Abstract
The gene regulatory networks underlying Ciona notochord fate specification and differentiation have been extensively investigated, but the regulatory basis for regionalized expression within the notochord is not understood. Here we identify three notochord-expressed genes, C11.331, C12.115 and C8.891, with strongly enriched expression in the secondary notochord cells at the posterior tip of the tail. C11.331 and C12.115 share a distinctive expression pattern that is highly enriched in the secondary notochord lineage but also graded within that lineage with the strongest expression at the posterior tip. Both genes show similar responses to pharmacological perturbations of Wnt and FGF signaling, consistent with an important role for Wnt and FGF ligands expressed at the tail tip. Reporter analysis indicates that the C11.331 cis-regulatory regions are extensively distributed, with multiple non-overlapping regions conferring posterior notochord-enriched expression. Fine-scale analysis of a minimal cis-regulatory module identifies discrete positive and negative elements including a strong silencer. Truncation of the silencer region leads to increased expression in the primary notochord, indicating that C11.331 expression is influenced by putative regulators of primary versus secondary notochord fate. The minimal CRM contains predicted ETS, GATA, LMX and Myb sites, all of which lead to reduced expression in secondary notochord when mutated. These results show that the posterior-enriched notochord expression of C11.331 depends on multiple inputs, including Wnt and FGF signals from the tip of the tail, multiple notochord-specific regulators, and yet-to-be identified regulators of regional identity within the notochord.
Introduction
The Ciona notochord consists of only 40 cells that intercalate to form a tapered, single-file column. Cell fate specification in the Ciona notochord has been extensively studied. In ascidians, the primary notochord cells are derived from blastomere pairs A7.3 and A7.7 and give rise to the anterior 32 cells in the intercalated notochord, whereas the secondary notochord cells are derived from B8.6 and give rise to the posterior 8 cells (Nishida, 1987). In Ciona, A7.3 and A7.7 become fate-restricted to notochord at the 64-cell stage downstream of both beta-catenin mediated mesendodermal specification and FGF signaling (Hudson et al., 2016, 2013; Yasuo and Hudson, 2007). B8.6 becomes fate-restricted at the 112-cell stage via different mechanisms that involve a Nodal/Delta2 signaling relay initiated by the lateral b6.5 cells (Hudson and Yasuo, 2006, 2005, Imai et al., 2006, 2002).
Both primary and secondary notochord cells express the key notochord-specific transcription factor Brachyury (Bra) (Corbo et al., 1997; Yasuo and Satoh, 1993), and the majority of notochord-enriched genes are expressed uniformly throughout the notochord (Hotta et al., 2000, 1999; José-Edwards et al., 2011; Kugler et al., 2008; Reeves et al., 2017). We have previously shown, however, that notochord cell sizes, shapes and behaviors vary depending on both lineage and anterior-posterior position (Carlson et al., 2015; Veeman and Smith, 2013). We have also shown that a subset of notochord-enriched genes are differentially expressed within subregions of the notochord (Reeves et al., 2014). The regulatory basis for regionalized notochord expression has received no previous attention.
Differential gene expression between primary and secondary notochord is of particular interest because secondary notochord cells are quite distinct from primary notochord cells in their morphology and behavior. Secondary notochord cells are more stereotyped in their patterns of intercalation than primary cells and give rise to a particularly distinct taper towards the tail’s distal tip (Carlson et al., 2015; Veeman and Smith, 2013). Unlike primary cells, they still intercalate effectively even in mutants for the planar cell polarity pathway gene Prickle (Jiang et al., 2005). Elements of differential gene expression have previously been noted between primary and secondary notochord cells, including a gene upregulated at both the anterior and posterior tips of the notochord, one expressed in a posterior to anterior gradient, and another expressed throughout the notochord but with visible enrichment in the secondary lineage (Reeves et al., 2014). Despite these indications of underlying patterning, markers that were strongly differentially expressed between primary and secondary notochord cells were previously elusive.
We recently used RNAseq on flow-sorted notochord cells to identify a large set of 1364 genes predicted to be enriched in the notochord. Extensive in situ validation of 151 genes showed that ~90% were indeed notochord enriched (Reeves et al., 2017 and unpublished data). In the course of this we identified several new genes showing regionalized notochord expression, including three that are strongly enriched at the notochord’s posterior tip. Here we characterize the expression patterns of these posterior notochord tip-specific genes with fine detail and investigate the molecular mechanisms controlling their regionalized expression.
Results
Secondary-enriched notochord expression
Although the majority of notochord enriched genes we identified by RNAseq are expressed uniformly across the notochord (Reeves et al., 2017), a subset display regionalized expression within the notochord. Fig. 1 shows in situ expression patterns for three genes with strongly differential expression between primary and secondary notochord. C11.331 has homology to the fibulin/hemicentin family of secreted extracellular matrix proteins (Timpl et al., 2003), but is not a clear ortholog of any vertebrate fibulin. C12.115 and C8.891 are both predicted SLC family solute carriers. We refer to all three here by their KH2012 gene model names (Brozovic et al., 2018; Satou et al., 2008) for unambiguity. During early notochord intercalation at mid neurula (Hotta stage 16) (Hotta et al., 2007), C11.331 is barely detectable (Fig. 1A), but is distinctly expressed at the notochord’s posterior tip late in intercalation at early tailbud stage 20 (Fig. 1B) and during tail elongation at late tailbud stage 23 (Fig. 1C). C12.115 has posterior-enriched notochord expression at all three stages (Fig. 1D–F) which resolves over time to be increasingly specific to the notochord’s posterior tip. C8.891 is qualitatively different than the other two genes and shows a more graded expression pattern at the first two stages (Fig. 1G,H) before developing a sharper transition between primary and secondary at stage 23 (Fig. 1I).
Fig. 1. Secondary-enriched notochord genes.
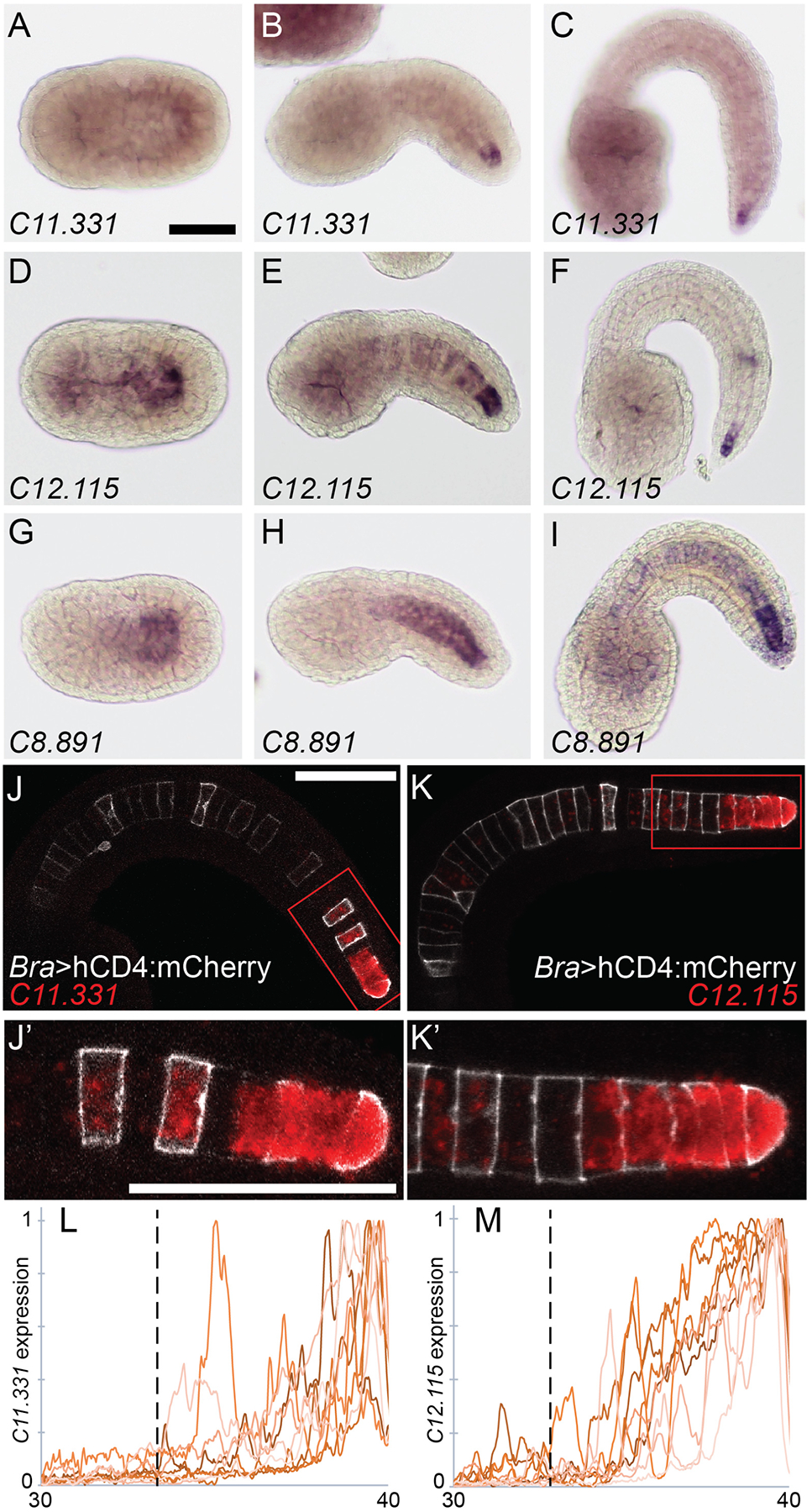
In situ hybridization of probes against (A-C) KH.C11.331, (D-F) KH.C12.115 and (G-I) KH.C8.891. Expression was tested during early intercalation at Hotta stage 16 (A, D, G), late in intercalation at Hotta stage 20 (B, E, H) and during notochord elongation at Hotta stage 23 (C, F, I). J-K) Fluorescent in situ hybridization against C11.331 (J) and C12.115 (K) in red at Hotta stage 22. Embryos were electroporated with Bra>hCD4:mCherry (white). J’-K’) Closeup of secondary notochord region indicated by red box in (J,K). L-M) Background subtracted and normalized expression of C11.331 (L) and C12.115 (M) in notochord cells 30–40. Nine embryos for each gene are shown. The location of the primary/secondary border is indicated by the vertical dotted line. Scale bars = 50 microns.
