Abstract
Neurons in the mammalian suprachiasmatic nucleus (SCN) contain a cell-autonomous circadian clock that is based on a transcriptional-translational feedback loop. The basic helix-loop-helix–PAS proteins CLOCK and BMAL1 are positive regulators and drive the expression of the negative regulators CRY1 and CRY2, as well as PER1, PER2, and PER3. To assess the role of mouse PER3 (mPER3) in the circadian timing system, we generated mice with a targeted disruption of the mPer3 gene. Western blot analysis confirmed the absence of mPER3-immunoreactive proteins in mice homozygous for the targeted allele. mPer1, mPer2, mCry1, and Bmal1 RNA rhythms in the SCN did not differ between mPER3-deficient and wild-type mice. Rhythmic expression of mPer1 and mPer2 RNAs in skeletal muscle also did not differ between mPER3-deficient and wild-type mice. mPer3 transcripts were rhythmically expressed in the SCN and skeletal muscle of mice homozygous for the targeted allele, but the level of expression of the mutant transcript was lower than that in wild-type controls. Locomotor activity rhythms in mPER3-deficient mice were grossly normal, but the circadian cycle length was significantly (0.5 h) shorter than that in controls. The results demonstrate that mPer3 is not necessary for circadian rhythms in mice.
A circadian clock in the mammalian suprachiasmatic nucleus (SCN) regulates rhythms in physiology and behavior (for reviews, see references 11 and 34). SCN neurons contain a cell-autonomous circadian clock (35). The SCN clock is based on a transcriptional-translational feedback loop in which the protein products of a set of genes feed back to inhibit their own transcription. The molecular components of the circadian clock mechanism appear to be well conserved within metozoan species, although the specific roles of the clock components differ between flies and mammals (4, 21).
In mammals, the basic helix-loop-helix (bHLH)–PAS proteins CLOCK and BMAL1 form heterodimers that drive the expression of responsive genes through E-box enhancer elements (7). The responsive genes include putative negative regulators as well as output genes (7, 9, 22). The mouse cryptochrome genes mCry1 and mCry2 play a critical role as negative regulators (12). Mice with targeted disruptions of both mCry1 and mCry2 are arrhythmic, while mice homozygous for targeted disruption of either of the two genes have an altered circadian cycle length (period) under constant conditions (29, 30, 32). The levels of expression of the mouse Period (mPer) genes are elevated in mCry1 mCry2 double-knockout mice, consistent with a defect in negative feedback (20, 32). The mCRYs facilitate the translocation of mPER proteins to the nucleus (12), but in vitro studies have demonstrated that the ability of mCRY proteins to inhibit transcription is independent of interactions with mPER proteins (26).
Understanding of the roles of the three mPer genes (mPer1, mPer2, and mPer3) in the mammalian clockwork continues to evolve. The structural homology of the mPER proteins to the Drosophila period gene product, which is an essential negative element in the fly feedback loop (4), led to the expectation that the mPER proteins would function as negative elements in the mammalian feedback loop. Indeed, each of the mPER proteins can inhibit CLOCK-BMAL1-mediated transcription in vitro (9, 12). The expression of the three mPer genes in the SCN is rhythmic, and the rhythms are temporally coordinated. Similarly, at the protein level, the rhythmic abundance of mPER1- and mPER2-containing nuclei in the SCN is virtually identical to and coincident with the rhythms of mCRY1 and mCRY2 (5, 8, 12). The timing of the peak levels of these proteins is synchronous with the decline in their mRNAs, suggestive of an autoregulatory mechanism. There is no direct evidence to indicate an important role of the mPER proteins in negative feedback within the clock loop, however.
Recent evidence suggests that the mPer genes have functionally distinct roles separate from their proposed role in negative feedback. A mutation of the mPer2 gene resulting in a deletion of 87 residues from the protein dimerization PAS domain (mPer2Brdm1) leads to circadian instability and delayed loss of rhythmicity in mice (37), indicating the importance of mPER2 in clock function. The primary role of mPER2 appears to be as a positive regulator of Bmal1 gene expression, rather than as a negative regulator of CLOCK-BMAL1-mediated transcription (26). mPer1 is thought to play a unique role in synchronizing the circadian clock to environmental stimuli (1, 2, 5, 25, 27, 28, 38). While these studies suggest that mPer1 and mPer2 have distinct roles in the mammalian clockwork, they provide little indication of a specific functional role of mPer3. To address this issue, we examined clock gene expression and behavioral rhythms in mice with a targeted disruption of the mPer3 gene.
MATERIALS AND METHODS
Generation of an mPer3 targeting construct.
Genomic DNA clones were isolated from a 129/sv mouse library (Stratagene) using a probe generated from a portion of the mPer3 cDNA corresponding to nucleotides (nt) 187 to 1139 of the sequence assigned GenBank accession number AF050182. The targeting construct was generated by inserting a 1.8-kb neomycin resistance cassette (PGK-NEO) in reverse orientation in place of a 1.6-kb EcoRI fragment of the genomic clone (Fig. 1A). The excised EcoRI fragment of the genomic clone began in intron 2, approximately 150 bp 5′ of exon 3, and contained exon 3, intron 3, and the 5′ portion of exon 4. The excised regions encode amino acid residues 92 to 154 of mPER3. The PGK-NEO cassette contains the phosphoglycerate kinase (PGK) promoter driving the expression of the neomycin (NEO) resistance gene and was provided by En Li (Massachusetts General Hospital, Boston).
