Significance
MicroRNAs (miRNAs) are a family of noncoding RNAs that regulate gene expression by posttranscriptional mechanisms. They are important for cell differentiation and were found to be misregulated in several immune disorders. However, the genes regulated by miRNAs during lymphocyte development remain largely unknown. Here, we show that miRNAs are conserved between early–B and early–T cells, but they target genes associated with natural killer, dendritic cell, and myeloid lineages in a cell type–specific manner. Knockdown and overexpression studies show miRNAs play an important role in the sustenance of lineage-specific gene expression programs and thereby enforce lymphoid cell-fate commitment. These studies provide insights into the role of miRNAs in the maintenance of lineage identity and regulation of cell-fate decisions.
Keywords: Ago2 RNA immunoprecipitation and sequencing, B- and T-cell development, lymphocyte commitment, microRNAs, lineage differentiation
Abstract
The commitment of hematopoietic multipotent progenitors (MPPs) toward a particular lineage involves activation of cell type–specific genes and silencing of genes that promote alternate cell fates. Although the gene expression programs of early–B and early–T lymphocyte development are mutually exclusive, we show that these cell types exhibit significantly correlated microRNA (miRNA) profiles. However, their corresponding miRNA targetomes are distinct and predominated by transcripts associated with natural killer, dendritic cell, and myeloid lineages, suggesting that miRNAs function in a cell-autonomous manner. The combinatorial expression of miRNAs miR-186-5p, miR-128-3p, and miR-330-5p in MPPs significantly attenuates their myeloid differentiation potential due to repression of myeloid-associated transcripts. Depletion of these miRNAs caused a pronounced de-repression of myeloid lineage targets in differentiating early–B and early–T cells, resulting in a mixed-lineage gene expression pattern. De novo motif analysis combined with an assay of promoter activities indicates that B as well as T lineage determinants drive the expression of these miRNAs in lymphoid lineages. Collectively, we present a paradigm that miRNAs are conserved between developing B and T lymphocytes, yet they target distinct sets of promiscuously expressed lineage-inappropriate genes to suppress the alternate cell-fate options. Thus, our studies provide a comprehensive compendium of miRNAs with functional implications for B and T lymphocyte development.
The differentiation of multipotent hematopoietic progenitors into mature cell types of the immune system has been shown to be under the control of gene regulatory networks involving transcription factors, cytokine signals, and other epigenetic mechanisms (1–5). The LMMPs (lymphoid-primed multipotent progenitors), which are the earliest predecessors of B and T lymphocytes (6, 7), predominantly differentiate toward T lineage upon entering thymus under the influence of Notch1 signaling in addition to TFs like TCF1, GATA-3, and BCL11B (8–11). Correspondingly, expression of active Notch1 in the bone marrow induces thymus-independent differentiation of early lymphoid progenitors into T cells at the expense of B lymphopoiesis (12). Studies from our laboratory and others have demonstrated that transcription factor EBF1 functions as the primary B lineage determinant and acts in conjunction with its downstream target, PAX5, to establish the B lineage identity of LMPPs (13–15). Targeted disruption of EBF1 results in a complete arrest of B cell development at CLP (pre–proB) stage, and the mutant Ebf1−/− progenitors exhibit developmental plasticity toward alternative lineages, including myeloid, dendritic cell (DC), natural killer (NK), and T cells, despite being maintained under B-lymphoid conditions (14, 16). Thus, development of B and T lymphocytes involves distinct transcriptional programs orchestrated and tightly regulated by key lineage determinants.
Multilineage transcriptional priming in progenitors has been shown to allow promiscuous expression of genes associated with divergent cell fates, thus allowing them to be “primed” for differentiation toward various hematopoietic lineages (17, 18). Besides driving the lineage-specific program, the primary cell-fate determinants also have an obligate role to repress the lineage-inappropriate genes in order to enable cell-fate commitment. Therefore, EBF1 and Notch1 may not only exert a direct regulatory control at transcriptional level but also employ an additional layer of control at the posttranscriptional level to effectively regulate gene expression during differentiation. To achieve this, we hypothesized that these primary determinants potentially induce microRNAs (miRNAs) to repress lineage-inappropriate genes and to fine-tune the random fluctuations in transcript abundance (19), thereby providing robustness to the gene expression programs that govern B and T lymphocyte differentiation.
miRNAs are small (22 nt) noncoding RNAs that bind to the target mRNAs (messenger RNAs) within the Ago2–RISC complexes. This interaction results in destabilization and subsequent degradation of the target mRNA molecules, thereby decreasing their translational output (20–22). Conditional deletion of Dicer or DGCR8 during early stages of B or T cell differentiation caused a developmental block at the pro–B stage or decreased survival of αβ-T cells, CD8+/CD4+ SP cells, and T-helper cells, respectively (23–26). Although several studies have earlier reported miRNA expression signatures of various hematopoietic lineages (27–31); the miRNAs which are expressed during lymphocyte differentiation and their ability to shape the cell-fate choice of multipotent progenitors has not yet been addressed comprehensively.
In this study, we have integrated high-throughput approaches (miRNA profiling and Ago2-RIPSeq [Ago2-RNA immunoprecipitation sequencing]) with functional studies (gain- and loss-of-function experiments) to show that miRNAs expressed in early–B and early–T cells are significantly correlated. Yet, these miRNAs target distinct sets of genes that are inappropriately expressed in these cell types but are associated with NK, DC, and myeloid lineages. Moreover, we identified a set of miRNAs: miR-186-5p, miR-128-3p, and miR-330-5p, which target myeloid lineage genes and thus attenuate the myeloid developmental potential of Ebf1−/− progenitors when expressed combinatorially. De novo motif analysis of commonly expressed lymphocyte miRNA promoters, as characterized by H3K4me3 ChIP-Seq (chromatin immunoprecipitation sequencing), revealed the presence of binding sites for B as well as T lineage determinants. Accordingly, the reporter assays indicate that promoters of commonly expressed miRNAs are active in B and T lineage cells. Collectively, our results provide a comprehensive understanding and functional insights on how lymphocyte miRNAs exert a combinatorial and synergistic control to attenuate the alternate cell-fate choices of developing B and T cells via suppression of lineage-inappropriate genes.
Results
miRNA Expression Pattern of Early–B Cells Extensively Overlaps with Early–T Cells.
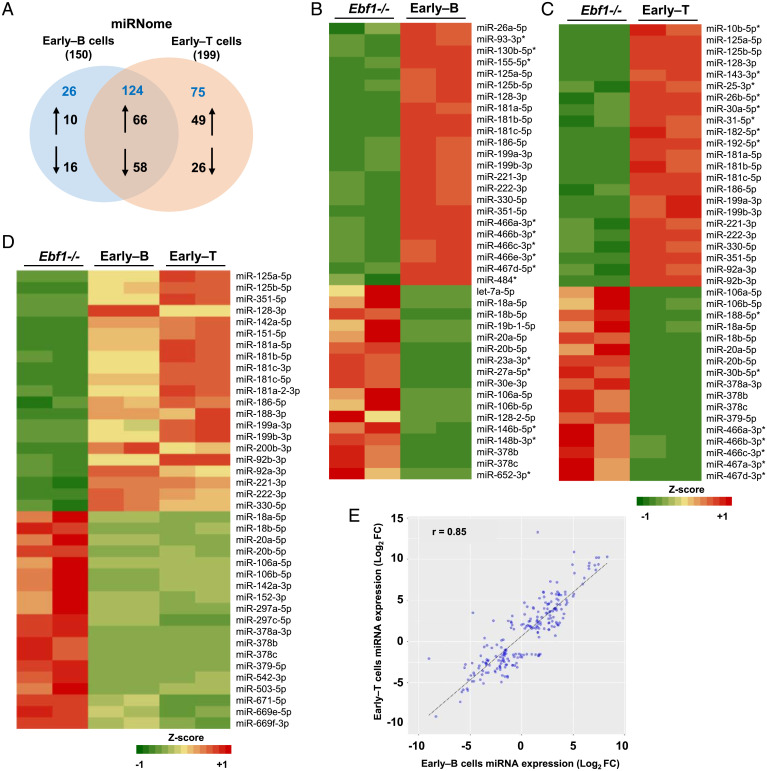
To probe the role of miRNAs during early stages of B and T lymphocyte development, we carried out genome-wide miRNA expression analysis upon differentiation of multipotent Ebf1−/− progenitors into “early–B” and “early–T” cells (SI Appendix, Methods and Fig. S1A). The Ebf1−/− progenitors were infected with a retrovirus encoding EBF1 fused to the ligand binding domain of Estrogen Receptor (MigR1-EBF1.ER) and treated with 4-hydroxytamoxifen (4-OHT) for 2 d on OP9 stroma to generate “early–B” cells (SI Appendix, Methods and Fig. S1 A and B). In parallel, the Ebf1−/− progenitors were cultured under T-lymphoid conditions (OP9-DL1 stromal cells) for 5 d to generate CD44+CD25+ (DN2-stage) “early–T” cells (SI Appendix, Methods and Fig. S1 A, C, and D) (14, 32). To profile the miRNAs expressed in Ebf1−/− progenitors, early–B cells and early–T cells, the small RNA libraries prepared from total RNA samples were deep-sequenced (SI Appendix, Methods and Fig. S1E) and miRNAs which exhibited a Log2-Fold difference, FC≥+1.5 or FC≤-1.5 (P ≤ 0.01) in each lineage, compared to Ebf1−/− progenitors were considered to be differentially “up-regulated” or “down-regulated,” respectively.
