V.M., R.A.L.B., Y.N., and A.J.M.D.
The authors declare no competing financial interest.
Supplementary Material
References
- Meyer V.; Basenko E. Y.; Benz J. P.; Braus G. H.; Caddick M. X.; Csukai M.; de Vries R. P.; Endy D.; Frisvad J. C.; Gunde-Cimerman N.; Haarmann T.; Hadar Y.; Hansen K.; Johnson R. I.; Keller N. P.; Kraševec N.; Mortensen U. H.; Perez R.; Ram A. F. J.; Record E.; Ross P.; Shapaval V.; Steiniger C.; van den Brink H.; van Munster J.; Yarden O.; Wösten H. A. B. Growing a Circular Economy with Fungal Biotechnology: A White Paper. Fungal Biol. Biotechnol. 2020, 7 (1), 5. 10.1186/s40694-020-00095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17 (3), 167–180. 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. C.; Grijseels S.; Prigent S.; Ji B.; Dainat J.; Nielsen K. F.; Frisvad J. C.; Workman M.; Nielsen J. Global Analysis of Biosynthetic Gene Clusters Reveals Vast Potential of Secondary Metabolite Production in Penicillium Species. Nat. Microbiol. 2017, 2, 17044. 10.1038/nmicrobiol.2017.44. [DOI] [PubMed] [Google Scholar]
- Caesar L. K.; Kelleher N. L.; Keller N. P. In the Fungus Where It Happens: History and Future Propelling Aspergillus nidulans as the Archetype of Natural Products Research. Fungal Genet. Biol. 2020, 144, 103477. 10.1016/j.fgb.2020.103477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanka F.; Cairns T.; Boecker S.; Berens C.; Happel A.; Zheng X.; Sun J.; Krappmann S.; Meyer V. Tet-on, or Tet-off, That Is the Question: Advanced Conditional Gene Expression in Aspergillus. Fungal Genet. Biol. 2016, 89, 72–83. 10.1016/j.fgb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Rantasalo A.; Landowski C. P.; Kuivanen J.; Korppoo A.; Reuter L.; Koivistoinen O.; Valkonen M.; Penttilä M.; Jäntti J.; Mojzita D. A Universal Gene Expression System for Fungi. Nucleic Acids Res. 2018, 46 (18), e111–e111. 10.1093/nar/gky558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mózsik L.; Büttel Z.; Bovenberg R. A. L.; Driessen A. J. M.; Nygård Y. Synthetic Control Devices for Gene Regulation in Penicillium chrysogenum. Microb. Cell Fact. 2019, 18 (1), 203. 10.1186/s12934-019-1253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K.; Bhabhra R.; Rhodes J. C.; Askew D. S. Doxycycline-Regulated Gene Expression in the Opportunistic Fungal Pathogen Aspergillus fumigatus. BMC Microbiol. 2005, 5 (1), 1. 10.1186/1471-2180-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derntl C.; Mach R. L.; Mach-Aigner A. R. Fusion Transcription Factors for Strong, Constitutive Expression of Cellulases and Xylanases in Trichoderma reesei. Biotechnol. Biofuels 2019, 12 (1), 231. 10.1186/s13068-019-1575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau M. F.; Entwistle R.; Chiang Y.-M.; Ahuja M.; Oakley C. E.; Akashi T.; Wang C. C. C.; Todd R. B.; Oakley B. R. Hybrid Transcription Factor Engineering Activates the Silent Secondary Metabolite Gene Cluster for (+)-Asperlin in Aspergillus nidulans. ACS Chem. Biol. 2018, 13 (11), 3193–3205. 10.1021/acschembio.8b00679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K.; Maurer S.; Kaffarnik F.; Ladendorf O.; Brachmann A.; Kämper J.; Feldbrügge M. Tetracycline-Regulated Gene Expression in the Pathogen Ustilago maydis. Fungal Genet. Biol. 2006, 43 (11), 727–738. 10.1016/j.fgb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Saunders G.; Picknett T. M.; Tuite M. F.; Ward M. Heterologous Gene Expression in Filamentous Fungi. Trends Biotechnol. 1989, 7 (10), 283–287. 10.1016/0167-7799(89)90048-6. [DOI] [Google Scholar]