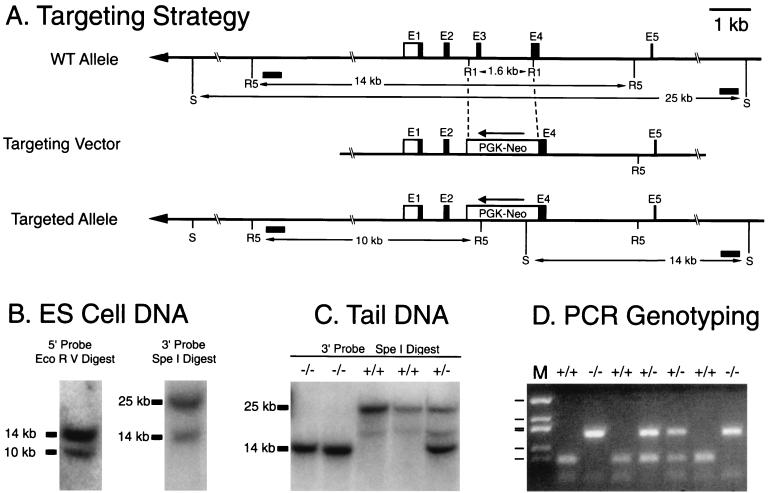
FIG. 1.
mPer3 targeting construct and genotyping strategies. (A) Schematic representation of the mPer3 gene, the targeting construct, and the targeted allele. A 1.6-kb portion of the mPer3 gene was excised with EcoRI (R1), and a PGK-NEO cassette was inserted in the reverse orientation. The 5′ and 3′ arms of the targeting construct were ca. 6 and 9 kb, respectively. Restriction sites introduced by the PGK-NEO cassette and used for Southern blot analysis were EcoRV (R5) and SpeI (S). Intron and exon structure is shown only for the first five exons (E1 to E5). Boxes represent exons, and the lines between the boxes indicate introns. Open boxes represent untranslated regions. Filled boxes represent putative coding regions based on translation from the first ATG with a Kozak consensus sequence at nt 358 of AF050182. (A second Kozak consensus sequence present in exon 2 could begin translation at nt 523). Horizontal filled boxes indicate the locations of probes used for Southern blotting. (B) Southern blot identification of the targeting event in ES cells. Representative autoradiograms are shown. The upper band in each case represents the wild-type allele, and the lower band represents the targeted allele (see panel A). (C) Southern blot analysis of DNA extracted from mouse tails. Genotypes are shown above lanes. (D) PCR genotyping of DNA extracted from mouse tails. Genotypes are shown above lanes. M, size marker (pUC19 DdeI with bands at 910, 540, 426, 409, 266, and 166 bp).
Generation of mPER3-deficient mice.
The targeting construct was linearized and then introduced into J1 embryonic stem (ES) cells by electroporation. Genomic DNA extracted from neomycin-resistant ES cell lines was digested with EcoRV, subjected to agarose gel electrophoresis, and hybridized with a 0.5-kb probe from the 5′ region flanking the targeting cassette. Four positive clones were identified out of 196 screened. Homologous recombination in these four positive clones was confirmed by digestion with SpeI and hybridization with a probe from the 3′-flanking region (Fig. 1). Two of the four positive ES cell lines (no. 12 and 135) were used for microinjection to generate chimeric founders.
Chimeric males were bred to females of the C57BL/6, C3H/HeJ, and isogenic (129/sv) strains. C57BL/6 and C3H/HeJ mice were purchased from Charles River Laboratories. Isogenic females were provided by En Li. For studies of gene expression, F1 heterozygotes generated from crosses to the C57BL/6 strain were crossed to produce F2 mice homozygous for the targeted allele (mPER3 deficient, or mPer3−/−) or homozygous for the wild-type allele (+/+). Behavioral studies were conducted with male mice from line 12 in a C57BL/6 background, from line 135 in a C3H background, and from both line 12 and line 135 in an isogenic (129/sv) background.
Genotyping.
Genotypes were determined by PCR and/or Southern blot analysis of tail biopsy DNA (Fig. 1C and D). Probes for Southern blots were the 5′- and 3′-flanking probes used to identify the ES cell lines (see above) radiolabeled with [32P]dCTP (New England Nuclear [NEN]) by use of random hexamer primers (Oligolabeling kit; Boehringer Mannheim Biochemicals).
The PCR method was done with a cocktail of three primers, a forward primer in intron 3 (3-43; 5′ TCTGTGAGTTCTTCCGTGTCTGTT) (present only in the wild-type [WT] allele), a primer located in the NEO cassette (Neo6-2; 5′ TGCCCCAAAGGCCTACCCGCTTCC), and a common reverse primer in exon 4 (3-41; 5′ GTCTTGAGGGGCAAGCAGGTCGAC). The presence of the WT allele led to the amplification of a ca. 200-bp band from primers 3-43 and 3-41, while the presence of the targeted allele was detected by amplification of a ca. 400-bp band with primers Neo6-2 and 3-41 (Fig. 1D). PCR was performed with Taq DNA polymerase (Fisher) in buffer H (Epicentre Technologies). The PCR protocol consisted of 3 min at 95°C, 30 cycles of amplification (each consisting of 30 s at 94°C, 30 s at 60°C, and 90 s at 72°C), and a final extension phase (10 min at 72°C). Products were separated on 1.5% agarose gels and viewed by UV transillumination with ethidium bromide.