We identified a total of 150 differentially expressed miRNAs in early–B cells, of which 76 were found to be up-regulated while 74 were down-regulated (Fig. 1 A and B and Dataset S1). In early–T cells, a total of 199 differentially expressed miRNAs were identified, of which 115 were up-regulated and 84 were down-regulated (Fig. 1 A and C and Dataset S1). A comparison of early–B and early–T cells miRNA profiles revealed that about 83% of miRNAs from early–B cells exhibited an expression trend similar to early–T cells (Fig. 1D, SI Appendix, Fig. S1F, and Dataset S1), indicating a significant correlation between their miRNA expression profiles (Pearson’s correlation coefficient, r = 0.85) (Fig. 1E). So, we categorized the differentially expressed early–B and early–T cells miRNAs into two groups: “common miRNAs” (Group-I) and “unique miRNAs” (Group-II). Group-I consists of 66 and 58 miRNAs that are “commonly up-regulated” or “commonly down-regulated,” respectively, in both cell types (Fig. 1 A and D and Dataset S1). Several members from families like miR-125, miR-199, miR-181, and miR-221/222 were commonly up-regulated; while members from miR-17/20/106, miR-378, and miR-297 families were commonly down-regulated (Fig. 1D and SI Appendix, Fig. S1G). Notably, of the “commonly up-regulated” miRNAs, ∼77% (51 out of 66) were found to exhibit relatively higher fold expression in early–T cells than early–B cells (SI Appendix, Fig. S1H). Within Group-II, 10 miRNAs were “uniquely up-regulated” in early–B cells compared to 49 in early–T cells; whereas, about 16 and 26 miRNAs were “uniquely down-regulated,” respectively (Fig. 1A and Dataset S1). Intriguingly, about 13 miRNAs from Group-II displayed an “inverse” expression pattern between early–B and early–T cells; however, the fold expression for the majority of these inversely expressed miRNAs was quite modest (within ± twofold) (SI Appendix, Fig. S1I and Dataset S1). The expression pattern of few miRNAs identified by miRNA-seq (microRNA sequencing) was validated using Stemloop qRT-PCR (SI Appendix, Fig. S1J).
Fig. 1.
miRNAs exhibit differential expression as multipotent Ebf1−/− progenitors differentiate into early–B or early–T cells. (A) Venn diagram depicting the distribution of miRNAs in early–B and early–T cells. The miRNAs that were up-regulated (≥+1.5-fold [Log2]) or down-regulated (≤−1.5-fold [Log2]) uniquely or commonly in early–B and early–T cells (compared to Ebf1−/− progenitors) are indicated by up/down arrows, respectively. The total number of miRNAs that are “common” or “unique” to each cell type are shown in blue while the total number of differentially expressed miRNAs in each cell type is indicated in parenthesis. (B and C) Heatmaps representative of miRNAs that are differentially expressed (by at least ±1.5-fold [Log2]) in early–B or early–T cells, respectively, compared to Ebf1−/− progenitors. Uniquely up- or down-regulated miRNAs in each cell type are indicated with (*). (D) Heatmap representing miRNAs that are “commonly” up- or down-regulated (by at least ±1.5-fold [Log2]) in early–B as well as early–T cells, compared to Ebf1−/− progenitors. (E) Correlation plot showing fold expression (Log2) of miRNAs in early–B versus early–T cells, indicating the Pearson’s Correlation coefficient (r) between the miRNA profiles of these cell types (complete list of differentially expressed, common, and unique miRNAs in early–B and early–T cells is provided in Dataset S1).
Interestingly, many of the up-regulated miRNAs discussed above were shown to be implicated in proliferation and survival of HSCs (hematopoietic stem cells) (e.g., miR-125b) and development of B and T cells (e.g., miR-181 family) (33–35). Contrastingly, myeloid-specific miRNAs like miR-146a/b-5p, miR-223-3p, and miR-23 cluster (miR-23a, miR-24, and miR-27a) (36–38); miR-126 which is predominantly expressed in CLPs (31); and miR-30b which antagonizes Notch (39) were all found to be significantly down-regulated during early–B or early–T cell differentiation. However, miR-17∼92 cluster, which is down-regulated during monocyte differentiation (40), was also repressed in early–B and early–T cells. Collectively, our genome-wide miRNA analyses indicate that despite distinct identity and gene expression pattern of early–B and early–T cells, their miRNA profiles are significantly correlated wherein both cell types exhibit a large fraction of commonly regulated miRNAs and fewer unique miRNAs. Nonetheless, we note that the early–T cells were found to exhibit a comparatively higher fold expression for a majority of commonly up-regulated miRNAs besides the higher total number of up-regulated miRNAs, compared to early–B cells (115 versus 76, respectively).
miRNAs Coexpressed in Early–B and Early–T Cells Target Distinct Sets of Genes in a Cell Type–Specific Manner.
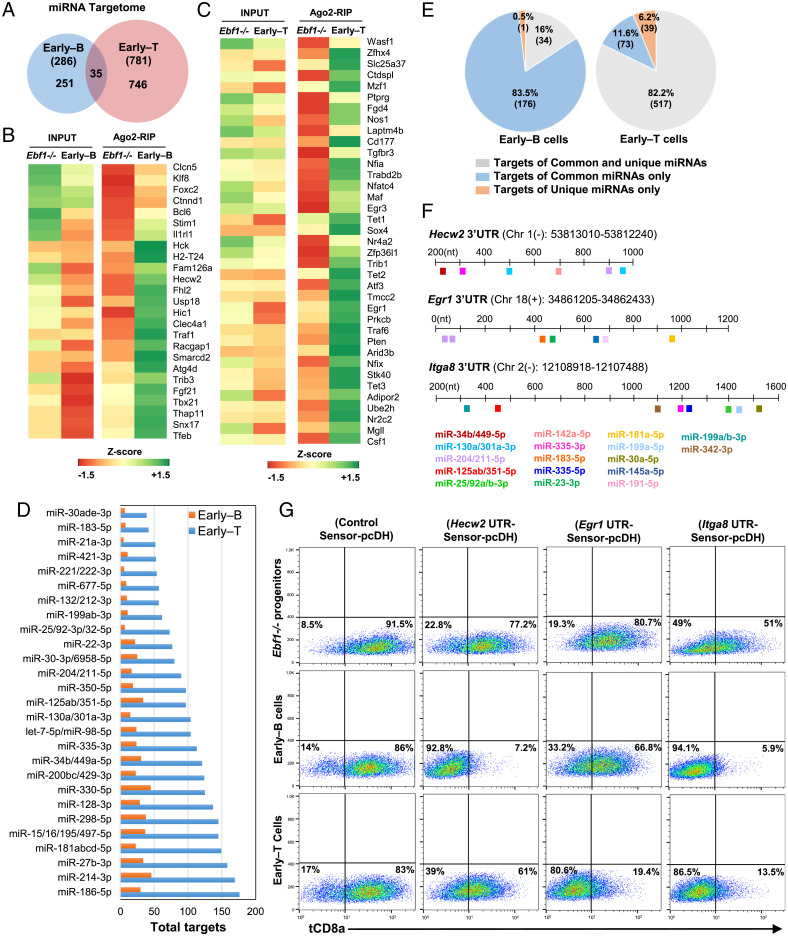
To identify the genome-wide targets of miRNAs (miRNA-targetome) during early–B and T cell differentiation, we performed Ago2-RIPSeq (Ago2-RNA Immunoprecipitation and Sequencing) in Ebf1−/− progenitors, early–B and early–T cells, to capture the transcripts targeted by miRNAs within the Ago2–RISC complexes. In parallel, we carried out total-mRNA-Seq (total messenger RNA sequencing, referred to as “Input”) to assess the expression levels of all mRNA transcripts in these cell types (SI Appendix, Methods and Fig. S2 A and B). The Principal Component Analysis (PCA) revealed a significant distinction between Input and Ago2-RIP profiles of Ebf1−/− progenitors versus early–B or early–T cells (SI Appendix, Fig. S2C). In total, we identified about 286 transcripts in early–B cells and 781 in early–T cells that were significantly Ago2-enriched (Ago2-RIP FC ≥ 2; P ≤ 0.05) compared to Ebf1−/− progenitors (Fig. 2A and SI Appendix, Methods and Fig. S2D). Of these, only 35 transcripts were commonly enriched in both cell types while the remaining transcripts were enriched in a cell type–specific manner (251 in early–B cells and 746 in early–T cells) (Fig. 2A). For instance, transcripts encoded by genes Foxc2, Sarm1, Klf8, Il10ra, and Itga8 were Ago2-enriched in both early–B and early–T cells (Dataset S2). However, transcripts encoded by genes like Hecw2, Fhl2, Eomes, Tbx21, and Bcl6 were enriched only in early–B cells while Egr1, Egr3, Ctdspl, Atf3, Trib1, and Maf were enriched only in early–T cells (Fig. 2 B and C, respectively). Notably, the miRNA-targetome of early–T cells was about three times larger compared to early–B cells (781 versus 286 transcripts, respectively) (Fig. 2A). Furthermore, the total mRNA (or Input) levels of the Ago2-enriched transcripts displayed an overall decrease in early–B or early–T cells compared to Ebf1−/− progenitors, suggesting miRNA-mediated destabilization and subsequent degradation of these transcripts during lineage differentiation (SI Appendix, Fig. S2E).
