Abstract
Panthothenate kinase-associated neurodegeneration (PKAN, OMIM 234200), is an inborn is an autosomal recessive inborn error of metabolism caused by pathogenic variants in PANK2. PANK2 encodes the enzyme pantothenate kinase 2 (EC 2.7.1.33), an essential regulatory enzyme in CoA biosynthesis. Clinical presentation includes dystonia, rigidity, bradykinesia, dysarthria, pigmentary retinopathy and dementia with variable age of onset ranging from childhood to adulthood. In order to provide an accurate incidence estimate of PKAN, we conducted a systematic review of the literature and databases for pathogenic mutations and constructed a bioinformatic profile for pathogenic missense variants in PANK2. We then studied the gnomAD cohort of ~140,000 unrelated adults from global populations to determine the allele frequency of the variants in PANK2 reported pathogenic for PKAN and for those additional variants identified in gnomAD that met bioinformatics criteria for being potentially pathogenic. Incidence was estimated based on three different models using the allele frequencies of pathogenic PKAN variants with or without those bioinformatically determined to be potentially pathogenic. Disease incidence calculations showed PKAN incidence ranging from 1:396,006 in Europeans, 1:1,526,982 in Africans, 1:480,826 in Latino, 1:523,551 in East Asians and 1:531,118 in South Asians. These results indicate PKAN is expected to occur in approximately 2 of every 1 million live births globally outside of Africa, and has a much lower incidence 1 in 1.5 million live births in the African population.
Keywords: PKAN, PANK2, NBIA, Hallervorden-Spatz, incidence, bioinformatics, population genetics, IEM, dystonia, movement disorder, retinopathy, eye of the tiger
Introduction
Panthothenate kinase-associated neurodegeneration (PKAN, OMIM 234200), also referred to as Neurodegeneration with brain iron accumulation 1 (NBIA1), is an autosomal recessive inborn error of metabolism (IEM) caused by pathogenic mutations in the PANK2 gene (OMIM 606157) [1–5]. Clinical presentation includes dystonia, rigidity, bradykinesia, dysarthria, pigmentary retinopathy and dementia [1, 2, 6]. Age of onset and progression for PKAN is variable: from early onset in the first decade of life with rapid progression to later onset with slower progression which may present as an adult-onset movement disorder[1, 2, 7].
PANK2 encodes the enzyme pantothenate kinase 2 (EC 2.7.1.33), an essential regulatory enzyme in CoA biosynthesis. CoA is the major acyl carrier, playing an integral role in intermediary and fatty acid metabolism. The gold standard for diagnosis of IEMs is typically the metabolic signature of either compromised enzyme activity or diminished or excessive metabolite levels. Contrary to most IEM’s, pantothenate kinase 2 enzyme activity studies have not shown a correlation between enzyme activity levels and PANK2 pathogenic mutations; activity level ranges from o to over 100 % of normal for pathogenic mutations [8]. Nearly all cases show a hallmark brain imaging phenotype resultant from iron deposits in the brain, called ‘eye of the tiger’; these are appreciated as a specific pattern of hypointensity in the pallidal nuclei with a central hyperintensity on T2-weighted MRI.
PKAN is rare but precise disease incidence for the general population has not been calculated. We have conducted a systematic, genetic approach to determine an accurate estimate of the incidence of PKAN in global populations. We conducted an exhaustive search for documented pathogenic PANK2 mutations and, further, we generated a bioinformatic profile for these known pathogenic variants to empirically determine a bioinformatic framework for prediction of pathogenicity of PANK2 variants of uncertain significance. We leveraged the Genome Aggregation Database (gnomAD) of 141,456 adults to determine the global population frequency of PANK2 deficiency-causing mutations. By determining the population frequency of PANK2 mutations that are documented pathogenic and those that are bioinformatically predicted to cause PANK2 deficiency we calculated a robust incidence for this disease.
Methods
Databases and genetic variants
Searches of medical literature and publicly available databases were conducted to identify variants reported as causing PKAN. PubMed http://www.ncbi.nlm.nih.gov/pubmed, ClinVar http://www.ncbi.nlm.nih.gov/clinvar, HGMD http://www.hgmd.cf.ac.uk/ac/index.php and Online Inheritance in Man (OMIM) http://www.omim.org were queried June 2019.
The Genome Aggregation Database (gnomAD) v2.1.1 was used to obtain allele frequencies of PANK2 variants in found in subjects with PKAN as well as a source of potentially pathogenic variants segregating in populations that may be predicted damaging to PANK2 function. gnomAD contains data from 125,748 exome sequences and 15,708 whole-genome sequences from unrelated individuals sequenced as part of various disease-specific and population genetic studies (https://gnomad.broadinstitute.org accessed July, 2019) [9].
Nomenclature for all variants follow HGVS standards and is reported in reference to HG19 and PANK2 NM_153638.3 and NP_705902.2. All genetic alleles studied that were not previously found in ClinVar have been submitted to ClinVar http://www.ncbi.nlm.nih.gov/clinvar/.
Bioinformatic analyses of variant potential pathogenicity
All pathogenic missense mutations in PANK2 were assessed by algorithms commonly used to predict pathogenicity of variants in order to generate a bioinformatic profile for these known pathogenic mutations: SIFT [10], PolyPhen2 HDIV [11], Genomic Evolutionary Rate Profiling (GERP)[12], and PhyloP [13]. Any value residing outside 1.5 times the inter-quartile range was labeled an outlier and removed for calculation of reported mean, standard deviation, boxplots and violin plots. SIFT uses a sequence alignment-based approach to predict and rank the effects of all possible substitutions at each position in a protein sequence. PolyPhen-2 HDIV uses a supervised machine-learning method and is trained on all damaging alleles with known effects on the molecular function causing human Mendelian diseases present in the UniProtKB database, together with differences between human proteins and their closely related mammalian homologs, assumed to be non-damaging. GERP detects evolutionary constraint while PhyloP considers both conservation and acceleration in the evolutionary rate of substitutions. The phyloP score reported here results from analysis of an alignment of primate sequences and the GERP score reported here is based on the alignment of 35 mammalian species. The average and range were generated for each predictor. The resulting bioinformatic profile was then applied to all missense PANK2 variants to predict their potential to be pathogenic.
