Abstract
We have shown recently that a stable hairpin preceded by a short upstream open reading frame (uORF) promotes nonlinear ribosome migration or ribosome shunt on a synthetic mRNA leader (M. Hemmings-Mieszczak and T. Hohn, RNA 5:1149–1157, 1999). We have now used the model mRNA leader to study further the mechanism of shunting in vivo and in vitro. We show that a full cycle of translation of the uORF, including initiation, elongation, and termination, is a precondition for the ribosome shunt across the stem structure to initiate translation downstream. Specifically, AUG recognition and the proper release of the nascent peptide are necessary and sufficient for shunting. Furthermore, the stop codon context must not impede downstream reinitiation. Translation of the main ORF was inhibited by replacement of the uORF by coding sequences repressing reinitiation but stimulated by the presence of the virus-specific translational transactivator of reinitiation (cauliflower mosaic virus pVI). Our results indicate reinitiation as the mechanism of translation initiation on the synthetic shunt-competent mRNA leader and suggest that uORF-dependent shunting is more prevalent than previously anticipated. Within the above constraints, uORF-dependent shunting is quite tolerant of uORF and stem sequences and operates in systems as diverse as plants and fungi.
The entire process of protein synthesis consists of three steps: translation initiation, elongation, and termination. During initiation, the translational apparatus selects an mRNA and forms a ribosome initiation complex on which the initiator Met-tRNAi interacts with the initiation codon, primarily via codon-anticodon base pairing. There are two well-recognized mechanisms leading to initiation in eukaryotes: 5′-end-dependent scanning (39) and internal initiation (reviewed in references 33 and 34), which occurs directly at or upstream of the initiation codon. According to the scanning model, the initiation complex can access the mRNA only from the 5′ end and can translate efficiently only the 5′-proximal cistron. However, many viral and a few cellular mRNAs have long, structured leaders with multiple upstream open reading frames (uORFs), which have critical regulatory functions (41, 42, 43). To accommodate such cases, the terms leaky scanning and reinitiation are used to describe the particular events leading to initiation at the main ORF of the mRNA. Leaky scanning occurs when some 40S ribosomal subunits bypass the first AUG codon, mostly due to an unfavorable context differing from the optimal A/GCCAUGG, and initiate at the next AUG codon downstream (36). The extent of leaky scanning can vary. Irrespective of context, the first AUG codon may be bypassed if it is too close to the 5′ end. In contrast, a weak context might be compensated for by the presence of a secondary structure located downstream, which may retard scanning and enhance initiation (40).
Reinitiation can occur after translation of a uORF if or when the 40S subunit remains bound to the mRNA, resumes scanning, and initiates at the downstream AUG codon (39). In principle, reinitiation is favored by long intercistronic distances (37) allowing ribosomes to reacquire some initiation factors, in particular the ternary complex of eIF-2, GTP, and Met-tRNAi (32). The ability of eukaryotic ribosomes to reinitiate is limited by the size of the uORF, with a cutoff length of around 30 codons (18, 46). In some cases, the frequency of reinitiation depends on the coding content of the short uORF (reviewed in references 21, 22, and 45). In the best-studied examples, the mammalian S-adenosylmethionine decarboxylase gene (AdoMetDc) (29, 30, 49), human cytomegalovirus gp48 (1, 7, 8, 59), the yeast CPA1 gene (13), its homologue in Neurospora crassa, arg-2 (65), and the bacterial cat (chloramphenicol acetyltransferase gene (45), the specific uORF peptide products inhibit reinitiation downstream. It is assumed that the nascent uORF-encoded peptide interacts with the components of the translation termination machinery and, by blocking peptidyl-tRNA hydrolysis, provokes ribosome stalling near its own termination codon. Indeed, in some cases, primer extension inhibition (toeprinting) allowed the detection of an arrested ribosome with the uORF termination codon at the ribosomal A site (9, 65, 66, 67). In another case, unreleased uORF-encoded peptide was found bound to ribosomes (45). In contrast to the aforementioned genes, GCN4 regulation in yeast (31) depends on 10-nucleotide (nt) sequences following termination codons. High reinitiation results from termination at uORF1, but repression follows termination at uORF4, indicating that reinitiation may be coupled to the efficiency of termination (23, 48). In a variety of organisms, termination efficiency and the fate of the ribosome after termination are influenced by the sequence context of the stop codon (47). In the case of the yeast Saccharomyces cerevisiae, upstream and downstream determinants act synergistically to influence the efficiency of translation termination (4, 6). In particular, there is base bias 3′ to the stop codon. Relative termination efficiencies for the respective stop codons are, for example, G > A > U > C for the UAA codon and A > U > C > G for the UAG codon. The influence of the termination codon context seems to be of particular significance for mRNAs bearing uORFs.
Translation initiation has been studied intensively on the cauliflower mosaic virus pregenomic mRNA (CaMV 35S RNA) leader, which contains several short uORFs and an elongated hairpin embodying a 480-nt sequence (25). Translation initiation on the CaMV 35S RNA leader occurs by a ribosome shunt mechanism (19) and requires the 5′ cap (60), a short, phylogenetically conserved uORF A (54), and an elongated hairpin structure (14, 26). Formation of this elongated hairpin allows exclusion of the internal part of the leader from the scanning process (26, 27). Recently, we showed that the excluded sequences and/or structure interact with the viral coat protein (24). Thus, the main function of shunting might be to ensure that translation and other processes essential for the viral life cycle can occur simultaneously. Details of the ribosome shunt model remain unclear and difficult to assess on the multifunctional viral pregenomic mRNA leader. Therefore, a simplified alternative mRNA leader was designed with a short but stable stem (Kozak stem [38]) replacing the elongated hairpin of the CaMV 35S RNA leader (27). This synthetic mRNA leader is shunt competent in translation assays both in vivo and in vitro, retaining all translational features of the natural viral counterpart with all the advantages of a simplified system (27). Hence, it seems suitable for analysis of the molecular mechanism of shunting.
Here we report the use of the model synthetic mRNA leader to study the mechanism of ribosome shunt in transfected plant protoplasts from Orychophragmus violaceus and in cell-free translation systems from wheat germ and S. cerevisiae. Our study reveals critical aspects of the mechanism of translation initiation occurring on the synthetic mRNA leader and suggests that uORF-dependent shunting is more prevalent than previously anticipated.
MATERIALS AND METHODS
Constructs and transcripts.
CAT reporter gene expression under the influence of artificial leader sequences was analyzed in vitro using pGEM-1 (Promega)-based constructs or in transiently transfected O. violaceus protoplasts, using pDH51-based constructs (53). Basic constructs S-A, S, and ΔS were previously described as pMH188, -191, and -212, respectively (27). Constructs L-A and L were derived from S-A and S, respectively, after deletion of the stem sequences by filling in and blunt-end ligation of XbaI and ClaI sites. Mutations in the A- and Y-series constructs (Fig. 4 and 7, respectively) were introduced by pairs of annealed oligonucleotides subcloned between the HindIII and XbaI restriction sites of the S-A construct. All constructs were sequenced; the sequences of oligonucleotides and constructs are available on request.