Fig. 2.
Ago2-RIPSeq identifies that miRNA targetomes of early–B and early–T cells are distinct and lineage-specific. (A) Venn diagram showing the size of miRNA-targetome (i.e., genome-wide miRNA targets) for early–B and early–T cells. The total number of transcripts that were Ago2-enriched in each of these cell types (Ago2-RIP Fold Change ≥2; P ≤ 0.05) compared to Ebf1−/− progenitors are represented in parenthesis. The number of transcripts that were Ago2-enriched “uniquely” in early–B and early–T cells are indicated within the circles. (B and C) Heatmaps representing transcripts that were “uniquely” Ago2-enriched either in early–B or early–T cells, respectively. (A complete list of transcripts that were Ago2-enriched in early–B and/or early–T cells is provided in Dataset S2). (D) Bar graph representing total number of transcripts targeted by commonly up-regulated miRNA(s) or their families in early–B or early–T cells (shown by orange and blue bars, respectively), identified using multiMiR. (E) Pie charts indicating the percentage of Ago2-enriched transcripts from early–B and early–T cells (Left and Right, respectively) that possess binding sites for common, unique, or both sets of miRNAs within their 3′UTRs. (F) Binding sites for miRNAs predicted within the 3′UTR regions of selected Ago2-enriched transcripts, Hecw2, Egr1, and Itga8, which were used for tCD8a reporter assays to study lineage-specific miRNA-mediated repression. (G) Flow cytometry analysis indicating surface levels of tCD8a reporter in Ebf1−/− progenitors, early–B or early–T cells, which expressed Control Sensor-pcDH or Sensor-pcDH vectors containing 3′UTR of Hecw2, Egr1, or Itga8 (SI Appendix, Fig. S3D).
Next, to identify the miRNAs that potentially target the Ago2-enriched transcripts in early–B and early–T cells, we mapped each of these transcripts to the up-regulated miRNAs from the corresponding lineages using an R-package called “multiMiR” (41) (Materials and Methods and Dataset S3). Subsequently, we sought to identify the transcripts that are regulated by the “common” or “unique” sets of miRNAs in each cell type. In early–B cells, a major fraction (∼83.5%) of the total Ago2-enriched transcripts were mapped specifically to the commonly up-regulated miRNAs, whereas a very small fraction (∼0.5%) were mapped only to unique miRNAs (Fig. 2 E, Left). Meanwhile, 16% of the Ago2-enriched transcripts in early–B cells were shown to be jointly targeted by both common and unique miRNAs. On the contrary, in early–T cells, only few of the total Ago2-enriched transcripts were targeted specifically by either common or unique sets of miRNAs (∼11.6% and 6.2%, respectively) while the majority of the enriched transcripts (∼82.2%) were found to be jointly targeted by both common and unique miRNAs (Fig. 2 E, Right). Since nearly all the Ago2-enriched transcripts in both these cell types possess binding sites for commonly up-regulated miRNAs (99%, 210 transcripts in early–B cells and 93.8%, 590 transcripts in early–T cells), it can be inferred that the commonly up-regulated miRNAs indeed target a large majority of Ago2-enriched genes in both early–B and early–T cells (Dataset S3). Accordingly, we noticed that the commonly up-regulated miRNAs like miR-186-5p, miR-214-3p, miR-27b-3p, miR-128-3p, and families like miR-181, miR-125/351, miR-132/212, and miR-204/211 were shown to target a large number of transcripts in early–B as well as early–T cells (Fig. 2D and Dataset S3). However, given the differences in the distribution of binding sites for common and unique miRNAs within the Ago2-enriched transcripts of early–B versus early–T cells (Fig. 2E), it is evident that besides the common miRNAs, the relatively larger number of unique miRNAs expressed in early–T cells (than early–B cells, i.e., 49 versus 10 miRNAs, respectively) partly accounts for the enlarged early–T cell target repertoire compared to early–B cells (781 versus 286 transcripts, respectively).
In order to validate the lineage-specific Ago2-enrichment of transcripts, we performed reporter assays using the 3′UTRs (3' untranslated regions) of three differentially enriched transcripts: Hecw2 (enriched only in early–B cells), Egr1 (enriched only in early–T cells), and Itga8 (enriched in early–B and early–T cells) (SI Appendix, Fig. S3A). The miRNAs that are predicted to bind to the 3′UTR regions of these transcripts are represented in Fig. 2F and SI Appendix, Fig. S3B. Each of these 3′UTRs or a control sequence (asEGFP) were inserted downstream of tCD8a (reporter gene) into the reporter construct “Empty Sensor-pcDH” (SI Appendix, Methods and Fig. S3C) and transduced into Ebf1−/− progenitors, which were subsequently differentiated into early–B or early–T cells. Analysis of tCD8a expression by flow cytometry (Fig. 2G and SI Appendix, Fig. S3D) and qRT-PCR (SI Appendix, Fig. S3E) revealed a lineage-specific decrease in tCD8a reporter levels, in consistence with the Ago2-RIPSeq findings. Since the majority of the miRNAs that putatively target Hecw2 and Egr1 transcripts were commonly expressed in both early–B and early–T cells (Fig. 2F and SI Appendix, Fig. S3B), the observed lineage-specific repression of these transcripts in the respective lineages suggests that miRNA-mediated regulation is cell type dependent and contingent not only upon the abundance of miRNAs and their targets but may also involve other complex cell-intrinsic mechanisms.
miRNAs Negatively Regulate the Abundance of Lineage-Inappropriate Transcripts during B and T Cell-Fate Commitment.
