Abstract
The simian virus 40 large T antigen (T antigen) inactivates tumor suppressor proteins and therefore has been used in numerous studies to probe the mechanisms that control cellular growth and to generate immortalized cell lines. Binding of T antigen to the Rb family of growth-regulatory proteins is necessary but not sufficient to cause transformation. The molecular mechanism underlying T-antigen inactivation of Rb function is poorly understood. In this study we show that T antigen associates with pRb and p130-E2F complexes in a stable manner. T antigen dissociates from a p130–E2F-4–DP-1 complex, coincident with the release of p130 from E2F-4–DP-1. The dissociation of this complex requires Hsc70, ATP, and a functional T-antigen J domain. We also report that the “released” E2F–DP-1 complex is competent to bind DNA containing an E2F consensus binding site. We propose that T antigen disrupts Rb-E2F family complexes through the action of its J domain and Hsc70. These findings indicate how Hsc70 supports T-antigen action and help to explain the cis requirement for a J domain and Rb binding motif in T-antigen-induced transformation. Furthermore, this is the first demonstration linking Hsc70 ATP hydrolysis to the release of E2F bound by Rb family members.
The tumor suppressor proteins retinoblastoma protein (pRb) and p53 have been the focus of intense study due to their crucial role in the regulation of normal cellular growth (reviewed in references 14 and 28). T antigen can inactivate both p53 and pRb and has been used as a tool to disable these cellular signaling pathways (3, 10, 17, 22).
The Rb family is composed of at least three proteins, pRb, p107, and p130, which are critical for cell cycle regulation and are thought to have overlapping functions in different stages of the cell cycle (31). pRb, p107, and p130 proteins bind to the E2F family of transcription factors. When an Rb family member binds to E2F, E2F-mediated transactivation is inhibited, and expression of E2F-responsive genes, such as cyclins A and E and dihydrofolate reductase, is decreased (1, 12, 24). When phosphorylated, pRb dissociates from E2F, inducing DNA synthesis and E2F-mediated gene expression, leading to advancement of the cell cycle (9, 46).
T antigen binds to the pRb proteins through an LXCXE motif (15) (see Fig. 1A for T-antigen domain map). This sequence is conserved in many pRb binding proteins, including cellular proteins such as BRG1 (13) and those expressed by other small DNA tumor viruses, such as E1A (adenovirus) and E7 (papillomavirus) (11, 15, 25). Mutation of the LXCXE motif renders T antigen defective for transformation (36, 42, 48). This suggests that T antigen's affinity for Rb family members displaces Rb from binding E2F, thus inducing transformation. However, the binding of pRb family members by T antigen is not sufficient to induce transformation (42).
FIG. 1.
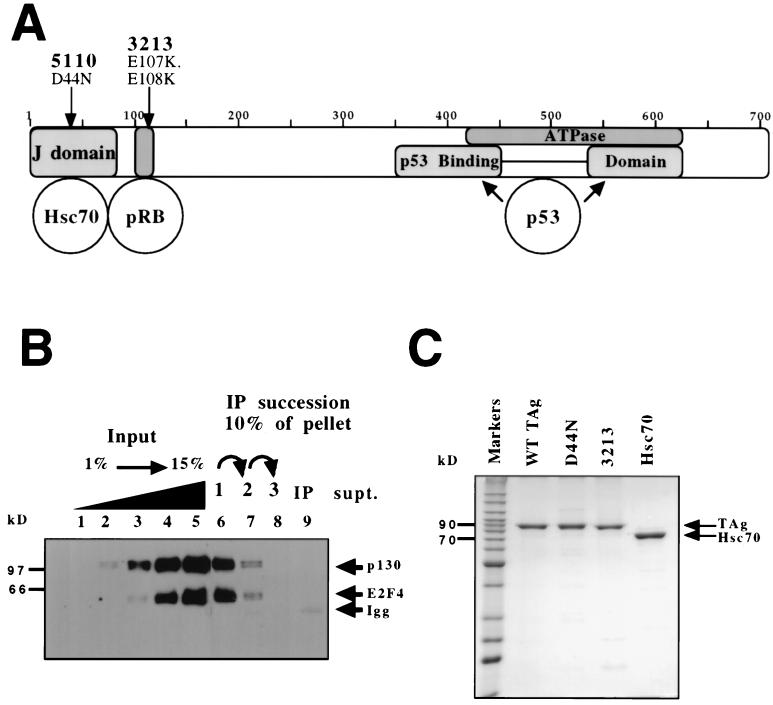
Structure of SV40 large T antigen. (A) T antigen comprises multiple domains, some of which are represented here as shaded boxes. The first 82 amino acids of T antigen have been shown to be a functional J domain that can bind to the DnaK homologue Hsc70. The pRb family of tumor suppressors bind to a conserved sequence of amino acids containing the motif LXCXE, which is found in many proteins that bind pRb. The bipartite binding site of the tumor suppressor p53 is represented by two separate boxes in the C terminus of T antigen. Overlapping this p53 binding region is the ATPase domain of T antigen. The positions and amino acid changes of each mutation used in this study are also shown. (B) E2F-4 is bound to p130 in insect cell lysate expressing p130, E2F-4, and DP-1. Increasing amounts of lysate (1, 2, 4, 8, and 15% input) are shown in lanes 1 to 5, respectively. Sequential immunoprecipitations (IP) for p130 were conducted, and steady-state levels of associated p130 and E2F-4 were determined using an SDS–8% polyacrylamide gel followed by immunoblot analysis for p130 and E2F-4 (10% of the pelleted complexes for the first, second, and third consecutive immunoprecipitation reactions are shown in lanes 6, 7, and 8, respectively). Immunoblot analysis for p130 and E2F-4 was conducted on the remaining supernatant (supt.) from the third immunoprecipitation (lane 9). Igg, band detected for the heavy chain of the immunoglobulin G antibody used in the immunoprecipitation reactions. (C) Wild-type and mutant T antigens (TAg) were purified from baculovirus-infected Sf9 cells as described in Materials and Methods. Hsc70 was purchased from Stress Gen Biotechnologies. Each lane contains 2 μg of protein. Samples were separated on an SDS–11% polyacrylamide gel and visualized by Coomassie brilliant blue staining. The migration of 90- and 70-kDa marker proteins is indicated.
Another region of T antigen required for transformation is the J domain (Fig. 1A) (for a review, see reference 2). J domains are a hallmark of a class of chaperones called J proteins or DnaJ chaperones that bear homology to Escherichia coli DnaJ. J proteins interact directly with DnaK-like proteins (such as Hsc70) and stimulate their ATPase activity. Stimulation of the ATPase activity of DnaK regulates its interaction with bound substrates (20). J proteins and DnaK-like chaperones are involved in numerous biological activities, including protein folding, protein transport across cellular membranes, protein degradation, and rearrangement of multiprotein complexes (4).
We have previously shown that T antigen requires a functional J domain and Rb binding motif in cis to transform REF52 and C3H10T1/2 cells (42). Both the J domain and pRb binding motif are also required for T-antigen-mediated inhibition of apoptosis (40). Furthermore, both the J domain and pRb binding motif are required for T antigen to alleviate pRb-mediated inhibition of E2F transactivation (19, 39, 47). Finally, cells expressing a mutant of T antigen defective for J domain or pRb binding functions contain a p130/E2F DNA-binding complex that is absent in cells expressing wild-type T antigen (47). Thus, it is likely that the J domain activity cooperates with the LXCXE motif of T antigen to disable Rb function, although the mechanism for how this occurs remains undetermined. This study seeks to better understand the mechanism by which T antigen inactivates the function of the Rb family of proteins. Specifically we show that T antigen requires its J domain function to cooperate with Hsc70 to release “free” E2F from association with Rb family members. The implications of these findings are discussed.
