Significance
We performed an in-cell screen of imatinib binding against a library of Abl kinase mutants derived from patients with imatinib-resistant chronic myeloid leukemia. The majority of mutations readily bind imatinib, posing the question of how these mutations cause resistance in patients. We identified several kinetic mutants, one of which binds imatinib with wild-type affinity but dissociates considerably faster from the mutant kinase. Using NMR and molecular dynamics, we found that this mutation increases the conformational dynamics of the mutant protein, linking conformational dynamics of the protein to drug dissociation. The results underline the importance of drug dissociation kinetics for drug efficacy and propose a kinetic resistance mechanism that may be targetable by altering drug treatment schedules.
Keywords: protein kinase, drug resistance, binding kinetics, drug binding, imatinib
Abstract
Protein kinase inhibitors are potent anticancer therapeutics. For example, the Bcr-Abl kinase inhibitor imatinib decreases mortality for chronic myeloid leukemia by 80%, but 22 to 41% of patients acquire resistance to imatinib. About 70% of relapsed patients harbor mutations in the Bcr-Abl kinase domain, where more than a hundred different mutations have been identified. Some mutations are located near the imatinib-binding site and cause resistance through altered interactions with the drug. However, many resistance mutations are located far from the drug-binding site, and it remains unclear how these mutations confer resistance. Additionally, earlier studies on small sets of patient-derived imatinib resistance mutations indicated that some of these mutant proteins were in fact sensitive to imatinib in cellular and biochemical studies. Here, we surveyed the resistance of 94 patient-derived Abl kinase domain mutations annotated as disease relevant or resistance causing using an engagement assay in live cells. We found that only two-thirds of mutations weaken imatinib affinity by more than twofold compared to Abl wild type. Surprisingly, one-third of mutations in the Abl kinase domain still remain sensitive to imatinib and bind with similar or higher affinity than wild type. Intriguingly, we identified three clinical Abl mutations that bind imatinib with wild type–like affinity but dissociate from imatinib considerably faster. Given the relevance of residence time for drug efficacy, mutations that alter binding kinetics could cause resistance in the nonequilibrium environment of the body where drug export and clearance play critical roles.
Imatinib, a selective inhibitor of Bcr-Abl kinase, reduced the mortality from chronic myeloid leukemia (CML) by 80% within a decade of its approval by the Food and Drug Administration in 2001 (1, 2). Of the patients treated with imatinib in the chronic phase, 95% achieve a complete hematologic remission and 60% a major cytogenetic response. However, most patients in blast crisis either fail to respond or quickly relapse following an initial response to imatinib. Mutations within the Bcr-Abl kinase domain (KD) are the most common cause for relapse (3). Over 1,000 substitution mutations throughout the KD have been identified in relapsed patients and are presumed to confer resistance to imatinib therapy (4, 5). Most of these mutations are located in areas of the KD that are involved in imatinib binding, including mutations at the imatinib-binding pocket, nucleotide-binding (P-loop) mutations, and activation loop (A-loop) mutations (6). Interestingly, more than 50% of the mutations in the Catalogue of Somatic Mutations in Cancer (COSMIC) (7) or Memorial Sloan Kettering's Precision Oncology Knowledge Base (OncoKB) (8) databases lie outside these areas. Early studies on small sets of KD mutants showed that some clinically observed imatinib resistance mutations lost sensitivity to imatinib, while others showed little or no change (9). Furthermore, for some of these mutants, the biochemical and cellular imatinib sensitivities do not correlate (10). For example, the Abl E355G mutant remains sensitive to imatinib in biochemical assays but only manifests its resistance in the context of cells (9). This observation could be explained based on several factors: 1) the purified mutant kinase constructs lack domains that facilitate resistance in the context of full-length Bcr-Abl expressed in cells, 2) mutations may increase kinase activity or biological stability in cells, or 3) mutations could increase cellular autophosphorylation of Bcr-Abl (11).
Here, we propose a general mechanism that mutations can cause resistance not by altering the equilibrium binding affinity but rather by altering binding and dissociation kinetics. To illuminate this mechanism, we characterize a clinically observed mutation with significantly accelerated dissociation kinetics associated with relapse from imatinib therapy.
In contrast to most biochemical and cellular assays that employ a constant drug concentration over time (4, 5, 9), the plasma drug concentration of patients fluctuates over time because of drug pharmacodynamics and pharmacokinetics (12, 13). Under constant drug concentration, the affinity of the drug for the receptor is the major contributor to drug efficacy; however, in the nonequilibrium environment of the body, drug binding and dissociation are coupled to other dynamic processes such as drug export and excretion that can limit drug efficacy. Drug–target residence time, defined as the inverse of the dissociation rate, has emerged as an excellent predictor of drug efficacy in certain contexts (14, 15, 16). Strategies aimed at prolonging drug–target residence time and minimizing off-target binding (i.e., kinetic selectivity) can drive drug efficacy (14, 15, 16). Indeed, many successful kinase inhibitors such as imatinib and lapatinib have long residence times during which they elicit a pharmacological effect (12, 17, 18–20). An earlier study on Bruton’s tyrosine kinase inhibitors showed that faster inhibitor dissociation correlated with lower efficacy (21). Based on these findings, we propose here that mutations can cause drug resistance by increasing dissociation rates, thereby decreasing drug residence times and reducing target inhibition in the nonequilibrium environment of the cell.
Here, we test how 94 patient-derived and resistance-associated Abl mutants affect imatinib affinity and binding kinetics in live cells. We use a cell-based assay technique (NanoBRET) that employs bioluminescence resonance energy transfer (BRET) to directly measure molecular interactions between inhibitors and their kinase target in real time (22, 23). This workflow represents a broad-spectrum analysis of the target engagement characteristics of the full range of mutations in a clinically relevant kinase. We envision that this will serve as a paradigm for further investigation of analogous mutations in other kinases such as epidermal growth factor receptor (EGFR).