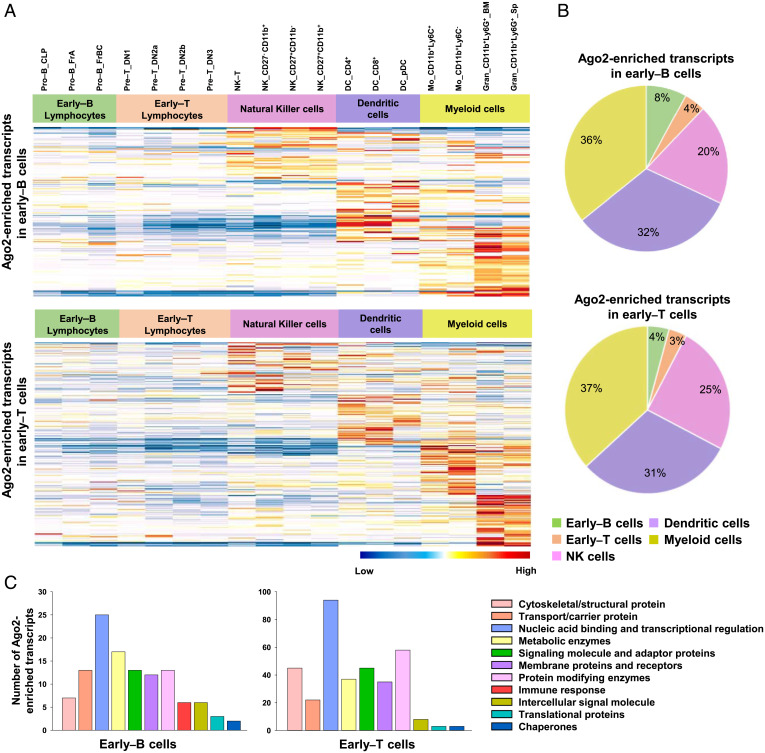
Next, we ascertained the lineage specificity of Ago2-enriched transcripts from early–B or early–T cells by comparing their expression levels across various hematopoietic lineages, using the RNA-Seq data available at ImmGen (Immunological Genome Project) (SI Appendix, Methods). Interestingly, a large majority of the total number of Ago2-enriched transcripts from early–B cells or early–T cells could be mapped to alternate lineages (i.e., NK cells, DCs and myeloid cells) and were found to exhibit a comparatively higher expression in these cell types than the early B or T lymphocytes (Fig. 3 A and B and Dataset S4). Using the Gene Ontology (GO) analysis, we identified that the Ago2-enriched transcripts encode proteins associated with diverse biological functions such as signal transducers (e.g., Ptprg and Map3k2), surface receptors (e.g., Tlr13, Tlr9, and Ciita), metabolic regulators (e.g., Ltf and Nos1), cytoskeletal proteins (e.g., Itga8), as well as regulators of transcription (e.g., TFs encoded by Tbx21, Eomes, Irf8, Tgfbr3, Nfatc2, or RNA-binding proteins [RBPs] encoded by Cpeb3) (Fig. 3C). Given these findings, we postulated, the developing B and T lymphocytes perhaps employ a common mechanism of miRNA-mediated posttranscriptional silencing to restrict the hitherto available alternate cell-fate options before undergoing commitment toward the respective lineages (42–48).
Fig. 3.
miRNAs expressed in early–B and early–T cells target lineage-inappropriate genes. (A) Heatmaps representing relative gene expression levels for Ago2-enriched transcripts from early–B cells (Top) or early–T cells (Bottom), analyzed in subsets of various immune cells (i.e., early–B lymphocytes, early–T lymphocytes, NK cells, DCs, and myeloid cells [monocytes and neutrophils]), using the RNA-Seq data available at ImmGen. (B) Pie chart showing percentages of Ago2-enriched transcripts from early–B cells (Top) or early–T cells (Bottom) that were mapped to various immune cell types using the RNA-Seq data available at ImmGen. (C) GO analysis of Ago2-enriched transcripts from early–B and early–T cells. The transcripts were classified into various categories based on their molecular function, using the annotation tool, PANTHER. The absolute number of genes under each category is represented using the colored bars.
To experimentally verify the findings obtained from ImmGen, we analyzed the mRNA levels of a few Ago2-enriched transcripts (those possibly associated with myeloid lineage) following differentiation of Ebf1−/− progenitors into myeloid cells. Ebf1−/− progenitors (maintained under lymphoid conditions) were directly differentiated into a heterogeneous population of Mac1+ (CD11b+) myeloid cells (shown as Mac1Lo, 52.2%) by culturing under myeloid-promoting cytokines (SCF, Flt3L, GMCSF, MSCF, and IL-3) (SI Appendix, Fig. S4 A and B). Alternatively, Ebf1−/− progenitors were first transduced with MigR1-PU1.ER vector (retrovirus encoding PU.1 fused to the ligand binding domain of Estrogen Receptor) and subsequently transferred to myeloid-promoting cytokines along with addition of 4-OHT to obtain myeloid cells expressing high levels of Mac1 (shown as Mac1Hi, 98.8%). Correspondingly, the transcript levels of PU.1 and its well-known targets like Csf1r and Cd11b (Mac1) were found to be comparatively higher in Mac1Hi cells than Mac1Lo cells, indicating a PU.1 dosage-dependent effect (SI Appendix, Fig. S4C). Consistent with the ImmGen analysis, the Ago2-enriched transcripts from early–B or early–T cells (e.g., Ctnnd1, Maf, Ptprg, Ctdspl, Tgfbr3, Gsn, Egr1, Zfhx4, Itga8, Fam126a, Fgr, Atg4d, Nfam1, and Il10ra) were expressed at higher levels in Mac1Lo myeloid cells and more so in Mac1Hi myeloid cells, compared to Ebf1−/− progenitors, suggesting their myeloid identity (SI Appendix, Fig. S4D).
Lymphocyte miRNAs Act Combinatorially to Suppress Myeloid Lineage Option of Multipotent Progenitors.
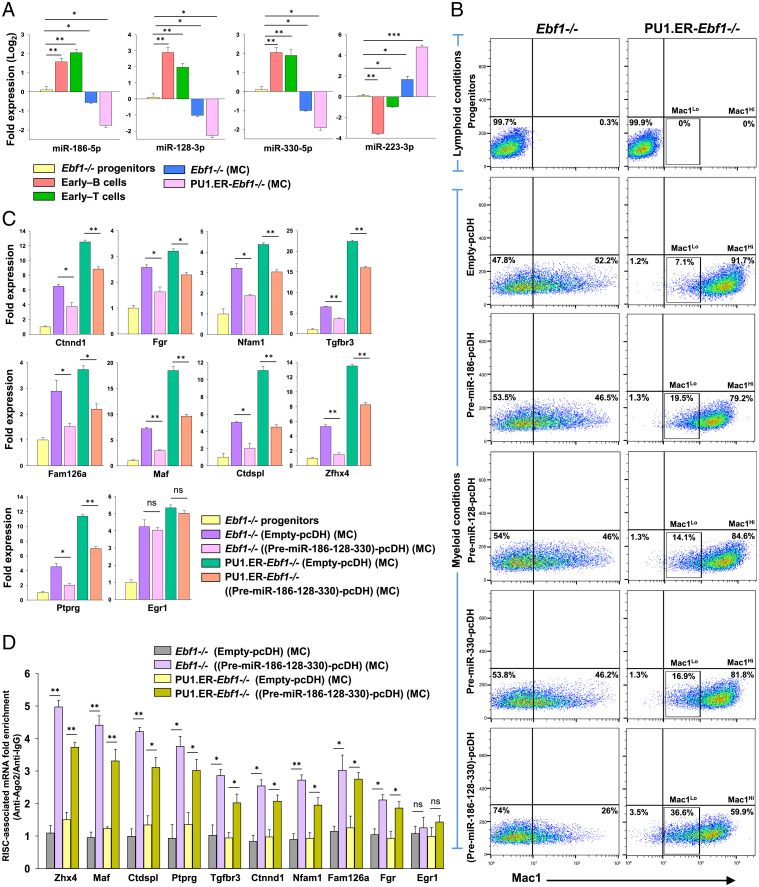
Next, to interrogate the functional significance of miRNA-mediated targeting of alternate lineage genes, we tested the ability of lymphocyte miRNAs to attenuate the myeloid cell-fate option of the multipotent Ebf1−/− progenitors. For this, we shortlisted three miRNAs: miR-186-5p, miR-128-3p, and miR-330-5p for ectopic expression studies based on their abundance, total number of myeloid-associated targets, and lymphocyte-specific expression pattern (Fig. 4A and Dataset S5). Precursors for each of these miRNAs were individually expressed (via the pcDH vector) into Ebf1−/− or PU1.ER-Ebf1−/− progenitors, which were subsequently analyzed for Mac1 expression following differentiation toward myeloid lineage (SI Appendix, Methods and Fig. S5 A–C). The levels of mature miRNAs (miR-186-5p, miR-128-3p, or miR-330-5p) were found to be approximately fivefold higher in cells transduced with the respective Pre-miRNA-pcDH vectors compared to cells transduced with Empty-pcDH (control vector), indicating effective processing of the precursor transcripts (SI Appendix, Fig. S5D). We found that expression of each of the precursor miRNAs, Pre-miR-186, Pre-miR-128, or Pre-miR-330, in Ebf1−/− progenitors resulted in a subtle yet significant decrease in the generation of heterogeneous Mac1+ myeloid cells (46.5%, 46%, or 46.2%, respectively) compared to Ebf1−/− progenitors that expressed Empty-pcDH vector (Mac1+, 52.2%) (Fig. 4B and SI Appendix, Fig. S6E). In parallel, PU1.ER-Ebf1−/− cells transduced with Pre-miR-186, Pre-miR-128, or Pre-miR-330 also exhibited a mild decrease in generation of Mac1Hi myeloid cells, leading to a corresponding increase in the Mac1Lo population (19.5%, 14.1%, and 16.9%, respectively), compared to the Mac1Lo population within the PU1.ER-Ebf1−/− cells expressing Empty-pcDH vector (Mac1Lo, 7.1%) (Fig. 4B and SI Appendix, Fig. S6E).