Western blots.
Mice used for analysis of proteins were housed in a light-dark cycle consisting of 12 h of light and 12 h of dark (12L:12D). Mice were euthanized by inhalation of carbon dioxide 30 min before lights were turned off. Tissues were removed, dissected, and frozen on dry ice. Tissue pools consisted of anterior hypothalamus containing SCN, whole hypothalamus, and brain (whole brain minus hypothalamus and cerebellum).
Tissue extracts were prepared as described previously (14). Briefly, tissues were homogenized at 4°C in 3 volumes of buffer 1 (0.4 M NaCl, 20 mM HEPES [pH 7.5], 1 mM EDTA, 5 mM NaF, 1 mM dithiothreitol, 0.3% Triton X-100, 5% glycerol, 0.25 mM phenylmethylsulfonyl fluoride [PMSF], 10 mg of aprotinin per ml, 5 mg of leupeptin per ml, 1 mg of pepstatin A per ml). Homogenates were cleared by centrifugation (twice, 12 min each, 12,000 × g). Supernatants were mixed with 2× sample buffer (14) and boiled. Proteins were separated by electrophoresis through sodium dodecyl sulfate (SDS)–6% polyacrylamide gels and then transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20 and then incubated with affinity-purified antisera to mPER3, mCRY1, or mCRY2 (Alpha Diagnostics International). Immunoreactive bands were visualized using anti-rabbit immunoglobulin G secondary antisera and enhanced chemiluminescence detection (NEN). Full-length mPer3 ligated into the pcDNA3.1-HA vector (12) was translated in vitro using a TnT T7 Quick kit (Promega), and approximately 0.03 fmol was loaded as a positive control.
Immunoprecipitation.
Frozen brain tissue was collected and homogenized as described above. After the second centrifugation step, supernatants were diluted with 2 volumes of buffer 2 (identical in composition to buffer 1 except that Triton X-100 was reduced to 0.05% and buffer 2 contains no NaCl). Extracts (500 mg of total protein) were incubated with 20 μl of protein G-Sepharose 4 Fastflow beads (Pharmacia) for 30 min at 4°C and then centrifuged. A slurry of antibody (1 μl of anti-PER3, 3 μl of anti-CRY1, or 3 μl of anti-CRY2) plus protein G-Sepharose 4 Fastflow beads was added to the clarified supernatants. After being mixed by gentle rotation for 2 h at 4°C, the beads were collected by centrifugation. Immune complexes were washed three times, mixed with sample buffer, boiled, and centrifuged. The final immune complexes in the supernatants were analyzed by Western blotting to detect mPER3 as described above. Blots were also analyzed for mCRY1 and mCRY2 to rule out the possibility that failure to immunoprecipitate mPER3 in mice homozygous for the targeted allele was due to an altered abundance of mCRY proteins; comparable amounts of mCRY1 and mCRY2 were immunoprecipitated in +/+ and mPer3−/− mice (data not shown).
Analysis of gene expression.
Mice used for analysis of gene expression were housed in 12L:12D for at least 10 days prior to analysis. The lighting timer was disabled on the day of study so that the animals remained in constant darkness (DD) on the day of tissue collection. Mice were euthanatized by carbon dioxide inhalation with the aid of dim-red illumination; tissues then were rapidly dissected and frozen (−80°C).
Gene expression in the SCN was examined by in situ hybridization using methods described previously (33). Coronal, 15-μm sections through the SCN were hybridized overnight with 35S-labeled cRNA probes generated by in vitro transcription (Promega). The templates for probe generation were PCR-generated fragments of cDNAs subcloned into the TA vector (Invitrogen). Probes were mPer1 (nt 340 to 761 of the sequence assigned GenBank accession number AF022992), mPer2 (nt 9 to 489, accession number AF035830), mPer3 (nt 108 to 622, accession number AF050182), mCry1 (nt 1081 to 1793, accession number AB000777), and Bmal1 (nt 864 to 1362, accession number AF015203).
Northern blot analysis was performed as previously described (38). RNA was extracted from skeletal muscle samples using Ultraspec reagent (Biotecx). Poly(A)+ RNA was isolated using oligo(dT) (Qiagen). RNA samples [1 μg of poly(A)+ RNA per lane] were separated by electrophoresis through agarose-formaldehyde gels and blotted to GeneScreen (NEN). Probes for Northern blots were labeled by the method of random priming (Oligolabeling kit) with [32P]dCTP (NEN). Templates were mPer3 5′ (nt 108 to 622, AF050182), mPer3 deletion-specific probe (nt 629 to 824, AF050182), mPer3 3′ (nt 1637 to 2223, AF050182), NEO (690-bp PstI fragment), mPer1 (nt 340 to 761, AF022992), and mPer2 (nt 1572 to 2338, AF035830). To verify equal loading across lanes, blots were stripped and reprobed with a probe generated from human β-actin cDNA (Clontech).
