Abstract
The FtsZ protein is a highly conserved bacterial tubulin homolog. In vivo, the functional form of FtsZ is the polymeric, ring-like structure (Z-ring) assembled at the future division site during cell division. While it is clear that the Z-ring plays an essential role in orchestrating cytokinesis, precisely what its functions are and how these functions are achieved remain elusive. In this article, we review what we have learned during the past decade about the Z-ring’s structure, function, and dynamics, with a particular focus on insights generated by recent high-resolution imaging and single-molecule analyses. We suggest that the major function of the Z-ring is to govern nascent cell pole morphogenesis by directing the spatiotemporal distribution of septal cell wall remodeling enzymes through the Z-ring’s GTP hydrolysis–dependent treadmilling dynamics. In this role, FtsZ functions in cell division as the counterpart of the cell shape–determining actin homolog MreB in cell elongation.
Keywords: FtsZ, bacterial cell division, cell wall constriction, cytoskeleton dynamics, septum synthesis, single-molecule imaging
INTRODUCTION
Bacteria cells, with their diverse cell cycles and shapes, have fascinated scientists since they were first seen by Antonie van Leeuwenhoek under his microscope in the late 1600s. Early microscopic analyses of bacteria cell cycles and shapes, even at low resolutions, provided a wealth of information setting the stage for modern-day genetic and molecular studies.
The minimal shape determinant of a bacterium is its rigid peptidoglycan (PG) cell wall, which surrounds the inner membrane and protects the cell from osmotic lysis (88, 218, 229). This gigantic mesh-like molecule (termed the sacculus) is composed of glycan chains cross-linked by short peptides (230). An isolated sacculus is able to maintain the cell shape in the absence of any other cellular components (233). During the cell cycle, the sacculus must accomplish both the insertion of new cell wall material and the degradation of old material while maintaining its structural integrity and cell shape at all times (48, 49, 218) (Figure 1). Disrupting the sacculus structure, either by mis-synthesis or enzymatic degradation, ultimately leads to cell lysis. Not surprisingly, a large number of antibiotics target the synthesis and/or integrity of the sacculus (188, 192).
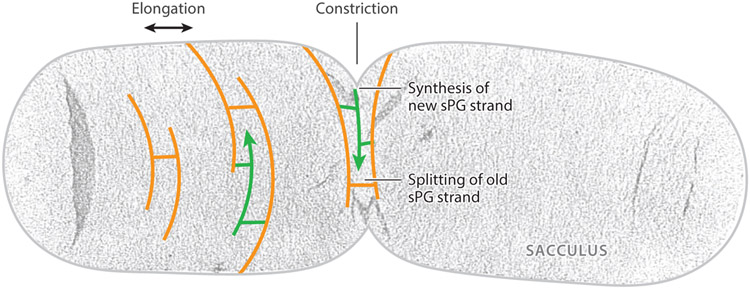
Figure 1.
Uranyl acetate–stained electron microscopy (EM) image of an isolated Escherichia coli sacculus with schematic drawing of the splitting of old (orange) and insertion of new (green) glycan strands in cell wall elongation and constriction. Figure adapted with permission from Reference 48. Abbreviation: sPG, septal peptidoglycan.
Successful bacterial cell division requires the synthesis of the septum and splitting of the existing sacculus, a process termed cell wall constriction. This process relies on the coordinated action of a multitude of septal cell wall enzymes and their regulators collectively. While many molecular players of this process have been identified, it remains unclear how various activities of these enzymes, often overlapping or redundant, are regulated and coordinated in time and space such that the cell wall constricts to produce two correctly shaped daughter cells without the danger of cell wall lesions or lysis.
We are still a long way from adequately answering this question, but it is clear from decades of research that a bacterial cell division protein, the tubulin homolog FtsZ, is at the center of this question. In this review, we attempt to illustrate what we know about FtsZ’s role in orchestrating the complicated task of bacterial cell wall constriction, with a focus on recent insights gained from high-resolution and high-sensitivity single-molecule imaging studies.
OVERVIEW OF FTSZ AND THE DIVISOME
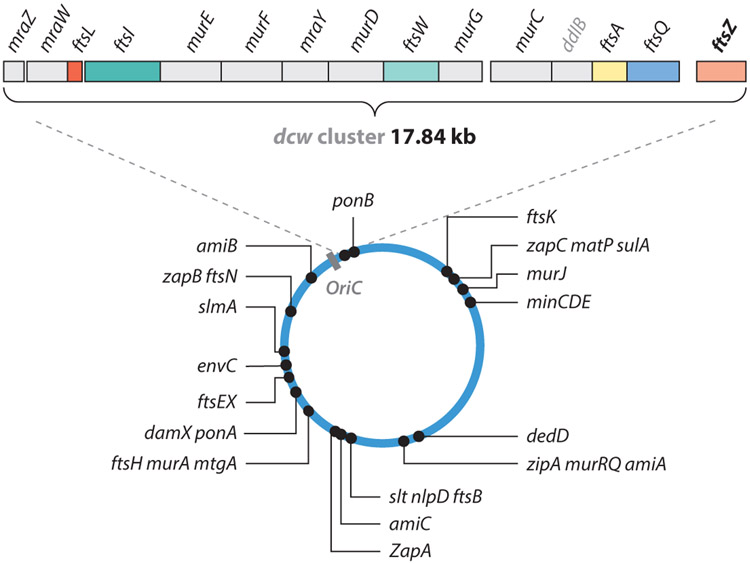
In 1968, Francis Jacob and colleagues (85) began probing Escherichia coli mutants that were able to divide at the permissive temperature of 30°C but not at 40°C. They identified that, in one mutant, PAT84, cells were able to segregate their newly replicated DNA but were unable to constrict the cell envelope at the incipient division site. The mutation in this strain was later mapped to the filamentous temperature sensitive gene Z (ftsZ) gene by Lutkenhaus and colleagues (17, 242). Interestingly, the ftsZ gene resides in the cell division and cell wall (dcw) cluster, which contains 12 cell wall biogenesis genes and four cell division genes that are likely transcribed as one single transcriptional unit (227) (Figure 2). The physical proximity of these genes within the cluster indicates that bacterial cell division is tightly coupled to cell wall constriction.
Figure 2.
Chromosomal locations and operon organizations of all divisome genes, including the dcw gene cluster, in Escherichia coli. Figure adapted with permission from Reference 56.
The FtsZ protein has been identified as a bacterial homolog of tubulin that hydrolyzes GTP and self-polymerizes in a GTP binding–dependent manner both in vitro and in vivo (25, 45, 58, 148, 155, 179). In most bacteria examined to date [except for Streptococcus pneumoniae (63)], FtsZ is the first protein to localize to the future division plane, where it polymerizes into a ring-like structure, termed the Z-ring. The Z-ring then recruits more than 30 proteins in a largely linear fashion to assemble into the macromolecular complex termed the divisome (5, 27, 70, 71). The fully assembled, or matured, divisome further forms the septal ring complex with other nonprotein components, including septal PG and inner membrane PG precursors, to carry out cytokinesis, which includes the complete segregation of the chromosome; inner membrane invagination; cell wall constriction; and, in gram negative bacteria, invagination of the outer membrane. As we show below, approximately 20 of the more than 30 divisome proteins are involved in septal PG (sPG) remodeling and regulation, suggesting again that the central task of bacterial cell division is cell wall constriction.
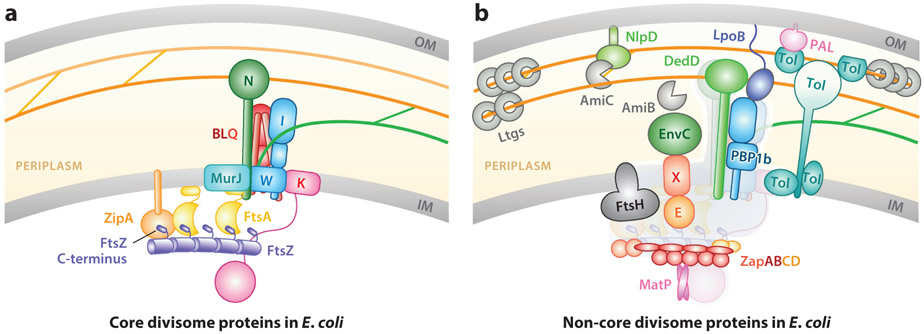
In E. coli, the divisome includes 10 core proteins (FtsA, B, I, K, L, N, Q, W, Z, and ZipA) (Figure 3a), which are essential for cell division and viability (55, 82). Among these core proteins are the division-specific sPG synthases: the PG glycosyltransferase (PGTase) FtsW (212a, 240) and transpeptidase (TPase) FtsI (153). The other core divisome proteins are involved in the assembly and stabilization of the Z-ring [FtsA (24, 172, 223) and ZipA (78, 147)], chromosome segregation [FtsK (14, 119)], or the regulation of sPG synthesis [FtsB, L, Q, and N (28, 118)]. MurJ, the conserved flippase for the PG synthesis precursor, lipid II (186, 193), has also recently been identified as a core divisome component in Staphylococcus aureus (145). The other more than 20 noncore proteins (Figure 3b) are individually dispensable for viability, but many play roles in important aspects of cell division (46, 127) such as Z-ring stability [Z-ring-associated proteins ZapA, B, C, and D (33, 53, 54, 76, 80, 178, 198, 235)], sPG enzyme activity control [FtsEX (141, 171, 191, 238), FtsH (156, 215, 216), LpoB (165, 219), NlpD (107, 217, 221), EnvC (220, 221), and DedD (117)], sPG synthesis and degradation [PBP1b (23, 39), AmiBC (84, 96, 226), and lytic transglycosylases (99, 231)], and outer membrane invagination [Tol-Pal (170)]. Both the core and noncore groups are critical for successful completion of normal cell division and correct cell pole shape morphogenesis (3, 46, 50, 127). Notably, all of these proteins’ abilities to assemble into the divisome [except SpMapZ (63)] are dependent on the localization of the Z-ring at the future division site (70, 71).
Figure 3.
(a) Core and (b) non-core proteins of the Escherichia coli divisome. Whenever possible, verified protein–protein, protein–membrane, and protein–sPG interactions are indicated by the spatial arrangement of respective components. Orange and green lines denote old and new sPG strands, respectively, as in Figure 1. In panel b, core proteins are colored gray in the background for simplicity. Abbreviations: IM, inner membrane; OM, outer membrane; sPG, septal peptidoglycan.
Z-RING STRUCTURE
Crucial to our understanding of the Z-ring’s function is a detailed knowledge of its structural organization in vivo. All known functions of FtsZ are fulfilled in its polymeric form (the Z-ring), and FtsZ can only hydrolyze GTP in this form, as the catalytic pocket is formed at the interface between two adjacent FtsZ monomers (158, 189). Additionally, all known protein and small-molecule antagonists of FtsZ [e.g., SulA (38, 95, 100), SlmA (16, 52), MinC (184), MciZ (19, 81), PC190723 (10, 83), and Compound 1 (206)] act to either promote or inhibit the polymerization of the Z-ring, highlighting the functional importance of FtsZ polymerization.
Z-Ring Structure Viewed by Conventional Imaging
The Z-ring is most certainly made up of a collection of FtsZ filaments, which could be individual single-stranded FtsZ protofilaments or units of multiple laterally associated protofilaments. Atomic structures of FtsZ monomers or dimers in complex with nucleotides from different bacterial species are available (111, 114, 138, 158, 159), but the in vivo arrangement of FtsZ protofilaments in the Z-ring remains elusive. In vitro structural studies showed that FtsZ protofilaments can adopt straight, curved, or circular conformation and can further form multistranded bundles, sheets, helices, and toroids depending on polymerization conditions such as GTP hydrolysis, metal ions, pH, and molecular crowding agents (57, 150, 182, 190). Such a high level of polymorphism is likely the key reason why it has been difficult to obtain a uniform polymeric species for crystallographic or electron microscopic studies.