Fig. 4.
Coexpression of multiple miRNAs with distinct seed sequences decreases myeloid differentiation potential of Ebf1−/− and PU1.ER-Ebf1−/−progenitors. (A) Expression levels of miR-186-5p, miR-128-3p, miR-330-5p, and miR-223-3p (a myeloid-specific miRNA) measured using stemloop qRT-PCR in Ebf1−/− progenitors, early–B cells, early–T cells, Mac1Lo, and Mac1Hi myeloid cells (generated from Ebf1−/− or PU1.ER-Ebf1−/− progenitors, respectively, by culturing under myeloid-promoting conditions [represented as “MC”; SI Appendix, Methods]). Data are represented with Ebf1−/− progenitors as control. (B) Analysis of myeloid differentiation potential of Ebf1−/− or PU1.ER-Ebf1−/− progenitors that were transduced with Empty-pcDH or pcDH vector expressing Pre-miR-186, Pre-miR-128, and Pre-miR-330, either individually or simultaneously. Transduced cells were cultured under myeloid-promoting conditions (represented as “MC”) and analyzed for Mac1 (CD11b) expression by flow cytometry. (SI Appendix, Fig. S6E). (C) qRT-PCR analysis of transcript levels of myeloid-associated genes targeted by miRNAs: miR-186-5p, miR-128-3p, or miR-330-5p, measured in Ebf1−/− or PU1.ER-Ebf1−/− progenitors that were transduced with Empty-pcDH or (Pre-miR-186-128-330)-pcDH construct and subsequently differentiated toward myeloid lineage (represented as “MC”). Egr1, a myeloid gene, not targeted by these miRNAs was used as negative control. Data are represented with Ebf1−/− progenitors as control sample. (D) RIP-qPCR analysis showing fold enrichment (Ago2-IP versus IgG-IP) of myeloid-associated transcripts targeted by miRNAs: miR-186-5p, miR-128-3p, or miR-330-5p, within the RISC-complexes of Ebf1−/− or PU1.ER-Ebf1−/− cells that were transduced with Empty-pcDH or (Pre-miR-186-128-330)-pcDH construct and subsequently differentiated toward myeloid lineage (represented as “MC”) (for A–D, two independent experiments were performed, each in duplicates, and data are shown as mean ± SD [*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ns = not significant]).
However, the efficiency and outcome of miRNA-mediated regulation of developmental processes has been shown to be defined not only by the expression levels, binding strength, or activity of individual miRNAs but also involves a combinatorial effect exerted by multiple miRNAs that coregulate several functionally related transcripts (49–51). Notably, each of the three miRNAs we had tested, miR-186-5p, miR-128-3p, and miR-330-5p, were predicted to target an average of 67 myeloid-associated transcripts, while in combination, they potentially target at least 148 myeloid-associated transcripts (Dataset S6). Considering this, we analyzed the effect of simultaneous expression of these miRNAs on myeloid differentiation capacity of Ebf1−/− and PU1.ER-Ebf1−/− progenitors. Strikingly, coexpression of Pre-miR-186, Pre-miR-128, and Pre-miR-330 in Ebf1−/− progenitors [via the construct (Pre-miR-186-128-330)-pcDH] (SI Appendix, Fig. S5A) resulted in a profound decrease in generation of heterogeneous Mac1+ myeloid population (Mac1+, 26%) (Fig. 4B) compared to the effect seen when each of these miRNAs were expressed individually (Mac1+, ∼46%). Similarly, PU1.ER-Ebf1−/− cells that coexpressed Pre-miR-186, Pre-miR-128, and Pre-miR-330 exhibited a substantial decrease in generation of Mac1Hi populations (Mac1Hi, 59.9%), leading to an increased accumulation of Mac1Lo population (Mac1Lo, 36.6%) (Fig. 4B and SI Appendix, Fig. S6E) compared to PU1.ER-Ebf1−/− cells that expressed these miRNAs individually (Mac1Lo, <20%). Although the expression levels of each of the mature/effector miRNAs (miR-186-5p, miR-128-3p, and miR-330-5p) generated from the corresponding precursors were similar between Ebf1−/− and PU1.ER-Ebf1−/− cells (SI Appendix, Fig. S5E), the myeloid differentiation capacity was abrogated to a lesser extent in PU1.ER-Ebf1−/− cells than Ebf1−/− cells (i.e., Mac1− population, 3.5% versus 74%, respectively). This could perhaps be due to the limited ability of miRNAs to antagonize the robust myeloid developmental program induced in PU1.ER-Ebf1−/− cells due to relatively high PU.1 abundance.
To determine whether the observed attenuation in myeloid differentiation capacity upon coexpression of miR-186-5p, miR-128-3p, or miR-330-5p is due to repression of myeloid-associated targets, we measured the transcript levels of few genes targeted by one or more of these miRNAs. Interestingly, previous reports suggest that many of these genes are involved in development and survival of myeloid cells (Dataset S6). As shown in Fig. 4C, myeloid-associated genes like Ctnnd1, Fgr, Nfam1, Fam126a, Maf, Tgfbr3, Ctdspl, Ptprg, and Zfhx4 underwent a decrease in their transcript levels in (Pre-miR-186-128-330)-pcDH vector expressing Ebf1−/− and PU1.ER-Ebf1−/− cells that were differentiated toward myeloid lineage, compared to their counterparts expressing Empty-pcDH vector (Egr1 is a nontarget control gene). Moreover, presence of binding sites for more than one of these miRNAs resulted in a more profound decrease in the transcript levels of the respective targets, indicating an additive effect (Fig. 4C). Furthermore, the RIP-qPCR analyses with Anti-Ago2 or Anti-IgG antibodies show a corresponding increase in Ago2-enrichment of these transcripts within the RISC complexes of myeloid-differentiating Ebf1−/− and PU1.ER-Ebf1−/− cells in presence of these miRNAs (Fig. 4D). Thus, coexpression of lymphocyte-specific miRNAs: miR-186-5p, miR-128-3p, and miR-330-5p, resulted in attenuation of myeloid differentiation capacity of multipotent progenitors due to simultaneous and cumulative repression of a large repertoire of myeloid-associated transcripts via their recruitment to cellular RISC complexes.
To further confirm whether the decrease in myeloid differentiation potential of Ebf1−/− and PU1.ER-Ebf1−/− progenitors observed in presence of (Pre-miR-186-128-330)-pcDH is a result of combinatorial and synergistic targeting of several myeloid-associated genes but not merely due to miRNA overexpression; we tested the myeloid lineage repression potential of a second set of lymphocyte-specific miRNAs: miR-125a-5p, miR-125b-5p, and miR-351-5p, which belong to miR-125 family and share an overlapping set of 62 myeloid targets due to their similar seed sequences (SI Appendix, Fig. S6A and Dataset S7). Interestingly, forced coexpression of miR-125 family precursors in Ebf1−/− or PU1.ER-Ebf1−/− progenitors [via (Pre-miR-125a-125b-351)-pcDH construct] caused a subtle decrease in their myeloid differentiation potential (SI Appendix, Fig. S6 B–E), despite a notable decline in the transcript levels of its gene targets (e.g., Gsn, Atg4d, Maf, Ctdspl, Itga8, and Il10ra) and a corresponding increase in their enrichment within the Ago2–RISC complexes (SI Appendix, Fig. S6 F and G, respectively). Notably, the mature miR-125 family members were generated from their precursors at similar levels as were miR-186-5p, miR-128-3p, and miR-330-5p (SI Appendix, Figs. S6H and S5E, respectively). These results suggest that coexpression of multiple lymphocyte miRNAs that share similar seed sequences could significantly lower the abundance of their respective myeloid-associated targets but is ineffective in attenuating the myeloid differentiation capacity of Ebf1−/− or PU1.ER-Ebf1−/− progenitors. Hence, effectual inhibition of a given lineage may require simultaneous repression of several lineage-associated genes, which is perhaps achieved by a concerted action of multiple miRNAs having distinct seed sequences.
To further assess the functional significance of the selected miRNAs in regulating lymphoid cell-fate decisions in vivo, we verified their expression patterns in ex vivo isolated pro–B cells (Fraction-B) and DN2 T cells as well as CD11b+ myeloid cells. Consistent with in vitro analyses, these miRNAs were significantly up-regulated in pro–B and DN2 T cells, compared to CD11b+ myeloid cells (SI Appendix, Fig. S7), suggesting an important role for these miRNAs in regulating lymphoid cell-fate choice.