RT-PCR.
Reverse transcription (RT)-PCR was performed to define the mPer3 transcripts present in mPER3-deficient mice. RNA was extracted from whole brain of WT mice and mice homozygous for the targeting construct (Ultraspec RNA isolation; Biotecx). RT with random hexamer primers was performed using an RT-PCR kit with a slight modification of the manufacturer's instructions (Perkin-Elmer Cetus). PCR was performed using Taq DNA polymerase and a PCR protocol consisting of 3 min at 95°C, 30 cycles of amplification (each consisting of 30 s at 94°C, 30 s at 60°C, and 90 s at 72°C), and a final extension phase (10 min at 72°C).
Primers for RT-PCR were as follows: exon 1 forward, 5′-GCAAACTGAGAGAAGCAGGCTGAG-3′; exon 2 forward, 5′-ACTAATCCCAGTACCCTAGATGCTCT-3′; Neo3, 5′-CGAGCTCATTCCTCCACTCATGATCTA-3′; Neo32147, 5′-CTGTCATCTCACCTTGCTCCTGCC-3′; Neo6-2, 5′-TGCCCCAAAGGCCTACCCGCTTCC-3′; exon 4 reverse, 5′-TTGGGTCCAGTTGTTCCAGAAAGG-3′; exon 4 reverse, 5′-GTCTTGAGGGGCAAGCAGGTCGAC-3′; and exon 5 reverse, 5′-TTCGCTGGTGCACATTCATACTGCG-3′.
PCR products were subcloned (TA vector), and nucleotide sequences were determined manually using the Sanger method (Sequenase; U.S. Biochemicals) or an ABI automated sequencer (Department of Molecular Biology, Massachusetts General Hospital).
Assessment of behavioral rhythms.
Mice used for behavioral analysis were housed individually within light-tight, ventilated environmental compartments in a temperature- and humidity-controlled facility. After 2 to 14 days of being monitored in 12L:12D, mice entered DD starting at the time when lights were normally turned off (2100 Eastern Standard Time). In most studies, dim-red light from fluorescent bulbs was present within each compartment at all times, including those designated as representing darkness. In study 5, the dim-red bulbs also were disabled.
For monitoring locomotor activity (spontaneous wheel running), male mice were housed individually in cages equipped with running wheels. Magnetic reed switches mounted near each running wheel detected the movement of a magnet mounted on the wheel. Switch closures were detected by a computer-based system (DataCol3; MiniMitter Corp.) and saved to a disk at 5-min intervals. Data were plotted in actogram format using the software within the DataCol package and were double plotted by photocopying. The free-running period was estimated without knowledge of the genotype of the animals. The period was determined from the slope of a hand-drawn line through activity onsets of each animal housed in DD. The first 5 days after discontinuation of the lighting cycle were excluded from the analysis to allow stabilization to constant conditions.
RESULTS
Generation of mice with a targeted disruption of mPer3.
The targeting construct resulted in the replacement of a 1.6-kb EcoRI genomic DNA fragment with a 1.8-kb neomycin resistance cassette (Fig. 1A). The excised EcoRI fragment includes exon 3 and the 5′ portion of exon 4. Insertion of the PGK-NEO cassette introduces stop codons in all three reading frames. The linearized construct was introduced into ES cells by electroporation. Two ES cell lines that gave the expected pattern of bands on Southern blot analysis with both 5′- and 3′-flanking probes (Fig. 1B) were selected for microinjection. Two independent lines of mice (no. 12 and 135) were generated. Genotypes of the offspring were determined by Southern blotting or PCR (Fig. 1C and D). Of the first 300 F2 offspring generated from crossing F1 heterozygotes of line 12, the distribution of genotypes was 74 homozygous wild-type mice, 149 heterozygotes, and 77 mice homozygous for the targeted allele, consistent with the expected Mendelian ratio of 1:2:1. mPER3 is not necessary for viability or fertility.
mPER3 protein is absent in mice homozygous for the targeted allele.
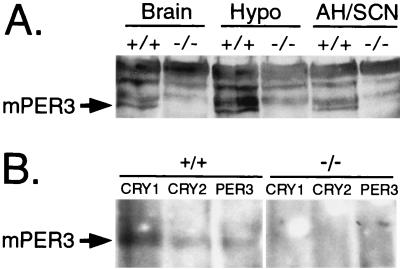
Western blot analysis confirmed the absence of mPER3 protein in mice with targeted disruption of the mPer3 gene (Fig. 2A). Proteins from brain tissue were extracted and separated by SDS-polyacrylamide gel electrophoresis, and the blots were incubated with an antiserum directed against the carboxy terminus of mPER3. The antiserum revealed several nonspecific bands that did not correspond in size to in vitro-translated mPER3 and that were present in samples from mice of either genotype. In vitro-translated mPER3 was apparent as a doublet at approximately 140 kDa. mPER3 immunoreactivity from brain extracts migrated as a single band, with a mobility similar to that of the in vitro-translated protein. mPER3 immunoreactivity was absent from mice homozygous for the targeted allele (Fig. 2A).
