Abstract
The expansion of a hexanucleotide repeat GGGGCC (G4C2) in the C9orf72 gene is the most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The G4C2 expansion leads to repeat-associated non-AUG (RAN) translation and the production of toxic dipeptide repeat (DPR) proteins, but the mechanisms of RAN translation remain enigmatic. Here, we report that the RNA helicase DHX36 is a robust positive regulator of C9orf72 RAN translation. DHX36 has a high affinity for the G4C2 repeat RNA, preferentially binds to the repeat RNA’s G-quadruplex conformation, and efficiently unwinds the G4C2 G-quadruplex structures. Native DHX36 interacts with the G4C2 repeat RNA and is essential for effective RAN translation in the cell. In induced pluripotent stem cells and differentiated motor neurons derived from C9orf72-linked ALS patients, reducing DHX36 significantly decreased the levels of endogenous DPR proteins. DHX36 is also aberrantly upregulated in tissues of C9orf72-linked ALS patients. These results indicate that DHX36 facilitates C9orf72 RAN translation by resolving repeat RNA G-quadruplex structures and may be a potential target for therapeutic intervention.
INTRODUCTION
The functions of nucleic acids are determined by both their linear sequences and their higher-order structures. G-quadruplexes are non-canonical four-stranded secondary structures formed in guanine-rich nucleic acids through Hoogsteen hydrogen bonding, and potential quadruplex-forming sequences are enriched in selected biologically significant regions of the human genome and RNAs, including telomeres, proto-oncogene promoters, replication origins, and untranslated regions of mRNA1–2. G-quadruplexes are thought to be involved in diverse biological phenomena, such as gene transcription and translation, DNA replication and genome instability, telomere regulation, and immunoglobulin gene class switch recombination3–5. Accordingly, G-quadruplexes are associated with pathogenic processes in many human diseases, including cancers and viral infections, making these unique structures potential drug targets for therapeutic intervention6–9.
Expansions of nucleotide repeats have been linked to approximately 40 different types of genetic disorders, primarily disorders affecting the neural and neuromuscular systems10–13. A hexanucleotide (GGGGCC) repeat expansion in the C9orf72 gene, ranging from hundreds to thousands of repeats, is the most frequent genetic cause of both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD)14–15. The genetics and pathologies shared by ALS, characterized by loss of motor neurons, and FTD, characterized by degeneration of the frontal and temporal lobes of the brain, suggest that these two conditions fall within the same continuous clinical spectrum16. The proposed mechanisms of pathogenesis induced by the C9orf72 repeat expansion include loss of C9orf72 function, repeat-associated RNA toxicity, and repeat-associated non-ATG-dependent (RAN) translation17.
RAN translation is a translational mechanism that does not require a start codon and can generate toxic repeat proteins from repeat expansions found in certain neurodegenerative disorders18. RAN translation of the C9orf72 repeat expansion occurs in all three frames in both the sense and anti-sense transcripts, yielding five types of poly-dipeptide repeats (DPR)17. Although several modifiers influencing the production of DPR have been reported, including those involving ribosomal functions, translation initiation, and RNA processing19–21, the mechanism of RAN translation is still poorly understood.
RNA structures play critical roles in biological processes that include translation, but the specific roles of RNA secondary structures in RAN translation remain enigmatic22. C9orf72 GGGGCC repeat RNAs have been found to adopt G-quadruplex structures with unusually high thermal stability23–26. However, the potential role of G-quadruplexes in the regulation of RAN translation has yet to be explored. Here, we report that an RNA helicase, DHX36, shows strong binding and unfolding activity in vitro towards the G-quadruplexes formed by C9orf72 repeat RNAs and that endogenous DHX36 interacts with C9orf72 repeat RNAs in the cell. Furthermore, we found that reducing DHX36 significantly decreases the levels of DPRs in C9orf72 ALS patient-derived induced pluripotent stem cells (iPSCs) and motor neurons. Our results indicate that DHX36 positively regulates the production of DPR by resolving C9orf72 repeat RNA G-quadruplexes, providing a mechanism for the pathogenesis of relevant neurological disorders.
MATERIALS AND METHODS
DNA constructs for translation reporters.
For cell-based translation reporters, a pcDNA3.1 backbone construct expressing EGFP under the CMV promoter and NeoR under the SV40 promoter was first generated. The EGFP coding sequence was amplified with a forward primer producing NheI, EcoRI, HindIII sites and a reverse primer producing an XhoI site. The resulting fragment was ligated into pcDNA3.1 (ThermoFisher) between the Nhe I and XhoI sites. The repeat sequence (GGGGCC)4, (GGGGCC)8, or their respective length-matched non-G-quadruplex-forming controls were synthesized (Integrated DNA Technologies) and ligated into the pcDNA3.1-EGFP backbone construct between the NheI and HindIII sites. The (GGGGCC)28 DNA fragment was generated using self-templating PCR as we previously described25 and inserted into the EcoRI site of pcDNA3.1-EGFP. A length-matched non-G-quadruplex-forming control for (GGGGCC)28 was amplified from a luciferase gene and ligated into pcDNA3.1-EGFP between the EcoRI and HindIII sites. For the in vitro translation assays, (GGGGCC)4-EGFP, (GGGGCC)8-EGFP, and their length-matched controls were amplified from the abovementioned pcDNA3.1 constructs and subcloned into pF3A WG(BYDY) (Promega) between the NcoI and SacI sites. The sequences of the control oligonucleotides are provided in Supplemental Table S1.
Protein purification and electrophoretic mobility shift assay.
DHX36 was purified as previously described27. Briefly, a variant of DHX36 (Δ1–56, EKK146AAA) containing a N-terminal GST-tag and a C-terminal poly-histidine tag was expressed in Escherichia coli LOBSTR (DE3) cells. The variant was used because it has wild-type-like G-quadruplex-binding and unwinding activities but shows enhanced solubility and resistance to proteolysis during protein purification. The cell cultures were induced by the addition of 1 mM IPTG and incubated overnight at 20 °C. The DHX36 variant was first purified using a Ni-NTA Superflow column (Qiagen), which was immediately followed by a GSTrap 4B (GE Healthcare) column. The eluted protein was incubated overnight at 4 °C with TEV protease for removal of the GST-tag. The following day, the protein sample was applied to a size-exclusion chromatography column (GE Healthcare) equilibrated with 25 mM HEPES-KOH pH 7.5, 150 mM KCl, 10% Glycerol, and 0.5 mM TCEP (tris(2-carboxyethyl)phosphine). Fractions corresponding to DHX36 were collected and stored at 4 °C. For electrophoretic mobility shift assays (EMSA), RNA oligonucleotides 3’-labelled with Cy5 (0.2 μM) were heated in annealing buffer (10 mM Tris-HCl pH 7.4, 50 mM KCl or LiCl, and 1 mM EDTA) at 95 °C for 5 min and slowly cooled to 25 °C. The annealed RNA oligonucleotides (20 nM) were then incubated with DHX36 at the indicated concentrations in binding reaction buffer [100 mM Tris-HCl pH 7.4, 100 mM KCl or LiCl, 1 mM EDTA, 1 mM DTT, 5% Glycerol, 2 ng/μl yeast tRNA, 0.01% BSA (w/v), and 5 U/μl RiboLock RNase Inhibitor (Thermo Scientific)] at 25 °C for 20 min. The mixture was then resolved on a 10% non-denaturing polyacrylamide gel (Acrylamide/Bis 37.5:1) in 1x TBE buffer with 75 mM KCl. DNA EMSAs follow the same protocol above without RNase inhibitors in the binding buffer or yeast tRNA but instead with 15 ng/ μl Sheared Salmon Sperm DNA (Ambion, AM9680). Images were captured with Amersham Typhoon Imager 9200 and analyzed with Image Studio version 5.2 (LI-COR Biosciences). The binding curve fittings were generated by GraphPad Prism 8 using nonlinear regression.
G-quadruplex-Hemin colorimetric Assay.
RNA oligonucleotides (5 μM) were annealed slowly in buffer containing 10 mM Tris-HCl (pH 7.4) and 100 mM KCl from 95 °C to 25 °C. 20 μl of the RNA oligonucleotide solution was then transferred to a final 50 μl reaction buffer (25 mM HEPES-NaOH pH 7.4, 100 mM KCl, 2 mM MgCl2, 0.05% Triton X-100, 1% DMSO and 2 μM RNA oligonucleotides) containing 5 μM Hemin. After an incubation at 25 °C for 30 min, another 50 μl of the reaction buffer containing 5 mM ABTS and 0.8 mM H2O2 was added to trigger the colorimetric reaction. The absorption spectra of the reaction were recorded by BioTek™ Synergy™ H4 Hybrid Microplate Reader. For the unwinding test, DHX36 or BSA was added together with the RNA in the reaction buffer to incubate for 15 min followed by the addition of 5 μM Hemin for another incubation at 25 °C for 15 min.
Single-molecule fluorescence resonance energy transfer data acquisition.
Single-molecule fluorescence resonance energy transfer (smFRET) data were acquired using a custom-built prism-type total internal reflection (TIR) inverted fluorescence microscope (Olympus IX 71) as described previously 28–29. The substrate was annealed in 10 mM Tris–HCl (pH 7.5) and 100 mM KCl to make a partial duplex RNA [5’-rUrGrGrCrGrArCrGrGrCrArGrCrGrArGrGrC(rGrGrGrGrCrC)4(rU)10/Cy3/−3′+5′-/Cy5/rGrCrCrUrCrGrCrUrGrCrCrGrUrCrGrCrCrA/biotin/−3] for immobilizing on the PEG-passivated surface coated with neutravidin (50 μg/ml). The smFRET measurements were carried out in an imaging buffer containing 10 mM Tris–HCl (pH 7.5), 100 mM KCl, 1 mM MgCl2, 10% glycerol, 0 or 1 mM ATP, and an oxygen scavenging system [1 mg/ml glucose oxidase, 0.5% (w/v) glucose, 4 μg/ml catalase, and 10 mM Trolox]. A solid-state 532 nm diode laser (Compass 315M, Coherent) was used to excite the Cy3 (donor). The fluorescence from Cy3 and Cy5 (acceptor) were simultaneously collected using a water immersion objective, projected onto the EMCCD camera (Andor) and donor and acceptor signals split through the dichroic mirror (cut off = 630 nm). Before and after addition of purified DHX36, all data were recorded with 100 ms frame integration time, processed by IDL script and analyzed by Matlab scripts30.
Cell culture and transfection.