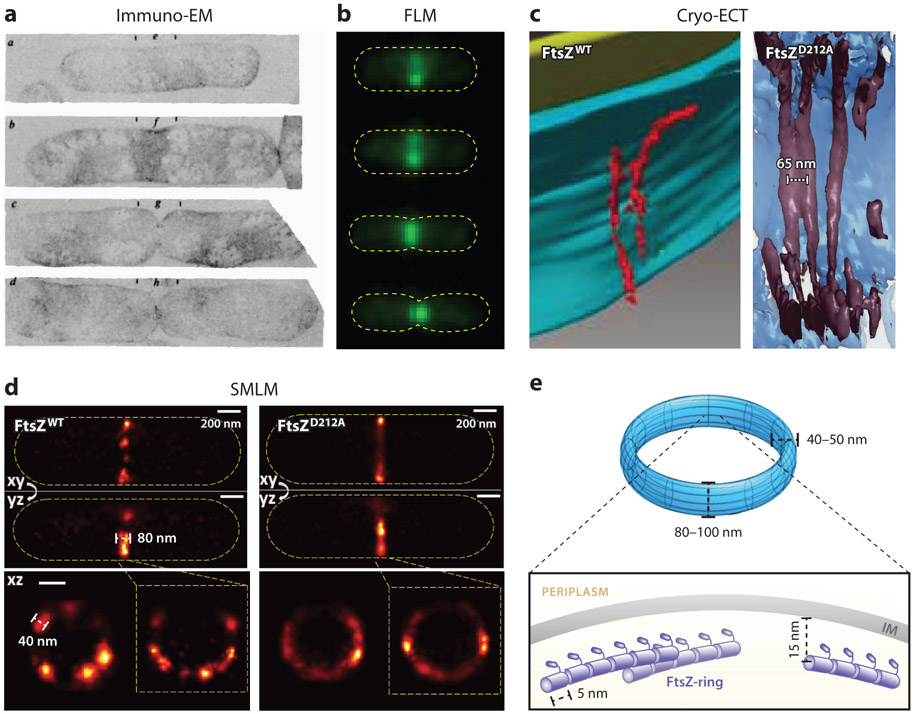
The first in vivo view of the ring-like arrangement of FtsZ was deduced by Bi & Lutkenhaus (18) in immunoelectron microscopy (ImmunoEM) sections of E. coli cells (Figure 4a). Using immuno-gold labeling, it was found that FtsZ always localizes to the invaginating division cleft (septum). Therefore, FtsZ must form a ring-like structure at the septum due to the symmetry of the rod-shaped cells, and the Z-ring must remodel itself over time to accommodate the inward growth of the septum. These observations were later confirmed and further complemented by fluorescence microscopy studies in which FtsZ was immunolabeled using dye-conjugated antibodies in fixed cells or tagged with a fluorescent protein (FP) in live cells (4, 113, 129) (Figure 4b). The smooth appearance of the Z-ring seen by epifluorescence microscopy led to the early proposal that the Z-ring could be composed of a single, long protofilament wrapped around the cell waist multiple times (61). However, calculations based on FtsZ’s GTP hydrolysis activity and the number of FtsZ monomers at midcell suggest that the Z-ring might instead be made of short overlapping and laterally interacting protofilaments (9, 61).
Figure 4.
Z-ring structure. (a) Immuno-gold EM images of constricting Escherichia coli cells show labeled FtsZ molecules (scattered black dots) at the leading edge of the invaginating septum (18). (b) Fluorescence images of FtsZ-GFP (green) in live E. coli cells outlined in yellow dashed lines (X. Yang, unpublished data). (c) Cryo-ECT images of FtsZ WT filaments in Caulobacter crescentus (left) (116) and reconstituted FtsZD212A mutant filaments in liposome (right) (212). (d) Three-dimensional fluorescence-based superresolution images of WT FtsZ-rings (left) and FtsZD212A mutant rings in E. coli (116). (e) Current model of the Z-ring depicting the spatial and dimensional features of the Z-ring (not to scale). The Z-ring is most likely composed of a single layer of short FtsZ protofilaments that randomly and heterogeneously associate underneath the inner membrane. Abbreviations: Cryo-ECT, cryo-electron tomography; EM, electron microscopy; FLM, fluorescence microscopy; GFP, green fluorescent protein; SMLM, single-molecule localization microscopy; WT, wild type.
These early pioneering works were among the first to demonstrate that bacterial proteins, like their eukaryotic counterparts, adopt specific subcellular localizations, and that their organizations are important for their functions. However, these early studies were unable provide detailed information on the spatial arrangement of FtsZ protofilaments in vivo, mainly due to the sparse labeling density in ImmunoEM and the diffraction limit of light microscopy experiments.
Z-Ring Structure by High-Resolution Imaging
In recent years, advances in new imaging technologies have pushed the detection limits of electron tomography and light microscopy to allow the construction of highly detailed molecular pictures of bacterial protein machineries both in vivo and in vitro (43, 143, 157, 236). These high-resolution images, coupled with genetic and biochemical studies, have proven powerful in dissecting the functions of the divisome in ways that were not possible before.
Cryo-electron tomography studies of Z-ring structure.
The first in vivo high-resolution structure of the Z-ring came from cryo-electron tomography (Cryo-ECT) imaging of Caulobacter crescentus (116). It was observed that individual, arc-like FtsZ filaments were positioned approximately 13 nm underneath the inner membrane at the site of midcell constriction (116) (Figure 4c). The filaments were approximately 40–160 nm long and approximately 5 nm in diameter, consistent with the dimensions of single FtsZ protofilaments based on previous calculations and in vitro EM studies (93, 183). Overexpression of wild-type (WT) FtsZ led to an increase in the number but not the length of filaments, whereas overexpression of a FtsZ GTPase mutant (FtsZG109S) led to a dramatic increase in the length of the arced filaments and bundling, presumably due to increased lateral interactions between filaments (116). These results demonstrate that the properties of FtsZ filaments inside the Z-ring are coupled to its FtsZ’s GTPase activity, similar to what has been seen in vitro (12, 144).
An interesting finding from this work is that, in all cells analyzed, there were only a few short, sparsely spaced FtsZ arcs and filaments and no complete Z-rings. Notably, the number of observed FtsZ filaments at midcell is significantly less than what would be expected from the expression level and midcell localization percentage of FtsZ in cells (102). It was proposed that, at any given time, only one or a few FtsZ protofilaments will come to the inner membrane and undergo a straight-to-bent conformation change to iteratively pinch the membrane to constrict the cell (116). An alternative explanation could be that FtsZ filaments in the Z-ring are not well organized or as well packed as what was seen for other cytoskeletons, such as ParA bundles (187). Therefore, most FtsZ filaments do not offer high enough contrast against the dense bacterial cytoplasm to be detected by Cryo-ECT. Nevertheless, this work was the first to suggest that that the Z-ring may not be a complete ring composed of long, continuous, and well-packed FtsZ filaments, as was previously proposed.
A later Cryo-ECT study using the same C. crescentus strain, however, observed long, continuous FtsZ filaments arranged in a single-layered, multistranded band with an interfilament distance of approximately 6–7 nm positioned approximately 15 nm underneath the inner membrane (212) (Figure 4c). A similar organization was seen in a skinny E. coli mutant strain, B/r H266 (212). The distance between FtsZ filaments and the inner membrane was demonstrated to be related to the length of FtsZ’s C-terminal linker and the size of FtsA: Extending the C-terminal linker length increased the distance to approximately 20 nm, whereas replacing FtsA with a direct membrane-targeting sequence (MTS) shortened the distance to approximately 10 nm (212).
The single-layered, multistranded configuration of the Z-ring proposed in this work is different from the short, scattered, sparsely arranged FtsZ filaments observed in the early Cryo-ECT study. If this proposal is true, then it would suggest that lateral interactions (direct or mediated by other factors) between FtsZ protofilaments play an important role in Z-ring organization. It was thought that the differences between the two studies could be due to new technical advances in Cryo-ECT imaging, and that the long, continuous FtsZ filaments observed in the later study could still be composed of multiple short, overlapping protofilaments due to the resolution limit. Other alternative sources of these differences could include but are not limited to cell cycle-dependent Z-ring reorganization, FtsZ expression level, GTPase activity, and cell growth conditions. For example, the long, multistranded configuration of FtsZ filaments was mainly observed in highly constricted, 2–3-fold WT FtsZ-overexpression cells, in GTPase mutant FtsZD212A overexpression cells, or in liposomes with reconstituted Z-rings using Thermotoga maritima FtsA and FtsZ (212).
Fluorescence superresolution studies of Z-ring structure.
In comparison to Cryo-ECT, fluorescence-based optical superresolution imaging has lower spatial resolution (approximately 20–50 nm) but offers advantages in specific labeling and the capacity for live-cell imaging (236). In particular, single-molecule localization microscopy (SMLM)-based superresolution imaging proves powerful for small bacterial cells due to its detection of single molecules, which allows quantitative characterizations of both the spatial features and molecular compositions of bacterial cellular structures (43, 236).
Early SMLM studies of the E. coli Z-ring revealed a discontinuous, heterogeneous organization of FtsZ clusters within the Z-ring (64) (Figure 4d). These FtsZ clusters, likely collections of FtsZ protofilaments unresolved at a spatial resolution of approximately 30 nm, are approximately200–800 nm in length, contain approximately 50–400 FtsZ monomers, and occupy a toroidal zone of approximately 80–100 nm in longitudinal width and approximately 40–60 nm in radial thickness at the midcell (31, 40, 64, 87, 128) (Figure 4e). The number of FtsZ protofilaments within the clusters likely ranges from a few to more than 10 (64) if the average length of a protofilament is assumed to be approximately 30 monomers long (9, 36, 205).
How FtsZ protofilaments associate with each other in the clusters remains unclear, but their association is likely mediated by protein factors instead of the intrinsic lateral affinity between FtsZ protofilaments (183). In vitro, it was shown that FtsZ protofilaments only form multistranded bundles in the presence of high concentrations of Ca2+ (51, 124), Mg2+ (37, 51), high-molecular-weight crowding agents (73, 174), or FtsZ bundling proteins (76, 80, 92,196, 198, 235). In vivo, the intracellular concentrations of Ca2+ [approximately 90 nM (67)] and Mg2+ [1–2 mM (7, 34)] are not high enough to promote FtsZ bundling, but the high level of molecular crowding [cytoplasmic protein concentration at approximately 200–300 g/L (34)], high concentrations of FtsZ bundling–promoting proteins [i.e., the Zaps, 2–10 μM (65, 235)], and the increased local concentration of FtsZ near the inner membrane due to membrane tethering could favor the lateral association of FtsZ protofilaments. In fact, removal of ZapA or ZapB, two Z-ring-associated proteins that promote the bundling of FtsZ protofilaments in vitro (65), leads to less coherent Z-rings made up of smaller, more dispersed FtsZ clusters (31, 235).
One interesting observation that emerged from these studies is that, upon overexpression of FtsZ (approximately 2–6-fold the WT level), the Z-ring width, thickness, and density (number of FtsZ molecules per occupied unit area) remain constant, while the occupied area within the Z-ring toroidal zone increases steadily and leads to smoother Z-rings (64, 128). These observations suggest that the Z-ring is not a tightly packed, regularly spaced polymeric structure, but instead is made up of heterogeneously organized, loosely associated FtsZ protofilaments that have large unoccupied spaces between them. If the same number of protofilaments (with a width of approximately 5 nm) were in tightly aligned clusters with no space in between, as was depicted by in vitro EM work, then the width of the corresponding FtsZ clusters and, consequently, the Z-ring would be as thin as approximately 30–50 nm, rather than approximately 80–100 nm, as was measured in vivo (64).
Although all of the fluorescence-based optical superresolution imaging studies of the Z-ring’s structure in vivo were consistent with each other, they were met with intense scrutiny. The central concern surrounding these studies is that, to generate a fluorescent signal, FtsZ must be tagged with a FP, which is prone to imaging artifacts and function disruption. However, to date, a fully functional FtsZ–FP fusion does not exist. A FtsZ–green FP (GFP) fusion in Bacillus subtilis (112) and a sandwich fusion of FtsZ–mNeonGreenSW in E. coli (146) have been shown to support WT-like cell growth and division as the sole cellular FtsZ copy, but the B. subtilis FtsZ–GFP fusion strain is temperature sensitive, and the FtsZ–mNeonGreenSW fusion in E. coli exhibits phenotypes similar to a mild FtsZ GTPase mutant (239). As such, almost all imaging studies were done in meriploid strains where the FtsZ–FP fusion was expressed exogenously at low levels to label the WT FtsZ-ring. A substantial amount of effort has been dedicated to ensuring that the information obtained from the partially labeled Z-ring represents the true underlying organization of the Z-ring. Specifically, the apparent heterogeneous morphology of the Z-ring has so far been observed with different FP tags, including Dendra (21, 87, 154, 235), mEos2 (40, 64), mEos3.2 (31, 128), Dronpa (31, 32), and PAmCherry (228); by different superresolution imaging techniques, including SMLM (21, 31, 32, 40, 64, 87, 128, 235), structured illumination microscopy (SIM) (185, 199, 204, 239), and stimulated emission depletion microscopy (98); by immuno-superresolution imaging targeting native FtsZ (31, 98, 185); by two-color immuno-superresolution imaging of the colocalization of FtsZ–GFP and native FtsZ clusters (40); by superresolution imaging of FtsZ-binding proteins (31, 185); and in both gram-negative and gram-positive bacteria, including E. coli (32, 40, 64, 128, 185, 199), B. subtilis (98, 204), C. crescentus (21, 87, 235), S. aureus (204), S. pneumoniae (97), and Laxus oneistus symbiont (167). In particular, nematode-associated L. oneistus symbiont γ-proteobacterial cells are large enough to resolve similar discontinuous, clustered distribution of the native Z-ring using ensemble epi-immuno-fluorescence (110, 167). These extensive characterizations demonstrate that the discontinuous, heterogeneous organization of the Z-ring is unlikely to be an imaging artifact, but rather reflects the true in vivo organization of the Z-ring. As we discuss below, such an organization turns out to be important for Z-ring function.
FtsZ’s GTPase activity modulates Z-ring structure.
What governs the Z-ring’s organization? Overexpression of FtsZ (approximately 10–15-fold WT level) in E. coli led to wider-than-WT Z-rings under ensemble epi-fluorescence imaging, but these wider Z-rings resolved into multiple thin bands resembling helical structures under superresolution imaging, and these thin bands have the same width and thickness as WT Z-rings (64, 128). Similar Z-ring dimensions were observed in E. coli cells where either a negative regulator (MinC) or positive regulators (ZapA, ZapB, or MatP) of Z-ring assembly were removed (31, 40). Furthermore, in other bacterial species where interacting partners and regulators of the Z-ring constitute distinct sets of proteins, Z-ring dimensions and organizations are also on par with what was measured in E. coli cells (21, 87, 97, 98, 204, 235). These observations suggest that the ability of FtsZ to assemble into a polymeric Z-ring with defined dimensions and organization appears to be independent of other proteins or the cell cycle and, instead, to be an intrinsic property of FtsZ’s polymerization.