To better quantify their expression patterns, we also imaged fluorescent in situ hybridizations against C11.331 and C12.115 by confocal microscopy. This allowed the boundary between primary and secondary notochord to be clearly identified. Both genes showed expression in secondary but not primary notochord (Fig. 1J,K). For both C11.331 and C12.115, this secondary-specific expression was stronger at the notochord’s posterior tip than at the boundary between primary and secondary notochord. Fig. 1L,M show quantitative analyses of these expression patterns across nine embryos each, confirming that both genes have graded expression in the secondary notochord that is strongest at the posterior tip.
Perturbation of Wnt and FGF signaling
These expression patterns suggested the potential involvement of a secreted signaling molecule expressed at the posterior tip of the tail. Candidate molecules for such a signal include Wnt5, FGF8/17/18 and FGF9/16/20, all of which are expressed in tail tip ectoderm or posterior tail muscles (Hudson et al., 2007; Hudson and Yasuo, n.d.; Ikuta et al., 2010; Imai et al., 2004). We tested their involvement pharmacologically using the FGF inhibitor U0126 and the canonical Wnt pathway activator BIO. Both of these drugs have been extensively used in Ciona and we used doses previously established to be specific and effective (Dumollard et al., 2013; Hudson et al., 2013; Racioppi et al., 2014; Sakabe et al., 2006). We treated the embryos with drugs starting at mid gastrula (Hotta stage 12). This is after notochord has been specified and has completed all cell divisions (Hotta et al., 2007), but before the earliest detectable expression of C11.331, C12.115 or C8.891 by in situ hybridization (data not shown). A Wnt pathway inhibitor would be conceptually appealing but no such reagents have been validated in Ciona embryos.
Both drugs caused mild defects in tail morphogenesis, as has been described previously for U0126 (Ikuta et al., 2010). As expected, the BIO and U0126 treatments had no effect on the expression of the key notochord transcriptional regulator Brachyury (Fig. 2A–D), but the drugs caused strong effects on the expression of the three posterior-specific notochord markers. C11.331 and C12.115 responded to both drugs in similar ways. BIO treatment led to the expansion of C11.331 and C12.115 expression throughout primary notochord (compare Fig. 2E and I to Fig. 2F and J). Interestingly, this ectopic expression was often strongest at the notochord’s anterior tip, giving rise to an overall expression pattern reminiscent of BCamL (KH.C2.209) (Reeves et al., 2014) in terms of being enriched at both the anterior and posterior tips of the notochord. FGF inhibition with U0126, however, led to a major decrease in the expression of both markers (Fig. 2G,K). This was particularly evident for C12.115 expression, which was virtually eliminated by U0126 treatment despite being much more robustly expressed than C11.331 in untreated embryos. C8.891 expression expanded throughout the notochord in response to Wnt pathway activation by BIO, but this ectopic expression was uniform and did not show the ‘both tips’ pattern usually seen with the other two genes (Fig. 2N). C8.891 also differed from C11.331 and C12.115 in that FGF inhibition by U0126 only caused a modest decrease in expression (Fig. 2O).
Fig. 2. Wnt and FGF signaling regulate posterior-enriched notochord expression.
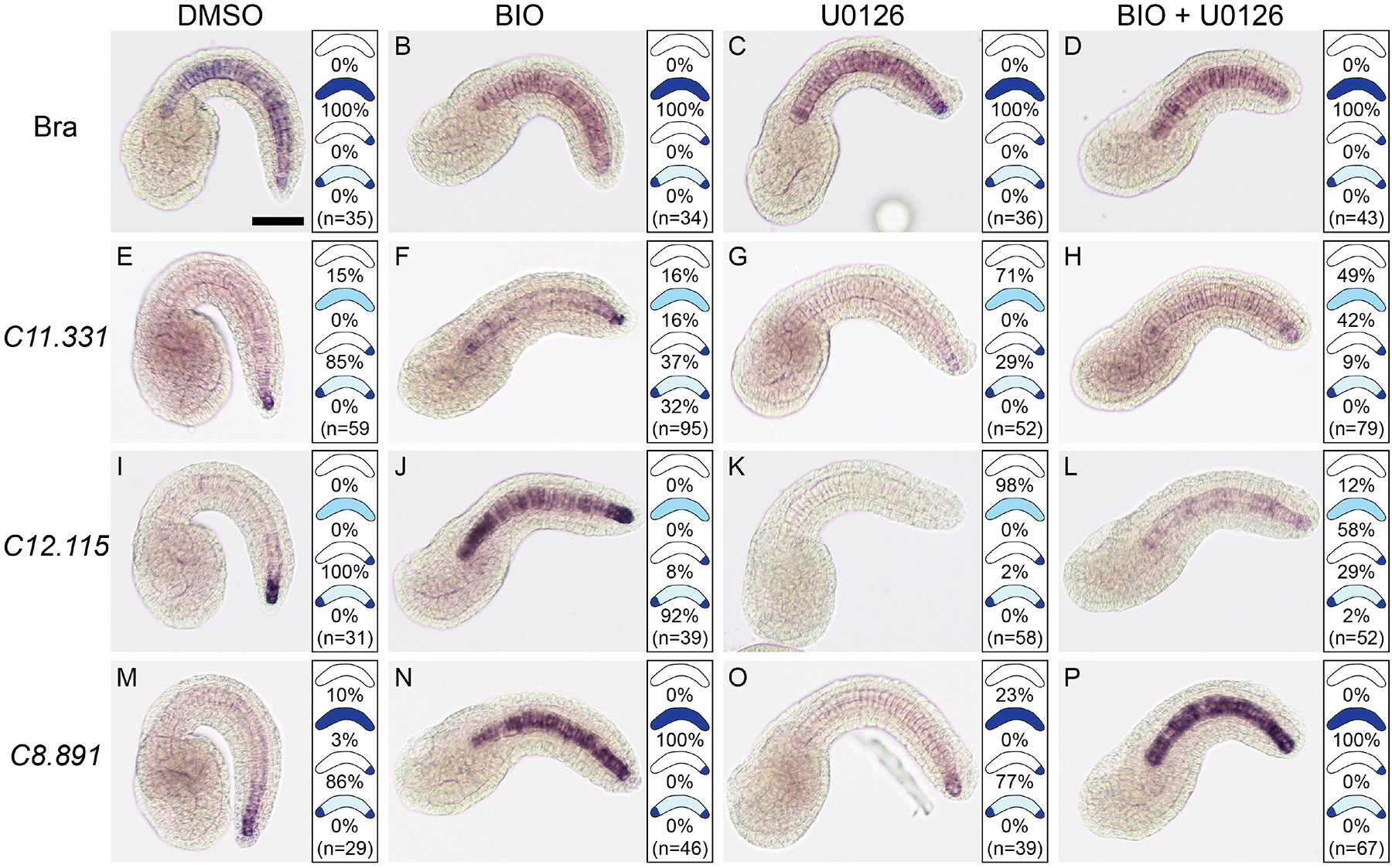
In situ hybridization against Brachyury (A-D), C11.331 (E-H), C12.115 (I-L) and C8.891 (M-P) on Hotta stage 23 embryos treated starting at mid gastrula (Hotta stage 12) with 1:1000 DMSO (A, E, I, M), 5 μM Wnt activator BIO (B, F, J, N), 4 μM FGF inhibitor U0126 (C, G, K, O) or BIO + U0126 (D, H, L, P). Representative images of the most common expression pattern for each gene/treatment are shown. Panels to the right of each image show the distribution of expression patterns observed. Scale bar = 50 microns.