Incidence estimation
The disease incidence estimates were conducted according to an analysis method we created for rare single gene disorders [14]. To determine carrier allele frequencies we queried gnomAD and the total carrier frequency was calculated as the number of individuals possessing a PKAN pathogenic or predicted pathogenic mutation divided by the total number of individuals ascertained. Disease incidence was calculated based on the total carrier frequency and Hardy-Weinberg principles under multiple models. One model was to calculate incidence using only the mutations reported pathogenic in the literature and alternate models used those mutations plus the variants predicted pathogenic.
Results
Determining a bioinformatic profile for PANK2 pathogenic missense variants
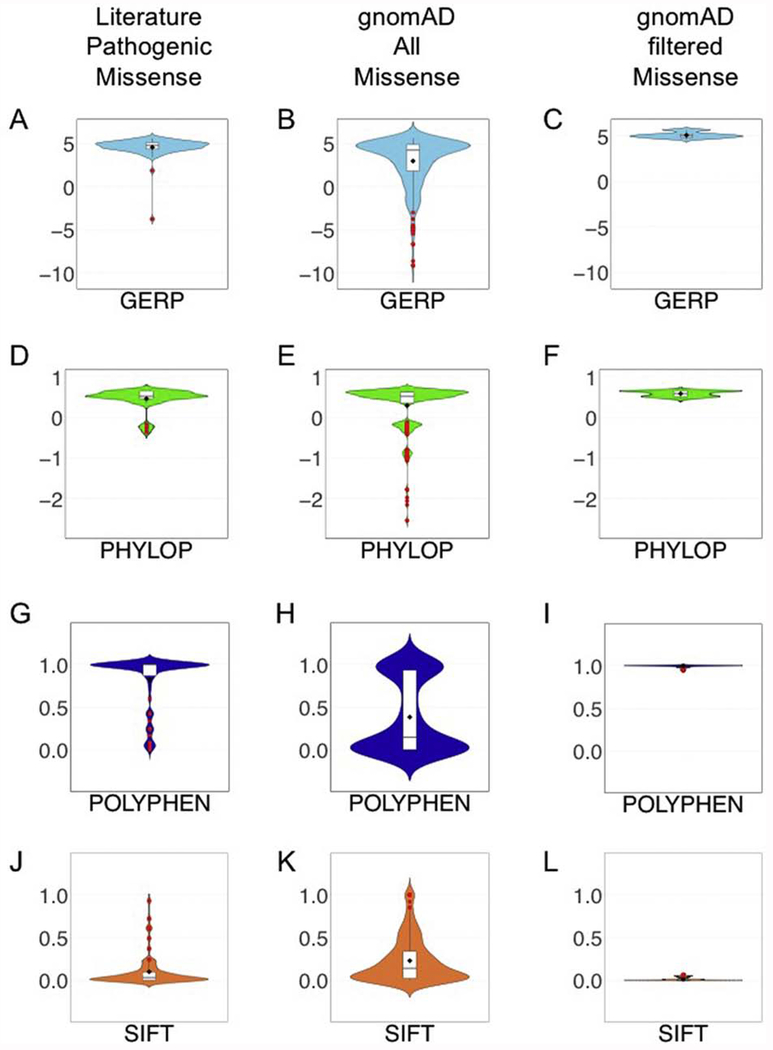
We conducted an exhaustive search for all PANK2 variants reported pathogenic for PKAN that included searches of public databases and medical literature. A total of 174 variants were identified: 123 missense, 16 suspected or shown to affect splicing and 36 small insertion/deletion variants (Table 1). All missense variants reported to cause PKAN were assessed for potential to perturb protein function using Sift, PolyPhen2 HDIV, GERP, and PhyloP Primate. SIFT and PolyPhen2 are widely utilized algorithms for predicting pathogenic effects of variants. We plotted the SIFT and PolyPhen2 HDIV scores, normalized from 0 to 1 with 1 being maximally damaging, for all missense PANK2 pathogenic variants. The average SIFT score for missense variants was 0.92 and the mean score for PolyPhen2 HDIV was 0.97 (Figure 1A and C). Evolutionary conservation was measured using PhyloP and GERP. The genome-wide PhyloP Primates score ranges from −2.525 to 0.71 and the average score for PKAN-causing missense variants was 0.53 (Figure 1E). GERP approximates evolutionary constraint at a locus by maximum likelihood estimation [12]. The genome-wide average GERP score is −0.125 [12], and the GERP score for PKAN-causing missense variants showed an elevated mean of 4.69 (Figure 1G).
Table 1.
PANK2 variants reported as pathogenic for PKAN.