The intrinsic property of FtsZ’s polymerization is most likely its GTPase activity. The polymerization of FtsZ is stimulated by nucleotide binding in a pocket formed at the monomer–monomer interface (158, 189), and stochastic hydrolysis of GTP leads to a high probability of the protofilament breaking at the site of hydrolysis (136), resulting in a discontinuous structure. Indeed, the Z-ring formed by a severe GTPase mutant D212A is much smoother and more homogenously distributed than WT Z-rings (128) (Figure 4d). This observation could be explained by the fact that a GTP binding–competent but hydrolysis-incompetent mutant (e.g., FtsZD212A) can form longer, more stable polymers than WT FtsZ (126). It is also consistent with the recent Cryo-ECT work, in which long, tightly aligned FtsZ filaments were mainly observed when the mutant FtsZD212A was overexpressed (212) (Figure 4c).
Single- versus multilayered arrangement of FtsZ filaments in the Z-ring.
One remaining question regarding the structural organization of the Z-ring is whether the Z-ring consists of a single layer or multiple layers of FtsZ filaments. It is unclear what biological implications such single- or multilayered filament arrangements might specify, but different arrangements could affect how FtsZ and its associated proteins interact with other periplasmic and cytoplasmic divisome proteins. Additionally, a single- or multilayered Z-ring may have different mechanical and/or dynamic properties that could impact the process of cell wall constriction.
To date, all available Cryo-ECT studies reported a single-layered configuration of FtsZ filaments in the Z-ring (26, 116, 212, 241). However, fluorescence-based superresolution imaging studies measured a radial thickness of the Z-ring at approximately 40–50 nm, thicker than what would be expected from a single layer of FtsZ filaments, even when the spatial resolution along the radial direction is taken into account [iPALM has a z-axis resolution of approximately 15–20 nm (40, 195)].
If the Z-ring were indeed multilayered, then FtsZ clusters observed by superresolution imaging would effectively be three-dimensional (3D) FtsZ bundles. FtsZ filaments facing the inner membrane would be anchored to the lipid bilayer by binding to its membrane tethers, and those inside and facing the cytoplasmic surface would associate with each other through the lateral interactions described above. With such an arrangement, one could reasonably expect that, when FtsZ is overexpressed, the bundle may gradually become thicker in all directions, as new FtsZ filaments could attach to the bundle from both the cytoplasmic and the membrane sides.
However, in all studies to date, when FtsZ was overexpressed, no increase in the Z-ring’s radial thickness was observed, even in the case of the FtsZD212A mutant (128). At the same overexpression levels (approximately 5–12-fold WT levels), ZD212A-rings are substantially wider (approximately 130–160 nm) compared to ZWT-rings, yet their radial thickness remains largely constant and similar to that of WT Z-rings (128). This observation suggests that the FtsZ filaments are preferentially assembled into the Z-ring along the membrane (via interaction with membrane tethers) rather than at the cytoplasmic side. This assembly preference would be most consistent with a single-layered arrangement of FtsZ filaments. A possible explanation of the observed thicker-than-a-single-layer arrangement of FtsZ filaments in fluorescence-based superresolution imaging could be that not all membrane-tethered FtsZ filaments are perfectly aligned in the same plane; instead, they have different curvatures and offsets from the membrane due to the stochastic GTP hydrolysis within the filaments and the flexible length of the disordered C-terminal linker of FtsZ (Figure 4e). Note that, if this were indeed the case, then such an irregular arrangement would also be difficult to detect by Cryo-ECT due to the low contrast that it would offer against the dense bacterial cytoplasm.
Current Z-Ring Model
In summary, existing high-resolution work suggests a common model for the in vivo structure of the Z-ring (Figure 4e). The Z-ring is composed of discontinuous, heterogeneously arranged FtsZ filaments confined in a toroidal zone of approximately 80–100 nm in width and positioned approximately 13–16 nm beneath the inner membrane. These FtsZ filaments are likely single-layered and loosely associated with each other via protein factors. Such an organization is largely independent of FtsZ’s cellular concentration or assembly regulators but is, instead, modulated by FtsZ’s GTPase activity, with FtsZ mutants with low GTPase activities forming more homogenously organized Z-rings.
Z-RING DYNAMICS
Assembly Dynamics
The Z-ring is not only heterogeneous, but also highly dynamic. Early fluorescent studies showed that nascent Z-rings in newborn E. coli cells exhibit rapid movement and oscillations in helix-like patterns prior to coalescing into stable assemblies at the midcell (207, 214). Similar dynamics were observed in B. subtilis cells during normal growth cycle and sporulation (113). These dynamics likely reflect the early polymerization and assembly dynamics of the Z-ring.
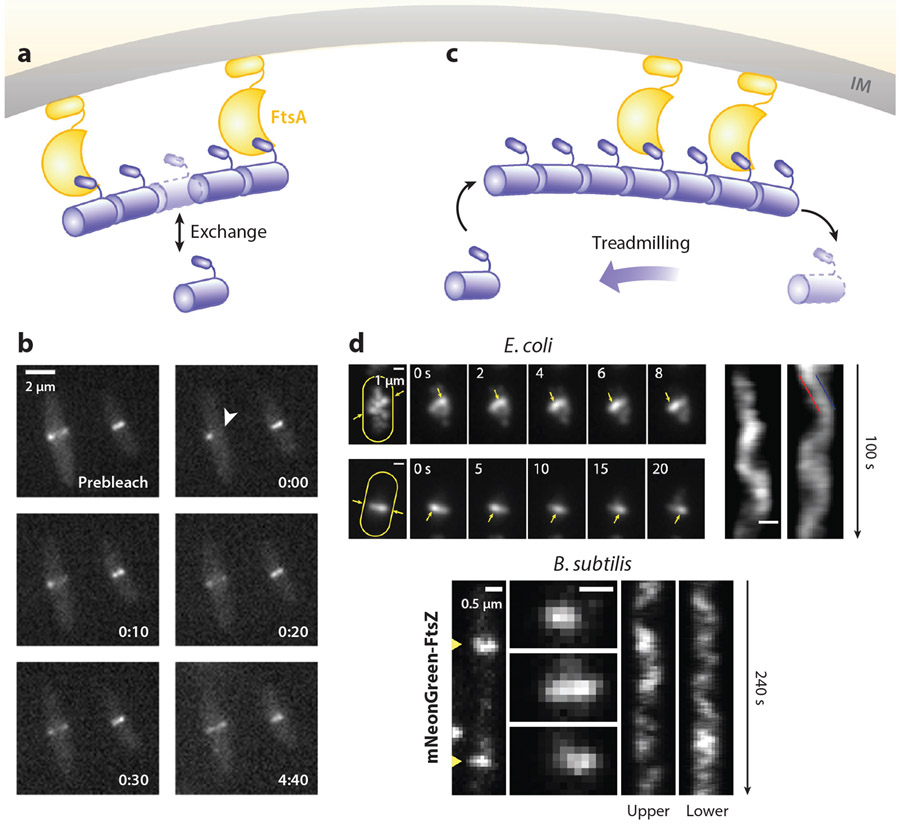
Subunit Exchange Dynamics
Even when it is stably assembled at the midcell, the Z-ring remains highly dynamic. Early in vivo fluorescence recovery after photobleaching (FRAP) experiments showed that the Z-ring undergoes rapid exchange with the cytoplasmic pool of FtsZ subunits on an approximately 10-s time scale (9, 205) (Figure 5a,b). This exchange rate is independent of the bacterial species examined (E. coli and B. subtilis), the presence or absence of Z-ring regulators (ZapA, EzrA, and MinC), and cell-cycle stage (early versus late constriction) but slows down approximately 10-fold when FtsZ’s GTPase activity is reduced to approximately 10% of the WT level using the FtsZG105S (FtsZ84) mutant. These dynamics are likely caused by the stochastic dissociation of FtsZ–GDP from the Z-ring and reassociation of cytoplasmic FtsZ–GTP. Specifically, upon GTP hydrolysis, FtsZ–GDP subunits in the Z-ring, if not directly attached to the membrane, may break the filament at the site of hydrolysis and dissociate into the cytoplasm, giving rise to the discontinuous, heterogeneous Z-ring structure discussed above. The reassociation of cytoplasmic FtsZ–GTP into the Z-ring maintains the Z-ring structure at a steady state. Interestingly, even with the fast exchange dynamics, single-molecule tracking (SMT) experiments revealed that individual FtsZ molecules within the Z-ring are stationary (31, 154), suggesting that the exchange dynamics do not involve direct movement of FtsZ monomers on the membrane.
Figure 5.
Z-ring dynamics. (a) Schematic of the subunit exchange dynamics of the Z-ring. (b) A FRAP imaging sequence showing the rapid recovery of fluorescence after the photobleaching of half of the Z-ring (white arrowhead) (205). (c) Schematic of the treadmilling dynamics of the Z-ring. (d) Treadmilling dynamics observed in Escherichia coli (top) (239) and Bacillus subtilis (bottom) (20). For E. coli, the maximum intensity projections (left), the montages (middle), and the corresponding kymographs (right) of Z-rings labeled with FtsZ–green fluorescent protein (GFP) in two cells are shown. For B. subtilis, two FtsZ–mNeonGreen-labeled Z-rings (left), montages at 8-s intervals (middle), and the corresponding kymographs (right) of two cells are shown. Abbreviations: FRAP, fluorescence recovery after photobleaching; IM, inner membrane.
Treadmilling Dynamics
Recently, treadmilling dynamics have been demonstrated in FtsZ polymers assembled in vitro on supported lipid bilayers via membrane tethers such as FtsA, ZipA, or an MTS (121, 177). Treadmilling is the apparent directional movement of a polymer by the continuous polymerization at the front of the polymer concurrent with depolymerization at the opposite end, while individual monomers in the polymer remain stationary (Figure 5c). In both E. coli and B. subtilis, it was also discovered that FtsZ clusters in the Z-ring exhibited apparent directional movement with a speed of approximately 25–30 nm/s at midcell (20, 239) (Figure 5d). This directional movement was independent of cell wall synthesis activity, Z-ring assembly regulators, and stabilizers but was tightly coupled to FtsZ’s GTPase activity (20, 239). Because individual FtsZ monomers are stationary in the Z-ring, and because the average speeds of the leading and trailing edges of moving FtsZ clusters are nearly identical, the directional movement of FtsZ clusters is most likely caused by treadmilling, as demonstrated in vitro. The treadmilling speed in vivo, however, is approximately 2–5-fold slower than what was observed in vitro, likely because the different labeling or membrane attachment schemes in vitro may affect FtsZ’s GTPase activity. Similar treadmilling dynamics in vivo were soon discovered in other bacteria such as S. aureus (145), S. pneumoniae (169), and Streptococcus mutans (115). Given the highly conserved GTPase activity of FtsZ, it is reasonable to expect that treadmilling Z-rings may be universal for all bacterial species. Note that the redistribution and reorganization dynamics of Z-rings in S. aureus and B. subtilis that were previously observed using SIM imaging are almost certainly the same treadmilling dynamics, although the spatial resolution of SIM imaging at that time was not high enough to resolve the directional treadmilling dynamics (185, 204).
FtsZ’s treadmilling dynamics must be related to subunit exchange dynamics because they both exhibit the same dependence on GTP hydrolysis. If one assumes that, on average, a single FtsZ protofilament treadmills at 25 nm/s, then the corresponding polymerization and depolymerization rates at the filament ends would be approximately 5 monomers per second [approximately 5 nm per monomer (123)]. This rate is approximately 50 times larger than what would be expected from the GTPase activity of FtsZ measured in vitro (approximately 0.1 s−1) (197). While it is possible that FtsZ’s GTPase activity in vivo could be higher than what is measured in vitro due to the presence of other regulator factors such as MinC or SlmA, the 10-s lifetime of individual FtsZ monomers in the Z-ring measured by FRAP and SMT is consistent with the 0.1 s−1 GTPase activity (i.e., every 10 s, a GTP molecule is hydrolyzed and, consequently, a FtsZ monomer dissociates; the dissociation and association rates should be equal on average to maintain a steady-state Z-ring). What could account for the discrepancy? One possibility is that the GTP hydrolysis activity may not be uniform along an FtsZ protofilament. The FtsZ subunits at the shrinking tip may hydrolyze GTP at a 50-fold faster rate than the ones in the middle; alternatively, the ones in the middle may hydrolyze GTP equally fast but not dissociate from the filament until the GDP-bound monomer moves to the tip. In such a scenario, one can calculate the average length of FtsZ filaments in vivo, which would be approximately 10 s × 25 nm/s = 250 nm. Further experiments and theoretical modeling are required to examine how the slow GTP hydrolysis rate measured in vitro gives rise to the fast treadmilling dynamics in vivo.