Both HEK293 cells and HeLa Flp-In cells were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum at 37 °C with 5% CO2. The C9orf72 RAN translation reporter assay was performed as described previously31. Briefly, HeLa Flp-In cells were plated in 6-well plates at 8 × 105 cells per well, followed by treatment with 2 μg/ml doxycycline for 24 h the following day. Then the cells were harvested to measure the NLuc and FLuc luciferase activity by The Nano-Glo Dual-Luciferase Reporter Assay System (Promega) on BioTek™ Synergy™ H1 Hybrid Microplate Reader. For transfection on HEK293 cells, cells were plated in 12-well plates at 4 × 105 cells per well and transfected with 500 ng of plasmid per well with 1 μl of Lipofectamine 2000 (Invitrogen). 24 h after the transfection, cells were harvested for quantitative RT-PCR and immunoblotting analyses.
Lentivirus-mediated gene knockdown.
For lentivirus production, the lentiviral vector pLKO.1 expressing shRNAs against DHX36 (clone ID: TRCN0000050704, TRCN0000050705; Broad Institute) or a control pLKO.1 vector expressing a non-targeting shRNA was co-transfected with the packing plasmid psPAX2 (Addgene #12260) and the envelope plasmid pMD2.G (Addgene #12259) into HEK293 cell at the molar ratio of 1:1:1. Fresh medium was changed 6 hours after the transfection and cells were allowed to continue to grow for 48 h. Supernatants containing the virus were then collected and filtered through 0.45 μm membrane (Millipore Sigma HVHP02500). The filtered supernatants were mixed with 4 x Lentivirus Concentrator Solution [1x PBS (pH 7.4) and 40% PEG-8000 (w/v)] and left at 4 °C overnight. The resulting solution was centrifuged at 1600 x g for 60 min at 4 °C, and the virus pellets were resuspended into cold PBS in 1/20 of the original volume and then aliquoted for storage. To generate stable DHX36 knockdown cell lines, HEK293 cells and HeLa Flp-In cells were transduced with 20 x concentrated lentiviruses 24 h after cell seeding in culture plates. Puromycin (2 μg/ml) was added at 48 h after transduction and cells were selected with the drug for 7 days before harvesting.
Reverse transcription and quantitative PCR.
RNA was isolated by RNeasy Plus Mini kit (Qiagen), followed by reverse transcription using QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacture’s protocol. Quantitative PCR was carried out with PowerUp SYBR Green Master Mix (Thermo Scientific) using a C1000 Touch thermal cycler with a CFX96 Real-Time System (BioRad). The primer sequences are provided in Supplemental Table S3.
Human tissues and western blotting.
Human spinal cord tissues used in this study were described in Supplementary Table S2. For immunoblot analysis, spinal cords were rinsed with cold PBS and homogenized with cold RIPA buffer using glass tissue grinders. The homogenates were then centrifuged at 4 °C at 1,000 g for 10 min, and the supernatants were centrifuged again at 16,000 g for 10 min, with the final supernatant used for immunoblot analysis.
Protein samples from tissues or cells were resolved on 12% SDS-PAGE gels and then transferred to nitrocellulose membranes (Bio-rad). The membranes were blocked in 5% milk in 1x TBST (Tris-Buffered Saline, 0.1% Tween-20) at room temperature for 1 h, followed by incubation with a primary antibody in 5% BSA in 1x TBST containing 0.03% sodium azide at 4 °C overnight. The primary antibodies include anti-DHX36 (Abcam, ab70269, 1:5000), anti-GFP (Invitrogen, GF28R, 1:500), anti-β-Tubulin (Cell Signaling, #2128, 1:1000), anti-GAPDH (Thermo Scientific, TAB1001, 1:2000) and anti-β-Actin (Santa Cruz, sc-47778, 1:1000). Membranes were washed three times with 1x TBST for 5 min at room temperature and then incubated with a secondary antibody diluted 1:1000 in blocking buffer at room temperature for 2 h. The secondary antibodies include goat anti-rabbit IgG IRDye (800 CW, 926–32211) and donkey anti-mouse IgG (680 LT, 926–68022). Membranes were again washed three times with 1x TBST at room temperature for 5 min and then imaged on the Odyssey system and analyzed with Image Studio version 5.2 (LI-COR).
In vitro transcription and translation.
The pF3A WG (BYDY) plasmids containing GGGGCC repeats or control sequences were linearized by XbaI and purified using the Wizard SV Gel and PCR Clean-Up system (Promega). The linearized plasmids were then used as templates for transcription by a HiScribe T7 transcription kit (NEB, E2030S). Following DNase treatments, the transcription products were purified using RNA Clean & Concentrator-25 (ZYMO RESEARCH) and examined by 5% denaturing urea polyacrylamide gel electrophoresis. In vitro translation assays were performed using a wheat germ extract kit (Promega, L4380). Briefly, reactions (50 μl) were carried out in 96-well black plates (Costar) containing 25 μl wheat germ extract, 1.25 μg template RNA, 0.08 mM amino acid mixture, 50 mM potassium acetate, and 0.8 U/μl RiboLock RNase Inhibitor (Thermo Scientific). The fluorescence signal was recorded by a BioTek Synergy H1 microplate reader with the following kinetic setting: temperature 25 °C, excitation 480 nm, emission 507 nm, running time 1.5 h, kinetic interval 2 min, shake linear for 1 s; read speed normal, optics position top, read height 5 mm.
RNA immunoprecipitation.
RNA immunoprecipitation was performed as described with modifications32. HeLa Flp-In cells were seeded in 10 cm dishes at 7 × 106 cells/dish and treated with 2 μg/ml doxycycline for 24 h the following day. Cells were then harvested and rinsed once with cold PBS. The cell pellets were resuspended in 0.5 ml cytoplasmic lysis buffer [25 mM HEPES pH 7.9, 5 mM KCl, 0.5 mM MgCl2, and 0.5% IGEPAL CA-630 (v/v)] supplemented with protease inhibitors (cOmplete, EDTA-free cocktail, Roche) and 1.6 U/μl of Ribolock RNase inhibitors (Thermo Scientific), left on ice for 20 min, and centrifuged at 2500 x g for 5 min at 4 °C. The supernatants were set aside on ice and the cell pellets were resuspended in 0.5 ml nuclear lysis buffer [25 mM Hepes, pH 7.9, 10% sucrose (w/v), 350 mM NaCl, and 0.01% IGEPAL CA-630 (v/v)] supplemented with protease inhibitors (cOmplete, EDTA-free cocktail, Roche) and 1.6 U/μl of Ribolock RNase inhibitors (Thermo Scientific). To lyse the nuclei, the tubes were vortexed for 30 s followed by 30-min incubation at 4 °C with end-over-end mixing. The nuclear and cytoplasmic fractions were combined, and the insoluble material was removed by centrifugation at 20,000 × g for 10 min at 4 °C. The cell lysates were then precleared with 10 μl equilibrated Pierce Protein A/G Magnetic Beads (Thermo Scientific) at 4 °C for 30 min. For 1 ml of the precleared cell lysate, 50 μl was taken for RNA input and 50 μl for protein input. For immunoprecipitations, 2.5 μg of anti-DHX36 antibody (ab70269, Abcam) or Normal Rabbit IgG (Cell Signaling Technology) was added to 450 μl cell lysate and incubated at 25 °C for 1 h with end-over-end mixing. Then 5 μl equilibrated Pierce Protein A/G Magnetic Beads was added to each sample for another 1-h mixing at 25 °C. Beads were washed 9 times with a 1:1 mixture of cytoplasmic and nuclear lysis buffers followed by RNA isolation with RNeasy Plus Mini Kit (Qiagen). The isolated RNA was then reverse transcribed by QuantiTect Reverse Transcription Kit (Qiagen) for qPCR using PowerUp SYBR Green Master Mix (Thermo Scientific) according to the manufacture’s protocols.
RNA fluorescence in situ hybridization.
RNA fluorescence in situ hybridization was performed as previously reported with modifications33. Briefly, cells grown on coverslips (Deckglaser, Germany) were fixed in 3.75% formaldehyde in PBS for 10 min at room temperature, followed by permeabilization in ice cold 70% ethanol for 30 min. After rehydration in wash buffer (40% deionized formamide in 2 x SSC) for 10 min, cells were prehybridized at 55 °C for 10 min in hybridization buffer (40% deionized formamide, 2 x SSC, 20 μg/mL BSA, 100 mg/mL dextran sulfate, 10 μg/mL yeast tRNA, and 2 mM Vanadyl Sulfate Ribonucleosides). Cells were then incubated at 55 °C for 2 h in hybridization buffer containing 125 nM (CCCCGG)4-Cy3 probe, which was denatured at 95 °C for 5 min before being added to the buffer, followed by washing three times with wash buffer at 55 °C for 10 min. Coverslips were then stained with 0.5 μg/mL DAPI (4’,6-diamidino-2-phenylindole) in PBS at room temperature for 10 min and mounted with ProLong Gold Antifade (Thermo Fisher Scientific). Images were obtained using a Leica SP8 confocal microscope with 0.5 μm z-step size and matched exposure settings. All solutions were made with DEPC-treated water and formamide was freshly used.
Human iPSC and motor neuron cultures.
iPSCs were grown in StemFlex medium (Thermo Scientific, A3349401) on matrigel (Corning, 354230) coated plates. To generate motor neurons, iPSCs were differentiated as previously described34. Briefly, iPSCs were first differentiated into neuroepithelial progenitor (NEP) cells using neural medium (1:1 DMEM/F12:neurobasal medium, glutamax, N2 supplement, B27 supplement, and ascorbic acid), supplemented with 3 μM CHIR99021, 2 μM SB431542, and 2 μM DMH-1 for 6 d. NEP cells were then split and grown in neural medium supplemented with 1 μM CHIR99021, 2 μM SB431542, 2 μM DMH-1, 0.1 μM retinoic acid (RA), and 0.5 μM purmorphamine to generate motor neuron progenitor (MNP) cells. To generate motor neuron-like cells, MNPs were disassociated by cell scraper and placed in suspension culture using neural medium supplemented with 0.5 μM RA and 0.1 μM purmorphamine. After 6 d, motor neuron-like cells were disassociated and plated onto Matrigel-coated plates. Mature motor neurons were generated in 12 d using neural medium supplemented with 0.5 μM RA, 0.1 μM purmorphamine, and 0.1 μM compound E, with medium change every other day. To generate stable shRNA-expressing iPSCs, 50 μl of concentrated lentivirus solution (shRNA clone ID: TRCN0000050704) was added to each well of iPSCs cultured in 6-well plates. Following 2 days of transduction, cells were cultured in selection medium containing 1 μg/ml puromycin for 7 days before analysis. For transduction of iMNs, lentiviruses were added at day 7 of the motor neuron maturing stage and incubated for 2 days. After another 8 days’ culturing, motor neurons were harvested for analysis. Human iPSCs used in this study were described in Supplementary Table S2.