MATERIALS AND METHODS
Cell lines and molecular methods.
Sf9 and High Five insect cells and their handling have been described previously (6).
Plasmids carrying the complete viral genomes of simian virus 40 (SV40) T-antigen mutants 5110 (pSV-5110) and 3213 (pSV-3213) have been described elsewhere (32, 42). Mutant 5110 harbors a single-amino-acid substitution in the conserved HPD loop of the J domain (D44N), and 3213 contains a double-amino-acid substitution in the Rb binding motif (E107K and E108K).
Wild-type baculovirus (Autographica californica nuclear polyhedrosis virus [AcNPV]), a recombinant baculovirus that expresses the wild-type T antigen (AcNPV941T), and a baculovirus transfer plasmid containing the wild-type T-antigen gene (pVL941T) were kindly provided by Robert Lanford (Southwest Foundation for Biomedical Research) and have been described previously (27). Baculovirus transfer plasmid pVL941-3213 was generated from pSV-3213 as described for pVL941T. Baculovirus transfer plasmid pVL1393-5110 was generated by cloning the StuI-BamHI fragment of pSV-5110 into the SmaI and BglII sites of pVL1393. Baculoviruses were constructed as previously described (7).
A baculovirus producing human pRb was provided by Robert Weinberg (Massachusetts Institute of Technology), and baculoviruses expressing human E2F-4 and a truncated version of human p130 (42) were provided by Peter Whyte (Institute for Molecular Biology and Biotechnology, McMaster University). Baculoviruses expressing human DP-1 and E2F-1 were provided by Helen Pinwica-Worms (Washington University).
Protein purification and lysate preparation.
Wild-type and mutant T antigens were purified essentially as described (6) except that the gel filtration step was eliminated. In brief, Sf9 cells were infected with recombinant baculovirus for 43 h. The cells were harvested, washed with phosphate-buffered saline–EDTA, and lysed in a buffer containing Nonidet P-40 (NP-40; Sigma). The cleared cellular lysate was filtered and passaged over a PAb419 immunoaffinity column. The column was washed two times at pH 8.0, once at pH 9.0, and eluted at pH 11.0. Fractions containing T antigen were pooled and dialyzed against a buffer containing 50% glycerol. Proteins were stored at −20°C. Purified bovine brain Hsc70 and the recombinant ATPase fragment of Hsc70, Hsc701–386, were purchased from StressGen Biotechnologies.
Lysates enriched for pRb-E2F family member complexes were generated by infecting Sf9 or High Five cells with multiple viruses encoding a pRb family member [either pRb or p130(Δ2–371)], E2F (either E2F-1 or E2F-4), and DP-1. Cells were incubated at 27°C for 43 h. Cells were resuspended with 10 times the packed cell volume in buffer B (400 mM KCl, 50 mM HEPES [pH 7.9], 0.5 mM EDTA, 10% glycerol, 0.1% NP-40) with 1 μg of pepstatin per ml and an EDTA-free protease inhibitor cocktail tablet, as recommended by the manufacturer (Boehringer Mannheim). The cells were lysed for 25 min on ice and centrifuged at 16,000 × g in microcentrifuge for 30 min at 4°C. The pellets were discarded, and the protein concentration in the supernatant was determined with the Bradford assay with bovine serum albumin as the standard (Bio-Rad). Expression of pRb, p130, E2F-1, E2F-4, and DP-1 was confirmed via immunoblot analysis as described below.
Immunochemical methods.
Antibodies against pRb (IF8), p130 (C20), E2F-1 (C20), E2F-1 (KH95), E2F-4 (C20), and DP-1 (K20) were purchased from Santa Cruz Biotechnologies. Anti-pRb-14001A was purchased from PharMingen International. Anti-Hsc70 (AB1) was purchased from StressGen Biotechnologies. The T-antigen-specific antibodies PAb416, which recognizes an epitope between amino acids 83 and 121, and PAb419, which recognizes an epitope between amino acids 1 and 82, have been described previously (18). Anti-T-antigen antibody 901 recognizes an epitope in the last C-terminal 15 amino acids of T antigen and was kindly provided by Judith Tevethia (The Pennsylvania State University Medical School, Hershey, Pa.).
A total of 10 μg of insect cell lysates expressing pRb-E2F family complexes were mixed in the presence or absence of 1 μg of T antigen or T-antigen mutants and in the presence or absence of an ATP regeneration system (50 μM GDP mannose, 40 μM creatine phosphate, 0.2 mg of creatine phosphokinase per ml) in a volume of 30 μl of 50 mM KCl–20 mM HEPES [pH 7.4]–8.5% glycerol–1 mM EDTA–1 mM MgCl2. The reactions were then immunoprecipitated for T antigen, pRb, p130, E2F-4, or E2F-1 by incubation with 1 to 2 μg of appropriate antibody for 25 min at 22°C and then adding 50 μl of a 50% slurry of protein A-Sepharose CL-4B beads (Pharmacia) in buffer I (40 mM KCl, 20 mM HEPES [pH 7.8], 6 mM MgCl2, 0.1% NP-40) and incubating for 30 min at 4°C. The complexes were captured via a 20-s spin in a microcentrifuge at 16,000 × g and washed three times with 1 ml of buffer M (150 mM NaCl, 50 mM HEPES [pH 7.4], 10% glycerol, 0.1% Tween 20). The final pellets were resuspended in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing the reductants dithiothreitol and β-mercaptoethanol and resolved by electrophoresis on an 8% gel. Gels were transferred to an Immobilon polyvinylidene difluoride membrane (Millipore) and immunoblotted using antibodies directed against appropriate proteins via the ECL+ protocol (Amersham). Where mentioned, autoradiography signal intensity was quantified using NIH image 2.1. Alternatively, some of the anti-T-antigen-immunoprecipitated pellets were assayed for the chaperone-induced release of bound complexes. This was accomplished by washing the pellets with buffer I (1 ml) three times and resuspending the pellets in 30 μl of buffer I in the presence or absence of Hsc70 (1 μg) and the presence or absence of the ATP regeneration system, followed by gel shift analysis (see below) of the E2F complexes released into the supernatant. Mock-infected insect lysate (9 μg/μl) was added back to some of the reactions.
Gel shift assay.
A total of 10 μg of insect cell lysate expressing p130, E2F-4, and DP-1 was incubated with 1 ng of 32P-, 5′-end-labeled (United States Biochemical T4 protocol) DNA probe that contains a consensus E2F binding site (5′-ATTTAAGTTTCGCGCCCTTTCTCAA-3′). Mutant competitor oligonucleotide had the following sequence with a 2-bp substitution (underlined): (5′-ATTTAAGTTTCGATCCCTTTCTCAA-3′). Reactions (20 μl) were incubated for 10 min on ice followed by 20 min at room temperature in 50 mM KCl–20 mM HEPES [pH 7.4]–8.5% glycerol–1 mM EDTA–1 mM MgCl2. Reactions were loaded onto a 0.25× Tris–borate–EDTA–4.5% acrylamide gel and run at 200 V for 3 h at 4°C. Antibodies were added at least 10 min after other reagents. Gels were dried and exposed to autoradiographic film (Biomax MR; Kodak) for 2 to 12 h or to the phosphorimager (Fuji) for quantification.
