Significance
Air pollution is ubiquitous and may increase neuropsychiatric risk, including for depression. However, the neural underpinnings and whether this environmental risk acts independently or interactively with genetic risk mechanisms are not well understood. In healthy individuals exposed to significant air pollution, we find that combined high air pollution exposures and relatively high polygenic risk for depression disproportionately augmented stress-related effects on brain circuitry. The coexpression of depression-associated genes across the brain tracked corresponding brain connectivity, driven by individuals with higher polygenic risk and with higher exposures to air pollution. These findings add to the mechanistic understanding of brain processes implicated in how individuals with high genetic risk for depression may be particularly vulnerable to the brain effects of air pollution.
Keywords: major depressive disorder, polygenic risk, fine particulate matter, PM2.5, gene–environment interaction
Abstract
Air pollution is a reversible cause of significant global mortality and morbidity. Epidemiological evidence suggests associations between air pollution exposure and impaired cognition and increased risk for major depressive disorders. However, the neural bases of these associations have been unclear. Here, in healthy human subjects exposed to relatively high air pollution and controlling for socioeconomic, genomic, and other confounders, we examine across multiple levels of brain network function the extent to which particulate matter (PM2.5) exposure influences putative genetic risk mechanisms associated with depression. Increased ambient PM2.5 exposure was associated with poorer reasoning and problem solving and higher-trait anxiety/depression. Working memory and stress-related information transfer (effective connectivity) across cortical and subcortical brain networks were influenced by PM2.5 exposure to differing extents depending on the polygenic risk for depression in gene-by-environment interactions. Effective connectivity patterns from individuals with higher polygenic risk for depression and higher exposures with PM2.5, but not from those with lower genetic risk or lower exposures, correlated spatially with the coexpression of depression-associated genes across corresponding brain regions in the Allen Brain Atlas. These converging data suggest that PM2.5 exposure affects brain network functions implicated in the genetic mechanisms of depression.
More than 90% of the world’s population live in places with atmospheric exposures exceeding World Health Organization air quality guidelines (1). Ambient air pollution is a major but potentially reversible cause of global morbidity and mortality (2). Empirical evidence suggests there may also be neurotoxic effects of air pollution, especially fine particulate matter (e.g., PM2.5) (3) that, over exposure periods of several months or more, is associated with increased risk for major depressive disorders (4, 5). These exposures have also been observed to affect cognition (6). Moreover, gene-by-environment interactions are implicated in air pollution and genetic risk for neurodegenerative disorders, potentially involving inflammatory processes (7, 8). Air pollution and inflammation may both affect mood regulation in major depression (9, 10), and indeed, inflammatory processes are implicated in putative risk genes associated with depression (11). These observations raise the possibility that air pollution may interact with depression-associated genes in influencing stress-related brain network function. While these associations at multiple levels of in vivo brain network function have yet to be reported, familial vulnerability to depression may be influenced by genetic interactions with environmental stressors (12, 13).
In this study, we examine the putative effects of recent months of relatively high air pollution exposures on cognitive (14) and emotional risk factors (trait anxiety/depression) of depressive illness (15–17) and, subsequently, pollution effects on underlying cognitive and stress-related brain network function in relation to genetic risk for major depression (11). In the latter, we focus on cognition during emotional stress as a paradigm to engage frontal and cortical–subcortical networks, which have been shown to be sensitive to disruption by stress and are also implicated in depressive disorders (18). Of note, the dysfunction of prefrontal and parietal cortex circuitry during working memory (WM) occurs in patients with depression and in healthy individuals with high polygenic risk for depression (14, 18, 19). Here, we aim to further define how exposure to recent months of air pollution may affect WM, social stress, and associated brain connectivity in the context of polygenic risk for depression.
We examine the behavioral risk factors for depression and associated brain networks engaged during WM under varying social stress levels in a community sample from Beijing, who were exposed to relatively high levels of air pollution (e.g., PM2.5). These exposures varied across the study period, during which pollution levels were moderated some 33% by policy interventions on industrial emissions (20), thus providing a unique opportunity to examine demographically and genomically well-matched individuals living in similar communities with varying (but still relatively high) pollution exposures. As poorer cognition was previously associated with higher levels of air pollution exposures over 3 mo to 3 y in an independent East Asian study (6) and adverse environments over recent months may affect emotional traits associated with depression (21) and indeed air pollution exposures over similar time periods have been associated with depression (4, 5), we examined the effects of recent 6 mo PM2.5 exposure on these behavioral characteristics, the effects of exposure on brain connectivity networks, and their interaction with polygenic risk for depression (11). We then leveraged recent developments linking live functional brain network data with postmortem gene expression data across the same brain regions (22, 23), made possible by the unique Allen Brain Atlas resource that has densely sampled genome-wide expression at multiple brain regions. Here, we surveyed spatial coexpression of depression-related genes across the human brain in the Allen Brain Atlas and the extent to which the spatial correlation of coexpression with cognitive and stress-related connectivity may differ in individuals based on levels of depression genetic risk and PM2.5 exposure.
Results
Effects of PM2.5 Exposure on Cognition and Trait Anxiety/Depression.