Z-RING FUNCTION
Z-Ring as a Divisome Scaffold
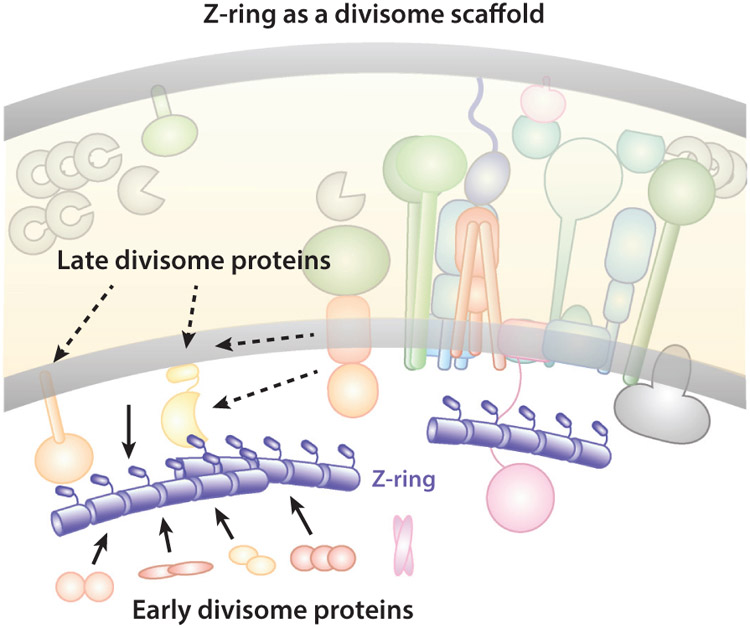
A well-acknowledged function of the Z-ring during cell division is that it acts as a scaffold for the assembly of all other division proteins into the divisome (Figure 6). FtsZ is the first protein (or among the first proteins) to localize to the future division site, and its localization is absolutely required for all of the other proteins to assemble into the divisome (70, 234). This scaffolding function is most likely achieved through the extensive protein–protein interaction network in the divisome (101, 104, 132, 166, 175). Protein–membrane and protein–PG interactions in the septal ring complex are certainly involved, too. In E. coli, direct interactions between FtsZ and the more than 30 divisome proteins mainly include proteins in the cytoplasm or at the inner membrane, such as Z-ring-associated proteins ZapA, B, C, D, and E (53, 54, 65, 76, 122, 135, 181); FtsZ’s membrane tethers FtsA (130, 173) and ZipA (78, 79, 120, 147); sPG remodeling regulators FtsQ (13, 175) and FtsE (44); and the N terminus of the chromosome segregation motor protein FtsK (15, 104). Most other periplasmic and outer membrane proteins, such as cell wall remodelers and their regulators, only make indirect contact with the Z-ring through other proteins. Additionally, the dynamic, discontinuous Z-ring organization makes it difficult to maintain stable protein–protein interactions. Therefore, it is possible that the scaffolding function of the Z-ring serves only to mark the division site for future septum synthesis, but not as a stabilizing force to hold all divisome proteins together.
Figure 6.
Schematic of the Z-ring functioning as a scaffold for the divisome assembly. For clarity, individual divisome proteins are not labeled, but instead are grouped according to their temporal assembly order (early or late). Most early divisome proteins reside in the cytoplasm or inner membrane and interact with FtsZ directly. Most late divisome proteins reside in the periplasm or outer membrane and interact with FtsZ indirectly. Additionally, some of the late divisome proteins’ location patterns do not exactly follow that of the Z-ring in mutants with perturbed Z-ring structures, indicating that the scaffolding function of the Z-ring is likely to mark the future division site and recruit other divisome proteins, but not necessarily to act as a scaffold for the stable assembly of the divisome.
Supporting this possibility, the assembly of the divisome is not instantaneous upon Z-ring localization, but rather temporally stepwise in E. coli (2), B. subtilis (66), and C. crescentus (72). Early division proteins localize to the future division site around the same time as the Z-ring and include mostly the ones interacting with the Z-ring directly. Late division proteins arrive approximately 10–20 min later. They are mainly periplasmic and outer membrane proteins that do not interact with the Z-ring directly, reflecting a protein-protein interaction relay of the divisome. Furthermore, in E. coli, overexpressing Z-ring antagonists such as MinC (22, 47), SulA (74, 95, 149), or SlmA (16) results in rapid disassembly of newly assembled Z-rings and slow dissociation of matured Z-rings (3, 100). Another study showed that, in ΔzapA or ΔzapB cells, where the Z-ring is destabilized and dispersed into multiple diffusive bands, the arrival times of all examined division proteins at the midcell were unaffected, but their localization patterns were clearly different (31). The early division protein ZipA followed the same diffusive, multibanded pattern as the destabilized Z-ring, but the late division proteins FtsK and FtsI still formed sharp single bands at the midcell (31). These observations suggest that late divisome proteins’ localization is not determined by the Z-ring scaffold. Indeed, recent superresolution imaging studies showed that, at least in E. coli, the late divisome protein FtsN (118) does not colocalize with the Z-ring, but instead gradually forms a separate N-ring approximately 40 nm away from the Z-ring in deeply constricted cells (199). Finally, the Z-ring was found to dissociate prior to the close of the inner membrane and the disassembly of all late division proteins (149, 201). These results are consistent with a limited scaffolding function of the Z-ring.
Z-Ring as a Cytokinesis Coordinator
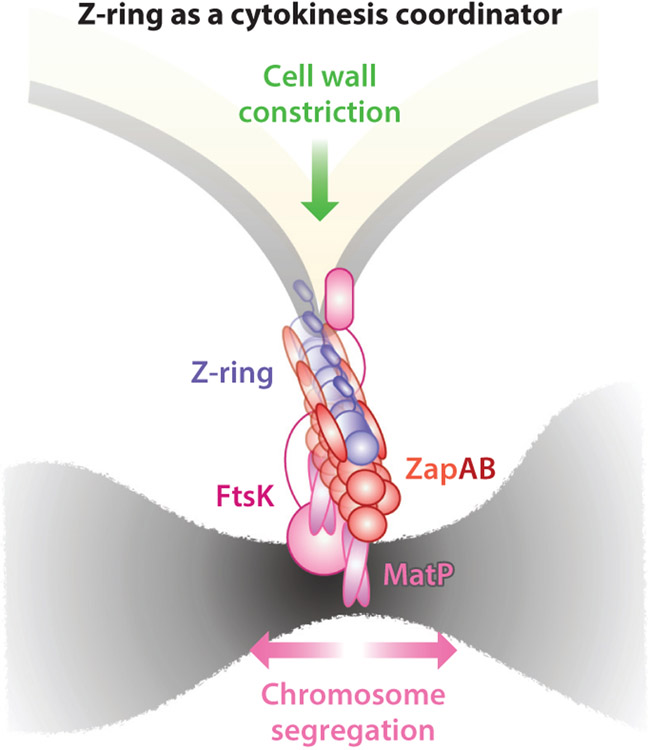
As discussed above, the Z-ring is positioned beneath the inner membrane and above the nucleoid. This subcellular location and the extensive divisome protein interaction network place the Z-ring at the prime position to bridge (or communicate with) divisome components from the cell envelope to the nucleoid and vice versa. In E. coli and C. crescentus, ZapA and ZapB in the cytoplasm stabilize the Z-ring by corralling heterogeneously distributed FtsZ clusters into the midcell assembly zone (31, 32, 235). Interestingly, in E. coli, ZapB binds not only to FtsZ [through ZapA (65)], but also to a DNA-binding protein, MatP (62). MatP binds to an array of matS sequences near the ter region of the chromosome and is involved in ter macrodomain condensation and segregation (32, 62, 134, 142). It was found that removing any one of the three proteins (ZapA, ZapB, or MatP) leads to the dispersion of the Z-ring into smaller FtsZ clusters occupying a wider region at the midcell (31). As revealed by 3D superresolution imaging, ZapA resides at similar level as the Z-ring, approximately 13 nm away from the inner membrane; ZapB forms large, contiguous polymeric structures approximately 40 nm beneath the Z-ring in the cytoplasm; and MatP forms one or two punctate clusters that are further displaced approximately 30 nm beneath ZapB (31). With these molecular-scale distances, a physical linkage of ZapA–ZapB–MatP could form and connect the Z-ring to the chromosomal ter macrodomain (Figure 7). This linkage could help stabilize the highly dynamic, discontinuous Z-ring at midcell, likely by anchoring the Z-ring to the stable nucleoid.
Figure 7.
Schematic of the Z-ring functioning as a cytokinesis coordinator. The Z-ring is sandwiched between the inner membrane and chromosome: FtsZ is attached to the inner membrane by FtsA, which makes further contacts with cell wall remodeling enzymes and regulators, and also attached to the chromosome by the ZapA–ZapB–MatP–DNA linkage. The presence of the linkage could coordinate the progression of cell wall constriction and chromosome segregation either mechanically or biochemically.
One interesting aspect of the presence of the membrane–FtsZ–ZapA–ZapB–MatP–DNA linkage is that it could present an elegant mechanism to coordinate the progression of cell wall constriction with chromosome segregation, either mechanically or biochemically. The Z-ring may act as sensor (or perhaps a brake) that relays the status of chromosome segregation and cell wall constriction between their respective machineries (31, 40, 42, 62). Supporting evidence for this idea came from recent studies showing that ΔmatP cells constrict at a faster rate than WT cells (32, 40) (Figure 8c). In the absence of the linkage, there is no coordination (or braking) between cell wall constriction and chromosome segregation; thus, new septum synthesis is able to proceed as fast as the corresponding enzymatic activities allow, and cells constrict over unsegregated chromosomes (133). Additionally, faster constriction under the condition of destabilized Z-rings also indicates that a stable Z-ring may not be essential for and could even hinder cell wall constriction, an important point on which we further elaborate below.
Figure 8.
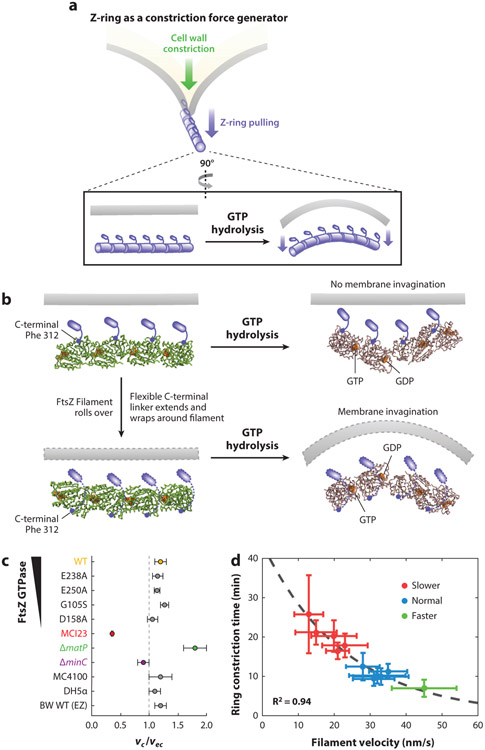
(a) Schematics of the Z-ring functioning as a constriction force generator through GTP hydrolysis-induced filament bending. (b) A recent crystallographic study (114) shows that GTP hydrolysis causes the FtsZ protofilament to bend, but the bending is toward the membrane (top) if the C-terminal tail of FtsZ (small blue rod) is directly attached to the membrane, which is opposite the direction of membrane invagination. To accommodate the bending conformation, the flexible linker between the globular domain and C-terminal tail of FtsZ has to wrap around the filament to attach to the membrane (dotted outlines, bottom). Note that the models shown are for visualization purposes only and are not based on the molecular dynamics simulations done in Reference 114. (c) Growth condition–normalized septum closure rate measurements made in wild-type (WT) BW25113 Escherichia coli cells in minimal media compared with different background strains including mutants affecting FtsZ’s GTPase activity (E238A, E250A, G105S and D158A), FtsI’s activity (MC123), chromosome segregation (ΔmatP), and Z-ring stability (ΔminC), in addition to different WT strain backgrounds (MC4100, DH5α) and WT BW25113 cells growing in rich defined growth medium (EZ). The FtsI mutant constricts significantly slower than WT cells, ΔmatP constricts faster, and all other perturbations of the Z-ring did not produce significant changes in septum closure rates (40, 239) (d) Constriction time measured in a series of Bacillus subtilis strains showed a high correlation with Z-ring treadmilling speed (20).
Note that, in addition to the Z-ring, ZapA and FtsK could also serve as coordinators between cell wall constriction and chromosome segregation. ZapA intriguingly interacts with a large number of divisome inner membrane proteins that do not interact directly with FtsZ, including FtsQ, FtsL, FtsB, FtsW, and FtsN (8, 132). These proteins are core divisome proteins involved in septum synthesis. FtsK interacts with FtsL, FtsQ, FtsI, and FtsN through its N-terminal transmembrane domain and with the chromosome through its C-terminal DNA motor domain (75). A recent study found that the ordered segregation of sister chromosomes by FtsK requires the presence of MatP, suggesting that these two proteins may indeed coordinate with each other (203).
Z-Ring as a Constriction Force Generator
Perhaps the most hotly debated function of the Z-ring in the past decade is whether it generates a mechanical force to drive the invagination of the inner membrane during cytokinesis (60, 61, 200, 237) (Figure 8a). One could easily rationalize that the Z-ring may convert the chemical energy released by GTP hydrolysis into mechanical energy to drive membrane constriction. Can FtsZ filaments indeed generate a mechanical force? Several in vitro liposome reconstitution studies have provided convincing evidence that this is the case. Specifically, when attached to membrane using a MTS, MTS–FtsZ was found to polymerize into circular or helical structures in liposomes and deform the liposome in a variety of geometries. In some cases, these filamentous MTS–FtsZ structures are also able to constrict the liposomes completely (160-163).
How might FtsZ filaments generate a force?