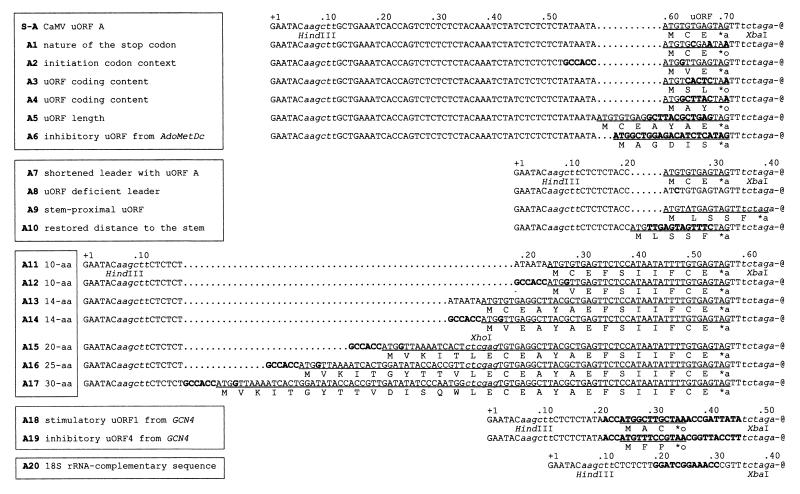
FIG. 4.
uORF mutations in the synthetic shunt-competent leader. Sequences are 3′-end aligned at the stem (represented by -@). Constructs with common 5′-proximal sequences are framed together, with dots introduced to facilitate the alignment. Restriction sites are in lowercase italics. uORF coding sequences are underlined, with the encoded peptides shown below. ∗a and ∗o indicate amber and ochre termination codons, respectively. Point mutations, insertions, and replacements are indicated in bold; Δ in A9 represents a 1-nt deletion. The uORF from AdoMetDc and GCN4 uORFs 1 and 4 (plus flanking sequences) are shown in bold in A6, A18, and A19, respectively. Sequence complementary to the 3′ end of the 18S rRNA is indicated in bold in A20.
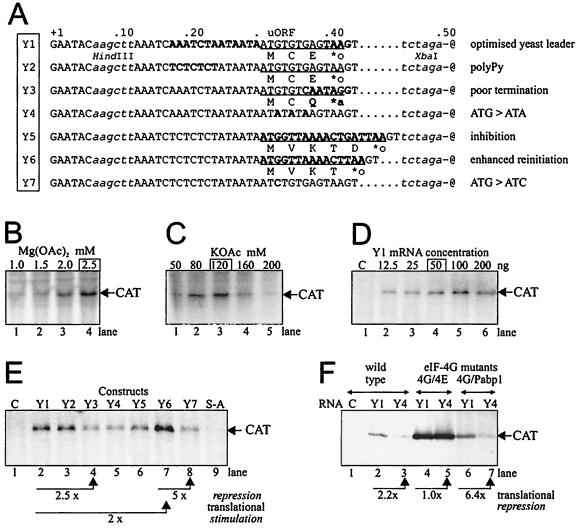
FIG. 7.
Expression of the CAT reporter gene under the control of the synthetic shunt-competent mRNA leader analyzed in an S. cerevisiae cell-free translation system. (A) uORF mutations shown in the context of sequences directly preceding the stem (represented by -@); dots are introduced to facilitate the alignment. Restriction sites are depicted with lowercase italics. uORF coding sequences are underlined, with encoded peptides shown below. ∗a and ∗o indicate amber and ochre termination codons, respectively. Point mutations, insertions, and replacements are distinguished in bold. Extracts were prepared from strains disrupted in both chromosomal eIF4G genes but bearing appropriate versions of the eIF4GI (TIF4631) gene on a plasmid. The strains harbored either wild-type eIF4GI (YAS 2069) or copies exhibiting mutations which weaken the interaction with eIF4E (YAS 2074) or with Pab1p (YAS 2075). Extracts prepared from YAS 2074 display strongly reduced cap effects on translation with concomitant activation of translation in cap-independent fashion, whereas extracts from YAS 2075 do not support poly(A)-promoted translation (63). Optimal concentrations of magnesium acetate (B), potassium acetate (C), and Y1 mRNA (D) are framed above the panels, showing typical resolution of in vitro-translated products separated by SDS-PAGE (12% gel). (E) Translation efficiency under the control of leader constructs shown in panel A analyzed in an extract derived from the wild-type yeast strain YAS 2069. C, no exogenous mRNA included. (F) Efficiency of translation of constructs Y1 and Y4 in extracts of wild-type (lanes 1 to 3) or mutant yeast strains YAS 2074 (lanes 4 and 5) and YAS 2075 (lanes 6 and 7). C, no exogenous mRNA included. Translational repression factors in panels E and F are calculated from translation efficiencies corresponding to the reactions represented by neighboring lanes and indicated by arrows.
SP6-directed transcripts were synthesized on templates digested with PstI or EcoRI, giving full-length or truncated CAT products, respectively. In vitro transcripts were capped with 7mGpppG and trace labeled with [32P]UTP. Transcription reactions were treated with RQ1 RNase-free DNase (Promega) and purified on Chroma-Spin-100 columns (Clontech).
Density centrifugation.
Translation initiation reactions were performed in wheat germ extract (Promega) as described by Kozak (38), using up to 100 ng of radiolabeled in vitro transcripts synthesized from templates digested with EcoRI. Reaction mixtures (10 μl) comprising 5 μl of extract, each amino acid at 1 mM (Promega), RNase inhibitor (1 U/μl; Promega), MgCl2 (2 mM), potassium acetate (100 mM), sparsomycin (200 μM; a gift from Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, National Cancer Institute, Bethesda, Md.), and cycloheximide (100 μg/ml; Sigma) were incubated for 10 min at 30°C, transferred to ice, and loaded on to precooled 5-ml 10 to 30% sucrose gradients prepared in 20 mM HEPES (pH 7.5)–2 mM MgCl2–100 mM potassium acetate–1 mM dithiothreitol–0.1 mM EDTA. Gradients were centrifuged for 90 min at 4°C in an SW60 rotor at 45,000 rpm. Following gradient fractionation, the absorbance profile at 260 nm was monitored to determine the size distribution and positions of ribosome-mRNA complexes. Quantification of radiolabeled transcript migration was performed by scintillation counting.
Primer extension inhibition (toeprinting).
Primer extension inhibition analysis was performed exactly as described by Wang and Sachs (65). Translation initiation reaction mixtures in wheat germ extract were assembled with 2 × 106 to 5 × 106 cpm of [γ-32P]ATP-labeled primer 1, 2, or 3 (Fig. 1A). Reverse transcription reactions (20 μl) were incubated with 100 U of Superscript II RNase H− reverse transcriptase (GibcoBRL) for 30 min at 30°C. Following phenol extraction and ethanol precipitation, cDNA products were separated by polyacrylamide gel electrophoresis (PAGE) on 6% polyacrylamide gels, which were then fixed, dried, and exposed for up to 1 week to X-ray films (Biomax-MR) or for up to 3 days in PhosphorImager (Molecular Dynamics) cassettes. Experiments were performed twice. Control reactions were prepared by omitting translation elongation inhibitors or wheat germ extract. Relevant plasmids were sequenced using radiolabeled primers.
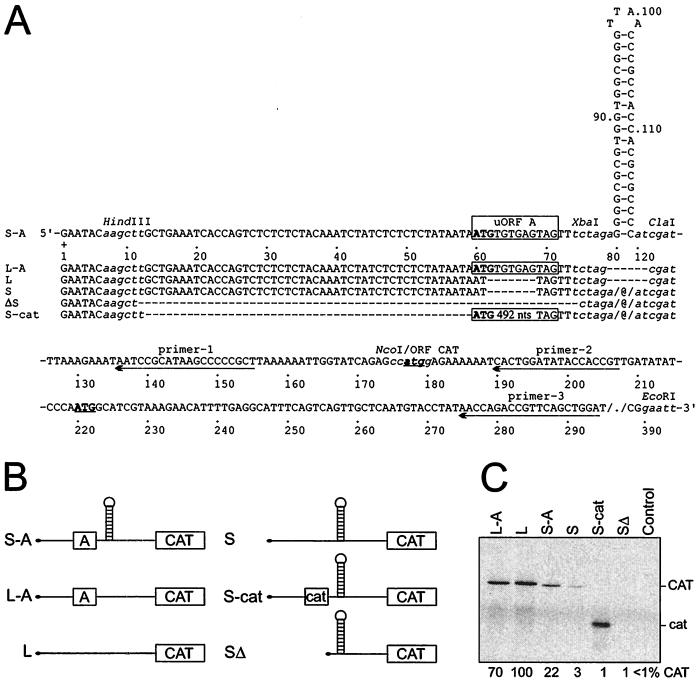
FIG. 1.