FIG. 2.
mPER3 protein is absent in mice with targeted disruption of the mPer3 gene. (A) Western blot incubated with antisera to mPER3 showing immunoreactive proteins from wild-type (+/+) and mPER3-deficient (−/−) mice. Tissues examined were whole brain minus cerebellum and hypothalamus (Brain), whole hypothalamus (Hypo), and anterior hypothalamus containing SCN (AH/SCN). The specific mPER3 band corresponds in mobility to in vitro-translated mPER3. Results shown are representative of three experiments. (B) Immunoprecipitation. Brain proteins extracted from +/+ or −/− mice were precipitated with antiserum to mCRY1, mCRY2, or mPER3 and separated by SDS-polyacrylamide gel electrophoresis, and the blot was incubated with antiserum to mPER3. The mPER3 band indicated by the arrow corresponds in mobility to in vitro-translated mPER3. Similar results were found in a replicate experiment.
Immunoprecipitation experiments confirmed the loss of mPER3 protein in mice homozygous for the targeted allele (Fig. 2B). Incubation of tissue extracts from wild-type mice with antiserum to mPER3, mCRY1, or mCRY2 led to precipitation of an mPER3-immunoreactive band of the expected size. This mPER3-immunoreactive band was absent from extracts from mice with targeted disruption of the mPer3 gene.
mPer3 transcripts are expressed in mPER3-deficient mice.
Preliminary experiments done to examine the rhythmicity of mPer3 gene expression in whole brain and skeletal muscle of mPER3-deficient mice revealed that mPer3 transcripts were present and rhythmically expressed in mice homozygous for the targeted allele. Northern blot analysis and RT-PCR were used to define the transcripts expressed from the targeted locus.
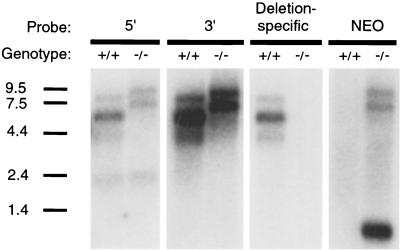
Wild-type mice expressed major mPer3 transcripts at ca. 6 and 7.5 kb (38) (Fig. 3A). In mPER3-deficient mice, the major transcripts were ca. 2 kb larger (Fig. 3A). This pattern is consistent with inclusion of the inserted PGK-NEO cassette in transcripts arising from the mPer3-targeted locus. Indeed, probes directed to regions of the mPer3 cDNA on the 5′ and 3′ sides of the PGK-NEO cassette gave a pattern of transcript sizes that was indistinguishable from the pattern seen when a probe directed to the PGK-NEO cassette itself was used (Fig. 3A); all three regions appeared to be present in the major transcripts of mPER3-deficient mice. A probe specific for exon 3 and the portion of exon 4 deleted in the targeted allele did not detect a transcript in mPER3-deficient mice, further confirming the targeting event. An intensely hybridizing 0.8-kb NEO transcript driven from the PGK promoter was noted in mPER3-deficient mice.
FIG. 3.
Northern blot analysis done to identify transcripts arising from the targeted mPer3 allele. Skeletal muscle RNA was hybridized with probes directed to the mPer3 cDNA sequence 5′ or 3′ of the PGK-NEO cassette, to the NEO cassette, and to the portion of mPer3 cDNA deleted by the targeting construct. RNA size standards (in kilobases) are shown on the left.
From the Northern blot analysis, it appeared that intronic splicing sites and/or splice sites within the PGK-NEO cassette were used to generate the transcripts found in the mPER3-deficient mice. RT-PCR with forward primers located in exon 1 or 2 and reverse primers located in the PGK-NEO cassette led to the detection of three transcript types. The first two transcript types resulted from splicing of exon 2 to cryptic splice sites in intron 2 located 39 and 36 bp upstream of the EcoRI site at which the PGK-NEO cassette began. The third type of transcript joined exon 2 directly to the PGK-NEO cassette, skipping over intron 2 and the first ca. 450 bp of the PGK-NEO cassette. Analysis of transcripts using primers located in the PGK-NEO cassette and reverse primers in exon 4 or 5 revealed that all transcripts detected consisted of the expected fusion of the PGK-NEO cassette with exon 4. No alternative transcripts were detected at this junction. Conceptual translation of the transcripts obtained revealed the presence of stop codons in all three frames in the portion of the PGK-NEO cassette included in transcripts arising from the targeted allele.
We cannot exclude the possibility that an amino-terminal fragment of mPER3 is generated from the mutant transcripts. Such a product would contain only the first 91 residues of mPER3 (of 1,113) fused to a sequence derived from translation of intronic or PGK-NEO cassette sequences. No portion of the protein dimerization PAS domain would be present.
Rhythmic gene expression in mPER3-deficient mice.