| PANK2 cDNA NM_153638.3 | PANK2 Protein NP_705902.2 | Reference |
|---|---|---|
| c.137A>T! | p.Asp46Val! | [15]! |
| c.332T>A | p.Leu111Gln | [16] |
| c.398T>A | p.Met133Lys | [6] |
| c.401A>G | p.Glu134Gly | [5] |
| NM_024960.5:c.419T>C | p.Phe140Ser | [17] |
| c.434C>A | p.Ser145* | [18] |
| c.445G>T | p.Glu149* | [19] |
| c.448C>T | p.Gln150* | [3] |
| c.533C>A | p.Ser178* | [20] |
| c.570C>G | p.Tyr190* | [3] |
| c.638G>A | p.Trp213* | [21] |
| c.649G>A | p.Asp217Asn | [2] |
| c.655G>A | p.Gly219Ser | [21] |
| c.656G>T | p.Gly219Val | [3] |
| c.680A>G | p.Tyr227Cys | [22, 23] |
| c.683T>C | p.Phe228Ser | [16] |
| c.695A>G | p.Asp232Gly | [24] |
| c.700A>C | p.Thr234Pro | [25] |
| c.700A>G | p.Thr234Ala | [3] |
| c.746G>C | p.Arg249Pro | [5] |
| c.745C>T | p.Arg249Trp | [6] |
| c.775G>A | p.Gly259Arg | [16] |
| c.787A>T | p.Ile263Phe | [26] |
| c.790C>T | p.Arg264Trp | [3] |
| c.791G>A | p.Arg264Gln | [2] |
| c.833G>T | p.Arg278Leu | [25] |
| c.832C>T | p.Arg278Cys | [3] |
| c.844C>G | p.Leu282Val | [3] |
| c.849C>G | p.His283Gln | [27] |
| c.847C>T | p.His283Tyr | [28] |
| c.852T>G | p.Phe284Leu | [29] |
| c.856C>T | p.Arg286Cys | [3] |
| c.894G>A | p.Met298Ile | [2] |
| c.893T>A | p.Met298Lys | [30] |
| c.898A>G | p.Arg300Gly | [31] |
| c.936T>A | p.Cys312* | [3] |
| c.965A>G | p.Glu322Gly | [16] |
| c.966G>T | p.Glu322Asp | [2] |
| c.970G>T | p.Asp324Tyr | [6] |
| c.980C>T | p.Thr327Ile | [3] |
| c.994C>T | p.Gln332* | [2] |
| c.1003A>G | p.Lys335Glu | [29] |
| c.1022G>C | p.Cys341Ser | [32] |
| c.1030A>T | p.Lys344* | [3] |
| c.1037T>G | p.Ile346Ser | [33] |
| c.1051T>C | p.Ser351Pro | [3] |
| c.1064A>G | p.Asn355Ser | [3] |
| c.1070G>C | p.Arg357Pro | [1] |
| c.1070G>A | p.Arg357Gln | [2] |
| c.1069C>T | p.Arg357Trp | [2] |
| c.1082A>G | p.Tyr361Cys | [32] |
| c.1130T>C | p.Phe377Ser | [34] |
| c.1133A>G | p.Asp378Gly | [35] |
| c.1148A>G | p.Tyr383Cys | [36] |
| c.1168A>T | p.Ile390Phe | [25] |
| c.1168A>T | p.Ile390Phe | [25] |
| c.1192G>A | p.Ala398Thr | [5] |
| c.1193C>T | p.Ala398Val | [6] |
| c.1208A>T | p.Asp403Val | [25] |
| c.1211A>T | p.Asn404Ile | [37] |
| c.1213T>G | p.Tyr405Asp | [1] |
| c.1215C>G | p.Tyr405* | [31] |
| c.1238T>C | p.Leu413Pro | [2] |
| c.1274T>A | p.Leu425His | [38] |
| c.1274T>C | p.Leu425Pro | [2] |
| c.1283G>A | p.Cys428Tyr | [5] |
| c.1301C>T | p.Ala434Val | [29] |
| c.1310T>C | p.Met437Thr | [39, 40] |
| c.1319G>C | p.Arg440Pro | [41] |
| c.1339G>A | p.Asp447Asn | [5] |
| c.1340A>C | p.Asp447Ala | [36] |
| c.1351C>T | p.Arg451* | [29] |
| c.1355A>G | p.D452G | [35] |
| c.1369G>T | p.Asp457Tyr | [42] |
| c.1391C>T | p.Pro464Arg | [31] |
| c.1391C>G | p.Pro464Leu | [43] |
| c.1405G>C | p.Ala469Pro | [4] |
| c.1412G>A | p.Ser471Asn | [3] |
| c.1442G>C | p.Arg481Pro | [27] |
| c.1442G>A | p.Arg481Gln | [2] |
| c.1441C>T | p.Arg481* | [44, 45] |
| c.1466T>C | p.Leu489Pro | [46] |
| c.1475C>T | p.Ala492Val | [27] |
| c.1490T>C | p.Ile497Thr | [3] |
| c.1499A>T | p.Asn500Ile | [16] |
| c.1502T>C | p.Ile501Thr | [5] |
| c.1502T>A | p.Ile501Asn | [29] |
| c.1510A>G | p.Ile504Val | [6] |
| c.1514C>A | p.Ala505Glu | [47] |
| c.1526C>T | p.Ala509Val | [5] |
| c.1531A>G | p.Asn511Asp | [5] |
| c.1555T>C | p.Phe519Leu | [48] |
| c.1561G>A | p.Gly521Arg | [2] |
| c.1583C>T | p.Thr528Met | [3] |
| c.1585A>G | p.Ile529Val | [31] |
| c.1594C>T | p.Arg532Trp | [25] |
| c.1607A>G | p.Tyr536Cys | [29] |
| c.1648T>C | p.Phe550Leu | [16] |
| c.1654G>A | p.Glu552Lys | [27] |
| c.1663G>A | p.Gly555Ser | [49] |
| c.1688T>C | p.Leu563Pro | [5] |
| c.1709C>T | p.Pro570Leu | [5] |
| c.628+2T>G | splice | [50, 51] |
| c.628+5G>C | splice | [1] |
| c.629-1G>A | splice | [34] |
| c.629-2A>T | splice | [5] |
| c.981+3A>G | splice | [5] |
| c.982-2A>C | splice | [52] |
| c.982-1G>A | splice | [16] |
| c.1235+5G>A | splice | [53] |
| c.1235+1G>T | splice | [54] |
| c.1236−2A>C | splice | [3, 5] |
| c.1413−58A>G | splice | [35] |
| c.1413−1G>T | splice | [5, 39, 40] |
| c.1537−1G>T | splice | [47] |
| c.1537−3C>G | splice | [3] |
| c.1663−1G>C | splice | [2] |
| c.1663−2A>G | splice | [5] |
| c.1153delinsTT | p.Leu385Phefs*11 | [41] |
| c.1017_1020delinsGCTTTGCAAATTCG | p.Asp340Leufs*14 | [55] |
| NM_024960.5:c.54_60delTGTCTTT | p.Phe20Profs*16 | [3] |
| c.391_418del | p.Ala131Profs*65 | [29] |
| c.515_527del | p.Val172Glyfs*29 | [53] |
| c.542_549del | p.Gly181Alafs*46 | [27] |
| c.573del | p.Ser191Argfs*14 | [2, 5] |
| c.615del | p.Lys207Serfs*46 | [5] |
| c.629_672del | p.Leu210Profs*5 | [34] |
| c.738del | p.Ser247Alafs*6 | [56] |
| c.749del | p.Lys250Serfs*3 | [36] |
| c.823_824delCT | p.Leu275Valfs*16 | [5] |
| c.828_829del | p.Cys276Trpfs*15 | [30] |
| c.927_933del | p.Phe311Profs*16 | [3] |
| c.944_952del | p.Gly315_Gly317del | [31] |
| c.1085_1088del | p.Tyr362Leufs*17 | [3] |
| c.1097del | p.Pro366Leufs*14 | [31] |
| c.1176_1177del | p.Val394* | [3] |
| c.1180_1181del | p.Val394* | [1] |
| c.1259del | p.Gly420Valfs*30 | [57] |
| c.1273_1275del | p.Leu425del | [5] |
| c.1317del | p.Arg440Valfs*10 | [31] |
| c.1323_1326del | p.Asp442Alafs*7 | [5] |
| c.1418_1424del | p.Asn474* | [2] |
| c.1426_1429delATGA | p.Met476Alafs | [58] |
| c.1442_1444delGAG | p.Arg481_Glu482delinsGln | [2, 20] |