RESULTS
In order to generate reactants to test the hypothesis that T antigen disrupts pRb-E2F family complexes, we expressed p130, E2F-4, and DP-1 in insect cells. From these cells, lysates were made and protein expression was confirmed as described in Materials and Methods. These lysates were sequentially immunoprecipitated with anti-p130 antibody and then probed for both p130 and E2F-4. Figure 1B (lanes 6 to 8) shows that greater than 90% of both p130 and E2F coprecipitated in the first immunoprecipitation. Furthermore, after two more consecutive immunoprecipitations, no E2F-4 or p130 was detectable in the supernatant (Fig. 1B, lane 9). Thus, we conclude that all of the E2F-4 present in this lysate is bound to p130. Similar immunodepletion experiments indicated that approximately 50% of the p130 present is complexed with E2F-4 (data not shown). Purified wild-type T antigen, mutants of T antigen, and Hsc70 were obtained as described in Materials and Methods. A Coomassie brilliant blue-stained gel of the purified proteins is shown in Fig. 1C.
T antigen associates with p130–E2F-4 complexes.
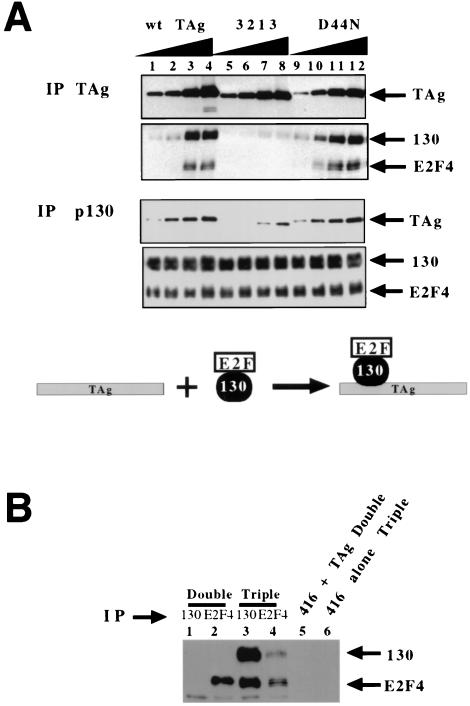
We asked if the association of T antigen with the p130 in a p130–E2F-4 complex dissociates p130 from E2F-4. Increasing amounts of purified T antigen were added to the lysate expressing the p130–E2F-4 complex, and T antigen was subsequently immunoprecipitated. Along with increasing amounts of T antigen, increasing amounts of p130 and E2F-4 also coprecipitated (Fig. 2A, top two panels, lanes 1 to 4). Immunoprecipitation of p130 from these T-antigen-treated lysates gave similar results; as greater amounts of T antigen were added, an increase in the amount of T antigen that coprecipitated with p130 was observed (Fig. 2A, bottom two panels, lanes 1 to 4). Adding increasing amounts of T antigen did not decrease the amount of E2F-4 associated with p130 (Fig. 2A, bottom panel, lanes 1 versus 4). Thus, T antigen binds to the p130–E2F-4 complex without dissociating p130 from E2F-4 (see diagram at bottom of Fig. 2A). Next, we tested the transformation-defective T-antigen pRb binding and J domain mutants (3213 and D44N, respectively) for the ability to associate with the p130–E2F-4 complex. The 3213 mutant contains an altered pRb binding motif and is defective for transformation and viral DNA replication (36). Immunoprecipitation of increasing amounts of 3213 fails to coprecipitate significant amounts of p130 and E2F-4 compared to immunoprecipitation of wild-type T antigen (Fig. 2A, top panel, lanes 5 to 8). The residual amount of p130 binding by 3213 is consistent with our previous results that showed that this mutant greatly reduces but does not eliminate T-antigen association with p130 (42).
FIG. 2.
T-antigen association with p130 and E2F-4. (A) Increasing amounts of wild-type (0.1, 0.3, 0.5, and 1.0 μg in lanes 1 to 4, respectively), Rb binding mutant 3213 (lanes 5 to 8), or J domain mutant D44N (lanes 9 to 12) T antigens were added to insect cell lysate expressing the p130–E2F-4–DP-1 complex. Immunoprecipitations for T antigen (IP TAg, top two panels) or p130 (bottom two panels) were conducted. Pelleted immunoprecipitates were assayed by SDS–8% PAGE and immunoblot analysis. The cartoon at the bottom of the figure diagrams the association of T antigen with the p130–E2F-4 complex. (B) Control reactions with anti-p130 antibody (lanes 1 and 3) or anti-E2F-4 antibody (lanes 3 and 4) were conducted in lysate made from cells doubly infected with E2F-4 and DP-1 viruses (lanes 1 and 2) or triply infected with p130, E2F-4, and DP-1 viruses (lanes 3 and 4). Lysate expressing E2F-4 and DP-1 was treated with T antigen and immunoprecipitated with anti-T-antigen pAB416 (lane 5). Lysate expressing p130, E2F-4, and DP-1 was immunoprecipitated with anti-T-antigen antibody without addition of T antigen.
The D44N point mutation lies in the strictly conserved HPD loop that is critical for J protein function and is conserved among T antigens of all polyomaviruses (33). We showed previously that this mutant is defective for replication, inhibition of apoptosis, and partially defective for transformation (32, 40, 42). When added to the lysate expressing the p130–E2F-4 complex, D44N coprecipitated p130 and E2F-4 as well as wild-type T antigen (Fig. 2A, top panel, lanes 9 to 12). Thus, a defective J domain does not inhibit the association of T antigen and p130–E2F-4 complex.
Next, increasing amounts of the T-antigen mutants 3213 and D44N were added to the p130–E2F-4 lysate, and anti-p130 antibody was also included. As with wild-type T antigen, a constant amount of E2F-4 coprecipitated with p130. D44N associated with p130–E2F-4 as well as wild-type T antigen, but a decreased association of 3213 with p130 was observed. We conclude that T antigen forms a stable complex with p130 and E2F-4 and that this association requires the T-antigen LXCXE motif but not the J domain. Furthermore, simple association of wild-type T antigen with the p130–E2F-4 complex does not dissociate p130 from E2F-4.
A control reaction in which T antigen was incubated with lysate expressing E2F-4–DP-1 showed that T antigen failed to coimmunoprecipitate any E2F-4 in the absence of p130 (Fig. 2B, lane 5). The addition of anti-T-antigen antibody (PAb416) in the absence of added T antigen failed to immunoprecipitate p130 or E2F-4 from the p130–E2F-4–DP-1 lysate (Fig. 2B, lane 6). Furthermore, the anti-p130 antibody is specific for p130, since it failed to precipitate any E2F-4 in the absence of p130 (Fig. 2B, lanes 1 and 3). We conclude that the E2F-4 that coprecipitates with T antigen does so by binding to p130.
ATP and Hsc70 induce T antigen to dissociate from p130–E2F-4.
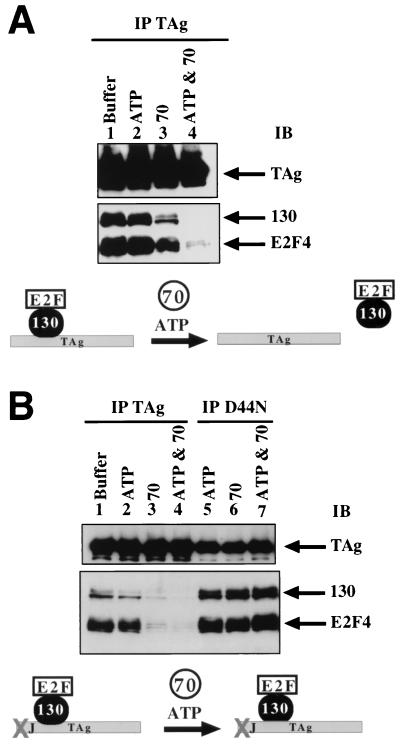
Given the evidence that the J domain of T antigen may act in conjunction with the Rb binding motif to alter Rb-E2F family complexes (see the introduction), we next tested the hypothesis that addition of a cochaperone DnaK affects the stability of the T-antigen–p130–E2F-4 complex. DnaK molecular chaperones interact with J proteins and are thought to change the conformation of bound protein substrates in an ATP-dependent manner (20). We chose to include the DnaK homologue Hsc70 in the reactions, since several studies have reported that T antigen interacts with Hsc70 and can stimulate its ATPase activity (5, 37, 42). We found that the addition of Hsc70 resulted in a modest decrease in the amount of p130 and E2F-4 which coprecipitated with T antigen. This decrease is further enhanced by the addition of an ATP regeneration system (Fig. 3A, lanes 3 and 4), although addition of an ATP regeneration system alone had little effect on the amount of p130 and E2F-4 that coprecipitated with T antigen (Fig. 3A, lane 2). We conclude that Hsc70 mediates release of p130–E2F-4 from T antigen and that this release is ATP dependent (diagram, bottom of Fig. 3A).