Nonredundant Role of miRNAs and Their Transcriptional Control during Lymphoid Cell Differentiation.
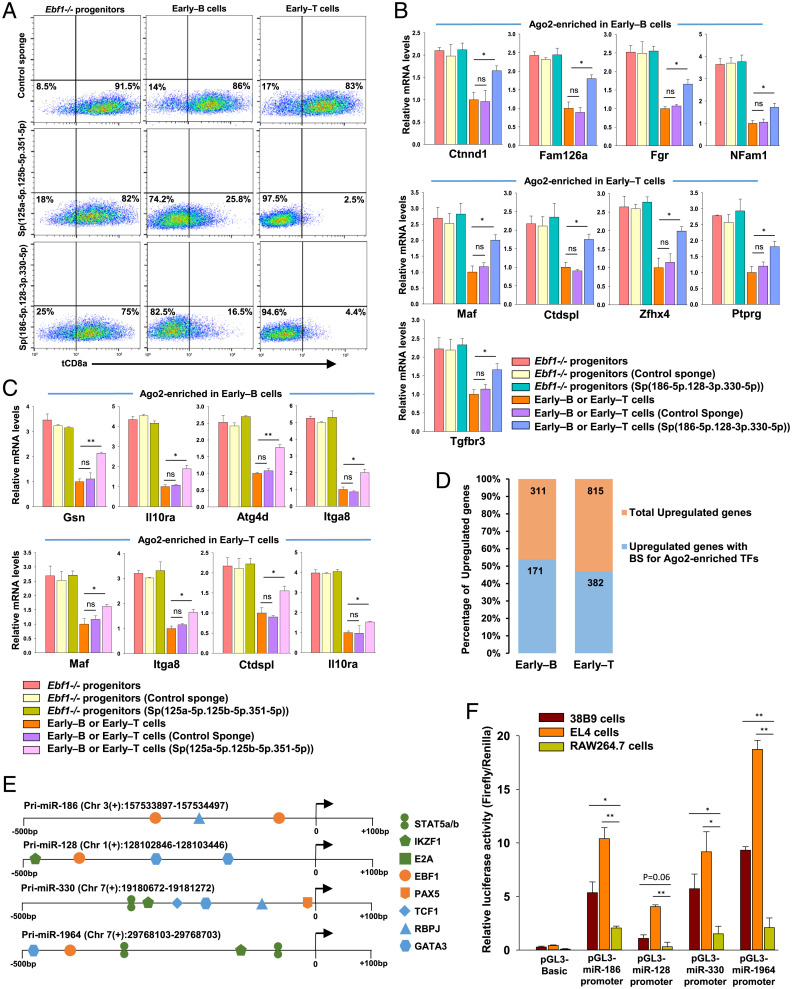
Next, we examined whether the early–B and early–T cell miRNAs repress lineage-inappropriate genes independently of B or T lineage determinants. For this, we used two sets of miRNA sponges, Sp(186-5p.128-3p.330-5p) and Sp(125a-5p.125b-5p.351-5p), to study the effect of knockdown of these lymphocyte miRNAs on their corresponding myeloid-associated gene targets during early–B and early–T cell differentiation (SI Appendix, Fig. S8A). The Sensor-pcDH vectors containing each of these miRNA sponges or a control sponge inserted downstream of tCD8a reporter were transduced into Ebf1−/− progenitors that were subsequently allowed to differentiate into early–B or early–T cells (SI Appendix, Fig. S8 B and C). We found a significantly decreased surface expression and transcript levels of tCD8a reporter in early–B and early–T cells expressing Sp(125a-5p.125b-3p.351-5p) or Sp(186-5p.128-3p.330-5p), indicating effective sponge-mediated knockdown of these miRNAs in both the cell types (Fig. 5A and SI Appendix, Fig. S8 D and E). More importantly, expression of miRNA sponges resulted in de-repression of myeloid genes targeted by these miRNAs (Fig. 5 B and C) but had no significant effect on the transcript levels of other nontarget genes that are expressed in early–B or early–T cells (e.g., Mb-1 and Pax5 or Tcf7 and Gata3, respectively) (SI Appendix, Fig. S8 F and G). However, for most of the tested myeloid transcripts, the extent of de-repression was found to be modest as they are potentially regulated by other miRNAs expressed in early–B or early–T cells. Thus, the contribution of early–B and early–T cell miRNAs toward repression of inappropriately expressed genes is nonredundant and independent of the contribution made by B and T lineage-specific TFs.
Fig. 5.
Knockdown of lymphocyte miRNAs in early–B and early–T cells rescues repression of myeloid-associated transcripts. (A) Flow cytometry analysis of tCD8a reporter levels in Ebf1−/− progenitors that were differentiated into early–B or early–T cells in presence of Control sponge or miRNA sponges, Sp(125a-5p.125b-5p.351-5p) or Sp(186-5p.128-3p.330-5p) (SI Appendix, Fig. S8D). (B) qRT-PCR analysis of transcript levels of myeloid-associated genes targeted by miRNAs: miR-186-5p, miR-128-3p, or miR-330-5p, measured in Ebf1−/− progenitors and early–B or early–T cells that expressed Control sponge or Sp(186-5p.128-3p.330-5p). Data are represented with early–B or early–T cells as control sample. (C) qRT-PCR analysis of transcript levels of myeloid-associated genes targeted by members of miR-125 family (miR-125a-5p, miR-125b-5p, and miR-351-5p), measured in Ebf1−/− progenitors and early–B or early–T cells that expressed Control sponge or Sp(125a-5p.125b-5p.351-5p). Data are represented with early–B or early–T cells as control sample (for A–C, two independent experiments were performed, each in duplicates, and data are shown as mean ± SD [*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ns = not significant]). (D) Bar graph showing the percentages of B or T lineage genes (i.e., genes up-regulated in early–B or early–T cells with RNAseq FC (Log2) ≥ 2) possessing binding sites for transcription factors encoded by transcripts that were Ago2-enriched in the respective cell type. (E) Schematic representation of binding sites for TFs within the promoter regions of miRNAs that were commonly expressed between early–B and early–T cells. TFs specific to early–B or early–T cells are represented in orange and blue, respectively, while TFs common to lymphocyte development are indicated in green. (F) Transcriptional activity of commonly expressed miRNA promoters analyzed by dual luciferase assay. Putative miRNA promoters represented in E were cloned into pGL3-basic vector and transfected into 38B9 (pro–B cells), EL4 (T cells), and RAW264.7 (macrophage) cells. After 48 h, cells were harvested and analyzed for luciferase reporter activity. Two independent experiments were performed, each in triplicates, and the data are shown as mean ± SD (*P ≤ 0.05; **P ≤ 0.01).
Subsequent to identifying the role for early–B and early–T cell miRNAs in repression of lineage-inappropriate genes, we speculated whether these inappropriately expressed, Ago2-enriched genes could possibly interfere with faithful expression of lineage-promoting genes in early–B and early–T cells and are thus subjected to repression by lymphocyte miRNAs. In this perspective, we probed the promoters of all the genes that were up-regulated during early–B and early–T cell differentiation for presence of binding sites for TFs encoded by Ago2-enriched genes. Interestingly, in early–B and early–T cells, 54% and 47% of up-regulated genes were found to possess binding sites for the Ago2-enriched TFs (Fig. 5D and Dataset S8). For example, promoters of B lineage-specific genes like Cd79a, Cd79b, Pax5, and Cd19 were shown to possess binding sites for Ago2-enriched TFs like IRF5, TBX21, EOMES, and FOXC2. Similarly, promoters of T lineage-specific genes like Bcl11b, Gata3, and Tcf7 possessed binding sites for Ago2-enriched TFs such as RUNX2, MAF, POU2F1, and NFIA. Therefore, miRNA-mediated repression of inappropriately expressed genes may allow for uninterrupted and sustained expression of B and T lineage-promoting genes to facilitate the progression of B or T cell differentiation program.