The BIO and U0126 treatments indicated that both Wnt and FGF signals may act as positive regulators of C11.331, C12.115 and C8.891 expression. As a preliminary test of the potential relationships between Wnt and FGF signaling in this context, we also treated embryos with both drugs simultaneously. Both C11.331 and C12.115 showed an intermediate phenotype in which expression expanded throughout the notochord but was much fainter than in response to BIO treatment alone (Fig. 2H,L). In addition, no enrichment in the anterior tip was seen and posterior enrichment was also lost in most embryos. This lack of a clear epistatic relationship suggests that Wnt and FGF signals act in parallel upon these two genes. C8.891 again behaved differently; treatment with combined BIO and U0126 led to expanded expression throughout the notochord, but this ectopic expression was strong and indistinguishable from BIO treatment alone (Fig. 2P). FGF signaling appears to play a less important role in the regulation of C8.891 compared to the other two posterior enriched genes.
Quantitative cis-regulatory analysis
To narrow in on the molecular mechanisms responsible for posterior-specific notochord expression, we developed a dual reporter strategy. Electroporated transgenes in Ciona are expressed mosaically, and it is common for individual embryos electroporated with reporter constructs for uniformly expressed notochord genes such as Brachyury to exhibit expression in secondary notochord but not primary and vice-versa (Carlson et al., 2015; Corbo et al., 1997). Different transgenes electroporated at the same time, however, typically show common patterns of mosaicism (Zeller et al., 2006). To control for differential reporter expression due to transgene mosaicism, as opposed to bona fide differences in regionalized expression, we co-electroporated HA-tagged Histone H2B under the control of the Bra enhancer/promoter together with Venus fluorescent protein downstream of candidate regulatory regions and a basal promoter (Fig. 3A).
Fig. 3. Cis-regulatory analysis of the C11.331 upstream intergenic region.
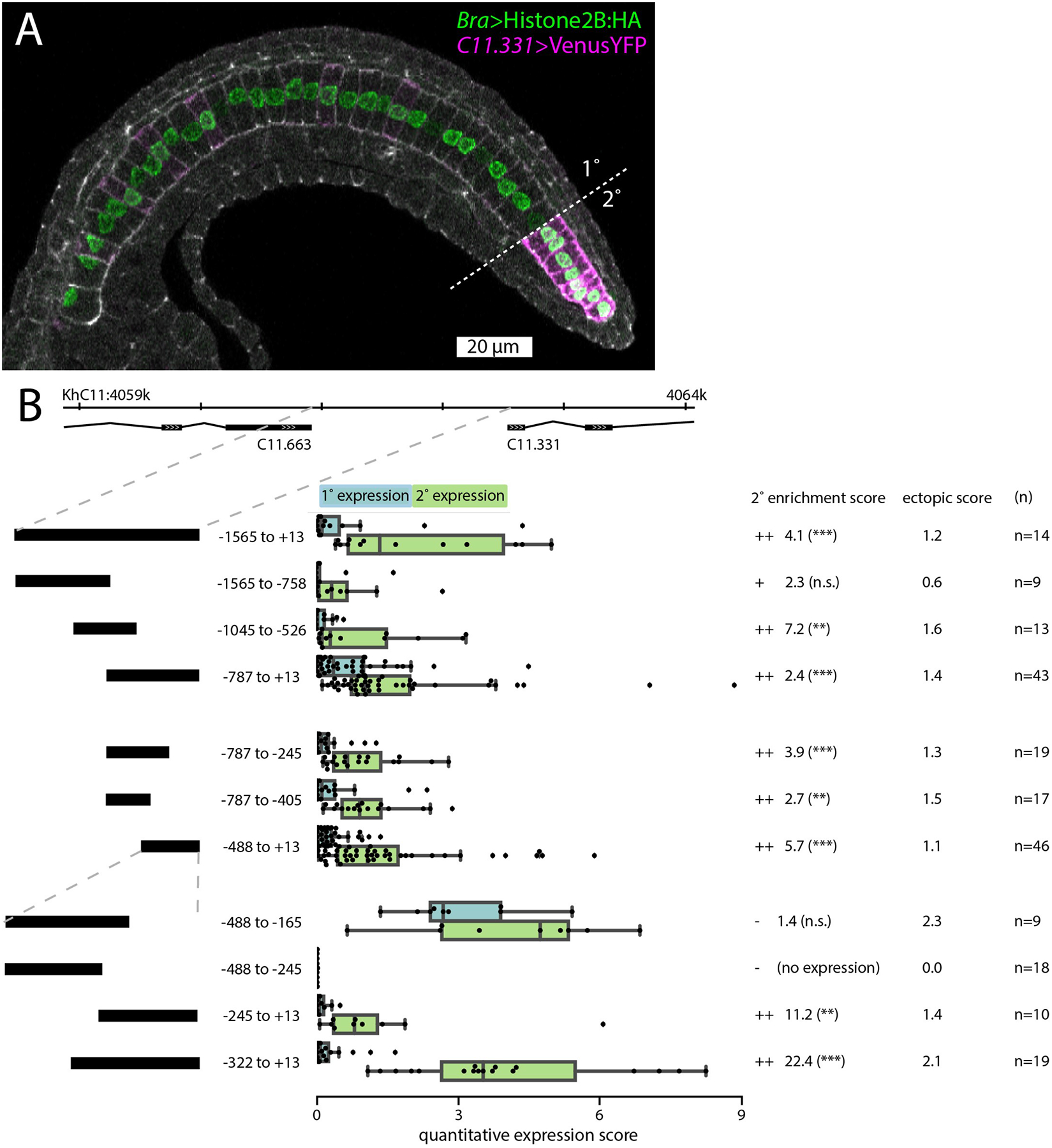
A: Representative embryo expressing C11.331 (−787 to +13)>Venus and Bra>Histone2B:HA fixed during tail elongation at Hotta stage 22. Cell cortices are labeled with Phalloidin-Alexa633 (white). Primary/secondary notochord boundary indicated with dashed line. Anterior is to the left and dorsal is to the top of the image. B: Summary statistics from reporter assays for the full-length C11.331 upstream region and a series of truncation mutants. Construct coordinates refer to the predicted transcriptional start site for C11.331. C11.331 quantitative expression score distributions are shown for primary (blue) and secondary (green) notochord. The superimposed box plots for each distribution show the median flanked by the interquartile range, with whiskers extending to the full range excluding outliers >1.5X IQR. Expression differences between notochord regions for each reporter are shown as enrichment scores, calculated as the mean secondary expression divided by the mean primary expression. ++ indicates that the construct was enriched in secondary notochord with statistical significance <0.05. + indicates that the construct showed signs of enrichment in secondary notochord but it was not statistically significant. – indicates that the construct was not enriched in secondary notochord. Secondary enrichment was tested by one-tailed Wilcoxon signed-rank tests (*, p<0.05; **, p<0.005; ***, p<0.0005). The ectopic expression scores represent the average expression in 5 non-notochord tissues using a 0–4 qualitative expression scale described in the methods. For a tissue-specific breakdown of the scores, see Tables S3,S4.
We used this dual reporter approach to investigate the cis-regulation of C11.331. All embryos were imaged by confocal microscopy using uniform imaging parameters. We developed a quantitative analysis approach based on measuring Venus and HA intensity along the AP axis of computationally straightened and flattened notochords. For each embryo analyzed, we measured the mean background-corrected Venus reporter signal for Bra>H2B:HA expressing cells in primary notochord (cells 1–32) and secondary notochord (cells 33–40), and normalized the results to the Bra>H2B:HA intensity in each region (Fig. 3B). We also scored a larger number of embryos with a qualitative system, ranging from 0 (no detectable expression) to 4 (oversaturated) (Tables S1,S2), which largely matched the quantitative analysis. An ectopic expression score was generated based on qualitative expression levels in several non-notochord tissues (Tables S3,S4). Full details of the scoring systems are given in the Methods section.
