To the Editor,
Measurement of allergen-specific IgE (sIgE), along with the patient’s clinical history and results of allergen provocation tests, are fundamental in diagnosing allergic disease1. The ImmunoCAP assay is used as a standard to quantify sIgE levels2. Although this method is sensitive (range, 0.1-100 kUA/L), it requires relatively large amount of plasma (which can be problematic when testing young children), and is also limited by its cost and the need for a specific instrument to analyze the test’s results. Therefore, further developments are needed to improve the sensitivity of sIgE detection methods, while markedly reducing the volume of sample required and the cost, without sacrificing assay robustness, reproducibility and accuracy.
Here, we report development of a highly sensitive method for quantifying sIgE levels using a luciferase-linked immunosorbent assay (LuLISA). The IgE LuLISA allows bioluminescent detection of sIgE using an anti-IgE nanobody (single variable heavy immunoglobulin domain [sdAb] or VHH) which recognizes the constant Cε3 region of human IgE3, and is expressed in tandem with the catalytic domain of the enzyme luciferase (nanoKAZ)4 (Figure 1A and Figure S1). The anti-IgE nanobody we used for this assay (sdAb026) has an affinity for IgE similar to that of the therapeutic anti-IgE antibody omalizumab (KD 1.4 nM vs. 2.6 nM, respectively3,5), and was reported to inhibit interactions between IgE and the two receptors FcεRI and CD233.
Figure 1. Sensitive and specific detection of allergen-specific IgE by LuLISA.
(A) Cartoon representation showing the anti-IgE nanobody-luciferase tandem (sdAb026-nanoKAZ) bound to the Fc portion of IgE (pink: nanoKAZ luciferase domain [PDB ID: 5B0U]3; blue: anti-IgE nanobody sdAb026; green: IgE Fc portion Cε3-4 domains [PDB ID: 5NQW])9. (B) Recombinant human anti-Der p 2 IgE, IgG1 and IgG4 were diluted in PBS at the indicated concentrations and incubated with plate-bound recombinant Der p 2. Bioluminescent detection of antibody levels was performed by LuLISA using the anti-IgE sdAb026-nanoKAZ. (C and D) Recombinant anti-OVA IgE (C) or plasma from a peanut allergic subject (D) were diluted in a pool of plasma from 30 healthy donors. Levels of OVA sIgE (C) or peanut sIgE (D) were assessed in aliquots from the same dilution sample using LuLISA or ImmunoCAP. All LuLISA data are from one experiment representative of three independent experiments. RLU: relative light unit.
To establish a proof-of-concept for the specific detection of sIgE using this method, we prepared dilution series in PBS of recombinant IgE, IgG1 (the major IgG subclass) or IgG4 (the main IgG subclass overproduced during allergen-specific immunotherapy) directed against the house dust mite allergen Der p 2 (Figure 1B). The 3 groups of samples were analyzed using IgE LuLISA. As expected, a concentration-dependent signal arose only for the sample containing anti-Der p 2 sIgE, with a detection limit of ~5x10−13 M sIgE (~1 pg/mL; ~0.0004 kUA/L) (Figure 1B). We also obtained high sensitivity with recombinant anti-ovalbumin (OVA) IgE, which was detectable by LuLISA at concentrations as low as 5 pg/mL (~0.002 kUA/L) (Figures S2 and S3). The sensitivity of LuLISA was also much higher than that of standard ELISA for the detection of sIgE with an extended dynamic range over 4 orders of magnitude instead of 2 (Figure S3).
Next, we compared the dynamic range and sensitivity of IgE LuLISA versus standard ImmunoCAP, using recombinant OVA sIgE diluted in plasma pooled from 30 healthy donors (Figure 1C). This head-to-head comparison revealed a markedly increased (≈250-fold) analytical sensitivity of LuLISA compared with ImmunoCAP (Figure 1C). We performed similar experiments with dilution series of a plasma sample from a highly peanut allergic subject, which was again diluted in a pool of plasma from 30 healthy donors (Figure 1D). ImmunoCAP allowed detection of peanut sIgE in plasma diluted up to 4,050 times, while peanut sIgE was still detected by LuLISA in allergic plasma diluted 100,000 to 300,000 times (Figure 1D). Dilution series of the anti-IgE nanobody-luciferase tandem gave a concentration-dependent signal at a fixed (1:50) dilution of this peanut allergic plasma sample, and confirmed the very low bioluminescent background signal of the IgE LuLISA (Figure S4).