Finally, we attempted to understand how early–B and early–T cells are able to express similar repertoire of miRNAs despite their distinct gene expression programs. By integrating the miRNA Transcription Start Sites (TSS) with H3K4me3 ChIP-Seq data from early–B and early–T cells, we connected miRNAs into the transcriptional circuitry to understand their regulation by transcription factors that are known to regulate early–B and T cell development. We found the promoter regions of almost all the early–B or early–T cells miRNAs were enriched in H3K4me3 mark, representing their active transcription in these cell types. Strikingly, de novo motif analysis of these miRNA promoters revealed high-confidence binding sites for TFs like E2A, IKAROS (Ikzf1), as well as STAT-5a/b (IL-7R signaling) that dictate the earliest stages of B as well as T lymphocyte development (15, 52, 53). Moreover, we found an increased cooccupancy of B lineage-specific factors (EBF1 and/or PAX5) as well as T lineage-specific factors (Notch1/RBPJ1, TCF1, and/or GATA3) within the promoters of several commonly expressed miRNAs (Fig. 5E and Dataset S9). To validate our hypothesis whether the promoters of commonly expressed miRNAs exhibit a lymphocyte-specific activity, we performed dual luciferase reporter assays wherein the putative miRNA promoter regions of miR-186, miR-128, miR-330, and miR-1964 were cloned into pGL3-basic luciferase reporter vector, and their activities were assessed following transfection into 38B9 cells (pro–B cells), EL4 cells (T cells), or RAW264.7 cells (myeloid cells). The outcome of these studies suggests that the selected miRNA promoters are transcriptionally more active in 38B9 as well as EL4 cells compared to RAW264.7 cells (Fig. 5F). The impaired activity of miRNA promoters in RAW264.7 cells, which lack primary lymphoid-specific determinants, indicates that expression of these miRNAs in early–B and early–T cells is critically dependent upon cell type–specific transcription factors.
Discussion
Despite the fact that B and T lineage options are mutually exclusive, our studies show that miRNAs expressed in early–B and early–T cells are significantly correlated; yet they target distinct sets of genes in these lineages. This cell type–specific targeting by commonly expressed miRNAs could be due to four possible reasons: Firstly, differences in the transcriptomes of early–B and early–T cells may result in differential Ago2 enrichment due to variation in the availability, specificity, frequency, and combination of miRNA binding sites within the 3′UTRs of differentially expressed transcripts. Quite possibly, the sites present in protein-coding sequences also contribute toward target repression, besides the sites within the 3′UTRs. Secondly, despite sharing similar seed sequences, each member of the miRNA families expressed in early–B and early–T cells may exhibit differential 3′-end (supplemental) base pairing, which compensates for seed-match imperfections and possibly contributes to diversity in target repertoires between the two lineages (54, 55). Thirdly, the relative abundance of commonly expressed miRNAs in early–T cells than early–B cells potentially permits targeting of transcripts that possess noncanonical BS (binding sites) (besides canonical BS), thereby further influencing the targeting specificity in early–T cells (56). Moreover, besides the commonly up-regulated miRNAs, the relatively higher number of unique miRNAs expressed in early–T cells also contribute significantly toward the increased targetome size of early–T cells. Lastly, the lineage-specific repression of transcripts as demonstrated by 3′UTR reporter assays suggests the existence of additional cell-intrinsic mechanisms that support cell type–specific miRNA targeting, independent of miRNA or mRNA abundance. These may include but are not limited to differentially expressed cellular RBPs that bind to and alter the local structure of target mRNAs, subsequently influencing the accessibility or affinity of binding sites for one or more miRNAs, thus altering the targeting efficiency in a cell type–specific manner (57–60). Given the differences in progression dynamics of lineage commitment during differentiation of LMPPs toward B versus T lineages (61), a possible implication of cell type–specific targeting demonstrated by miRNAs is that it perhaps allows differential regulation of genes, which act as rate-limiting factors for commitment and are thus required to be maintained at different thresholds in developing B versus T cells.
Interestingly, a vast majority of miRNA-targeted genes in early–B or early–T cells are associated with alternate cell fates like NK, DC, and myeloid and are thus aptly suppressed to enable commitment toward B or T lineages. In this perspective, we suggest that the developmental arrest seen upon absolute depletion of miRNAs in B and T cells (by abrogation of DGCR8 or Dicer) prior to lineage commitment (23–25) could possibly be a consequence of accumulation of lineage-inappropriate genes, which in turn prevents the transition of lineage-specified cells into a committed stage. Since a large fraction of miRNA-targeted genes in both early–B and early–T cells encode proteins from signaling cascades (like cell surface receptors and signal transducers), minor changes in their threshold levels holds a potential to greatly impact the cellular output (62). Perhaps, this highlights the inconspicuous yet significant role for miRNAs in maintaining optimal levels of such important modulators of cell development and function. Notably, TF PU.1, which is expressed in both lymphoid and myeloid lineages (albeit at different dosages) (63), was not targeted by miRNAs in early–B or early–T cells, whereas several PU.1-dependent, myeloid-associated transcripts were repressed by miRNAs in both cell types.
Our study also highlights the functional significance of combinatorial action of miRNAs, which is often not captured in single miRNA studies. For instance, miRNAs like miR-186-5p, miR-128-3p, and miR-330-5p having distinct seed sequences, when coexpressed caused a synergistic and simultaneous repression of several myeloid-associated genes, resulting in a more-efficient inhibition of myeloid differentiation capacity compared to single miRNAs or miRNAs sharing the same seed (e.g., miR-125 family). However, besides these three miRNAs, other combinations of lymphocyte miRNAs can quite possibly generate such functional outcomes. We show that combinatorial targeting can impact cell-fate decisions, thus highlighting the fact that complex processes like lineage differentiation may involve a multitude of individual miRNA–target interactions. Given the significance of miRNAs demonstrated by this study, we propose a lymphocyte developmental network wherein the early–B or T lineage determinants induce miRNAs as an additional tier of regulators during differentiation of LMPPs into committed B and T lymphocytes (SI Appendix, Fig. S9). Upon activation, the miRNAs act hand-in-hand with lineage-specific factors to antagonize inappropriately expressed genes in order to block available alternate cell-fate options, leading to establishment of lineage identity. Collectively, this study establishes miRNAs as significant and nonredundant regulatory components of B and T lymphocyte developmental networks.
Materials and Methods
Ago2-RIPSeq.
Cell lysates were prepared for Ebf1−/− progenitors, early–B, and early–T cells. The cells were washed once with 1× PBS (phosphate buffered saline, pH = 7.4), and lysed in Cell Lysis buffer (CLB) (50 mM Tris·HCl [pH = 7.4], 150 mM NaCl, 5 mM MgCl2, and 1.0% Nonidet P-40), supplemented with 1 mM DTT (dithiothreitol), 1× Protease inhibitors, and RNase inhibitor (at 200 U/mL). The lysates were centrifuged at 4 °C for 15 min to remove debris and then precleared with Protein-G Dynabeads at 4 °C for 30 min. For each RIP sample, about 4 mg of cleared protein lysate from the respective cell type was immunoprecipitated with anti-Ago2 antibody (Abcam ab32381) for 7 to 8 h at 4 °C, followed by pull-down using Protein-G Dynabeads to capture the Ago2-RISC complexes. The beads were washed five times with CLB and resuspended in TRIzol for RNA isolation. For Input samples, cleared protein lysates were directly used for RNA isolation using TRIzol. The Input and IP RNA samples were subsequently used for preparing mRNA libraries using NEBNext Ultra-II Directional RNA Library Prep kit (NEB, New England Biolabs), as per the manufacturer’s instructions. The libraries were sequenced on Illumina’s HiSeq 2500. Ago2-RIPSeq data analysis is described in detail under SI Appendix, Methods.
Target Prediction.
The miRNAs up-regulated in early–B and early–T cells were mapped with Ago2-enriched transcripts from the respective cell type, using R-package multiMiR (41). The user-defined cut off for target strength was set to 40%, in order to select only the top 40% predictions ranked by the primary score for each of the enlisted databases in multiMiR, including MicroCosm, miRanda, miRDB, PicTar, TargetScan, DIANAmicroT, PITA, ElMMo, miRecords, and miRTarbase. Subsequently, the miRNA–target interactions that were shown by at least two of the prediction tools were selected for further studies.
GO Analysis.
GO analysis was performed by submitting the list of Ago2-enriched genes to the protein classification tool, Protein ANalysis THrough Evolutionary Relationships (PANTHER, version 14.0) for analysis based on molecular function (64). Genes that play functionally related molecular roles were grouped together into broad categories for representation.
Ago2-Enriched TF Binding Analysis.
To identify the active promoters for B or T lineage-specific genes (i.e., genes up-regulated in early–B or early–T cells with RNA-seq FC (Log2)≥2), the TSS of the genes were mapped with the respective H3K4me3 ChIP-Seq datasets from early–B cells (SI Appendix, Methods) or early–T cells (65). For each cell type, the H3K4me3 peaks located within ±2.5 kb region from the TSS of each gene were probed for presence of binding sites for TFs encoded by Ago2-enriched genes from the respective lineages, using the position weight matrices from JASPAR and the Transcription Element Search System (TESS).
miRNA Promoter Analysis.