Poly-GP ELISA.
Cells were lysed in RIPA buffer (Sigma, R0278) containing 1 x proteinase inhibitor (Roche, cOmplete, EDTA-free) and centrifuged at 16000 x g for 20 min at 4 °C. The supernatants were collected, and protein concentrations were quantified with the Pierce BCA protein assay (Thermo Scientific). The samples were then diluted to the concentration of 1 mg/ml for ELISA as previously described21. Briefly, 0.375 μg/mL biotinylated rabbit anti-GP antibody was incubated in 96-well small spot streptavidin coated plates for 1 h at room temperature. Following three PBST washes, 35 μL cell lysate was added per well in duplicate and incubated for a 3 h at room temperature. After three PBST washes, sulfo-tagged detection antibody was then added at 1 μg/mL and incubated for 1 h. Following another set of PBST washes, 150 μL read buffer was added and samples immediately imaged by MESO QuickPlex SQ 120. Specificity was verified using lysates of HEK293 cells overexpressing GFP-tagged dipeptide repeat proteins. All reagents were from Meso Scale Discovery (MSD).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism 8. Quantitative analysis was performed using two-tailed Student’s t-test. P value less than to 0.05 was taken as statistically significant. The n values and other details of statistical analysis are described in the figure legends.
RESULTS
DHX36 helicase has high affinity for G-quadruplexes formed by C9orf72 repeat RNA.
C9orf72 GGGGCC repeat RNA can form parallel G-quadruplexes that exhibit high thermal stability23, 25–26, 35. We hypothesized that a helicase that resolves the RNA G-quadruplex structure may play a regulatory role in the cellular functions associated with the C9orf72 GGGGCC repeat RNAs. As a member of the DEAH/RHA family of helicases, DHX36 shows an extremely high affinity for DNA and RNA G-quadruplexes, and it preferentially unwinds parallel G-quadruplexes27, 36. To examine whether DHX36 could potentially act on r(GGGGCC)n G-quadruplexes, we first performed EMSAs to determine whether the various structures of C9orf72 repeat RNAs can be recognized by DHX36. We have previously shown that four repeats of the G4C2 RNA are sufficient to form stable G-quadruplexes25. Since K+ stabilizes G-quadruplex structures better than does Li+ 37, the formation of r(GGGGCC)4 G-quadruplexes is more likely induced by the presence of K+, whereas the formation of hairpins is more likely induced by r(GGGGCC)4 Li+ (Figure S1). In the EMSA assay, DHX36 preferentially bound the G-quadruplex conformation of r(GGGGCC)4 or r(GGGGCC)8, since it showed a much higher affinity for the RNAs under K+ conditions than under Li+ conditions (Figure 1A–D). Notably, r(GGGGCC)8 formed more stable multiple G-quadruplexes than did r(GGGGCC)4, as indicated on gel electrophoresis by the smeared bands and the relatively high proportion of upper-shifted bands for r(GGGGCC)8 in the presence of K+ when compared to Li+ (Figure 1A, C), consistent with the concept that the longer C9orf72 repeat RNA can adopt multiple G-quadruplex conformations that increase its stability and complexity23. To confirm the specificity of DHX36 binding for r(GGGGCC)4 G-quadruplexes, we introduced mutations into the repeat RNA that disabled the formation of the G-quadruplex structure. The mutated repeat RNA, r(GTGTCC)4, did not exhibit any K+-dependent binding to DHX36 (Figure 1E, F), confirming our conclusion that the interaction between the helicase and the repeat RNAs was mediated by the G-quadruplex. In addition, we found that another control, the C9orf72 antisense repeat RNA r(CCCCGG)4, which cannot form G-quadruplexes38, showed no interaction with DHX36 (Figure 1G, H). Notably, unlike parallel RNA G-quadruplexes, single strand RNAs can be bound by DHX36 but with a much lower affinity as shown in Figure 1E. Furthermore, DHX36 has little preferential binding toward the d(GGGGCC)4 DNA G-quadruplexes (Figure S2), which we and other have found to adopt antiparallel oritention25, 39, in agreement with the concept that DHX36 favors to bind parallel G-quadruplexes36. Taken together, our results demonstrate that DHX36 can specifically bind G-quadruplexes of C9orf72 G4C2 repeat RNA.
Figure 1.
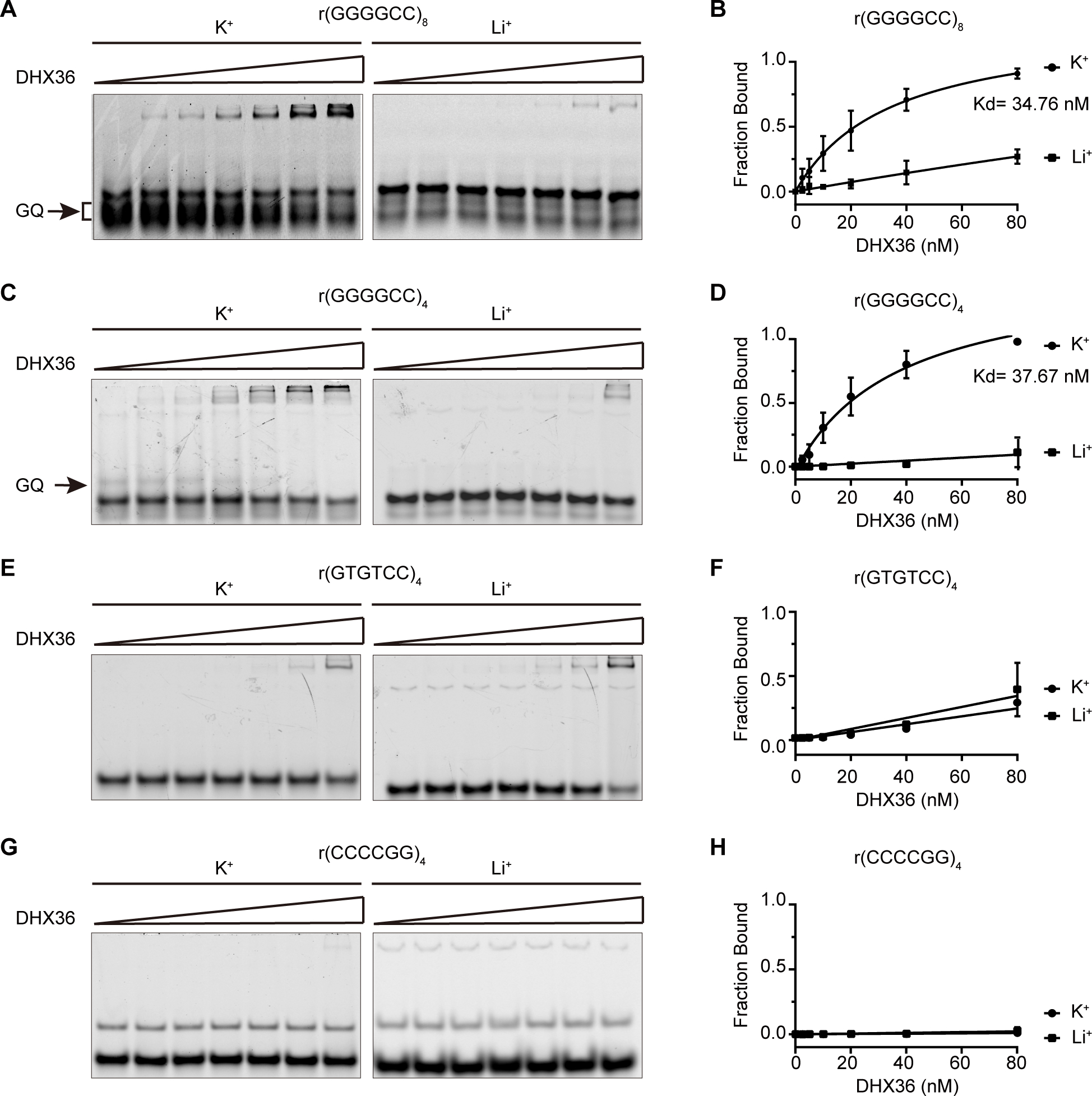
DHX36 protein shows high affinity for G-quadruplex structures formed by GGGGCC RNA repeats. (A), (C), (E), (G) EMSA experiments were performed by annealing 20 nM of four Cy5-labeled RNA probes, r(GGGGCC)8, r(GGGGCC)4, r(GTGTCC)4, and r(CCCCGG)4, gradually from 95 °C to 25 °C and then incubating them with DHX36 protein in increasing concentrations (0, 2.5, 5, 10, 20, 40, and 80 nM) in the presence of 100 mM KCl or LiCl. The G-quadruplexes (GQ) that specifically appeared on the gels in the presence of K+ are indicated by arrows. (B), (D), (F), (H) Quantification was performed to generate the binding curves of DHX36 protein to various RNA probes in the presence of K+ or Li+ (N = 3), as indicated. Data are given as means ± SD of three independent experiments.
DHX36 unwinds the G-quadruplex structure formed by C9orf72 repeat RNA.
After establishing the binding of DHX36 to the C9orf72 repeat RNA G-quadruplex, we investigated the helicase activity of DHX36 on this G-quadruplex structure. First, we used a G-quadruplex-hemin RNAzyme system to monitor the integrity of the G-quadruplex structure. G-quadruplexes are known to be able to associate with hemin (iron(III)-protoporphyrin IX) to form peroxidase-mimicking enzymes, whose activity is dependent on an intact G-quadruplex structure40–41. To test the ability of C9orf72 repeat RNA to catalyze peroxidase-mimic reactions, we used ABTS (2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) as a substrate. The G-quadruplexes, once folded from C9orf72 repeat RNAs, bind hemin with high affinity and form RNAzyme complexes to catalyze the oxidation of ABTS, yielding a peroxidation product, ABTS·+, which has a maximum absorption around 412–422 nm (Figure 2A). As expected, r(GGGGCC)10 showed strong G-quadruplex-mediated production of ABTS·+, exhibiting an absorption maximum around 420 nm (Figure 2B). In contrast, the antisense repeat r(CCCCGG)10, known to form intramolecular A-form-like helices38, showed a significantly lower signal identical to that of the negative buffer control, indicating a lack of peroxidase activity (Figure 2B). Using the G-quadruplex-hemin RNAzyme system, we examined the effect of DHX36 on the G-quadruplexes formed by r(GGGGCC)10. When DHX36 was added to the reaction mixture, the amount of ABTS·+ decreased significantly as the concentration of DHX36 increased (Figure 2C), suggesting that DHX36 bound to the G-quadruplex-hemin complex and altered its conformation and activity. As a control for the DHX36 protein, the addition of bovine serum albumin (BSA) protein to the reaction had no major effect on the activity of the G-quadruplex-hemin RNAzyme, confirming the specificity of DHX36 activity. In addition, we tested the effects of DHX36 on the RNAzyme activities of r(GGGGCC)4 and r(GGGGCC)8, which showed similar results as those of r(GGGGCC)10 (Figure S3).