This technique has been recently used to study libraries of inhibitors against 178 full-length kinases (24). We present a study in which a library of resistance-associated mutant kinases is screened for target engagement and residence time in live cells. We find that two-thirds of the tested clinically observed imatinib resistance mutations in Abl kinase significantly reduce drug affinity, whereas one-third bind imatinib with similar or tighter affinity than Abl wild type (wt). These values correspond with previous measurements of imatinib’s effect on Bcr-Abl autophosphorylation and cell viability in resistance-associated mutations (25). We identified three imatinib resistance mutations, N368S, V299L, and G251E, that have similar imatinib affinity as Abl wt but increased imatinib dissociation rates. We propose here that clinical Abl mutations may cause drug resistance by suppressing the protracted residence time of imatinib in a pathophysiological setting.
Results
To measure kinase engagement in live cells, we used a recently described technique that employs BRET (NanoBRET) as a proximity-based measure of imatinib binding (26). Previously, this technique has successfully been applied to quantify compound engagement over a number of intracellular target proteins (22, 23). Briefly, a reporter complex forms inside live cells when a cell-permeable fluorescent probe molecule (tracer) binds to the ATP-binding site of a luciferase-tagged full-length target protein. In the presence of a cell-permeable luciferase substrate, the reporter complex emits a BRET signal. The addition of an unlabeled test compound (e.g., imatinib or dasatinib) disrupts the reporter complex and decreases the BRET signal. Our NanoBRET tracers were based on adenosine triphosphate (ATP)–competitive kinase inhibitors that directly compete with the tested inhibitors for the same binding site on the kinase (24).
Two-Thirds of Imatinib Resistance–Associated Mutations Bind Imatinib with Weaker Affinity than Abl wt.
We selected 94 mutations in Abl KD mutations from the COSMIC (7) or OncoKB (8) databases that were annotated as disease relevant (OncoKB) or resistance causing (COSMIC) (SI Appendix, Table S1). Some mutations are located in the ATP-binding site where they are likely to interfere with imatinib binding; however, many mutations are distal to the imatinib-binding site, and their resistance mechanism is unclear (Fig. 1A). We introduced individual point mutations to full-length Abl kinase fused to the C terminus of NanoLuc luciferase (Abl1-Nluc) for expression in human embryonic kidney (HEK293T) cells.
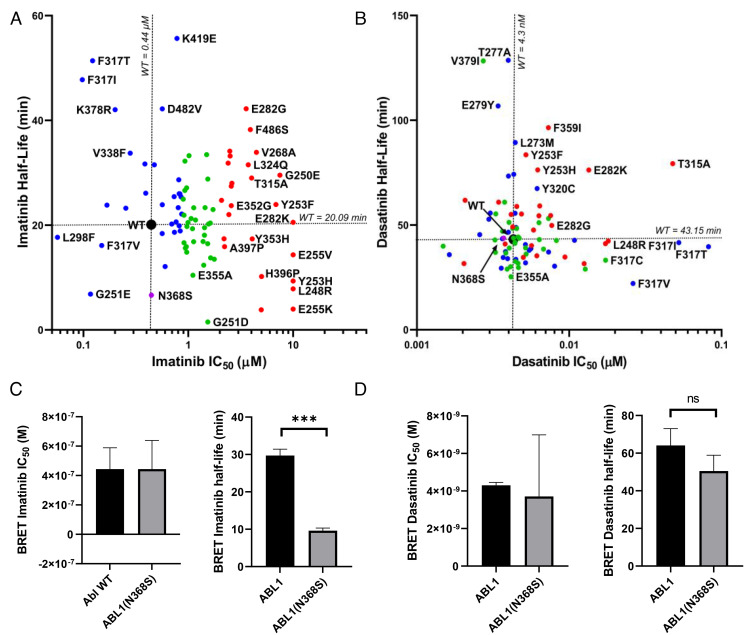
Fig. 1.
In-cell inhibitor–binding affinities of mutant panel. (A) Structure of the Abl KD (PDB entry 2HYY). The locations of 94 patient-derived Abl mutants associated with resistance are indicated as spheres. The color of the spheres indicates the level of imatinib resistance conferred by the mutation: blue, affinity of mutant is less than twofold weaker than wt; green, affinity is between two- and fourfold weaker than wt; red, affinity is more than eightfold weaker than wt. (B) Fraction of mutations in different resistance classes. The colors correspond to A. (C) Imatinib and dasatinib resistance distribution of Abl mutants determined by in-cell NanoBRET target engagement assay. Colors correspond to A. Confidence of values is ± 23% based off of wt SD, n = 2.
Imatinib bound to Abl wt with a cellular affinity (IC50) of 0.44 ± 0.10 μM at equilibrium. For two-thirds of resistance-associated mutants (63 out of 94), the affinity for imatinib was weakened at least twofold (IC50 ≥ 0.8 μM) (Fig. 1 B and C). At the standard dose of 400 mg daily, imatinib’s free serum concentration fluctuates from a mean maximum concentration of 4.4 ± 1.3 μM to a mean plasma trough concentration of 2 ± 1.4 μM (27). This indicates that in a population of patients, the trough plasma concentration would fall as low as 0.6 μM, which is below the IC50 of one-third of the tested mutants. The mutants that most drastically compromised imatinib affinity included the gatekeeper mutant and P-loop mutations, which had more than eightfold higher imatinib IC50 values (SI Appendix, Table S2). We were surprised to find that one-third of imatinib resistance mutations bound imatinib equally or more potently than wt. The mutation that most drastically increased imatinib affinity was Abl L298F, which increased imatinib affinity fourfold over Abl wt (IC50 = 0.1 μM).
About 85% of mutants (80 out of 94) bound to dasatinib with similar affinity as Abl wt (cellular IC50 of 0.0043 μM) (Fig. 1B). Only 15% (14 out of 94) of all the mutations tested had twofold weaker dasatinib affinity than Abl wt, consistent with the clinical use of dasatinib as a second line therapeutic following emergence of imatinib resistance. Furthermore, mutations with weaker dasatinib affinity also showed weaker imatinib affinity. Exceptions to this were the mutations at F317, with F317I/T/V all remaining sensitive to imatinib, despite compromising dasatinib affinity (Fig. 1C).