| c.1500_1501delCA | p.Ile501TrpfsX10 | [59] |
| c.569dup | p.Tyr190* | [2] |
| c.569dup | p.Tyr190* | [3] |
| c.544dup | p.Thr182Asnfs*48 | [3] |
| c.545_546insA | p.Arg183Glufs*47 | [5] |
| c.1047dup | p.Asp350* | [60] |
| c.1152dup | p.Leu385Serfs*11 | [61] |
| c.1413-9_1413-10insTTCCCC | splice | [53] |
| c.1501_1504dup | p.Gly502Aspfs*11 | [3] |
| c.1632dupG | p.Gln545Alafs*21 | [2] |
this variant scored below the first quartile in 3 out of 4 bioinformatic predictors and is predicted benign
Figure 1. Bioinformatic analyses of PANK2 variants.
Missense PANK2 variants reported pathogenic in PKAN patients were assessed bioinformatically and the resulting scores were plotted as violin plots. Outliers are plotted as red dots. The metric studied are GERP, PhyloP, PolyPhen, and SIFT. A, D, G, and J show the bioinformatic results for the missense variants reported pathogenic in the literature. B, E, H, and K show the results for all missense variants in gnomAD. C, F, I and L show the results for the gnomAD missense variants that pass the filters derived from study of the PANK2 variants reported pathogenic in the literature.
One variant had scores that were outliers for three of the four bioinformatic metrics. PANK2 NM_153638.3:c.137A>T;p.Asp4.6Val was reported in one paper as potentially causing PKAN [15]; however, it scores PolyPhen2 HDIV = 0, PhyloP Primate = −0.17 and GERPRS = −3.75. These scores are so low we considered this variant benign and did not include it in further analyses.
Bioinformatically predicting pathogenic variants for PKAN
We hypothesized that not all variants that can cause PKAN have yet been reported and that there may be individuals who are carriers of pathogenic PKAN variants in gnomAD database; which holds sequence data from approximately 141,000 individuals, none of whom are reported to carry a diagnosis of NBIA. These individuals represent global populations and include 64,603 Non-Finnish Europeans, 12,562 Finnish Europeans, 12,487 Africans, 17,720 Latinos, 9,977 East Asians, and 15,308 South Asians. All gnomAD variants present in PANK2 were analyzed for Sift, PolyPhen HDIV, GERP, and PhyloP Primate (Figure 1B, 1D, 1F, 1G). Using the bioinformatics profile generated for PKAN-causing missense variants as a benchmark, we determined thresholds for classifying variants not previously reported as pathogenic as ‘potentially pathogenic’. Missense variants were considered potentially pathogenic if they have a value for Sift, PolyPhen, GERP, and PhyloP that is equal to or higher than the mean value for three out of four of these metrics with the all scores being higher than the 1st quartile in the PKAN causing missense variant group (Table 2). All nonsense, frameshift and canonical splice site variants in gnomAD were considered pathogenic.
Table 2.
Bioinformatic predictors of pathogenicity for variants reported to cause PKAN
|
|
||
|---|---|---|
| MISSENSE | ||
| Mean | Std.Dev | |
|
|
||
| SIFT | 0.05 | 0.08 |
| PolyPhen2 | 0.94 | 0.15 |
| PhyloP | 0.53 | 0.10 |
| GERP | 4.69 | 0.56 |
A total of 46 missense variants in gnomAD met the bioinformatics profile of PKAN-causing variants. Of these 46 missense variants present in gnomAD, 33 had not previously been reported as causing PKAN. Conversely, of the 123 missense variants reported in the literature as causing PKAN, 52 were observed in gnomAD and only 25% of these passed the bioinformatic selection criteria we devised, illustrating the conservative nature of this filtering strategy. An additional 45 variants were present that were either stop gain, splice donor/acceptor, or frameshift and these were all considered pathogenic; 16 of these loss of function variants have been reported in PKAN patients. In frame insertions and deletions in gnomAD that were not already reported pathogenic in the literature were not considered pathogenic despite their potential to be, in keeping with our desire to set conservative criteria for prediction of pathogenicity.
Global allele frequencies of disease-causing PANK2 variants
The allele frequency distribution of PANK2 variants in the gnomAD cohort of approximately 141,000 individuals was similar for the pathogenic and ‘potentially pathogenic’ groups of variants. In both groups, most variants were observed in < 0.01% in all populations (Figure 2) and there was no significant difference in allele frequency distribution between the two groups. We posited that the variants reported in the literature as pathogenic may have had higher frequency than those not yet reported but observed in gnomAD. We did not detect this pattern in the data, although it may be this study was not powered to detect a pattern such as this for a rare disease.
Figure 2. Population allele frequency distributions of PANK2 variants reported pathogenic in PKAN patients and those predicted pathogenic.
The allele frequency of every variant found through literature is shown in A and the allele frequency for every variant identified in gnomAD passing bioinformatics criteria is shown in B. Each population is represented individually: EUR is Non-Finnish European, FIN is Finnish, AFR is African, LAT is Latino, EAS is East Asian, and SAS is Southeast Asian.