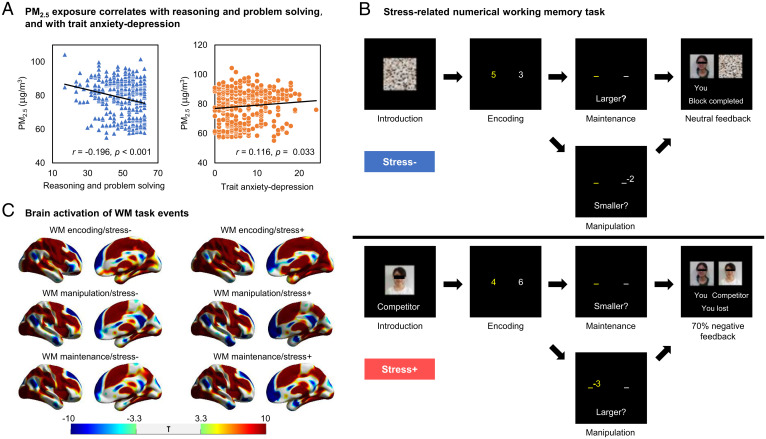
We recruited 352 healthy adults living in Beijing whose demographics are summarized in SI Appendix, Table S1. Subjects performed the MATRICS Consensus Cognitive Battery (MCCB) (24, 25); we assessed personality characteristics associated with risk for depression (15, 16, 26) using a validated translation of the trait anxiety–depression (neuroticism) subscale of the Eysenck personality questionnaire (26–28). Subjects underwent genome-wide genotyping to derive each individual’s polygenic risk score for depression (see SI Appendix for the details of methods). We estimated each subject’s PM2.5 exposure in the 6 mo immediately prior to study participation using data from the nearest air monitoring station to their residence address. The mean distance from the residence of each subject to the nearest air quality monitoring station was 4.49 ± 4.8 kilometers. The 12 monitoring stations were well distributed across Beijing (SI Appendix, Fig. S1 and Table S2). Controlling age, sex, and education that may confound the association between cognition and exposure to air pollution (6), we found that PM2.5 exposure was associated with reduced reasoning and problem solving (r = −0.196, P < 0.001, and P < 0.05 with Bonferroni correction across the seven cognitive domains of the MCCB) and nominally associated with poorer social cognition (r = −0.106 and P = 0.05) as well as with increased trait anxiety/depression (r = 0.116 and P = 0.033) (Fig. 1A). On the other hand, there was an absence of gene–environment correlation in that PM2.5 exposure was not correlated with polygenic risk for depression (r = −0.005 and P = 0.932). Given the epidemiological links between similar periods of PM2.5 exposure and risk for major depression (4, 5), we then followed up the above associations between exposure and behavioral risks of depression, by interrogating across multiple levels of cognitive and emotional brain network function, and genes associated with depression.
Fig. 1.
Cognitive and emotional behavioral tasks. (A) Correlations between PM2.5 exposure and poorer reasoning and problem solving (blue, n = 352) and higher trait anxiety/depression (orange). (B) WM task across stress contexts: In stress+ trials, subjects were shown their photograph alongside that of a similar age and sex competitor. Feedback at the end of each WM maintenance or manipulation task was such that the competitor tended to perform better (“You lost” 70% of the time versus “You won”). In the stress− trials, a scrambled competitor image was shown instead, and no relative performance feedback was provided. In the WM tasks, subjects first encoded a pair of numbers that they maintained or manipulated in WM. In the maintenance task, they then responded to a probe as to which number was larger or smaller. In the manipulation task, they performed a subtraction of 2 or 3 from one number before responding to a probe as to which result was larger or smaller. (C) Brain activation engaged by the WM tasks with lateral and medial views of right hemisphere (n = 352, P < 0.05 family-wise error corrected at the whole-brain voxel level). Color bars: green to blue denotes deactivation T-values, while green to red denotes activation T-values.
Behavioral Performance during Functional MRI WM Task under Varying Stress Contexts and PM2.5 Exposure.
Subjects performed an event-related functional MRI (fMRI) WM task under varying social threat stress contexts in a 3-Tesla MRI scanner (29, 30) (Fig. 1B). At each trial in the WM task, subjects encoded a pair of integer numbers, followed by a jittered maintenance interval between 3 and 5 s. In the WM maintenance trials, subjects then responded to a probe as to which number was larger or smaller. In the WM manipulation trials, subjects performed a subtraction of 2 or 3 from one number before responding to a probe as to which result was larger or smaller. In social stress contexts (stress+), subjects were shown a photograph of an age- and gender-matched “competitor” who they were not acquainted with but against whom their WM task performance would be compared. About 70% of stress+ trials were subsequently fed back with “You lost” relative to “You won.” In noncompetition contexts (stress−), subjects were shown an equivalent scrambled photograph representing no competitor, and the feedback provided was “Block completed.”
During WM maintenance and manipulation across stress contexts (SI Appendix, Table S1), there were significant interactions between stress and WM task in relation to accuracy [F(1, 351) = 45.96 and P < 0.001] and reaction time [F(1, 351) = 5.31 and P = 0.022]. Here, WM maintenance at stress+ had higher accuracy than maintenance at stress− (P < 0.001) (SI Appendix, Fig. S2), while the stress-induced advantage was moderated in WM manipulation. A similar pattern of stress-induced advantage at WM maintenance but less so at WM manipulation was observed for reaction time. These stress-induced biases against WM manipulation in favor of maintenance are consistent with canonical disease and stress-related effects on these constituent WM tasks (14, 31–33).
In relation to PM2.5 exposure, and consistent with the association between pollution and cognition (above), PM2.5 exposure was associated with a slower reaction time across the WM tasks [F(1, 350) = 5.14 and P < 0.05; SI Appendix, Fig. S3]. The exposure effects appeared more prominent under stress+ than at stress−, at least at WM manipulation [exposure-by-stress interaction, F(1, 350) = 3.87 and P = 0.05; SI Appendix, Fig. S3]. The interaction effect was not significant at WM maintenance.
Effective Connectivity Networks Engaged during WM Tasks across Stress Contexts.
As there were behavioral associations with PM2.5 exposure across cognitive and stress-related information processing (Fig. 1A and SI Appendix, Fig. S3), we then examined their potential network connectivity underpinnings in fMRI. During WM encoding, maintenance, or manipulation events under stress+ or stress− contexts, bilateral dorsolateral prefrontal cortex, inferior prefrontal cortex, parietal cortex, temporal cortex, striatum, thalamus, and hippocampus were robustly engaged, accompanied by a deactivation of bilateral medial cortical regions (whole brain P < 0.05 voxel-wise, family-wise-error correction; Fig. 1C and SI Appendix, Table S3).
To elucidate how the information transfer (effective connectivity) across these brain regions were influenced by the constituent WM tasks (encoding/stress+, encoding/stress−, maintenance/stress+, maintenance/stress−, manipulation/stress+, and manipulation/stress−), time series extracted from 20 cortical and three subcortical regions of interest (ROIs) were modeled with deterministic bilinear dynamic causal models (DCMs; Fig. 2A) (31, 34). Herein, all six tasks robustly modulated effective connectivity across the network of 23 brain regions (P < 0.05 with false discovery rate [FDR] correction, SI Appendix, Table S4).