- Roux I.; Woodcraft C.; Hu J.; Wolters R.; Gilchrist C. L. M.; Chooi Y.-H. CRISPR-Mediated Activation of Biosynthetic Gene Clusters for Bioactive Molecule Discovery in Filamentous Fungi. ACS Synth. Biol. 2020, 9 (7), 1843–1854. 10.1021/acssynbio.0c00197. [DOI] [PubMed] [Google Scholar]
- Mózsik L.; Hoekzema M.; de Kok N. A. W.; Bovenberg R. A. L.; Nygård Y.; Driessen A. J. M. CRISPR-Based Transcriptional Activation Tool for Silent Genes in Filamentous Fungi. Sci. Rep. 2021, 11 (1), 1118. 10.1038/s41598-020-80864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Huang L.; Pan L.; Wang B.; Pan L. CRISPR/DCas9-Mediated Epigenetic Modification Reveals Differential Regulation of Histone Acetylation on Aspergillus niger Secondary Metabolite. Microbiol. Res. 2021, 245, 126694. 10.1016/j.micres.2020.126694. [DOI] [PubMed] [Google Scholar]
- Snoek I. S. I.; van der Krogt Z. A.; Touw H.; Kerkman R.; Pronk J. T.; Bovenberg R. A. L.; van den Berg M. A.; Daran J. M. Construction of an HdfA Penicillium chrysogenum Strain Impaired in Non-Homologous End-Joining and Analysis of Its Potential for Functional Analysis Studies. Fungal Genet. Biol. 2009, 46 (5), 418–426. 10.1016/j.fgb.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Aleksenko A.; Clutterbuck A. J. Autonomous Plasmid Replication in Aspergillus nidulans: AMA1 and MATE Elements. Fungal Genet. Biol. 1997, 21 (3), 373–387. 10.1006/fgbi.1997.0980. [DOI] [PubMed] [Google Scholar]
- Deng H.; Gao R.; Liao X.; Cai Y. CRISPR System in Filamentous Fungi: Current Achievements and Future Directions. Gene 2017, 627, 212–221. 10.1016/j.gene.2017.06.019. [DOI] [PubMed] [Google Scholar]
- Weber E.; Engler C.; Gruetzner R.; Werner S.; Marillonnet S. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS One 2011, 6 (2), e16765 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. J.; Lai H.-E.; Kelwick R. J. R.; Chee S. M.; Bell D. J.; Polizzi K. M.; Freemont P. S. EcoFlex: A Multifunctional MoClo Kit for E. coli Synthetic Biology. ACS Synth. Biol. 2016, 5 (10), 1059–1069. 10.1021/acssynbio.6b00031. [DOI] [PubMed] [Google Scholar]
- Lee M. E.; DeLoache W. C.; Cervantes B.; Dueber J. E. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 2015, 4 (9), 975–986. 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- Obst U.; Lu T. K.; Sieber V. A Modular Toolkit for Generating Pichia pastoris Secretion Libraries. ACS Synth. Biol. 2017, 6 (6), 1016–1025. 10.1021/acssynbio.6b00337. [DOI] [PubMed] [Google Scholar]
- Engler C.; Youles M.; Gruetzner R.; Ehnert T.-M.; Werner S.; Jones J. D. G.; Patron N. J.; Marillonnet S. A Golden Gate Modular Cloning Toolbox for Plants. ACS Synth. Biol. 2014, 3 (11), 839–843. 10.1021/sb4001504. [DOI] [PubMed] [Google Scholar]