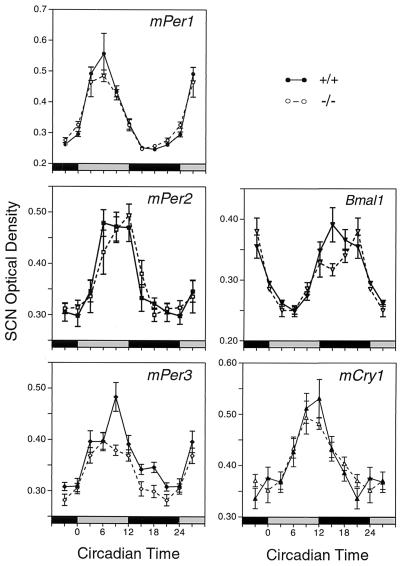
In situ hybridization was used to examine the expression of putative clock genes in the SCN. Mice were entrained to 12L:12D, and then tissue samples were collected at eight times on the first day in DD. The mPer1, mPer2, Bmal1, and mCry1 rhythms were unaffected in mPer3-deficient mice (Fig. 4). For each of these four genes, two-way analysis of variance revealed a significant effect of time (P < 0.0001) but no significant effect of genotype and no significant interaction of genotype and time (P > 0.05). The mPer3 rhythm was altered in mice with targeted disruption of the mPer3 gene, however. Two-way analysis of variance revealed significant effects of time and genotype (P = 0.0001) and a significant interaction (P = 0.025). The peak levels of mPer3 expression, at circadian time 9, were significantly lower in the mPER3-deficient mice than in +/+ controls (t test, P < 0.01). These studies demonstrate a minor molecular phenotype in the central circadian clock.
FIG. 4.
Rhythmic gene expression in the SCN, as assessed by in situ hybridization. Significant rhythms of expression of mPer1, mPer2, mCry1, and Bmal1 were detected in both wild-type and mPER3-deficient mice, and there were no differences between the genotypes. mPer3 RNA levels were significantly reduced in the SCN of mPer3-deficient mice. Each value represents the mean ± standard error of the mean for six to eight mice. Adjacent sections from the same group of mice were used for all probes. Tissues were collected on the first day in DD. The horizontal bar at the bottom of each panel represents the lighting cycle that the animals experienced prior to entry into DD: subjective day is gray, and subjective night is black. (Circadian times 0 and 12 represent the times at which the lights would have been turned on and off had the animals remained in LD.) Data from circadian times 3, 21, and 24 are double plotted.
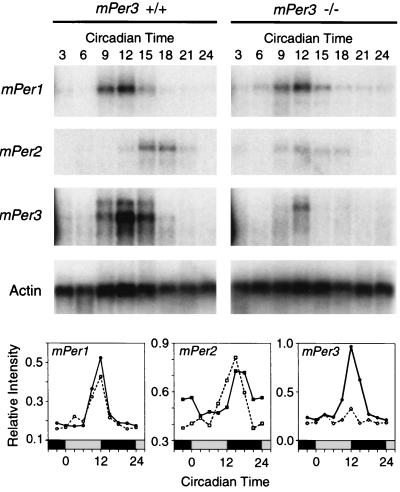
Northern blot analysis was used to examine the temporal pattern of expression of mPer genes in skeletal muscle. The expression of mPer1, mPer2, and mPer3 was rhythmic in both wild-type and mPER3-deficient mice (Fig. 5). Peak levels of mPer3 RNA were consistently lower in mPER3-deficient mice than in wild-type mice. Rhythms of mPer1 in the same RNA samples did not differ between genotypes. The level of expression of mPer2 was variable across blots; on one blot, the phase of the mPer2 rhythm was advanced (Fig. 5). Overall, however, there was no consistent change in the mPer2 RNA rhythm in mPER3-deficient mice.
FIG. 5.
Rhythmic gene expression in skeletal muscle. Autoradiograms from Northern blots illustrate rhythmic expression of mPer1, mPer2, and mPer3 in −/− and +/+ mice on the first day in DD. Hybridization of the actin control probe verified equal loading of the lanes (bottom autoradiogram). Graphs illustrate temporal profiles of mPer gene expression in skeletal muscle. Values are expressed relative to those for actin, and each value represents the mean of two blots. See the legend to Fig. 4 for plotting conventions.
Circadian behavior in mPER3-deficient mice.
Preliminary studies were conducted on small groups of mPER3-deficient and control mice in three different genetic backgrounds (Table 1). In all cases, there was evidence of entrainment to the light-dark cycle as well as long-term persistence of circadian rhythms under constant conditions. Upon release into DD, animals of both genotypes showed free running rhythms, with initial activity onsets occurring close to the time when the lights were turned off at the end of the previous lighting cycle. Several mice were exposed to a 30-min light pulse at circadian time 14, 2 h after activity onset. This procedure produced the expected (23) phase delay of the locomotor activity rhythm in mice of both genotypes. Gross aspects of circadian rhythmicity were thus normal in mPER3-deficient mice.
TABLE 1.
Period length of locomotor activity rhythms in DD
| Study | Line | Background | Genotypea | No. of mice tested | Period (h), mean ± SEM | Pb |
|---|---|---|---|---|---|---|
| 1 | 12 | C57BL/6 F2 | +/+ | 2 | 23.24 ± 0.38 | |
| −/− | 4 | 22.35 ± 0.32 | NS | |||
| 2 | 135 | C3H/He F2 | +/+ | 1 | 23.60 | |
| −/− | 3 | 23.32 ± 0.12 | NS | |||
| 3 | 135 | 129/sv | +/+ | 6 | 23.66 ± 0.10 | |
| −/− | 6 | 23.22 ± 0.13 | <0.021 | |||
| 4 | 135 | C3H/He F2 | +/+ | 3 | 23.77 ± 0.06 | |
| −/− | 5 | 23.54 ± 0.21 | NS | |||
| 5 | 12 | 129/sv | +/+ | 12 | 23.77 ± 0.09 | |
| −/− | 12 | 23.27 ± 0.18 | <0.025 |
+/+, wild type; −/−, mPER3 deficient.