FIG. 3.
ATP and Hsc70 disrupt T antigen from p130. (A) Purified T antigen (TAg, 1 μg) was incubated with lysate (10 μg) containing the p130–E2F-4–DP-1 complex in buffer alone (lane 1), in the presence of an ATP regeneration system (lane 2), in the presence of 3 μg of Hsc70 (lane 3), and in the presence of an ATP regeneration system and Hsc70 (lane 4). The cartoon at the bottom of the figure shows that the T-antigen–p130–E2F-4 complex dissociates in the presence of Hsc70 and ATP. (B) Purified D44N (1 μg) was incubated with lysate (10 μg) containing the p130–E2F-4–DP-1 complex in the presence of an ATP regeneration system (lane 5), in the presence of 3 μg of Hsc70 (lane 6), and in the presence of an ATP regeneration system and Hsc70 (lane 7). Reactions identical to those in panel A are shown for comparison (lanes 1 to 4). The cartoon at the bottom of the figure shows that D44N, unlike wild-type T antigen, does not release the p130–E2F-4 complex upon incubation with Hsc70 and ATP.
We next examined if a functional T-antigen J domain is required to dissociate p130–E2F-4 from T antigen. By incubating the J domain mutant D44N with the lysate expressing the p130–E2F-4 complex, we found that D44N remained associated with p130 and E2F-4, even in the presence of Hsc70 and ATP (Fig. 3B, lanes 5 to 7). Thus, mutation of the J domain abolishes the ability of T antigen to release p130–E2F-4 upon incubation with Hsc70 and ATP (diagram, bottom of Fig. 3B).
T antigen requires Hsc70 and ATP to release p130 from E2F-4.
Next we sought to determine if E2F-4 remains associated with p130 after release from T antigen. The ability of T antigen to dissociate p130 from E2F-4 was examined by immunoprecipitating E2F-4 and examining for the release of p130 via immunoblot analysis. Upon incubation with T antigen, Hsc70, and an ATP regeneration system, the amount of p130 as well as the amount of T antigen associated with E2F-4 decreased by approximately 90% (Fig. 4A, lane 3). Incubation with Hsc70 and the ATP regeneration system without T antigen had no effect on the levels of p130 associated with E2F-4 (Fig. 4C, lane 3). In control reactions, a slight decrease in the amount of p130 associated with E2F-4 is observed if T antigen is incubated with either Hsc70 or an ATP regeneration system alone (Fig. 4B, lanes 3 and 4). The addition of T antigen without Hsc70 and ATP did not decrease the amount of p130 associated with E2F-4; in fact, a consistent increase in p130 was observed (Fig. 4A, lane 2). Similar results were obtained when p130 was immunoprecipitated from T antigen treated extracts and the amount of associated E2F-4 was examined (Fig. 4A, right). A fragment of Hsc70 comprising the ATPase binding domain, Hsc701–386, that fails to bind T antigen (unpublished data) was unable to stimulate the T-antigen-induced release of p130 from E2F-4 (Fig. 4C, lane 5). We conclude that T antigen mediates the disruption of E2F from p130 in a reaction that requires Hsc70 and ATP.
FIG. 4.
T-antigen-dependent p130 dissociation from E2F-4. (A) Lysate containing the p130–E2F-4–DP-1 complex was incubated with T antigen (TAg) in the presence or absence of Hsc70 and ATP. Reactions were then immunoprecipitated (IP) with antibody that recognizes E2F-4 (C20, lanes 1 to 3) or p130 (C20, lanes 4 and 5) and immunoblotted (IB) for T antigen or p130 and E2F-4. Lysates were untreated (lane 1) or treated with 1 μg of T antigen (lanes 2 and 4) or T antigen, ATP, and 3 μg of Hsc70 (lanes 3 and 5). The cartoon to the right shows that p130–E2F-4 complex dissociates in the presence of T antigen, Hsc70, and ATP. (B) The J domain and Rb binding motif are required for the disruption of p130–E2F-4 complex. Lysates were untreated (lane 1), treated with 1 μg of T antigen (lane 2), treated with T antigen and ATP (lane 3), or treated with T antigen and 3 μg of Hsc70 (lane 4); lanes 5 to 7 are treated with ATP, Hsc70, and the indicated T antigen. The cartoon to the right shows that unlike wild-type T antigen, D44N does not dissociate p130 from E2F-4 in the presence of Hsc70 and ATP (top). In the presence of T antigen, Hsc70, and ATP, p130 dissociates from E2F-4–DP-1 (bottom). (C) ADP, ATPγS, and Hsc701–386 do not foster release of p130 from E2F-4. Lysates were untreated (lane 1), treated with 1 μg of T antigen (lane 2), treated with ATP and 3 μg of Hsc70 (lane 3), treated with T antigen, ATP, and Hsc70 (lane 4), treated with T antigen, ATP and 3 μg of Hsc701–386 (denoted with asterisk) (lane 5), treated with T antigen, ADP, and Hsc70 (lane 6), or treated with T antigen, ATPγS, and 3 μg of Hsc70 (lane 7). (D) Steady-state levels of p130 and E2F-4. Immunoblot steady-state analysis was conducted on 10 μg of insect cell lysate expressing the p130–E2F-4–DP-1 complex. Lysate was untreated (lane 1) or treated with T antigen (1 μg), Hsc70 (3 μg), and an ATP regeneration system (lane 2).
We explored the nucleotide dependence of this dissociation reaction. ADP (Fig. 4C, lane 6) and the ATP hydrolysis-defective analog ATPγS (Fig. 4C, lane 7) were included in the release reaction instead of the ATP regeneration system. Neither could cooperate with Hsc70 and T antigen to induce the disruption of p130 from E2F-4. We conclude that hydrolysis of ATP is required for the Hsc70- and T-antigen-induced disruption of E2F-4 from p130.
T antigen requires a functional J domain and Rb binding motif to release p130–E2F-4 complexes.
Since the J domain is required to disengage T antigen from the p130–E2F-4 complex (Fig. 3B), we tested whether a functional J domain is required for the T-antigen-induced release of p130 from E2F-4. Either purified wild-type T antigen or the J domain mutant D44N was incubated with Hsc70, the ATP regeneration system, and the lysate expressing p130–E2F-4–DP-1 followed by immunoprecipitation with antibody against E2F-4. As shown before, incubation with T antigen, Hsc70, and ATP resulted in a decrease in the p130 that coprecipitated with E2F-4 (Fig. 4B, lane 5). However, with D44N, no change in the amount of p130 that coprecipitated with E2F-4 was observed (Fig. 4B, lane 7). Also, D44N was found in the E2F-4 immunoprecipitates even though wild-type T antigen is released in the presence of Hsc70 and ATP (Fig. 4B, lanes 5 and 7). We conclude that T antigen requires a functional J domain to mediate disruption of p130 from E2F-4 (see diagram at right of Fig. 4B).