CAT reporter gene-expressing constructs with structured and linear leaders. (A) Structured leaders: S-A, synthetic shunt-competent leader shown with numbering every 10 nt. Restriction sites used for further modifications are represented by lowercase italics. S and ΔS have uORF A or the 5′-proximal sequence, respectively, deleted. S-cat has an uORF extended by the insertion of 492 nt derived from the C-terminal part of CAT. Linear leaders: L-A and L, derivatives of S-A and S, respectively, with deletion of the stem sequences (nt 79 to 120). All uORFs are boxed, ATG codons are indicated in bold, and those in CAT are underlined. Dashes represent deletions, and @ indicates the stem structures. Arrows depict sequences targeted with the antisense oligonucleotides used in the primer extension assay (primers 1 to 3). (B) Schematic representation of constructs. (C) CAT expression in wheat germ in vitro translation system; resolution by SDS-PAGE (15% gel). Translation efficiency, migration of CAT, and its cat derivative are indicated. The control has no exogenous RNA added to the system.
In vitro translation.
In vitro translation in wheat germ extract was carried out in a 10-μl reaction volume as instructed by the manufacturer (Promega) in the presence of 2 mM MgCl2 and 100 mM potassium acetate using equal molar amounts (100 fmol) of in vitro transcripts synthesized from templates digested with PstI. Yeast translation extracts were prepared as described previously (55, 62). In vitro translation in yeast extracts was carried out in 15-μl reaction volume as described elsewhere (56) in the optimized conditions of 2.5 mM MgCl2–120 mM potassium acetate, using equal amounts (50 or 100 ng) of in vitro transcripts. Translation products labeled with [35S]Met were separated by sodium dodecyl sulfate-PAGE on 12 or 15% (Fig. 1C) polyacrylamide gels, which were fixed, dried, and exposed to X-ray films (Fuji) or quantified using a PhosphorImager (Molecular Dynamics). Experiments and calculations were repeated three times.
Transient expression assays.
Protoplasts prepared from O. violaceus cell suspension cultures were transfected by electroporation (18), using 15 μg of plasmid DNA per transformation, including 10 μg of CAT-expressing construct supplemented with 5 μg of HELP7 expressing CaMV pVI (5) or empty vector pDH51 (53). An internal control plasmid expressing β-glucuronidase was cotransformed in all experiments. After an overnight incubation, a protein extract was prepared and CAT enzyme-linked immunosorbent assays were carried out as instructed by the manufacturer (Roche-Boehringer). CAT expression was always calculated relative to the β-glucuronidase activity of the internal control measured as described by Fütterer and Hohn (17). Activities cited are the means from three independent transformations.
RESULTS
Effects of a short uORF on linear scanning and shunting.
To analyze the role of the uORF/hairpin element in shunting, constructs with structured (S-series) or unstructured linear (L-series) leaders were prepared (see Materials and Methods and Fig. 1A) based on the model structured leader construct S-A (construct pMH188 in reference 27). The influence of these leader mutants on expression of the CAT reporter gene was analyzed in a commercial wheat germ cell-free translation system, in which mRNAs stabilities were measured (60) and shown to be unaffected by mutations in the leader (14). Compared with linear scanning, translation on the structured leader was rather inefficient (20 to 30% efficiency; Fig. 1C, S-A versus L-A) due to a very stable hairpin (Kozak stem [38]). This hairpin effectively blocked translation when placed near or at a distance from the 5′ end (Fig. 1C, constructs ΔS and S, respectively). Its effect was partially suppressed by a short uORF (Fig. 1C, construct S-A).
The 3-amino-acid (aa)-uORF A in constructs L-A and S-A (Fig. 1A) is derived from the CaMV 35S RNA shunt-competent leader (CaMV strain S [16]). Its effect on translation depended strictly on the structure of the leader. In the linear context, uORF A slightly diminished translation at the main ORF (maximum, 30%; Fig. 1C, compare L-A and L). Thus, either it was not efficiently recognized or reinitiation was very efficient. On the structured leader, however, uORF A stimulated translation downstream of the stem at least sevenfold (Fig. 1C, compare S-A and S). A strong stem can stall or slow down scanning ribosomes, thus prolonging the ribosome-mRNA interaction (40). Here, this may have enhanced recognition of the short uORF. Efficient recognition of the uORF may be a precondition for stem skipping to allow translation at the main ORF.
uORF A is very short, and the corresponding translation product is therefore difficult to detect. Extension of the uORF length by in-frame insertion of sequences derived from the C-terminal part of the CAT ORF (165-aa uORF of S-cat) abolished translation of the full-length CAT ORF located downstream (Fig. 1C). Due to the problem of assessing the level of uORF A translation, we set out to directly examine the initiation complex formation at uORF A.
Ribosome assembly at the initiation codon of a short uORF.
Ribosome assembly at the initiation codon of the uORF A was analyzed by sucrose density gradient centrifugation (Fig. 2) and by toeprinting (Fig. 3). Translation initiation in wheat germ extract was performed in the presence of two translation elongation inhibitors: cycloheximide, which blocks the translocation of peptidyl-tRNA from the A site to the P site (65), and sparsomycin, a highly specific ribosomal peptidyltransferase inhibitor that blocks binding of substrates to the A site while strongly stimulating interaction at the P site (3). Under these conditions, CAT expression on both linear and structured leaders was effectively abolished (data not shown).
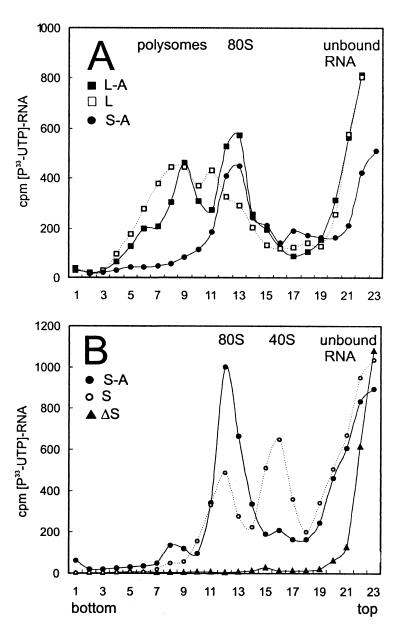
FIG. 2.
Effect of a short uORF A in the linear and structured leader context on the formation of translation initiation complexes. Labeled transcripts were incubated briefly in wheat germ extract in the presence of translation elongation inhibitors (cycloheximide [100 μg/ml] and sparsomycin [200 μM]). Initiation complexes formed with full-length mRNAs were assayed by density centrifugation in 10 to 30% sucrose gradients. Sedimentation was from right to left. Positions of unbound RNA, 40S subunits, and 80S and high-molecular-weight polysomal complexes are indicated. (A) Migration of the L-A, L, and S-A transcripts; (B) migration of the S-A, S, and ΔS transcripts.