NS, not significant.
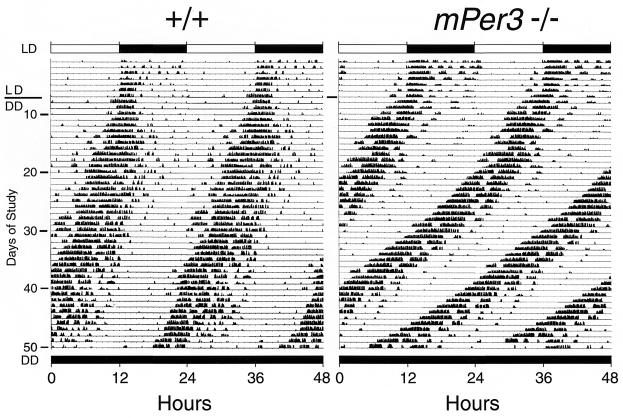
Analysis of the free-running period of mice housed in DD indicated a consistent trend toward a shorter circadian period among the mPER3-deficient mice, but the sample sizes were insufficient to detect a difference of this magnitude in three of the four studies (Table 1, studies 1 to 4). A larger group of isogenic male mice was examined in study 5 (Fig. 6). Wild-type mice in this study showed free running rhythms in DD with an average period of 23.77 ± 0.09 h (mean ± standard error of the mean). mPER3-deficient mice had a significantly shorter free-running period (23.27 ± 0.18 h; P < 0.025). A subset of mPER3-deficient mice showed slight, progressive shortening of the period length with extended time in DD (Fig. 6).
FIG. 6.
Representative actograms illustrating rhythmic wheel-running behavior in a wild-type mouse (left) and an mPER3-deficient mouse (right). Each horizontal line represents a 48-h period; the second 24-h period is double plotted, appearing both to the right of and below the first 24-h period. Vertical deflections represent periods of wheel running. Animals were maintained in 12L:12D for the first 8 days of the record and then entered DD. The timing of the lighting cycle is indicated by the horizontal bar at the top. Records shown are from isogenic male mice from study 5 (Table 1).
DISCUSSION
Mice with a targeted disruption of mPer3 have a subtle circadian phenotype, consisting of a shortening of the circadian period in DD. The only molecular phenotype detected was a reduction in the peak levels of mutant mPer3 transcripts in SCN and skeletal muscle. The mutant mPer3 transcript may be less stable than the transcript expressed from the endogenous mPer3 gene. No major up- or down-regulation of other clock components was noted for either tissue, suggesting that the lack of a robust phenotype is not due to molecular compensation involving up-regulation of a related gene with overlapping function. These results indicate that mPER3 is not essential for a functioning circadian clock. Thus, mPER3-mPER1 interactions that influence the subcellular distribution of these proteins in vitro (12, 36) are not critical for circadian function.
The present results are in striking contrast to the effects of targeted disruption of mPer2. A mutation of mPer2 that leads to deletion of a portion of the PAS domain leads to severely altered rhythms and a reduction in the levels of mPer1 and mPer2 RNA transcripts in the SCN and peripheral tissues (37). If this mutation is a loss-of-function mutation, then the phenotype suggests that mPer2 plays a necessary role in the maintenance of circadian rhythms that cannot be fulfilled by other members of the mPer gene family (37). Recent evidence indeed suggests that mPer2 is a critical component of an interlocking, positive loop within the circadian system (26) and that neither mPER2 nor mPER3 is critical for negative feedback in vivo.
What, then, is the role of mPER3 that accounts for the subtle alteration in circadian period in mPer3-deficient mice? The most likely explanation is that mPER3 plays a role in buffering protein-protein interactions among essential clock proteins within SCN neurons. mPER proteins interact with each other and with mCRY proteins (12, 26, 39). This capacity of mPER3 to interact with both positive and negative components of the interlocking circadian loops may provide fine-tuning of the cycle length. The phosphorylation state of the mPER proteins also may play an important regulatory role by altering the affinity for dimerization partners in a partner-specific manner (10, 17, 31).
In addition to this proposed buffering role, mPER3 may also function as an “output” protein, transducing the oscillation of the central circadian clock to effector genes. In this role, mPER3 would not be involved in the core molecular feedback loop. An example of a mechanism by which mPER3 could influence behavioral rhythms is through regulating the levels of transcription factors or cofactors outside of the core oscillator. Recent studies on the albumin D-element binding protein (DBP) provide a precedent. DBP is rhythmically expressed in the SCN and is transcriptionally regulated by CLOCK-BMAL1 (16, 19, 22). Targeted disruption of the DBP gene leads to a behavioral phenotype that is almost identical to the phenotype of mice with targeted disruption of the mPer3 gene, e.g., the circadian period in DBP−/− mice is 0.5 h shorter than that in isogenic control mice (16). One important distinction between mPER3 and DBP, however, is that DBP has a recognized role as a transcriptional regulator (6, 13), while mPER3 does not contain domains known to regulate transcription (38). PAS-mediated interactions with transcription factors of the bHLH-PAS family seem the most likely mechanism by which mPER3 could influence gene expression (3, 15, 18). Several members of the bHLH-PAS family are expressed in the SCN (24).