We next determined if the Rb binding mutant of T antigen (3213) was capable of dissociating p130 from E2F-4. 3213, Hsc70, and ATP were added to the p130–E2F-4 lysate and immunoprecipitated with antibody against E2F-4. No decrease in the amount of p130 coprecipitating with E2F-4 was observed (Fig. 4B, lane 6). Thus, disruption of the p130–E2F-4 complex requires a functional LXCXE motif.
Even though E2F-4 is released from p130 following treatment with T antigen, Hsc70, and ATP, it still remains associated with its heterodimeric partner DP-1 (Fig. 4B). Therefore, treatment with T antigen and Hsc70 does not disrupt all protein complexes but rather is specific for the release of p130 from E2F-4–DP-1.
Finally, T antigen induces the degradation of p130 in vivo (43), and we have observed a reduction in the steady-state levels of p130 and E2F-4 upon SV40 infection (unpublished results). These data raise the possibility that the dissociation of p130 from E2F-4 could be due to their degradation. To test this, mock immunoprecipitation assays were performed, and the steady-state levels of p130 and E2F-4 were determined via immunoblot analysis. No change in the level of either E2F-4 or p130 was detected upon treatment with T antigen, Hsc70, and an ATP regeneration system (Fig. 4D, lanes 1 and 2). Therefore, with our in vitro results, degradation cannot account for the disassociation of p130 from E2F-4.
Free E2F-4 and p130–E2F-4 released from T antigen are capable of binding DNA.
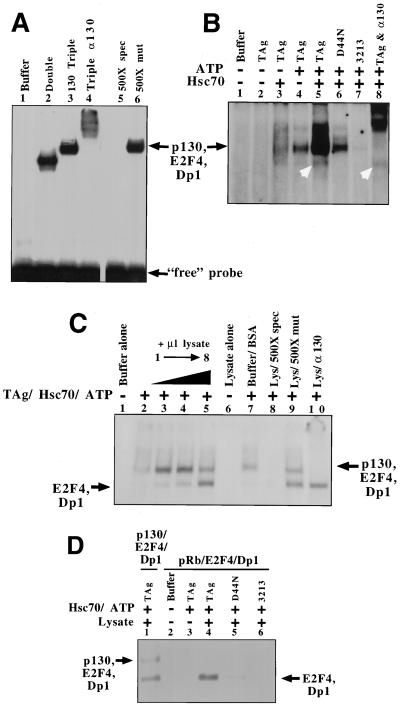
Electrophoretic mobility shift analysis shows that the p130–E2F-4–DP-1 complex from lysates containing p130, E2F-4, and DP-1 binds to DNA containing an E2F consensus site. Incubation of lysate that contains only E2F-4 and DP-1 showed a free E2F complex that migrates as a single band (Fig. 5A, lane 2). A slower-migrating complex is observed when lysates from cells expressing p130–E2F-4–DP-1 are incubated in the gel shift reaction (Fig. 5A, lane 3). Incubation with mock-infected Sf9 cell lysate showed no complex, indicating that endogenous insect E2F DNA binding activity is below the level of detection of this assay (Fig. 5A, lane 1). Incubation of the lysate containing the p130–E2F-4–DP-1 complex with antibody to p130 supershifted the entire complex, indicating that all of the active E2F is complexed to p130 (Fig. 5A, lane 4). The p130–E2F-4–DP-1–DNA complex is specific, since including a 500-fold molar excess of specific competitor oligonucleotide abolished formation of the complex; however, incubation with a 500-fold molar excess of mutant competitor oligonucleotide had no effect (Fig. 5A, lanes 5 and 6). Thus, the p130–E2F-4–DP-1 complex present in the lysate specifically binds to DNA containing an E2F consensus binding site.
FIG. 5.
p130–E2F-4 complexes disrupted by T antigen can bind to DNA. Gels shifts were conducted using insect cell lysates and radiolabeled DNA oligonucleotide containing an E2F consensus binding site as the probe. (A) Control reactions. Uninfected insect cell lysate (lane 1), E2F-4 and DP-1 double-infected insect cell lysate (lane 2), p130, E2F-4, and DP-1 triple-infected insect cell lysate (lane 3) or p130–E2F-4–DP-1-expressing lysate was incubated with anti-p130 antibody (1 μg, lane 4), a 500-fold molar excess of specific (spec., lane 5) or mutant (mut., lane 6) nucleotide competitor. (B) E2F-containing DNA-binding complexes are released from T antigen (TAg). T antigen or mutants of T antigen were incubated with lysate containing p130–E2F-4–DP-1 and immunoprecipitated for T antigen. Complexes released from these pellets were assayed via gel shift analysis using the E2F probe. The following treatments were applied: no T antigen in the immunoprecipitation, treated with buffer (lane 1), T antigen immunoprecipitation treated with buffer (lane 2), T antigen immunoprecipitation treated with Hsc70 (lane 3), T antigen immunoprecipitation treated with an ATP regeneration system (lane 4), T antigen immunoprecipitation treated with Hsc70 and an ATP regeneration system (lane 5), D44N immunoprecipitation treated with Hsc70 and an ATP regeneration system (lane 6), 3213 immunoprecipitation treated with Hsc70 and an ATP regeneration system (lane 7), T antigen immunoprecipitation treated with Hsc70, an ATP regeneration system, and anti-p130 antibody included in the gel shift reaction (lane 8). White arrows, free E2F. (C) Treatment with lysate increases the efficiency of p130–E2F-4 release. T antigen was incubated with p130–E2F-4 complexes and immunoprecipitated for T antigen. Pellets were treated and assayed for E2F DNA-binding activity. T-antigen immunoprecipitation was treated with buffer alone (lane 1), Hsc70 and an ATP regeneration system (lane 2), Hsc70 and an ATP regeneration system and 1, 3, or 8 μl of mock lysate (lanes 3 to 5, respectively), 8 μl of mock buffer and BSA (lane 7), Hsc70, an ATP regeneration system, 8 μl of mock lysate, and a 500-fold excess of unlabeled specific (lane 8) or mutant (lane 9) E2F probe competitor, or Hsc70, an ATP regeneration system, 8 μl of mock lysate, and anti-p130 antibody (lane 10). Mock lysate alone (8 μl) was run to demonstrate that no E2F binding activity was detectable in this lysate (lane 6). (D) Treatment with Hsc70 and ATP releases free E2F-4 from T-antigen-bound pRb–E2F-4 complexes. T antigen was incubated with pRb-E2F complexes and immunoprecipitated for T antigen. T-antigen immunoprecipitation was treated with buffer alone (lane 2), Hsc70 and an ATP regeneration system (lane 2), or lysate, Hsc70, and an ATP regeneration system (lane 4). Alternatively, T-antigen mutants D44N (lane 5) and 3213 (lane 6) were immunoprecipitated and treated with lysate, ATP regeneration system, and Hsc70. Free E2F-4 and p130–E2F-4 complexes (released from T antigen as described for panel C) are shown for comparison (lane 1).
We next investigated whether the p130–E2F-4 that is released from T antigen is active for binding to DNA containing an E2F consensus binding site. Lysate containing the p130–E2F-4–DP-1 complex was incubated with T antigen and then immunoprecipitated using a T-antigen-specific antibody (PAb416). The pelleted complexes were incubated with various treatments, and the complexes that were released into the supernatant were assayed for the ability to bind to DNA containing the E2F consensus binding site. We observed that incubation of the pelleted complexes with Hsc70 and ATP releases only a small amount of free E2F-4–DP-1 complex as well as a p130–E2F-4–DP-1 complex (Fig. 5B, lane 5).