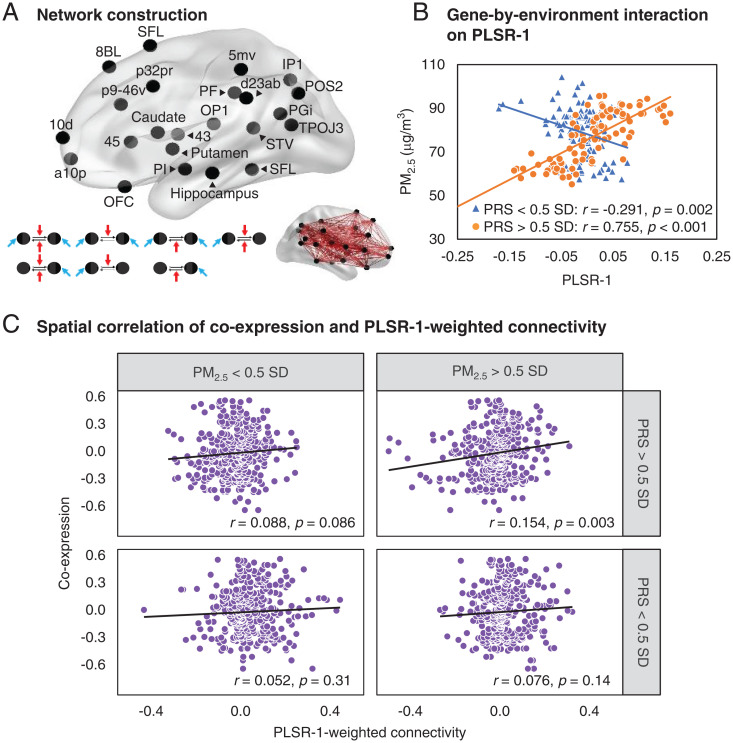
Fig. 2.
Brain connectivity networks engaged by the WM tasks and in the gene-by-environment interaction. (A) Brain rendering of the locations of the subcortical ROIs (caudate, putamen, and hippocampus) from the Automated Anatomical Labeling atlas and 20 cortical ROIs from the Human Connectome Project parcellation (POS2: dorsal part of parieto-occipital sulcus area, SFL: superior frontal language area, STV: superior temporal visual area, d23ab: dorsal part of posterior cingulate cortex, 5mv: ventral part of mid-cingulate cortex, p32pr: dorsal part of anterior cingulate cortex, 8BL: lateral part of frontal eye fields, 10d: dorsal part of frontopolar prefrontal cortex, 45: anterior part of frontopolar prefrontal cortex, p9-46v: central part of dorsolateral prefrontal cortex, a10p: anterior part of frontopolar prefrontal cortex, OFC: orbitofrontal cortex, 43:supplementary and cingulate eye field, OP1:posterior part of posterior opercular cortex, TE1p: posterior part of middle temporal gyrus, TPOJ3: part of temporo-parieto-occipital junction, IP1: posterior part of inferior parietal cortex, PF: anterior part of inferior parietal cortex, PGi: postero-inferior part of inferior parietal cortex, and PI: parainsular area). For each pair of ROIs, we constructed seven task-modulated DCM models, from which we derived task-modulated effective connectivity in each direction with Bayesian model averaging: black arrow denotes intrinsic effective connectivity, blue arrow denotes task input, and red arrow denotes modulatory effect of task stimuli on effective connectivity. (B) PLSR component across WM tasks with the largest variance explained and with a significant gene-by-environment interaction (n = 352 and P < 0.0001). The engagement of brain networks in this component was disproportionately accentuated by PM2.5 exposure in those with higher polygenic risk for depression. While significant in the entire sample, this interaction is shown in a subset of individuals with relatively higher polygenic risk for depression (orange, >0.5 SD and n = 98) and a subset of individuals with relatively lower polygenic risk (blue, <0.5 SD and n = 111). (C) To the extent the WM networks engaged in the PLSR component was implicated in the effects of gene-by-environment interactions, we found that the coexpression of depression-related genes in the orthogonal Allen Brain dataset spatially correlated with the corresponding connectivity weighted according to this PLSR component only in the group with relatively higher (>0.5 SD) polygenic risk score (PRS) and higher-PM2.5 exposure. Spatial correlations between coexpression and connectivity were not significant in groups with relatively lower (<0.5 SD) PRS or lower-PM2.5 exposure.
Interaction between PM2.5 Exposure and Polygenic Risk for Depression on Effective Connectivity.
We used partial least square regression (PLSR) as a dimensionality reduction strategy to define orthogonal effective connectivity networks across the WM tasks (encoding, maintenance, and manipulation) and stress contexts (stress− versus stress+), in which these networks were influenced by PM2.5 exposure, polygenic risk for depression, and their interaction (35). The top four PLSR components explained 45.76% variance of PM2.5 exposure, polygenic risk for depression, and their interaction [P < 0.001 nonparametric permutation test (35)]. These PLSR components, each comprising orthogonal connectivity networks engaged across the WM tasks, were associated with the interaction between PM2.5 exposure and polygenic risk for depression (P < 10−4; SI Appendix, Fig. S4; see SI Appendix, Fig. S5 for factor loadings on each stress-related connectivity). Here, component WM network responses to PM2.5 exposure diverged in relation to polygenic risk for depression. The largest PLSR component, contributing 12.8% to the explained variance, was associated with opposing stress-related effects during WM manipulation and maintenance (SI Appendix, Fig. S5), and these effects were disproportionately accentuated by PM2.5 exposure in those with higher polygenic risk for depression (interaction P < 10−5; Fig. 2B and SI Appendix, Fig. S4).