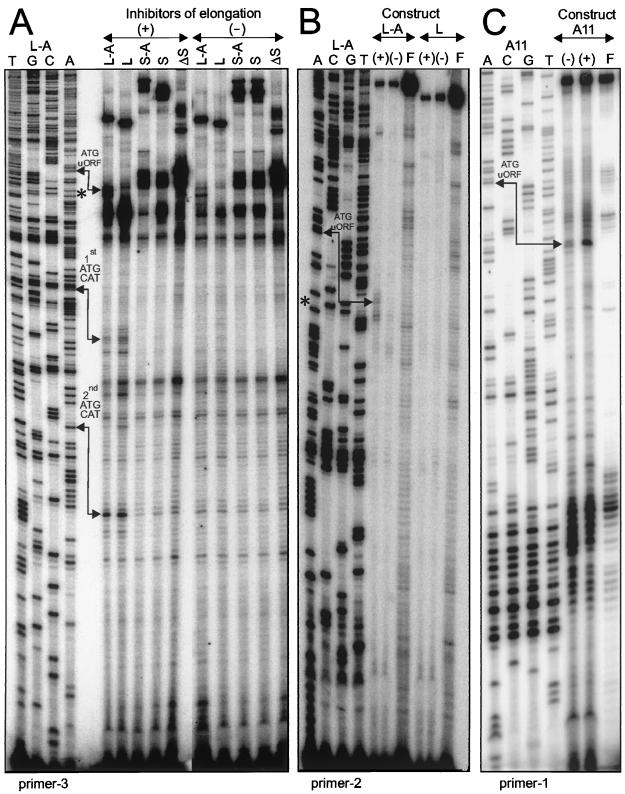
FIG. 3.
Toeprinting of translation initiation complexes. Translation initiation reactions were performed in the presence (+) or absence (−) of translation elongation inhibitors on transcripts with linear (L-series) or structured (S-series and A11) leaders, as depicted in Fig. 1A and 4 (A11). Reverse transcription reactions performed with antisense primer 3, 2, or 1 resulted in extended cDNA products analyzed by PAGE (6% gel). Formation of a functional 80S ribosome complex at the initiation ATG codon resulted in premature reverse transcription termination 17 to 18 nt downstream from the A base. Toeprints corresponding to translation initiation complexes located at the first and the second, out-of-frame ATG codons of CAT were detected with primer 3 on both linear transcripts (A). Note that primer 2 did not detect any toeprint band at the ATG of CAT on L and L-A mRNAs, as its target site partially overlaps the region protected by the initiating ribosome. An initiation complex at uORF A was detected with both primer 3 and primer 2 on transcript L-A (A and B) and with primer 1 on transcript A11 (C). Lanes F in panels B and C contain products of the control translation initiation reactions performed in the absence of the wheat germ extracts; lanes A, C, G, and T contain products of the sequencing reactions performed on the corresponding plasmids, as indicated. Positions of the hairpin are shown within the L-A sequence (A and B) by asterisks corresponding to cytosine 122 (Fig. 1A).
Comparative analysis of translation initiation on two linear leaders showed that both L-A and L transcripts accumulated in the 80S and polysomal fractions (Fig. 2A). Toeprinting analysis of translation initiation complexes confirmed extensive ribosome binding to both linear leaders (Fig. 3A and B). Two common toeprints detected within the CAT ORF correspond to the first (Fig. 3A, 1st ATG CAT; Fig. 1A, nt 177 to 179) and to the second, out-of-frame ATG codons (Fig. 3A, 2nd ATG CAT; Fig. 1A, nt 220 to 222). An additional signal detected on the L-A transcript in the presence of inhibitors mapped to the ATG codon of uORF A (Fig. 3A, ATG uORF; Fig. 1A, nt 60 to 62). This particular toeprint was detected by the reverse transcription reaction primed with two different oligonucleotides: primer 3 was successfully used to analyze complexes formed at the CAT ORF but gave rise to multiple signals of dubious significance among longer products on both linear and structured leaders (Fig. 3A). Hence, 5′-proximal primer 2 was used on the L-A leader to map precisely the ribosome complex positioned at the initiation codon of uORF A (Fig. 3B, ATG uORF). The corresponding signal was missing in the absence of uORF A (Fig. 3B, L). In this case, the most 5′-proximal ATG codon on the L mRNA, i.e., the initiation codon of the CAT ORF, was more efficiently used, and the toeprint signal detected with primer 3 was stronger on L than on L-A (Fig. 3A).
Our results show directly that 80S ribosome assembly occurs at the initiation codon of the 3-aa uORF A, indicating that uORF A can be properly recognized and translated despite its shortness and suboptimal initiation context. In the presence of the translation elongation inhibitors, both initiation codons present on the L-A transcript can be used independently for initiation complex assembly. This is consistent with a leaky scanning mechanism allowing initiation at the upstream or the downstream ATG codon, in contrast to the reinitiation mechanism, which assumes that arrest at the uATG excludes ribosome binding downstream. Consequently, we consider leaky scanning as the operating mechanism during translation initiation on the linear leader containing a very short uORF (see also the effect of CaMV pVI on translation, below).
Translational arrest at the uORF blocks shunting.
Analysis of initiation complexes by density gradient centrifugation showed that the S-A shunt-competent transcript accumulates exclusively in the 80S ribosomal fraction (Fig. 2, S-A), suggesting an equimolar binding to ribosomes. Deletion of the uORF on the S leader results in strongly diminished binding to 80S complexes and comigration with 40S ribosomal subunits (Fig. 2B). Thus, the Kozak stem (−50 kcal/mol) provokes 40S subunit stalling, correlating with inefficient translation on this transcript (Fig. 1C). A small amount of S transcript comigrating with the 80S fraction probably results from the residual alternative initiation at the non-AUG start codon (Fig. 1A, ATT codon created by 7-nt deletion [20]) that correlates with a very low CAT ORF translation (Fig. 1C). This supposition was further confirmed by a single-point mutation creating uORF-deficient leader A8 (Fig. 4) with undetectable CAT ORF expression (see below). Near complete deletion of the sequences preceding the stem on the ΔS leader totally precludes binding to ribosomes (Fig. 2B) and correlates with abolished translation (Fig. 1C).
Toeprinting analysis failed to detect any specific signal downstream of the Kozak stem on the structured leaders (Fig. 3A, gel area below the asterisks). Thus, the 80S complex identified by density centrifugation on the S-A leader likely forms at the uORF ATG codon. Toeprinting analysis of the initiation complexes upstream of the strongly base-paired Kozak stem resulted in multiple signals which are difficult to interpret (Fig. 3A; e.g., intense nonspecific signals above the asterisks on all structured leaders). Only when the GC-rich primer 1 complementary to the stem-proximal sequence (Fig. 1A) was used in the reaction on a slightly modified construct could a specific signal, located at the correct distance from the ATG codon of the uORF, be detected (Fig. 3C). The modified construct used in the latter experiment contains an N-terminally enlarged uORF; thus, the distance from the ATG codon to the stem is extended (Fig. 4, A11). This modification influenced neither ribosome shunt efficiency (see below) nor ribosome binding (data not shown) but improved the quality of toeprinting and allowed detection of the translation initiation complexes formed at the uORF.
Altogether, these results demonstrate that on the structured leader, the primary translation initiation event occurs exclusively at the uATG, i.e., upstream of the stem. Both deletion of the uORF and translational arrest at the uATG in the presence of translation elongation inhibitors abolished initiation at the main ORF, downstream of the stem. These results underline the link between translation of a short uORF and stem skipping to allow translation downstream and indicate reinitiation as a possible mechanism of translation initiation on the synthetic shunt-competent mRNA leader. Consequently, we propose and discuss below that the very same ribosome which translates the short uORF later resumes translation downstream of the stem.
Effects of uORF mutations on shunting in vitro.