Figure 2.
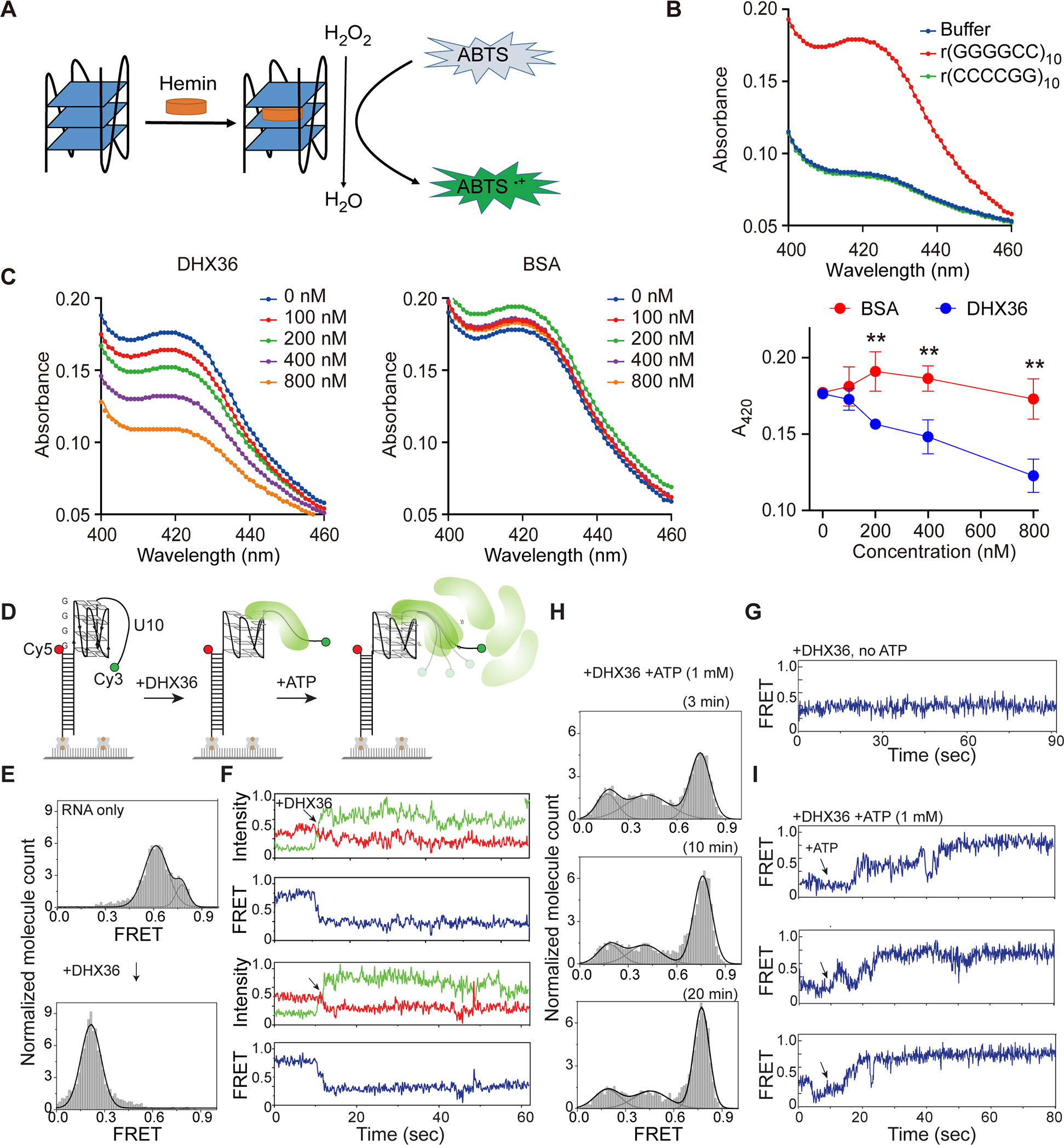
DHX36 efficiently unwinds G-quadruplexes formed by GGGGCC RNA repeats. (A) Schematic representation of the G-quadruplex-hemin RNAzyme system. (B) Absorption spectra of r(GGGGCC)10, r(CCCCGG)10, or the buffer control in the colorimetric reactions. [RNA]=1 μM, [Hemin]=2.5 μM, [ABTS]=2.5 μM, and [H2O2]=0.4 mM. (C) DHX36 or BSA proteins were added to the r(GGGGCC)10 RNAzyme system at increasing concentrations (0, 100, 200, 400, or 800 nM) before triggering the colorimetric reaction. Maximum absorbance at 420 nm for each group was used to compare the RNAzyme activities. Data are given as means ± SD of four independent experiments. **P <0.01. (D) Schematic of smFRET using RNA r(GGGGCC)4-U10. (E) FRET histograms before and after addition of the DHX36 protein. FRET values were collected from >4000 molecules in 20 different fields of view. (F) Two sets of representative smFRET traces upon addition of DHX36 (arrows). The smFRET trace shows fluorescence intensities observed for Cy3 (green) and Cy5 (red) and the calculated FRET efficiency (blue) when 10 nM DHX36 protein was added. (G) The smFRET trace taken after DHX36 was added to the G4C2-U10 substrate. (H) FRET histograms taken 3, 10, and 20 minutes after 1 mM ATP was added to the DHX36 and G4C2-U10 substrate mix. (I) Three sets of representative smFRET traces after ATP was added.
To obtain further insight into DHX36 helicase activity toward the C9orf72 repeat RNA G-quadruplexes, we performed single-molecule fluorescence resonance energy transfer (smFRET) experiments. Here, we designed a GGGGCC-containing RNA construct labelled with Cy3 and Cy5 for smFRET imaging. The RNA consisted of four GGGGCC repeats and a poly rU10 tail for DHX36 loading at the 3’ end (Figure 2D). FRET values were collected from >4000 molecules in 20 different fields of view to build the FRET histogram. The G4C2-containing RNA alone yielded the main FRET peak at ∼0.6, consistent with the expected distance between Cy3 and Cy5 separated by the G4C2 repeat and U10 tail (Figure 2E). Upon addition of 10 nM DHX36, the mid-FRET peak immediately shifted to low FRET at ~0.2, indicating that all RNA molecules are occupied by DHX36 (Figure 2e). The representative smFRET traces displayed a decrease in FRET signals, suggesting the partial disruption of G-quadruplex structures by DHX36 binding (Figure 2F). The DHX36 remains bound as reflected by a steady low FRET state (Figure 2G). When 1 mM ATP was added to the DHX36-bound G4C2-U10 substrate, the FRET histogram gradually shifted back to high values (Figure 2H), and the smFRET traces displayed slow FRET fluctuations that transitioned to the high-FRET state (Figure 2I). This pattern represents ATP-dependent repetitive unfolding of G-quadruplexes by DHX36 followed by dislodging of DHX36, in accordance with our previous observations of the action of DHX36 on another model G-quadruplex substrates42–43. In summary, our data indicate that DHX36 binds G-quadruplex structure formed by C9orf72 repeat RNA with high affinity and unfolds the G-quadruplex structure by hydrolyzing ATP.
DHX36 positively regulates C9orf72 repeat-associated RAN translation.
Given the strong binding of DHX36 protein to the C9orf72 repeat RNA and its effects on the RNA structures, we asked whether DHX36 plays a role in repeat RNA-mediated cellular processes. One of the unique consequences of C9orf72 hexanucleotide repeat expansion is repeat-associated non-AUG-dependent (RAN) translation. To investigate a potential role for DHX36 in this non-canonical translation, we employed an established system using inducible dual-luciferase-based reporter cell lines designed for monitoring C9orf72 RAN translation (Figure 3A)31. Briefly, a (GGGGCC)70 repeat was inserted upstream of the ATG-less coding sequence of the NanoLuc luciferase (NLuc) in the poly-GP frame to induce RAN translation, and the firefly luciferase (FLuc) with an ATG start codon was cloned in the same construct as an internal control for canonical translation. The reporter cassette, with or without the (GGGGCC)70 repeat, was integrated into the unique Flp-In site in the stable reporter HeLa cell lines31. Using an RNA fluorescence in situ hybridization (FISH) assay with the (CCCCGG)4 probe, we confirmed the expression of the GGGGCC repeat RNA in the cells harboring the (GGGGCC)70 repeats (Figure 3B), as evidenced by the repeat RNA foci in the nucleus — a pathological hallmark in C9orf72-related ALS patients44. As expected, the (GGGGCC)70 repeat induced significant RAN translation, as measured by the NLuc expression in the poly-GP frame relative to the FLuc expression, whereas the no-insert control gave only a minimal background signal (Figure 3C). To test the role of DHX36 in RAN translation, we stably knocked down the helicase in the (GGGGCC)70 reporter cell line by using two different lentivirus-mediated shRNAs. In accordance with the reduction of the DHX36 protein, the level of RAN translation, as indicated by the NLuc signal relative to FLuc, was significantly reduced (Figure 3C), demonstrating that DHX36 positively regulates C9orf72 RAN translation.
Figure 3.