Initial inspection of the number of pathogenic variants in each individual population showed noticeable differences between populations (Figure 2A). Based solely on the variants reported pathogenic in the literature, the European population had more than twice as many variants as other populations. However, this elevated number in Europeans reflects likely reflects ascertainment bias in two ways. First, ascertainment of the patient population reported in the literature is commonly skewed togward European and European derived populations. Second, the number of European chromosomes in the gnomAD database is higher than other populations with −114,000 chromosomes ascertained for most of the variants in this study. Bioinformatic prediction of pathogenic variants was included to help overcome ascertainment bias that may exist in the population of published patients, and indeed the ratio of pathogenic and predicted pathogenic variants in each population per number of chromosomes was similar across most populations (Figure 2B). For example, the European population had 3x as many variants as other populations, however, the European population had roughly 3x as many chromosomes while the other populations had roughly one-third the pathogenic and ‘predicted pathogenic’ variants (Figure 2B).
Estimation of PKAN disease incidence reveals global population variation
We sought to generate an accurate estimate of PKAN disease incidence based on carrier frequencies in adult populations who do not carry a diagnosis of PKAN. Studying carrier frequencies, using both the pathogenic and ‘potentially pathogenic’ variants in a large cohort, provides a disease estimate that should less susceptible to ascertainment bias; similar to the observations on the number of variants (Figure 2). Disease incidence was estimated under three different models: 1. Variants reported in the literature only 2. Variants reported in the literature plus all loss-of-function variants in gnomAD 3. The variants described in condition 2 plus the missense variants in gnomAD predicted pathogenic. As expected these models show progressively increasing incidence (Table 3). The most conservative model (literature only) results in incidences of 1:825,974 in Europeans, 1:5,423,073 in Africans, 1:619,935 in Latino, 1:1,181,521 in East Asians and 1:6,509,160 in South Asians. However, we show there is ascertainment bias in the literature and consequently this estimate of incidence based on the literature is less accurate (Figure 2). The more robust estimate of incidence includes the variants in gnomAD not reported in the literature. This model resulted in incidence of: 1:396,006 in Europeans, 1:1,526,982 in Africans, 1:480,826 in Latino, 1:523,551 in East Asians and 1:531,118 in South Asians. These estimates show an even distribution of disease incidence across Europe, Asia and the Latin population of roughly 1 in 400,000-500,000. A notable exception is the African population incidence which remains relatively low despite bioinformatic interpretation. One possibility is that this is a sign of selective pressure against PANK2 dysfunctional alleles in this population or else African PANK2 pathogenic alleles remain unascertained.
Table 3.
PKAN disease incidence based on known and predicted pathogenic variants
|
|
||||||
|---|---|---|---|---|---|---|
| MODEL | EUR | FIN | AFR | LAT | EAS | SAS |
|
|
||||||
| Literature | 825,974 | 2,804,955 | 5,423,073 | 619,935 | 1,181,521 | 6,509,160 |
| Literature + g-LOF | 534,440 | 1,708,905 | 1,996,795 | 567,126 | 641,618 | 1,005,929 |
| Literature + g-LOF + g-missense | 396,006 | 1,708,905 | 1,526,982 | 480,826 | 523,551 | 531,118 |
Discussion
We created a bioinformatic framework to assess the potential pathogenicity of genetic alleles segregating in a large cohort of unrelated adults and subsequently generate an unbiased estimate of disease incidence [14] and applied this to PKAN. Through our bioinformatic approach we identified 28 missense variants in gnomAD that are likely pathogenic for PKAN that were not in the literature and an additional 30 loss of function variants in gnomAD that have not been reported in a PKAN patient. Disease incidence calculations based on these PANK2 variants plus those in the literature show PKAN incidence ranging from 1:396,006 in Europeans, 1:1,526,982 in Africans, 1:480,826 in Latino, 1:523,551 in East Asians and 1:531,118 in South Asians. This is the largest population-based study of the incidence of PKAN and consequently the most robust estimates for this disease.
Figure 3. Number of variants in PANK2 reported pathogenic and predicted pathogenic.
A Literature B. Literature plus those predicted pathogenic in gnomAD.
The black bars indicate the number of PANK2 missense variants and the white bars indicate the number of loss of function variants. EUR is Non-Finnish European, FIN is Finnish, AFR is African, LAT is Latino, EAS is East Asian, and SAS is Southeast Asian.