We next determined via gel shift analysis whether a functional J domain and Rb binding motif are required to release E2F complexes. D44N or 3213 was immunoprecipitated with anti-T-antigen antibody after incubation with the p130–E2F-4–DP-1 complex. Treatment of the pelleted complexes with ATP and Hsc70 releases a greatly diminished amount of E2F-containing complexes compared to wild-type complexes (Fig. 5B, lanes 6 and 7). As shown in Fig. 2, 3213 cannot bind to a significant amount of the p130-E2F complex. Thus, we do not detect any release of E2F complexes. This corresponds well with the observation that an identical immunoprecipitation-release reaction that does not include T antigen also fails to release any E2F complexes (Fig. 5B, lane 1). Finally, a functional J domain is required to obtain optimal release of the p130-E2F complex from T antigen, since D44N is defective for release (Fig. 5B, lane 6).
It should be noted that significantly fewer E2F complexes are detected upon incubation of the pellets with ATP or Hsc70 alone (Fig. 5B, lanes 3 and 4) compared to incubation with both Hsc70 and ATP (Fig. 5B, lane 5). Incubation with buffer alone did not release any E2F complexes (Fig. 5B, lanes 1 and 2). A large portion of the E2F-containing complex released by treatment with ATP and Hsc70 is supershifted with antibody to p130 (Fig. 5B, lane 8). However, some free E2F activity is released from T antigen, since approximately 10% of the released complex migrates appropriately and is unaffected by treatment with anti-p130 antibody (Fig. 5B, lane 8, white arrow). We conclude that the p130–E2F-4 complex that binds to T antigen releases in the presence of ATP and Hsc70 and that a small fraction of the p130–E2F-4 complex dissociates into free E2F-4. In addition, the E2F complexes released from T antigen can bind to DNA.
These data suggested that our immunoprecipitation Western blot assay (Fig. 3), which was done in the context of cellular lysate, is more efficient than our semi-immunopurified gel shift release assay, in which the lysate was washed away before treatment with Hsc70 and ATP (90% compared to 10%). To determine if lysate increased the ratio of free E2F-4 released from the anti-T-antigen-immunoprecipitated pellets, increasing amounts of mock-infected insect cell lysate was incubated with Hsc70 and ATP in the release assay (described above), and gel shift analysis was conducted (Fig. 5C, lanes 3 to 5). As shown in Fig. 5B, without addition of lysate to the semipure pellet, treatment with Hsc70 and ATP produces only about 10% of the total E2F signal bound to DNA as free. However, if 8 μl (63 μg of total protein) of mock lysate was included in the reaction with ATP and Hsc70, 60% (approximately sixfold more than without lysate) of the total E2F signal observed is free E2F (Fig. 5C, lane 5). Furthermore, the amount of total E2F signal detected is increased in a dose-dependent manner (Fig. 5C, lanes 2 to 5). Treatment with buffer and bovine serum albumin (8 μl, 65 μg) (Fig. 5C, lane 7), or heat-killed mock lysate (8 μl, 95°C, 10 min) (data not shown) did not increase the ratio of free E2F signal released by Hsc70 and ATP. As a control, mock lysate was incubated in a direct gel shift assay, and no shift was detected (Fig. 5C, lane 6). Therefore, the increase in released free E2F signal induced by coincubation with mock lysate that we observed cannot be due to any endogenous E2F complexes present in the mock lysate. We incubated anti-p130 antibody in the released E2F complexes and, as expected, observed a supershift of the top p130-containing band with no effect on the bottom free E2F-4 band (Fig. 5C, lane 10). Furthermore, the released E2F-4 complexes are specific, since incubation with a 500-fold excess of unlabeled specific (Fig. 5C, lane 8) but not mutant (Fig. 5C, lane 9) competitor inhibits detection of gel-shifted bands. We conclude that the release assay is most efficient in the context of the cellular lysate.
T antigen requires Hsc70 and ATP to disrupt pRb from E2F.
We next tested if T antigen could dissociate E2F from an Rb family member other than p130. Insect cell lysates overexpressing pRb were mixed with lysates overexpressing E2F-4 and DP-1 to allow a complex to form. Subsequent immunoprecipitation and gel shift analysis confirmed that pRb bound to E2F-4 (data not shown). These mixed lysates were then incubated with T antigen and immunoprecipitated with anti-T-antigen antibody. We then treated these pellets as in the p130 release assay described above. Treatment with lysate, an ATP regeneration system, and Hsc70 releases free E2F-4, as assayed by gel shift analysis (Fig. 5D, lane 4). Surprisingly, unlike the previous experiments with p130, very little (less than 5%) of the released E2F complexes is bound to pRB. As one might expect from the p130–E2F-4 results, mutants of T antigen in the J domain and Rb binding motif are defective for this activity (Fig. 5D, lanes 5 and 6; 60 and 85% defective, respectively). Therefore, T antigen induces the disruption of multiple pRb family members from E2F in an Hsc70-, ATP-, and J domain-dependent manner.
DISCUSSION
SV40 T antigen is a multifunctional, multidomain protein that can induce neoplastic transformation and is required for viral DNA replication, gene transactivation, and virion assembly. Previous studies demonstrated that the J domain of T antigen is essential for both viral replication and transformation (32, 42). In some circumstances, transformation by T antigen requires that the J domain act in cis with the pRb binding motif (42). Furthermore, T antigen requires a functional J domain to alleviate pRb-mediated repression of E2F transactivation (19, 39, 43). This evidence is consistent with the hypothesis that T antigen's effect on pRb family complexes requires the action of both its J domain and Rb binding motif to inhibit pRb function and drive the cells to divide. These data have led to a model in which T antigen chaperones the rearrangement of multiprotein complexes to elicit its growth-promoting effects on the cell (39, 42). However, this model has not been proven, and the mechanism by which the J domain modulates the activity of pRb family complexes remains undetermined. In this work we show that T antigen binds to pRb family members and dissociates E2F by a mechanism that requires ATP hydrolysis and the DnaJ molecular chaperone function of T antigen.
T antigen does not disrupt Rb-E2F complexes by affinity displacement.
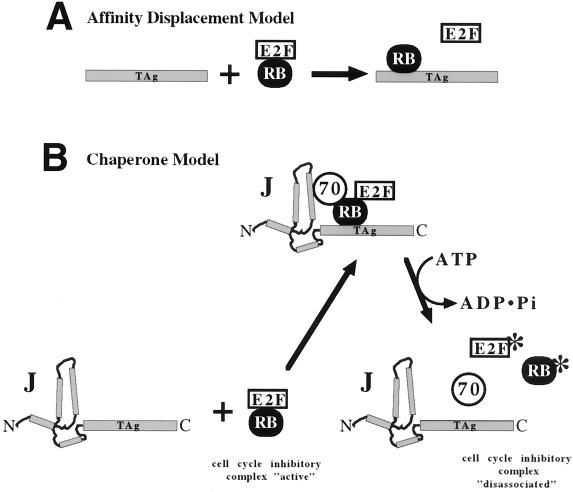
One model to explain how T antigen frees E2F from its association with Rb is that T antigen and E2F compete for the same binding site on Rb but that T antigen has a higher affinity for this site than E2F (8). Thus, the addition of T antigen to the Rb-E2F complex induces the displacement of free E2F by affinity displacement (Fig. 6A). This is consistent with observations that T-antigen binding to pRb is required to inactivate Rb's growth-suppressive functions. However, recent evidence indicates that in addition to the Rb binding motif, a functional J domain is required for T antigen to block Rb function (32, 39, 47). Consequently, a chaperone-based model was proposed in which the J domain recruits Hsc70 into association with the Rb-E2F complex and E2F is released due to the action of Hsc70 on the complex (42).
FIG. 6.