How FtsZ filaments would generate the mechanical force to deform or constrict liposomes is unclear. Three basic mechanisms could be at play: (a) the intrinsic persistence length or bending rigidity of FtsZ filaments, (b) GTP hydrolysis–induced filament conformational change (41, 57, 59-61, 125, 161, 183), and (c) FtsZ polymerization dynamics. Other, more complex mechanisms stemming from the basic three have also been proposed, such as immediate reannealing of FtsZ protofilaments upon GTP hydrolysis–induced subunit loss (211), condensation of FtsZ protofilaments caused by lateral affinity (105), sliding of FtsZ protofilaments by continuous polymerization or depolymerization (212), or combinations of these mechanisms (69, 89, 164, 211). For excellent reviews on different force generation mechanisms, the readers are referred to References 59, 61, and 137.
In the first basic mechanism, any polymer with a given persistence length will be able to deform or bend an attached membrane if the membrane is softer than the polymer. In relation to FtsZ, this mechanism could be independent of GTP hydrolysis, as formation of the polymeric Z-ring only requires the binding of GTP by FtsZ. Consistent with this idea, in vitro liposome reconstitution experiments have shown that purified FtsZ can induce visible liposome constrictions that are independent of GTP hydrolysis, as Z-rings assembled in the presence of a slowly hydrolyzable GTP analog, GMPCPP, deformed liposomes in a similar manner as those formed with GTP (160). In another reconstitution experiment, the persistence length of FtsZ filaments (likely FtsZ bundles made of multiple protofilaments) was estimated to be approximately 0.7–1.4 μm based on the bending modulus of the attached soft lipid tubes (176). This persistence length is orders of magnitude smaller than that of microtubules [approximately 1.5 mm (176)] or actin filaments [approximately 20 μm (176)] and leads to softer FtsZ filaments [Young’s modulus of approximately 8–16 MPa (105)] compared to the E. coli membrane and cell wall [Young’s modulus of approximately 20–25 MPa (224)]. Therefore, although it appears that FtsZ filaments could indeed deform and constrict membranes in vitro, it is unlikely that, in vivo, the rigidity of FtsZ filaments will be able to exert a sufficient force to deform the membrane and cell wall to the extent of driving cell wall constriction.
The second mechanism relies on a straight-to-curved conformational change of FtsZ filaments upon GTP hydrolysis (Figure 8a). The straight-to-curved conformation change has been proposed for some years; both straight and curved FtsZ filaments have been observed in vivo by Cryo-ECT studies (116, 212, 241) and in vitro by EM studies (94, 125, 183). However, the curved or bent confirmation of a FtsZ dimer was only recently observed at the atomic level (114, 139). Li et al. (114) found that a GDP-bound Mycobacterium tuberculosis FtsZ protofilament exhibits a curved conformation, in contrast to the straight conformation that was invariably observed in nearly all previous FtsZ structures from other bacterial species (108, 111, 139, 158, 180, 213). Surprisingly, the GDP-bound open conformation causes the C-terminal surface of the Mtb FtsZ protofilament to bend toward but not away from the membrane (Figure 8b). Such a conformation is at odds with the known membrane-facing geometry of the C terminus of FtsZ filaments (90, 161). An early study has convincingly demonstrated that, when an MTS is placed at FtsZ’s C terminus, the filaments cause concaved depressions on liposomes; conversely, when the MTS is switched to the N terminus, only concaved bulges are observed (161). It was proposed that the flexible, disordered C-terminal linker (29, 30, 68) could possibly wrap around the filament to allow the C-terminal tail to reach over to the membrane so that the both the N and C termini of FtsZ protofilament would face the membrane (Figure 8b). It is unclear what would prevent the protofilament from simply rolling over and adopting the opposite curvature toward the membrane.
Finally, recent computational simulations suggest that, in order for the filament-bending model to work, one critical requirement must be satisfied: The linker between FtsZ and the membrane should be rigid enough to prevent the filament from rolling over to curve on the surface of the membrane instead of bending away from the membrane (152). As such, while it remains a formal possibility that FtsZ protofilaments exhibit GTP hydrolysis–dependent conformational change, such a change may not necessarily be responsible for membrane invagination in vivo.
In the third basic mechanism, treadmilling dynamics (i.e., the continuous polymerization at one end and depolymerization at the other end) of FtsZ filaments could also be responsible for generating a constrictive force. In a recent in vitro work, soft lipid tubes were pulled from giant FtsZ-decorated lipid vesicles by optical tweezers (177). As FtsZ filaments treadmilled on the pulled tubes, dynamic helical deformations of the tubes were observed. The helical deformations were interpreted as the lipid tube (modeled as an elastic rod) being twisted by a constant force generated by the treadmilling FtsZ filaments. A FtsZ GTPase mutant that is able to assemble into ring-like structures but unable to treadmill also caused a helical twist of the pulled tubes; however, the development of the twist was delayed, and the pitches of the twists were much larger than those caused by WT FtsZ. The difference in the helical compression of the lipid tubes between the WT and mutant FtsZ was used to deduce that the treadmilling activity of FtsZ could indeed generate a torsional force on the order of 1–2 pN (contingent on the size of the treadmilling FtsZ filament). This force is small compared to what is thought to be required to deform the inner membrane or cell wall of bacterial cells (on the order of nN) (106, 237). Note that the force generated by treadmilling dynamics is not dependent on, but rather is only enhanced by, GTP hydrolysis. Further theoretical work needs to be developed to explain the mechanism by which treadmilling could generate a mechanical force perpendicular to the filament’s travel direction to invaginate the inner membrane during cytokinesis.
Is a force required for cell wall constriction?
Regardless of what mechanism might govern FtsZ’s force generation, an important question to ask is whether such a force is required for cell wall constriction in vivo. Recent computational simulation work suggests that a mechanical constrictive force might be necessary to bias the synthesis of new septum toward the inner face of the cell at the site of the septum (151). In this work, Nguyen et al. (151) showed that, in order for the septal cell wall to constrict (i.e., reduce in diameter), a newly inserted sPG hoop must be smaller than the previous one. This requirement could be satisfied if a sufficient force against the turgor pressure is available to pull (or push) the newly synthesized sPG strand inward to skip one potential cross-linking site on the old sPG strand and cross-link with the next available site instead. Thus, the number of cross-links made in the new hoop of sPG strands will be less than the previous one, and in turn, the septum diameter will reduce gradually as more new sPG strands are inserted. The same work also suggests that septal cell wall growth in the absence of such a biasing force would not lead to constriction, but instead only to cell elongation, as newly inserted PG strands will match one-to-one with the existing strands without tightening them. The estimated force required for this mechanism to work is approximately 15 nN (151); such a force could theoretically be generated by the Z-ring if 100% of the energy released by GTP were harnessed for this purpose (91, 237).
In other works, it was suggested that the periplasm and cytoplasm of gram-negative bacteria may be isosmotic, and therefore there is no need to have a force to overcome the turgor pressure at the inner membrane (35, 60, 163, 202). Supporting this hypothesis, there are Cryo-ECT images showing that the inner membrane is far ahead of the cell wall and outer membrane during cell division (11). In such a scenario, the Z-ring does not need to rely on a force generation mechanism to invaginate the inner membrane (99a). However, it is commonly acknowledged that, in gram-negative bacteria such as E. coli and C. crescentus, the invagination of the three layers of cell envelope (inner membrane, cell wall, and outer membrane) is concurrent at all constriction stages with no apparent changes in the distances in between (35, 202). It has also been pointed out that, even though the periplasm and cytoplasm may be isosmotic, upon cell wall constriction, the volumes of both periplasm and cytoplasm will be reduced (131). Therefore, the Z-ring (or some other components of the divisome) will still need to overcome turgor pressure at the constriction site to invaginate the inner membrane (131).
Does a force from the Z-ring drive cell wall constriction?
Finally, while plenty of evidence has demonstrated that FtsZ filaments can indeed generate a constrictive force through a variety of potential mechanisms, it is unknown whether such a force is relevant in vivo, or if it is sufficient to drive membrane invagination and/or cell wall constriction. In two comprehensive investigations of whether the Z-ring generates a constrictive force to limit the progression of cell wall constriction, Coltharp et al. (40) and Yang et al. (239) found that the septum closure rate (i.e., or how fast a cell constricts) was insensitive to substantial changes in all Z-ring properties proposed to generate a constrictive force (Figure 8c). These perturbations include changes in FtsZ’s GTPase activity, Z-ring density, and the timing of Z-ring assembly and disassembly. Instead, the constriction rate was limited by the activity of FtsI, the essential sPG transpeptidase. There is also no correlation between FtsZ’s treadmilling dynamics and the corresponding septum closure rate in E. coli (239). Consistent with these observations, the septum synthesis activity, estimated by the incorporation of fluorescent D-Ala-D-Ala analogs, remained the same across all of the FtsZ mutant backgrounds (239).
These results, while surprising in the context of the Z-ring-centric force generation model, are consistent with known observations that severe FtsZ GTPase mutants, such as FtsZ84 and D212A in E. coli, are viable and grow with minimal cell division defects at permissive temperatures (86). This suggests that, even though the Z-ring may indeed generate a constrictive force in vivo, this force is not dependent on GTP hydrolysis, nor is it limiting for the cell wall constriction rate. Instead, the septum cell wall synthesis rate, perhaps limited by available precursor levels (11), sets the limit for how fast the cell wall constricts (40). Interestingly, in B. subtilis, the opposite was observed: The faster FtsZ treadmills, the faster a cell constricts (20) (Figure 8d). Thus, in gram-positive bacteria, cell wall constriction may not be limited by cell wall precursor levels, but instead by FtsZ dynamics or the corresponding force generation. It has yet to be determined whether placing E. coli cells under rich growth conditions, where cell wall precursors are unlimited, or placing B. subtilis cells under poor growth conditions, where cell wall precursors are limited, would lead to opposite results. Additionally, a new theoretical study suggests that, depending on the diffusive dynamics of cell wall synthesis enzymes in different bacterial species, the same treadmilling dynamics of FtsZ could produce differential effects in modulating the level of enzymes available for cell wall constriction, thus altering the cell wall constriction rate (140). We further elaborate on this point in the next section.
Z-ring as a Cell Pole Shape Determinant
While many studies in the past decade have focused on the force generation function of the Z-ring, it is important to emphasize that there is a long history of studies documenting the significant involvement of the Z-ring in septal cell wall synthesis and cell pole morphogenesis. Early fluorescence microscopy and EM studies in E. coli found that the shape of FtsZ filaments determines the shape of the constricting septum: Cells with WT Z-rings produced smooth, symmetric septa, while cells with arcs or spirals of mutant FtsZ filaments displayed asymmetric, incomplete, or twisted septa (6). This role is attributed to the ability of the Z-ring to recruit all of the essential and nonessential septal cell wall remodelers and localize them to the division plane (46, 77, 127). Indeed, it was subsequently observed that new sPG is only inserted into the cell wall where FtsZ polymerizes in E. coli (225), C. crescentus (1), and S. aureus (168). It is possible that the major function of the Z-ring in bacterial cell division is not to divide the cell but to govern the cell pole morphogenesis through its participation in septal cell wall synthesis. In fact, in wall-less L-form bacteria, FtsZ becomes dispensable and is no longer required for cell proliferation or division (109).
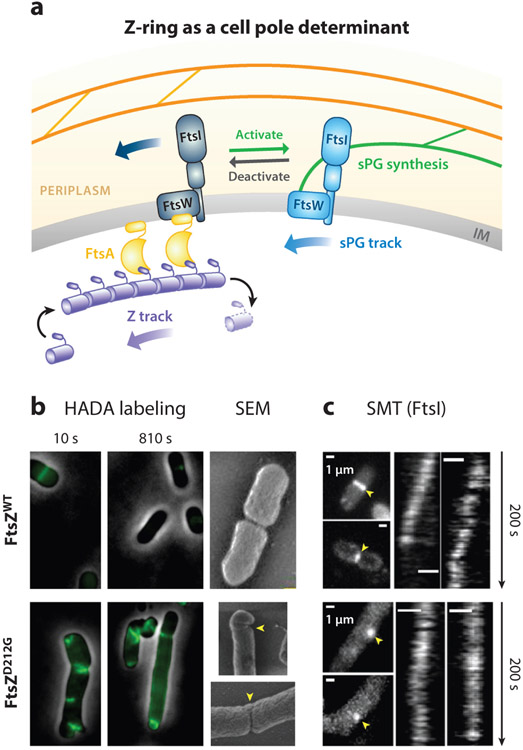
How does the Z-ring function as a cell pole shape determinant? Given our current understanding of Z-ring structure and dynamics, the mechanism of determining septum shape simply by recruitment and localization does not appear to suffice. For example, if new sPG were only inserted where FtsZ clusters are located, one should expect that the septa of the GTPase mutant FtsZD212A cells should be much smoother and more symmetric than those of WT cells because the mutant ZD212A-rings are much more continuous and homogenous than the WT Z-rings (128) (Figure 4d). However, the opposite was observed experimentally (239) (Figure 9).
Figure 9.