Altogether, these results indicate that the IgE LuLISA has a very high sensitivity and specificity, and could thus potentially be used to quantify IgE in samples from patients with very low sIgE. However, the cut-off level commonly used in clinical practice to define IgE positivity is 0.35 kUA/L, which can be measured by ImmunoCAP and is much higher than the sensitivity of the IgE LuLISA. Thus, the main advantage of the IgE LuLISA over ImmunoCAP is that it requires extremely low volume of sample. In the case of the sample from the peanut-allergic patient used in Figure 1D, peanut sIgE could still be detected using less than 1 nanoliter of the initial patient’s sample. Thus, very large screens of sIgE against arrays of potential allergens can be envisioned using IgE LuLISA, even when patient’s sample sizes are limited, automatable in 96 and 384-well plates.
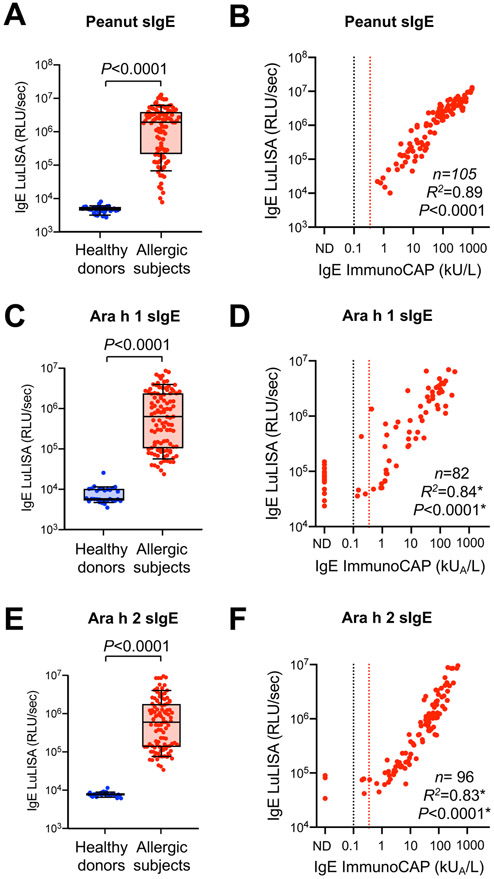
We then sought to further validate this approach by measuring sIgE against total peanut extract, or against the major peanut allergens Ara h 1 and Ara h 2 using 1 μL of plasma from 31 healthy donors (obtained from the French blood bank EFS with unknown allergic status) and 82-105 peanut-allergic subjects (collected upon their enrollment into the institutional review board–approved peanut oral immunotherapy study: safety, efficacy and discovery trial; ClinicalTrials.gov identifier: NCT02103270)6. Dilution series from reference samples with titrated high peanut sIgE were used for assay calibration, to ensure that all plasma samples were analyzed within the linear range of detection of our method (Figure S5). As expected, significantly higher levels of peanut sIgE, Ara h 1 sIgE and Ara h 2 sIgE were detected in plasma samples from peanut allergic subjects as compared to healthy donors (Figure 2A,C,E). Head-to-head comparison between LuLISA and ImmunoCAP in allergic patients showed a high correlation between both methods (R2=0.89, 0.84 and 0.83 for peanut sIgE, Ara h 1 sIgE and Ara h 2 sIgE, respectively) (Figure 2B,D,F). These correlations were calculated using all plasma samples for which sIgE levels were above the detection cut-off of ImmunoCAP (0.1 kUA/L). This was the case for all samples for peanut sIgE. However, 17 out of 82 samples (19.7%) for Ara h 1 sIgE and 3 out of 96 samples (3.1%) for Ara h 2 sIgE were below the detection limit of ImmunoCAP (Figure 2B,D,F). However, all these subjects had clear clinical reactivity to peanut, as assessed by performing double-blind, placebo-controlled food challenge (DBPCFC) and skin prick tests (Table S1). Altogether, these results demonstrate that the IgE LuLISA is highly sensitive and accurate for the clinical detection of sIgE, and requires very low volumes of plasma.
Figure 2. Detection of sIgE levels by LuLISA in 1 μL plasma samples from healthy donors and peanut allergic subjects.
(A, C, E) Levels of peanut sIgE (A), Ara h 1 sIgE (C) or Ara h 2 sIgE (E) by LuLISA in 1 μL plasma samples from 31 healthy donors and 82-105 peanut allergic subjects. Data in A, C and E are shown as box and whisker plots (10th and 90th percentiles), and each circle represents an individual patient. P values were calculated by nonparametric Mann-Whitney test (2-tailed). (B, D, F) Correlation between peanut sIgE (B), Ara h 1 sIgE (D) or Ara h 2 sIgE (F) by LuLISA vs. ImmunoCAP. Black dashed line indicates ImmunoCAP cut-off level (0.1 kUA/L); Red dashed line indicates cut-off level commonly used in clinical practice (0.35 kUA /L). RLU: relative light unit. *Pearson’s R2 correlation coefficients and P values (two-tailed) were calculated using all samples above the ImmunoCAP cut-off level (0.1 kUA/L).