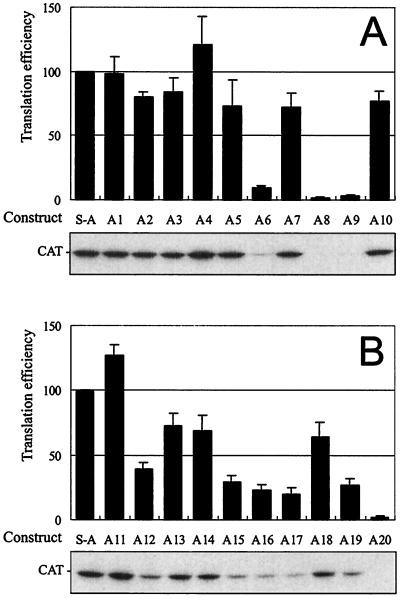
To characterize requirements for stem skipping, uORF mutants A1 to A20 (Fig. 4) were tested in the wheat germ cell-free translation system (Fig. 5). In the plant extract, ribosome shunt did not depend on the sequences preceding the uORF (A2 and A7) or on the type of the termination codon (A1). Replacement of the viral uORF A by a different 3-aa coding sequence (A2 to A4) or still longer, arbitrarily chosen ORFs of 5 (A10), 7 (A5), 10 (A11), and 14 (A13 and 14) aa resulted in efficient shunting. However, further enlargement of the uORF gradually reduced translation efficiency (A15 to A17). In general, improving the initiation context of the uORF had no effect on translation at the main ORF in case of constructs containing 3-aa (A2), 14-aa (A14), or still longer (A15 to A17 [not shown]) uORFs, except for the A12 construct. Decreased CAT expression on the latter can be attributed to occlusion of its uATG codon due to a peculiar structural effect not studied further.
FIG. 5.
In vitro expression of the CAT reporter gene under control of uORF mutations in the synthetic shunt-competent leader. Constructs are as depicted in Fig. 4. Average expression levels are represented by bars. Typical resolution of in vitro-translated products by SDS-PAGE (12% gel) is shown in the lower panel.
Five of the uORF mutants manifested low shunting efficiency. Removal of the start codon by a G-to-C point mutation resulted in 40-times-lower CAT expression (A8). Due to a single-nucleotide deletion, another uORF frameshift mutant acquired a new 5-aa uORF that terminated just at the stem structure and abolished translation downstream (A9). The effect of the A9 mutation was position dependent and reversed by relocation of the uORF to recreate the proper 8-nt distance to the stem (A10). Taking into account that uORF-dependent shunting on the synthetic mRNA is unaffected by introducing ATG codons into the stem (27), the latter result suggests that the structure in the synthetic shunt-competent leader is very tight and resists melting even by translating ribosomes. This, in turn, indicates the importance of proper stop codon recognition and termination of the uORF translation for shunting. This point was further analyzed with the help of two mutants in which uORF A is replaced by inhibitory uORFs affecting translation termination in heterologous systems.
The 6-aa uORF from mammalian AdoMetDc, which inhibits reinitiation in cis by blocking proper termination and peptide release at its own stop codon (22), was found in our system to repress CAT ORF expression in A6. Compared with uORF A, another heterologous uORF, the inhibitory uORF4 from yeast GCN4 (23), reduced CAT ORF expression in A19. In contrast, the stimulatory uORF1 from GCN4 was found to reverse the latter effect in A18, similarly as shown in the yeast system (23). Both yeast uORFs are flanked by the original sequences required for translational regulation of GCN4 (23, 48), thus doubling the distance from the termination codon of the uORF to the stem (16 versus the original 8 nt). This change should not influence shunting, as in the viral shunt-competent leader even 30-nt spacing between cis-acting elements is tolerated (14). Finally, uORF A was replaced by the sequence complementary to the 3′-proximal sequences from the 18S rRNA suspected to stimulate uORF-independent shunting in the adenovirus tripartite leader (A20) (68). This replacement, however, did not support translation in our system, probably due to the suboptimal choice or location of the involved sequences required for uORF-independent shunting in adenovirus (69).
In summary, translation on the synthetic shunt-competent leader depends strongly on the length and position of the uORF. Further, it is sensitive to the coding content of the uORF and sequences following it. In our system only uORFs of 14 aa or less and terminating 8 nt in front of the stem structure were able to support stem skipping and efficient translation at the main ORF. Substitution for another uORF is possible, provided that it allows proper termination and release of the uORF-encoded peptide. Again this indicates reinitiation as the mechanism of uORF-dependent shunting.
Effects of uORF mutations on shunting in vivo in the presence of CaMV pVI, the viral transactivator of reinitiation.
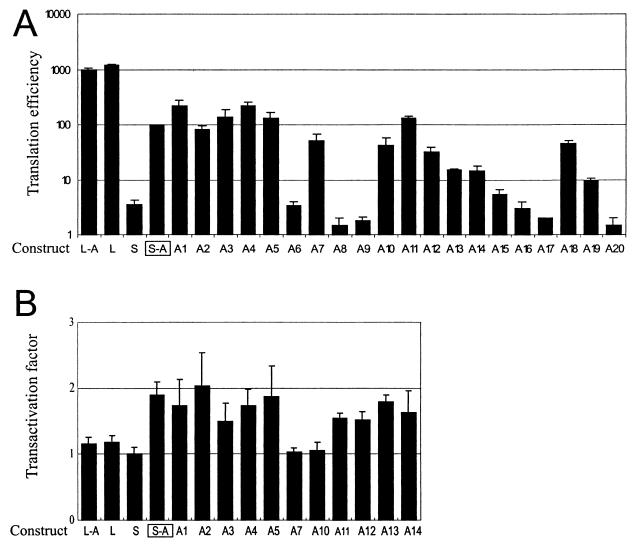
Expression of the CAT reporter gene under control of linear and structured leaders was tested in vivo in transfected plant protoplasts. In this system, mRNA stabilities are unaffected by mutations introduced into the leader sequences (26, 27). Generally, translation levels in vivo paralleled translation data in vitro, indicating that our results do reflect differences in translation efficiencies (Fig. 6A). In vivo as in vitro, the effect of the uORF on translation was strong in the structured but negligible in the linear leader context (Fig. 6A, compare S-A and S with L-A and L). All uORF mutants blocking translation on the structured leader in vitro resulted in 10% or lower CAT expression in vivo. As before, they fall into the three categories of deleted (A8 and A20), improperly terminated (A6, A9, and A19), and overextended (A15 to A17) uORFs. Expression of several constructs manifesting more than 10% efficiency of S-A expression (Fig. 6A) was analyzed in vivo in the presence of the CaMV pVI or transactivator (5). It is well documented that the virus-encoded RNA-binding protein pVI stimulates reinitiation on polycistronic mRNAs (5, 17), as a result of its interaction with subunit 5 of eIF3 and/or ribosomal protein L24 (2). When tested in the presence of the CaMV pVI, CAT expression under the control of the linear leader was unaffected (Fig. 6B, L-A and L). This result is consistent with the leaky scanning mechanism during translation initiation on the linear leader proposed above. In the structured leader context, however, the uORF effect on CAT expression was enhanced 1.5- to 2.5-fold in the presence of the pVI protein (Fig. 6B, S-A and following constructs). These results speak in favor of a reinitiation-type mechanism during translation initiation on the structured leader.
FIG. 6.
In vivo expression of the CAT reporter gene under control of mutations in the leader. (A) Effect of uORF mutations in the context of linear and structured shunt-competent mRNA leaders. Note that translation efficiencies represented by bars are plotted in a logarithmic scale. (B) Effect of the CaMV-specific translational transactivator of reinitiation on translation. See Fig. 4 for constructs.
Interestingly, the positive effect of CaMV pVI was more pronounced on constructs with a functional uORF being placed within original viral 5′-proximal sequences (Fig. 6B, A1 to A5). Significant shortening of these sequences did not affect shunting (Fig. 6A, A7 to A10) but abolished transactivation by pVI (Fig. 6B, A7 and A10). Extension from 21 to 24 nt partially reversed the latter effect (Fig. 6B, A11 to A14), but the results were not as consistent as in the presence of the 59-nt original CaMV sequence. These results indicate that transactivation of reinitiation on the synthetic shunt-competent leader might act in a sequence-dependent manner, being more sensitive to the virus-derived sequences.
