Significance
Double-stranded RNA (dsRNA) is the strongest-known “molecular signature” of viral infections that activates antiviral interferon responses and inflammation in human cells. Due to this activity, dsRNA-sensing pathways belong to the core interests of academic laboratories and pharma working on antiviral defenses and inflammatory diseases. dsRNAs do not always arise from viral infections. Mammalian cells can also produce their own dsRNAs. Precisely what are the endogenous dsRNAs that activate innate immune responses in human cells, and which receptors of the innate immune system sense endo-dsRNAs? Here, we find that human endo-dsRNAs can arise from jammed decay of intronic and intergenic transcripts folded as RNA duplexes. We show that the receptors OAS3/RNase L and PKR are important sensors of these dsRNAs.
Keywords: retrotransposon, dsRNA, RNase L, phosphorothioate, interferon
Abstract
Double-stranded RNA (dsRNA), a hallmark viral material that activates antiviral interferon (IFN) responses, can appear in human cells also in the absence of viruses. We identify phosphorothioate DNAs (PS DNAs) as triggers of such endogenous dsRNA (endo-dsRNA). PS DNAs inhibit decay of nuclear RNAs and induce endo-dsRNA via accumulation of high levels of intronic and intergenic inverted retroelements (IIIR). IIIRs activate endo-dsRNA responses distinct from antiviral defense programs. IIIRs do not turn on transcriptional RIG-I/MDA5/IFN signaling, but they trigger the dsRNA-sensing pathways of OAS3/RNase L and PKR. Thus, nuclear RNA decay and nuclear-cytosolic RNA sorting actively protect from these innate immune responses to self. Our data suggest that the OAS3/RNase L and PKR arms of innate immunity diverge from antiviral IFN responses and monitor nuclear RNA decay by sensing cytosolic escape of IIIRs. OAS3 provides a receptor for IIIRs, whereas RNase L cleaves IIIR-carrying introns and intergenic RNAs.
Sensing of double-stranded RNA (dsRNA) is an important mechanism of antiviral defense in cells of all higher vertebrates. Apart from viral infections, molecules of dsRNA can be produced endogenously as sources of natural small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs) involved in retrotransposon silencing in oocytes (1) and as stress signals inducing innate immune responses in somatic cells (2–5). Mechanisms of endo-dsRNA biogenesis remain unclear. Mammals lack RNA-dependent RNA polymerase (RdRp) and must use RdRp-independent routes (1). Reported mechanisms range from bidirectional DNA transcription (6) and transcriptional derepression of genomic repeat elements (4, 7) to formation of secondary structures within 3′-untranslated regions of messenger RNAs (mRNAs) due to in cis base pairing of inverted Alu elements present in more than 500 human mRNAs (8).
The normal levels of endo-dsRNAs are low, but they increase sufficiently to activate antiviral responses upon stress caused by cell treatment with DNA methyltransferase inhibitor 5-Aza-CdR, a US Food and Drug Administration (FDA)–approved drug used against hematologic neoplasms (4, 7). The 5-Aza-CdR inhibits DNA methylation and, after several days of treatment, leads to derepression of short interspersed nuclear elements and bidirectional transcription of near-centromeric satellite DNA repeats (9). Base pairing of the resulting complementary RNAs forms endogenous dsRNAs, which have been shown to suppress cancerous cells through an apoptotic interferon (IFN) response (9).
In human, intracellular dsRNAs are recognized via three major pathways. The first involves the receptors MDA5 and RIG-I, which activate type I and type III IFN responses (10, 11). Two additional pathways involve the protein kinase PKR, which arrests global translation initiation (12), and the dsRNA-binding enzymes OAS1, OAS2, and OAS3 (OASs), which activate cytosolic mRNA decay (13, 14). The OASs are structurally related to the antiviral dsDNA sensor cGAS (15), but they function as specific receptors for dsRNA. Upon binding dsRNA, the OASs synthesize 2′,5′-linked iso-oligoadenylates (2–5A) (2, 16–18), which represent high-affinity activators of the 2–5A receptor endonuclease, RNase L (19, 20). The RNase L•2–5A complex cleaves intracellular single-stranded RNAs (ssRNAs) at consensus UN^N sites (N is A, G, C, or U) (21, 22), leading to 2–5A-mediated mRNA decay (2-5AMD) that tilts translation toward preferential synthesis of IFNs (13, 14). The antiviral roles of the dsRNA-sensing pathways have been investigated in detail, whereas their roles in sensing endo-dsRNAs are unclear. Emerging evidence suggests that endo-dsRNA responses are critically important and can be lethal to cells as well as whole embryos (23, 24).
Endo-dsRNAs are continuously removed by the adenosine-to-inosine editing enzyme adenosine deaminase 1 (ADAR1) (25). ADAR1 targets predominantly Alu elements in polymerase II (pol II) transcripts (3, 26) and converts AU base pairs into single-stranded bulges (Fig. 1A). ADAR1 is an essential enzyme that protects from Aicardi-Goutieres autoimmune syndrome (25) as well as from aberrant IFN signaling via the dsRNA sensor MDA5, which causes embryonic death (23). Endo-dsRNAs emerge as components of mammalian cells, for which we lack sufficient knowledge of biogenesis, regulation, and recognition by the innate immune system. Here, we describe a cellular mechanism protecting from accumulation of excessively high levels of endo-dsRNAs in human cells. We describe the precise molecular origin and innate immune recognition of these endo-dsRNAs.
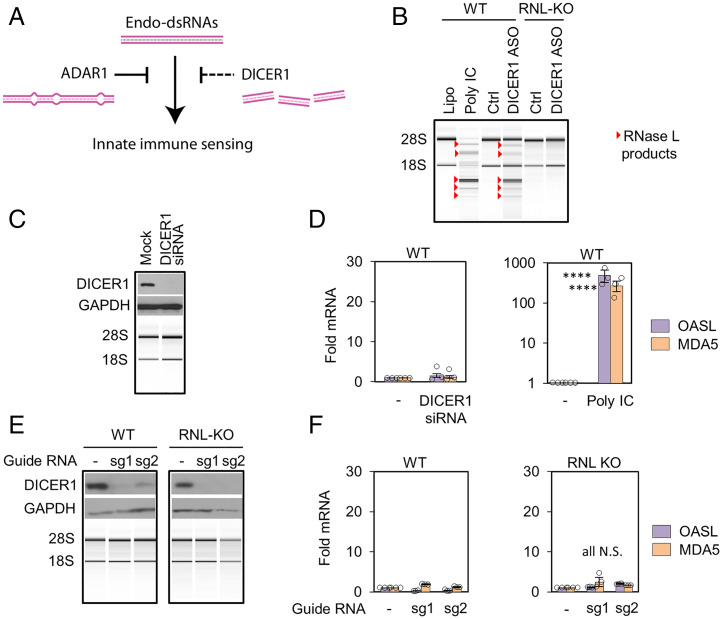
Fig. 1.
