Highlights
-
•
Diazotrophic bacteria were screened and characterized from long-term organic rice fields of Sikkim
-
•
Screened the diazotrophic bacteria for nitrogen fixing ability and plant growth promoting traits
-
•
Enterobacter was commonly found in paddy soils of Sikkim
-
•
Enterobacter species were significantly enhanced the above and below ground responses in rice plants
Keywords: Diazotroph, 16S rRNA gene, Rice, Nitrogen dynamics, Soil, Rhizosphere
Abstract
This study addresses the plant beneficial enterobacteria present in rice rhizosphere and their efficiency for enhancing nitrogen uptake in rice plant. Using culturable approaches, the population of total diazotrophs present in rhizosphere samples collected from different organic rice fields of Sikkim were studied and recorded in the range between 4.62 to 4.97 log10 CFU/g soil. All the isolated commonly occurred diazotrophic bacterial isolates were screened based on their ability to fix nitrogen in milligram per gram of sugar consumed under in-vitro condition with the reference check. In addition to nitrogen fixation, plant growth promoting traits such as production of indole-3-acetic acid and gibberellic acid were estimated using spectrophotometric approaches and compared against Bacillus subtilis as reference multi-potent plant growth promoting strain. In-vivo evaluation of these diazotrophic species in rice found improvement in both above and below ground responses in rice plant evaluated by estimating changes in chlorophyll concentration, plant biomass, root architecture, nitrogen uptake, microbial biomass and associated biochemical activity of soil. Further, the selected isolates were identified through DNA targeted analysis of 16S rRNA gene present in diazotrophs and which identified that the isolates belonged to the Enterobacter genus. Statistical models were prepared for deciphering the dynamics of plant growth improvement due to selective enrichment of rhizosphere bacteria and found significant (p<0.05) correlation between soil and plant parameters. This study concludes that Enterobacter spp. present in organic paddy soils of Sikkim having good nitrogen fixing abilities and whose selective enrichment in rhizosphere improved nitrogen uptake and plant growth promotion in rice plant.
Graphical abstract
Introduction
In plant and soil interactions, rhizosphere play important roles for plant nutrition, growth promotion, and biotic and abiotic stress management. Rhizosphere is an important site for residents of most of the microorganisms, of which bacteria play a major role for soil and plant health management. The soil which is nearest to the plant root is called rhizosphere, wherein plant root releases different metabolites (Nihorimbere et al., 2011) and it decides the colonization of microorganism. Sikkim an agricultural state located at the Himalayan mountain range in the North Eastern India cultivates rice as the major cereal crop under organic farming practice. As hill farming becomes the predominant farming practice in mountainous regions, crops cultivated are vulnerable towards edaphic stresses among which soil nutrients have been given importance due to their role in plant growth and development. Among the different macronutrients present in soil, nitrogen (N) has been regarded as one of the primary growth limiting nutrient required excessively by rice plant for biosynthesis of chlorophyll and enzymes, as compared to other macronutrients (Ladha and Reddy, 2000). Hence, rice cultivation greatly depends upon the soil N content and has been estimated in terms of chemical N fertilizers to require atleast 100 kg N ha−1 to maintain crop productivity (Fierer et al., 2012). N deficiency in soil have resulted in decrease of photosynthetic ability of leaf apparently due to reduction of total chlorophyll content (Huang et al., 2004). Moreover, N deficiency in soil have been found responsible for disturbing the root architecture by reduction of root growth (Hsieh et al., 2018). Therefore, stringent N management practices is required in organic farming for alleviating N stress in plant and increasing rice production. For preserving soil N content under organic farming practices, application of animal manures, green manures, and compost have been traditionally practiced in Sikkim. In addition to organic soil amendments, application of heterotrophic diazotrophs have been found to play important role towards improvement of soil N content by participating in soil biological nitrogen fixation (BNF) (Fierer et al., 2012). In rice cultivation, BNF have played pivotal role in coping with crop N demand (Ladha and Reddy, 2003) by fixing 180 × 106 metric tons/year N globally (Tilak et al., 2005) and has the potential to substitute chemical N fertilizer, (Choudhury et al., 2004). Therefore, BNF in soil provides an effective measure for sustainable management of soil N, which is in compliances with the principles of ecological security and resource conservation (Sofi and Wani, 2007). Most of the work on rice BNF has been carried using Azotobacter and Azospirillum strains (Tilak et al., 2005). Comparatively there are limited descriptions of other rice rhizosphere associated bacteria such as Enterobacter oryziphilus, E. oryzendophyticus (Hardoim et al., 2013), E. cloacae (Shankar et al., 2011), E. oryzae (Peng et al., 2009) and E. agglomerans (Achouak et al., 1994) which are capable of tolerating anaerobic growth environment as found in case of wetland rice cultivation (Hardoim et al., 2013) and also have been reported to benefit rice plant growth particularly by stimulating root growth by bacterial phytohormones, and improvement of nutrient availability in rhizosphere by fixing atmospheric nitrogen (Lima et al., 2018; Shankar et al., 2011). As organic farming has been the only form of agriculture judiciously practiced in Sikkim, application of nitrogenous biofertilizers shall provide viable solution to combat soil N depletion and improve plant health. Moreover, as soil management in this region has been maintained by organic methods, the soil microbial diversity could have been well preserved as compared to regions where rice cultivation is carried by conventional farming practices. Therefore, in order to provide sustainable measures for counteracting N loss from soil, the diazotrophic bacterial diversity was studied and indigenous diazotrophic isolates were assayed for their competency as plant growth regulators in rice plants cultivated under organic farming practices at Sikkim, India.
Material and methods
Soil sampling and isolation of diazotrophic bacteria
Soil samples were collected at depth of 15–2 cm from surface of soil in five organic rice (Oryza sativa L.) growing areas located at East Sikkim (27.3084°N and 88.6724°E) (Panneerselvam et al., 2019). In each field five representative soil samples were collected. From the soil samples, roots and stones were removed and samples were stored at 4°C. For isolation of diazotrophic bacteria, N free Jensen's medium was used containing 20.0 g sucrose, 1.0 g K2HPO4, 0.5 g MgSO4.7H2O, 0.5 g NaCl, 0.1 g K2SO4, 0.005 g Na2MoO4, 18 g agar, per liter distilled water, pH 6.9; serial dilutions of the rhizospheric soils was done up to 10−6 dilutions and then 100µl of diluted samples were inoculated on N free Jensen's medium plates and the colonies developed after 24 hours of incubation were isolated, purified and preserved using Jensen's agar medium.
Estimating nitrogen assimilation by diazotrophs under in-vitro condition
Nitrogen fixed by free living bacteria was evaluated from the amount of nitrogen present in N free Jensen's broth medium after fifteen days of incubation and expressed as mg of N fixed per g of sucrose consumed. Nitrogen assimilated in broth from atmosphere by the diazotrophs was estimated using Kjeldahl method (Kirk, 1950) from the broth residues after subjecting to centrifugation at 5000 rpm for 20 min. Total sugar present in the supernatant was estimated using anthrone method (Morse, 1947) and was used to calculate nitrogen fixed per gram of carbon.