Abl N368S Binds Imatinib with High Affinity but Dissociates More Rapidly.
Since a large fraction (31 out of 94) of imatinib resistance mutations still bound imatinib with affinity similar to wt Abl, we determined the effect of mutation on cellular imatinib dissociation kinetics. Our rationale was that mutations might cause resistance by altering drug affinity or dissociation kinetics. We used NanoBRET assays to measure the real-time imatinib and dasatinib dissociation kinetics of mutants with wt-like imatinib affinities in live cells (Fig. 2 A and B and SI Appendix, Table S6). In brief, Abl kinase was expressed in HEK293T cells preloaded with imatinib or dasatinib, and excess drug was washed out. An excess of fluorescent tracer was added that bound to Abl as imatinib/dasatinib dissociated, resulting in an increased BRET signal.
Fig. 2.
In-cell dissociation kinetics. (A and B) Abl mutants were characterized for imatinib (A) and dasatinib (B) affinity and dissociation kinetics using NanoBRET assay in live cells. Mutants were colored according to imatinib affinity as in Fig. 1. The novel Abl N368S mutant was characterized for (C) imatinib- and (D) dasatinib-binding affinities and dissociation kinetics by NanoBRET in live cells. ns: P > 0.05, ***P < 0.001.
Imatinib and dasatinib dissociated slowly from Abl wt with half-lives of 20.1 and 43.2 min, respectively. When comparing the correlation between drug half-life and affinity (Fig. 2 A and B), we found that the mutant panel varied more from wt when treated with imatinib (average distance normalized to wt values: 3.94) than dasatinib (distance: 1.1). This is consistent with mutations identified in imatinib-resistant CML and not selected for dasatinib resistance. We found that seven mutants dissociated at least twice as fast from imatinib as Abl wt. Of those, Abl G251E exhibits both an increased imatinib affinity and faster dissociation (0.12 μM and 6.8 min). A mutation at the same site, G251D, has reduced imatinib affinity (1.5 μM) compared to wt (0.44 μM), but its dissociation from imatinib is more than 15 times faster (1.2 versus 20.1 min). Abl N368S, a mutation that has only been reported once (28), dissociated three times faster (half-life 6.6 ± 0.7 min) but bound imatinib with similar affinity as wt (0.44 ± 0.2 μM, 0.44 ± 0.14 μM respectively) (Fig. 2C). This effect was specific to imatinib, as dasatinib dissociated from Abl N368S and Abl wt with near-identical kinetics (43.8 and 43.2 min, respectively) (Fig. 2D). The sites of the kinetic mutations are frequently altered in different kinases and various cancer types with 13 mutations across eight kinases reported for G251 and nine mutations in four kinases reported for N368 (29).
Enzymatic Activity of N368S KD Requires Activation Loop Phosphorylation.
The asparagine at position 368 in Abl wt is nearly universally conserved among kinases (Fig. 3A) (30). The sidechain of N368 forms hydrogen bonds with D381 and the catalytic aspartate D363 when the kinase is in the active conformation and the aspartate sidechain of the Asp-Phe-Gly (DFG) motif faces into the active site (DFG-Asp-in conformation) (Protein Data Base [PDB] entry 2G2I) (Fig. 3B). Both D381 and D363 coordinate Mg2+/ATP, and they are required for catalysis (31, 32). Therefore, we speculated that N368 could be critical for enzyme activity as well.
Fig. 3.
Characterization of the Abl N368S mutation. (A) Sequence alignment of human protein kinases represented as weblogo. The size of the letters reflects the degree of conservation, and numbering is according to Abl1A numbering. Asparagine 368 is as highly conserved as the catalytic His-Arg-Asp (HRD) and the DFG motif. (B) Structural overview of Abl N368 in the active conformation (PDB entry 2G2I). The sidechain of N368 is near the nucleotide and is positioned to form hydrogen bonds with the catalytic aspartate D363 and D381 in the DFG motif. (C) Kinase activity of purified Abl N368S is undetectable in the absence of activation loop phosphorylation (designated as pAbl). (D) The Michaelis–Menten constant for ATP of purified pAbl N368S is twofold higher than that of pAbl wt. (E) The affinity of purified Abl N368S for imatinib determined by a spectroscopic-binding assay is slightly weaker than for Abl wt. (F) BaF3 cells expressing Bcr-Abl wt, T315I, or N368S. BaF3 cells expressing N368S are equally sensitive to imatinib as cells expressing Bcr-Abl wt. (G) BaF3 cells expressing imatinib resistance mutations T315I or N368S exhibit a faster doubling time in the absence of imatinib. ns: P > 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We expressed and purified unphosphorylated Abl KD to characterize the N368S mutant biochemically. Abl N368S was predicted to be stable according to FoldX calculations, and the experimental melting temperature of Abl N368S (46.05 ± 0.13 °C) was virtually identical to that of Abl wt (45.69 ± 0.07 °C) (SI Appendix, Fig. S1). Next, we determined how the N368S mutation affected the affinity of Abl KD for imatinib. The intrinsic protein fluorescence of Abl decreases upon imatinib binding and therefore can be used as a readout for imatinib binding (33). Consistent with our cellular NanoBRET assays, we found that the dissociation constant (Kd) of imatinib for purified Abl N386S (Kd = 11.8 ± 0.9 nM) was only 1.5-fold higher than that of Abl wt (8.4 ± 0.8 nM), making it unlikely that this difference contributes to inhibitor efficacy in vivo (Fig. 3E).