Testing endo-dsRNA suppressor function of DICER1. (A) Illustration of endo-dsRNA sensing and removal routes. (B) Total RNA profiling by BioAnalyzer in A549 cells. ASO (50 nM for 32 h) or poly IC control (1 µg/mL for 6 h) was used. (C) DICER1 knockdown for 48 h with siRNA. (D) Analysis of dsRNA-inducible antiviral genes OASL and MDA5 by qPCR. (E) DICER1 KO characterization by Western blotting (WB) and BioAnalyzer. (F) Profiling OASL and MDA5 in DICER1 KO cells. Two experiments were done for each of the two guide RNAs and three for remaining figures. Statistical significance (P) from Welch’s two-tailed unpaired t test: P ≤ 0.05*, 0.01**, 0.001***, 0.0001****; and N.S., nonsignificant.
Results
DICER1 as Endo-dsRNA Suppressor Candidate.
RNase L activation upon loss of ADAR1 (5) inspired us to use RNase L for testing the hypothesis that the endonuclease DICER1 acts as a second direct suppressor of endo-dsRNAs. It has been reported that DICER1 fragments endo-dsRNAs formed by convergent bidirectional pol II transcription to prevent immune responses and that it degrades Alu RNA to protect retinal cells from endo-dsRNA toxicity and death (6, 27). DICER1 and ADAR1 may enable two parallel strategies of endo-dsRNA neutralization (Fig. 1A). To test this model, we knocked down DICER1 in human A549 cells using the published DICER1 antisense oligonucleotide (ASO) (27) (SI Appendix, Table S1). DICER1 ASO activated RNase L as revealed by specific RNA fragments derived from cleavage of 18S and 28S ribosomal RNA (rRNA). The same pattern of cleaved fragments forms in response to exogenously supplied viral dsRNA mimic (polyinosinic polycytidylic acid [poly IC]; Fig. 1B). The rRNA cleavage was absent in RNase L knockout (KO) cells (Fig. 1B and SI Appendix, Fig. S1A), confirming activation of the OAS/RNase L pathway and demonstrating that DICER1 ASO induces endo-dsRNAs.
To independently show the role of DICER1 in RNase L regulation, we depleted DICER1 by RNA interference (RNAi) knockdown (Fig. 1C and SI Appendix, Fig. S1B). In surprising discord with the DICER1 ASO data, DICER1 RNAi failed to activate RNase L, suggesting that loss of DICER1 does not induce endo-dsRNAs (Fig. 1C and SI Appendix, Fig. S1B). The absence of dsRNA accumulation with DICER1 RNAi treatment was further supported by unchanged expression of dsRNA-inducible antiviral genes, MDA5 and OASL, which exhibit >100-fold increase in the control treatment with poly IC (Fig. 1D).
To reconcile the discrepancy between RNAi and ASO, we used a third approach and knocked out DICER1 with CRISPR/Cas9 using lentiviral delivery of two different guide RNAs (sg1 and sg2; Fig. 1E and SI Appendix, Table S2 and Fig. S1C). In contrast to ADAR1 KO that could not be generated in A549 cells due to endo-dsRNA toxicity (5), DICER1 KOs were viable, indicating that DICER1 loss does not cause endo-dsRNA buildup. Neither RNase L activation nor IFN response were detected in DICER1 KO cells (Fig. 1 E and F and SI Appendix, Fig. S1C), consistent with RNAi experiments but contradicting the DICER1 ASO data. To reconcile the opposing outcomes of different techniques to ablate DICER1, we proposed that DICER1 ASO induces endo-dsRNAs via a DICER1-independent mechanism.
Phosphorothioate Oligonucleotides Are Potent Inducers of Endo-dsRNAs.
To elucidate the mechanism of endo-dsRNA induction by DICER1 ASO, we designed an ASO targeting a nonessential gene unrelated to dsRNA biogenesis and innate immune dsRNA sensing. We selected clathrin adaptor protein GGA2 involved in Golgi protein trafficking. Transfection of GGA2 ASO activated RNase L, revealing that DICER1 sequence is not required for triggering endo-dsRNAs (Fig. 2 A and B and SI Appendix, Fig. S2 A and B). Both oligonucleotides that induced endo-dsRNAs, DICER1 ASO and GGA2 ASO, have 2′-OMe and phosphorothioate (PS) modifications (Fig. 2 A and B), which distinguish them from control DNA (Fig. 2 and SI Appendix, Table S1). To determine which of these two modifications cause endo-dsRNA accumulation, we synthesized ASOs modified individually with 2’-OMe versus PS groups (SI Appendix, Table S1). The 2′-OMe DICER1 ASO did not activate RNase L, whereas PS DICER1 ASO did (Fig. 2B and SI Appendix, Fig. S3A). To verify that PS modification is sufficient for endo-dsRNA induction, we designed NON-DICER1 PS DNA, which contains PS modifications but does not pair with DICER1 and other human mRNAs (SI Appendix, Table S1). NON-DICER1 PS DNA activated RNase L (SI Appendix, Fig. S3B).
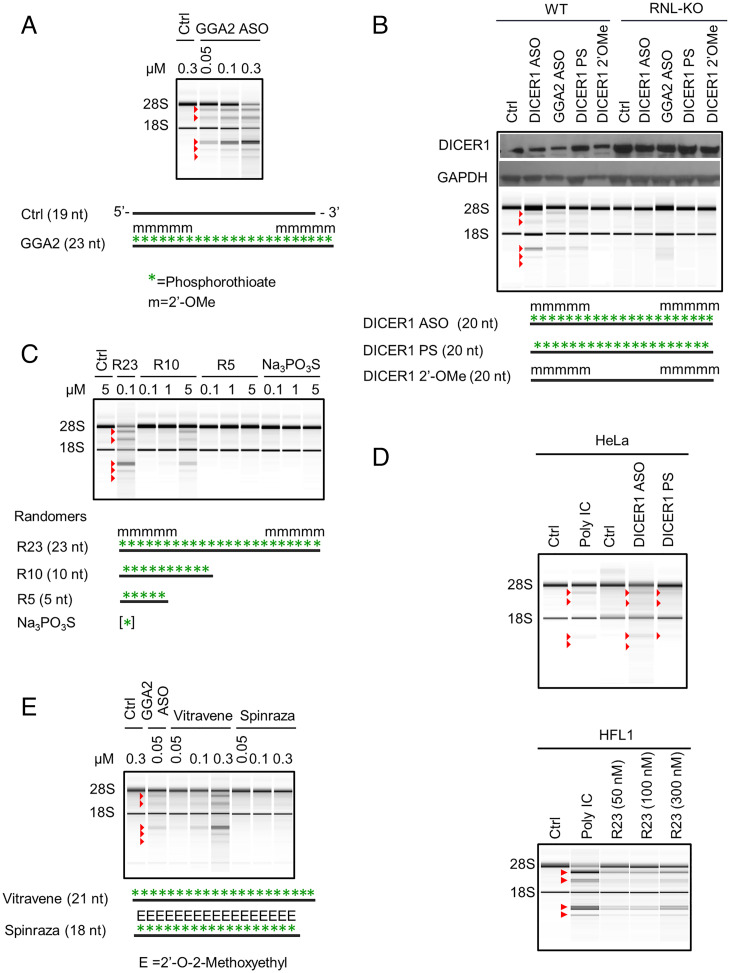
Fig. 2.