Spectrophotometric estimation of phytohormone production by bacteria
Indole-3-acetic acid (IAA) production: IAA was determined as per the standard method (Gordon and Weber, 1951). Bacteria cultured in 5mM L-tryptophan amended nutrient broth for 5 days at 30 °C was centrifuged at 5000 rpm for 20 min. Salkowaski reagent (2% 0.5M FeCl3 in 35% perchloric acid) 2 mL was added to 1 mL of culture supernatant and the colour change was monitored after 30 min at 535 nm in spectrophotometer. Standard graph was prepared using IAA (Sigma) and concentration of IAA expressed in μg ml−1. Gibberellic acid (GA3) production: GA3 was determined as per the spectrophotometric method (Borrow et al., 1995). Bacteria cultured in nutrient broth for 7 days was centrifuged at 5000 rpm for 20 min. Equal volume of zinc acetate solution was added to 2 mL supernatant followed by addition of equal volume of potassium ferrocyanide and centrifuged at 5000 rpm for 20 min. GA3 was extracted from supernatant using equal volume of 30% hydrochloric acid upon incubation at 27 °C for 75 min and intensity measured at 254 nm in a UV-VIS spectrophotometer. Standard graph was prepared using GA3 (Sigma) and concentration of GA3 expressed in μg ml−1.
Bacterial identification using 16S rRNA molecular probes and DNA sequence analysis
Genomic DNA isolation from bacteria isolates were carried using Bacterial DNA isolation kit manufactured by ZymoTM, USA. Approximately 5 mL of 24 h old bacterial culture grown on nutrient broth was centrifuged at 8000 rpm for 15 min and the DNA extraction from cell pellet was carried as per protocol available in the kit. The quantity and purity of isolated DNA was determined by UV spectrophotometer (Nanodrop; Thermo Scientific). PCR amplification of 16S rRNA gene from bacteria isolates was carried using the primers 27f (5′-AGAGTTTGATCCTGGCTCAG) and 1492r (5′-GGTTACCTTGTTACGACTT) (Suzuki and Giovannoni, 1996), following which the gene sequencing of the purified PCR product was conducted through Sanger sequencing by Eurofins, Bengaluru, India. Contigs were generated from the resulting forward and reverse read sequences using CAP3 assembly program (Huang and Madan, 1999) following which presence of chimera was checked using DECIPHER v2.0 (Wright, 2016) and the sequences were submitted to NCBI database after identifying the bacteria using nBLAST. The sequence data was aligned with MEGAX-ClustalW and the phylogenetic tree analyses were conducted in MEGAX using neighbour-joining method. The topology of phylogenetic dendrogram was ascertained using 1000 bootstrap resampling method.
Seed inoculation with bacteria and description of experimental control
Seeds of Oryza sativa var. Naveen were used for the experiment under glass house conditions, which were surface sterilized in 2% sodium hypochlorite for 5 min and thoroughly washed five times with sterile double distilled water, prior to bacterial treatment procedure. There were total nine treatments with four replication, which included five enterobacterial isolates as test strains viz. E. asburiae strain PANW4 (MK318650), Enterobacter sp. strain PANW4/1 (MK318651), Enterobacter sp. strain PANW5 (MK318652), E. kobei strain PANW6 (MK318653) and Enterobacter sp. strain PANW9 (MK318654), three positive experimental controls out of which one was Gram negative diazotrophs belonging to A. tropicalis AT and one Gram positive diazotroph belonging to Bacillus genera i.e. B. subtilis, and one negative control which included seed treatment and soil drenching with sterile Jensen's broth medium. Seed bacterization process which involved thorough mixing of inoculum and seeds through agitation for 30 min was carried with an inoculum dose of 5 mL inoculum per 1 kg of rice seeds. The bacterial cultures used as inoculants with minimum of 5 × 1012 CFU/ml bacteria cell density maintained colorimetrically after repeated sedimentation of broth suspensions by centrifugation at 5000 rpm for 20 min and dilution using sterile distilled water. A. tropicalis AT (NAIMCC-B-01336), and B. subtilis BioCWB (MG490133.1) used as potential standard nitrogen fixing and phytohormone producing strains, were collected from Crop Production Division, ICAR-NRRI among which the strain BioCWB was previously isolated from Sikkim soil and has proven multifunctional plant growth promoting traits (Panneerselvam et al., 2019). Five bacterized seeds were sown per pot (19.43 cm diameter x 30.48 cm depth) filled with 5 kg sterile soil. Following seed germination, two seedlings per pot was maintained. Sterile distilled water was used for maintaining moisture content at field capacity throughout the experiment. Parameters measured for plant included agronomic properties and total plant nitrogen content in plant, whereas for soil the parameters included soil available nitrogen content and soil biochemical and microbiological properties as described in the following sections.
Plant sample analysis
Plant growth parameters were monitored after 60 days after sowing seeds, which includes shoot length (SL), root length (RL), shoot weight, root weight and total dry plant biomass (BIOM) which was the cumulating measure from dry shoot weight and root weight. Non destructive analysis of leaf chlorophyll content was measured using SPAD-502 (Konica-Minolta, Japan) meter. Root samples were scanned at 600 dpi (Epson V 700) and analyzed using WinRhizo v.2007d software. Nitrogen content of plant was analyzed by hot acid digestion and estimating of nitrogen content using Kjeladhl method (Tkachuk, 1969).
Soil sample analysis
Soil samples collected from each experimental pot after 60 days after sowing were used for analysis purposes. Soil parameters pH and electrical conductivity (EC) were measured using digital sensors after appropriate dilution in distilled water (Black et al., 1965). Kjeladhl method was used for estimation of soil available nitrogen (AN) as per standard procedure (Subbiah and Asija, 1956). Dichromate titrimetric method was used for estimation of soil organic carbon (SOC) (Jackson 1967). Spectrophotometric analysis of Bray's extract was used for estimating available phosphorus (AP) in soil (Bray and Kurtz, 1945). Flame spectroscopy of ammonium acetate extract was used for estimation of available potassium (AK) in soil. Soil microbial biomass carbon (MBC) was estimated using chloroform fumigation method using 0.5 M potassium sulfate solution (K2SO4) as extractant (Kumar et al., 2017). Fluorescein diacetate (FDA) hydrolase activity was estimated spectrophotometrically by extracting soil enzymes particularly esterase (Mahapatra et al., 2017). Reduction of triphenyl tetrazolium chloride (TTC) by soil enzymes was used for estimating dehydrogenase activity (DHA) spectrophotometrically (Chatterjee et al., 2019).
Data analysis
R v3.4.1 was used for performing data analysis of the results pertaining to plant and soil analysis. Correlation was made using the package ‘igraph’ and linear regression model was prepared using different parameters, following which ‘ols_step’ function of package olsrr was used for calculating regression coefficients of parameters in step modeling. Spearman's p correlation was carried using the function ‘cor’ and the correlation network was visualized using the function ‘qgraph’. Welch's t-test was used to determine the variances between treatments. For linear regression models the function ‘lm’ was used to predict nitrogen uptake (Nup) values i.e. Y based on different plant and soil parameters estimated i.e. X. Supervised learning was performed using the linear models followed the generalized mathematical equation: Y = β1 + β2X + ε, where the regression coefficients β1 is the intercept and β2 is the slope with ε representing the error of regression model.