Acknowledgements
Research reported in this publication was supported by National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS083726 to PEB. PEB served as a consultant to Retrophin during the period of time it attempted to develop a treatment for PKAN. A conflict management plan was followed ensuring commitment to transparency and research integrity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pellecchia MT, Valente EM, Cif L, Salvi S, Albanese A, Scarano V, Bonuccelli U, Bentivoglio AR, D’Amico A, Marelli C, Di Giorgio A, Coubes P, Barone P, Dallapiccola B The diverse phenotype and genotype of pantothenate kinase-associated neurodegeneration Neurology, 64 (2005), pp. 1810–1812 [DOI] [PubMed] [Google Scholar]
- [2].Hartig MB, Hortnagel K, Garavaglia B, Zorzi G, Kmiec T, Klopstock T, Rostasy K, Svetel M, Kostic VS, Schuelke M, Botz E, Weindl A, Novakovic I, Nardocci N, Prokisch H, Meitinger T Genotypic and phenotypic spectrum of PANK2 mutations in patients with neurodegeneration with brain iron accumulation Ann Neurol, 59 (2006), pp. 248–256 [DOI] [PubMed] [Google Scholar]
- [3].Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome Nat Genet, 28 (2001), pp. 345–349 [DOI] [PubMed] [Google Scholar]
- [4].Morales-Briceno H, Chacon-Camacho OF, Perez-Gonzalez EA, Arteaga-Vazquez J, Rodriguez-Violante M, Cervantes-Arriaga A, Perez-Rodriguez L, Zenteno JC, Mutchinick OM Clinical, imaging, and molecular findings in a sample of Mexican families with pantothenate kinase-associated neurodegeneration Clin Genet, 87 (2015), pp. 259–265 [DOI] [PubMed] [Google Scholar]
- [5].Hayflick SJ, Westaway SK, Levinson B, Zhou B, Johnson MA, Ching KH, Gitschier J Genetic, clinical, and radiographic delineation of Hallervorden-Spatz syndrome N Engl J Med, 348 (2003), pp. 33–40 [DOI] [PubMed] [Google Scholar]
- [6].Ma LY, Wang L, Yang YM, Lu Y, Cheng FB, Wan XH Novel gene mutations and clinical features in patients with pantothenate kinase-associated neurodegeneration Clin Genet, 87 (2015), pp. 93–95 [DOI] [PubMed] [Google Scholar]
- [7].Thomas M, Hayflick SJ, Jankovic J Clinical heterogeneity of neurodegeneration with brain iron accumulation (Hallervorden-Spatz syndrome) and pantothenate kinase-associated neurodegeneration Mov Disord, 19 (2004), pp. 36–42 [DOI] [PubMed] [Google Scholar]
- [8].Zhang YM, Rock CO, Jackowski S Biochemical properties of human pantothenate kinase 2 isoforms and mutations linked to pantothenate kinase-associated neurodegeneration J Biol Chem, 281 (2006), pp. 107–114 [DOI] [PubMed] [Google Scholar]
- [9].Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Vallabh Minikel E, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Neale BM, Daly MJ, MacArthur DG Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes bioRxiv (2019), pp. 531210 [Google Scholar]
- [10].Ng PC, Henikoff S Predicting deleterious amino acid substitutions Genome Res, 11 (2001), pp. 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR A method and server for predicting damaging missense mutations Nat. Methods, 7 (2010), pp. 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol, 6 (2010), pp. e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A Detection of nonneutral substitution rates on mammalian phylogenies Genome Res, 20 (2010), pp. 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Appadurai V, DeBarber A, Chiang PW, Patel SB, Steiner RD, Tyler C, Bonnen PE Apparent underdiagnosis of Cerebrotendinous Xanthomatosis revealed by analysis of ~60,000 human exomes Mol Genet Metab, 116 (2015), pp. 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].DaRe JT, Vasta V, Penn J, Tran NT, Hahn SH Targeted exome sequencing for mitochondrial disorders reveals high genetic heterogeneity BMC Med Genet, 14 (2013), pp. 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Leoni V, Strittmatter L, Zorzi G, Zibordi F, Dusi S, Garavaglia B, Venco P, Caccia C, Souza AL, Deik A, Clish CB, Rimoldi M, Ciusani E, Bertini E, Nardocci N, Mootha VK, Tiranti V Metabolic consequences of mitochondrial coenzyme A deficiency in patients with PANK2 mutations Mol Genet Metab, 105 (2012), pp. 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Beheshtian M, Saee Rad S, Babanejad M, Mohseni M, Hashemi H, Eshghabadi A, Hajizadeh F, Akbari MR, Kahrizi K, Riazi Esfahani M, Najmabadi H Impact of whole exome sequencing among Iranian patients with autosomal recessive retinitis pigmentosa Arch Iran Med, 18 (2015), pp. 776–785 [PubMed] [Google Scholar]
- [18].Zolkipli Z, Dahmoush H, Saunders DE, Chong WK, Surtees R Pantothenate kinase 2 mutation with classic pantothenate-kinase-associated neurodegeneration without ‘eye-of-the-tiger’ sign on MRI in a pair of siblings Pediatr Radiol, 36 (2006), pp. 884–886 [DOI] [PubMed] [Google Scholar]
- [19].Song XW, Wang YL, Shi YW, Deng WY, Chen SQ, Lin H, Yi YH, Liao WP [Clinical manifestations and detection of pantothenate kinase 2 gene mutation in a patient with Hallervorden-Spatz syndrome] Zhonghua Yi Xue Za Zhi, 89 (2009), pp. 3320–3323 [PubMed] [Google Scholar]
- [20].Rump P, Lemmink HH, Verschuuren-Bemelmans CC, Grootscholten PM, Fock JM, Hayflick SJ, Westaway SK, Vos YJ, van Essen AJ A novel 3-bp deletion in the PANK2 gene of Dutch patients with pantothenate kinase-associated neurodegeneration: evidence for a founder effect Neurogenetics, 6 (2005), pp. 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gatto E, Etcheverry JL, Converso DP, Bidinost C, Rosa A Pantothenate kinase-associated neurodegeneration: novel mutations in the PANK2 gene in an Argentinean young woman Mov Disord, 25 (2010), pp. 2262–2264 [DOI] [PubMed] [Google Scholar]