PS DNAs induce endo-dsRNAs that activate RNase L. (A) Activation of RNase L pathway by GGA2 ASO. A549 cells were treated with the ASO for 24 h. (B) Profiling endo-dsRNA/RNase L activation by ASOs in WT versus RNase L-KO cells, conducted as in A. (C) Endo-dsRNA/RNase L activation by PS randomers and sodium PS. (D) PS-induced activation of RNase L pathway in HeLa and normal lung fibroblast cells (HFL1). Cells were treated with 50 nM DICER1 ASOs and with indicated doses of R23 for 24 h. Controls with poly IC (1 μg/mL) were done for 7 h in HeLa cells and 4 h in HFL1. (E) Endo-dsRNA/RNase L activation effect of Vitravene and Spinraza. Work was conducted as in A.
These results indicate that PS DNAs induce endo-dsRNA by a sequence-independent mechanism. To validate this conclusion, we synthesized a series of randomer PS DNAs comprised of equal mixtures of A, G, C, or T at each position (Fig. 2C and SI Appendix, Table S1). Random 23-mer PS DNA activated RNase L at 100 nM concentrations (Fig. 2C) and exhibited improved activity when PS modification was combined with 2′-OMe group (SI Appendix, Fig. S3C). These data show that 2′-OMe group is not essential, but it enhances R23 potency presumably by stabilizing PS DNA against intracellular degradation. Random PS DNA with 10 nucleotides (R10) was still capable of inducing endo-dsRNA and activating RNase L (Fig. 2C and SI Appendix, Fig. S3D), although only at 10 to 50-fold higher concentrations compared to the 23-mer. The pentamer R5 and sodium thiophosphate were inactive at any concentration tested (Fig. 2C and SI Appendix, Fig. S3 D and E). PS DNAs induce endo-dsRNAs within hours compared to days needed with 5-Aza-CdR (4, 7), indicating that PS DNAs and 5-Aza-CdR use different mechanisms. The mechanism engaged by PS DNAs is not limited to A549 cells. RNase L activation takes place in HeLa cells (less pronounced rRNA cleavage reflects low endogenous RNase L levels in HeLa) and in noncancer lung epithelial cells HFL1 (Fig. 2D and SI Appendix, Fig. S3F).
PS Drugs Vitravene and Spinraza Show Contrasting Ability to Induce Endo-dsRNAs.
A number of PS oligonucleotides have been subject to clinical trials and several were approved by the FDA (28–30). Our data predict that these oligonucleotides may induce endo-dsRNAs and RNase L activity as an unknown part of their therapeutic mechanism or as a side effect. We obtained the oligonucleotide drugs Vitravene and Spinraza and tested their ability to activate RNase L. Vitravene, an antiviral drug for patients with AIDS, readily activated RNase L (Fig. 2E and SI Appendix, Fig. S3G), whereas Spinraza, a drug used to treat spinal muscular atrophy, did not (Fig. 2E and SI Appendix, Fig. S3G). Both oligos contain PS substitutions throughout the backbone, indicating that the absence of endo-dsRNA induction by Spinraza is due to its 2′-O-methoxyethyl groups. Therefore, therapeutic oligonucleotides containing PS modifications can induce endo-dsRNAs preventable by sugar chemical modifications.
Endo-dsRNAs Are Sensed via OAS3/RNase L and Are Undetected by Any dsRNA Receptors that Induce IFN Response.
PS DNAs activate RNase L comparably to poly IC treatment (i.e., leading to robust 28S rRNA cleavage), suggesting that endo-dsRNAs accumulate to high levels and should activate all relevant innate immune pathways involved in endo-dsRNA sensing. To test these pathways, we examined the activity of the major cytosolic dsRNA sensors: PKR, OAS1, OAS2 and OAS3, and MDA5/RIG-I. PS DNA induced time-dependent phosphorylation of the PKR substrate, translation initiation factor eIF2α (Fig. 3A and SI Appendix, Fig. S4A). Phosphorylation of eIF2α started at the same time as activation of RNase L, consistent with endo-dsRNA being the common activator of both pathways. As expected, the time-dependent increase in eIF2α phosphorylation no longer occurred in PKR-KO cells (Fig. 3A and SI Appendix, Fig. S4A).
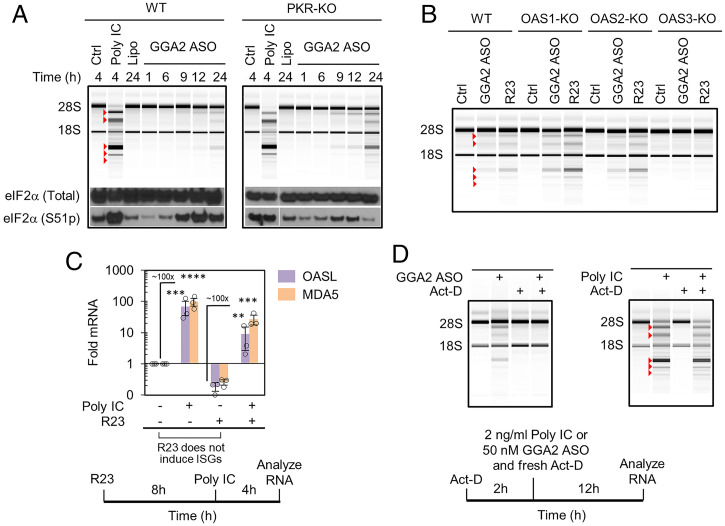
Fig. 3.