C11.331 is separated from the 3’ end of the closest upstream gene by ~1.5kb, and we found that this intergenic region (−1565 to +13) gave rise to strong expression in secondary notochord, much weaker expression in primary notochord, and relatively little ectopic expression (Fig. 3B). We dissected this region with a series of truncations and found evidence for extensive functional redundancy. On a coarse scale, both the distal −1565 to −758 and the proximal −787 to +13 region gave rise to secondary-enriched notochord expression, with the distal region showing somewhat less ectopic expression but the proximal region showing stronger notochord expression and greater statistical evidence for enrichment in the secondary notochord. We further dissected the −787 to +13 region and again found aspects of redundancy, with both the −787 to −405 and the minimally overlapping −488 to +13 region showing secondary-enriched notochord expression.
Finer-scale analysis of the −488 to +13 region allowed us to identify distinct regulatory regions. A construct spanning the −488 to −165 region was still strongly expressed in secondary notochord. Unlike the previous constructs, however, it was strongly expressed in primary notochord as well, indicating the presence of a silencer element for primary notochord expression proximal to −165. A further truncation to give the −488 to −245 construct eliminated expression altogether, indicating the presence of essential enhancer elements in the −245 to −165 interval. Truncating from the other side, the −322 to +13 construct was highly expressed and strongly enriched in secondary notochord, whereas the −245 to +13 construct was enriched in secondary notochord, but much more weakly expressed overall, indicating that there are additional enhancer elements between −322 and −245. Our subsequent efforts therefore focused on the −322 to +13 interval.
Fig. 4A shows normalized C11.331(−322 to +13)>Venus expression as a function of AP position for 19 imaged notochords. The normalized mean intensity (green line) reveals a sharp transition from low to high expression precisely at the boundary between primary and secondary notochord. This is in contrast to the Bra>H2B:HA control that is expressed uniformly along the notochord’s AP axis (Fig. 4B).
Fig. 4. Quantitative analysis of the C11.331(−322 to +13) CRM.
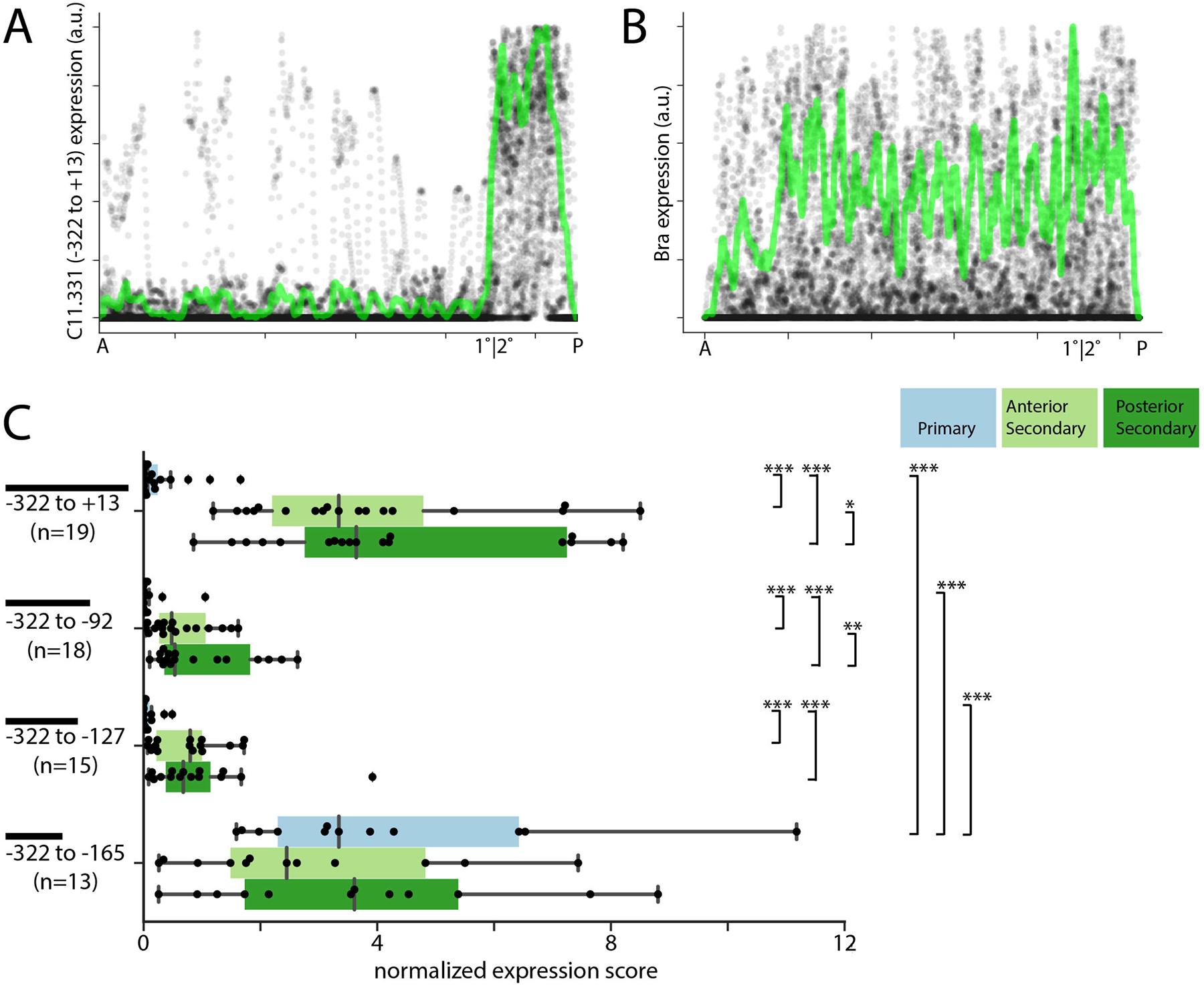
A-B) Quantitative traces of C11.331 (−322 to +13) (A) and Bra (B) expression from 19 embryos as a function of anterior-posterior position. Expression data from each embryo was scaled in intensity to {0 1}, scaled to a common length, aligned by the primary/secondary notochord boundary and plotted as transparent gray to help visualize overlapping data points. Normalized mean intensity is overlaid in green. C) C11.331 quantitative expression score distributions are shown for primary (blue) anterior secondary (light green) and posterior secondary (dark green) notochord. The superimposed box plots for each distribution show the median flanked by the interquartile range, with whiskers extending to the full range excluding outliers >1.5X IQR. Expression differences between notochord regions for a given reporter were tested by one-tailed Wilcoxon signed-rank tests. Expression differences for each notochord region between the different reporter constructs were tested by ANOVA followed by Tukey’s Honest Significant Difference post-hoc test (*, p<0.05; **, p<0.005; ***, p<0.0005).
For these finer-scale dissections, we quantified reporter expression separately for the anterior (cells 33–36) and posterior (cells 37–40) secondary notochord. For the −322 to +13 construct, the sample mean of this quantitative metric of reporter expression was 19.7 fold higher in anterior secondary notochord as compared to primary notochord, and 26.6 fold higher in posterior secondary notochord as compared to primary notochord. These differences were highly significant as measured by the Wilcoxon sign-rank test. Two truncated constructs, −322 to −92 and −322 to −127, retained significant enrichment in secondary notochord but were less highly expressed overall, indicating that there are enhancer elements promoter-proximal to −92. A further truncation to give the −322 to −165 construct was significantly derepressed in primary notochord, confirming the presence of important silencer elements promoter-proximal to −165. Combined with the previous constructs (Fig. 3B), this indicates that the primary notochord silencer has essential elements between −165 and −127.
Identification and testing of candidate TFBSs
We searched the −165 to −127 interval and flanking regions for putative transcription factor binding sites (TFBSs) using both TRANSFAC position weight matrices (PWMs) (Matys et al., 2006) via the LASAGNA web tool (Lee and Huang, 2013) and also using SELEX-seq-derived (Slattery et al., 2011) consensus sequences for known Ciona notochord TFs downloaded from the ANISEED community database (Brozovic et al., 2018). We found several predicted sites of interest, including an ETS site just outside of the silencer in a region required for strong expression (Fig. 5A). This was of particular interest given that ETS family transcription factors are key mediators of ascidian FGF signaling (Gainous et al., 2015; Miya and Nishida, 2003). We did not identify any predicted LEF/TCF sites (Brannon et al., 1997), suggesting that any Wnt inputs into this particular cis-regulatory module are likely to be indirect. Other sites of interest included a GATA site, an LMX site and a Myb site that were all in very close proximity within the putative silencer region. An LMX ortholog (KH.C9.485) is known to be expressed in the Ciona notochord (José-Edwards et al., 2011), and functionally important Myb sites have been identified in several Ciona notochord CRMs (José-Edwards et al., 2015). GATA TFs have not been implicated in notochord-specific gene expression, but immunostaining suggests there may be GATA.a expression in the posterior notochord (Oda-Ishii et al., 2016).