(a) Schematic of the Z-ring functioning as a cell pole shape determinant. The directional treadmilling dynamics of the Z-ring provides a spatial cue to attract sPG synthase molecules to follow the directional movement, thus distributing them evenly along the septum. (b) HADA labeling of Escherichia coli septum synthesis showed that, at a short time scale (10 s), both WT and the D212G mutant had punctate incorporation. At a long time scale (810 s), WT cells showed homogenous incorporation of HADA intensity, corresponding to smooth septum in the scanning electron microscopy image on the right, while the D212G mutant showed incomplete and asymmetric HADA incorporation and deformed septa (yellow arrowheads) (239). (c) SMT of FtsI molecules with maximal intensity projections (left, yellow arrowheads) and corresponding kymographs (right) in the WT (top) and D212G (bottom) FtsZ mutant E. coli cells showed correlated directional movement with FtsZ’s treadmilling speeds (239). Abbreviation: SMT, single-molecule tracking; sPG, septal peptidoglycan; WT, wild-type.
Another example is that, in C. crescentus, disrupting the C-terminal linker between FtsZ’s GTPase domain and extreme C-terminal tail caused a cell wall bulging and lysis phenotype at future division sites (209). The cell wall composition was also altered—there was lower cross-linking and longer glycan strands in the mutant cells. Nevertheless, the recruitment and localization patterns of many divisome proteins remained the same. These proteins include Z-ring-associated proteins, proteins involved in PG precursor synthesis, and proteins involved in sPG synthesis and regulation (209). It was later found that FtsZ’s linker modulates filament structure and dynamics in vitro (208, 210), suggesting FtsZ’s dynamics are important for proper septal cell wall remodeling and cell pole shape morphogenesis.
The discovery of Z-ring’s treadmilling dynamics provides an important clue as to how the Z-ring functions to govern septal morphogenesis. It is possible that the dynamic remodeling of the Z-ring, rather than its localization pattern, is required for a symmetric, smooth cell wall constriction. The treadmilling dynamics would allow the Z-ring to sample the surface of the growing septum evenly over time, thereby ensuring a uniform distribution of sPG synthesis along the septum (Figure 9a). Specifically, because each round of FtsZ filament treadmilling along the septum only takes approximately 100 s (assuming a 3 μm circumference), whereas the constriction period is usually orders of magnitude longer (>10 min under most lab growth conditions), the time scale separation allows treadmilling Z-rings to average out any possible stochastic fluctuations in the septal distribution of sPG enzymes to ensure an even synthesis of the septum from all around.
Supporting this hypothesis, when new sPG synthesis was pulse-labeled using fluorescent D-amino acid analogues (FDAAs) (103) in E. coli (Figure 9b), WT cells showed dot-like punctate incorporation of FDAAs at very short time scales (<10 s) but homogenous incorporation at long time scales (>100 s) (239). In contrast, FtsZ GTPase mutant cells showed only partial, incomplete FDAA incorporation along the septa even at very long time scales (>800 s), indicating that although the structure of the mutant Z-rings are more continuous and homogenous, the lack of treadmilling dynamics renders an uneven distribution of sPG synthesis activity, which gives rise to the abnormal septum morphology (239).
Most excitingly, SMT found that individual septum-specific cell wall enzyme molecules, such as TPases [PBP2b in B. subtilis (20) and PBP3 in E. coli (239)] and PGTase [FtsW in E. coli (241)], move directionally along the septum, and that their movements were driven by FtsZ’s treadmilling dynamics (Figure 9c). How stationary FtsZ monomers in treadmilling FtsZ polymers in the cytoplasm drive the directional movement of single sPG enzyme molecules in the periplasm is unknown. However, a recent study indicated that a Brownian rachet model could be at play (13, 140). In this model, sPG enzyme molecules continuously track with the shrinking end of a treadmilling FtsZ filament via local diffusion coupled with repeated binding and unbinding (140). As such, while bacterial cells do not have linear motors akin to kinesin or myosin, FtsZ could use GTP hydrolysis–driven treadmilling to function as a motor and deliver cargo (sPG enzyme molecules) directionally along the septum.
It is important to note that, although the influence of FtsZ’s treadmilling dynamics on total sPG synthesis activity is different between B. subtilis [faster Z-ring treadmilling speed corresponds to higher total sPG synthesis activity (20) (Figure 8d)] and other bacteria [total sPG synthesis activity independent of FtsZ’s treadmilling speed in E. coli, S. pneumoniae, and S. aureus (20, 145, 169, 239) (Figure 8c)], it is difficult to envision a molecular mechanism in which, at the single-molecule level, a sPG synthase molecule would gain higher sPG synthesis activity if it moves faster on the FtsZ’s treadmilling track.
In fact, a new study in E. coli found that the population of FtsW molecules driven by FtsZ’s treadmilling dynamics is not active in sPG synthesis (240). This study observed that two populations of processively moving FtsW molecules exist at the septum (240). A fast-moving population of FtsW is driven by the treadmilling dynamics of FtsZ and independent of sPG synthesis, as previously shown on FtsI in E. coli (239, 240). A slow-moving population of FtsW is driven by active sPG synthesis and independent of FtsZ treadmilling dynamics (240). In other words, FtsZ’s treadmilling dynamics only influence the speed of the fast-moving, inactive population but not that of the slow-moving, active population of FtsW in E. coli. Furthermore, FtsN, a late divisome protein and potential sPG synthesis activator, was found to promote the slow-moving, sPG synthesis–dependent population. Based on these results, a two-track model was proposed (Figure 9a). Inactive sPG synthase molecules follow the fast treadmilling Z-track to be distributed along the septum; FtsN promotes their release from the Z-track to become active in sPG synthesis on the slow sPG-track (Figure 9a). This model integrates spatial information into the regulation of sPG synthesis activity and could serve as a mechanism for the spatiotemporal coordination of bacterial cell wall constriction.
Supporting the existence of a FtsZ-independent sPG-track, in S. pneumoniae, the septum-specific TPase, PBP2x, and FtsW were also found to move directionally, but their moving speeds were independent of FtsZ treadmilling and only dependent on active sPG synthesis (169). In S. aureus, septum constriction continues even in the absence of FtsZ treadmilling in highly constricted cells (145), indicating that FtsZ’s treadmilling dynamics are no longer necessary once cells pass the initial constriction stage.
Such a two-track model (240), coupled with the Brownian rachet model described above (140), could also potentially explain the differential dependence of total sPG synthesis activity on FtsZ’s treadmilling speed in different bacterial species. A recent theoretical study shows that, when FtsZ treadmills too fast, a sPG enzyme molecule will not be able to keep up with the treadmilling FtsZ polymer if it diffuses slowly between consecutive end-tracking steps. Therefore, the faster FtsZ treadmills, the more sPG enzyme molecules will be released from the Z-track to become available for active sPG synthesis, increasing the total sPG synthesis activity to that observed in B. subtilis (20). If sPG enzyme molecules diffuse relatively fast, so that they can always keep persistent end-tracking of treadmilling FtsZ polymers (as in E. coli, where FtsW and FtsI molecules diffuse approximately 10-fold faster than their counterparts in B. subtilis), or extremely slow to become completely independent of FtsZ’s treadmilling, as in S. pneumoniae and S. aureus, then the total sPG synthesis activity would be insensitive to FtsZ’s treadmilling speed. In the latter scenario, Z-ring treadmilling could perhaps function to distribute other sPG regulators that indirectly influence the spatial distribution of sPG synthases.
Taken together, these newer studies suggest that the Z-ring mainly works as a shuttle to transport sPG enzymes to different sites along the septum but does not dictate how fast these enzymes work. It is possible that, in this role, FtsZ in cell division governs the cell pole shape morphogenesis, as MreB does in cell elongation: MreB is an actin homolog (222) that determines the rod-like cell shape (194, 232); it guides the directional movement of the Rod system for lateral cell wall synthesis during cell elongation to maintain the rod cell shape (194).
CONCLUSIONS AND OUTLOOK
Since the discovery of FtsZ half a century ago, this essential, highly conserved bacterial tubulin homolog has stayed center stage in the study of bacterial cell division and sparked intense scientific interest. From the initial genetic and cytological investigations in bulk cultures, to biochemical and biophysical characterizations in vitro, and finally to single-molecule studies in single cells with unprecedented resolution and sensitivity, our knowledge of the structure, function, and dynamics of the Z-ring has advanced substantially.
We now understand that the Z-ring is not a static or regularly packed structure, but is instead highly dynamic and discontinuous; the treadmilling dynamics and heterogeneous organization of the Z-ring are direct consequences of FtsZ’s GTPase activity and have little to do with other known Z-ring regulators. Such dynamics and structure are also essential to carry out the functions of the Z-ring in directing the spatiotemporal distribution of septal cell wall remodelers to ensure correct cell pole morphogenesis. Dynamic treadmilling may also allow the Z-ring to constantly survey the leading edge of the new septum, perhaps mending unevenly constricted septum as needed. A static, homogenously organized Z-ring such as that of FtsZD212A would lack this critical ability to adjust and thus lead to highly deformed and incomplete septa.
It remains unknown whether the Z-ring generates a mechanical force in vivo, but if it does, then the force does not limit how fast the cell wall constricts. The most likely scenario is that the Z-ring may bias the direction of new sPG strand insertion by deforming the inner membrane. How this spatial cue is transmitted to the sPG synthesis direction is unknown. The two-track and Brownian ratchet models also need to be further examined under different cell division scenarios and in different bacterial species. Finally, we do not know how sPG synthesis activity is spatiotemporally coordinated with sPG hydrolase activity to avoid cell wall lesion during constriction, which is perhaps one of the most important and dangerous tasks a bacterial cell has to accomplish.
It is an exciting time for the field of bacterial cell division. New discoveries in the past few years only begin to scratch the surface of the complex spatiotemporal regulation mechanism in bacterial cell division. With the substantial knowledge foundation built in the past half century and new technical advances in single-cell and single-molecule analyses, the field is poised to make breakthroughs for many years to come.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Xinxing Yang for providing unpublished FtsZ-GFP images for illustration purposes in this article; Josh McCausland, Xinxing Yang, Jason Z. Lyu, Jian Liu, David Weiss, and Piet de Boer for their collaborative work that made new perspectives in the article possible; and participants of the Super Z Group meeting from S. Holden, E. Garner, E. Goley, and H. Erickson’s groups for inspiring discussions. This work was supported in part by National Institutes of Health (NIH) grant T32 GM008403 (to R.M.), NIH grants R01GM086447 (to J.X.) and GM125656 (subcontract to J.X.), and a Hamilton Smith Innovative Research Award (to J.X.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol 64:938–52 [DOI] [PubMed] [Google Scholar]
- 2.Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Distèche M, den Blaauwen T. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol 55:1631–45 [DOI] [PubMed] [Google Scholar]
- 3.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol 7:642–53 [DOI] [PubMed] [Google Scholar]
- 4.Addinall S, Bi E, Lutkenhaus J. 1996. FtsZ ring formation in fts mutants. J. Bacteriol 178:3877–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Addinall S, Lutkenhaus J. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol 178:7167–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Addinall SG, Lutkenhaus J. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol 22:231–37 [DOI] [PubMed] [Google Scholar]
- 7.Alatossava T, Jütte H, Kuhn A, Kellenberger E. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol 162:413–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexeeva S, Gadella TW, Verheul J, Verhoeven GS, den Blaauwen T. 2010. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol. Microbiol 77:384–98 [DOI] [PubMed] [Google Scholar]
- 9.Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol 186:5775–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreu JM, et al. 2010. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J. Biol. Chem 285:14239–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arjes HA, Lai B, Emelue E, Steinbach A, Levin P. 2015. Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function. BMC Microbiol. 15:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auer GK, Weibel DB. 2017. Bacterial cell mechanics. Biochemistry 56:3710–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranova N, Radler P, Hernández-Rocamora VM, Alfonso C, López-Pelegrín M, et al. 2020. Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nat. Microbiol 5:407–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Begg K, Dewar S, Donachie W. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol 177:6211–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berezuk AM, Glavota S, Roach EJ, Goodyear MC, Krieger JR, Khursigara CM. 2018. Outer membrane lipoprotein RlpA is a novel periplasmic interaction partner of the cell division protein FtsK in Escherichia coli. Sci. Rep 8:12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernhardt TG, de Boer P. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi E, Lutkenhaus J. 1990. FtsZ regulates frequency of cell division in Escherichia coli. J. Bacteriol 172:2765–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi E, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–64 [DOI] [PubMed] [Google Scholar]
- 19.Bisson-Filho AW, Discola KF, Castellen P, Blasios V, Martins A, et al. 2015. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. PNAS 112:E2130–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, et al. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biteen JS, Goley ED, Shapiro L, Moerner W. 2012. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. ChemPhysChem 13:1007–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boer DP, Crossley R, Rothfield L. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol 174:63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boes A, Olatunji S, Breukink E, Terrak M. 2019. Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. mBio 10:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bork P, Sander C, Valencia A. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. PNAS 89:7290–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bramhill D, Thompson C. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. PNAS 91:5813–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol. Microbiol 62:5–14 [DOI] [PubMed] [Google Scholar]
- 27.Buddelmeijer N, Beckwith J. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol 5:553–57 [DOI] [PubMed] [Google Scholar]
- 28.Buddelmeijer N, Beckwith J. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol 52:1315–27 [DOI] [PubMed] [Google Scholar]
- 29.Buske P, Levin P 2013. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol 89:249–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buske PJ, Mittal A, Pappu RV, Levin P. 2015. An intrinsically disordered linker plays a critical role in bacterial cell division. Semin. Cell Dev. Biol 37:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, et al. 2013. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol 89:1099–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J. 2015. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLOS Genet. 11:e1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buss JA, Peters NT, Xiao J, Bernhardt TG. 2017. ZapA and ZapB form an FtsZ-independent structure at midcell. Mol. Microbiol 104:652–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cayley S, Lewis BA, Guttman HJ, Record MT. 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity: implications for protein-DNA interactions in vivo. J. Mol. Biol 222:281–300 [DOI] [PubMed] [Google Scholar]