Transcriptional dependence and innate immune receptor profile of PS DNA–induced endo-dsRNA. (A) The effect of GGA2 ASO on phosphorylation of eIF2α in WT and PKR-KO cells. A549 cells were treated with 1 µg/mL poly IC or 50 nM GGA2 ASO. (B) Profiling RNase L activation by GGA2 ASO (50 nM for 24 h) in A549 cells with individual KO of each OAS. (C) Profiling IFN response by qPCR of IFN-stimulated genes MDA5 and OASL. A549 cells were treated with 50 nM R23 and 1 µg/mL poly IC. (D) Act-D (1 μg/mL) does not inhibit RNase L activation by externally supplied dsRNA poly IC (2 ng/mL) but blocks endo-dsRNA induction by GGA2 ASO (50 nM) in A549 cells.
To test the role of the OAS receptors, we examined cells with individual OAS proteins knocked out by CRISPR (31). Cells lacking OAS1 or OAS2 retained normal RNase L activation. In contrast, loss of OAS3 disabled RNase L (Fig. 3B and SI Appendix, Fig. S4B). We have shown previously that OAS1 requires relatively short dsRNAs with ≥17 bp for optimal binding but has weak dsRNA affinity, whereas OAS3 senses longer dsRNAs (≥50 bp) and has strong dsRNA affinity (2). The identification of OAS3 as the key sensor driving RNase L indicates that endo-dsRNAs that appear in human cells during our experiments have length ≥ 50 bps.
To test the engagement of MDA5/RIG-I, which transcriptionally up-regulate IFNs and IFN-stimulated genes (ISGs) in the presence of cytosolic dsRNAs, we measured changes in expression of two bona fide ISGs: OASL and MDA5 itself. Exogenously supplied dsRNA (poly IC) activated both RNase L and a robust IFN response revealed by ∼100-fold up-regulation of ISGs. In contrast, PS DNA (R23) did not trigger IFN response (Fig. 3C and SI Appendix, Figs. S5 A–D and S6), while activating RNase L as expected (SI Appendix, Fig. S5 A–D). We confirmed that R23 does not block transcriptional dsRNA-induced IFN signaling by combining poly IC and R23 treatments (Fig. 3C and SI Appendix, Fig. S5B). Analogous to R23, the FDA-approved phosphorothioate drugs Vitravene and Spinraza too did not activate IFN signaling (SI Appendix, Fig. S5E).
Our observations using the OASL/MDA5 readout were in line with cell-wide analysis of IFN responses by RNA sequencing (RNA-seq). Under conditions producing robust RNase L activation, treatments with R23 did not induce IFNs α, β, λ, or γ and did not up-regulate ISGs (SI Appendix, Fig. S5F and Dataset S1; Gene Expression Omnibus [GEO] ID GSE143638). These data suggest that all cytosolic dsRNA receptors driving transcriptional IFN responses remain mute in the presence of endo-dsRNAs.
Endo-dsRNAs Accumulate via Actively Ongoing Transcription.
To define whether endo-dsRNAs arise from premade cellular RNAs or from new transcripts that are produced after PS DNA addition, we treated the cells with the transcriptional inhibitor actinomycin D (Act-D). We established conditions that block transcription by using RT-qPCR analysis of pol II-transcribed proto-oncogene cMYC (SI Appendix, Fig. S7A). Under these conditions cells treated with Act-D stopped producing endo-dsRNA in response to PS DNAs, as indicated by the absence of rRNA cleavage (Fig. 3D and SI Appendix, Fig. S7B). In contrast, Act-D did not inhibit RNase L activity when dsRNA was supplied exogenously as poly IC (Fig. 3D and SI Appendix, Fig. S7C). These data indicate that PS DNAs activate the RNase L pathway via de novo transcribed endo-dsRNAs, which arise subsequent to the addition of PS DNA.
PS DNAs Block Decay of Introns and Intergenic RNAs.
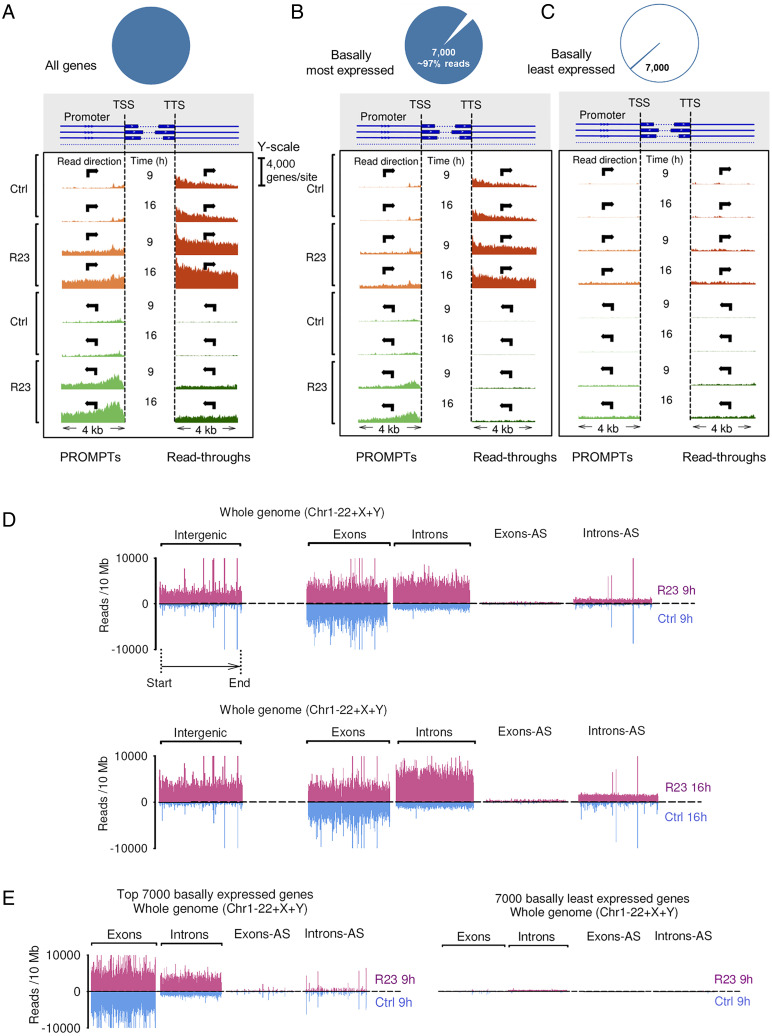
To identify the endo-dsRNAs and elucidate their mechanism of production, we analyzed the transcriptome of cells treated with R23 by ribo-zero RNA-seq. R23 treatment caused accumulation of promoter upstream region RNAs (PROMPTs) transcribed in both directions as well as RNA polymerase read-through transcripts (Fig. 4A). The accumulation of PROMPTs has been previously observed upon disruption of nuclear decay (32), suggesting that R23 inhibits nuclear RNA decay. Further analysis supported this mechanism. PROMPTs and read-through RNAs arise from genes that are actively expressed in normal cells (Fig. 4B) but not from genes that are basally repressed (Fig. 4C). These data indicate that PS DNAs and 5-Aza-CdR indeed work via different mechanisms. In contrast to 5-Aza-CdR, PS DNAs do not cause gene derepression, but they disrupt RNA homeostasis by inhibiting nuclear decay of nonexonic RNAs, which leads to accumulation of rogue RNAs as a putative source of endo-dsRNAs.