When evaluating the enzymatic activity of Abl N368S KD, we were surprised that unphosphorylated Abl N368S had no detectable catalytic activity (kcat unmeasurable) compared to Abl wt (kcat = 129.0 ± 31.5 min−1). However, the activation loop phosphorylation of N368S by Hck increased kinase activity (kcat = 36.9 ± 17.6 min−1). This phosphorylation activity was still sevenfold lower than the activity of Abl wt phosphorylated at the activation loop by Hck (kcat 258.0 ± 52.2 min−1) (Fig. 3C). Next, we determined how the N368S mutation affected the Michaelis–Menten constant (Km) of Abl KD for ATP. We found that activation loop–phosphorylated Abl (pAbl) N368S (KmATP = 110.3 ± 1.9 μM) had greater than twofold Km for ATP as pAbl wt and nonphosphorylated Abl wt (KmATP = 53.2 ± 8.9 μM) (Fig. 3 D and F). This suggested that the mutation had a twofold lower ATP affinity, and therefore, increased competition between ATP and imatinib was not the mechanism behind N368S’s resistance phenotype.
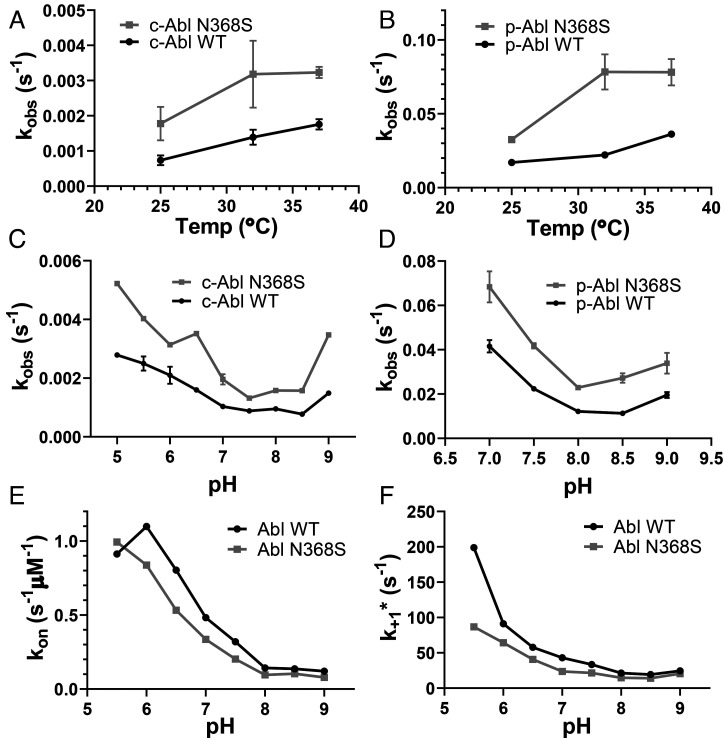
N368S Affects Drug Dissociation Kinetics of Isolated KD.
It is conceivable that regulatory domains, interactions with cellular proteins or ligands, or biological processes such as protein turnover facilitate the faster dissociation of imatinib from Abl N368S in cells. Thus, we determined whether purified Abl N368S KD also showed an increased rate of imatinib dissociation using a competition assay between the two ATP-competitive drugs bosutinib and imatinib. Upon binding to Abl kinase, bosutinib fluorescence at 460 nm increases, making it a useful probe to monitor inhibitor binding to kinases (34, 35). Briefly, after the formation of Abl–imatinib complexes, we add a 25-fold molar excess of bosutinib. Bosutinib competes with imatinib for the ATP-binding site, and upon binding, there is an increase in fluorescence at 460 nm. We found that imatinib dissociated twofold faster from Abl N368S than from Abl wt at 25, 32, and 37 °C (Fig. 4A). Similarly, imatinib dissociated twofold faster from phosphorylated Abl N368S than from Abl wt at 25, 32, and 37 °C (Fig. 4B). These results suggested that faster imatinib dissociation from full-length Abl N368S in cells is directly caused by an increased inhibitor off rate from the KD.
Fig. 4.
In vitro kinetics of imatinib binding and dissociation from purified Abl KD. (A and B) Dissociation kinetics of imatinib from unphosphorylated (A) and activation loop–phosphorylated (B) Abl wt and N368S at various temperatures. (C and D) Dissociation of imatinib from unphosphorylated (C) and activation loop–phosphorylated (D) Abl wt and N368S at various pH values. (E and F) Binding kinetics of imatinib to Abl wt and N368S at various pH values yielding the rate constant of imatinib binding (E) and the rate constant of the DFG-flip (F).
Faster Imatinib Dissociation From Abl N368S Is pH Independent.
We showed previously that the rate of imatinib binding to Abl KD is pH dependent. For imatinib to bind, the sidechain of Asp381 in the DFG motif must rotate from a position where it faces into the active site (DFG-Asp-in) to an orientation where it faces the solvent (DFG-Asp-out), a process referred to as the DFG-flip. The protonation of Asp381 reduces the energetic barrier for the 180° crankshaft-like rotation of the DFG motif and increases the imatinib-binding rate without changing the affinity (36). This indicates that the DFG-flip may be the rate-limiting step for the imatinib-binding process. The kinetics of the DFG-flip and further conformational exchange between Abl’s active conformation and the inactive binding-competent conformation are acutely sensitive to changes in pH (36, 37). Given the proximity of the N368 residue to D381, we wanted to investigate whether the N368S substitution subtly altered D381’s protonation state, thereby affecting the rate-limiting DFG-flip.
We determined how pH affected the kinetics of imatinib binding to Abl wt and N368S using our previously described fast transient stopped-flow assay (33). We found that the rate constants for imatinib binding (kon) and the DFG-flip (k+1*) were about 30% slower for N368S (kon = 0.096 ± 0.006 μM−1 ⋅ s−1 and k+1* = 14.6 s−1, respectively, at pH 8.0) than for Abl wt (kon = 0.143 ± 0.076 μM−1 ⋅ s−1 and k+1* = 21.4 s−1, respectively, at pH 8.0) across the pH range (Fig. 4 E and F) (36).