Results
Physicochemical and microbiological characteristics of soil from different organic rice cultivating areas in Sikkim
Soil collected from five different locations were found to be acidic in nature with values ranging from pH 4.54 to 6.04. As represented in Table 1, among the five different sampling locations, Nandok had highly acidic soil followed by Lingzey, Chhota Singtam, Saurini and Saramsa. Measurements of soil EC and SOC showed variations between 32.4 uS/cm to 64.38 uS/cm and 1.9 % to 2.8 %, respectively. The concentration of major plant nutrients in soil were determined: AN (185.04 kg/ha to 274.63 kg/ha), AP (36.7 kg/ha to 41.22 kg/ha) and AK (322.70 kg/ha to 328.19 kg/ha), which showed soil samples collected from Nandok were optimally rich in major plant nutrients. Compelling differences in MBC (432.5 µg/g soil to 1182.5 µg/g soil) were observed in soil samples from Chhota Singtam demonstrating the richness in soil microflora at that location. Heterotrophic diazotrophs cultured from soils of Saramsa demonstrated higher diazotropic population log10 4.97 CFU as compared to other rice cultivating locations. The functional attributes of soil microflora were characterized by monitoring FDA and DHA activities, which revealed higher functionality of microflora in Saramsa as determined through FDA activity which ranged from 7.27 µg fluorescein/g soil to 18.22 µg fluorescein/g soil and DHA activity which ranged from 262.04 µg TPF/kg/24 h to 621.13 µg TPF/kg/24 h. Soil samples from Saramsa had higher AP content and heterotrophic diazotropic population in soil as compared to other soil sampling areas.
Table 1.
Physicochemical properties of surveyed soil samples from different parts of Sikkim (pH, EC: electrical conductivity, SOC: soil organic carbon, AN: available nitrogen, AP: available phosphorus and AK: available potassium) and soil microbiological properties estimates (MBC: microbial biomass carbon, DHA: soil dehydrogenase active, FDA: Fluorescein diacetate hydrolysis.
| Parameters | Saurini | Chhota Singtam | Nandok | Saramsa | Lingzey |
| pH | 5.55±0.31 | 5.3±0.26 | 4.54±0.48 | 6.04±0.52 | 5.09±0.27 |
| EC (µS/cm) | 44.37±0.79 | 39.88±0.29 | 32.4±0.54 | 64.38±1.04 | 38.13±0.87 |
| SOC (%) | 2.2±0.23 | 1.9±0.29 | 2.8±0.41 | 2.40±0.25 | 1.60±0.42 |
| AN (kg/ha) | 268.21±0.73 | 210.1±0.16 | 274.63±1.00 | 271.31±1.43 | 185.04±1.46 |
| AP (kg/ha) | 38.43±0.32 | 37.11±1.08 | 40.19±0.69 | 41.22±1.29 | 36.70±1.31 |
| AK (kg/ha) | 325.12±0.52 | 322.7±0.70 | 325.5±0.50 | 328.19±0.82 | 326.09±0.66 |
| MBC (µg/g soil) | 783.83±1.51 | 1182.5±1.32 | 981.5±1.20 | 432.5±1.37 | 863.86±0.29 |
| FDA (µg fluorescein /g) | 12.36±0.51 | 7.27±0.86 | 16.93±0.60 | 18.22±0.26 | 11.84±0.99 |
| DHA (µg TPF/kg/24h) | 402.23±1.68 | 279.10±1.16 | 436.42±0.98 | 621.13±1.09 | 262.04±1.07 |
| Diazotrophs (log10CFU/gsoil) | 4.85±0.52 | 4.67±0.50 | 4.60±0.51 | 4.97±0.54 | 4.62±0.50 |
Characterization of nitrogen fixing and phytohormones production potential in diazotrophic isolates
A total of 63 heterotrophic diazotrophs were isolated from Sikkim soils among which 15 isolates had abilities to assimilate >1 mg N/g of sucrose utilized in N-free Jensen's medium (Table 2). Preliminary determination of N-fixing potentiality was carried by repeated sub-culturing of isolates in N-free Jensesn's agar medium for 10 times, which showed similar growth pattern for all isolates in subsequent sub-culturing. Out of fifteen isolates, following six isolates viz., PANW4/1 (10.16 mgN/gC), PANW4 (9.3 mgN/gC), PANW6 (9.1 mgN/gC), PANW5 (7.19 mgN/gC) and PANW2/1 (6.72 mgN/gC) and PANW9 (6.3 mgN/gC) had higher N assimilating potential. Phytohormones production ability by selected diazotrophic isolates were measured and the results are given in (Table 2). From the spectrophotometric estimation of phytohormones production, it was inferred that among the 15 diazotrophic isolates, IAA production was found to be highest in PANW4/1 (45.1 ug/mL), followed by PANW5 (37.01 ug/mL), PANW4 (36.8 ug/mL), PANW9 (29.83 ug/mL) and PANW6 (27.8 ug/mL). Whereas GA production, PANW9 (8.12 ug/mL) and PANW4/1 (7.63 ug/mL) recorded higher than other isolates (Table 2).
Table 2.
Screening of diazotrophs based on nitrogen (N) fixing ability, indole acetic acid (IAA) and gibberellic acid (GA3) production. Similar letter found across each parameter shows no difference between the respective values at 5% level of significance. Responses undetected are represented with ‘nd’.
| Diazotrophs | N fixation | IAA | GA3 |
| (mg N/g sugar consumed) | (ug/ml) | (ug/ml) | |
| PANW1 | 5.11i | 12.82g | 1.14jk |
| PANW2 | 3.25k | 9.60i | 2.30g |
| PANW2/1 | 6.72f | Nd | 0.56mn |
| PANW4 | 9.3c | 36.80b | 4.13e |
| PANW4/1 | 10.16b | 45.20a | 7.63c |
| PANW5 | 7.19e | 37.01b | 2.05h |
| PANW5/2 | 4.33j | 10.19i | 0.22nop |
| PANW6 | 9.10d | 27.80d | 6.43d |
| PANW7 | 6.14g | Nd | Nd |
| PANW9 | 6.30g | 29.83c | 8.12d |
| PANW9/1 | 1.07m | 12.11h | 0.84op |
| PANW9/2 | 0.84m | 8.51j | 1.51b |
| PANW9/3 | 0.72m | Nd | 0.93lm |
| PANW11 | 5.43h | 12.13h | Nd |
| PANW12/1 | 2.19l | 15.40f | 1.60kl |
| +ve Control AT | 12.31a | 23.28e | 8.60a |
| +ve Control BioCWB | 4.51j | 45.05a | 8.47a |
| CD(0.05) | 0.32 | 0.72 | 0.31 |
16SrRNA. molecular identification of phytohormones producing diazotrophic isolates
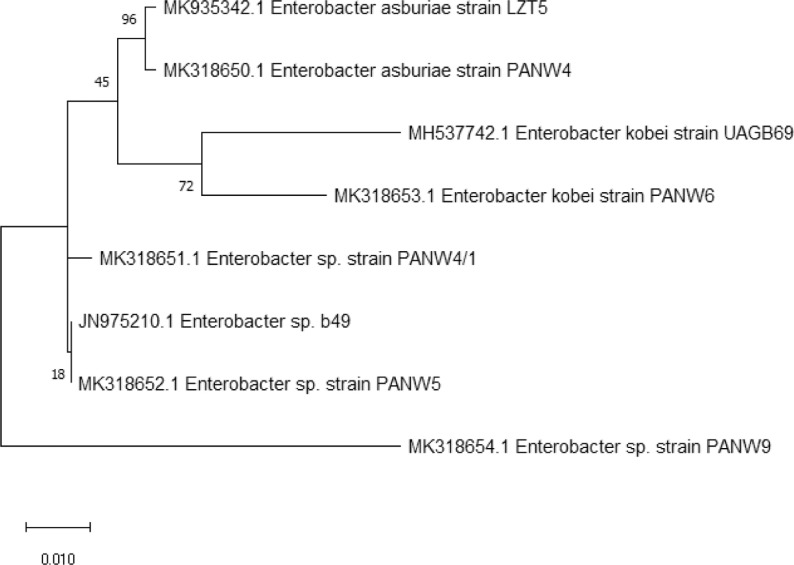
Phylogenetic dendrogram constructed based on molecular distance matrix showed more than maximum similarity in species level. Fig. 1 shows all of the isolates belong to the heterobacterial diazotrophs and Enterobacter genera i.e. E. asburiae strain PANW4 (MK318650), Enterobacter sp. strain PANW4/1 (MK318651), Enterobacter sp. strain PANW5 (MK318652), E. kobei strain PANW6 (MK318653), Enterobacter sp. strain PANW9 (MK318654).