Fig. 4.
RNA expression profiling by RNA-seq. (A) Whole-genome RNA-seq profiles of promoter upstream regions (PROMPTs) and transcription read-throughs. TSS marks transcription start site. y axis shows count of genes with experimentally observed reads, calculated individually for every nucleotide position. (B) Data for 7,000 most abundant RNAs (∼97% of RNA-seq reads). (C) Data for 7,000 least abundant RNAs (<0.1% of RNA-seq reads). (D) Whole-genome expression profiles for exons, introns, and intergenic regions. Blue: naïve A549 cells, magenta: cells treated with R23 for 9 and 16 h. Mapped reads from ribo-zero RNA-seq are plotted. (E) Data in A for 7,000 most highly abundant and 7,000 least basally abundant genes.
Further analysis of the RNA-seq data revealed that R23 activates genome-wide accumulation of introns and intergenic transcripts (Fig. 4D and SI Appendix, Fig. S8). As we observed with PROMPTs and read-throughs, the up-regulated intronic and intergenic transcripts originated from basally actively expressed genes (Fig. 4E). This analysis confirms our conclusion that R23 does not derepress genomic locations but blocks degradation of normally transcribed but rapidly cleared RNAs (Fig. 4E). To test whether the activity of PS DNAs could be recapitulated by targeted disruption of known nuclear decay machines, we knocked down nuclear exosomal proteins (32) and nuclear exonuclease involved in RNA decay, XRN2 (33). The knockdowns were insufficient for activation of RNase L and recapitulation of the effect of PS DNAs (SI Appendix, Figs. S9–S11). Our current data suggest that PS DNAs may inhibit different, perhaps unknown, components of nuclear RNA decay machine or inhibit proteins responsible for nuclear retention and sorting of introns and intergenic transcripts, leading to their escape from nuclear decay to the cytosol. Thus, the RNA-binding protein target (or targets) of PS DNAs responsible for endo-dsRNA up-regulation remains to be identified, and based on our data, the target protein must 1) require more than 5 but less than 11 nucleotides for binding, 2) have a long oligonucleotide binding tract such that 23-mers bind better than 10-mers, 3) bind RNA without forming important interactions with 2′-OH groups, and 4) not tolerate the bulky 2′ groups present in Spinraza.
Introns Leak to the Cytosol and Encode Endo-dsRNAs That Activate RNase L Without Inducing Transcriptional IFN Response.
Introns represent the largest RNA pool up-regulated by PS DNAs (Fig. 4D). If introns are the source of endo-dsRNAs in our study, nuclear compartmentalization could physically separate them from the cytosolic receptors RIG-I/MDA5 and explain the absence of IFN responses to endo-dsRNAs. Activation of cytosolic RNase L would still be possible via the nuclear pool of OAS3 (34) and nuclear-cytosolic diffusion of 2–5A (47). To elucidate subcellular localization of the accumulating introns, we designed a series of qPCR primers that detect unspliced, spliced, intronic, and total forms of mRNA encoding alpha subunit in integrin M (ITGAM; Fig. 5A). ITGAM belongs to the group of mRNAs with the strongest up-regulated introns (>500-fold increase with R23, GEO GSE143638 and Dataset S1). Subcellular fractionation revealed intron-containing forms of ITGAM both in the nucleus and in the cytosol (Fig. 5A and SI Appendix, Fig. S12 A and B). The strongest induction was observed for the cytosolic ITGAM intron, indicating intron leakage to the cytoplasm. Fractionation data were confirmed using confocal microscopy with cell staining by dsRNA-specific J2 antibody (35). R23 treatments promoted dsRNA accumulation both in the nucleus and in the cytosol (SI Appendix, Fig. S12C). These results suggest that introns can become accessible to the cytosolic receptors MDA5 and RIG-I and that IFN response could be absent for reasons other than subcellular compartmentalization, such as a lack of intronic endo-dsRNA recognition by RIG-I and MDA5.
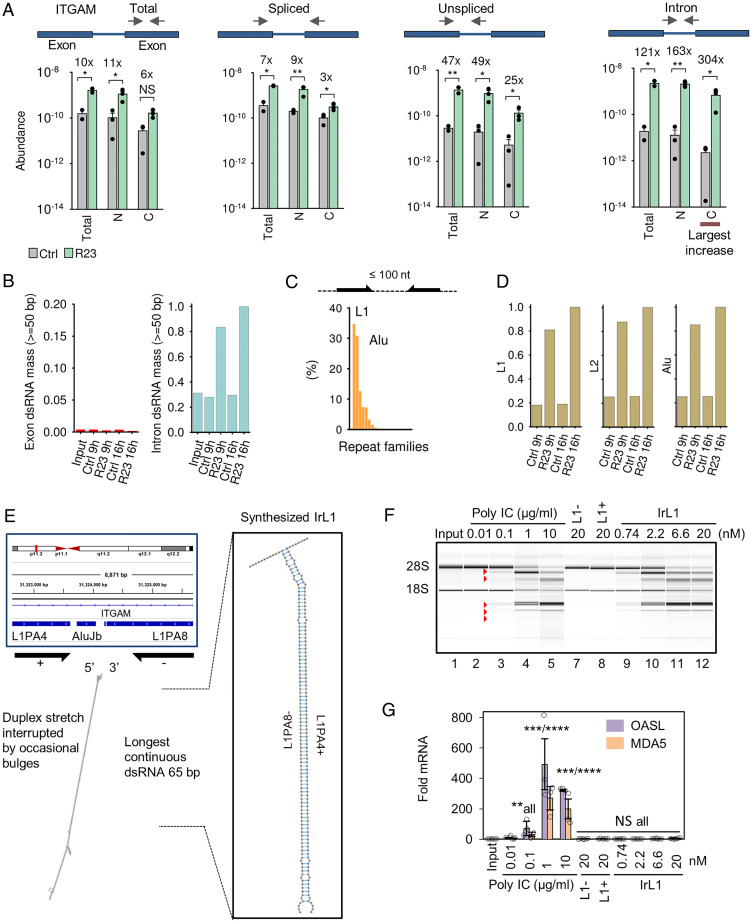
Fig. 5.