Fig. 5. Mapping TFBSs in the C11.331 (−322 to +13) CRM.
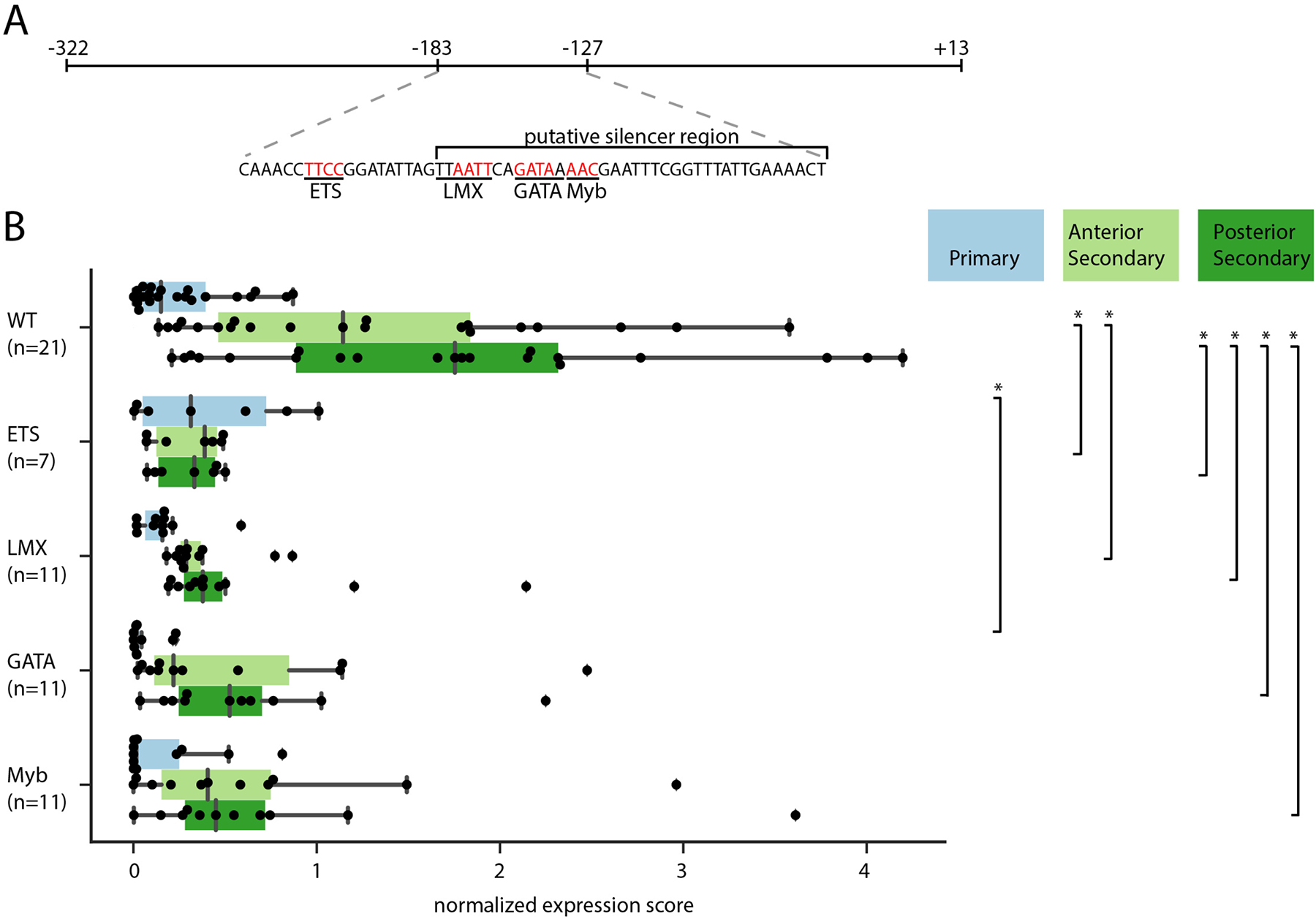
A) Schematic of C11.331 (−322 to +13) with the sequence of the −183 to −127 region shown and the −165 to −127 putative silencer region indicated. Core binding sites for the transcription factors ETS, LMX, GATA, and Myb are underlined, with the specific bases mutated in derivative constructs shown in red. B) Quantitative analysis of regionalized notochord expression for the C11.331(−322 to +13) construct and derivatives in which each of these predicted TFBSs have been mutated. Quantitative expression scores for individual embryos are shown for primary notochord (blue) anterior secondary (light green) and posterior secondary (dark green). The superimposed box plots for each distribution show the median flanked by the interquartile range, with whiskers extending to the full range excluding outliers >1.5X IQR. Between-reporter differences in expression for each notochord region were tested by ANOVA followed by Tukey’s Honest Significant Difference post-hoc test (*, p<0.05).
We individually mutated the core motif for each of these TFBSs in the −322 to +13 construct. The GATA and Myb core sequences are almost overlapping, and the LMX site is also nearby, so we took care to mutate these core sequences without affecting the adjacent core sites (Fig. 5A). We cannot exclude, however, that these mutations may have promiscuous effects via important but uncharacterized flanking sequences. Our hypothesis was that one or more of these mutations would have a phenotype similar to the −322 to −165 sequence in which expression in primary notochord is derepressed. We instead found that all 4 mutations led to decreased expression overall, particularly in secondary notochord cells (Fig. 5B). This was particularly evident for the ETS mutation, which is not unexpected given its location outside of the silencer region, the U0126 phenotype and the expression of FGF8/17/18 and FGF9/16/20 in the tip of the tail. It remains unclear, however, whether repression in the primary notochord involves cryptic combinatorial effects of LMX, Myb and/or GATA, or whether there are essential binding sites for unknown factors that have yet to be identified in the −164 to −127 region.
Discussion and Conclusions
Here we characterized the expression patterns and dependencies on Wnt and FGF signaling for three newly identified genes with strongly enriched expression in the posterior notochord. These fell into two classes: C11.331 and C12.115 were consistently differentially expressed between primary and secondary notochord, whereas C8.891 showed a graded expression pattern at several stages extending past the primary/secondary boundary; C11.331 and C12.115 were strongly and consistently affected by manipulations of both Wnt and FGF signaling, whereas C8.891 was strongly affected by ectopic Wnt activation but only modestly affected by FGF inhibition. This indicates that there may be at least two functionally distinct mechanisms of posterior-specific notochord gene regulation involving multiple signaling molecules expressed in the posterior tip of the tail.
Both C11.331 and C12.115 showed a surprisingly complex expression pattern when the canonical Wnt pathway was ectopically activated by BIO treatment. In both cases, the ectopic expression induced by BIO treatment is highly specific to notochord and is not seen in other tissues, confirming that there must be distinct tissue-specific inputs into the expression of these genes. Both expression patterns expanded throughout the notochord upon BIO treatment, but with distinct enrichment in most embryos at both the anterior and posterior tips. This is reminiscent of the wildtype expression pattern for BCamL which is similarly enriched in the anterior and posterior notochord. FGFs are expressed in the trunk as well as the posterior tail tip (Hudson and Yasuo, n.d.; Imai et al., 2004), suggesting that they might play a key role in generating these bipolar expression patterns. This is supported by the weak uniform notochord expression of C11.331 and C12.115 seen in embryos treated with both BIO and U0126, which shows that induction of tip specific expression by ectopic Wnt pathway activation requires functional FGF signaling. We predict that the combination of both Wnt and FGF signaling at the tail tip is essential for the robust expression of C11.331 and C12.115 in posterior notochord, but that BCamL likely responds to FGF alone. If this hypothesis is correct, BCamL expression should be reduced or eliminated by U0126 treatment but not affected by BIO.
We have also extensively probed the cis-regulatory architecture underlying the posterior-specific expression of C11.331 and find evidence for extensively distributed enhancer function. There are multiple non-overlapping or minimally overlapping regions in the C11.331 upstream intergenic region that confer posterior notochord-enriched expression in reporter assays. Relatively few ‘shadow/redundant/distributed’ enhancers have been identified to date in Ciona (Farley et al., 2016), but they are extremely common in other model systems (Barolo, 2012; Cannavò et al., 2016; Perry et al., 2011) and easily missed in studies focused on rapidly identifying minimal CRM regions. A recent study using ATAC-seq to identify candidate enhancer regions genome-wide suggests that they may be more common in Ciona as well (Madgwick et al., 2018).