Fig. 1.
Phylogenetic analysis of hypervariation within 16S rRNA region of diazotrophic isolates.
Effect of bacterial inoculation on upper and below ground responses in rice plants
A total of 12 parameters were studied for understanding the upper and below ground responses in bacteria inoculated plants. The changes associated with the agronomic characteristics (Table 3) showed that plants inoculated with Enterobacter sp. strain PANW4/1 demonstrated results at par with positive control of Azotobacter strains AT. The Enterobacter sp. strain PANW4/1 inoculated plants registered an increase in leaf chlorophyll content (34.230 SPAD value), root length (RL) 38.279 cm, projected root area (PRA) 26.84 mm2, root surface area (RSA) 27.81 mm2, root average diameter (RAD) 0.716 mm and dry biomass weight (BIOM) 2.809 g/plant. The changes associated with soil nitrogen content and soil microbial properties viz. MBC, DHA and FDA in relation to seed bacterization are given in (Table 4), from which it was deciphered the available nitrogen (AN) content in soil significantly (p < 0.05) varied in rice plants treated with diazotrophic heterobacteria as compared to negative control with the highest amount of soil nitrogen availability recorded at 357.6 kg/ha for AT treated rice seeds which was significantly at par with Enterobacter sp. strain PANW4/1 and Enterobacter sp. strain PANW5 treatments recorded at 378.3 kg/ha and 376.5 kg/ha AN, respectively. There was significant (p < 0.05) variance in the soil microbial properties including MBC, DHA and FDA of treated plants as compared with untreated negative control.
Table 3.
Comparative analysis of above and below ground responses of rice plants (60 days after sowing) inoculated with selected diazotropic bacteria isolated from Sikkim soils. Above ground responses: shoot length (SL) and leaf chlorophyll concentrations (SPAD) calculated using ODD (optical density difference) which is the ratio between reflection and absorption of incident light by SPAD meter. Below ground responses: root length (RL), projected root area (PRA), root surface area (RSA) and root average diameter (RAD). BIOM: Biomass calculated using shoot and root dry weights. PN: total plant nitrogen. Similar alphabets found across each parameter shows no difference between the respective values at 5% level of significance.
| Treatment | SPAD | SL | RL | PRA | RSA | RAD | BIOM | PN |
| (ODD) | (cm) | (cm) | (mm2) | (mm2) | (mm) | (g/plant) | (%/plant) | |
| PANW4 | 33.416c | 34.23a | 36.221c | 26.246b | 27.866a | 0.53bc | 2.23c | 0.386a |
| PANW4/1 | 34.23a | 34.123ab | 38.279a | 26.84a | 27.81ab | 0.716a | 2.809a | 0.315d |
| PANW5 | 34.13a | 33.247d | 38.038b | 26.39b | 27.62b | 0.669a | 2.722b | 0.327c |
| PANW6 | 33.27cd | 33.177d | 35.226e | 25.52c | 26.98c | 0.515c | 2.195c | 0.386a |
| PANW9 | 33.162d | 33.167d | 35.027f | 25.172d | 26.482d | 0.475c | 2.015b | 0.33c |
| +ve Control AT | 33.716b | 33.707bc | 36.108d | 26.196b | 27.806ab | 0.669a | 2.797a | 0.31d |
| +ve Control BioCWB | 33.406c | 33.413cd | 31.299g | 19.706e | 19.556e | 0.595b | 2.167c | 0.352b |
| -ve Control N broth | 32.686e | 32.183e | 23.516h | 13.286f | 16.516f | 0.382d | 1.789e | 0.36b |
| CD (0.05) | 0.242 | 0.459 | 0.036 | 0.242 | 0.24 | 0.068 | 0.068 | 0.009 |
Table 4.
Estimating the changes associated with rhizospheric soil nitrogen content and microbial properties of rice plants inoculated with diazotrophic bacteria (60 days after sowing). AN: soil available nitrogen, soil MBC: microbial biomass carbon, DHA: soil dehydrogenase active, FDA: Fluorescein diacetate hydrolysis. Similar alphabets found across one parameter shows no difference between the respective values at 5% level of significance.
| Treatment | AN | MBC | DHA | FDA |
| (kg/ha) | (µg/g soil) | (µg TPF/kg/24h) | (µg fluorescein/g soil) | |
| PANW4 | 359.8abc | 356.92bc | 648.77bcd | 3.63bc |
| PANW4/1 | 378.3ab | 366.12ab | 660.56ab | 3.656bc |
| PANW5 | 376.5ab | 373.71ab | 663.52ab | 3.909a |
| PANW6 | 340.9cd | 343.82c | 638.55cd | 3.685b |
| PANW9 | 329.4d | 319.74d | 636.14d | 3.525d |
| +ve Control AT | 357.6bc | 386.93a | 672.99a | 3.629bc |
| +ve Control BioCWB | 322.9de | 368.15ab | 653.36abcd | 3.574cd |
| -ve Control | 314.3e | 281.84e | 642.42bcd | 2.816e |
| CD (0.05) | 22.52 | 22.183 | 21.581 | 0.093 |
Statistical models for analysing responses of bacteria with respect to changes in plant nitrogen uptake
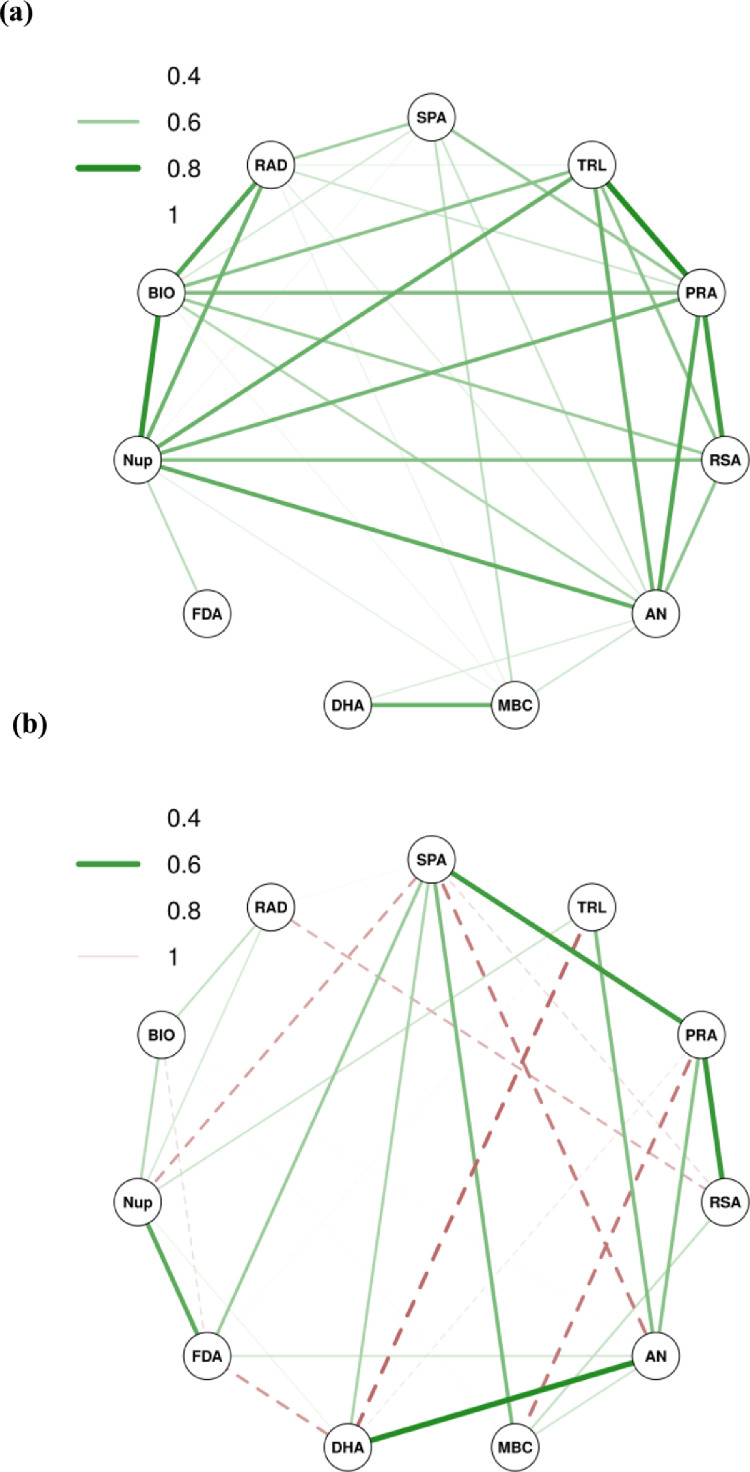
Step regression analysis was performed and the two parameters viz. shoot length (SL) and total plant nitrogen (PN) were removed, which improved the regression values in linear models. Figure 2a and 2b illustrates the correlation developed using Spearman's p statistic which was used to measure the variances through rank-based associations. Figure 2a provided the complete correlation network from which it was evident that highest correlations i.e. >90% were found among total root length (TRL) and projected root area (PRA) with 0.948, followed by plant biomass (BIOM) and nitrogen uptake (Nup) with 0.935, and PRA and root surface area (RSA) with 0.922 regression p values, respectively. Relatively, lower degree of correlations i.e. >80% were found between Nup, (root area diameter) RAD, TRL and available nitrogen (AN). Furthermore, Figure 2b provided the partial correlation network which showed there was higher significant difference (p < 0.05) with correlations greater than 75% between the parameters of DHA and AN, followed by correlations between leaf chlorophyll content (SPAD), PRA and RSA. Following step regression analysis, a linear regression model was prepared with 11 parameters leaving SL as the only parameter, from which the following regression equation was obtained:
Fig. 2.
Understanding the interaction between plant and soil parameters through network modelling of Spearman's p correlation co-efficient (n = 45). Fig. 2a: representation of complete correlation network. Fig. 2b: representation of partial correlation network. Thickness of line is proportional to the correlation values. Green line (bold) indicates significant positive correlation and red line (dashed) indicates non-significant positive correlation.
Nup = 1.204 + 0.018 * SPAD – 0.001 * TRL – 0.358 * PRA + 0.063 * RSA + 0.377 * RAD + 0.001 * AN + 0.003 * MBC – 0.004 * DHA – 0.051 * FDA + 0.041 * BIOM
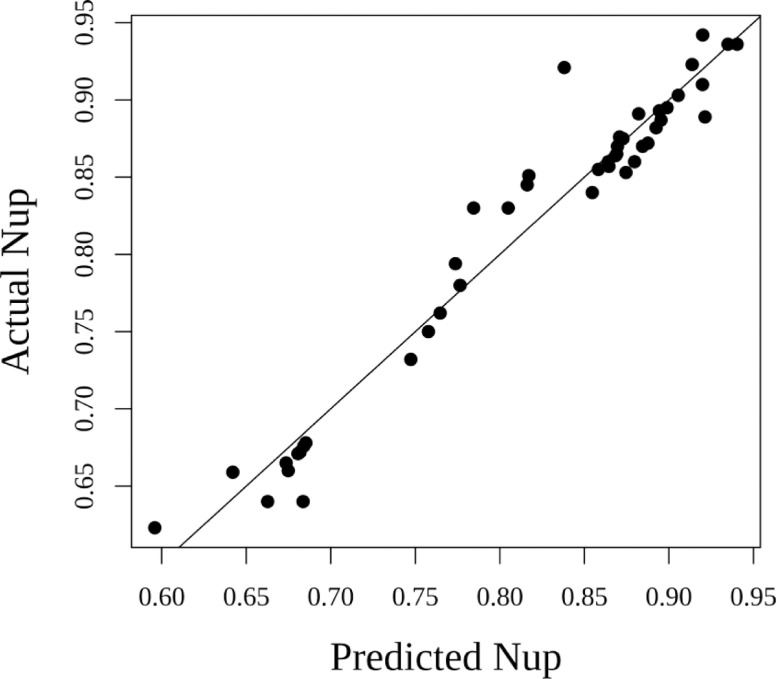
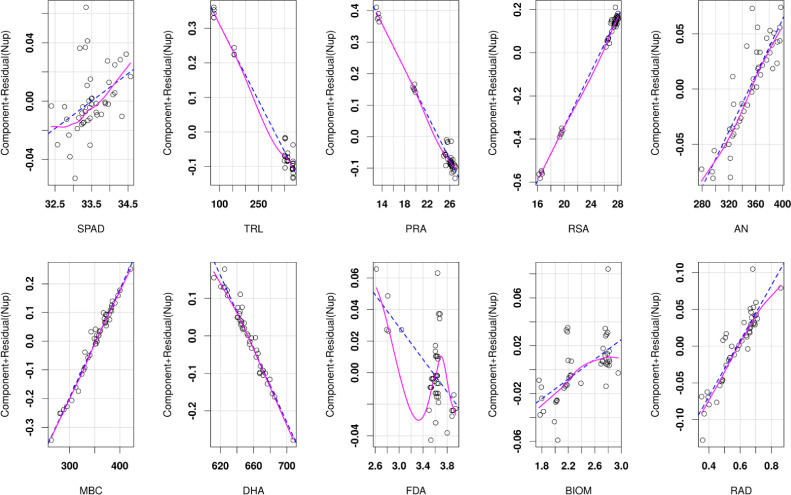
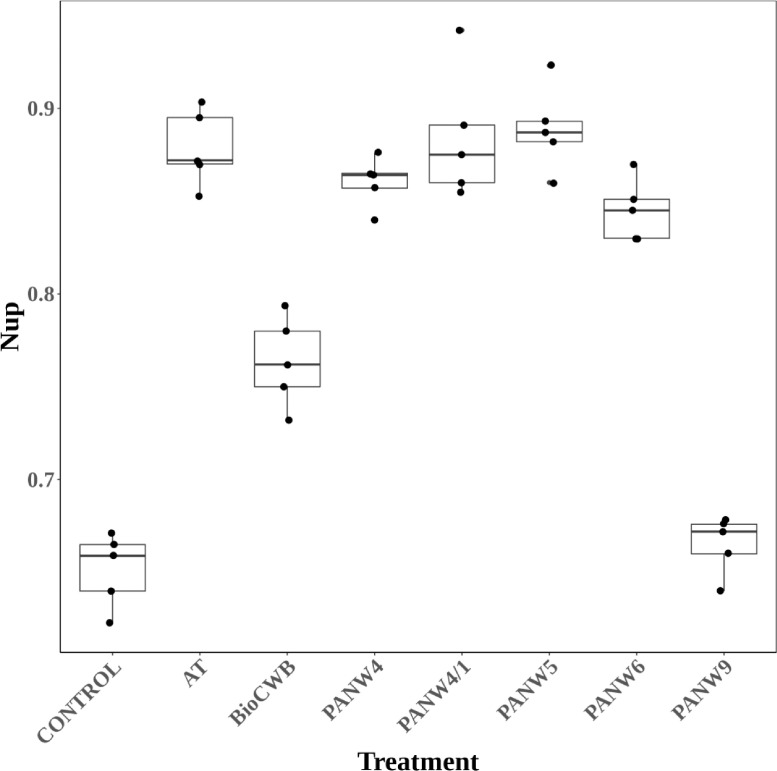
The regression equation had R2 value of 0.965 and p value of < 0.001. The F-statistic was 86.77 on 10 with 34 degrees of freedom (df) having total 44 df measuring five replication values from nine treatments. Using the regression equation, the nitrogen uptake (Nup) value was predicted, whose comparisons with actual Nup has been illustrated in Fig. 3 from which it could be depicted that there was no requirement of shifting the responses in the model as the levels of maximum Nup estimated was minimum and the predictions were found precise as the observed values were closer to the predicted values particularly when the Nup values were higher than 0.85 g/pot. The linear relationships between nitrogen uptakes (Nup) with other parameters are given in component and residual plot as illustrated in Fig. 4, wherein the dotted lines show the fitted least square values and smooth lines represent original distributions. Nitrogen uptake (Nup) calculated based on total nitrogen content present in plant biomass (Fig. 5) shows PANW4, PANW4/1, PANW5 and PANNW6 performed better role on improvement of nitrogen uptake as compared to positive controls AT strain with the maximum Nup recorded in rice, whereas in test isolates PANW5 recorded maximum Nup at 0.890 g/pot. To further understand the assumption regarding correlation between nitrogen uptake (Nup) and plant biomass (BIOM), linear regression model was used which computed the estimated nitrogen uptake and the significance of its relationship with biomass. The results of the linear regression model have been mentioned in Table 5, which showed the highly significant relationship among Nup and BIOM. Additionally, the model showed, the impact of Enterobacter species on plant growth was at par with the test strains.
Fig. 3.
Analysing the precision of linear model by comparing nitrogen uptake (Nup) derived from actual and predicted values.
Fig. 4.
Representation of linear relationships by nitrogen uptake with different plant and soil parameters using modelled component and residual values. Pink (bold) lines represent smoothened regression curve whereas blue (dashed) lines represent fitted linear regression curve.
Fig. 5.
Estimating the efficacy of diazotrophs on improvement of plant nitrogen uptake estimated as g/pot. The comparisons between isolates and positive controls have been performed using Welch t-test.
Table 5.
Estimating the nitrogen uptake (Nup) by plants in response to bacterial inoculation and analysing their relationship with plant biomass (BIOM) using linear models. Intercept represents the estimated Nup. ** represent significance of the interaction at 0.01 and *** represent significance of the interaction at 0.001 level.
| Treatment | Intercept | p-value | Model R2 | Pr(>|t|) |
| PANW4 | 0.932 | 0.032 | 0.89 | 0.002** |
| PANW4/1 | 0.964 | 0.033 | 0.972 | 0.004** |
| PANW5 | 0.901 | 0.029 | 0.795 | <0.001*** |
| PANW6 | 0.865 | 0.032 | 0.875 | <0.001*** |
| PANW9 | 0.7 | 0.022 | 0.648 | 0.001*** |
| +ve Control AT | 0.713 | 0.026 | 0.763 | <0.001*** |
| +ve Control BioCWB | 1.071 | 0.044 | 0.661 | 0.003** |
| -ve Control | 0.82 | 0.033 | 0.988 | <0.001*** |
Discussion