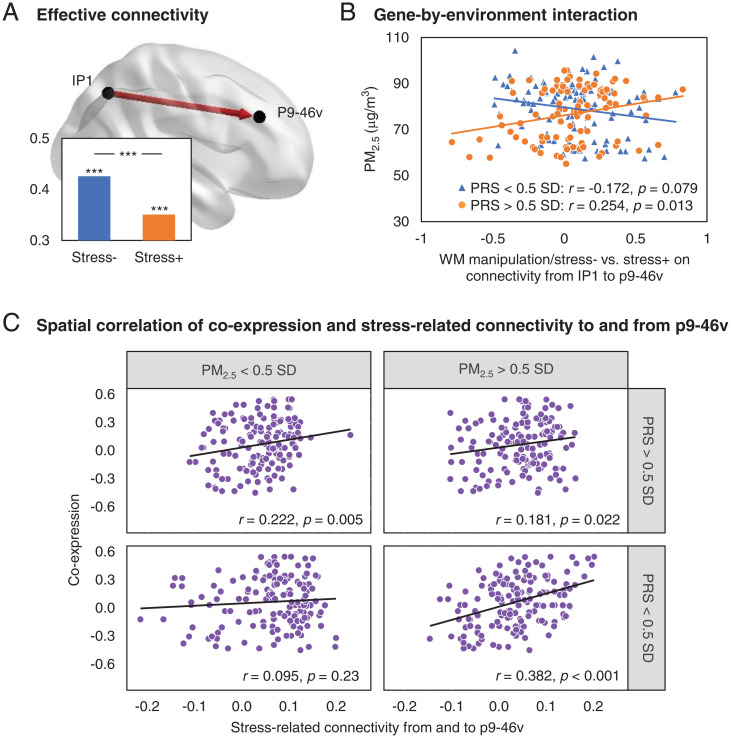
In analyses complementary to broad connectivity networks across WM tasks in the gene-by-environment interactions above, we asked if there were also discernable interaction effects in specific task-related connectivity across specific brain regions. We engaged a principled search by initially ranking the relative importance of individual brain regions across the top four PLSR components in each WM task and subsequently testing for interaction effects in effective connectivity engaged by the most important nodes to and from the other brain regions. Specifically, we considered the weighted degree of each node (brain region) as weighted by their task-specific effective connectivity to and from all the other 22 brain regions and the respective task-specific loadings on connectivity across the top four components. Herein, p9-46v (central part of the dorsolateral prefrontal cortex) during WM manipulation was associated with the highest weighted degree (SI Appendix, Table S6). Across task-modulated effective connectivity engaging p9-46v to and from other brain regions, PM2.5 exposure interacted with polygenic risk for depression in augmenting the stress-related changes in effective connectivity during WM manipulation from IP1 to p9-46v (P < 0.001, with P < 0.05 FDR correction across connectivity engaging p9-46v; Fig. 3 A and B). Similar effects were observed at POS2 to p9-46v effective connectivity (P < 0.001 and P < 0.05 FDR corrected; SI Appendix, Fig. S6). Other nominally significant interaction effects (P < 0.05) implicated in all brain regions are detailed in SI Appendix, Table S7).
Fig. 3.
Specific WM task-related effective connectivity engaged in the gene-by-environment interaction. (A) Effective connectivity from IP1 to p9-46v during WM manipulation/stress− (mean ± SD) and manipulation/stress+ (mean ± SD) were significant and differed across stress (***P < 0.001). IP1: posterior part of inferior parietal cortex; p9-46v: central part of dorsolateral prefrontal cortex. (B) PM2.5 exposure disproportionately increased the stress-related effect (stress− versus stress+) on connectivity from IP1 to p9-46v in individuals with higher polygenic risk for depression in a significant gene-by-environment interaction (P < 0.001, in the entire sample n = 352) and plotted in a subsample of individuals with relatively higher polygenic risk of depression (>0.5 SD and n = 98, orange), compared to those with relatively lower polygenic risk (<0.5 SD and n = 111, blue). (C) To explore the extent to which the interaction effects could also correspond to the coexpression of depression-associated genes, in the context of p9-46v connectivity with other brain regions, we examined if the spatial coexpression correlated differently with corresponding stress-related connectivity in vivo depending on polygenic risk score (PRS) and PM2.5 exposure levels. Across subjects with relatively higher (>0.5 SD) exposures or higher PRS, the coexpression of depression genes in these brain regions spatially correlated with connectivity. However, this spatial correlation was not present in subjects with lower PRS for depression and lower PM2.5 exposure.
Validation Tests.
While we regressed out the effects of age, sex, and education to control for potential socioeconomic confounders influencing PM2.5 exposure (6), we are cognizant that there could be confounding by other unmeasured geographic factors associated with subjects’ residence location and by the spatial imprecision of each individual’s exposure estimate (36). We therefore performed several further sensitivity tests to examine these potential confounders. First, we replaced our pollution exposure variable with average pollution exposure measured in the day, week, and month after experimental participation. We reasoned that unmeasured factors associated with each subject’s residence, behavior, or social situations would nevertheless remain stable in this time frame and that if these were driving our findings rather than the PM2.5 exposure prior to participation, then this test would still yield positive findings. However, we found that the deleterious effects of PM2.5 exposure on behavior and their brain network connectivity interactions with polygenic risk for depression were no longer significant after replacing the exposure variable with future exposures (SI Appendix, Fig. S7). Further adding a regressor of each subject’s time living in Beijing prior to the study resulted in similar behavioral and brain network effects of PM2.5 exposure (SI Appendix, Supplementary Results and Discussion). We therefore suggest that our findings more likely relate with PM2.5 exposure rather than behavioral, social, or geographic factors potentially linked to PM2.5 exposures.
Next, we considered that because government policy appeared to reduce PM2.5 over the years we performed this study (20) and PM2.5 is therefore negatively correlated with the date when subjects participated in our study (r = −0.153 and P = 0.004; SI Appendix, Fig. S9), it could be possible our findings were driven instead by date of study enrollment and other unmeasured variables associated with an earlier or later date of participation. To test this, we replaced the PM2.5 exposure with an index of the participation date of each individual and found that the effects of participation date on cognition and the connectivity interactions were not significant (SI Appendix, Fig. S7).
Spatial Correlation between WM Network Connectivity and Coexpression of Depression Genes.