Next, we tested whether the dissociation rate of imatinib from Abl is pH dependent, which would indicate that the DFG-flip was rate limiting for imatinib dissociation. We measured imatinib dissociation kinetics at different pH conditions ranging from pH 5.0 to 9.0. We found that for both Abl wt and N368S, imatinib dissociation kinetics were independent of pH. This makes it unlikely that the protonation-sensitive DFG-flip was a rate-limiting step in imatinib dissociation. Imatinib dissociated from Abl N368S twofold faster than from Abl wt across the tested pH range (Fig. 4C). Likewise, imatinib dissociated twofold faster from phosphorylated Abl N368S than from wt across the tested pH range (Fig. 4D).
Taken together, these data show that compared to Abl wt, faster imatinib dissociation from N368S is not facilitated by a faster DFG-flip, which in fact is slightly slower in N368S. Instead, the N368S mutation mainly alters the dissociation process.
Ba/F3 Cells Expressing Bcr-Abl N368S Proliferate and Remain Sensitive to Imatinib under Constant Drug Concentration.
Next, we determined whether the Abl N368S mutation had any effect on cellular proliferation. To show the relevance of the N368S mutations for CML, we tested N368S in the context of Bcr-Abl using Ba/F3 cells. These cells are widely used in kinase drug discovery because they can only survive in the absence of IL-3 if they constitutively express an enzymatically active tyrosine kinase (38). Upon expression of enzymatically active human Bcr-Abl, murine hematopoietic Ba/F3 cells proliferate independently of IL-3, and inhibition of Bcr-Abl kinase activity prevents proliferation (1). Ba/F3 cells expressing the Bcr-Abl N368S mutant proliferated in the absence of IL-3, which is further evidence that the Bcr-Abl N368S mutant is active in a cellular context. We determined the imatinib cellular IC50 for three cell lines expressing Bcr-Abl wt or resistance-associated mutations (wt, T315I, N368S). Imatinib inhibited the proliferation of cells expressing Bcr-Abl wt and N368S with IC50 values of 1.23 ± 0.47 and 1.78 ± 0.60 μM, respectively (Fig. 3F). We used a cell line expressing the T315I gatekeeper mutation as a control and found that it was highly resistant to imatinib as expected (IC50 = 36.6 ± 12.0 μM). These results were in excellent agreement with the NanoBRET target engagement results for wt, N368S, and T315I (IC50 values of 0.44 ± 0.14 μM, 0.44 ± 0.2 μM, >10 μM, respectively), considering the use of different cell lines and Abl construct in the different assays.
We also determined whether the presence of imatinib resistance–associated mutations affects cell doubling time. We found that doubling times were statistically significantly shorter for Ba/F3 cells expressing Abl T315I (33.7 ± 2.0 h) and N368S (32.9 ± 1.7 h) than for cells expressing Abl wt (51.9 ± 1.1 h) (Fig. 3G). Because of the exponential growth of cells, the 1.6-fold faster doubling times results in 10 times more cells expressing N368S than wt after 300 h. Taken together, Bcr-Abl N368S accelerates cell proliferation and is inhibited by imatinib under constant drug concentration similarly to wt.
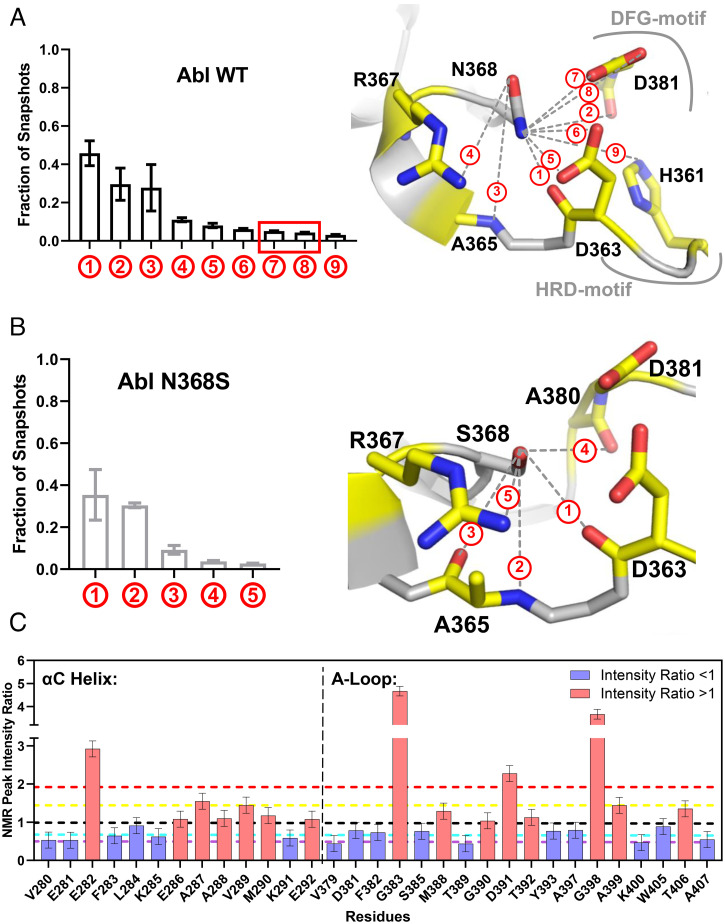
The H-Bond Interaction between N368 and D381 Is Only Observed in the DFG-in Conformation.
Since drug binding and dissociation is a dynamic process, we speculated that protein dynamics could be affected by the N368S mutation. Therefore, we performed 12 500-ns molecular dynamics (MD) simulations with a self-adjusted mixture sampling (SAMS)-enhanced sampling scheme (39) that accelerates sampling of DFG-flip events. We chose two different starting structures: Abl in the DFG-out conformation bound to imatinib (PDB entry 2HYY) and Abl in the DFG-in conformation bound to dasatinib (PDB entry 2GQG). Simulations were run for each of these starting models in the presence and absence of the corresponding ligand and with and without the N368S mutation. We employed two slightly different SAMS biasing strategies, which we describe in Materials and Methods.