Tracing the molecular origin of endo-dsRNAs. (A) Subcellular fractionation of ITGAM in R23-treated A549 cells. C, cytosolic; N, nuclear. (B) Calculated abundance of self-dsRNA in controls versus R23-treated A549 cells (Dataset S1). Scale is relative to introns in R23-treated cells at 16 h. (C) Calculated abundance of ≥50 bp dsRNAs formed by inverted repeats separated by ≤100 nt in the human genome. (D) Observed expression of introns carrying L1, L2, and Alu elements. (E) Double-stranded region within a strongly induced intron mapping to ITGAM and structure of IrL1. (F, G) IrL1 activates dsRNA/RNase L but not IFN response. Equal amounts of RNA by OD260 were used for qPCR normalization.
To assess dsRNA content within introns, we identified ≥50 bp duplexes required for OAS3 activation by mapping sequence complementarity across the human transcriptome (Methods; Dataset S2). We used these data and RNA-seq to estimate total dsRNA levels in A549 cells treated with R23. Our analysis showed that exons encode relatively few endo-dsRNAs, which do not increase with R23 (Fig. 5B). In contrast, introns contain vast amounts of endo-dsRNAs that become up-regulated by R23. The ≥50 bp dsRNA duplexes within introns are formed by inverted pairs of homologous retroelements (repeats). Using University of California, Santa Cruz (UCSC) repeat data (36), we identified L1 and Alu as predominant repeats (SI Appendix, Fig. S12D) and also as major types of retroelements present as proximal inverted pairs (Fig. 5C). Transcripts with intronic repeats are induced by R23 similar to dsRNA levels, consistent with these repeats being the source of endo-dsRNAs (Fig. 5D versus 5B). Examination of the largest ITGAM intron identified repeats from L1, L2, and Alu families (SI Appendix, Fig. S13), including an inverted L1 pair forming an extended endo-dsRNA protrusion with 65 bp uninterrupted duplex (Fig. 5E). This length should be sufficient for optimal activation of OAS3 (2), suggesting that the L1 duplex is representative of endo-dsRNA activating RNase L. A key prediction of this model is that such inverted intronic repeats should activate RNase L without inducing IFN responses.
To test this prediction, we obtained inverted intronic L1 repeat (IrL1) via total chemical synthesis of each L1 element with single-stranded overhangs, followed by annealing (Fig. 5E). We have shown previously that chemical RNA synthesis is necessary for interpretable analysis of innate immune responses to dsRNA (2). The common method of RNA production via transcription with T7 RNA polymerase generates spurious dsRNA byproducts, which contaminate RNA samples and trigger unintentional dsRNA responses. Chemically synthesized RNAs do not show this spurious activity (2). The individual single-stranded L1 elements did not activate RNase L and did not induce IFN response, as expected (Fig. 5F, lanes 7 to 8; SI Appendix, Fig. S14A). Duplex IrL1 activated RNase L at nanomolar concentrations and exhibited the same potency as a high dose of poly IC (lanes 9 to 12 versus 2 to 5). However, in contrast to poly IC, IrL1 did not induce IFN response (Fig. 5G).
To confirm that the size of L1 elements in our test was sufficient, in principle, for IFN induction, when L1 forms a perfect (unnatural) duplex, we transcribed L1 with T7 RNA polymerase as a control. The resulting T7-dsL1 transcript forms a perfectly base-paired dsRNA with 112 bps, which no longer mimics intronic inverted retroelements but resembles viral dsRNAs. T7-dsL1 activated both RNase L and strong IFN response, as expected (SI Appendix, Fig. S14B). These results validate the RNase L pathway as a sensor of inverted retroelements that are ubiquitous within introns and intergenic RNAs, and confirm that these intronic and intergenic inverted retroelements (IIIRs) are undetected by antiviral IFN signaling.
RNase L Can Access and Cleave Introns and Intergenic Transcripts.
The presence of an exclusively dsRNA-specific sensor (OAS3) and an exclusively ssRNA-specific processor (RNase L) within the RNase L pathway suggests that this pathway could recognize RNA molecules that combine dsRNA and ssRNA (37), which matches the composition of introns and intergenic regions packed with retroelements (SI Appendix, Fig. S15A). Intronic and intergenic transcripts that escape into the cytosol therefore could serve not only as activators, but also as targets of the RNase L pathway. This model can explain the long-paradoxical UN^N (22) sequence preference of RNase L by active site complementarity to distinct prevalence of U within introns, particularly within introns that encode endo-dsRNAs (Fig. 6A). U/A enrichment is also observed for endo-dsRNA–rich intergenic transcripts (SI Appendix, Fig. S15B).
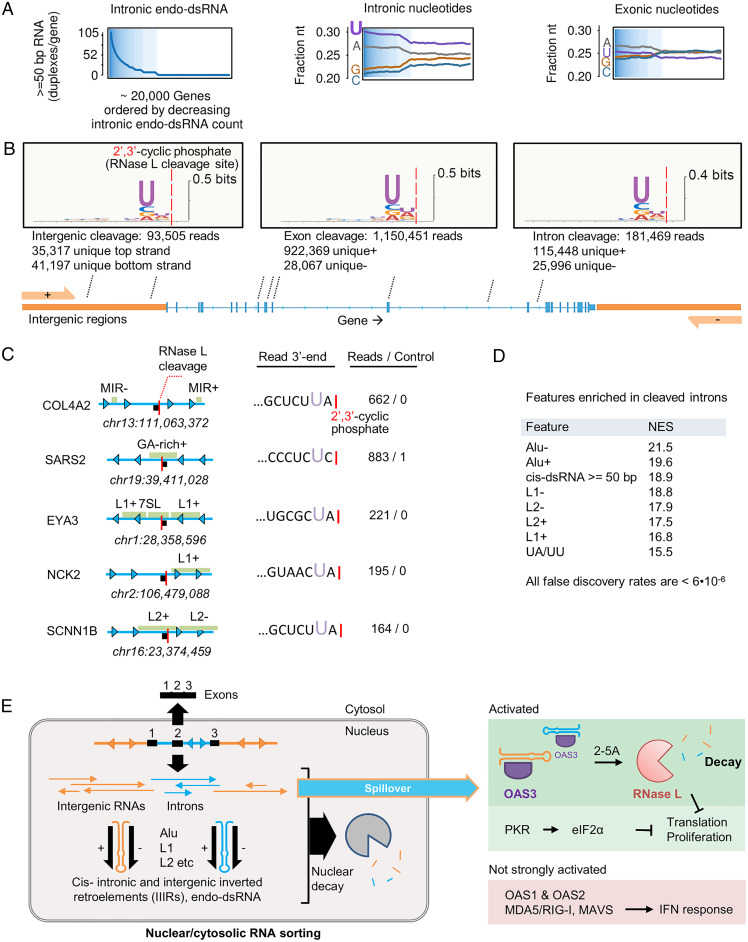
Fig. 6.