Brain connectivity networks may reflect transcriptomic coexpression in the same networks (23, 35). We thus reasoned that if PM2.5 exposure and polygenic risk for depression interacted to putatively influence depression-related connectivity networks, these networks, at least in networks engaged by the first PLSR component, might be reflected in the spatial coexpression of depression-associated genes. Specifically, we tested if indeed the spatial coexpression of depression risk–associated genes across the same brain networks in the Allen Brain Atlas correlated differently with the connectivity network architectures depending on levels of genetic risk and PM2.5 exposure in our research volunteers. We examined the coexpression of 49 genome-wide association study (GWAS) associated depression genes (11) with expression data in the Allen Brain Atlas across the same cortical brain regions examined in the fMRI effective connectivity analysis. Across pairs of these brain regions, we then examined the spatial correlation between coexpression and effective connectivity weighted according to the first PLSR component. This weighted connectivity comprised the sum of the WM task-related effective connectivities across the paired regions, weighted by the corresponding WM task loading from the first PLSR component. The weighted connectivity thus represented the composite WM effective connectivity associated with the first PLSR component of any given group of individuals. We examined four groups of individuals according to relatively higher (>0.5 SD) or lower (<0.5 SD) polygenic depression risk and PM2.5 exposures (see SI Appendix, Table S8 for the sample size and demographics of the four subgroups) and, accordingly, the spatial correlation between coexpression and weighted effective connectivity in each group. To the extent the WM networks engaged in the PLSR component was implicated in the effects of gene-by-environment interactions, we found that the coexpression of depression-related genes in the Allen Brain dataset spatially correlated with the corresponding connectivity weighted according to this PLSR component but only in the group with relatively higher polygenic risk and higher-PM2.5 exposure (380 coexpression–connectivity data points, r = 0.15, and P < 0.004; Fig. 2C). The spatial correlation between coexpression and connectivity was not significant in groups with relatively lower polygenic risk or lower-PM2.5 exposure (difference in correlations between the former and the latter groups, P < 0.027, using Fisher’s r-to-z transformation).
In complementary analyses focused on specific WM task-related connectivity, p9-46v engagement during WM manipulation was associated with the highest weighted degree, wherein PM2.5 exposure and polygenic risk for depression interacted to putatively influence stress-related IP1 to p9-46v effective connectivity (Fig. 3A). We then sought to explore the extent to which these effects could also correspond to coexpression of depression-associated genes in the context of p9-46v connectivity with other brain regions. Specifically, we examined if the spatial coexpression correlated differently with corresponding stress-related connectivity in vivo depending on polygenic risk and PM2.5 exposure levels. Here, we found that across subjects with relatively higher (>0.5 SD) exposures or higher polygenic risk, coexpression of depression genes in these brain regions spatially correlated with connectivity [subjects with lower polygenic risk for depression and higher-PM2.5 exposure (161 coexpression–connectivity data points, r = 0.382, and P < 0.001), subjects with higher polygenic risk for depression and lower-PM2.5 exposure (r = 0.222 and P = 0.005), and subjects with higher polygenic risk for depression and higher-PM2.5 exposure (r = 0.181 and P = 0.022)]. However, this spatial correlation was not present in subjects with lower polygenic risk for depression and lower PM2.5 exposure (r = 0.095 and P = 0.23; difference in correlations between the former and the latter groups, P < 0.001, using Fisher’s r-to-z transformation) (Fig. 3C). In a follow-up exploratory analysis, we further clustered the depression-associated genes to define subsets of genes enriched in driving the connectivity spatial coexpression patterns across genetic and environmental risk profiles (SI Appendix, Fig. S8). These enrichment analyses suggest that a subset of genes including RBFOX1, ENOX1, and others (SI Appendix, Supplementary Results and Discussion) implicated in mitochondrial and neuroinflammatory processes may be particularly salient in driving the connectivity–coexpression patterns across genetic and air pollution risks.
Discussion
In a healthy community sample in Beijing exposed to relatively high levels of PM2.5, we found that PM2.5 exposure over 6 mo was associated with higher trait anxiety/depression, with relatively reduced reasoning and problem solving, and with behavioral effects during WM tasks across social stress contexts. These behavioral associations were then interrogated at the level of brain network connectivity and genetic risk for depression. Individuals with higher-PM2.5 exposure and higher polygenic risk for depression had disproportionately augmented effects on network connectivity engaged across WM and social stress contexts in significant gene-by-environment interactions. Dorsolateral prefrontal cortex effective connectivity was critically implicated in these processes. These gene-by-environment interaction findings are independent of those in previous work from our group on the effects of childhood urbanicity (29) (SI Appendix, Supplementary Results and Discussion). Moreover, higher-PM2.5 exposures and higher polygenic risk for depression were selectively associated with network connectivity that were correlated spatially with the coexpression of depression-associated genes across similar human brain regions in the orthogonal Allen Brain Atlas dataset. This spatial correlation of gene coexpression and connectivity was less apparent in individuals with lower PM2.5 exposures or lower polygenic risk.
Extending previous associations between months or more of air pollution and depressive illness (4, 5), our findings suggest links between the deleterious effects of PM2.5 exposure on cognitive and emotional brain function that are also implicated in the genetic mechanisms of depressive illness. Consistent with significant negative life stress over a period of months having deleterious effects on measures of trait anxiety/depression (21), we found that PM2.5 exposures over 6 mo may potentiate the higher emotional reactivity manifest in higher trait anxiety. Consistent with the association between air pollution and cognitive deficits (6), we found that PM2.5 exposure was associated with effects on reasoning, problem solving, and WM. Moreover, PM2.5 exposure selectively slowed the reaction time of WM manipulation under stress, consistent with the greater vulnerability of WM manipulation, relative to maintenance processes, to disruption under stress and disease processes (14, 31, 32).
Building on the behavioral associations, we examined biophysical models of effective connectivity (34) engaged during WM (31, 37). WM has been shown to be sensitive to stress and depression risk (14, 18, 38) and also to air pollution (6). However, the putative mechanistic links across these disease and environmental associations on human brain networks have been unclear, especially in terms of how these networks may be influenced by the interaction between depression genetic risk and air pollution. In examining the overlapping environmental, genetic, and brain–behavior relationships, we engaged a PLSR strategy to address interrelationships across connectivity networks engaged in the constituent WM tasks across social stress contexts (35). We canvassed using PLSR, effective connectivity networks across these WM processes, to identify orthogonal brain networks through which the genetic risk for depression putatively influenced the behavioral and emotional impact of air pollution. Within brain networks with these gene-by-environment interactions, we found divergent loadings at WM manipulation relative to maintenance processes (SI Appendix, Fig. S5), consistent with our behavioral data, and other data, suggesting differing effects of physiological stress on these WM processes (14, 29, 31). In the context of these component patterns of network engagement, increased genetic risk for depression disproportionately augmented the effects of PM2.5 exposure on these networks (Fig. 2B), suggesting that these networks may be physiologically susceptible to PM2.5 effects, at least in part through genetic mechanisms implicated in depression.