Among the 12 simulations, we observed five DFG flips in the apo simulations and none in the presence of ligands (SI Appendix, Table S3). In four simulations of Abl wt and N368S, the aspartate of the DFG motif flipped from a solvent-exposed, outward orientation to an orientation facing into the active site of the kinase (out-to-in flip). In one additional simulation of N368S, the DFG motif flipped in the reverse direction (in to out). This indicated that N368S could perform the DFG flip in both directions.
Next, we investigated the H-bonding of Abl wt and N368S within the active site. We found that the sidechain of N368 formed an H-bond with DFG-Asp only in the DFG-Asp–in conformation (Fig. 5 A and B), and those interactions were absent in the N368S mutant. The H-bond between N368 and D381 likely stabilized the DFG-in (active) conformation and acted as a kinetic barrier for the DFG flip. Conversely, loss of the H-bond in Abl N368S would be expected to destabilize the active conformation, consistent with the low specific activity of N368S. Next, we investigated how the N368S mutation affected the solution dynamics of the activation loop. Based on the hydrogen-bonding pattern of Asn368, we hypothesized that Asn368 could anchor the amino-terminal end of the activation loop and helix αC. We therefore compared 1H-15N NMR peak intensities for wt and N368S (Fig. 5C and SI Appendix, Fig. S3). Sharper NMR peaks leading to higher signal intensities correspond to increased solution dynamics. We found that several residues within the activation loop of N368S showed increased dynamics compared to wt. In particular, G383 in the DFG motif exhibited a fivefold stronger signal relative to wt, indicating a sharp increase in dynamics. Similarly, all residues in helix αC that face the DFG motif showed increased dynamics in N368S. This observation is consistent with the notion that in the N368S mutant, the activation loop and helix αC become more dynamic. In a separate study, we have performed MD simulations of the dissociation process of imatinib from Abl wt and N368S (40). These simulations quantitatively reproduce the experimentally observed imatinib dissociation rates for wt and N368S. The MD studies indicated that the flexibility of the activation loop and helix αC are crucial for imatinib dissociation, consistent with our experimental observations.
Fig. 5.
MD and NMR analyses of the N368S mutant protein. (A) Fraction of snapshots from MD simulation in which the sidechain of N368 engages in a hydrogen bond with neighboring atoms (Left) illustrated in the structure of Abl kinase in the active conformation (PDB entry 2GQG) (Right). (B) Fraction of snapshots from MD simulations in which the sidechain of S368 engages with neighboring atoms (Left) illustrated on the model of S368 in the structure of active Abl wt (PDB entry 2GQG). (C) NMR 1H-15N peak intensity ratios (IN368S/IWT) for residues in helix αC and the activation loop. Black line: ratio of 1, yellow line: mean of all values larger than 1, red line: 1 SD from mean of values larger than 1, cyan line: mean of all values smaller than 1, purple line: 1 SD from mean of values smaller than 1.
Discussion
In this study, we determined the target engagement of 94 resistance-associated mutations of Abl kinase for the cancer drugs imatinib and dasatinib in cells. Surprisingly, we found that one-third of imatinib resistance–associated mutations remain sensitive to imatinib and bind with similar affinity as Abl wt. One explanation could be that some mutations may not cause resistance to imatinib, but they exist on the background of other resistance mechanisms such as up-regulation of drug exporters, down-regulation of drug importers, increased serum protein binding, or overexpression of Src kinase (41–43). Given that Bcr-Abl activity underlies CML and imatinib still inhibits Bcr-Abl, in theory, the drug should still be an effective therapeutic. Importantly, this would indicate that approximately one-third of mutations in the relevant databases are incorrectly assigned. If identified in patient samples, the presence of these mutations should not necessarily alter the clinical standard of care. Imatinib could remain the first-line therapy for patients harboring these mutations unless other clinical metrics indicate disease resistance to imatinib.
Another potential explanation could be that these distal mutations may destabilize the imatinib-binding competent conformation of Abl as demonstrated by a recent NMR investigation of key allosteric mutants (37, 44). A further dissection of the remaining mutations in this context could further illuminate Abl’s allosteric regulation.
Potential limitations of this Proof of Principle pilot study include the use of full-length Abl instead of Bcr-Abl and the use of HEK293T cells instead of a more disease-relevant hematopoietic cell line. Differences in the concentration, activity, and localization of other proteins and ligands in Bcr-Abl–positive leukemic cells could affect drug–target engagement. Therefore, we cannot exclude the possibility that some mutations may be more resistant to imatinib in the Bcr-Abl protein and in the context of leukemic cells. For example, mutations might sensitize Abl to tetramerization via the Bcr region or to activation loop phosphorylation through other kinases, leading to increased Abl activation loop phosphorylation and reduced imatinib affinity (45).
To explore novel molecular resistance mechanisms, we determined the cellular drug dissociation kinetics for 34 Abl mutants. Given that drug residence time is a better predictor for drug efficacy than affinity alone (12, 14, 17), we predicted that, conversely, mutations could reduce drug efficacy by reducing drug residence time. We found that Abl N368S, V299L, and G251E had similar imatinib affinity compared to wt, but their imatinib dissociation was faster than wt. This observation posed the possibility of kinetic drug resistance caused by mutations. Interestingly, mutations at the analogous N368 position are also present in BLK, BRAF, EGFR, and PTK6 in patient cancer samples (29, 46), and BRAF N581S has been described previously as a sorafenib resistance mutation (47). Given the high sequence conservation at the N368 position, this suggests that this kinetic resistance mechanism may not be limited to Abl alone. Such kinetic mutations may not be detectable in assays with constant drug concentration, and their resistance phenotype may vary from patient to patient by other pharmacodynamic and pharmacokinetic parameters. Importantly, kinetic resistance mechanisms may be addressable by altering the treatment schedule, for example, from a single daily dose to multiple doses.