- 35.Cayley SD, Guttman HJ, Record TM. 2000. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys. J 78:1748–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Erickson HP. 2005. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem 280:22549–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Erickson HP. 2009. FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry 48:6664–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Milam SL, Erickson HP. 2012. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51:3100–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, et al. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol 1:16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coltharp C, Buss J, Plumer TM, Xiao J. 2016. Defining the rate-limiting processes of bacterial cytokinesis. PNAS 113:E1044–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coltharp C, Xiao J. 2016. Beyond force generation: Why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? Bioessays 39:1600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coltharp C, Xiao J. 2016. How do bacteria divide and multiply? Atlas of Science, May 2. https://atlasofscience.org/how-do-bacteria-divide-and-multiply/ [Google Scholar]
- 43.Coltharp C, Yang X, Xiao J. 2014. Quantitative analysis of single-molecule superresolution images. Curr. Opin. Struc. Biol 28:112–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corbin BD, Wang Y, Beuria TK, Margolin W. 2007. Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol 189:3026–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Boer P, Crossley R, Rothfield L. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–56 [DOI] [PubMed] [Google Scholar]
- 46.de Boer PA. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol 13:730–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–49 [DOI] [PubMed] [Google Scholar]
- 48.de Pedro M, Quintela J, Höltje J, Schwarz H. 1997. Murein segregation in Escherichia coli. J. Bacteriol 179:2823–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Pedro MA, Cava F. 2015. Structural constraints and dynamics of bacterial cell wall architecture. Front. Microbiol 6:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev 32:321–44 [DOI] [PubMed] [Google Scholar]
- 51.Dhaked H, Ray S, Battaje R, Banerjee A, Panda D. 2019. Regulation of Streptococcus pneumoniae FtsZ assembly by divalent cations: paradoxical effects of Ca2+ on the nucleation and bundling of FtsZ polymers. FEBS J. 286:3629–46 [DOI] [PubMed] [Google Scholar]
- 52.Du S, Lutkenhaus J. 2014. SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLOS Genet. 10:e1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. 2012. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol 194:3189–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol 68:720–35 [DOI] [PubMed] [Google Scholar]
- 55.Egan AJ, Vollmer W. 2013. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci 1277:8–28 [DOI] [PubMed] [Google Scholar]
- 56.Eraso JM, Markillie LM, Mitchell HD, Taylor RC, Orr G, Margolin WM. 2014. The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli. J. Bacteriol 196(11):2053–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erickson H, Taylor D, Taylor K, Bramhill D. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. PNAS 93:519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erickson HP. 1995. FtsZ, a prokaryotic homolog of tubulin? Cell 80:367–70 [DOI] [PubMed] [Google Scholar]
- 59.Erickson HP. 2009. Modeling the physics of FtsZ assembly and force generation. PNAS 106:9238–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erickson HP. 2017. How bacterial cell division might cheat turgor pressure: a unified mechanism of septal division in Gram-positive and Gram-negative bacteria. Bioessays 39(8). 10.1002/bies.201700045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev 74:504–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espéli O, Borne R, Dupaigne P, Thiel A, Gigant E, et al. 2012. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31:3198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, et al. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516:259–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLOS ONE 5:e12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galli E, Gerdes K. 2012. FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol 194:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol 191:4186–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gangola P, Rosen B. 1987. Maintenance of intracellular calcium in Escherichia coli. J. Biol. Chem 262:12570–74 [PubMed] [Google Scholar]
- 68.Gardner KA, Moore DA, Erickson HP. 2013. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol 89:264–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghosh B, Sain A. 2008. Origin of contractile force during cell division of bacteria. Phys. Rev. Lett 101:178101. [DOI] [PubMed] [Google Scholar]
- 70.Goehring NW, Beckwith J. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol 15:R514–26 [DOI] [PubMed] [Google Scholar]
- 71.Goehring NW, Gueiros-Filho F, Beckwith J. 2005. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Gene Dev. 19:127–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, et al. 2011. Assembly of the Caulobacter cell division machine. Mol. Microbiol 80:1680–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.González JM, Jiménez M, Vélez M, Mingorance J, Andreu JM, et al. 2003. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem 278:37664–71 [DOI] [PubMed] [Google Scholar]
- 74.Gottesman S, Halpern E, Trisler P. 1981. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J. Bacteriol 148:265–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grenga L, Luzi G, Paolozzi L, Ghelardini P. 2008. The Escherichia coli FtsK functional domains involved in its interaction with its divisome protein partners. FEMS Microbiol. Lett 287:163–67 [DOI] [PubMed] [Google Scholar]
- 76.Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Gene Dev. 16:2544–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haeusser DP, Margolin W. 2016. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol 14:305–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hale CA, de Boer P. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–85 [DOI] [PubMed] [Google Scholar]
- 79.Hale CA, Rhee AC, de Boer PA. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol 182:5153–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hale CA, Shiomi D, Liu B, Bernhardt TG, Marggolin W, et al. 2011. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol 193:1393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Handler AA, Lim J, Losick R. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol. Microbiol 68:588–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harry E, Monahan L, Thompson L. 2006. Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol 253:27–94 [DOI] [PubMed] [Google Scholar]
- 83.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, et al. 2008. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321:1673–75 [DOI] [PubMed] [Google Scholar]
- 84.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, et al. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol 41:167–78 [DOI] [PubMed] [Google Scholar]
- 85.Hirota Y, Ryter A, Jacob F. 1968. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol 33:677–93 [DOI] [PubMed] [Google Scholar]
- 86.Holden S 2018. Probing the mechanistic principles of bacterial cell division with super-resolution microscopy. Curr. Opin. Microbiol 43:84–91 [DOI] [PubMed] [Google Scholar]
- 87.Holden SJ, Pengo T, Meibom KL, Fernandez C, Collier J, Manley S. 2014. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. PNAS 111:4566–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Höltje J 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev 62:181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hörger I, Velasco E, Mingorance J, Rivas G, Tarazona P, Vélez M. 2008. Langevin computer simulations of bacterial protein filaments and the force-generating mechanism during cell division. Phys. Rev. E 77:011902. [DOI] [PubMed] [Google Scholar]
- 90.Housman M, Milam SL, Moore DA, Osawa M, Erickson HP. 2016. FtsZ protofilament curvature is the opposite of tubulin rings. Biochemistry 55:4085–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsin J, Gopinathan A, Huang KC. 2012. Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. PNAS 109:9432–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang KH, Durand-Heredia J, Janakiraman A. 2013. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J. Bacteriol 195:1859–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huecas S, Llorca O, Boskovic J, Martín-Benito J, Valpuesta JM, Andreu JM. 2008. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys. J 94:1796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huecas S, Ramírez-Aportela E, Vergoñós A, Núñez-Ramírez R, Llorca O, et al. 2017. Self-organization of FtsZ polymers in solution reveals spacer role of the disordered C-terminal tail. Biophys. J 113:1831–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huisman O, D’Ari R, Gottesman S. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. PNAS 81:4490–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol 48:1171–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jacq M, Adam V, Bourgeois D, Moriscot C, Di Guilmi A-M, et al. 2015. Remodeling of the Z-ring nanostructure during the Streptococcus pneumoniae cell cycle revealed by photoactivated localization microscopy. mBio 6:e01108–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jennings PC, Cox GC, Monahan LG, Harry EJ. 2011. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron 42:336–41 [DOI] [PubMed] [Google Scholar]
- 99.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann. N. Y.Acad. Sci 1277:54–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99a.Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. 2015. Distinct constrictive processes, separated in time and space, divide caulobacter inner and outer membranes. J. Bacteriol 187:6874–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Justice SS, García-Lara J, Rothfield LI. 2000. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol 37:410–23 [DOI] [PubMed] [Google Scholar]
- 101.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol 187:2233–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV. 1998. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Gene Dev. 12:880–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuru E, Tekkam N, Hall E, Brun YV, Nieuwenhze MS. 2015. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc 10:33–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lallo DG, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353–59 [DOI] [PubMed] [Google Scholar]
- 105.Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. 2009. Condensation of FtsZ filaments can drive bacterial cell division. PNAS 106:121–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lan G, Wolgemuth CW, Sun SX. 2007. Z-ring force and cell shape during division in rod-like bacteria. PNAS 104:16110–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lange R, Hengge-Aronis R. 1994. The nIpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol 13:733–43 [DOI] [PubMed] [Google Scholar]
- 108.Läppchen T, Pinas VA, Hartog AF, Koomen GJ, Schaffner-Barbero C, et al. 2008. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem. Biol 15:189–99 [DOI] [PubMed] [Google Scholar]
- 109.Leaver M, Domínguez-Cuevas P, Coxhead J, Daniel R, Errington J. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849–53 [DOI] [PubMed] [Google Scholar]
- 110.Leisch N, Verheul J, Heindl NR, Gruber-Vodicka HR, Pende N, et al. 2012. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr. Biol 22:R831–32 [DOI] [PubMed] [Google Scholar]
- 111.Leung AK, Lucile White E, Ross LJ, Reynolds RC, DeVito JA, Borhani DW. 2004. Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J. Mol. Biol 342:953–70 [DOI] [PubMed] [Google Scholar]
- 112.Levin P, Kurtser IG, Grossman AD. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. PNAS 96:9642–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Levin P, Losick R. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Gene Dev. 10:478–88 [DOI] [PubMed] [Google Scholar]
- 114.Li Y, Hsin J, Zhao L, Cheng Y, Shang W, et al. 2013. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science 341:392–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y, Shao S, Xu X, Su X, Sun Y, Wei S. 2018. MapZ forms a stable ring structure that acts as a nanotrack for FtsZ treadmilling in Streptococcus mutans. ACS Nano 12:6137–46 [DOI] [PubMed] [Google Scholar]
- 116.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu B, Hale CA, Persons L, Phillips-Mason PJ, de Boer PA. 2019. Roles of the DedD protein in Escherichia coli cell constriction. J. Bacteriol 201:e00698–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu B, Persons L, Lee L, de Boer PA. 2015. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol 95:945–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu G, Draper CG, Donachie W. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol 29:893–903 [DOI] [PubMed] [Google Scholar]
- 120.Liu Z, Mukherjee A, Lutkenhaus J. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol 31:1853–61 [DOI] [PubMed] [Google Scholar]
- 121.Loose M, Mitchison TJ. 2013. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol 16:38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Low HH, Moncrieffe MC, Löwe J. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol 341:839–52 [DOI] [PubMed] [Google Scholar]
- 123.Löwe J, Amos LA. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–6 [DOI] [PubMed] [Google Scholar]
- 124.Löwe J, Amos LA. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu C, Reedy M, Erickson HP. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol 182:164–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lu C, Stricker J, Erickson HP. 2001. Site-specific mutations of FtsZ: effects on GTPase and in vitro assembly. BMC Microbiol. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lutkenhaus J, Du S. 2013. E. coli cell cycle machinery. Sub-Cell Biochem. 84:27–65 [DOI] [PubMed] [Google Scholar]
- 128.Lyu Z, Coltharp C, Yang X, Xiao J. 2016. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional superresolution imaging. Biopolymers 105:725–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. PNAS 93:12998–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ma X, Margolin W. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol 181:7531–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.MacCain WJ, Kannan S, Jameel DZ, Troutman JM, Young KD. 2018. A defective undecaprenyl pyrophosphate synthase induces growth and morphological defects that are suppressed by mutations in the isoprenoid pathway of Escherichia coli. J. Bacteriol 200:e00255–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maggi S, Massidda O, Luzi G, Fadda D, Paolozzi L, Ghelardini P. 2008. Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology 154:3042–52 [DOI] [PubMed] [Google Scholar]
- 133.Männik J, Bailey MW, O’Neill JC, Männik J. 2017. Kinetics of large-scale chromosomal movement during asymmetric cell division in Escherichia coli. PLOS Genet. 13:e1006638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Männik J, Castillo DE, Yang D, Siopsis G, Männik J. 2016. The role of MatP, ZapA and ZapB in chromosomal organization and dynamics in Escherichia coli. Nucleic Acids Res. 44:1216–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, et al. 2014. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 5:e00022–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mateos-Gil P, Paez A, Hörger I, Rivas G, Vicente M, et al. 2012. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. PNAS 109:8133–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mateos-Gil P, Tarazona P, Vélez M. 2018. Bacterial cell division: modeling FtsZ assembly and force generation from single filament experimental data. FEMS Microbiol. Rev 43:73–87 [DOI] [PubMed] [Google Scholar]
- 138.Matsui T, Han X, Yu J, Yao M, Tanaka I. 2014. Structural change in FtsZ induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem 289:3501–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matsui T, Yamane J, Mogi N, Yamaguchi H, Takemoto H, et al. 2012. Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Crystallogr. D 68:1175–88 [DOI] [PubMed] [Google Scholar]