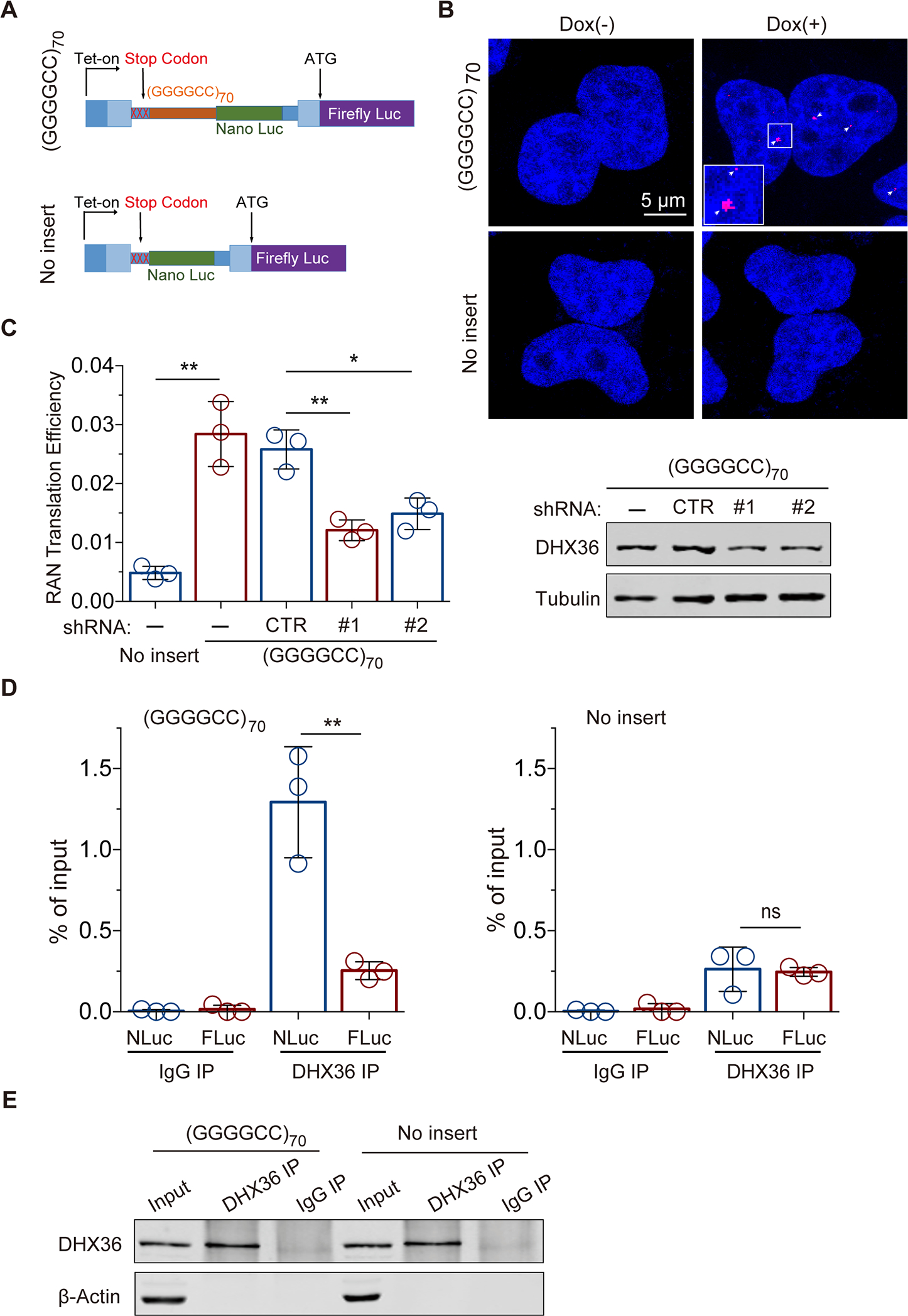
DHX36 binding to C9orf72 repeat RNA is required for efficient RAN translation in cells. (A) Schematic of the inducible luciferase-based C9orf72 RAN translation reporter system in HeLa Flp-In cells. The wide and narrow rectangles represent exons and introns, respectively. (B) RNA FISH with a (CCCCGG)4-Cy3 probe demonstrated repeat RNA foci in the cells expressing the (GGGGCC)70. (C) DHX36 knockdown significantly decreased C9orf72 RAN translation efficiency, measured as the ratio of Nano luciferase signal to firefly luciferase signal, in the HeLa Flp-In cells (left, N = 3). The knockdown of DHX36 was confirmed by immunoblot analysis of the cells expressing (GGGGCC)70 (right). (D) RNA immunoprecipitation experiments in HeLa Flp-In cells with or without (GGGGCC)70 demonstrated the enrichment of (GGGGCC)70-containing transcripts (Nano Luc RNAs) but not the internal control firefly Luc transcripts upon pulldown of DHX36 (N = 3). (E) Immunoblotting confirmed the pulldown of DHX36 in the RNA immunoprecipitation experiments in HeLa Flp-In cells with and without (GGGGCC)70. Data are given as means ± SD of three independent experiments. *P < 0.05, **P < 0.01.
To test the hypothesis that DHX36 regulates C9orf72 RAN translation by interacting with repeat RNA and further validate the in vitro observation of direct binding of DHX36 to GGGGCC repeat RNAs, we performed native RNA immunoprecipitation experiments in the cell-based system. Endogenous DHX36 was pulled down by incubating a specific antibody with whole cell lysates from the (GGGGCC)70 or no-repeat reporter lines, and RNA associated with DHX36 was co-immunoprecipitated for further quantitative analysis. When compared with a non-specific IgG control, there was an enrichment of DHX36 protein from the pull-down samples, indicating the specific immunoprecipitation of DHX36 protein (Figure 3E). Quantitative reverse transcription and PCR analysis showed that the NLuc RNA fused with (GGGGCC)70 RNA was significantly enriched in the pull-down samples when compared to the FLuc RNA control (Figure 3D). In contrast, the NLuc RNA without the (GGGGCC)70 flanking sequence from the no-insert control cells had only a minimal background signal, comparable to that of FLuc RNA. The nearly 5-fold enrichment of (GGGGCC)70 repeat-containing RNA confirmed that DHX36 binds the C9orf72 repeat RNA in vivo (Figure 3D). Taken together, our results indicate that DHX36 interacts with C9orf72 repeat RNA and positively regulates its RAN translation.
DHX36 releases translational repression caused by C9orf72 repeat RNA G-quadruplexes.
Given the reported role of G-quadruplexes in inhibiting translation elongation45, we hypothesized that DHX36 regulates C9orf72 RAN translation by resolving the G-quadruplexes in the repeat RNA and thereby relieving the translational repression. To test this hypothesis, we engineered a series of reporter constructs in which GGGGCC repeats were placed between the ATG start codon and the EGFP coding sequence (Figure 4A). Since C9orf72 hexanucleotide repeat expansion can impair transcription in a G-quadruplex-dependent manner25, we tested different lengths of GGGGCC repeats in order to minimize the effect of impaired transcription on the translation of the reporter gene. For example, when a segment of 28 hexanucleotide repeats, (GGGGCC)28, was engineered into the reporter, the level of EGFP mRNA as measured by quantitative RT-PCR was significantly reduced when compared to those with shorter repeats; however, when the repeats were replaced with the non-G-quadruplex-forming sequence, the EGFP mRNA signal reverted to normal levels (Figure 4B), suggesting that the G-quadruplex structures formed by (GGGGCC)28 led to a reduction in the transcription of the reporter gene. By comparison, for the reporters containing (GGGGCC)4 or (GGGGCC)8, the levels of EGFP mRNA did not change when the repeats were replaced with the non-G-quadruplex-forming sequences (Figure 4B), indicating that the transcription of the reporter gene was not affected by the secondary structures of these shorter hexanucleotide repeats. Therefore, we employed the reporters containing (GGGGCC)4 or (GGGGCC)8 to examine the effects of G-quadruplexes and DHX36 protein on translation in the absence of the confounding effects of G-quadruplex structures on transcription.
Figure 4.
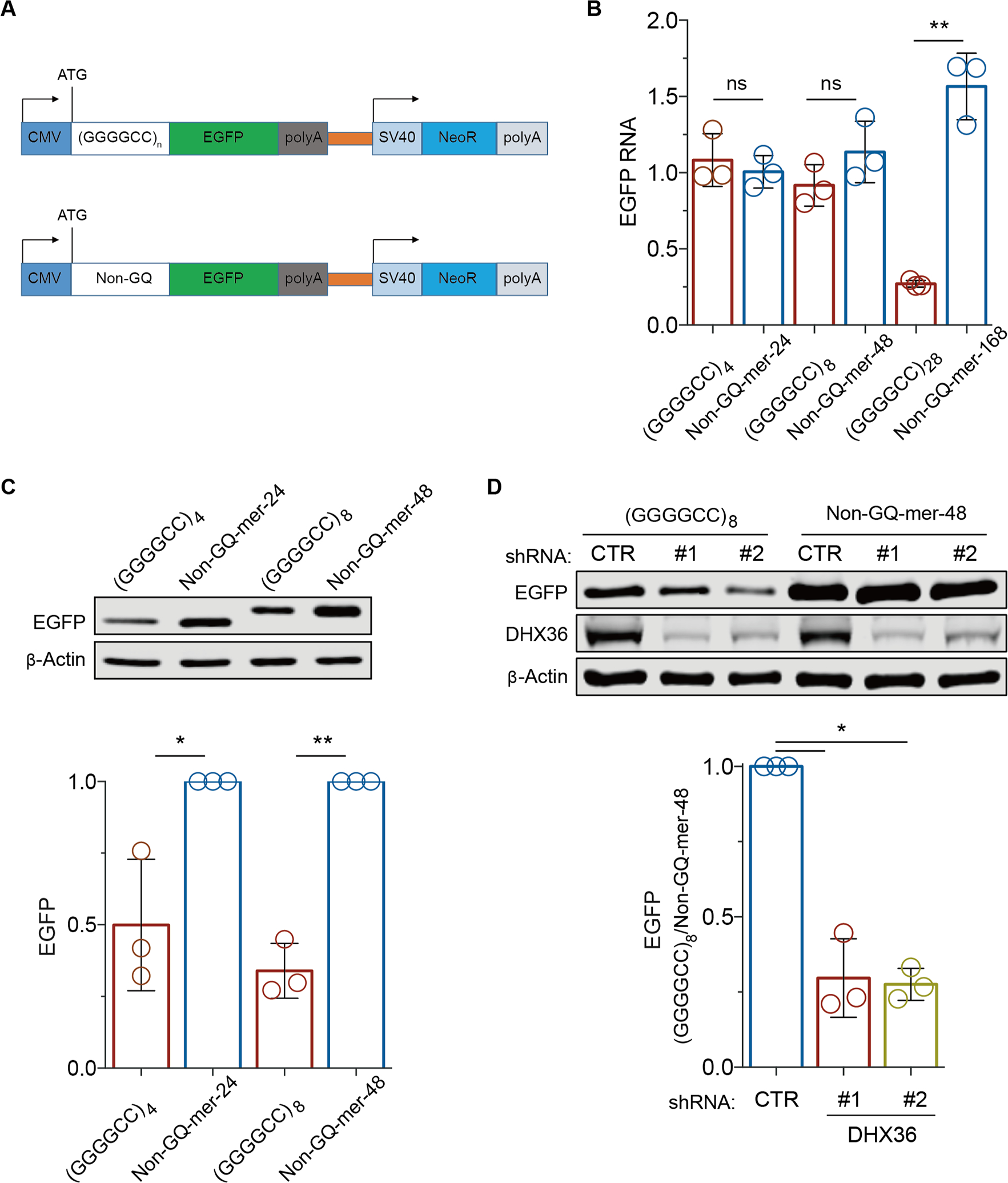
DHX36 is required for efficient translation elongation through GGGGCC repeat RNAs. (A) Schematic of the reporter constructs used to monitor the effects of (GGGGCC)n repeats on translation elongation. (B) The effects of (GGGGCC)n repeats on transcription were compared by RT-PCR analyses of EGFP RNA levels in HEK293 cells expressing reporter constructs containing various GGGGCC repeats and non-G-quadruplex-forming control sequences (non-GQ-mers) of the same sizes. The level of EGFP RNA was normalized to that of NeoR RNA, which is independently expressed on the same reporter construct. (C) Immunoblot analysis of EGFP from HEK293 cells expressing the reporter and control constructs. β-actin was used as the loading control. (D) Immunoblot analysis of EGFP from HEK293 cells expressing the reporter and control constructs after the cells were treated with shRNAs against DHX36 or non-targeting control shRNAs. Data are given as means ± SD of three independent experiments. *P < 0.05, **P < 0.01.