Besides ImmunoCAP, several other methods have been reported for the detection of sIgE, including IMMULITE and, more recently, isotype-specific agglutination-PCR (ISAP)7,8. IMMULITE appears to be the closest method to LuLISA as it uses a chemiluminescent approach to detect sIgE. However, the reported detection limit for sIgE with IMMULITE is the same as for ImmunoCAP (0.1 kUA/L)7 Similarly to LuLISA, detection of sIgE by ISAP can be performed using 1 μl of clinical sample. Moreover, the two tests are based on different approaches as ISAP requires chemically-synthesized allergen-DNA (for each type of allergen) and secondary anti-IgE antibody-DNA conjugate for the detection of sIgE by quantitative PCR.
In summary, the IgE LuLISA is a new method for the detection of sIgE of ultra-high sensitivity requiring only very small (1 μL or less) plasma sample volumes. The use of bioluminescence offers markedly increased sensitivity and extended dynamic range over classical colorimetric (ELISA) or fluorescent (ImmunoCAP) IgE detection methods. The method is fully automatable and uses commercialized plates and a standard luminometer for the bioluminescent detection of IgE. Thus, IgE LuLISA should be very cost-effective over conventional ImmunoCAP. Further tests will be performed to extend the potential use of IgE LuLISA for multiplexed detection of sIgE against arrays of allergens.
Supplementary Material
Funding source
This work was supported by the Institut Pasteur initiative for valorizing the applications of research (ValoExpress 2016-2017, Innov-IARP Pasteur-Carnot 2019-2020 S.G., Y.L.J., P.P., T.R.) and NIH/NIAID U19AI104209 (S.J.G, R.S.C, S-C.L., and K.C.N). B.B. acknowledges support from the Pasteur - Paris University (PPU) International PhD program and a fellowship from the French “Fondation pour la Recherche Médicale FRM”. L.L. Reber acknowledges support from the INSERM and an ATIP-Avenir grant.
Footnotes
Conflicts of interest
The authors declare no competing interests.
References
- 1.Canonica GW, Ansotegui IJ, Pawankar R, et al. A WAO - ARIA - GA(2)LEN consensus document on molecular-based allergy diagnostics. World Allergy Organ J. 2013;6(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Hage M, Hamsten C, Valenta R. ImmunoCAP assays: Pros and cons in allergology. J Allergy Clin Immunol. 2017;140(4):974–977. [DOI] [PubMed] [Google Scholar]
- 3.Jabs F, Plum M, Laursen NS, et al. Trapping IgE in a closed conformation by mimicking CD23 binding prevents and disrupts FcepsilonRI interaction. Nat Commun. 2018;9(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inouye S, Sato J, Sahara-Miura Y, Yoshida S, Hosoya T. Luminescence enhancement of the catalytic 19 kDa protein (KAZ) of Oplophorus luciferase by three amino acid substitutions. Biochem Biophys Res Commun. 2014;445(1):157–162. [DOI] [PubMed] [Google Scholar]
- 5.Gasser P, Tarchevskaya SS, Guntern P, et al. The mechanistic and functional profile of the therapeutic anti-IgE antibody ligelizumab differs from omalizumab. Nat Commun. 2020;11(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukai K, Gaudenzio N, Gupta S, et al. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 hours before analysis. J Allergy Clin Immunol. 2017;139(3):889–899 e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton RG, Franklin Adkinson N Jr. In vitro assays for the diagnosis of IgE-mediated disorders. J Allergy Clin Immunol. 2004;114(2):213–225; quiz 226. [DOI] [PubMed] [Google Scholar]
- 8.Tsai CT, Mukai K, Robinson PV, et al. Isotype-specific agglutination-PCR (ISAP): A sensitive and multiplex method for measuring allergen-specific IgE. J Allergy Clin Immunol. 2018;141(5):1901–1904 e1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomabechi Y, Hosoya T, Ehara H, Sekine SI, Shirouzu M, Inouye S. Crystal structure of nanoKAZ: The mutated 19 kDa component of Oplophorus luciferase catalyzing the bioluminescent reaction with coelenterazine. Biochem Biophys Res Commun. 2016;470(1):88–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.