Direct detection of intron and intergenic cleavage by RNase L in A549 cells. (A) Intronic nucleotide composition versus dsRNA content. Exons are shown for comparison. (B) LRtcB-seq analysis of A549 cells treated with poly IC for 1 h reveals RNase L-mediated RNA cleavage within exons, introns, and intergenic regions. The UN^N consensus of RNase L is observed for all transcript types shown. (C) Examples of strongly cleaved sites mapping to human introns. (D) GSEA enrichment analysis demonstrating preferential cleavage of introns enriched in repeats (L1, L2, Alu), dsRNA, and UA/UU sequences. (E) Proposed scheme for IIIR regulation and innate immune sensing.
To test whether RNase L can, in principle, access introns and intergenic RNAs and whether it can cleave these RNAs, we used long RtcB RNA-seq (LRtcB RNA-seq) (13). Previously, we applied LRtcB RNAs-seq to map RNase L cleavage sites within human 18S and 28S rRNA. Here, we extended our analysis to find reads mapping to exons, introns, and intergenic regions and generated by RNase L during response to poly IC. The advantage of using poly IC for activating RNase L during this experiment is that, compared to PS DNA, poly IC does not disrupt RNA homeostasis and preserves naturally low levels of cytosolic intronic/intergenic transcripts inside the cells to better approximate nonabundant physiological RNA targets of RNase L. We identified more than a million nonribosomal reads with cleavage sites exhibiting global UN^N consensus and mapping to exons, introns, and intergenic regions, including PROMPTs and read-throughs (Fig. 6B and SI Appendix, Fig. S16). Nearly 20% of all pol II reads mapped to intronic and intergenic RNAs, with cleavage sites located near retroelements (Fig. 6C). Using functional enrichment analysis (38, 39), we established that introns with mapped cleavage sites have elevated counts of L1, L2, and Alu retroelements, endo-dsRNA duplexes, and UA/UU dinucleotides that are the cognate cleavage sites of RNase L (Fig. 6D and SI Appendix, Fig. S17). These data show that intronic and intergenic transcripts serve as activators of OAS3 and as substrates of the downstream receptor RNase L. Supporting a role of OAS3/RNase L in intron homeostasis and decay, both OAS3 and RNase L are basally expressed in human cell lines and in human tissues (SI Appendix, Fig. S18), whereas our fractionation RNA-seq found that RNase L KO cells accumulate cytosolic introns (SI Appendix, Fig. S19).
Discussion
We show that nuclear RNA decay and the associated nuclear-cytosolic RNA sorting suppress endo-dsRNAs and protect human cells from rogue innate immune signaling. This protective barrier can be disrupted by PS oligonucleotides, leading to activation of endo-dsRNA surveillance mechanisms. Inhibition of RNA sorting is observed with a broad range of PS DNAs longer than ∼10 nts, including randomers and the antiviral drug Vitravene, but not with Spinraza bearing a bulky 2′ modification. PS DNAs induce endo-dsRNA via a different mechanism than that of the previously described endo-dsRNA–inducing drug, 5-Aza-CdR. Whereas 5-Aza-CdR alters transcription and requires several days for detectable endo-dsRNA production, PS DNAs within several hours induce introns and intergenic transcripts to levels that exceed the total mass of all cellular mRNAs.
Reported studies of endo-dsRNAs that used 5-Aza-CdR treatments or ablation of ADAR1 often described activation of IFN signaling (7, 9, 23), whereas our data suggest that IFN response is not the major mechanism activated by endo-dsRNAs, at least with respect to IIIRs in human A549 cells, where IIIRs are sensed primarily via PKR and OAS3/RNase L. Based on these findings, we propose that PKR and OAS3/RNase L represent a specialized part of the innate immune system tuned for sensing introns and intergenic transcripts that have escaped decay in the nucleus (Fig. 6E).
The lack of IFN production in response to IIIRs is in line with antiviral purposes of IFNs. IFN signaling serves to alert neighboring cells of an upcoming virus. Endo-dsRNAs do not pose the risk of spreading infectious material to nearby tissues, eliminating the need for paracrine defense mechanisms provided by IFNs. Mechanistically, the absence of IFN response could reflect structural properties of IIIRs. Cytosolic dsRNAs up-regulate IFNs via two related helicases: RIG-I and MDA5. RIG-I recognizes blunt-end dsRNAs with 5′-triphosphate (41), whereas MDA5 recognizes long ≥500 bp dsRNAs (42). IIIRs lack blunt ends and 5′-terminal triphosphate, and form smaller than 500 bp dsRNA stretches. Endo-dsRNA length is sufficient for optimal activation of OAS3 (2) but not for activation of MDA5. We must note that our model dsRNA IrL1 is removed out of the context of full introns, which could limit its potency to activate MDA5 and alternatively explain the absence of MDA5 signaling with IrL1 (40). Nevertheless, the absence of IFN response in cells filled with full introns (Fig. 3 and SI Appendix, Fig. S5) argues against this alternative. Our data suggest that transcription-activating dsRNA receptors evade endo-dsRNA, perhaps as a mechanism to minimize paracrine signaling via IFNs in the absence of genuine viruses.
The two pathways activated by IIIRs (Fig. 6E) inhibit global translation as well as reprogram protein synthesis (13, 14, 43), suggesting that translational control is a desired outcome in cells that fail to resolve intronic and intergenic transcripts. We propose that IIIRs are the physiological drivers of RNase L activity, which may account for many roles of this protein in the absence of viral infections. RNase L stimulates adipocyte differentiation, impedes cellular growth and proliferation, as well as acts as a potent inhibitor of cell migration and metastasis (39, 44–46). Our work suggests that these processes may involve imbalances with nuclear decay and changes in cytosolic acessibility of IIIRs. By directly detecting RNase L cleavage sites within human introns and intergenic RNAs, we show that the pathway of RNase L serves not only as a sensor but also as a cleavage system for these transcripts that carry IIIRs.