Although (GGGGCC)4 or (GGGGCC)8 did not affect the mRNA levels of the reporter gene, these repeats significantly reduced the levels of the EGFP reporter protein, when compared to the non-G-quadruplex-forming control sequence (Figure 4C, D). Since these hexanucleotide repeats are positioned downstream of the start codon, this finding suggests that the G-quadruplex structures formed by these repeats inhibits translation elongation, leading to a reduced level of protein product. We also developed a wheat germ extract-based in vitro system for monitoring translation in real time by placing the EGFP coding sequence immediately following the GGGGCC repeats and then measuring the translation rates based on the EGFP fluorescence. Consistent with the observations in HEK293 cells, the reporters containing either (GGGGCC)4 or (GGGGCC)8 showed lower translation rates than did the controls with the GGGGCC repeats replaced by non-G-quadruplex-forming sequences of equal lengths (Figure S4), further confirming that the GGGGCC repeats impaired translation.
Next, to examine whether DHX36 plays a role in the hexanucleotide repeat-dependent regulation of translation, we generated stable DHX36 knockdown HEK293 cells using shRNA-expressing lentivirus. We found that the levels of EGFP protein expressed from the reporter containing (GGGGCC)8 were significantly reduced after DHX36 knockdown when compared to the EGFP protein levels in a non-targeting shRNA control (Figure 4D). Since EGFP mRNA levels were not changed (Figure S5), this result indicates that endogenous DHX36 promotes translation elongation over C9orf72 repeat RNA. Furthermore, when (GGGGCC)8 was replaced with a randomized, non-G-quadruplex-forming control sequence of the same length, no effect of DHX36 knockdown on the reporter gene translation was not detected (Figure 4D), consistent with the observation that DHX36 recognizes and resolves the G-quadruplex structure of GGGGCC repeat RNA. Taken together, these data suggest that DHX36 unwinds the G-quadruplexes of C9orf72 repeat RNA and promotes translation elongation through the repeat RNA sequences.
DHX36 facilitates native RAN translation and is upregulated in patient tissues.
We next asked whether DHX36 regulates endogenous DPR protein production in cells from C9orf72-linked ALS patients. We utilized an enzyme-linked immunosorbent assay (ELISA) that has been optimized for quantification of the levels of poly-GP, one of the five DPRs generated in the patients. The proteins extracted from cells of patients and control individuals, together with biotinylated anti-GP antibodies, were applied to a multi-spot immunoassay platform with high sensitivity21. As expected, the levels of poly-GP detected in both the C9orf72 ALS patient-derived iPSCs and motor neurons were significantly higher than those in healthy control cells, confirming the sensitivity and specificity of the poly-GP detection assay (Figure 5A and 5B). To test the role of DHX36 in the regulation of poly-GP production, we knocked down DHX36 in the iPSC lines from the C9orf72-linked ALS patients via lentivirus-mediated stable expression of DHX36-specific shRNAs. We found that the poly-GP proteins were significantly decreased in the patient iPSCs upon the DHX36 knockdown when compared to the non-targeting control shRNA treatment (Figure 5A), indicating that DHX36 is an important positive regulator of the endogenous poly-GP production in the cells of patients. We also induced differentiation of the iPSCs into motor neurons (iMNs) and then tested the effects of DHX36 knockdown on poly-GP production. The iMNs were transduced with lentiviruses expressing either DHX36-specific shRNAs or non-targeting control shRNAs for 2 days, followed by culturing for another 8 days before the ELISA analysis. The DHX36 knockdown produced a result similar to that obtained with the iPSCs with a decrease in the levels of poly-GP in the iMNs derived from the C9orf72-linked ALS patients (Figure 5B), indicating that DHX36 positively regulates RAN translation in the patients’ motor neurons. Furthermore, to determine whether DHX36 is pathologically relevant to C9orf72-linked ALS, we analyzed the expression levels of DHX36 protein in spinal cord tissues from five C9orf72-linked ALS patients and six control individuals. We found that the average protein levels of DHX36 in the ALS patients’ spinal cord tissues were higher than those in the control tissues (Figure 5C), indicating that DHX36 is abnormally upregulated in the ALS patients.
Figure 5.
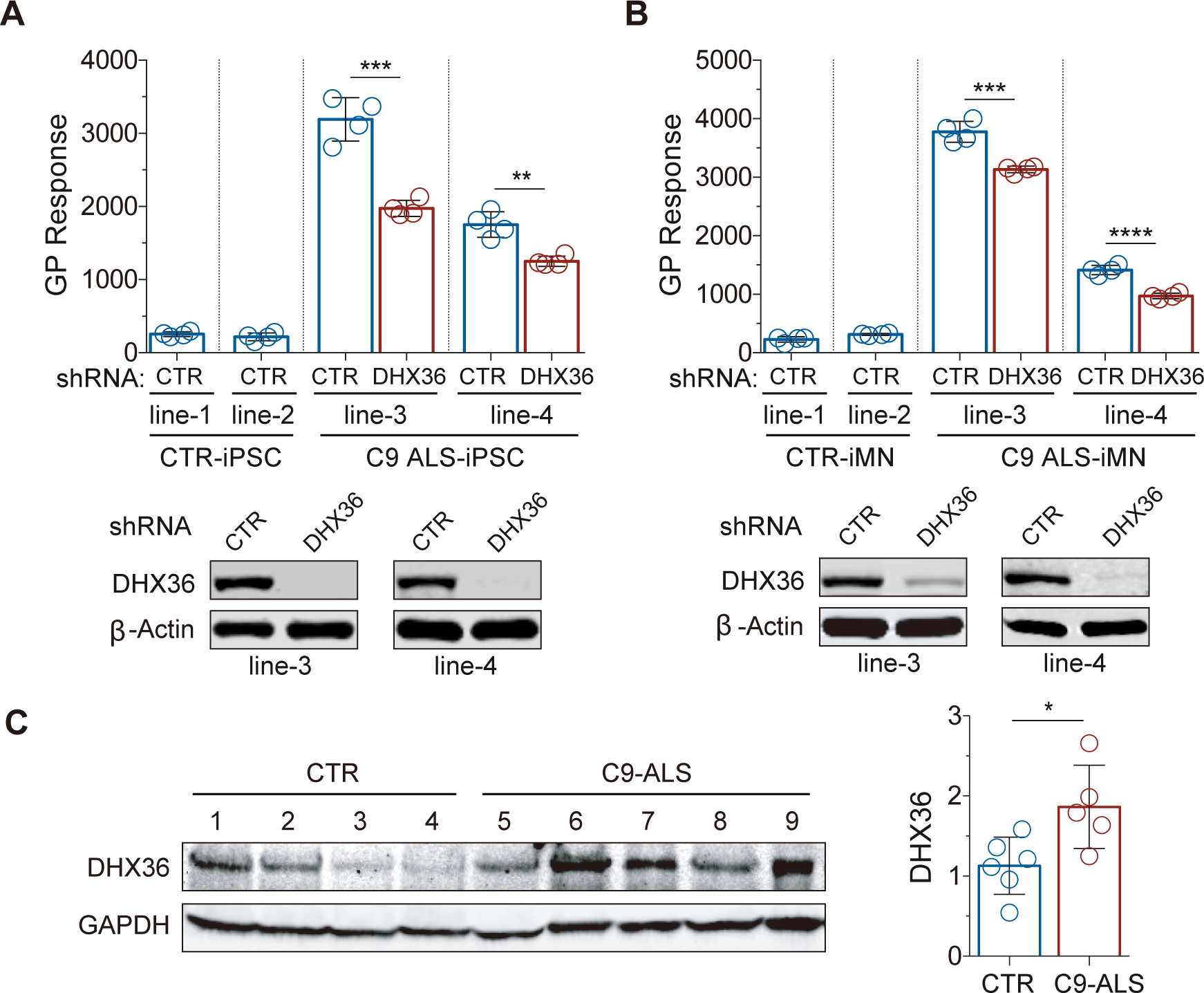
Reduction in poly-GP by knockdown of DHX36 in patient-derived iPSCs and iMNs and upregulation of DHX36 in the spinal cords of C9orf72-linked ALS patients. (A) C9orf72-linked ALS patient-derived iPSCs were analyzed for poly-GP levels by ELISA, with or without the stable knockdown of DHX36. Data are given as means ± SD of four replicates from two independent experiments. **P < 0.01, ***P < 0.001. (B) C9orf72-linked ALS patient-derived iMNs were analyzed for poly-GP levels by ELISA, with or without the stable knockdown of DHX36. Data are given as means ± SD of four replicates from two independent experiments. ***P < 0.001, ****P < 0.0001. (C) DHX36 protein expression levels are higher in the spinal cord tissues from C9orf72-linked ALS patients than in the controls. Data are given as means ± SD of samples from five patients or five controls. *P = 0.0335.
DISCUSSION
In the present study, we have identified a regulatory mechanism for RAN translation that is mediated by the nucleic acid structure G-quadruplex and its resolving helicase (Figure S6). RNA helicases constitute a large family of RNA structure-modulating proteins that are involved in nearly all aspects of RNA metabolism46. The DHX36 helicase has been shown to bind to RNA G-quadruplexes with high affinity and to be the major source of RNA quadruplex resolving activity in HeLa cell lysates47. Moreover, our previous study has demonstrated that DHX36 is capable of binding to several RNA G-quadruplexes in living bacterial cells48. A high-throughput study probing RNA structures reported that RNA G-quadruplexes are globally unfolded in eukaryotic cells49, reflecting the effects of endogenous RNA helicases such as DHX36 on resolving these structures. Indeed, several studies have shown that DHX36 is required for the effective translation of G-quadruplex-containing mRNAs50–53. In the present study, we found that DHX36 plays a critical role in the RAN translation occurring at the C9orf72 GGGGCC repeats that form stable G-quadruplexes. The finding clearly implicates the helicase function of DHX36 in the regulation of the non-canonical translation associated with repeat expansions.