The functional significance of shadow/redundant/distributed enhancers remains unclear, but multiple non-exclusive hypotheses involving the strength, robustness, tissue-specificity and evolvability of gene expression have been proposed (Barolo, 2012). While not extensively investigated here, our data are consistent with distributed enhancer function being important for both the strength and the tissue-specificity of expression. The smaller constructs that recapitulated the secondary enriched expression seen with the full length −1565 to +13 region tended to have greater ectopic expression and/or weaker expression overall (Fig. 3B).
While the C11.331 upstream regions showed extensively redundant cis-regulatory activity, further dissections of the minimal −488 to +13 regulatory module identified discrete regions with predominant roles in positive and negative regulation. These include a region between −165 and −127 essential for repressing expression in primary notochord, and a region between −245 and −165 inferred to contain essential elements required for both notochord-specific and ectopic expression. The −165 to −127 silencer region is of particular interest, because it implies that there is a key role for negatively-acting factors that prevent C11.331 expression in primary notochord. We accordingly searched the −165 to −127 region for TFBSs of interest, but mutations in several predicted sites led to decreased expression in secondary notochord as opposed to increased expression in primary notochord. It remains unclear whether there are yet to be identified sites with clear repressive function, or whether some more complex regulatory scheme might be at work.
It was not clear from the initial in situ hybridization experiments whether C11.331 and C12.115 are truly specific to secondary notochord, or whether they are induced in the posterior notochord by FGFs and Wnts expressed in the posterior tail tip and coincidentally have an anterior limit of detectable expression that is closely aligned with the primary/secondary boundary. Both the chromogenic and fluorescent in situs made it clear that there is a graded quality to C11.331 and C12.115 expression patterns, which are consistently strongest in the posteriormost notochord cells and drop off rapidly towards the primary/secondary boundary. It was unclear, however, whether there was a sharp transition in expression levels at the primary/secondary boundary given the limited sensitivity and modest dynamic range of traditional (non-single molecule) in situ hybridization. Quantitative analysis of C11.331 reporter constructs revealed, however, that although there is graded expression within the secondary notochord, there is a sharp and abrupt transition between very low expression levels in the primary notochord and much higher levels in secondary notochord (Fig. 4A). This suggests that there are inputs into C11.331 expression based on primary vs secondary notochord fate and that it is not solely based on proximity to Wnt and FGF signals in the tip of the tail.
Fig. 6 shows a provisional model for C11.331 expression in the secondary notochord. FGF and Wnt signals from a signaling center at the tip of the tail help to induce regionalized C11.331 expression, which is also dependent on notochord-specific transcriptional regulators (possibly including LMX1 and potential Myb family members) and an as-yet unidentified transcriptional repressor that is predicted to be differentially expressed in primary but not secondary notochord. The identification of this putative primary notochord repressor is of considerable interest, but it will also be important to determine if similar mechanisms are at work in other C11.331 CRMs as well as in the regulatory regions for other regionally expressed notochord genes. The 1kb upstream regions of C12.115 and C8.891 both contain predicted Ets, LMX, GATA and Myb sites that define candidate regions of interest.
Fig. 6. Regulatory network model for C11.331 expression.
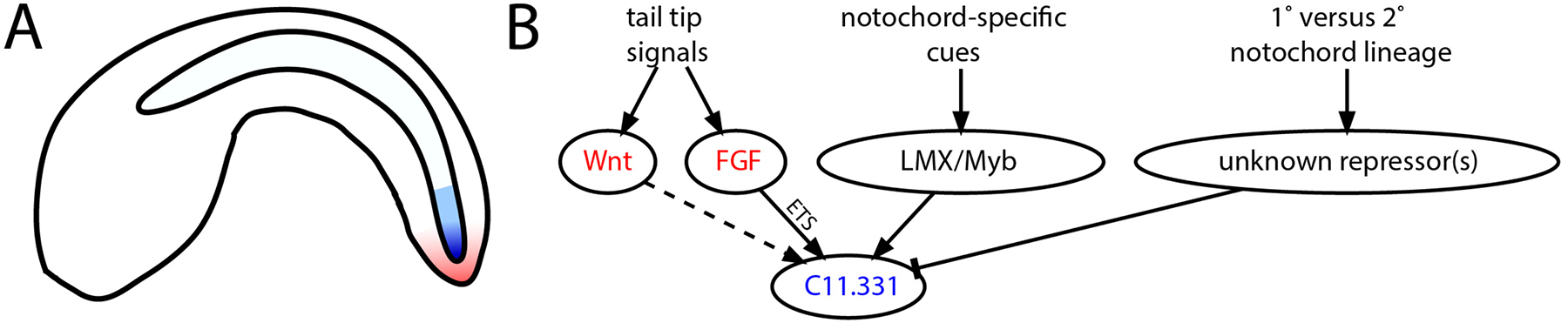
A) Schematic of a Ciona embryo showing Wnt and FGF expression in the posterior tail tip (red) and C11.331 expression in blue. C11.331 expression is both highly enriched in secondary notochord compared to primary notochord and also graded within the secondary notochord. B) Simple network model of inferred inputs into C11.331 expression.
Methods:
Ciona husbandry and embryology
Ciona robusta (formerly known as Ciona intestinalis type A) (Pennati et al., 2015) were collected in San Diego and shipped to KSU by Marine Research and Educational Products Inc. (M-REP, San Diego, CA). Adult Ciona were maintained in a recirculating aquarium. Standard fertilization, dechorionation and electroporation protocols were used (Veeman et al., 2011). Staging is based upon the series of Hotta (Hotta et al., 2007).
Drug treatments: BIO (GSK inhibitor IX, CAS 667463-62-9, Sigma-Aldrich) was dissolved in DMSO at 5 mM and used at a final concentration of 5 micromolar. MEK1/MEK2 inhibitor U0126 (CAS109511–58, Sigma-Aldrich) was dissolved in DMSO at 4 mM and used at a final concentration of 4 micromolar. DMSO alone (at 1:1000 dilution) was used as a control. Drugs were added to artificial seawater (ASW) at Hotta stage 12 and embryos were grown in drug-treated ASW until fixation at Hotta stage 23.
In situ hybridization
Probe synthesis, embryo collection, in situ hybridization and imaging were performed essentially as described previously (Reeves 2014), except that embryos for fluorescent in situs were electroporated with 60 micrograms Bra>hCD4:mCherry (Gline et al., 2015) to visualize notochord cell boundaries post-hybridization. mCherry antibody (Biovision, #5993) was included at 1:500 during the anti-DIG-AP incubation. After SIGMA-FAST FastRed staining (Sigma-Aldrich; F4648) was stopped by washing 5 times in PBS-Tween20, embryos were incubated overnight at 4°C with 1:1000 anti-rabbit AlexaFluor 488 (Invitrogen, A-11034). Embryos were washed 5 times in PBS-Tween20, then mounted, cleared, imaged and fluorescence was quantified in cells 30–40 as described previously (Reeves et al., 2014). All fluorescence intensities were background subtracted and then normalized to a 0–1 scale. Expression data was centered on the primary/secondary boundary and scaled to a common length.
The Brachyury in situ probe was previously described (Reeves et al., 2014). Regions of C11.331, C12.115 and C8.891 were amplified from cDNA and cloned into pBSII-SK(−), using the following primers: C11.331 (993bp, Forward: ACGGGACTCACACAACTTCC, Reverse: GTCTCCAATCGCTTGCTGTT); C12.115 (865bp, Forward: ACGCCATTAACACCGGTTC, Reverse: CAAATGTTTAGAAAACTGATTTTGAC), C8.891 (900bp, Forward: GTGCTGATGCCAAGAATGC, Reverse: GTTTCACACAGCTGGTAGGC).
Reporter cloning and mutagenesis
The genomic region spanning from −1565 to +13 of KH2012:KH.C11.331.v1.A.SL2–1 was PCR amplified from genomic DNA (Forward primer: TAAAATGGCGCGCCCAGGTGCCACAAATAAACC; Reverse primer: CCTCCGTCTAGACCTATTTGTCCTTCTGAAATAACAG and cloned into the AscI and XbaI sites of pX2+bpFOG>UNC76:Venus (Stolfi et al., 2015). All subsequent reporters were subcloned from this original plasmid (see Table S5 for primers and genomic coordinates). TFBS single mutants were generated through the Q5 Site-Directed Mutagenesis Kit (E0554S, NEB), following manufacturer directions. Mutagenesis primers are listed in Table S5. The Bra-H2B:HA reporter was generated by standard Gateway cloning of pENTR-H2B into pSP72BSSPE-SwaI::RFA-HA (Roure et al., 2007) A 2.2kb Brachyury enhancer was cloned into the plasmid’s XhoI/HindIII sites.