- Martella A.; Matjusaitis M.; Auxillos J.; Pollard S. M.; Cai Y. EMMA: An Extensible Mammalian Modular Assembly Toolkit for the Rapid Design and Production of Diverse Expression Vectors. ACS Synth. Biol. 2017, 6 (7), 1380–1392. 10.1021/acssynbio.7b00016. [DOI] [PubMed] [Google Scholar]
- Sarkari P.; Marx H.; Blumhoff M. L.; Mattanovich D.; Sauer M.; Steiger M. G. An Efficient Tool for Metabolic Pathway Construction and Gene Integration for Aspergillus niger. Bioresour. Technol. 2017, 245, 1327–1333. 10.1016/j.biortech.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Dahlmann T. A.; Terfehr D.; Becker K.; Teichert I. Golden Gate Vectors for Efficient Gene Fusion and Gene Deletion in Diverse Filamentous Fungi. Curr. Genet. 2021, 67 (2), 317–330. 10.1007/s00294-020-01143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polli F.; Meijrink B.; Bovenberg R. A. L.; Driessen A. J. M. New Promoters for Strain Engineering of Penicillium chrysogenum. Fungal Genet. Biol. 2016, 89, 62–71. 10.1016/j.fgb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Pohl C.; Kiel J. A. K. W.; Driessen A. J. M.; Bovenberg R. A. L.; Nygård Y. CRISPR/Cas9 Based Genome Editing of Penicillium chrysogenum. ACS Synth. Biol. 2016, 5 (7), 754–764. 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- Kolar M.; Punt P. J.; van den Hondel C. A. M. J. J.; Schwab H. Transformation of Penicillium Chrysogenum Using Dominant Selection Markers and Expression of an Escherichia coli LacZ Fusion Gene. Gene 1988, 62 (1), 127–134. 10.1016/0378-1119(88)90586-0. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T.; Kadonaga J. T. Regulation of Gene Expression via the Core Promoter and the Basal Transcriptional Machinery. Dev. Biol. 2010, 339 (2), 225–229. 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L.; Ouedraogo J.-P.; Kolbusz M.; Nguyen T. T. M.; Tsang A. Efficient Genome Editing Using TRNA Promoter-Driven CRISPR/Cas9 GRNA in Aspergillus niger. PLoS One 2018, 13 (8), e0202868 10.1371/journal.pone.0202868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nødvig C. S.; Nielsen J. B.; Kogle M. E.; Mortensen U. H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS One 2015, 10 (7), e0133085. 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmaier F.; Philippsen P. Identification of Two Genes Coding for the Translation Elongation Factor EF-1 Alpha of S. cerevisiae. EMBO J. 1984, 3 (13), 3311–3315. 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo L.; Lamas-Maceiras M.; Ullán R. V.; Campoy S.; Teijeira F.; Casqueiro J.; Martín J. F. Characterization of the Oat1 Gene of Penicillium chrysogenum Encoding an ω-Aminotransferase: Induction by l-Lysine, l-Ornithine and l-Arginine and Repression by Ammonium. Mol. Genet. Genomics 2005, 274 (3), 283–294. 10.1007/s00438-005-0019-2. [DOI] [PubMed] [Google Scholar]
- Orejas M.; MacCabe A. P.; Pérez González J. A.; Kumar S.; Ramón D. Carbon Catabolite Repression of the Aspergillus nidulans XlnA Gene. Mol. Microbiol. 1999, 31 (1), 177–184. 10.1046/j.1365-2958.1999.01157.x. [DOI] [PubMed] [Google Scholar]
- Bovenberg R. A. L.; Kiel J. A. K. W.; Wenzel T. J.; Los A. P.. Vector-Host System. WO 2012123429, 2012.