Models for T-antigen-induced dissociation of pRb-E2F family complexes. Disruption of growth-inhibitory complexes (labeled Rb/E2F) drives cells to enter the cycle. (A) Affinity displacement model. T antigen (TAg) binds to pRb with a higher affinity for E2F than pRb's affinity for E2F, thus sequestering pRb from E2F. (B) Chaperone model. T antigen, with cochaperone Hsc70 and ATP hydrolysis, elicits a conformational change in the pRb-E2F complex or T antigen, thereby rearranging the complex and decreasing the amount of pRb associated with E2F. Note that T antigen does not remain associated with pRb. Some other modification (denoted with an asterisk) of pRb or E2F may be required to keep proteins from reassociating. To underscore the role of the J domain, it is depicted as four interacting rods which represent the four alpha helices of the J domain (21) at the amino terminus (N) of T antigen.
To explore these two hypotheses, we performed experiments in which T antigen was added to a preformed p130–E2F-4–DP-1 complex. An affinity displacement model predicts that as T antigen is added to the complex, E2F-4 should be released. However, we observed that no E2F-4 was released from p130 when T antigen was added to the complex; rather, T antigen formed a stable association with the complex (Fig. 2). Thus, T antigen and E2F do not compete for the same binding site on p130. The fact that T antigen associates with the complex but does not release free E2F-4, combined with previous results that show that the J domain is required in cis with the Rb binding motif to inactivate Rb-mediated growth suppression (42), strongly argues against affinity displacement as the mechanism for inactivation of Rb function.
Disruption of Rb-E2F complexes requires the molecular chaperone activity of T antigen.
Previous work has shown that T antigen binds Hsc70 through its J domain and that a functional J domain is required for T antigen inactivation of Rb function (5, 39, 42, 47). We proposed a model in which disruption of the Rb-E2F complex requires energy that is derived from Hsc70-mediated ATP hydrolysis. In this model, T antigen serves as a scaffold that brings the Rb-E2F complex in contact with the Hsc70 chaperone machine.
In this study we found that T antigen could mediate the disruption of Rb-E2F complexes in a reaction that requires exogenous Hsc70 and ATP. Mutants of T antigen that cannot bind to Rb or that contain a nonfunctional J domain fail to disrupt Rb-E2F complexes. As expected, a mutant T antigen that is defective for Rb binding failed to either associate with p130-E2F complexes or mediate the release of E2F from p130 in the presence of ATP and Hsc70. A T-antigen mutant containing a defective J domain associated with p130-E2F complexes as well as wild-type T antigen, but was defective for the Hsc70- and ATP-dependent dissociation of the p130-E2F complexes. An Hsc70 mutant that fails to bind to T antigen does not disrupt p130 from E2F-4. The nucleotide dependence of this reaction is specific to ATP, since ADP and ATPγS fail to induce the disruption of p130 from E2F-4. Both T antigen and Hsc70 have robust ATPase activities. However, ATPase-defective T-antigen mutants are still capable of disrupting p130–E2F-4 in the presence of Hsc70 and ATP (data not shown). Therefore, we conclude that the reaction requires Hsc70-mediated ATP hydrolysis.
The above results are consistent with a chaperone-based model (Fig. 6B) in which T antigen binds to p130-E2F complexes through its LXCXE motif. In this model, the J domain of T antigen recruits Hsc70 to the Rb-E2F complex, stimulating the ATPase activity of Hsc70. This in turn releases the p130–E2F-4 complex from T antigen, inducing the dissociation of p130 from E2F. Presumably, T antigen and Hsc70 are then free to recycle and act on additional p130-E2F complexes. In the context of the cell, this action of T antigen could remove a cellular growth transcription-inhibitory p130–E2F-4–DP-1 complex and/or induce a growth-promoting complex composed of free E2F-4–DP-1.
Once the p130–E2F-4 complex is disrupted by T antigen, some modification to p130 or E2F-4 may be required to prevent their reassociation. In the context of the cell, this modification may target p130 or E2F-4 for degradation, since the J domain of T antigen is required to decrease the half-life of p130 (43) and SV40 infection decreases the steady-state levels of p130 and E2F-4 (unpublished results). It is also possible that the action of T antigen causes changes in the association of Hsc70 with p130 and that this renders the p130 complex incapable of reassociating with E2F-4. It has been shown that pRb can bind to Hsc70 in tissue cell culture and in vitro (23). We have shown that p130 can form a stable complex with Hsc70 in vitro (unpublished observation), but the in vivo relevance of this interaction or whether this can modify the function of p130 remains to be determined. We have shown that a semipurified p130-E2F complex released from a T-antigen-immunoprecipitation pellet can bind to DNA, but only approximately 10% is unbound to p130. If, however, we add back lysate to the immunoprecipitated T-antigen–p130–E2F-4 complex, we obtain a sixfold increase in the amount of free E2F-4 observed. Additions of buffer alone, buffer and BSA, or heat-treated lysate all fail to induce this effect. This evidence corresponds well with our immunoprecipitation Western blot data (Fig. 3), in which we are able to detect up to 90% of the p130-E24F complex being disrupted. For these experiments, Hsc70 and ATP are added directly to the insect lysate expressing the p130–E2F-4 complex, and then immunoprecipitation is conducted. Thus, any factors present in the lysate that increase the efficiency of the chaperone-mediated reaction are present. If, however, we first immunoprecipitate the T-antigen–p130–E2F-4 complex and then treat with Hsc70 and ATP, it is difficult to detect any release of the p130 from E2F-4 by Western blot analysis (data not shown). Since the factor is heat sensitive, we conclude that it is most likely proteinaceous. This factor increases the efficiency of the T-antigen-mediated chaperone-dependent release of p130–E2F-4 from T antigen as well as the release of p130 from E2F-4. Future experimentation is required to identify the components of this factor.
T antigen directs the rearrangement of multiprotein complexes.
Recently it has been reported that the J domain of T antigen is required for synergistic transactivation and binding with the transcription factor complex Tst-1–Oct6–SCIP (41). Others have shown that expression of a J domain-containing fragment of T antigen is sufficient to downregulate the Her-2 promoter, which is hyperactivated in certain breast cancers (26). Mutations in the J domain of T antigen render SV40 defective for viral DNA replication and virion assembly as well as transformation (32, 42). Perhaps recruiting different multiprotein complexes into association with the Hsc70 chaperone machine is a common theme linking T antigen's diverse functions (2).
Chaperone-mediated rearrangement of multiprotein complexes could be a common viral strategy, as phage lambda uses host E. coli DnaJ and DnaK to rearrange the replication machinery proteins essential for phage replication (45). Since T antigen disrupts Rb-E2F family complexes via a molecular chaperone mechanism, we wonder if adenovirus E1A or papillomavirus E7, which also contain an LXCXE Rb binding motif, utilize a chaperone mechanism to disrupt Rb-E2F complexes. The papillomavirus protein E1 utilizes the cellular chaperones Hsc70 and DnaJ to stimulate viral genome replication (29). The papillomavirus E7 protein interacts with a mitochondrial J protein involved in apoptosis regulation (38, 44). Since E1A and E7 contain no homology to J proteins, it is unlikely that they directly stimulate the ATPase activity of Hsc70 to disrupt Rb-E2F complexes. However, it is possible that E1A or E7 indirectly uses cellular chaperones to assist in disrupting Rb-E2F complexes. It has been shown that a peptide of the conserved region 1 (CR1) of E1A may compete with E2F for the same binding site on Rb (15a, 22a). Our experiments do not rule out a role for the T antigen CR1 motif in the disruption of Rb-E2F complexes. However, we observe a stable association of T antigen with Rb-E2F complexes, indicating that E2F and T antigen can exist in the same complex in the presence of CR1. Thus, the presence of an intact amino terminus (including the CR1 motif and J domain) is not sufficient to disrupt Rb from E2F unless exogenous Hsc70 and ATP are included in the reaction. Furthermore, the J domain mutant D44N, which contains a wild-type CR1, is defective for disrupting Rb from E2F even in the presence of Hsc70 and ATP. Therefore, the disruption of Rb-E2F complexes that we observe is truly a chaperone-dependent phenomenon that cannot be accounted for simply by competition for Rb binding between E2F and the CR1 motif of T antigen.