This study demonstrated the nitrogen fixing ability of free living diazotrophs such as Enterobacter species and their involvement in plant growth promotion in rice plants. Although the taxonomic relatedness of Enterobacter isolates is poorly understood due to their phenotypic resemblances among species type, for which molecular identification based on DNA-DNA relatedness has been generally preferred for identification of Enterobacter spp. In this study partial DNA sequences of the 16S rRNA gene were aligned Based on the 16S rRNA sequencing results, the isolates were found to be different at species level. The Enterobacter strains were found in organic paddy soils of Sikkim, some of which were found to produce copious amount of exopolysaccharides (data not shown), which was a positive trait responsible for biofilm formation that is partially responsible for attachment of soil microbes with plant. Among these exopolysaccharides producing isolates, production by E. asburaei was higher. The contribution of Enterobacter via BNF was at par to those observed by Azotobacter species confirms their desirability in use as biofertilizer for rice production. Although the developments of Enterobacter inoculums are questionable due to issues regarding pathogenicity against human beings however, their potent nitrogen assimilation ability in soil will definitely seek assistance for compensating soil nitrogen loss in long run. As result of which their pathogenicity should be addressed in future studies.
Enterobacter are also known to produce phytohormones such as IAA (Nutaratat et al., 2017). In this study we emphasised both phytohormones production and nitrogen fixing abilities of isolates. Enterobacter is proposed among the potent culturable bacteria to be used as biofertilizer in rice ecosystem because of its potency on growth improvement of rice plants (de Souza et al., 2018; Lima et al., 2018; Hardoim et al., 2013). Previous works supports the facts the Enterobacter species have preferential colonization to plant root surfaces preferably on root hairs as observed in E. cloacae GS1 and E. agglomerans using epifluorescence micrographs (Shankar et al., 2011). The microcolonies develop at the tip of root and the biofilm developed around the root tip, which is basically due to the exopolysaccharides secreted by some Enterobacter assist other microbes to survive in the rhizosphere (Shankar et al., 2011). In this study, the rhizospheric association of Enterobacter strains was found to improve root architecture as evident from the models which proposes improvement in the following growth parameters such as root length, surface area and root volume which were found to be significantly correlated with the improvement of nitrogen uptake in rice plants. Based on the supervised learning models provided by linear regression algorithms, we speculated that soil microbial activity induced by Enterobacter strains were chiefly responsible for plant growth improvement and found Enterobacter species to be K-strategist rather than R-strategist. Based upon the r/K selection theory, K-strategist have been found to perform better than r-strategist in terms of competition for resource since K-strategist have higher population close to carrying capacity as compared to r-strategist (Vadstein et al. 2018). Linear models have been used for identifying the type of most influencing variable based upon the distribution of responses, wherein the main challenge has been the selection of smoothing parameters (Gertheiss et al., 2013). In this work the parameter screening strategies have provided more realistic prediction of changes in microbial densities of individual bacterization treatments by analyzing their biomass and enzymatic activity in soil. Even, the increase in soil enzymatic activities as depicted through DHA and FDA activities represent higher functionality of soil microbiome from which it was revealed that bio-inoculation of soil through bacterized rice seeds has effect on improving soil microbiome. This strategy may be followed to preserve soil fertility, by artificially intensifying soil biogeochemical cycles. Owing to the innate abilities of Enterobacter species for growth promotion in plants, their long-term association with humans is also intriguing. Soil made rich with beneficial Enterobacter species will add benefit to cropping plants participating in the food chain. The nitrogen assimilation by Enterobacter diazotrophs which were found in Sikkim soil microflora shows promising results on sustainable agriculture by fulfilling plant nitrogen demands. Similar findings have shown E. ludwigii isolated from rice soil ecosystem particularly have multiple physiological traits including phytohormones production, silicon and phosphate solubilization revealing the applicability of Enterobacter as potential biofertilizer in rice ecosystem (Lee et al., 2019). Furthermore, zinc solubilizing Enterobacter species identified as E. cloacae have been found to plat active role in bioremediation of hexavalent chromium and also possess several other plant growth promoting traits for improving crop productivity in rice ecosystems having heavy metal contamination (Pattnaik et al., 2020). At end, the clinical consequences of Enterobacter isolates should never be forgotten since these strains have been shown by earlier research workers to share similarity to clinical isolates (Brenner et al., 1986). Additionally, previous research work have indicate that most of r-strategist species are pathogens due to rapid growth rate but have slower competition ability at low substrate concentration (Vadstein et al., 2018). As the Enterobacter species used in this research share similarities with K-strategist, it may be inferred these strains to be non-pathogenic in nature.