Our biochemical and biophysical studies indicate that imatinib dissociation is not a simple reversal of the binding process. Consistent with such a mechanism, a recent computational study using adaptive sampling between conformational “milestones” proposes a dissociation pathway of imatinib from Abl where the kinase lingers in the DFG-out conformation via a temporary deformation of the N-lobe (48). To address the mechanism of N368S, we performed extensive simulations of imatinib dissociation from wt and N368S kinase using infrequent metadynamics (40, 49). This method captured qualitatively and quantitatively the off rates for both systems and demonstrated how the faster off rate for the mutant is a direct consequence of enhanced protein flexibility in the DFG region.
The work presented here is a study that systematically evaluates a large panel of clinical mutations for drug engagement and changes in residence time in live cells. This workflow provides a template for broader evaluations of disease-relevant mutations and how such mutations impact drug affinity and binding and dissociation kinetics.
With more clinical sequencing efforts of patient samples, we expect that databases on disease-related mutations will grow rapidly in size. Thus, our results indicate that the experimental characterization of mutant phenotypes in cells remains essential to distinguish mutations that are causal for observed clinical phenotypes versus those observed in the background of other biological processes.
Materials and Methods
Drug-Binding Studies.
The KD of human c-Abl (UniProtID: P00519, residues 248 to 531), human Abl 1a numbering, was expressed in Escherichia coli and purified with a Tobacco Etch Virus-cleavable His-tag as described previously (50). NanoBRET assay for target engagement was performed in HEK293 cells according to manufacturer’s conditions using a human Abl1a full-length construct with an N-terminal fusion of NanoLuc luciferase.
Additional materials and methods are provided in SI Appendix, including detailed information on protein expression and purification, NMR spectroscopy, tissue culture, Ba/F3 cell viability assays, NanoBRET assay for target engagement and residence time, molecular simulations and drug affinity, and drug-binding kinetics experiments with purified protein.
Acknowledgments
We acknowledge support for this work by NIH R35 GM119437 (M.A.S.), NIH S10 OD028478 (M.A.S.), NIH F30 CA225172 (A.L.), NIH F30 CA260771 (A.M.R.), NIH T32 GM008444 (A.L., A.M.R.), NIH P30 CA008748 (J.D.C.), NIH R01 GM121505 (J.D.C.), and the Sloan Kettering Institute and Cycle for Survival (J.D.C.). The content is solely our responsibility and does not necessarily represent the official views of the NIH. We are grateful to Brooks Bond-Watts and Chris Jeans of the QB3 MacroLab at the University of California, Berkeley for their work on the high-throughput expression of the Abl mutants. S.K. and B.-T.B. are grateful for support by the Structural Genomics Consortium, a registered charity (no. 1097737) that receives funds from AbbVie, Bayer AG, Boehringer Ingelheim, the Canada Foundation for Innovation, Eshelman Institute for Innovation, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond, Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN Grant 875510], Janssen, Merck KGaA (EMD in Canada and the United States), Pfizer, and Takeda as well as support from the German translational cancer network (DKTK), the Deutsche Forschungsgemeinschaft network grant SFB1399, and the Frankfurt Cancer Institute. We would like to thank Barbara P. Craddock, Sultan Ahmed, and W. Todd Miller for helpful discussions and troubleshooting during cellular experiments. We are grateful to Victoria R. Mingione for her insightful comments during manuscript editing and preparation.
Footnotes
Author contributions: B.-T.B., A.M.R., J.G., S.K.A., S.K., J.D.C., and M.A.S. designed research; A.L., B.-T.B., A.M.R., Y.P., J.T., H.P., J.G., S.K.A., S.K., J.D.C., and M.A.S. performed research; M.B.R. and S.K. contributed new reagents/analytic tools; A.L., B.-T.B., A.M.R., Y.P., J.G., S.K.A., M.B.R., S.K., J.D.C., and M.A.S. analyzed data; and A.L., B.-T.B., A.M.R., Y.P., J.G., S.K.A., M.B.R., S.K., J.D.C., and M.A.S. wrote the paper.
Competing interest statement: J.D.C. is a current member of the Scientific Advisory Board of OpenEye Scientific Software, Redesign Science, and Interline Therapeutics and has equity interests in Redesign Science and Interline Therapeutics. The J.D.C. laboratory receives or has received funding from multiple sources, including the NIH, the NSF, the Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Entasis Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biotechnology, Bayer, XtalPi, Interline Therapeutics, the Molecular Sciences Software Institute, the Starr Cancer Consortium, the Open Force Field Consortium, Cycle for Survival, a Louis V. Gerstner Young Investigator Award, and the Sloan Kettering Institute.
This article is a PNAS Direct Submission.
1A.L., B.-T.B., and A.M.R. contributed equally to this work.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111451118/-/DCSupplemental.
Data Availability
MD simulation data/codes have been deposited in GitHub (https://github.com/choderalab/Abl_kinase_N368S). All other study data are included in the article and/or SI Appendix.