Reporter expression, staining, and imaging
Fertilized dechorionated eggs were co-electroporated with 30 μg Bra > H2B:HA plasmid and 60 μg C11.331 reporter plasmid (region of interest-bpFOG>Venus). At least three separate replicates were performed for each reporter. Embryos were fixed at Hotta stage 21–22, stained and prepared for imaging as previously described (Carlson et al., 2015). GFP polyclonal antibody (Invitrogen, A-11122) was used at 1:000 and HA-Tag mouse monoclonal antibody (Cell Signaling Technology, 2367S) at 1:750. Secondary antibodies (Invitrogen; anti-mouse AlexaFluor 488, A-11029 and anti-rabbit AlexaFluor 555, A-21420) were used at 1:1000, and AlexaFluor 633 Phalloidin (Invitrogen, A22284) was used at 1:150. Embryo identities were blinded prior to mounting and clearing embryos. Mounted embryos were imaged on a Zeiss 700 laser scanning confocal microscope, using a 40× 1.3NA objective. Z-stacks of each embryo were collected at a slice interval of 0.5 μm and a pixel size of 0.24 μm, using consistent settings for laser power, PMT gain and scan speed. Embryos were selected for imaging based on expression of the Bra>H2B:HA control in both primary and secondary notochord, but without respect for the expression of the C11.331 reporter plasmid.
Quantitative reporter analysis
Confocal stacks of embryos electroporated with the indicated reporter plasmids were analyzed interactively using FIJI/ImageJ (Schindelin et al., 2012). The tail is rarely flat enough or sufficiently parallel to the coverslip to visualize the entire notochord in one 2D image. To allow a simple 2D analysis of reporter intensity, the notochord was first computationally flattened in the Z axis to bring all the Bra>H2B:HA labeled nuclei into a single plane. This involved reslicing the image to view the image volume as XZ or YZ slices, manually tracing a polyline that followed the notochord in Z, and then reslicing again along the polyline to reconstruct a flattened plane through the full length of the notochord. Embryos that could not be cleanly resliced in this manner to generate a clear image of the embryo were rejected from quantitative analysis. A 40 pixel wide polyline was then traced along the notochord’s curvature in X and Y to capture the full width of each labeled nucleus from cell 1 to cell 40. Intensity data from the Venus and HA reporter channels was collected along that line using the ‘plot profile’ function. All of these steps used standard FIJI/ImageJ tools.
We then derived a normalized expression metric implemented as a MATLAB function. Inputs to the function include the AP intensity profiles of Venus and HA, manually estimated values for Venus and HA background levels, and the inferred positions of the primary/secondary notochord boundary and the cell 36/37 boundary (anterior versus posterior secondary notochord). The intensity profiles were segmented into the three regions of interest. For each region, a normalized expression score was generated by dividing background-subtracted reporter>Venus intensity by control background-subtracted Bra>HA intensity, but only for the positions with greater than threshold levels of Bra>HA expression.
Wilcoxon signed-rank tests were used to compare expression levels between notochord regions for the quantitative analysis. ANOVA followed by Tukey’s Honest Significant Difference post-hoc test was used to compare differences in expression between different reporter constructs for each notochord subregion to better control for multiple comparisons. All statistical tests used standard MATLAB functions. We used the Python Pandas, Numpy, Matplotlib and Seaborn packages for plotting and data visualization.
Qualitative reporter analysis
Confocal stacks of embryos from the reporter dissections were scored for expression in the primary notochord, anterior four cells of the secondary notochord, and posterior four cells of the secondary notochord on a scale from zero (no visible expression) to four (oversaturated expression). Scores were assigned based on the cell in the notochord region with the strongest Venus expression that also expressed Bra>H2B:HA. Scores were also given for ectopic expression in CNS, epidermis, endoderm, muscle, and mesenchyme, but without the requirement for Bra>H2B:HA expression in the same cell. Little evidence was seen for distinct tissue-specific silencers outside the notochord, so a combined ectopic expression score was derived by averaging the scores across the 5 tissues examined. Scoring was performed separately and without knowledge of the specific secondary reporter plasmid by both MH and CB prior to reconciling any score differences and recording consensus scores for each embryo. For simpler presentation, we averaged the anterior and posterior secondary notochord scores into a single combined value.
TFBS Analysis
The predicted ETS, GATA and Myb sites were identified using TRANSFAC PWMs (Matys et al., 2006) via the LASAGNA web tool (http://biogrid-lasagna.engr.uconn.edu/lasagna_search/)(Lee and Huang, 2013). The LMX site was identified by manually searching the minimal region of interest for matches to the core sequence identified by SELEX against Ciona Lmx1A (Brozovic et al., 2018). We also searched for matches to SELEX-derived motifs for Brachyury (shared by Tbx2/3) and FoxAa but did not locate any within this interval.
Resources
Information on key resources are provided in the KRT table.
KEY RESOURCES TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-mCherry | Biovision | Cat#5993 |
| Goat anti-rabbit polyclonal AlexaFluor488 | Invitrogen | Cat#A11034 |
| Goat anti-mouse polyclonal AlexaFluor488 | Invitrogen | Cat#A11029 |
| Goat anti-rabbit polyclonal AlexaFluor555 | Invitrogen | Cat#A22420 |
| Rabbit polyclonal anti-GFP | Invitrogen | Cat#A11122 |
| Mouse monoclonal anti-HA | Cell Signaling Technology | Cat#2367S |
| Bacterial and Virus Strains | ||
| n/a | ||
| Biological Samples | ||
| n/a | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| GSK inhibitor IX (BIO) | Sigma-Aldrich | Cat#361550; CAS667463-62-9 |
| U1026 | Sigma-Aldrich | Cat#662005; CAS109511-58-2 |
| SIGMA-FAST FastRed | Sigma-Aldrich | Cat#F4648 |
| AlexaFluor633 Phalloidin | Invitrogen | Cat#A22284 |
| Critical Commercial Assays | ||
| n/a | ||
| Deposited Data | ||
| Ciona SELEX data | ANISEED | https://www.aniseed.cnrs.fr/ |
| Experimental Models: Cell Lines | ||
| n/a | ||
| Experimental Models: Organisms/Strains | ||
| Wild Ciona robustus | M-REP, San Diego, CA | N/A |
| Oligonucleotides | ||
| See Table S1 | ||
| Recombinant DNA | ||
| Bra>hCD4:mCherry | Gline et al, 2015 | N/A |
| All C11.331>UNC76:Venus reporter plasmids | This paper | N/A |
| Bra>H2B:HA | This paper | N/A |
| Software and Algorithms | ||
| MATLAB | The Mathworks, Inc | Release 2012b |
| FIJI/ImageJ | Schindelin et al, 2012 | https://fiji.sc/ |
| LASAGNA-GRID | Lee & Huang, 2013 | (http://biogrid-lasagna.engr.uconn.edu/lasagna_search |
| Other | ||
Supplementary Material
Acknowledgements
We gratefully acknowledge support from the KSU CVM confocal microscopy core.
Funding
This work was supported by NIH 1R01HD085909 and NSF IOS 1456555 (MV).