G-quadruplex sequence motifs are frequently found throughout the genome. The fact that they are enriched in the promoters or non-coding regions but rarely found in the coding sequences is consistent with the view that exons may have evolved to avoid the stable higher-order structures that impede ribosome scanning54. We have also observed that the GGGGCC repeats reduce translation efficiency in our experimental systems; however, translation can still be completed and DPR can be produced in the cell, suggesting that cellular machinery exists to unwind G-quadruplexes or lock them in unfolded states to allow for productive ribosome scanning. Our observation that knockdown of DHX36 suppresses translation elongation at the GGGGCC repeats and decreases the production of poly-GP suggests that endogenous DHX36 is actively engaged in resolving stable G-quadruplexes to enhance ribosome processivity.
The cellular effects of DHX36 on C9orf72 repeat-associated translation are in accord with our biochemical and biophysical analyses of the recognition and resolution of the r(GGGGCC)n repeats by the helicase. DHX36 binds to the GGGGCC repeat RNA G-quadruplexes with specificity and high affinity, and it resolves the GGGGCC repeat RNA G-quadruplexes in an ATP-dependent repetitive unfolding and refolding maneuver. We and others have previously reported that GGGGCC repeat RNAs form parallel G-quadruplex structures whereas its DNA repeats form anti-parallel G-quadruplexes23, 25–26, 35. DHX36 is known to preferentially binds to the parallel form of G-quadruplexes36. A recent co-crystal structure of DHX36 bound to a parallel DNA G-quadruplex has revealed intimate contact between DHX36 and the quadruplex as well as contact with an overhanging single-stranded DNA. Upon engaging with the substrate, DHX36 resolves the quadruplex structure by repetitively pulling a single guanine base out of the folded quadruplex structure27, 42. A similar repetitive unfolding activity has also been observed for RNA G-quadruplexes43. Collectively, the evidence leads us to propose that the DHX36 protein actively and repetitively resolves G-quadruplexes on GGGGCC repeat RNAs with its binding and catalytic domains as part of a dynamic regulation that allows RAN translation to take place.
Quadruplex nucleic acids have long been studied as targets for anticancer and antiviral therapeutics, and more recently for developmental disorders such as ATR-X syndrome55–58. Consistent with our observation of an inhibitory role for G-quadruplexes in translation, a set of small molecules stabilizing r(GGGGCC)n quadruplexes has been found to reduce the levels of DPR products in fly and mammalian cell models59. Separately, a small molecule binding the hairpin form of the r(GGGGCC)n repeat and the helicase DDX3X recognizing the same hairpin form of the repeat have been found to negatively regulate the DPR production21, 60. Thus, the evidence obtained so far indicates that, while hairpins seem to promote the RAN translation, G-quadruplexes obstruct the RAN translation occurring at the GGGGCC repeats. Our identification of DHX36 as a facilitator of RAN translation and the resolving activity of DHX36 in promoting conformational transition from G-quadruplexes to hairpins further support the differential roles of various secondary RNA structures in C9orf72 repeat-associated RAN translation. In addition, it remains possible that DHX36 may influence other steps of the GGGGCC repeat RNA metabolism, from transcription to trafficking and decay, through its structure-modulating activities.
We observed the abnormal accumulation of DHX36 protein in the spinal cord tissues of C9orf72-linked ALS patients, suggesting that the upregulation of DHX36 may contribute to both the RAN translation and the relevant pathologies observed in the patients carrying C9orf72 repeat expansions. Considering that G-quadruplexes formed on the C9orf72 repeat RNAs are also involved in other cellular processes such as nucleocytoplasmic transport, alternative splicing, phase separation, and assembly of membraneless organelles such as paraspeckles61–64, it is possible that DHX36 is involved in these related pathological events as well. It should be noted that there have been reports that DHX36 protein prevents the accumulation of translationally inactive mRNAs with G-quadruplexes and is associated with, and affects, the formation of stress granules50, 65, which are stress-induced organelles implicated in ALS/FTD pathogenesis66. Together with our findings concerning the critical role of DHX36 in RAN translation, an increasing understanding of the cellular functions of this helicase may enable the design of effective interventions for the pathogenesis related to the C9orf72 nucleotide repeat expansions.
CONCLUSION
We have uncovered a critical role for G-quadruplex and its partnering helicase DHX36 in the regulation of the non-canonical RAN translation associated with the GGGGCC repeat expansion in C9orf72-linked ALS/FTD. The DHX36 helicase acts by unwinding G-quadruplexes formed by GGGGCC repeat RNAs and eliminating this structural barrier to C9orf72 RAN translation. Given this newly recognized role of DHX36 in positively regulating RAN translation and the observation of its aberrant upregulation in patients’ pathological tissues, our results suggest that DHX36 participates in C9orf72 RAN translation and may serve as a target for alleviating RAN-associated toxicity in relevant neurodegenerative diseases.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Shuying Sun for kindly providing the luciferase reporter cell lines, Lin Xue for helping with the ELISA assay, Yang Liu for helping with the iPSC culturing, Target ALS for patient tissues, and members of Wang laboratory for discussion.
Funding Sources
This work was supported by grants from NIH (NS074324, NS089616, NS110098, GM115631, NS113636), the U.S. Department of Defense, Packard Center for ALS Research at Johns Hopkins, and the Muscular Dystrophy Association, and the intramural program of the National Heart, Lung and Blood Institute (NHLBI). M.T.B. is a Lenfant Fellow of the NHLBI.
Footnotes
Notes
All authors declare no competing interests.
ASSOCIATED CONTENT
Supporting Information.
The supporting information is available free of charge via the Internet at http://pubs.acs.org. Oligonucleotide sequences, human iPSCs and tissues.
REFERENCES
- 1.Tran PL; De Cian A; Gros J; Moriyama R; Mergny JL, Tetramolecular quadruplex stability and assembly. Topics in current chemistry 2013, 330, 243–73. [DOI] [PubMed] [Google Scholar]
- 2.Yang D, G-Quadruplex DNA and RNA. Methods Mol Biol 2019, 2035, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes D; Lipps HJ, G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 2015, 43 (18), 8627–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C; Liu HH; Zheng KW; Hao YH; Tan Z, DNA G-quadruplex formation in response to remote downstream transcription activity: long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res 2013, 41 (14), 7144–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerner LK; Holzer S; Kilkenny ML; Svikovic S; Murat P; Schiavone D; Eldridge CB; Bittleston A; Maman JD; Branzei D; Stott K; Pellegrini L; Sale JE, Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication. The EMBO journal 2020, 39 (18), e104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharel P; Balaratnam S; Beals N; Basu S, The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley interdisciplinary reviews. RNA 2020, 11 (1), e1568. [DOI] [PubMed] [Google Scholar]
- 7.Spiegel J; Adhikari S; Balasubramanian S, The Structure and Function of DNA G-Quadruplexes. Trends Chem 2020, 2 (2), 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cammas A; Millevoi S, RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res 2017, 45 (4), 1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asamitsu S; Obata S; Yu Z; Bando T; Sugiyama H, Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirkin SM, Expandable DNA repeats and human disease. Nature 2007, 447 (7147), 932–40. [DOI] [PubMed] [Google Scholar]
- 11.Orr HT; Zoghbi HY, Trinucleotide repeat disorders. Annu Rev Neurosci 2007, 30, 575–621. [DOI] [PubMed] [Google Scholar]
- 12.La Spada AR; Taylor JP, Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature reviews. Genetics 2010, 11 (4), 247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd PK; Paulson HL, RNA-mediated neurodegeneration in repeat expansion disorders. Annals of neurology 2010, 67 (3), 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeJesus-Hernandez M; Mackenzie IR; Boeve BF; Boxer AL; Baker M; Rutherford NJ; Nicholson AM; Finch NA; Flynn H; Adamson J; Kouri N; Wojtas A; Sengdy P; Hsiung GY; Karydas A; Seeley WW; Josephs KA; Coppola G; Geschwind DH; Wszolek ZK; Feldman H; Knopman DS; Petersen RC; Miller BL; Dickson DW; Boylan KB; Graff-Radford NR; Rademakers R, Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72 (2), 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renton AE; Majounie E; Waite A; Simon-Sanchez J; Rollinson S; Gibbs JR; Schymick JC; Laaksovirta H; van Swieten JC; Myllykangas L; Kalimo H; Paetau A; Abramzon Y; Remes AM; Kaganovich A; Scholz SW; Duckworth J; Ding J; Harmer DW; Hernandez DG; Johnson JO; Mok K; Ryten M; Trabzuni D; Guerreiro RJ; Orrell RW; Neal J; Murray A; Pearson J; Jansen IE; Sondervan D; Seelaar H; Blake D; Young K; Halliwell N; Callister JB; Toulson G; Richardson A; Gerhard A; Snowden J; Mann D; Neary D; Nalls MA; Peuralinna T; Jansson L; Isoviita VM; Kaivorinne AL; Holtta-Vuori M; Ikonen E; Sulkava R; Benatar M; Wuu J; Chio A; Restagno G; Borghero G; Sabatelli M; Consortium I; Heckerman D; Rogaeva E; Zinman L; Rothstein JD; Sendtner M; Drepper C; Eichler EE; Alkan C; Abdullaev Z; Pack SD; Dutra A; Pak E; Hardy J; Singleton A; Williams NM; Heutink P; Pickering-Brown S; Morris HR; Tienari PJ; Traynor BJ, A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72 (2), 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Langenhove T; van der Zee J; Van Broeckhoven C, The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann Med 2012, 44 (8), 817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitler AD; Tsuiji H, There has been an awakening: Emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain research 2016, 1647, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen L; Cleary JD; Ranum LPW, Repeat-Associated Non-ATG Translation: Molecular Mechanisms and Contribution to Neurological Disease. Annu Rev Neurosci 2019, 42, 227–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman LD; Prudencio M; Srinivasan AR; Rifai OM; Lee VM; Petrucelli L; Bonini NM, eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. Acta Neuropathol Commun 2019, 7 (1), 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada SB; Gendron TF; Niccoli T; Genuth NR; Grosely R; Shi Y; Glaria I; Kramer NJ; Nakayama L; Fang S; Dinger TJI; Thoeng A; Rocha G; Barna M; Puglisi JD; Partridge L; Ichida JK; Isaacs AM; Petrucelli L; Gitler AD, RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat Neurosci 2019, 22 (9), 1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng W; Wang S; Zhang Z; Morgens DW; Hayes LR; Lee S; Portz B; Xie Y; Nguyen BV; Haney MS; Yan S; Dong D; Coyne AN; Yang J; Xian F; Cleveland DW; Qiu Z; Rothstein JD; Shorter J; Gao FB; Bassik MC; Sun S, CRISPR-Cas9 Screens Identify the RNA Helicase DDX3X as a Repressor of C9ORF72 (GGGGCC)n Repeat-Associated Non-AUG Translation. Neuron 2019, 104 (5), 885–898 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciesiolka A; Jazurek M; Drazkowska K; Krzyzosiak WJ, Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions. Front Cell Neurosci 2017, 11, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy K; Zamiri B; Stanley SY; Macgregor RB Jr.; Pearson CE, The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem 2013, 288 (14), 9860–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conlon EG; Lu L; Sharma A; Yamazaki T; Tang T; Shneider NA; Manley JL, The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeusler AR; Donnelly CJ; Periz G; Simko EA; Shaw PG; Kim MS; Maragakis NJ; Troncoso JC; Pandey A; Sattler R; Rothstein JD; Wang J, C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507 (7491), 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Z; Zhang Y; Gendron TF; Bauer PO; Chew J; Yang WY; Fostvedt E; Jansen-West K; Belzil VV; Desaro P; Johnston A; Overstreet K; Oh SY; Todd PK; Berry JD; Cudkowicz ME; Boeve BF; Dickson D; Floeter MK; Traynor BJ; Morelli C; Ratti A; Silani V; Rademakers R; Brown RH; Rothstein JD; Boylan KB; Petrucelli L; Disney MD, Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 2014, 83 (5), 1043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen MC; Tippana R; Demeshkina NA; Murat P; Balasubramanian S; Myong S; Ferre-D’Amare AR, Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558 (7710), 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HT; Sanford S; Paul T; Choe J; Bose A; Opresko PL; Myong S, Position-Dependent Effect of Guanine Base Damage and Mutations on Telomeric G-Quadruplex and Telomerase Extension. Biochemistry 2020, 59 (28), 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul T; Voter AF; Cueny RR; Gavrilov M; Ha T; Keck JL; Myong S, E. coli Rep helicase and RecA recombinase unwind G4 DNA and are important for resistance to G4-stabilizing ligands. Nucleic Acids Res 2020, 48 (12), 6640–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy R; Hohng S; Ha T, A practical guide to single-molecule FRET. Nature methods 2008, 5 (6), 507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng W; Wang S; Mestre AA; Fu C; Makarem A; Xian F; Hayes LR; Lopez-Gonzalez R; Drenner K; Jiang J; Cleveland DW; Sun S, C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation. Nature communications 2018, 9 (1), 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booy EP; Meier M; Okun N; Novakowski SK; Xiong S; Stetefeld J; McKenna SA, The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res 2012, 40 (9), 4110–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zu T; Liu Y; Banez-Coronel M; Reid T; Pletnikova O; Lewis J; Miller TM; Harms MB; Falchook AE; Subramony SH; Ostrow LW; Rothstein JD; Troncoso JC; Ranum LP, RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A 2013, 110 (51), E4968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y; Wang T; Ji YJ; Johnson K; Liu H; Johnson K; Bailey S; Suk Y; Lu YN; Liu M; Wang J, A C9orf72-CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes & development 2018, 32 (21–22), 1380–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fratta P; Mizielinska S; Nicoll AJ; Zloh M; Fisher EM; Parkinson G; Isaacs AM, C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Scientific reports 2012, 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heddi B; Cheong VV; Martadinata H; Phan AT, Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc Natl Acad Sci U S A 2015, 112 (31), 9608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya D; Mirihana Arachchilage G; Basu S, Metal Cations in G-Quadruplex Folding and Stability. Front Chem 2016, 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodd DW; Tomchick DR; Corey DR; Gagnon KT, Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches. Biochemistry 2016, 55 (9), 1283–6. [DOI] [PubMed] [Google Scholar]
- 39.Sket P; Pohleven J; Kovanda A; Stalekar M; Zupunski V; Zalar M; Plavec J; Rogelj B, Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiology of aging 2015, 36 (2), 1091–6. [DOI] [PubMed] [Google Scholar]
- 40.Adeoye RI; Osalaye DS; Ralebitso TK; Boddis A; Reid AJ; Fatokun AA; Powell AK; Malomo SO; Olorunniji FJ, Catalytic Activities of Multimeric G-Quadruplex DNAzymes. Catalysts 2019, 9 (7). [Google Scholar]
- 41.Li W; Li Y; Liu Z; Lin B; Yi H; Xu F; Nie Z; Yao S, Insight into G-quadruplex-hemin DNAzyme/RNAzyme: adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res 2016, 44 (15), 7373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tippana R; Hwang H; Opresko PL; Bohr VA; Myong S, Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc Natl Acad Sci U S A 2016, 113 (30), 8448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tippana R; Chen MC; Demeshkina NA; Ferre-D’Amare AR; Myong S, RNA G-quadruplex is resolved by repetitive and ATP-dependent mechanism of DHX36. Nature communications 2019, 10 (1), 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagier-Tourenne C; Baughn M; Rigo F; Sun S; Liu P; Li HR; Jiang J; Watt AT; Chun S; Katz M; Qiu J; Sun Y; Ling SC; Zhu Q; Polymenidou M; Drenner K; Artates JW; McAlonis-Downes M; Markmiller S; Hutt KR; Pizzo DP; Cady J; Harms MB; Baloh RH; Vandenberg SR; Yeo GW; Fu XD; Bennett CF; Cleveland DW; Ravits J, Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 2013, 110 (47), E4530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endoh T; Kawasaki Y; Sugimoto N, Stability of RNA quadruplex in open reading frame determines proteolysis of human estrogen receptor alpha. Nucleic Acids Res 2013, 41 (12), 6222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourgeois CF; Mortreux F; Auboeuf D, The multiple functions of RNA helicases as drivers and regulators of gene expression. Nature reviews. Molecular cell biology 2016, 17 (7), 426–38. [DOI] [PubMed] [Google Scholar]
- 47.Creacy SD; Routh ED; Iwamoto F; Nagamine Y; Akman SA; Vaughn JP, G4 Resolvase 1 Binds Both DNA and RNA Tetramolecular Quadruplex with High Affinity and Is the Major Source of Tetramolecular Quadruplex G4-DNA and G4-RNA Resolving Activity in HeLa Cell Lysates. J Biol Chem 2008, 283 (50), 34626–34634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu HH; Zheng KW; He YD; Chen Q; Hao YH; Tan Z, RNA G-quadruplex formation in defined sequence in living cells detected by bimolecular fluorescence complementation. Chemical Science 2016, 7 (7), 4573–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo JU; Bartel DP, RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353 (6306). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauer M; Juranek SA; Marks J; De Magis A; Kazemier HG; Hilbig D; Benhalevy D; Wang X; Hafner M; Paeschke K, DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nature communications 2019, 10 (1), 2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murat P; Marsico G; Herdy B; Ghanbarian A; Portella G; Balasubramanian S, RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome biology 2018, 19 (1), 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martone J; Mariani D; Santini T; Setti A; Shamloo S; Colantoni A; Capparelli F; Paiardini A; Dimartino D; Morlando M; Bozzoni I, SMaRT lncRNA controls translation of a G-quadruplex-containing mRNA antagonizing the DHX36 helicase. EMBO reports 2020, 21 (6), e49942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vester K; Eravci M; Serikawa T; Schutze T; Weise C; Kurreck J, RNAi-mediated knockdown of the Rhau helicase preferentially depletes proteins with a Guanine-quadruplex motif in the 5’-UTR of their mRNA. Biochemical and biophysical research communications 2019, 508 (3), 756–761. [DOI] [PubMed] [Google Scholar]
- 54.Huppert JL; Balasubramanian S, Prevalence of quadruplexes in the human genome. Nucleic Acids Res 2005, 33 (9), 2908–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neidle S, Quadruplex nucleic acids as targets for anticancer therapeutics. Nature Reviews Chemistry 2017, 1 (5), 0041. [Google Scholar]
- 56.Ruggiero E; Richter SN, G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res 2018, 46 (7), 3270–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shioda N; Yabuki Y; Yamaguchi K; Onozato M; Li Y; Kurosawa K; Tanabe H; Okamoto N; Era T; Sugiyama H; Wada T; Fukunaga K, Targeting G-quadruplex DNA as cognitive function therapy for ATR-X syndrome. Nat Med 2018, 24 (6), 802–813. [DOI] [PubMed] [Google Scholar]
- 58.Butovskaya E; Solda P; Scalabrin M; Nadai M; Richter SN, HIV-1 Nucleocapsid Protein Unfolds Stable RNA G-Quadruplexes in the Viral Genome and Is Inhibited by G-Quadruplex Ligands. ACS Infect Dis 2019, 5 (12), 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simone R; Balendra R; Moens TG; Preza E; Wilson KM; Heslegrave A; Woodling NS; Niccoli T; Gilbert-Jaramillo J; Abdelkarim S; Clayton EL; Clarke M; Konrad MT; Nicoll AJ; Mitchell JS; Calvo A; Chio A; Houlden H; Polke JM; Ismail MA; Stephens CE; Vo T; Farahat AA; Wilson WD; Boykin DW; Zetterberg H; Partridge L; Wray S; Parkinson G; Neidle S; Patani R; Fratta P; Isaacs AM, G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol Med 2018, 10 (1), 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang ZF; Ursu A; Childs-Disney JL; Guertler R; Yang WY; Bernat V; Rzuczek SG; Fuerst R; Zhang YJ; Gendron TF; Yildirim I; Dwyer BG; Rice JE; Petrucelli L; Disney MD, The Hairpin Form of r(G4C2)(exp) in c9ALS/FTD Is Repeat-Associated Non-ATG Translated and a Target for Bioactive Small Molecules. Cell Chem Biol 2019, 26 (2), 179–190 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar V; Hasan GM; Hassan MI, Unraveling the Role of RNA Mediated Toxicity of C9orf72 Repeats in C9-FTD/ALS. Front Neurosci 2017, 11, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K; Donnelly CJ; Haeusler AR; Grima JC; Machamer JB; Steinwald P; Daley EL; Miller SJ; Cunningham KM; Vidensky S; Gupta S; Thomas MA; Hong I; Chiu SL; Huganir RL; Ostrow LW; Matunis MJ; Wang J; Sattler R; Lloyd TE; Rothstein JD, The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015, 525 (7567), 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fay MM; Anderson PJ; Ivanov P, ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell reports 2017, 21 (12), 3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simko EAJ; Liu H; Zhang T; Velasquez A; Teli S; Haeusler AR; Wang J, G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res 2020, 48 (13), 7421–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chalupnikova K; Lattmann S; Selak N; Iwamoto F; Fujiki Y; Nagamine Y, Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem 2008, 283 (50), 35186–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li YR; King OD; Shorter J; Gitler AD, Stress granules as crucibles of ALS pathogenesis. The Journal of cell biology 2013, 201 (3), 361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