- Fiedler M. R. M.; Gensheimer T.; Kubisch C.; Meyer V. HisB as Novel Selection Marker for Gene Targeting Approaches in Aspergillus niger. BMC Microbiol. 2017, 17 (1), 57. 10.1186/s12866-017-0960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hartingsveldt W.; Mattern I. E.; van Zeijl C. M. J.; Pouwels P. H.; van den Hondel C. A. M. J. J. Development of a Homologous Transformation System for Aspergillus niger Based on the PyrG Gene. Mol. Gen. Genet. 1987, 206 (1), 71–75. 10.1007/BF00326538. [DOI] [PubMed] [Google Scholar]
- Panozzo C.; Capuano V.; Fillinger S.; Felenbok B. The Zinc Binuclear Cluster Activator AlcR Is Able to Bind to Single Sites but Requires Multiple Repeated Sites for Synergistic Activation of the AlcA Gene in Aspergillus nidulans. J. Biol. Chem. 1997, 272 (36), 22859–22865. 10.1074/jbc.272.36.22859. [DOI] [PubMed] [Google Scholar]
- Riabinina O.; Luginbuhl D.; Marr E.; Liu S.; Wu M. N.; Luo L.; Potter C. J. Improved and Expanded Q-System Reagents for Genetic Manipulations. Nat. Methods 2015, 12 (3), 219–222. 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossani Z. Y.; Reider Apel A.; Szmidt-Middleton H.; Hillson N. J.; Deutsch S.; Keasling J. D.; Mukhopadhyay A. A Combinatorial Approach to Synthetic Transcription Factor-Promoter Combinations for Yeast Strain Engineering. Yeast 2018, 35 (3), 273–280. 10.1002/yea.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigl C.; Handler M.; Sprenger G.; Kürnsteiner H.; Zadra I. A Novel Homologous Dominant Selection Marker for Genetic Transformation of Penicillium chrysogenum: Overexpression of Squalene Epoxidase-Encoding ErgA. J. Biotechnol. 2010, 150 (3), 307–311. 10.1016/j.jbiotec.2010.09.941. [DOI] [PubMed] [Google Scholar]
- Punt P. J.; van den Hondel C. A. M. J. J. Transformation of Filamentous Fungi Based on Hygromycin B and Phleomycin Resistance Markers. Methods Enzymol. 1992, 216, 447–457. 10.1016/0076-6879(92)16041-H. [DOI] [PubMed] [Google Scholar]
- Fraaije B. A.; Bayon C.; Atkins S.; Cools H. J.; Lucas J. A.; Fraaije M. W. Risk Assessment Studies on Succinate Dehydrogenase Inhibitors, the New Weapons in the Battle to Control Septoria Leaf Blotch in Wheat. Mol. Plant Pathol. 2012, 13 (3), 263–275. 10.1111/j.1364-3703.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C.; Polli F.; Schütze T.; Viggiano A.; Mózsik L.; Jung S.; de Vries M.; Bovenberg R. A. L.; Meyer V.; Driessen A. J. M. A Penicillium rubens Platform Strain for Secondary Metabolite Production. Sci. Rep. 2020, 10 (1), 7630. 10.1038/s41598-020-64893-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M. R. M.; Cairns T. C.; Koch O.; Kubisch C.; Meyer V. Conditional Expression of the Small GTPase ArfA Impacts Secretion, Morphology, Growth, and Actin Ring Position in Aspergillus niger. Front. Microbiol. 2018, 9, 878. 10.3389/fmicb.2018.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A.; Scheiman J.; Vora S.; Pruitt B. W.; Tuttle M.; P R Iyer E.; Lin S.; Kiani S.; Guzman C. D.; Wiegand D. J.; Ter-Ovanesyan D.; Braff J. L.; Davidsohn N.; Housden B. E.; Perrimon N.; Weiss R.; Aach J.; Collins J. J.; Church G. M. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 2015, 12 (4), 326–328. 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoz D. S. M.; Rudolf F.; Stelling J. Inducible, Tightly Regulated and Growth Condition-Independent Transcription Factor in Saccharomyces cerevisiae. Nucleic Acids Res. 2014, 42 (17), e130–e130. 10.1093/nar/gku616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvion J.-F.; Havaux-Copf B.; Picard D. Fusion of GAL4-VP16 to a Steroid-Binding Domain Provides a Tool for Gratuitous Induction of Galactose-Responsive Genes in Yeast. Gene 1993, 131 (1), 129–134. 10.1016/0378-1119(93)90681-R. [DOI] [PubMed] [Google Scholar]
- Hilton I. B.; D’Ippolito A. M.; Vockley C. M.; Thakore P. I.; Crawford G. E.; Reddy T. E.; Gersbach C. A. Epigenome Editing by a CRISPR-Cas9-Based Acetyltransferase Activates Genes from Promoters and Enhancers. Nat. Biotechnol. 2015, 33 (5), 510–517. 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Holbert M. A.; Wurtele H.; Meeth K.; Rocha W.; Gharib M.; Jiang E.; Thibault P.; Verreault A.; Cole P. A.; Marmorstein R. Fungal Rtt109 Histone Acetyltransferase Is an Unexpected Structural Homolog of Metazoan P300/CBP. Nat. Struct. Mol. Biol. 2008, 15 (7), 738–745. 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.