References
- Barolo S, 2012. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34, 135–41. 10.1002/bies.201100121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D, 1997. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev 11, 2359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozovic M, Dantec C, Dardaillon J, Dauga D, Faure E, Gineste M, Louis A, Naville M, Nitta KR, Piette J, Reeves W, Scornavacca C, Simion P, Vincentelli R, Bellec M, Ben Aicha S, Fagotto M, Guéroult-Bellone M, Haeussler M, Jacox E, Lowe EK, Mendez M, Roberge A, Stolfi A, Yokomori R, Brown CT, Cambillau C, Christiaen L, Delsuc F, Douzery E, Dumollard R, Kusakabe T, Nakai K, Nishida H, Satou Y, Swalla B, Veeman M, Volff J-N, Lemaire P, 2018. ANISEED 2017: extending the integrated ascidian database to the exploration and evolutionary comparison of genome-scale datasets. Nucleic Acids Res 46, D718–D725. 10.1093/nar/gkx1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E, Khoueiry P, Garfield DA, Geeleher P, Zichner T, Gustafson EH, Ciglar L, Korbel JO, Furlong EEM, 2016. Shadow Enhancers Are Pervasive Features of Developmental Regulatory Networks. Curr. Biol 26, 38–51. 10.1016/j.cub.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Reeves W, Veeman M, 2015. Stochasticity and stereotypy in the Ciona notochord. Dev. Biol 397, 248–256. 10.1016/j.ydbio.2014.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW, 1997. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Hebras C, Besnardeau L, McDougall A, 2013. Beta-catenin patterns the cell cycle during maternal-to-zygotic transition in urochordate embryos. Dev. Biol 384, 331–42. 10.1016/j.ydbio.2013.10.007 [DOI] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS, 2016. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc. Natl. Acad. Sci. U. S. A 113, 6508–13. 10.1073/pnas.1605085113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainous TB, Wagner E, Levine M, 2015. Diverse ETS transcription factors mediate FGF signaling in the Ciona anterior neural plate. Dev. Biol 399, 218–25. 10.1016/j.ydbio.2014.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gline S, Kaplan N, Bernadskaya Y, Abdu Y, Christiaen L, 2015. Surrounding tissues canalize motile cardiopharyngeal progenitors towards collective polarity and directed migration. Development 142, 544–554. 10.1242/dev.115444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K, 2007. SPECIAL ISSUE RESEARCH ARTICLE A Web-Based Interactive Developmental Table for the Ascidian Ciona intestinalis, Including 3D Real-Image Embryo Reconstructions : I. From Fertilized Egg to Hatching Larva. Dev. Biol 236, 1790–1805. 10.1002/dvdy.21188 [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Asakura T, Saitoh B, Takatori N, Satou Y, Satoh N, 2000. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev. Biol 224, 69–80. 10.1006/dbio.2000.9765 [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Erives A, Levine M, Satoh N, 1999. Temporal expression patterns of 39 Brachyury-downstream genes associated with notochord formation in the Ciona intestinalis embryo. Dev. Growth Differ 41, 657–664. 10.1046/j.1440-169X.1999.00467.x [DOI] [PubMed] [Google Scholar]
- Hudson C, Kawai N, Negishi T, Yasuo H, 2013. β-Catenin-driven binary fate specification segregates germ layers in ascidian embryos. Curr. Biol 23, 491–5. 10.1016/j.cub.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Hudson C, Lotito S, Yasuo H, 2007. Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development 134, 3527–37. 10.1242/dev.002352 [DOI] [PubMed] [Google Scholar]
- Hudson C, Sirour C, Yasuo H, 2016. Co-expression of Foxa.a, Foxd and Fgf9/16/20 defines a transient mesendoderm regulatory state in ascidian embryos. Elife 5. 10.7554/eLife.14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Yasuo H, 2006. A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development 133, 2855–64. 10.1242/dev.02466 [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H, 2005. Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development 132, 1199–210. 10.1242/dev.01688 [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H, n.d. C2.125 in situ hybridization [WWW Document]. ANISEED. URL https://www.aniseed.cnrs.fr/aniseed/gene/show_expression?unique_id=Cirobu.g00004295&offset=60 (accessed 5.31.18). [Google Scholar]
- Ikuta T, Satoh N, Saiga H, 2010. Limited functions of Hox genes in the larval development of the ascidian Ciona intestinalis. Development 137, 1505–13. 10.1242/dev.046938 [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y, 2004. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047–4058. 10.1242/dev.01270 [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y, 2006. Regulatory blueprint for a chordate embryo. Science 312, 1183–7. 10.1126/science.1123404 [DOI] [PubMed] [Google Scholar]
- Imai KS, Satou Y, Satoh N, 2002. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development 129, 2723–32. [DOI] [PubMed] [Google Scholar]
- Jiang D, Munro EM, Smith WC, 2005. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr. Biol 15, 79–85. [DOI] [PubMed] [Google Scholar]
- José-Edwards DS, Kerner P, Kugler JE, Deng W, Jiang D, Di Gregorio A, 2011. The identification of transcription factors expressed in the notochord of Ciona intestinalis adds new potential players to the brachyury gene regulatory network. Dev. Dyn 240, 1793–1805. 10.1002/dvdy.22656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Edwards DS, Oda-Ishii I, Kugler JE, Passamaneck YJ, Katikala L, Nibu Y, Di Gregorio A, 2015. Brachyury, Foxa2 and the cis-Regulatory Origins of the Notochord. PLoS Genet 11, 1–16. 10.1371/journal.pgen.1005730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JE, Passamaneck YJ, Feldman TJ, Beh J, Regnier TW, Di Gregorio A, 2008. Evolutionary conservation of vertebrate notochord genes in the ascidian Ciona intestinalis. Genesis 46, 697–710. 10.1002/dvg.20403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Huang C-H, 2013. LASAGNA-Search: an integrated web tool for transcription factor binding site search and visualization. Biotechniques 54. 10.2144/000113999 [DOI] [PubMed] [Google Scholar]
- Madgwick A, Magri MS, Dantec C, Gailly D, Fiuza U-M, Guignard L, Hettinger S, Gomez-skarmeta JL, Lemaire P, 2018. Evolution of the embryonic cis-regulatory landscapes between divergent Phallusia and Ciona ascidians. bioRxiv doi 10.1101/123456. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E, 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34, D108–10. 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya T, Nishida H, 2003. An Ets transcription factor, HrEts, is target of FGF signaling and involved in induction of notochord, mesenchyme, and brain in ascidian embryos. Dev. Biol 261, 25–38. [DOI] [PubMed] [Google Scholar]
- Nishida H, 1987. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev. Biol 121, 526–41. [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I, Kubo A, Kari W, Suzuki N, Rothbächer U, Satou Y, 2016. A Maternal System Initiating the Zygotic Developmental Program through Combinatorial Repression in the Ascidian Embryo. PLoS Genet 12, e1006045. 10.1371/journal.pgen.1006045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennati R, Ficetola GF, Brunetti R, Caicci F, Gasparini F, Griggio F, Sato A, Stach T, Kaul-Strehlow S, Gissi C, Manni L, 2015. Morphological Differences between Larvae of the Ciona intestinalis Species Complex: Hints for a Valid Taxonomic Definition of Distinct Species. PLoS One 10, e0122879. 10.1371/journal.pone.0122879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M, 2011. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A 108, 13570–5. 10.1073/pnas.1109873108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racioppi C, Kamal AK, Razy-Krajka F, Gambardella G, Zanetti L, di Bernardo D, Sanges R, Christiaen LA, Ristoratore F, 2014. Fibroblast growth factor signalling controls nervous system patterning and pigment cell formation in Ciona intestinalis. Nat. Commun 5, 4830. 10.1038/ncomms5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves W, Thayer R, Veeman M, 2014. Anterior-posterior regionalized gene expression in the Ciona notochord. Dev. Dyn 243, 612–620. 10.1002/dvdy.24101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Wu Y, Harder MJ, Veeman MT, 2017. Functional and evolutionary insights from the Ciona notochord transcriptome. Development 144, 3375–3387. 10.1242/dev.156174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure A, Rothbächer U, Robin F, Kalmar E, Ferone G, Lamy C, Missero C, Mueller F, Lemaire P, 2007. A multicassette Gateway vector set for high throughput and comparative analyses in ciona and vertebrate embryos. PLoS One 2, e916. 10.1371/journal.pone.0000916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe E, Tanaka N, Shimozono N, Gojobori T, Fujiwara S, 2006. Effects of U0126 and fibroblast growth factor on gene expression profile in Ciona intestinalis embryos as revealed by microarray analysis. Dev. Growth Differ 48, 391–400. 10.1111/j.1440-169X.2006.00877.x [DOI] [PubMed] [Google Scholar]
- Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, Wiley GB, Macmil SL, Roe BA, Zeller RW, Hastings KEM, Lemaire P, Lindquist E, Endo T, Hotta K, Inaba K, 2008. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol 9, R152. 10.1186/gb-2008-9-10-r152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–82. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, Mann RS, 2011. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–82. 10.1016/j.cell.2011.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L, 2015. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527, 371–374. 10.1038/nature15758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R, Sasaki T, Kostka G, Chu ML, 2003. Fibulins: A versatile family of extracellular matrix proteins. Nat. Rev. Mol. Cell Biol 4, 479–489. 10.1038/nrm1130 [DOI] [PubMed] [Google Scholar]
- Veeman MT, Chiba S, Smith WC, 2011. Ciona Genetics, in: Vertebrate Embryogenesis pp. 401–422. 10.1007/978-1-61779-210-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Smith WC, 2013. Whole-organ cell shape analysis reveals the developmental basis of ascidian notochord taper. Dev. Biol 373, 281–9. 10.1016/j.ydbio.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo H, Hudson C, 2007. FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev. Biol 302, 92–103. 10.1016/j.ydbio.2006.08.075 [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N, 1993. Function of vertebrate T gene. Nature 364, 582–583. 10.1038/364582b0 [DOI] [PubMed] [Google Scholar]
- Zeller RW, Virata MJ, Cone AC, 2006. Predictable mosaic transgene expression in ascidian embryos produced with a simple electroporation device. Dev. Dyn 235, 1921–32. 10.1002/dvdy.20815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