References
- 1.Jemal A., et al., Cancer statistics, 2007. CA Cancer J. Clin. 57, 43–66 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Parker S. L., Tong T., Bolden S., Wingo P. A., Cancer statistics, 1997. CA Cancer J. Clin. 47, 5–27 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Gorre M. E., et al., Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Azam M., Latek R. R., Daley G. Q., Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell 112, 831–843 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Shah N. P., et al., Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2, 117–125 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Lamontanara A. J., Gencer E. B., Kuzyk O., Hantschel O., Mechanisms of resistance to BCR-ABL and other kinase inhibitors. Biochim. Biophys. Acta 1834, 1449–1459 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Tate J. G., et al., COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravarty D., et al., OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 2017, 2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbin A. S., La Rosée P., Stoffregen E. P., Druker B. J., Deininger M. W., Several Bcr-Abl kinase domain mutants associated with imatinib mesylate resistance remain sensitive to imatinib. Blood 101, 4611–4614 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Eck M. J., Manley P. W., The interplay of structural information and functional studies in kinase drug design: Insights from BCR-Abl. Curr. Opin. Cell Biol. 21, 288–295 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Patel A. B., O’Hare T., Deininger M. W., Mechanisms of resistance to ABL kinase inhibition in chronic myeloid leukemia and the development of next generation ABL kinase inhibitors. Hematol. Oncol. Clin. North Am. 31, 589–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copeland R. A., Pompliano D. L., Meek T. D., Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5, 730–739 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Dahl G., Akerud T., Pharmacokinetics and the drug-target residence time concept. Drug Discov. Today 18, 697–707 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Walkup G. K., et al., Translating slow-binding inhibition kinetics into cellular and in vivo effects. Nat. Chem. Biol. 11, 416–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.V. Georgiet al., . Binding Kinetics Survey of the Drugged Kinome. J. Am. Chem. Soc. 140, 15774–15782 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Holdgate G. A., Gill A. L., Kinetic efficiency: The missing metric for enhancing compound quality? Drug Discov. Today 16, 910–913 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Lu H., Tonge P. J., Drug-target residence time: Critical information for lead optimization. Curr. Opin. Chem. Biol. 14, 467–474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swinney D. C., The role of binding kinetics in therapeutically useful drug action. Curr. Opin. Drug Discov. Devel. 12, 31–39 (2009). [PubMed] [Google Scholar]
- 19.Wood E. R., et al., A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 64, 6652–6659 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Willemsen-Seegers N., et al., Compound selectivity and target residence time of kinase inhibitors studied with surface plasmon resonance. J. Mol. Biol. 429, 574–586 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Bradshaw J. M., et al., Prolonged and tunable residence time using reversible covalent kinase inhibitors. Nat. Chem. Biol. 11, 525–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robers M. B., et al., Target engagement and drug residence time can be observed in living cells with BRET. Nat. Commun. 6, 10091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robers M. B., et al., Quantifying target occupancy of small molecules within living cells. Annu. Rev. Biochem. 89, 557–581 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Vasta J. D., et al., Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem. Biol. 25, 206–214.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisberg E., et al., AMN107 (nilotinib): A novel and selective inhibitor of BCR-ABL. Br. J. Cancer 94, 1765–1769 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robers M. B., et al., Quantitative, real-time measurements of intracellular target engagement using energy transfer. Methods Mol. Biol. 1888, 45–71 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Peng B., et al., Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J. Clin. Oncol. 22, 935–942 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Elias M. H., et al., BCR-ABL kinase domain mutations, including 2 novel mutations in imatinib resistant Malaysian chronic myeloid leukemia patients-Frequency and clinical outcome. Leuk. Res. 38, 454–459 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Gauthier N. P., et al., MutationAligner: A resource of recurrent mutation hotspots in protein domains in cancer. Nucleic Acids Res. 44, D986–D991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McSkimming D. I., et al., ProKinO: A unified resource for mining the cancer kinome. Hum. Mutat. 36, 175–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J., et al., Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry 32, 2154–2161 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Bossemeyer D., Engh R. A., Kinzel V., Ponstingl H., Huber R., Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24). EMBO J. 12, 849–859 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeliger M. A., et al., c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure 15, 299–311 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Levinson N. M., Boxer S. G., Structural and spectroscopic analysis of the kinase inhibitor bosutinib and an isomer of bosutinib binding to the Abl tyrosine kinase domain. PLoS One 7, e29828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albanese S. K., et al., An open library of human kinase domain constructs for automated bacterial expression. Biochemistry 57, 4675–4689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan Y., et al., A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc. Natl. Acad. Sci. U.S.A. 106, 139–144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie T., Saleh T., Rossi P., Kalodimos C. G., Conformational states dynamically populated by a kinase determine its function. Science 370, eabc2754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warmuth M., Kim S., Gu X. J., Xia G., Adrián F., Ba/F3 cells and their use in kinase drug discovery. Curr. Opin. Oncol. 19, 55–60 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Tan Z., Optimally adjusted mixture sampling and locally weighted histogram analysis. J. Comput. Graph. Stat. 26, 54–65 (2017). [Google Scholar]
- 40.Shekhar M., Smith Z., Seeliger M. A., Tiwary P., Protein flexibility and dissociation pathways differentiation can explain onset of resistance mutations: Abl kinase and Gleevec case study. bioRxiv [Preprint] (2021).10.1101/2021.07.02.450932. Accessed 5 July 2021.
- 41.Gambacorti-Passerini C., et al., Alpha1 acid glycoprotein binds to imatinib (STI571) and substantially alters its pharmacokinetics in chronic myeloid leukemia patients. Clin. Cancer Res. 9, 625–632 (2003). [PubMed] [Google Scholar]
- 42.Pene-Dumitrescu T., Smithgall T. E., Expression of a Src family kinase in chronic myelogenous leukemia cells induces resistance to imatinib in a kinase-dependent manner. J. Biol. Chem. 285, 21446–21457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shukla S., Sauna Z. E., Ambudkar S. V., Evidence for the interaction of imatinib at the transport-substrate site(s) of the multidrug-resistance-linked ABC drug transporters ABCB1 (P-glycoprotein) and ABCG2. Leukemia 22, 445–447 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Saleh T., Rossi P., Kalodimos C. G., Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat. Struct. Mol. Biol. 24, 893–901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindler T., et al., Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289, 1938–1942 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Miller M. L., et al., Pan-cancer analysis of mutation hotspots in protein domains. Cell Syst. 1, 197–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan P. T., et al.; Cancer Genome Project, Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Narayan B., Buchete N. V., Elber R., Computer simulations of the dissociation mechanism of Gleevec from Abl Kinase with milestoning. J. Phys. Chem. B 125, 5706–5715 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiwary P., Mondal J., Berne B. J., How and when does an anticancer drug leave its binding site? Sci. Adv. 3, e1700014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeliger M. A., et al., High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 14, 3135–3139 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
MD simulation data/codes have been deposited in GitHub (https://github.com/choderalab/Abl_kinase_N368S). All other study data are included in the article and/or SI Appendix.