The nitrogen movement in the soil has been an important attribute of BNF (Ladha et al., 2005). One of the major advantages of bacteria mediated nitrogen fixation over addition of FYM was that by reducing nitrous oxide greenhouse gas (GHG) emission which was often found in soils applied with farm yard manure (Pathak et al., 2002). Therefore, application of diazotrophs may be one of the alternatives for enriching nitrogen content in soil without emitting nitrous oxide gas. Another advantage of bacteria mediated nitrogen fixation is production of bacterial phytohormones that induce root growth which subsequently improves nitrogen uptake by enhancing the root surface area (El-Sharkawi, 2012). The root architectural characteristics were highly influenced by seed bacterization using E. asburiae and B. subtilis which favoured the growth of rice seedlings which plausibly was due to the higher amount of phytohormones produced by these diazotrophic species (Mehnaz et al., 2001; Ambreetha et al., 2018). Similar findings also support the fact that phytohormone producing bacteria play an important role in modification of root architecture which in turn improves plant growth and development (Martinez-Rodriguez et al., 2019; Panneerselvam et al., 2019). Moreover, significant correlations were obtained between plant nitrogen uptake and root parameters demonstrating the additional advantages of phytohormones production by Enterobacter species that have played an active role in improving the root surface area as evident from root scanning data.
Conclusions
Based on presumptive models generated by combining soil and plant parameters through supervised learning approaches, this study provided evidence for the associative nature of Enterobacter in rice plants and participation in crop improvement through soil nitrogen assimilation from atmosphere. Contribution of Enterobacter species towards BNF in soil was compared with other free living heterotrophic diazotrophs. This study clearly demonstrated the ability of Enterobacter species to alleviate nitrogen stress in rice plant under organic farming practices by improving plant nitrogen uptake through cumulative contribution of nitrogen fixing and phytohormone production traits of heterotrophic bacterial diazotrophs. This study indicates that different Enterobacter spp. may play some role for nitrogen uptake of rice plants from atmosphere under organic farming practices in Sikkim.
Declaration of Competing Interest
The authors have declared no conflict of interest.
Acknowledgments
Funding
We are grateful to the Department of Biotechnology, Government of India for promotion of Biotechnology in the North Eastern Region of India through Biotech Consortium India Limited and giving grants (BT/PR16291/NER/95/185/2015) for this research work.
ORCID
Periyasamy Panneerselvam (0000-0001-7144-2947)
Debasis Mitra (0000-0003-0525-8812)
Intellectual property
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
Research ethics
We further confirm that any aspect of the work covered in this manuscript that has involved human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
Author Contributions
Conception and design of study: P. Panneerselvam Acquisition of data: A. Senapati, L. Sharma, A Kumar, U Kumar, S. R. Prabhukarthikeyan, D Mitra and M S Sagarika Analysis and/or interpretation of data: P. Panneerselvam, A. Senapati, D Mitra Drafting the manuscript: P. Panneerselvam, A. Senapati, D Mitra Revising the manuscript critically for important intellectual content: P. Panneerselvam, U Kumar, S. R. Prabhukarthikeyan, A K Nayak, L. Sharma Approval of the version of the manuscript to be published: P. Panneerselvam, A. Senapati, L. Sharma, A K Nayak, A Kumar, U Kumar, S. R. Prabhukarthikeyan, D Mitra and M S Sagarika.
Acknowledgments
We are grateful to Director, ICAR-National Rice Research Institute, Odisha, India for providing all the support for carrying out this research work and special thanks to DBT, India for financial support. We thankful to Prof. Sergio de los Santos Villalobos and Dr. Fannie I. Parra Cota, Laboratorio de Biotecnología del Recurso Microbiano Instituto Tecnológico de Sonora, Sonora, México and Guest Editors of the Special Issue “Current trends in plant growth-promoting microorganisms research for sustainable food security” in Current Research in Microbial Sciences for the invitation.
Reference
- Borrow A., Brian P.W., Chester V.E., Curtis P.J., Hemming H.G., Henehan C., Jeffreys E.G., Lloyd P.B., Nixon I.S., Norris G.L.F., Radley M. Gibberellic acid, a metabolic product of the fungus Gibberella fujikuroi: some observations on its production and isolation. J. Sci. Food Agric. 1955;6(6):340–348. [Google Scholar]
- Martinez-Rodriguez A., Macedo-Raygoza G., Huerta-Robles A.X., Reyes-Sepulveda I., Lozano-Lopez J., et al. Seed Endophytes. Springer; Cham: 2019. Agave seed endophytes: ecology and impacts on root architecture, nutrient acquisition, and cold stress tolerance; pp. 139–170. [Google Scholar]
- Choudhury A.T., Kennedy I.R. Prospects and potentials for systems of biological nitrogen fixation in sustainable rice production. Biol. Fert. Soils. 2004;39(4):219–227. [Google Scholar]
- Mahapatra B., Adak T., Patil N.K., Gowda G.B., Jambhulkar N.N., Yadav M.K., et al. Imidacloprid application changes microbial dynamics and enzymes in rice soil. Ecotoxicol. Environ. Safe. 2017;144:123–130. doi: 10.1016/j.ecoenv.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Subbiah B.V., Asija G.L. A rapid procedure for the determination of available nitrogen in soil. Curr. Sci. 1956;25:259–260. [Google Scholar]
- Black C.A., Evans D.D., White J.L., Ensminger L.E., Clark F.E. American Society of Agronomy. Inc. Publisher; Madison, Winconsin, USA: 1965. Methods of Soıl Analysıs. Part I. [Google Scholar]
- Chatterjee D., Kuotsu R., Ao M., Saha S., Ray S.K., Ngachan S.V. Does rise in temperature adversely affect soil fertility, carbon fractions, microbial biomass and enzyme activities under different land uses? Curr. Sci. 2019;116(12):2044. [Google Scholar]
- Brenner D.J., McWhorter A.C., Kai A., Steigerwalt A.G., Farmer J.J. Enterobacter asburiae sp. nov., a new species found in clinical specimens, and reassignment of Erwinia dissolvens and Erwinia nimipressuralis to the genus Enterobacter as Enterobacter dissolvens comb. nov. and Enterobacter nimipressuralis comb. nov. J. Clin. Microbiol. 1986;23(6):1114–1120. doi: 10.1128/jcm.23.6.1114-1120.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima D.R., Santos I.B., Oliveira J.T., Barbosa J.G., Diniz W.P., Farias A.R., Freire F.J., Sobral J.K. Tolerance of potentially diazotrophic bacteria to adverse environmental conditions and plant growth-promotion in sugarcane. Arch. Agron. Soil Sci. 2018;64(11):1534–1548. [Google Scholar]