The TSS of mouse primary miRNAs available at ref. 66 were overlaid with H3K4me3 ChIP-Seq datasets obtained from early–B cells (SI Appendix, Methods) or early–T cells (65) to identify the active promoters for common and unique miRNAs expressed in early–B and early–T cells (Mouse genome assembly GRCm38.p6). The region spanning −500bp to +100bp across the TSS of each primary miRNA was scanned for the presence of binding sites for transcription factors (E2A, IKAROS, STAT5a/b, EBF1, PAX5, FOXO1, RBPJ, TCF1, and GATA3) that are crucial for development of early–B or early–T cells. Position weight matrices obtained from JASPAR and TESS were used to predict the TF binding sites within the miRNA promoters.
Statistical Analysis.
Statistical analyses were performed using Microsoft Excel (2013), SigmaPlot (version 12.3) and GraphPad prism (version 7). The data are expressed as mean ± SD. Wherever indicated, two or three independent experiments (in duplicates or triplicates) were performed, and statistical significance was determined by unpaired two-sample t test. A value of P ≤ 0.05 was considered statistically significant (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ns = not significant).
Additional Methods.
Additional details on vectors and constructs, cell culture procedures, viral transductions, in vitro differentiation of progenitors into early–B/T or myeloid cells, quantification of miRNAs and mRNA transcripts, miRNA library preparation, data analysis for miRNA-seq and Ago2-RIPSeq, Ago2-RIP-qPCR, 3′UTR reporter assays, H3K4me3 ChIP-Seq, ImmGen analysis, dual luciferase assays for miRNA promoter activity, and flow cytometry analyses are described in SI Appendix, Methods.
Acknowledgments
We thank the Next-Generation Genomics Facility at National Centre for Biological Sciences (Bangalore) for NGS services and Dr. Ullas Kolthur-Seetharam (Tata Institute of Fundamental Research, Mumbai) for his help in procuring miRNA sponge constructs. We acknowledge the following funding sources: Department of Biotechnology, Government of India (BT/PR1825/AGR/36/679/2011) and (BT/PR32456/BRB/10/1785/2019); Department of Science and Technology, Government of India (DST/CRG/2018/002753). S.N., A.D.Y., A.P., and P.K.N. are recipients of fellowships from University Grants Commission, Council for Scientific and Industrial Research, or Rajiv Gandhi National Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104297118/-/DCSupplemental.
Data Availability
The NGS data generated for this manuscript were uploaded to the Gene Expression Omnibus under one superseries with accession number GSE153544. The datasets used for analysis from studies published earlier (65) can be accessed using accession numbers GSM2418356, GSM2418357, and GSM2418369 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90958).
References
- 1.Lara-Astiaso D., et al., Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Javierre B. M., et al., Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu G., et al., Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T-cells. Immunity 48, 227–242.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magilnick N., Boldin M. P., Molecular Moirai: Long noncoding RNAs - mediators of HSC fate. Curr. Stem Cell Rep. 4, 158–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pongubala J. M. R., Murre C., Spatial organization of chromatin: Transcriptional control of adaptive immune cell development. Front. Immunol. 12, 633825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai A. Y., Kondo M., T and B lymphocyte differentiation from hematopoietic stem cell. Semin. Immunol. 20, 207–212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazaki K., Miyazaki M., Murre C., The establishment of B versus T cell identity. Trends Immunol. 35, 205–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambandam A., et al., Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 6, 663–670 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Pai S. Y., et al., Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Weber B. N., et al., A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikawa T., et al., An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Pui J. C., et al., Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Lin H., Grosschedl R., Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376, 263–267 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Pongubala J. M. R., et al., Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9, 203–215 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Boller S., Grosschedl R., The regulatory network of B-cell differentiation: A focused view of early B-cell factor 1 function. Immunol. Rev. 261, 102–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nechanitzky R., et al., Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat. Immunol. 14, 867–875 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Hu M., et al., Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto T., et al., Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Ebert M. S., Sharp P. A., Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel D. P., Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo H., Ingolia N. T., Weissman J. S., Bartel D. P., Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichhorn S. W., et al., mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koralov S. B., et al., Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860–874 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Brandl A., et al., The microprocessor component, DGCR8, is essential for early B-cell development in mice. Eur. J. Immunol. 46, 2710–2718 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Cobb B. S., et al., T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201, 1367–1373 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muljo S. A., et al., Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 202, 261–269 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landgraf P., et al., A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neilson J. R., Zheng G. X. Y., Burge C. B., Sharp P. A., Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21, 578–589 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., et al., Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 113, 4586–4594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchen S., et al., Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petriv O. I., et al., Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc. Natl. Acad. Sci. U.S.A. 107, 15443–15448 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt T. M., Zúñiga-Pflücker J. C., Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Ooi A. G. L., et al., MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. U.S.A. 107, 21505–21510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C. Z., Li L., Lodish H. F., Bartel D. P., MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Li Q. J., et al., miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129, 147–161 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Khalaj M., Tavakkoli M., Stranahan A. W., Park C. Y., Pathogenic microRNA’s in myeloid malignancies. Front. Genet. 5, 361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurkewich J. L., et al., The miR-23a∼27a∼24-2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 13, e1006887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajasekhar M., et al., Identifying microRNA determinants of human myelopoiesis. Sci. Rep. 8, 7264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su X., et al., miRNomes of haematopoietic stem cells and dendritic cells identify miR-30b as a regulator of Notch1. Nat. Commun. 4, 2903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontana L., et al., MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 9, 775–787 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Ru Y., et al., The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 42, e133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikawa T., Kawamoto H., Fujimoto S., Katsura Y., Commitment of common T/Natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J. Exp. Med. 190, 1617–1626 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izon D., et al., A common pathway for dendritic cell and early B cell development. J. Immunol. 167, 1387–1392 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Manz M. G., Traver D., Miyamoto T., Weissman I. L., Akashi K., Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood 97, 3333–3341 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Rumfelt L. L., Zhou Y., Rowley B. M., Shinton S. A., Hardy R. R., Lineage specification and plasticity in CD19- early B cell precursors. J. Exp. Med. 203, 675–687 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wada H., et al., Adult T-cell progenitors retain myeloid potential. Nature 452, 768–772 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Rothenberg E. V., T cell lineage commitment: Identity and renunciation. J. Immunol. 186, 6649–6655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zandi S., et al., Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, 15871–15876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pons-Espinal M., et al., Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Reports 8, 1046–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cursons J., et al., Combinatorial targeting by MicroRNAs co-ordinates post-transcriptional control of EMT. Cell Syst. 7, 77–91.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Bhaskaran V., et al., The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat. Commun. 10, 442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh H., Medina K. L., Pongubala J. M. R., Contingent gene regulatory networks and B cell fate specification. Proc. Natl. Acad. Sci. U.S.A. 102, 4949–4953 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothenberg E. V., et al., Transcriptional establishment of cell-type identity: Dynamics and causal mechanisms of T-cell lineage commitment. Cold Spring Harb. Symp. Quant. Biol. 78, 31–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Broughton J. P., Lovci M. T., Huang J. L., Yeo G. W., Pasquinelli A. E., Pairing beyond the seed supports microRNA targeting specificity. Mol. Cell 64, 320–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore M. J., et al., miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 6, 8864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brancati G., Großhans H., An interplay of miRNA abundance and target site architecture determines miRNA activity and specificity. Nucleic Acids Res. 46, 3259–3269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacobsen A., Wen J., Marks D. S., Krogh A., Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 20, 1010–1019 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.HafezQorani S., et al., Modeling the combined effect of RNA-binding proteins and microRNAs in post-transcriptional regulation. Nucleic Acids Res. 44, e83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doyle F., Tenenbaum S. A., Trans-regulation of RNA-binding protein motifs by microRNA. Front. Genet. 5, 79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Min K. W., et al., AUF1 facilitates microRNA-mediated gene silencing. Nucleic Acids Res. 45, 6064–6073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rothenberg E. V., Zhang J., Li L., Multilayered specification of the T-cell lineage fate. Immunol. Rev. 238, 150–168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inui M., Martello G., Piccolo S., MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 11, 252–263 (2010). [DOI] [PubMed] [Google Scholar]
- 63.DeKoter R. P., Singh H., Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288, 1439–1441 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isoda T., et al., Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171, 103–119.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Rie D., et al., An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 35, 872–878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The NGS data generated for this manuscript were uploaded to the Gene Expression Omnibus under one superseries with accession number GSE153544. The datasets used for analysis from studies published earlier (65) can be accessed using accession numbers GSM2418356, GSM2418357, and GSM2418369 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90958).