To further test the extent of how WM networks, as defined by the largest PLSR component, were implicated in the effects of gene-by-environment interactions, we showed that connectivity metrics from groups of individuals with higher genetic risk and higher exposures, but not others from lower risk groups, selectively predicted the coexpression of depression-related genes across the same brain regions in the Allen Brain dataset. These findings extend recent observations that interrelated networks across levels of neuroimaging investigation reflect transcriptomic coexpression relationships across corresponding brain regions in the human and primate brains (23, 35). We adapted this approach to study a more specific set of genome-wide associated depression genes, analogous to earlier work on Alzheimer’s disease (39). The spatial correlations between connectivity and coexpression linking these orthogonal datasets thus provide converging evidence supportive of our hypothesis that genetic risk mechanisms of depression interacted with air pollution exposures to affect stress-related information processing brain networks.
To the extent brain networks across WM tasks were implicated in the gene-by-environment interactions, we then asked if there were also discernable interaction effects within specific task-related connectivity in specific brain regions. Using a principled search through brain regions with the highest weighted network degree, p9-46v during WM manipulation emerged as a key node, and therein, the strongest interaction occurred in which stress-induced effects on effective connectivity from IP1 to p9-46v were disproportionately augmented in individuals with higher polygenic risk for depression and higher-PM2.5 exposure (Fig. 3A). These results, implicating the rapid updating of new information in WM manipulation, are consistent with their stronger engagement in prefrontal circuits than simpler WM maintenance processes (30, 40). These higher-order WM processes have also been postulated to be vulnerable to disruption by neuroinflammation (41) associated with stress, depression (14, 42), and genetic risk for depression (11).
In the context of connectivity and coexpression involving p9-46v with other cortical regions, we found that the spatial coexpression of depression-associated genes predicted prefrontal connectivity at higher PM2.5 exposures or genetic risk but not connectivity when exposures and genetic risk were both low. The spatial correlation between stress-related connectivity with coexpression of depression-associated genes is remarkably consistent with the sensitivity of these prefrontal-associated networks to chronic stress and depression (38) and, indeed, consistent with the mapping of these genome-wide signals to the expression of genes enriched in the prefrontal cortex (11). Moreover, genes such as RBFOX1, ENOX1, and others implicated in neuroinflammatory processes in depression (11) appear enriched in these spatial correlations (SI Appendix, Supplementary Results and Discussion). These findings thus provide converging evidence supportive of the conceptualization that air pollution exposures interact with prefrontal cortical genetic risk mechanisms of depression and may engage neuroinflammatory processes.
In the present study, we have controlled for age, sex, education, and genetic background that may confound the associations across cognition, stress, and exposure to air pollution (6). We are also cognizant that gene–environment correlations may cause artifactual interaction effects (12). However, a feature of the study population is the geographically extensive air pollution exposures they experienced, which may minimize socioeconomic (and related genetic) relationships with exposure. Indeed, there were no correlations between pollution exposure and depression genetic risk or between pollution and socioeconomic factors. Furthermore, we suggest through several validation tests that the effects of unmeasured geographic and personal factors (36) may not be driving our findings. While our findings appear consistent with the impact of air pollution on stress-related prefrontal networks implicated in depression genetic risk, this may be specific to the higher exposures in our study population, and it remains to be determined if similar findings might generalize to other populations and at lower levels of air pollution exposures. We also cannot exclude the likelihood that more spatially specific measurements (e.g., using portable monitors) could yield more individually specific results.
A number of limitations surrounding the complex genetics of depression are pertinent in this study. Firstly, we have endeavored to use extant genome-wide significant signals from large GWAS of depression (11) in calculating the polygenic risk for depression that converges subsequently in the definition of cognate genes studied in the spatial correlation between coexpression and connectivity. Inevitably, there is significant missing heritability not accounted for by these rarified signals, as the cognate genes whose coexpression we have studied likely represent only a fraction of that implicated in depressive illness. Future bioinformatic advances accounting for epigenetic and tissue-specific regulatory mechanisms could augment the principled definition of cognate genes, including from selected, subthreshold genetic signals (43). Genes and genetic signals from functional networks associated with genes enriched in the differential spatial correlation across genetic and environmental risks may also be considered. Our power to define ancestry-specific effects is also necessarily limited (44) because we have used genetic signals from a European sample (11) in the absence of similarly powered East Asian data. Nevertheless, the significant findings we report herein might, at least in part, suggest the explanatory power of these large GWAS and the cognate gene-level associations they implicate, extending beyond their original ancestral populations (45, 46). The large environmental exposure effects we examine could also have mitigated the reduced genetic power of association. Moreover, including signals from smaller East Asian GWAS (47) in our data yielded consistent findings (SI Appendix, Supplementary Results and Discussion). Still, the limited sets of genes we have studied, and the limited samples in the Allen Brain Atlas, could have constrained the brain networks we have implicated, albeit to prefrontal networks also enriched in the expression of the same genes we study (11), but possibly less so other key regions and mechanisms implicated in depression (e.g., at hippocampus). Extending our assay of brain networks across multiple cognitive, resting state, and other neuroimaging modalities could further augment the definition of these genetic and environmental brain mechanisms. Finally, we have studied the genes and brain networks within the context of recent air pollution exposures over 6 mo, a time frame suggested to affect risk for depression (4, 5) and cognition (6). Differing exposure durations in samples across neurodevelopment (e.g., different stages of prenatal life, childhood, and aging), as well as long-term cumulative effects, might well yield additional disease mechanisms that should be targets of future work.
In conclusion, we find that in an otherwise healthy and homogeneous volunteer sample in Beijing exposed to higher levels of PM2.5 than their Western counterparts, PM2.5 exposure affects cognition and levels of trait anxiety/depression. These behavioral associations were reflected in changes at brain network connectivity in which polygenic risk for depression nonadditively augmented the neural impact of PM2.5 exposure. Spatial correlation between brain network connectivity and the coexpression of depression-associated genes was found in relation to higher exposures and higher genetic risk but not in relation to lower exposures or lower genetic risk. This study adds to the mechanistic understanding of brain processes implicated in how individuals with a relatively high genetic risk for depression may be particularly vulnerable to the brain effects of air pollution. While our unique study context has minimized gene–environment correlations, we might anticipate that correlations between genetic and environmental risk factors (e.g., poverty, pollution, and genetic risk) could nevertheless, in other study contexts, augment these interactions to cascade upon cognitive and mood dysfunction.