In conclusion, the subtle circadian phenotype of mPER3-deficient mice further supports the specialization of function among the three Period genes in mammals and indicates that mPER3 is not a necessary component of the mouse circadian clock.
ACKNOWLEDGMENTS
L.P.S. and X.J. contributed equally to this work.
We thank Camala Capodice, Kurtis Gray, Kathryn Picanso, and Aditi Chavda for technical assistance and En Li and Hong Lei for generation of the chimeric founder mice.
This work was supported by NIH grants HD14227 and NS39303 to S.M.R. L.P.S. was supported in part by a fellowship from the Program for Training in Sleep, Circadian and Respiratory Neurobiology at Harvard Medical School (NHLBI HL07901). X.J. was supported in part by NRSA grant MH12067.
REFERENCES
- 1.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 3.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 4.Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 5.Field M D, Maywood E S, O'Brien J A, Weaver D R, Reppert S M, Hastings M H. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 6.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 7.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 8.Hastings M H, Field M D, Maywood E S, Weaver D R, Reppert S M. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:1–7. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin X, Shearman L P, Weaver D R, Zylka M J, de Vries G J, Reppert S M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 10.Keesler G A, Camacho F, Guo Y, Virshup D, Mondadori C, Yau Z. Phosphorylation and destabilization of human period 1 clock protein by human casein kinase 1ɛ. NeuroReport. 2000;11:951–955. doi: 10.1097/00001756-200004070-00011. [DOI] [PubMed] [Google Scholar]
- 11.Klein D C, Reppert S M, Moore R Y, editors. Suprachiasmatic nucleus: the mind's clock. New York, N.Y: Oxford University Press; 1991. [Google Scholar]
- 12.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X, Maywood E S, Hastings M H, Reppert S M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 13.Lavery D J, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C. Circadian expression of the steroid 15 alpha-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 15.Lindebro M C, Poellinger L, Whitelaw M L. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowrey P L, Shimomura Z, Antoch M P, Yamazaki S, Zemenideds P D, Ralph M R, Menaker M, Takahashi J S. Positional syntenic cloning and functional characterization of a mammalian circadian mutation tau. Science. 2000;288:483–491. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moffett P, Reece M, Pelletier J. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol Cell Biol. 1997;17:4933–4947. doi: 10.1128/mcb.17.9.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oishi K, Fukui H, Ishida N. Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem Biophys Res Commun. 2000;268:164–171. doi: 10.1006/bbrc.1999.2054. [DOI] [PubMed] [Google Scholar]
- 20.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasia A, Muijtjens M, Hoeijmakers J H J, van der Horst G T J. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 21.Reppert S M. A clockwork explosion! Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 22.Ripperger J A, Shearman L P, Reppert S M, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz W J, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shearman L P, Zylka M J, Reppert S M, Weaver D R. Expression of basic helix-loop-helix-PAS genes in the mouse suprachiasmatic nucleus. Neuroscience. 1999;89:387–397. doi: 10.1016/s0306-4522(98)00325-x. [DOI] [PubMed] [Google Scholar]
- 25.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Two period homologues: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 26.Shearman L P, Sriram S, Weaver D R, Maywood E S, Chaves I, Zheng B, Kume K, Lee C C, van der Horst G T J, Hastings M H, Reppert S M. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 27.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J L, Dunlap J C, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 28.Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, Yagita K, Yan L, Young M W, Okamura H. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thresher R J, Vitaterna M H, Miyamoto Y, Kazantsev A, Hsu D S, Petit C, Selby C P, Dawut L, Smithies O, Takahashi J S, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 30.van der Horst G T J, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker A P, van Leenen D, Buijs R, Bootsma D, Hoeijmakers J H, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 31.Vielhaber E, Eidi E, Rivers A, Gao Z-H, Virshup D M. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase 1 ɛ. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitaterna M H, Selby C P, Todo T, Niwa H, Thompson C, Fruechte E M, Hitomi K, Thresher R J, Ishikawa T, Miyazaki J, Takahashi J S, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver D R. A2a adenosine receptor gene expression in developing rat brain. Mol Brain Res. 1993;20:313–327. doi: 10.1016/0169-328x(93)90058-w. [DOI] [PubMed] [Google Scholar]
- 34.Weaver D R. The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms. 1998;13:100–112. doi: 10.1177/074873098128999952. [DOI] [PubMed] [Google Scholar]
- 35.Welsh D K, Logothetis D E, Meister M, Reppert S M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 36.Yagita K, Yamaguchi S, Tamanini F, van der Horst G T J, Hoeijmakers J H J, Yasui A, Loros J J, Dunlap J C, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. The mPer2 gene encodes a functional component of the mammalian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 38.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Three period homologues in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 39.Zylka M J, Shearman L P, Levine J D, Jin X, Weaver D R, Reppert S M. Molecular analysis of mammalian Timeless. Neuron. 1998;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]