- [22].Saleheen D, Ali T, Aly Z, Khealani B, Frossard PM Novel mutation in the PANK2 gene leads to pantothenate kinase-associated neurodegeneration in a Pakistani family Pediatr Neurol, 37 (2007), pp. 296–298 [DOI] [PubMed] [Google Scholar]
- [23].Schiessl-Weyer J, Roa P, Laccone F, Kluge B, Tichy A, De Almeida Ribeiro E, Prohaska R, Stoeter P, Siegl C, Salzer U Acanthocytosis and the c.680 A>G Mutation in the PANK2 Gene: A Study Enrolling a Cohort of PKAN Patients from the Dominican Republic PLoS One, 10 (2015), pp. e0125861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tanteles GA, Spanou-Aristidou E, Antoniou C, Christophidou-Anastasiadou V, Kleopa KA Novel homozygous PANK2 mutation causing atypical pantothenate kinase-associated neurodegeneration (PKAN) in a Cypriot family J Neurol Sci, 340 (2014), pp. 233–236 [DOI] [PubMed] [Google Scholar]
- [25].Dezfouli MA, Alavi A, Rohani M, Rezvani M, Nekuie T, Klotzle B, Tonekaboni SH, Shahidi GA, Elahi E PANK2 and C19orf12 mutations are common causes of neurodegeneration with brain iron accumulation Mov Disord, 28 (2013), pp. 228–232 [DOI] [PubMed] [Google Scholar]
- [26].Assami S, Azzedine H, Nouioua S, Mundwiller E, Mahoui S, Makri S, Djemai M, Grid D, Brice A, Hamadouche T, Stevanin G, Tazir M Pantothenate kinase-associated neurodegeneration: clinical description of 10 patients and identification of new mutations Mov Disord, 26 (2011), pp. 1777–1779 [DOI] [PubMed] [Google Scholar]
- [27].Westaway SK, Ching KH, Levinson B, Gitschier J, Hayflick SJ Gene symbol: PANK2. Disease: pantothenate kinase-associated neurodegeneration (PKAN) Hum Genet, 119 (2006), pp. 678. [PubMed] [Google Scholar]
- [28].Saleheen D, Nazir A, Khanum S, Haider SR, Frossard P A novel mutation in a patient with pantothenate kinase-associated neurodegeneration Cmaj, 173 (2005), pp. 578–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lim BC, Ki CS, Cho A, Hwang H, Kim KJ, Hwang YS, Kim YE, Yun JY, Jeon BS, Lim YH, Paek SH, Chae JH Pantothenate kinase-associated neurodegeneration in Korea: recurrent R440P mutation in PANK2 and outcome of deep brain stimulation Eur J Neurol, 19 (2012), pp. 556–561 [DOI] [PubMed] [Google Scholar]
- [30].Kojovic M, Kuoppamaki M, Quinn N, Bhatia KP “Progressive delayed-onset postanoxic dystonia” diagnosed with PANK2 mutations 26 years after onset-an update Mov Disord, 25 (2010), pp. 2889–2891 [DOI] [PubMed] [Google Scholar]
- [31].Egan RA, Weleber RG, Hogarth P, Gregory A, Coryell J, Westaway SK, Gitschier J, Das S, Hayflick SJ Neuro-ophthalmologic and electroretinographic findings in pantothenate kinase-associated neurodegeneration (formerly Hallervorden-Spatz syndrome) Am J Ophthalmol, 140 (2005), pp. 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vasconcelos OM, Harter DH, Duffy C, McDonough B, Seidman JG, Seidman CE, Campbell WW Adult Hallervorden-Spatz syndrome simulating amyotrophic lateral sclerosis Muscle Nerve, 28 (2003), pp. 118–122 [DOI] [PubMed] [Google Scholar]
- [33].Doi H, Koyano S, Miyatake S, Matsumoto N, Kameda T, Tomita A, Miyaji Y, Suzuki Y, Sawaishi Y, Kuroiwa Y Siblings with the adult-onset slowly progressive type of pantothenate kinase-associated neurodegeneration and a novel mutation, Ile346Ser, in PANK2: clinical features and (99m)Tc-ECD brain perfusion SPECT findings J Neurol Sci, 290 (2010), pp. 172–176 [DOI] [PubMed] [Google Scholar]
- [34].Shan J, Wen B, Zhu J, Lin P, Zheng J, Yan C Novel PANK2 gene mutations in two Chinese siblings with atypical pantothenate kinase-associated neurodegeneration Neurol Sci, 34 (2013), pp. 561–563 [DOI] [PubMed] [Google Scholar]
- [35].Wu YR, Chen CM, Chao CY, Lyu RK, Lee-Chen GJ Pantothenate kinase-associated neurodegeneration in two Taiwanese siblings: identification of a novel PANK2 gene mutation Mov Disord, 24 (2009), pp. 940–941 [DOI] [PubMed] [Google Scholar]
- [36].Aggarwal A, Schneider SA, Houlden H, Silverdale M, Paudel R, Paisan-Ruiz C, Desai S, Munshi M, Sanghvi D, Hardy J, Bhatia KP, Bhatt M Indian-subcontinent NBIA: unusual phenotypes, novel PANK2 mutations, and undetermined genetic forms Mov Disord, 25 (2010), pp. 1424–1431 [DOI] [PubMed] [Google Scholar]
- [37].Camargos ST, Gurgel-Giannetti J, Lees A, Hardy J, Singleton A, Cardoso F Low prevalence of PANK2 mutations in Brazilian patients with early onset generalised dystonia and basal ganglia abnormalities on MRI J Neurol Neurosurg Psychiatry, 82 (2011), pp. 1059–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kazek B, Jamroz E, Gencik M, Jezela Stanek A, Marszal E, Wojaczynska-Stanek K A novel PANK2 gene mutation: clinical and molecular characteristics of patients short communication J Child Neurol, 22 (2007), pp. 1256–1259 [DOI] [PubMed] [Google Scholar]
- [39].Houlden H, Lincoln S, Farrer M, Cleland PG, Hardy J, Orrell RW Compound heterozygous PANK2 mutations confirm HARP and Hallervorden-Spatz syndromes are allelic Neurology, 61 (2003), pp. 1423–1426 [DOI] [PubMed] [Google Scholar]
- [40].Orrell RW, Amrolia PJ, Heald A, Cleland PG, Owen JS, Morgan-Hughes JA, Harding AE, Marsden CD Acanthocytosis, retinitis pigmentosa, and pallidal degeneration: a report of three patients, including the second reported case with hypoprebetalipoproteinemia (HARP syndrome) Neurology, 45 (1995), pp. 487–492 [DOI] [PubMed] [Google Scholar]
- [41].Yoon WT, Lee WY, Shin HY, Lee ST, Ki CS Novel PANK2 gene mutations in korean patient with pantothenate kinase-associated neurodegeneration presenting unilateral dystonic tremor Mov Disord, 25 (2010), pp. 245–247 [DOI] [PubMed] [Google Scholar]