Materials and Methods
Subjects.
Subjects were part of a Peking University–Lieber Institute for Brain Development research partnership on genes and the environment (29). A total of 352 healthy adults with 3-Tesla MRI and air pollution exposure data were included in this study, which was approved by the Institutional Review Boards of the Peking University Institute of Mental Health and the Johns Hopkins University School of Medicine. Informed consent was given by each subject. Details of the mood and cognition measurements, the stress-related numerical WM task, the MRI sequences, the estimates of individual PM2.5 exposure, genotyping quality control, and the calculation of the polygenic risk score for depression are in SI Appendix.
Neuroimaging Data Analyses.
Brain imaging data were processed and analyzed with SPM12 (Statistical Parametric Mapping 12, https://www.fil.ion.ucl.ac.uk/spm). We examined biophysical models of effective connectivity using DCM (34) as implemented in SPM12 (DCM10) to estimate the modulatory effects of WM task stimuli on effective connectivity. We focused on DCMs across a representative set of 20 cortical and 3 subcortical ROIs (SI Appendix). To identify potential interactions through which PM2.5 exposure and polygenic risk for depression influenced effective connectivity networks across WM tasks (encoding, maintenance, and manipulation) and stress contexts (stress− versus stress+), we performed PLSR in MATLAB 2018b across all 1,518 cortical–subcortical task-modulated connections during these tasks (35). To define interaction effects at specific task-related effective connectivity across specific brain regions, we ranked the relative importance of individual brain regions based on the weighted degree of each brain region’s connectivity and PLSR loadings; subsequently, from each of these brain regions, we examined the effective connectivity to and from all the other brain regions in terms of gene-by-environment interactions using generalized linear models. We visualized interaction effects by plotting PM2.5 exposure and connectivity across varying higher (>0.5 SD) or lower polygenic risks for depression. We also utilized validation tests, replacing the exposure variable with variables sensitive to geography and time, to putatively exclude their confounding effects on our gene-by-environment interaction findings. Additional details of data analysis are in SI Appendix.
Spatial Correlation across Stress-Related Connectivity and the Coexpression of Depression Genes.
To the extent PM2.5 exposure may interact with depression genetic risk on brain connectivity, we then examined if spatial coexpression of depression-related genes across the human brain in Allen Brain Atlas (https://human.brain-map.org/) may couple differently with corresponding connectivity across polygenic risk for depression and PM2.5 exposure. Specifically, we examined the coexpression of the 49 GWAS-associated depression genes (11) that also had quality-controlled expression data in the Allen Brain Atlas (SI Appendix) across the same cortical brain regions examined in the fMRI data. Across pairs of these brain regions, we then examined the spatial correlation between coexpression and effective connectivity weighted according to the first PLSR component. We examined four groups of individuals according to relatively higher (>0.5 SD) or lower (<0.5 SD) polygenic depression risk and PM2.5 exposures and, accordingly, the spatial correlation between coexpression and weighted effective connectivity in each group.
In complementary analyses on specific WM task-related connectivity, we focused on coexpression and stress-related WM manipulation connectivity across the brain related to the p9-46v, implicated in our findings (see Results). Effective connectivity involving p9-46v and 106 other ROIs that were engaged by the task and sensitive to stress (161 connections, P < 0.05 FDR corrected) were examined. The coexpression of 49 depression-associated genes (11) across the 106 pairs of ROIs from human gene expression data in Allen Brain Atlas (23) were computed. The spatial correlation across the same pairs of brain regions in the coexpression and stress-related connectivity data were then computed. To examine the extent to which this spatial correlation may vary with polygenic risk for depression and PM2.5 exposure, subjects were divided into four groups with relatively higher (>0.5 SD) or lower (<0.5 SD) exposures (SI Appendix, Table S8) and polygenic risk. Further details are in SI Appendix.
Acknowledgments
This work was supported by the US NIH (R01MH101053, H.Y.T.), the National Natural Science Foundation of China (81361120395, D.Z.; 81825009, W.Y.), and the Lieber Institute for Brain Development.
Footnotes
Author contributions: Z.L. and H.Y.T. conceived research on air pollution and brain connectivity; Z.L., H.Y., S.H., D.Z., D.R.W., W.Y., and H.Y.T. designed research; Z.L., H.Y., X.Z., D.Z., W.Y., and H.Y.T. performed or supervised data collection; Z.L., S.S., G.Y., Q.C., S.H., W.Y., and H.Y.T. contributed new reagents/ analytic tools; Z.L. and H.Y.T. analyzed data; Z.L. and H.Y.T. wrote the paper with contributions from S.H. and D.R.W.
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109310118/-/DCSupplemental.
Data Availability
All study data in support of our findings are included in the article and/or SI Appendix. Computing code and source data are available on GitHub (https://github.com/psychlizhi/Air-pollution-interacts-with-genetic-risk-to-influence-cortical-networks-implicated-in-depression.git).