Ribosome shunt in a heterologous cell-free translation system.
In S. cerevisiae, requirements for translation termination are well characterized and appear to be strict (6, 47, 64). Also, cap-dependent initiation of translation in yeast is particularly sensitive to hairpin elements in the 5′ leader (47). Thus, to study further the effect of uORF termination on shunting and to explore the potential of a uORF/hairpin shunting element across species, we chose to use a yeast cell-free translation system (55). This system displays characteristic responses to the presence of a 5′ cap structure and a 3′ poly(A) tail on the mRNA: initiation in micrococcal nuclease-treated extracts is strongly stimulated by the presence of a single end modification, while untreated extracts require the synergistic cooperation of both cap structure and poly(A) tail for efficient translation (55). Conveniently, reporter mRNA stability in this system does not depend significantly on the presence of a cap structure or poly(A) tail (55) and is not sensitive to the insertion of uORF sequences in the leader (56).
We prepared a series of seven uORF mutants incorporated into the synthetic mRNA leader dedicated for use in yeast (Fig. 7A, Y series). After determining favorable magnesium, potassium, and mRNA concentrations for the capped reference mRNA Y1 (Fig. 7B to D), we used these optimized conditions for a comparison of capped mRNAs Y1 to Y7 in a wild-type yeast extract treated with micrococcal nuclease (YAS 2069 [63]). CAT expression under the control of the basic S-A leader (Fig. 1A) was found to be less efficient (Fig. 7E, lane 9) than Y1 with its shortened 5′-proximal sequence and the optimal yeast-type uORF termination signal. Insertion of a pyrimidine-rich sequence had no effect (Y2), but substitutions leading to poor termination context (Y3) or to non-uAUG codons (Y4 and Y7) resulted in translational repression two- to threefold relative to Y1. Collectively, these results indicate that CAT expression under the control of an optimized Y1 leader occurs by a uORF-dependent ribosome shunt mechanism. Replacement of the CaMV-derived uORF A by a specific short ORF blocking peptidyltransferase activity of yeast ribosomes in cis (45) affected shunting in our system: a uORF encoding the peptide MVKTD provoked twofold inhibition on Y5 compared with Y1, but as expected, a single codon deletion in Y6 resulting in MVKT stimulated translation by a factor of 2 compared with Y1 (and by a factor of 5 compared with Y7). This demonstrates that, similar to the plant system, shunting on a uORF/hairpin element in yeast depends on proper termination at the uORF and substantiates the notion that translation at the main ORF occurs by a reinitiation-type mechanism.
Ribosome shunt on the model mRNA leader was further tested using capped and polyadenylated transcripts in untreated extracts derived from yeast strain YAS 2074 or YAS 2075 (63), bearing mutations in the eIF4G1 gene that affect binding to eIF4E or Pabp1p, respectively (Fig. 7F). This allowed assessment of the influence of poly(A) tail function on shunting. In conditions of abolished cap-dependent and stimulated poly(A)-dependent translation (YAS 2074 [Fig. 7F, lanes 4 and 5), the regulatory effect of the uORF on shunting was occluded by strong 5′-independent initiation that can bypass steric blocks in the leader (55). This equal translatability of Y1 and Y4 mRNA in YAS 2074 extract confirms the similar stabilities of these mRNAs in the translation extracts (56). In contrast, in conditions of abolished poly(A)-dependent and intact cap-dependent translation (YAS 2075), the uORF stimulated downstream CAT translation about sixfold (Fig. 7F, lanes 6 and 7). These results are consistent with a cap-dependent shunting phenomenon in yeast, and they demonstrate that it does not require participation of a poly(A)-tail stimulated process.
DISCUSSION
Accumulated data suggest a regulatory function for uORFs during translation initiation in eukaryotes (reviewed in references 21, 22, and 45). There are many examples of uORFs in the leaders of viral and cellular genes involved in control of growth and differentiation (41). Diverse uORFs can be categorized in different ways (22), e.g., overlapping and autonomous uORFs, depending on position of the initiator and termination codons with respect to the downstream ORF; sequence-dependent and -independent uORFs, reflecting the character of the encoded peptide; and stimulatory, neutral, and repressive uORFs, taking into account their effect on translation at the main ORF. Translation can be also regulated by metabolic conditions that counteract inhibitory uORFs by, e.g., modifying the structure or usage of the leader (68) or allowing reinitiation after termination at the end of the uORF (32). In contrast to repressive uORFs which occur frequently, only a few cases of positive regulators are known, including uORF1 of the GCN4 mRNA (31), uORFs from Rous sarcoma virus (15), and uORF A from CaMV (19, 27, 54). These stimulatory uORFs can alleviate the effects of cis-acting inhibitory elements, other uORFs, or structural motifs. The positive effect of ORF A on translation in the synthetic shunt-competent mRNA leader has been also well documented (27). In the current work, we explored the translational potential and regulatory function of the 3-aa uORF A in both linear and structured leader contexts. Further, detailed mutational analysis of the short uORF was carried out to study the mechanism of shunting in vivo and in vitro.
Our data indicate that despite being short and in the unfavorable initiation context, uORF A is recognized by the translational machinery and thus potentially translated. Its regulatory effect on translation depends on the leader context: it is negligible in the unstructured but stimulatory in the structured leader. We have shown that translational arrest at the uATG codon abolished translation at the main ORF in the context of the synthetic shunt-competent mRNA leader. This result suggests that translation of uORF A is absolutely required for shunting and excludes the leaky scanning mechanism. Replacement of the 3-aa uORF A by other short coding sequences seems possible, except those uORFs whose products repress reinitiation by blocking peptidyltransferase and their proper release from the ribosome. Two such heterologous uORFs were used in our replacement experiments: both the 6-aa mammalian uORF from AdoMetDc (22) and the 5-aa bacterial cat leader peptide (45) affected uORF-dependent shunting on the synthetic mRNA leader, when analyzed in plant and yeast systems, respectively. In a parallel study performed in our laboratory, it was shown also that the former uORF prevents shunting by provoking ribosome stalling when replacing uORF A in the CaMV 35S RNA leader context (58). The character of the nascent peptide affects the translating ribosome and precludes shunting, thus proving that in the context of the shunt-competent leaders, a short uORF is indeed efficiently translated. Theoretically, translation of the uORF might have been required for the direct transfer of initiating or elongating ribosomes to the main ORF, as happens during hopping in prokaryotes (28). However, we can rule out this possibility, as the extended CAT reporter gene products were never observed in our experiments.
The results of our replacement experiments point out that a full cycle of a short uORF translation including initiation, elongation, and termination is a precondition for stem skipping to allow translation at the main ORF. In particular, proper termination at the uORF stop codon appears to be absolutely essential for shunting, as we demonstrated in different ways using diverse expression systems. We have shown that the proper location and availability of the uORF termination codon, as well as the proper release of the uORF-encoded peptide, stimulate shunting in the plant system. Moreover, efficient termination of translation, due to an optimized uORF stop codon context, enhanced shunting in yeast. Interestingly, the well-known effect of the extended termination sequence on translation termination has been interpreted to mean that the fourth-base context influences the recognition of the triplet stop codons by modulating direct interaction with a release factor (64). The recently solved crystal structure of the human eRF1 protein displays the overall structural mimicry with the tRNA molecule that allowed a conserved groove present on domain 1 to be identified as the stop codon interaction site—the anticodon site of eRF1 (61). The codon recognition site on eRF1 is quite long compared with the anticodon on the tRNA, consistent with an extended stop codon recognition site capable of interacting with four or more bases (61; David Barford, personal communication). Thus, conclusions drawn from recent structural studies are in line with earlier data indicating the importance of the sequences flanking the stop codon in translation termination and indirectly corroborate their significance for termination at the uORF and shunting.