- 140.McCausland JW, Yang X, Lyu Z, Söderström B, Xiao J, Liu J. 2019. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. bioRxiv 857813. 10.1101/857813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meier EL, Daitch AK, Yao Q, Bhargava A, Jensen GJ, Goley ED. 2017. FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLOS Genet. 13:e1006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, et al. 2008. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135:475–85 [DOI] [PubMed] [Google Scholar]
- 143.Milne J, Subramaniam R. 2009. Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat. Rev. Microbiol 7:666–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mohammadi T, Ploeger GE, Verheul J, Comvalius AD, Martos A, et al. 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48:11056–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, et al. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554:528–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moore DA, Whatley ZN, Joshi CP, Osawa M, Erickson HP. 2017. Probing for binding regions of the FtsZ protein surface through site-directed insertions: discovery of fully functional FtsZ-fluorescent proteins. J. Bacteriol 199:e00553–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, et al. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mukherjee A, Dai K, Lutkenhaus J. 1993. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. PNAS 90:1053–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mukherjee A, Cao C, Lutkenhaus J. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. PNAS 95:2885–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mukherjee A, Lutkenhaus J. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol 176:2754–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nguyen LT, Oikonomou CM, Ding HJ, Kaplan M, Yao Q, et al. 2019. Simulations suggest a constrictive force is required for Gram-negative bacterial cell division. Nat. Commun 10:1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nguyen LT, Oikonomou CM, Jensen GJ. 2019. Simulations of proposed mechanisms of FtsZ-driven cell constriction. bioRxiv 737189. 10.1101/737189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nguyen-Distèche M, Fraipont C, Buddelmeijer N, Nanninga N. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol. Life Sci 54:309–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Niu L, Yu J. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys. J 95:2009–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nogales E, Downing KH, Amos LA, Löwe J. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol 5:451–58 [DOI] [PubMed] [Google Scholar]
- 156.Ogura T, Tomoyasu T, Yuki T, Morimura S, Begg KJ, et al. 1991. Structure and function of the ftsH gene in Escherichia coli. Res. Microbiol 142:279–82 [DOI] [PubMed] [Google Scholar]
- 157.Oikonomou CM, Jensen GJ. 2017. The development of cryo-EM and how it has advanced microbiology. Nat. Microbiol 2:1577–79 [DOI] [PubMed] [Google Scholar]
- 158.Oliva MA, Cordell SC, Löwe J. 2004. Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol 11:1243–50 [DOI] [PubMed] [Google Scholar]
- 159.Oliva MA, Trambaiolo D, Löwe J. 2007. Structural insights into the conformational variabilityof FtsZ. J. Mol. Biol 373:1229–42 [DOI] [PubMed] [Google Scholar]
- 160.Osawa M, Anderson DE, Erickson HP. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Osawa M, Anderson DE, Erickson HP. 2009. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28:3476–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Osawa M, Erickson HP. 2011. Inside-out Z rings: constriction with and without GTP hydrolysis. Mol. Microbiol 81:571–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Osawa M, Erickson HP. 2018. Turgor pressure and possible constriction mechanisms in bacterial division. Front. Microbiol 9:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Paez A, Mateos-Gil P, Hörger I, Mingorance J, Rivas G, et al. 2009. Simple modeling of FtsZ polymers on flat and curved surfaces: correlation with experimental in vitro observations. PMC Biophys. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, et al. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pazos M, Natale P, Margolin W, Vicente M. 2013. Interactions among the early Escherichia coli divisome proteins revealed by bimolecular fluorescence complementation. Environ. Microbiol 15:3282–91 [DOI] [PubMed] [Google Scholar]
- 167.Pende N, Wang J, Weber PM, Verheul J, Kuru E, et al. 2018. Host-polarized cell growth in animal symbionts. Curr. Biol 28:1039–51.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pereira AR, Hsin J, Król E, Tavares AC, Flores P, et al. 2016. FtsZ-dependent elongation of a coccoid bacterium. mBio 7:e00908–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Perez AJ, Cesbron Y, Shaw BL, Bazan Villicana J, Tsui HT, et al. 2019. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. PNAS 116:3211–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Petiti M, Serrano B, Faure L, Lloubes R, Mignot T, Duché D. 2019. Tol energy-driven localization of Pal and anchoring to the peptidoglycan promote outer-membrane constriction. J. Mol. Biol 431:3275–88 [DOI] [PubMed] [Google Scholar]
- 171.Pichoff S, Du S, Lutkenhaus J. 2019. Roles of FtsEX in cell division. Res. Microbiol 170:374–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol 55:1722–34 [DOI] [PubMed] [Google Scholar]
- 173.Pichoff S, Lutkenhaus J. 2007. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol 64:1129–38 [DOI] [PubMed] [Google Scholar]
- 174.Popp D, Iwasa M, Narita A, Erickson HP, Maéda Y. 2009. FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91:340–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rajagopala SV, Sikorski P, Kumar A, Mosca R, Vlasblom J, et al. 2014. The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol 32:285–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ramirez-Diaz D, Merino-Salomon A, Heymann M, Schwille P. 2019. Bidirectional FtsZ filament treadmilling promotes membrane constriction via torsional stress. bioRxiv 587790. 10.1101/587790 [DOI] [Google Scholar]
- 177.Ramirez-Diaz DA, García-Soriano DA, Raso A, Mücksch J, Feingold M, et al. 2018. Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLOS Biol. 16:e2004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rand-Heredia J, Yu HH, Carlo S, Lesser CF, Janakiraman A. 2011. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol 193:1405–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.RayChaudhuri D, Park JT. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–54 [DOI] [PubMed] [Google Scholar]
- 180.Raymond A, Lovell S, Lorimer D, Walchli J, Mixon M,et al. 2009. Combined protein constructand synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer. BMC Biotechnol. 9:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Roach EJ, Kimber MS, Khursigara CM. 2014. Crystal structure and site-directed mutational analysis reveals key residues involved in Escherichia coli ZapA function. J. Biol. Chem 289:23276–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Romberg L, Levin P 2003. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol 57:125–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Romberg L, Simon M, Erickson HP. 2001. Polymerization of FtsZ, a bacterial homolog of tubulin: Is assembly cooperative? J. Biol. Chem 276:11743–53 [DOI] [PubMed] [Google Scholar]
- 184.Rowlett V, Margolin W 2013. The bacterial Min system. Curr. Biol 23:R553–56 [DOI] [PubMed] [Google Scholar]
- 185.Rowlett V, Margolin W. 2014. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys. J 107:L17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ruiz N 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. PNAS 105:15553–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Salje J, Zuber B, Löwe J. 2009. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science 323:509–12 [DOI] [PubMed] [Google Scholar]
- 188.Sauvage E, Terrak M. 2016. Glycosyltransferases and transpeptidases/penicillin-binding proteins: valuable targets for new antibacterials. Antibiotics 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJM. 2002. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41:521–29 [DOI] [PubMed] [Google Scholar]
- 190.Scheffers DJ, Driessen A. 2001. The polymerization mechanism of the bacterial cell division protein FtsZ. FEBS Lett. 506:6–10 [DOI] [PubMed] [Google Scholar]
- 191.Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, et al. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol 186:785–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schneider T, Sahl H-G. 2010. An oldie but a goodie: cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol 300:161–69 [DOI] [PubMed] [Google Scholar]
- 193.Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. 2014. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345:220–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shi H, Bratton BP, Gitai Z, Huang K. 2018. How to build a bacterial cell: MreB as the foreman of E. coli construction. Cell 172:1294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, et al. 2009. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. PNAS 106:3125–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Singh J, Makde RD, Kumar V, Panda D. 2008. SepF increases the assembly and bundling of FtsZ polymers and stabilizes FtsZ protofilaments by binding along its length. J. Biol. Chem 283:31116–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Small E, Addinall SG. 2003. Dynamic FtsZ polymerization is sensitive to the GTP to GDP ratio and can be maintained at steady state using a GTP-regeneration system. Microbiology 149:2235–42 [DOI] [PubMed] [Google Scholar]
- 198.Small E, Marrington R, Rodger A, Scott DJ, Sloan K, et al. 2007. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J. Mol. Biol 369:210–21 [DOI] [PubMed] [Google Scholar]
- 199.Söderström B, Chan H, Shilling PJ, Skoglund U, Daley DO. 2018. Spatial separation of FtsZ and FtsN during cell division. Mol. Microbiol 107:387–401 [DOI] [PubMed] [Google Scholar]
- 200.Söderström B, Mirzadeh K, Toddo S, van Heijne G, Skoglund U, Daley DO. 2016. Coordinated disassembly of the divisome complex in Escherichia coli. Mol. Microbiol 101:425–38 [DOI] [PubMed] [Google Scholar]
- 201.Söderström B, Skoog K, Blom H, Weiss DS, von Heijne G, Daley DO. 2014. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol. Microbiol 92:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stock J, Rauch B, Roseman S. 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem 252:7850–61 [PubMed] [Google Scholar]
- 203.Stouf M, Meile J-C, Cornet F. 2013. FtsK actively segregates sister chromosomes in Escherichia coli. PNAS 110:11157–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 2012. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLOS Biol. 10:e1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Stricker J, Maddox P, Salmon E, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. PNAS 99:3171–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sun N, Lu YJ, Chan FY, Du RL, Zheng YY, et al. 2017. A thiazole orange derivative targeting the bacterial protein FtsZ shows potent antibacterial activity. Front. Microbiol 8:855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sun Q, Margolin W 1998. FtsZ dynamics during the division cycle of live Escherichia coli cells. J. Bacteriol 180:2050–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sundararajan K, Goley ED. 2017. The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro. J. Biol. Chem 292:20509–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Huang KC, Goley ED. 2015. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun 6:7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sundararajan K, Vecchiarelli A, Mizuuchi K, Goley ED. 2018. Species- and C-terminal linker-dependent variations in the dynamic behavior of FtsZ on membranes in vitro. Mol. Microbiol 110:47–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Surovtsev IV, Morgan JJ, Lindahl PA. 2008. Kinetic modeling of the assembly, dynamic steady state, and contraction of the FtsZ ring in prokaryotic cytokinesis. PLOS Comput. Biol 4:e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Szwedziak P, Wang Q, Bharat TA, Tsim M, Löwe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3:e04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212a.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, et al. 2019. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol 4:587–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tan CM, Therien AG, Lu J, Lee SH, Caron A, et al. 2012. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci. Transl. Med 4:126ra35. [DOI] [PubMed] [Google Scholar]
- 214.Thanedar S, Margolin W. 2004. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol 14:1167–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, et al. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 14:2551–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tomoyasu T, Yamanaka K, Murata K, Suzaki T, Bouloc P, et al. 1993. Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol 175:1352–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tsang MJ, Yakhnina AA, Bernhardt TG. 2017. NlpD links cell wall remodeling and outer membrane invagination during cytokinesis in Escherichia coli. PLOS Genet. 13:e1006888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol 10:123–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, et al. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Uehara T, Dinh T, Bernhardt TG. 2009. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol 191:5094–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Uehara T, Parzych KR, Dinh T, Bernhardt TG. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29:1412–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.van den Ent F, Amos LA, Löwe J. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44 [DOI] [PubMed] [Google Scholar]
- 223.van den Ent F, Löwe J. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.van Mameren J, Vermeulen KC, Gittes F, Schmidt CF. 2009. Leveraging single protein polymers to measure flexural rigidity. J. Phys. Chem. B 113:3837–44 [DOI] [PubMed] [Google Scholar]
- 225.Varma A, de Pedro MA, Young KD. 2007. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol 189:5692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. 2019. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol 10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Vicente M, Gomez M, Ayala J. 1998. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell Mol. Life Sci 54:317–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Virant D, Turkowyd B, Balinovic A, Endesfelder U. 2017. Combining primed photoconversion and UV-photoactivation for aberration-free, live-cell compliant multi-color single-molecule localization microscopy imaging. Int. J. Mol. Sci 18:1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Vollmer W 2007. Structure and biosynthesis of the murein (peptidoglycan) sacculus. In The Periplasm, ed. Ehrmann M, pp. 198–213. Sterling, VA: ASM Press [Google Scholar]
- 230.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev 32:149–67 [DOI] [PubMed] [Google Scholar]
- 231.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev 32:259–86 [DOI] [PubMed] [Google Scholar]
- 232.Wachi M, Okada IY, Matsuhashi M. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol 171:6511–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Weidel W, Pelzer H. 1964. Bagshaped macromolecules: a new outlook on bacterial cell. Adv. Enzymol 26:193–232 [DOI] [PubMed] [Google Scholar]
- 234.Weiss DS. 2004. Bacterial cell division and the septal ring. Mol. Microbiol 54:588–97 [DOI] [PubMed] [Google Scholar]
- 235.Woldemeskel S, McQuillen R, Hessel AM, Xiao J, Goley ED. 2017. A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus. Mol. Microbiol 105:721–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xiao J, Dufrêne YF. 2016. Optical and force nanoscopy in microbiology. Nat. Microbiol 1:16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xiaoc J, Goley ED. 2016. Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr. Opin. Microbiol 34:90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. 2011. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. PNAS 108:E1052–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2017. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355:744–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yang X, McQuillen R, Lyu Z, Phillips-Mason P, De La Cruz A, et al. 2019. FtsW exhibits distinct processive movements driven by either septal cell wall synthesis or FtsZ treadmilling in E. coli. bioRxiv 850073. 10.1101/850073 [DOI] [Google Scholar]
- 241.Yao Q, Jewett AI, Chang YW, Oikonomou CM, Beeby M, et al. 2017. Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J. 36:1577–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yi Q-M, Lutkenhaus J. 1985. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene 36:241–47 [DOI] [PubMed] [Google Scholar]