Methods
Chemically modified DNA oligonucleotides used in this study were purchased from IDT and are listed in SI Appendix, Table S1. Human cells were grown using American Type Culture Collection or provider-recommended conditions in minimum essential media + 10% fetal bovine serum (FBS) (HeLa) or Roswell Park Memorial Institute (RPMI) media + 10% FBS (A549), F12K medium + 10% FBS (HFL1). All media were purchased from Gibco, Life Technologies. HeLa and HFL1 cells were a gift from the laboratory of Yibin Kang (Princeton University). Wild type (WT), RNase L KO, PKR-KO, and OAS KOs A549 were a gift from the laboratory of Susan Weiss (University of Pennsylvania). RNA-seq experiments and data processing were conducted as described previously by our group (47). Briefly, total RNA was extracted using RNeasy kit (Qiagen). RNA integrity was verified by an RNA 6000 Nano Chip using BioAnalyzer and 2100 Expert software (Agilent Technologies). rRNA was depleted from the total RNA by hybridization to bead bound rRNA probes. Subsequent steps involved fragmentation, adapter ligation, PCR amplification, and sequencing on Illumina HiSeq 2000 platform. The methodology is described in detail in SI Appendix, Methods.
Acknowledgments
We thank Prof. Susan Weiss (University of Pennsylvania School of Medicine) for the gift of KO A549 cells (RNase L, PKR, OAS1/2/3). We thank Dr. Wei Wang at Princeton University High Throughput Sequencing and MicroArray Facility and facility staff and all members of the A.K. laboratory for helpful comments on the manuscript. We would like to also thank Dr. Gary Laevsky at the Confocal Imaging Facility and Nikon Center of Excellence at the Department of Molecular Biology at Princeton University. This study was funded by NIH Grant 1R01GM110161-01 (to A.K.), Sidney Kimmel Foundation Grant AWD1004002 (to A.K.), Burroughs Wellcome Foundation Grant 1013579 (to A.K.), The Vallee Foundation (A.K.), National Institute of General Medical Services (NIGMS) Training Grant 5T32GM007388 (to S.R. and K.S.-K.), and F99 CA212468-01 (to S.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102134118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) (GSE143638, GSE171296). Previously published data were used for this work (RNA-seq datasets from GEO database: GSE131130, GSE123034).
References
- 1.Watanabe T., et al. , Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Donovan J., Whitney G., Rath S., Korennykh A., Structural mechanism of sensing long dsRNA via a noncatalytic domain in human oligoadenylate synthetase 3. Proc. Natl. Acad. Sci. U.S.A. 112, 3949–3954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung H., et al. , Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiappinelli K. B., et al. , Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., et al. , Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. eLife 6, e25687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White E., Schlackow M., Kamieniarz-Gdula K., Proudfoot N. J., Gullerova M., Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat. Struct. Mol. Biol. 21, 552–559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roulois D., et al. , DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad S., et al. , Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonova K. I., et al. , p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 110, E89–E98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneyama M., et al. , The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Kawai T., et al. , IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Dar A. C., Dever T. E., Sicheri F., Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122, 887–900 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Rath S., et al. , Concerted 2-5A-mediated mRNA decay and transcription reprogram protein synthesis in the dsRNA response. Mol. Cell 75, 1218–1228.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke J. M., Moon S. L., Matheny T., Parker R., RNase L reprograms translation by widespread mRNA turnover escaped by antiviral mRNAs. Mol. Cell 75, 1203–1217.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Civril F., et al. , Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann R., Justesen J., Sarkar S. N., Sen G. C., Yee V. C., Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase. Mol. Cell 12, 1173–1185 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Donovan J., Dufner M., Korennykh A., Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Natl. Acad. Sci. U.S.A. 110, 1652–1657 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Floyd-Smith G., Slattery E., Lengyel P., Interferon action: RNA cleavage pattern of a (2'-5')oligoadenylate--dependent endonuclease. Science 212, 1030–1032 (1981). [DOI] [PubMed] [Google Scholar]
- 19.Silverman R. H., et al. , Purification and analysis of murine 2-5A-dependent RNase. J. Biol. Chem. 263, 7336–7341 (1988). [PubMed] [Google Scholar]
- 20.Dong B., et al. , Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J. Biol. Chem. 269, 14153–14158 (1994). [PubMed] [Google Scholar]
- 21.Donovan J., Rath S., Kolet-Mandrikov D., Korennykh A., Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Y., et al. , Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343, 1244–1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liddicoat B. J., et al. , RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee S., et al. , OAS-RNase L innate immune pathway mediates the cytotoxicity of a DNA-demethylating drug. Proc. Natl. Acad. Sci. U.S.A. 116, 5071–5076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice G. I., et al. , Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswami G., Li J. B., RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42, D109–D113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko H., et al. , DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansoor M., Melendez A. J., Advances in antisense oligonucleotide development for target identification, validation, and as novel therapeutics. Gene Regul. Syst. Bio. 2, 275–295 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein C. A., Castanotto D., FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 25, 1069–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laina A., Gatsiou A., Georgiopoulos G., Stamatelopoulos K., Stellos K., RNA Therapeutics in cardiovascular precision medicine. Front. Physiol. 9, 953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., et al. , Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 2241–2246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preker P., et al. , RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Schmid M., Jensen T. H., Controlling nuclear RNA levels. Nat. Rev. Genet. 19, 518–529 (2018). [DOI] [PubMed] [Google Scholar]
- 34.S. Besse, D. Rebouillat, I. Marie, F. Puvion-Dutilleul, A. G. Hovanessian, Ultrastructural localization of interferon-inducible double-stranded RNA-activated enzymes in human cells. Exp. Cell Res. 239, 379–392 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Dhir A., et al. , Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kent W. J., et al. , The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsen T. W., Baglioni C., Mechanism for discrimination between viral and host mRNA in interferon-treated cells. Proc. Natl. Acad. Sci. U.S.A. 76, 2600–2604 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rath S., et al. , Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proc. Natl. Acad. Sci. U.S.A. 112, 15916–15921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G., Gack M. U., Distinct and orchestrated functions of RNA sensors in innate immunity. Immunity 53, 26–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Schleeet al., . Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31, 25–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. Katoet al., . Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205, 1601–1610 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starck S. R., et al. , Translation from the 5' untranslated region shapes the integrated stress response. Science 351, aad3867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabre O., et al. , RNase L controls terminal adipocyte differentiation, lipids storage and insulin sensitivity via CHOP10 mRNA regulation. Cell Death Differ. 19, 1470–1481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee S., et al. , RNase L is a negative regulator of cell migration. Oncotarget 6, 44360–44372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou A., et al. , Interferon action and apoptosis are defective in mice devoid of 2',5'-oligoadenylate-dependent RNase L. EMBO J. 16, 6355–6363 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chitrakar A., et al. , Real-time 2-5A kinetics suggest that interferons β and λ evade global arrest of translation by RNase L. Proc. Natl. Acad. Sci. U.S.A. 116, 2103–2111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) (GSE143638, GSE171296). Previously published data were used for this work (RNA-seq datasets from GEO database: GSE131130, GSE123034).