Since efficient termination at the short uORF precedes shunting, we favor the view that translation initiation at the main ORF occurs by a reinitiation-type mechanism. Our results support reinitiation in several ways. First, as in other systems (46), reinitiation via shunting requires a short uORF A. Second, replacement of uORF A is in principle possible, indicating that shunting does not depend directly on the sequence of the nascent peptide encoded by the uORF. Third, both uORFs from GCN4 incorporated in the synthetic mRNA leader affect shunting similarly to the way they are known to regulate reinitiation in yeast (23). Fourth, the CaMV-derived translational transactivator known to enhance reinitiation on polycistronic transcripts (5) was found to promote translation downstream of the Kozak stem in our system. Altogether, our data point to a single ribosome being involved in all three steps of translation initiation on the synthetic shunt-competent leader, i.e., translation of the uORF, shunting, and reinitiation at the main ORF.
An alternative is a two-ribosome model with the first ribosome translating the uORF and preparing the internal binding site for the second ribosome to initiate downstream. Note, however, that shunting has been studied extensively in the viral and synthetic leader context (26, 27). The two leaders differ substantially from each other with respect to primary and overall secondary structure. Therefore, it seems improbable that translation of the uORF would lead in both cases to the creation of a similar, however special, internal ribosome entry site. In addition, shifting the uORF termination codon sequences backward (14) and forward (58) with respect to the stem is tolerated to a certain extent, which excludes the possibility that only a special location of the termination codon, as part of the internal ribosome entry site, would activate shunting. Rather, stop codon availability for recognition and proper termination is critical because its proximity to the very strong stem which is not melted by scanning preinitiation complexes (27) precludes shunting on the synthetic leader (A9). We should further mention that in all systems and conditions tested, uORF-dependent shunting is cap dependent and not active when tested in the dicistronic constructs with the shunt-competent leader introduced between two reporter genes (Fig. 1 [S-cat] and reference 14). This allows us to exclude the typical or an induced internal initiation model in shunting and points rather to reinitiation as the operating mechanism of translation initiation on the synthetic shunt-competent mRNA leader.
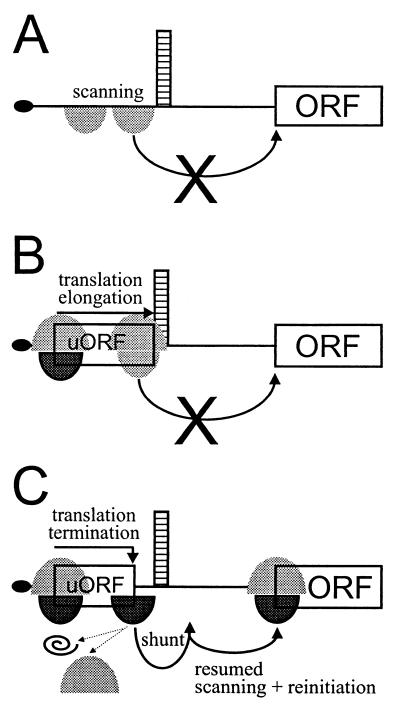
We demonstrated that neither scanning 40S subunits nor elongating ribosomes are capable of overcoming the repressive effect imposed by the presence of the strong stem in the leader (Fig. 8A and B, respectively). However, immediately after translation termination at the uORF, ribosomes might be in a state that allows somewhat more relaxed movement on the mRNA. The recently published crystal structure of prokaryotic 70S ribosome functional complexes (11) allows an interpretation of our data at the molecular level. Details of the interactions between the ribosome, mRNA, and P- and A-site-bound tRNAs can be seen in the X-ray structures. At the P site, the ribosome grips the tRNA and mRNA with six fingers of electron density to maintain the translational reading frame. Although ribosomal components of these fingers have not yet been identified, they seem to hold the mRNA being translated very tightly. In contrast to the P site, the A site makes only weak contacts with the tRNA-mRNA complex. In the absence of bound tRNA, the A-site codon is disordered in electron density, suggesting minimal if any A-site contacts between mRNA and the ribosome in the absence of an A-site tRNA. Taking into account structural constraints imposed during translation elongation, proper termination at the stop codon of the uORF enhances flexibility of the ribosomes required for shunting across the stem to allow translation at the main ORF (Fig. 8C).
FIG. 8.
Model for ribosome shunt on the artificial shunt-competent mRNA leader. The structured leader is shown schematically with the black oval representing the cap, scanning 40S subunits and elongating 80S ribosomes represented by gray shadows, initiation- and reinitiation-competent 40S subunits shown in black, and migration of subunits and 80S ribosomal complexes indicated by arrows. Scanning 40S subunits (A) and elongating 80S ribosomes (B) stall in front of a stable stem structure, unfit for translation downstream. (C) Translation of a short uORF culminating in proper termination promotes reinitiation. Upon release of the uORF-encoded peptide and dissociation of the 60S (indicated by dotted-line arrows on the left), the released reinitiation-competent 40S subunit skips over the stem sequences and resumes scanning downstream.
Following termination at the uORF, shunting ribosomes must regain initiation capacity, but how they reach the main ORF remains to be determined. As described here, nonlinear ribosome migration or shunting allows skipping of the stem and resumed progressive scanning in the conventional 5′-to-3′ direction. Another special case of translation reinitiation is known in the case of overlapping ORFs, when the termination codon of an uORF is situated downstream of but close to the initiation codon of the downstream ORF; here ribosomes can scan backwards up to 50 nt to engage a suitable initiation site (51). Thus, in contrast to forward scanning, shunting as well as backward scanning can be seen as examples of eukaryotic ribosome migration occurring by lateral diffusion rather than unidirectional migration. For reinitiation, some initiation factors, including eIF1 and eIF1A, which help to locate initiation codons (52), and eIF2, which delivers Met-tRNAi (32), should be reacquired, but others, e.g., the helicase complex including eIF4A (35), may be lacking. In the case of GCN4, reinitiation is distance dependent and inversely correlated to the concentration of active eIF2 (32, 50). In the case of the adenovirus major late mRNA tripartite leader, shunting is uORF independent and correlates with reduced activity of the RNA cap-binding complex, including eIF4A (68).
Clearly, ribosome shunt on the structured mRNA leader constitutes a very particular type of translation initiation in eukaryotes. Until now, only a few shunt-competent leaders equipped with uORFs have been characterized (12, 19, 20, 44, 57). However, the frequency of uORF occurrence in eukaryotic transcripts demonstrates that complex polycistronic mRNA structures are reasonably common (10, 41). Consequently, translation initiation via ribosome shunt might occur much more frequently than previously anticipated. Our results demonstrate that uORF-dependent shunting occurs in both plant and yeast cell-free translation systems in a cap-dependent fashion which does not require poly(A)-stimulated processes. Within defined limits it is tolerant of uORF and stem sequences and operates via a reinitiation-type mechanism to allow translation at the main ORF. As a consequence, the synthetic shunt-competent mRNA leader may be invaluable as a universal substrate in further studies of the uORF-dependent shunting, as well as in studying other steps of eukaryotic translation, including translation termination and reinitiation.
ACKNOWLEDGMENTS
We thank Matthias Müller and Dave Kirk for technical assistance, Alan Sachs for yeast strains, Matthias Hentze for helpful discussions, and Pat King and Witek Filipowicz for comments on the manuscript.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (PR616/1-1 and PR616/1-2) to T.P.