- Morse E.E. Anthrone in estimating low concentrations of sucrose. Anal Chem. 1947;19(12):1012–1013. [Google Scholar]
- Wright E.S. Using DECIPHER v2. 0 to analyze big biological sequence data. R Journal. 2016;8(1):352–359. [Google Scholar]
- Peng G., Zhang W., Luo H., Xie H., Lai W., Tan Z. Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia. Int. J. Syst. Evol. Micr. 2009;59(7):1650–1655. doi: 10.1099/ijs.0.65484-0. [DOI] [PubMed] [Google Scholar]
- El-Sharkawi H.M. Effect of nitrogen sources on microbial biomass nitrogen under different soil types. ISRN Soil Sci. 2012:1–7. [Google Scholar]
- Pathak H., Bhatia A., Prasad S., Singh S., Kumar S., Jain M.C., et al. Emission of nitrous oxide from rice-wheat systems of Indo-Gangetic plains of India. Environ. Monit. Assess. 2002;77(2):163–178. doi: 10.1023/a:1015823919405. [DOI] [PubMed] [Google Scholar]
- Gertheiss J., Maity A., Staicu A.M. Variable selection in generalized functional linear models. Stat. 2013;2(1):86–101. doi: 10.1002/sta4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladha J.K., Pathak H., Krupnik T.J., Six J., van Kessel C. Efficiency of fertilizer nitrogen in cereal production: retrospects and prospects. Adv. Agron. 2005;87:85–156. [Google Scholar]
- Ladha J.K., Reddy P.M. Nitrogen fixation in rice systems: state of knowledge and future prospects. Plant soil. 2003;252(1):151–167. [Google Scholar]
- Ladha J.K., Reddy P.M. In: Proceedings of the Third Working Group Meeting of Assessing Opportunities for Nitrogen Fixation in Rice, IRRI. Ladha J.K., Reddy P.M., editors. Vol. 354. Manila, Philippines; 2000. Steps towards nitrogen fixation in rice. The quest for nitrogen fixation in rice; p. 2000. 33-46. [Google Scholar]
- Lee K.E., Adhikari A., Kang S.M., You Y.H., Joo G.J., Kim J.H., Kim S.J., Lee I.J. Isolation and characterization of the high silicate and phosphate solubilizing novel strain Enterobacter ludwigii GAK2 that promotes growth in rice plants. Agronomy. 2019;9(3):144. [Google Scholar]
- Tilak K.V., Ranganayaki N., Pal K.K., De R., Saxena A.K., Nautiyal C.S., et al. Diversity of plant growth and soil health supporting bacteria. Curr. Sci. 2005:136–150. [Google Scholar]
- Shankar M., Ponraj P., Ilakkiam D., Gunasekaran P. Root colonization of a rice growth promoting strain of Enterobacter cloacae. J Basic Microbiol. 2011;51(5):523–530. doi: 10.1002/jobm.201000342. [DOI] [PubMed] [Google Scholar]
- Suzuki M.T., Giovannoni S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M.L. Jackson, Prentice Hall of India. Pvt. Ltd., New Delhi, pp. 498, 1967.
- Fierer N., Lauber C.L., Ramirez K.S., Zaneveld J., Bradford M.A., Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6(5):1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadstein O., Attramadal K.J., Bakke I., Olsen Y. K-selection as microbial community management strategy: a method for improved viability of larvae in aquaculture. Front. Microbiol. 2018;9:2730. doi: 10.3389/fmicb.2018.02730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutaratat P., Monprasit A., Srisuk N. High-yield production of indole-3-acetic acid by Enterobacter sp. DMKU-RP206, a rice phyllosphere bacterium that possesses plant growth-promoting traits. 3 Biotech. 2017;7(5):305. doi: 10.1007/s13205-017-0937-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P., Senapati A., Kumar U., Sharma L., Lepcha P., Prabhukarthikeyan S.R., Jahan A., Parameshwaran C., Govindharaj G.P.P., Lenka S., Nayak P.K., Mitra D., Sagarika M.S., Thangappan S., Sivakumar U. Antagonistic and plant-growth promoting novel Bacillus species from long-term organic farming soils from Sikkim, India. 3 Biotech. 2019;9(11):416. doi: 10.1007/s13205-019-1938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofi P., Wani S. Prospects of nitrogen fixation in rice. Asian J. Plant Sci. 2007;6(1):203–213. [Google Scholar]
- Hsieh P.H., Kan C.C., Wu H.Y., Yang H.C., Hsieh M.H. Early molecular events associated with nitrogen deficiency in rice seedling roots. Sci. Rep. 2018;8(1):1–23. doi: 10.1038/s41598-018-30632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk P.L. Kjeldahl method for total nitrogen. Anal. Chem. 1950;22(2):354–358. [Google Scholar]
- Hardoim P.R., Nazir R., Sessitsch A., Elhottová D., Korenblum E., van Overbeek L.S., et al. The new species Enterobacter oryziphilus sp. nov. and Enterobacter oryzendophyticus sp. nov. are key inhabitants of the endosphere of rice. BMC Microbiol. 2013;13(1):164. doi: 10.1186/1471-2180-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachuk R. Nitrogen-to-protein conversion factors for cereals and oilseed meals. Cereal. Chem. 1969;46:419–424. [Google Scholar]
- Bray R.H., Kurtz L.T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945;59(1):39–46. [Google Scholar]
- Ambreetha S., Chinnadurai C., Marimuthu P., Balachandar D. Plant-associated Bacillus modulates the expression of auxin-responsive genes of rice and modifies the root architecture. Rhizosphere. 2018;5:57–66. [Google Scholar]
- Mehnaz S., Mirza M.S., Haurat J., Bally R., Normand P., Bano A., Malik K.A. Isolation and 16S rRNA sequence analysis of the beneficial bacteria from the rhizosphere of rice. Can. J. Microbiol. 2001;47(2):110–117. doi: 10.1139/w00-132. [DOI] [PubMed] [Google Scholar]
- Pattnaik S., Dash D., Mohapatra S., Pattnaik M., Marandi A.K., Das S., Samantaray D.P. Improvement of rice plant productivity by native Cr (VI) reducing and plant growth promoting soil bacteria Enterobacter cloacae. Chemosphere. 2020;240 doi: 10.1016/j.chemosphere.2019.124895. [DOI] [PubMed] [Google Scholar]
- Gordon S.A., Weber R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951;26(1):192. doi: 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar U., Shahid M., Tripathi R., Mohanty S., Kumar A., Bhattacharyya P., et al. Variation of functional diversity of soil microbial community in sub-humid tropical rice-rice cropping system under long-term organic and inorganic fertilization. Ecol. Indic. 2017;73:536–543. [Google Scholar]
- Nihorimbere V., M. O., Smargiassi M., Thonart P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnologie, Agronomie, Société et Environnement. 2011;15(2):327–337. [Google Scholar]
- Achouak W., Heulin T, Villemin G., Balandreau J. Root colonization by symplasmata-forming Enterobacter agglomerans. FEMS Microbiol. Ecol. 1994;13(4):287–294. [Google Scholar]
- Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.A., Jiang D.A., Yang Y., Sun J.W., Jin S.H. Effects of nitrogen deficiency on gas exchange, chlorophyll fluorescence, and antioxidant enzymes in leaves of rice plants. Photosynthetica. 2004;42(3):357–364. [Google Scholar]