Our results suggest several new functions of Hsc70. For the first time we demonstrate that Hsc70 assists in the release of p130 from E2F-4 as well as the release of p130 from T antigen (Fig. 3 and 4). The tumor suppressor proteins p53 and pRb both associate with the molecular chaperone Hsc70 (16, 23, 30), although the function of these interactions is unclear. One possibility is that the normal cellular role of these tumor suppressors is modified by Hsc70, similar to the way T antigen induces Hsc70 to affect Rb. Similarly, chaperones may be involved in regulating the function of the p53 tumor suppressor. T antigen binds and inactivates p53, and it is possible that the J domain plays a role in this process (34, 35). The work presented in this paper suggests that the cellular chaperone machinery is essential for the virus-induced disruption of pRb-E2F family complexes and that this accounts for the J domain requirement for T-antigen-induced transformation. Future experiments will test if a similar mechanism is utilized in nonvirally induced cell cycle progression.
ACKNOWLEDGMENTS
This work was supported by NIH grant CA40586.
We thank J. Brodsky, K. Sachsenmeier, R. Hendrix, A. McClellan, A. Slinskey, and L. Engler for their critical reading of the manuscript. We thank T. Harper for assistance with the figures.
REFERENCES
- 1.Blake M C, Azizkhan J C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodsky J L, Pipas J M. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan T M, Reddel R R. SV40-induced immortalization of human cells. Crit Rev Oncog. 1994;5:331–357. doi: 10.1615/critrevoncog.v5.i4.10. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 5.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 6.Cantalupo P, Saenz-Robles M T, Pipas J M. Expression of SV40 large T antigen in baculovirus systems and purification by immunoaffinity chromatography. Methods Enzymol. 1999;306:297–307. doi: 10.1016/s0076-6879(99)06019-x. [DOI] [PubMed] [Google Scholar]
- 7.Castellino A M, Cantalupo P, Marks I M, Vartikar J V, Peden K W, Pipas J M. trans-Dominant and non-trans-dominant mutant simian virus 40 large T antigens show distinct responses to ATP. J Virol. 1997;71:7549–7559. doi: 10.1128/jvi.71.10.7549-7559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellappan S, Kraus V B, Kroger B, Munger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 10.Chen J G, Tobin J, Pipas J M, Van Dyke T. T-antigen mutant activities in vivo: roles of p53 and pRB binding in tumorigenesis of the choroid plexus. Oncogene. 1992;7:1167–1175. [PubMed] [Google Scholar]
- 11.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 12.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. . (Erratum, 15:5846–5847.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Fattey A R, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fourie A M, Hupp T R, Lane D P, Sang B C, Barbosa M S, Sambrook J F, Gething M J. HSP70 binding sites in the tumor suppressor protein p53. J Biol Chem. 1997;272:19471–19479. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- 17.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris K F, Christensen J B, Radany E H, Imperiale M J. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 21.Hill R B, Flanagan J M, Prestegard J H. 1H and 15N magnetic resonance assignments, secondary structure, and tertiary fold of Escherichia coli DnaJ(1–78) Biochemistry. 1995;34:5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- 22.Ibaraki N, Chen S C, Lin L R, Okamoto H, Pipas J M, Reddy V N. Human lens epithelial cell line. Exp Eye Res. 1998;67:577–585. doi: 10.1006/exer.1998.0551. [DOI] [PubMed] [Google Scholar]
- 22a.Ikeda M A, Nevins J R. Identification of distinct roles for separate E1A domains in the disruption of E2F complexes. Mol Cell Biol. 1993;13:7029–7035. doi: 10.1128/mcb.13.11.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue A, Torigoe T, Sogahata K, Kamiguchi K, Takahashi S, Sawada Y, Saijo M, Taya Y, Ishii S, Sato N, et al. 70-kDa heat shock cognate protein interacts directly with the N-terminal region of the retinoblastoma gene product pRb: identification of a novel region of pRb-mediating protein interaction. J Biol Chem. 1995;270:22571–22576. doi: 10.1074/jbc.270.38.22571. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 25.Kaelin W G, Jr, Ewen M E, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao M C, Liu G Y, Chuang T C, Lin Y S, Wuu J A, Law S L. The N-terminal 178-amino-acid domain only of the SV40 large T antigen acts as a transforming suppressor of the HER-2/neu oncogene. Oncogene. 1998;16:547–554. doi: 10.1038/sj.onc.1201513. [DOI] [PubMed] [Google Scholar]
- 27.Lanford R E. Expression of simian virus 40 T antigen in insect cells using a baculovirus expression vector. Virology. 1988;167:72–81. doi: 10.1016/0042-6822(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 28.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 29.Liu J S, Kuo S R, Makhov A M, Cyr D M, Griffith J D, Broker T R, Chow L T. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 30.Merrick B A, He C, Witcher L L, Patterson R M, Reid J J, Pence-Pawlowski P M, Selkirk J K. HSP binding and mitochondrial localization of p53 protein in human HT1080 and mouse C3H10T1/2 cell lines. Biochim Biophys Acta. 1996;1297:57–68. doi: 10.1016/0167-4838(96)00089-1. [DOI] [PubMed] [Google Scholar]
- 31.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 32.Peden K W, Pipas J M. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes. 1992;6:107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- 33.Pipas J M. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quartin R S, Cole C N, Pipas J M, Levine A J. The amino-terminal functions of the simian virus 40 large T antigen are required to overcome wild-type p53-mediated growth arrest of cells. J Virol. 1994;68:1334–1341. doi: 10.1128/jvi.68.3.1334-1341.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rushton J J, Jiang D, Srinivasan A, Pipas J M, Robbins P D. Simian virus 40 T antigen can regulate p53-mediated transcription independent of binding p53. J Virol. 1997;71:5620–5623. doi: 10.1128/jvi.71.7.5620-5623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutila J E, Imperiale M J, Brockman W W. Replication and transformation functions of in vitro-generated simian virus 40 large T antigen mutants. J Virol. 1986;58:526–535. doi: 10.1128/jvi.58.2.526-535.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawai E T, Butel J S. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schilling B, De-Medina T, Syken J, Vidal M, Munger K. A novel human DnaJ protein, hTid-1, a homolog of the Drosophila tumor suppressor protein Tid56, can interact with the human papillomavirus type 16 E7 oncoprotein. Virology. 1998;247:74–85. doi: 10.1006/viro.1998.9220. [DOI] [PubMed] [Google Scholar]
- 39.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slinskey A, Barnes D, Pipas J M. Simian virus 40 large T antigen J domain and Rb-binding motif are sufficient to block apoptosis induced by growth factor withdrawal in a neural stem cell line. J Virol. 1999;73:6791–6799. doi: 10.1128/jvi.73.8.6791-6799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sock E, Enderich J, Wegner M. The J domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol Cell Biol. 1999;19:2455–2464. doi: 10.1128/mcb.19.4.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Syken J, De-Medina T, Munger K. TID1, a human homolog of the Drosophila tumor suppressor 1(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci USA. 1999;96:8499–8504. doi: 10.1073/pnas.96.15.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 46.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 47.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate Rb family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J, Rice P W, Gorsch L, Abate M, Cole C N. Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J Virol. 1992;66:2780–2791. doi: 10.1128/jvi.66.5.2780-2791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]