- [42].Dusek P, Tovar Martinez EM, Madai VI, Jech R, Sobesky J, Paul F, Niendorf T, Wuerfel J, Schneider SA 7-Tesla Magnetic Resonance Imaging for Brain Iron Quantification in Homozygous and Heterozygous PANK2 Mutation Carriers Mov Disord Clin Pract, 1 (2014), pp. 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chan KY, Lam CW, Lee LP, Tong SF, Yuen YP Pantothenate kinase-associated neurodegeneration in two Chinese children: identification of a novel PANK2 gene mutation Hong Kong Med J, 14 (2008), pp. 70–73 [PubMed] [Google Scholar]
- [44].Ching KH, Westaway SK, Gitschier J, Higgins JJ, Hayflick SJ HARP syndrome is allelic with pantothenate kinase-associated neurodegeneration Neurology, 58 (2002), pp. 1673–1674 [DOI] [PubMed] [Google Scholar]
- [45].Higgins JJ, Patterson MC, Papadopoulos NM, Brady RO, Pentchev PG, Barton NW Hypoprebetalipoproteinemia, acanthocytosis, retinitis pigmentosa, and pallidal degeneration (HARP syndrome) Neurology, 42 (1992), pp. 194–198 [DOI] [PubMed] [Google Scholar]
- [46].Yapici Z, Akcakaya NH, Tekturk P, Iseri SA, Ozbek U A novel gene mutation in PANK2 in a patient with severe jaw-opening dystonia Brain Dev, 38 (2016), pp. 755–758 [DOI] [PubMed] [Google Scholar]
- [47].Matarin MM, Singleton AB, Houlden H PANK2 gene analysis confirms genetic heterogeneity in neurodegeneration with brain iron accumulation (NBIA) but mutations are rare in other types of adult neurodegenerative disease Neurosci Lett, 407 (2006), pp. 162–165 [DOI] [PubMed] [Google Scholar]
- [48].Pan LS, Yu LH, Yin YY, Xu YM A novel PANK2 mutation in a 12-year-old Chinese boy with pantothenate kinase-associated neurodegeneration Neurol India, 61 (2013), pp. 175–176 [DOI] [PubMed] [Google Scholar]
- [49].Perez-Gonzalez EA, Chacon-Camacho OF, Arteaga-Vazquez J, Zenteno JC, Mutchinick OM A novel gene mutation in PANK2 in a patient with an atypical form of pantothenate kinase-associated neurodegeneration Eur J Med Genet, 56 (2013), pp. 606–608 [DOI] [PubMed] [Google Scholar]
- [50].Tanrikulu B, Ozen A, Gunal DI, Turkdogan D, Bayrakli F, Bayri Y, Dagcinar A, Seker A Deep brain stimulation as treatment for dystonic storm in pantothenate kinase-associated neurodegeneration syndrome: case report of a patient with homozygous C.628 2 T > G mutation of the PANK2 gene Acta Neurochir (Wien), 157 (2015), pp. 1513–1516; discussion 1516–1517 [DOI] [PubMed] [Google Scholar]
- [51].Siegl C, Hamminger P, Jank H, Ahting U, Bader B, Danek A, Gregory A, Hartig M, Hayflick S, Hermann A, Prokisch H, Sammler EM, Yapici Z, Prohaska R, Salzer U Alterations of red cell membrane properties in neuroacanthocytosis PLoS One, 8 (2013), pp. e76715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wieczorek SW, Epplen JT Gene symbol: PANK2. Disease: Pantothenate kinase associated neurodegeneration (PKAN) Hum Genet, 116 (2005), pp. 545. [PubMed] [Google Scholar]
- [53].Hajek M, Adamovicova M, Herynek V, Skoch A, Jiru F, Krepelova A, Dezortova M MR relaxometry and 1H MR spectroscopy for the determination of iron and metabolite concentrations in PKAN patients Eur Radiol, 15 (2005), pp. 1060–1068 [DOI] [PubMed] [Google Scholar]
- [54].Cherot E, Keren B, Dubourg C, Carre W, Fradin M, Lavillaureix A, Afenjar A, Burglen L, Whalen S, Charles P, Marey I, Heide S, Jacquette A, Heron D, Doummar D, Rodriguez D, Billette de Villemeur T, Moutard ML, Guet A, Xavier J, Perisse D, Cohen D, Demurger F, Quelin C, Depienne C, Odent S, Nava C, David V, Pasquier L, Mignot C Using medical exome sequencing to identify the causes of neurodevelopmental disorders: Experience of 2 clinical units and 216 patients Clin Genet, 93 (2018), pp. 567–576 [DOI] [PubMed] [Google Scholar]
- [55].Aryani O, Houshmand M, Fatehi F A novel PANK2 gene mutation in a Persian boy: the first report from Iran Clin Neurol Neurosurg, 115 (2013), pp. 1170–1172 [DOI] [PubMed] [Google Scholar]
- [56].Sohn EH, Michaelides M, Bird AC, Roberts CJ, Moore AT, Smyth D, Brady AF, Hungerford JL Novel mutation in PANK2 associated with retinal telangiectasis Br J Ophthalmol, 95 (2011), pp. 149–150 [DOI] [PubMed] [Google Scholar]
- [57].Zorzi G, Zibordi F, Chiapparini L, Bertini E, Russo L, Piga A, Longo F, Garavaglia B, Aquino D, Savoiardo M, Solari A, Nardocci N Iron-related MRI images in patients with pantothenate kinase-associated neurodegeneration (PKAN) treated with deferiprone: results of a phase II pilot trial Mov Disord, 26 (2011), pp. 1756–1759 [DOI] [PubMed] [Google Scholar]
- [58].Dastsooz H, Nemati H, Fard MAF, Fardaei M, Faghihi MA Novel mutations in PANK2 and PLA2G6 genes in patients with neurodegenerative disorders: two case reports BMC Med Genet, 18 (2017), pp. 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim J, Shin H, Youn J, Ki CS, Cho AR, Cho JW A novel PANK2 gene mutation with sudden-onset dystonia Can J Neurol Sci, 39 (2012), pp. 395–397 [DOI] [PubMed] [Google Scholar]
- [60].Lee CH, Lu CS, Chuang WL, Yeh TH, Jung SM, Huang CL, Lai SC Phenotypes and genotypes of patients with pantothenate kinase-associated neurodegeneration in Asian and Caucasian populations: 2 cases and literature review Scientific World Journal, 2013 (2013), pp. 860539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Seo JH, Song SK, Lee PH A Novel PANK2 Mutation in a Patient with Atypical Pantothenate-Kinase-Associated Neurodegeneration Presenting with Adult-Onset Parkinsonism J Clin Neurol, 5 (2009), pp. 192–194 [DOI] [PMC free article] [PubMed] [Google Scholar]