References
- 1.World Health Organization, Health topics: Air pollution. https://www.who.int/health-topics/air-pollution. Accessed 8 October 2021. [Google Scholar]
- 2.Burnett R. T., et al., An integrated risk function for estimating the global burden of disease attributable to ambient fine particulate matter exposure. Environ. Health Perspect. 122, 397–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genc S., Zadeoglulari Z., Fuss S. H., Genc K., The adverse effects of air pollution on the nervous system. J. Toxicol. 2012, 782462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan S. J., et al., Ambient air pollution and depression: A systematic review with meta-analysis up to 2019. Sci. Total Environ. 701, 134721 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Braithwaite I., Zhang S., Kirkbride J. B., Osborn D. P. J., Hayes J. F., Air pollution (particulate matter) exposure and associations with depression, anxiety, bipolar, psychosis and suicide risk: A systematic review and meta-analysis. Environ. Health Perspect. 127, 126002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X., Chen X., Zhang X., The impact of exposure to air pollution on cognitive performance. Proc. Natl. Acad. Sci. U.S.A. 115, 9193–9197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eid A., Mhatre I., Richardson J. R., Gene-environment interactions in Alzheimer’s disease: A potential path to precision medicine. Pharmacol. Ther. 199, 173–187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee P. C., et al., Gene-environment interactions linking air pollution and inflammation in Parkinson’s disease. Environ. Res. 151, 713–720 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Thomson E. M., Air pollution, stress, and allostatic load: Linking systemic and central nervous system impacts. J. Alzheimers Dis. 69, 597–614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dantzer R., O’Connor J. C., Freund G. G., Johnson R. W., Kelley K. W., From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wray N. R., et al.; eQTLGen; 23andMe; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutter M., Silberg J., Gene-environment interplay in relation to emotional and behavioral disturbance. Annu. Rev. Psychol. 53, 463–490 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Kendler K. S., Karkowski-Shuman L., Stressful life events and genetic liability to major depression: Genetic control of exposure to the environment? Psychol. Med. 27, 539–547 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Joormann J., Gotlib I. H., Updating the contents of working memory in depression: Interference from irrelevant negative material. J. Abnorm. Psychol. 117, 182–192 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Kendler K. S., Kuhn J., Prescott C. A., The interrelationship of neuroticism, sex, and stressful life events in the prediction of episodes of major depression. Am. J. Psychiatry 161, 631–636 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Duggan C., Sham P., Lee A., Minne C., Murray R., Neuroticism: A vulnerability marker for depression evidence from a family study. J. Affect. Disord. 35, 139–143 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Nagel M., et al.; 23andMe Research Team, Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 50, 920–927 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Garrett A., et al., Aberrant brain activation during a working memory task in psychotic major depression. Am. J. Psychiatry 168, 173–182 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Yüksel D., et al., Polygenic risk for depression and the neural correlates of working memory in healthy subjects. Prog. Neuropsychopharmacol. Biol. Psychiatry 79, 67–76 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q., et al., Drivers of improved PM2.5 air quality in China from 2013 to 2017. Proc. Natl. Acad. Sci. U.S.A. 116, 24463–24469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeronimus B. F., Ormel J., Aleman A., Penninx B. W. J. H., Riese H., Negative and positive life events are associated with small but lasting change in neuroticism. Psychol. Med. 43, 2403–2415 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Burt J. B., et al., Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat. Neurosci. 21, 1251–1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawrylycz M., et al., Canonical genetic signatures of the adult human brain. Nat. Neurosci. 18, 1832–1844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuechterlein K. H., et al., The MATRICS consensus cognitive battery, part 1: Test selection, reliability, and validity. Am. J. Psychiatry 165, 203–213 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Shi C., et al., The MATRICS consensus cognitive battery (MCCB): Co-norming and standardization in China. Schizophr. Res. 169, 109–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J., et al., The relationship between neuroticism, major depressive disorder and comorbid disorders in Chinese women. J. Affect. Disord. 135, 100–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eysenck H. J., Eysenck S. B. G., Manual of the Eysenck Personality Questionnaire (Junior and Adult) (Hodder and Stoughton, London, 1975). [Google Scholar]
- 28.Gong Y. X., Revised Eysenck Personality Questionnaire (EPQ-R) (Map Publishing House, Changsha, 1992). [Google Scholar]
- 29.Zhang X., et al., Childhood urbanicity interacts with polygenic risk for depression to affect stress-related medial prefrontal function. Transl. Psychiatry11, 522 (2021). [DOI] [PMC free article] [PubMed]
- 30.Tan H. Y., et al., Catechol-O-methyltransferase Val158Met modulation of prefrontal-parietal-striatal brain systems during arithmetic and temporal transformations in working memory. J. Neurosci. 27, 13393–13401 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenman D. L. B., et al., Parietal–prefrontal feedforward connectivity in association with schizophrenia genetic risk and delusions. Am. J. Psychiatry 177, 1151–1158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oei N. Y., Everaerd W. T., Elzinga B. M., van Well S., Bermond B., Psychosocial stress impairs working memory at high loads: An association with cortisol levels and memory retrieval. Stress 9, 133–141 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Cannon T. D., et al., Dorsolateral prefrontal cortex activity during maintenance and manipulation of information in working memory in patients with schizophrenia. Arch. Gen. Psychiatry 62, 1071–1080 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Friston K. J., Harrison L., Penny W., Dynamic causal modelling. Neuroimage 19, 1273–1302 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Seidlitz J., et al.; NSPN Consortium, Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron 97, 231–247.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheppard L., et al., Confounding and exposure measurement error in air pollution epidemiology. Air Qual. Atmos. Health 5, 203–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Esposito M., Postle B. R., Ballard D., Lease J., Maintenance versus manipulation of information held in working memory: An event-related fMRI study. Brain Cogn. 41, 66–86 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Arnsten A. F. T., Stress signalling pathways that impair prefrontal cortex structure and function. Nat. Rev. Neurosci. 10, 410–422 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grothe M. J., et al.; Alzheimer’s Disease Neuroimaging Initiative, Molecular properties underlying regional vulnerability to Alzheimer’s disease pathology. Brain 141, 2755–2771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray S., Almas R., Arnold A. E. G. F., Iaria G., MacQueen G., Intraparietal sulcus activity and functional connectivity supporting spatial working memory manipulation. Cereb. Cortex 25, 1252–1264 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Levesque S., et al., Diesel exhaust activates and primes microglia: Air pollution, neuroinflammation, and regulation of dopaminergic neurotoxicity. Environ. Health Perspect. 119, 1149–1155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moghaddam B., Stress activation of glutamate neurotransmission in the prefrontal cortex: Implications for dopamine-associated psychiatric disorders. Biol. Psychiatry 51, 775–787 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Sey N. Y. A., et al., A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 23, 583–593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin A. R., et al., Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levey D. F., et al.; 23andMe Research Team; Million Veteran Program, Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 24, 954–963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam M., et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Indonesia Schizophrenia Consortium; Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN), Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai N., et al.; CONVERGE consortium, Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data in support of our findings are included in the article and/or SI Appendix. Computing code and source data are available on GitHub (https://github.com/psychlizhi/Air-pollution-interacts-with-genetic-risk-to-influence-cortical-networks-implicated-in-depression.git).