REFERENCES
- 1.Alderete J P, Jarrahian S, Geballe A P. Translation effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J Virol. 1999;73:8330–8337. doi: 10.1128/jvi.73.10.8330-8337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey-Serres J, Rochaix J-D, Wassenegger M, Filipowicz W. Plants, their organelles, viruses and transgenes reveal the mechanisms and relevance of posttranscriptional processes. EMBO workshop report. EMBO J. 1999;18:5153–5158. doi: 10.1093/emboj/18.19.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballesta J P G, Lazaro E. Peptidyltransferase inhibitors: structure-activity relationship analysis by chemical modification. In: Hill W E, Dalberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome. Washington, D.C.: American Society for Microbiology; 1990. pp. 502–510. [Google Scholar]
- 4.Bonetti R, Fu L, Moon J, Bedwell D M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 5.Bonneville J-M, Sanfaçon H, Fütterer J, Hohn T. Posttranscriptional transactivation in cauliflower mosaic virus. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- 6.Brown C M, Dalphin M E, Stockwell P A, Tate W P. The translation termination signal database. Nucleic Acids Res. 1993;21:3119–3123. doi: 10.1093/nar/21.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, Geballe A P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, Geballe A P. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA. 1998;4:181–188. [PMC free article] [PubMed] [Google Scholar]
- 10.Carter P S, Jarquin-Pardo M, De Benedetti A. Differential expression of Myc1 and Myc2 isoforms in cells transformed by eIF4E: evidence for internal ribosome repositioning in the human c-myc 5′UTR. Oncogene. 1999;18:4326–4335. doi: 10.1038/sj.onc.1202890. [DOI] [PubMed] [Google Scholar]
- 11.Cate J H, Yusupov M M, Yusupova G Z, Earnest T N, Noller H F. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 12.Curran J, Kolakofsky D. Scanning independent ribosomal initiation of the Sendai virus X protein. EMBO J. 1988;7:2869–2874. doi: 10.1002/j.1460-2075.1988.tb03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delbecq P, Werner M, Feller A, Filipkowski R K, Messenguy F, Piérard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez D I, Ryabova L A, Pooggin M M, Schmidt-Puchta W, Fütterer J, Hohn T. Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J Biol Chem. 1998;273:3669–3678. doi: 10.1074/jbc.273.6.3669. [DOI] [PubMed] [Google Scholar]
- 15.Douzé O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franck A, Guilley H, Jonard G, Richards K, Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980;21:285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- 17.Fütterer J, Hohn T. Translation of the polycistronic mRNA in presence of the CaMV transactivator protein. EMBO J. 1991;10:3887–3896. doi: 10.1002/j.1460-2075.1991.tb04958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fütterer J, Hohn T. Role of an upstream open reading frame in the translation of polycistronic mRNA in plant cells. Nucleic Acids Res. 1992;20:3851–3857. doi: 10.1093/nar/20.15.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fütterer J, Kiss-László Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 20.Fütterer J, Potrykus I, Bao Y, Li L, Burns T M, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geballe A P, Morris D R. Initiation codons within 5′-leaders of mRNA as regulators of translation. Trends Biochem Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 22.Geballe A P. Translational control mediated by upstream AUG codons. In: Hershey J, Matthews M, Sonnenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 173–197. [Google Scholar]
- 23.Grant C M, Hinnebusch A G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerra-Peraza O, de Tapia M, Hohn T, Hemmings-Mieszczak M. Interaction of the cauliflower mosaic virus coat protein with the pregenomic RNA leader. J Virol. 1999;74:2067–2072. doi: 10.1128/jvi.74.5.2067-2072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemmings-Mieszczak M, Steger G, Hohn T. Alternative structures of the cauliflower mosaic virus 35 S RNA leader: implications for viral expression and replication. J Mol Biol. 1997;267:1075–1088. doi: 10.1006/jmbi.1997.0929. [DOI] [PubMed] [Google Scholar]
- 26.Hemmings-Mieszczak M, Steger G, Hohn T. Regulation of CaMV 35 S RNA translation is mediated by a stable hairpin in the leader. RNA. 1998;4:101–111. [PMC free article] [PubMed] [Google Scholar]
- 27.Hemmings-Mieszczak M, Hohn T. A stable hairpin preceded by a short open reading frame promotes nonlinear ribosome migration on a synthetic mRNA leader. RNA. 1999;5:1149–1157. doi: 10.1017/s1355838299990325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbst K L, Nichols L M, Gesteland R F, Weiss R B. A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc Natl Acad Sci USA. 1994;91:12525–12529. doi: 10.1073/pnas.91.26.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill J R, Morris D R. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA. J Biol Chem. 1992;267:21886–21893. [PubMed] [Google Scholar]
- 30.Hill J R, Morris D R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. J Biol Chem. 1993;268:726–731. [PubMed] [Google Scholar]
- 31.Hinnebusch A G. Translational control of GCN4: an in vivo barometer of initiation-factory activity. Trends Biochem Sci. 1994;19:409–414. doi: 10.1016/0968-0004(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 32.Hinnebusch A G. Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 33.Jackson R J, Hunt S L, Reynolds J E, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr Top Microbiol Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 34.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 35.Jaramillo M, Dever T E, Merrick W C, Sonnenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF4B. Mol Cell Biol. 1991;11:5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eukaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. Circumstances and mechanism of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 43.Kozak M. A consideration of alternative models for the initiation of translation in eukaryotes. Crit Rev Biochem Mol Biol. 1992;27:385–402. doi: 10.3109/10409239209082567. [DOI] [PubMed] [Google Scholar]
- 44.Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luukkonen B G, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarthy J E G. Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller P F, Hinnebusch A G. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 1989;3:1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- 49.Mize G J, Ruan H, Low J J, Morris D R. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- 50.Müller P P, Grüter P, Hinnebusch A G, Trachsel H. A ribosomal protein is required for translational regulation of GCN4 mRNA. Evidence for involvement of the ribosome in eIF2 recycling. J Biol Chem. 1998;273:32870–32877. doi: 10.1074/jbc.273.49.32870. [DOI] [PubMed] [Google Scholar]
- 51.Peabody D S, Subramani S, Berg P. Effect of upstream reading frames on translational efficiency in simian virus 40 recombinants. Mol Cell Biol. 1986;6:2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pestova T V, Borukhov S I, Hellen C U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 53.Pietrzak M, Shillito M, Hohn T, Potrykus I. Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res. 1986;14:5857–5868. doi: 10.1093/nar/14.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pooggin M M, Hohn T, Fütterer J. Forced evolution reveals the importance of short open reading frame A and secondary structure in the cauliflower mosaic virus 35S RNA leader. J Virol. 1998;72:4157–4169. doi: 10.1128/jvi.72.5.4157-4169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preiss T, Hentze M. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 56.Preiss T, Muckenthalter M, Hentze M W. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Remm M, Remm A, Ustav M. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J Virol. 1999;73:3062–3070. doi: 10.1128/jvi.73.4.3062-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryabova L, Hohn T. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev. 2000;14:817–829. [PMC free article] [PubMed] [Google Scholar]
- 59.Schleiss M R, Degnin C R, Geballe A P. Translational control of human cytomegalovirus gp48 expression. J Virol. 1991;65:6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt-Puchta W, Dominguez D, Lewetag D, Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854–2860. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song H, Mugnier P, Das A K, Webb H M, Evans D R, Tuite M F, Hemmings B A, Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 62.Tarun S, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 63.Tarun S, Sachs A B. Binding of eukaryotic initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tate W P, Poole E S, Mannering S A. Hidden infidelities of the translational stop signal. Prog Nucleic Acid Res Mol Biol. 1996;52:293–333. doi: 10.1016/s0079-6603(08)60970-8. [DOI] [PubMed] [Google Scholar]
- 65.Wang Z, Sachs M S. Ribosomal stalling is responsible for arginine-specific translation attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, Fang P, Sachs M S. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol. 1998;18:7528–7536. doi: 10.1128/mcb.18.12.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z, Gaba A, Sachs M S. A highly conserved mechanism of regulated ribosomal stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J Biol Chem. 1999;274:37565–37574. doi: 10.1074/jbc.274.53.37565. [DOI] [PubMed] [Google Scholar]
- 68.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 69.Yueh A, Schneider R J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]