Highlights
-
•
Equatorial lakes Olbolosat and Oloiden are rich in prokaryotic diversity.
-
•
Prokaryotic diversity differ across samples between the two lakes
-
•
Environmental factors shape the prokaryotic community structures
-
•
Alpha Proteobacteria are the most abundant taxa in both lakes Olbolosat and Oloiden
Keywords: Prokaryotic, 16S rRNA gene, Environmental factors, Illumina sequencing, Equatorial lakes
Abstract
Total community 16S rDNA was used to determine the diversity and composition of bacteria and archaea within lakes Olbolosat and Oloiden in Kenya. The V3-V4 hypervariable region of the 16S rRNA gene was targeted since it's highly conserved and has a higher resolution for lower rank taxa. High throughput sequencing was performed on 15 samples obtained from the two lakes using the Illumina Miseq platform. Lakes Olbolosat and Oloiden shared 280 of 10,523 Amplicon Sequence Variants (ASVs) recovered while the four sample types (water, microbial mats, dry and wet sediments) shared 4 ASVs. The composition of ASVs in lake Olbolosat was highly dependent on Cu+, Fe2+, NH4+, and Mn2+, while L. Oloiden was dependent on Mg2+, Na+, Ca2+, and K+. All the alpha diversity indices except Simpson were highest in the dry sediment sample (EC1 and 2) both from lake Oloiden. The abundant phyla included Proteobacteria (33.8%), Firmicutes (27.3%), Actinobacteriota (21.2%), Chloroflexi (6.8%), Cyanobacteria (3.8%), Acidobacteriota (2.8%), Planctomycetota (1.9%) and Bacteroidota (1.1%). Analysis of similarity (ANOSIM) revealed a significant difference in ASV composition between the two lakes (r = 0.191, p = 0.048), and between the sample types (r = 0.6667, p = 0.001). The interaction network for prokaryotic communities within the two lakes displayed Proteobacteria to be highly positively connected with other microbes. PERMANOVA results suggest that temperature controls the functioning of the two ecosystems.
1. Introduction
Lake Olbolosat is an endangered freshwater lake and the only lake in Central Kenya, Africa. It is a drainage wetland lake that was formed by down warping and it's among the lakes in Kenya outside the rift valley. The freshwater lake supports the livelihoods of communities, wildlife and livestock and it forms the headwaters for Ewaso Nyiro river to Thomson's falls in Nyahururu town. However, the lake has been experiencing massive shrinking attributed to anthropogenic activities (Wafula and Murunga, 2020). Lake Oloiden is a satellite of L. Naivasha (Maina et al., 2018). This lake is hydrologically closed and the evaporation in the lake leads to water loss while the rise in the lake is brought by the rainfall and via a peninsula from lake Naivasha (Luo et al., 2017). The two endorheic (terminal) lakes are ecosystems that are highly rich in the resources and conditions available for microbial growth (Kiama et al., 2021).
Studies on bacterial/archaeal communities within endorheic lake ecosystems are important in acquiring knowledge about the bacterial/archaeal diversity and composition of microbial genetic resources, patterns of microorganisms, the functional role of microbial diversity and the regulation of microbial diversity (Yang et al., 2016). Numerous studies of prokaryotic communities have been done on different natural habitats in Kenya but despite this, exploitable microbial diversity is not exhaustive, and microorganisms represent the largest reservoir of untapped biodiversity (Mwirichia et al., 2011; Kambura et al., 2016; Ghilamicael et al., 2017). Bacteria and archaea are unique microbial communities of prokaryotic taxon groups that are important in the functioning of a lake ecosystem (Silveira et al., 2020). Prokaryotes play an important role in the decomposition of organic material into nutrients taken as food by other organisms and controlling the quality of water in the lake (Zhang et al., 2019). They are also important in balancing between respiration and photosynthesis in the natural cycles of oxygen, carbon, sulfur and metals (Silveira et al., 2020). However, a large range of abiotic and biotic factors influences microbial communities which affect their diversity, abundance, composition, distribution, processes and existence within these ecosystems. Environmental factors have been reported to promote the diversity of microorganisms. Endorheic lakes of the tropics exhibit considerable variation in their hydrogeology due to climate change. These lakes experience phases of being flooded with freshwater to salinity or even dried out caused by evaporation and anthropogenic activities (Luo et al., 2017).
Prokaryotic communities obtained from water, mats, wet and dry sediments play a very important role within saline-alkaline lakes. However, due to the complexity of the culture-dependent techniques, only a limited number of prokaryotes have been extensively studied (Kambura et al., 2016). Marker gene sequencing is fast and obtains community/ taxonomic distribution profile or fingerprinting using PCR amplification and sequencing of evolutionarily conserved marker genes. The taxonomic distribution can be associated with metadata derived from the sampling sites under investigation (Paul et al., 2016). High-throughput sequencing is used widely to study the diversity and composition of prokaryotic communities. Taxonomic analysis of bacteria and archaea is regularly performed using Illumina sequencing (Oulas et al., 2015). The higher resolution resulting from Amplicon Sequence Variants (ASVs) also known as Exact Sequence Variant (ESVs) are inferred unique sequences present in the original sample, after correcting for sequencing and PCR errors. The ASVs method was adopted in this study since the method has combined the benefits for subsequent analysis of closed-referencing and de novo Operational Taxonomic Units (OTUs). Replacing the traditional OTUs with ASVs makes marker gene sequencing reusable, more precise, reproducible, high resolution and comprehensive. (Callahan et al., 2017). Callahan et al. (2017) reported that the ASV method deduces the biological sequences in the sample before the introduction of amplification and sequencing errors, and distinguishing sequence variants. This is the first culture-independent study for the microbial diversity and composition within lakes Olbolosat and Oloiden. In addition, we sought to evaluate the environmental factors shaping characteristics of microbial taxa within these ecosystems.
2. Material and methods
2.1. Study sites
Lake Olbolosat a freshwater body is 43 Km2. The lake is located at a longitude of 36° 26′E and a latitude of 0° 09′S in the central part of Kenya, Nyandarua county. The lake is wedged in shape and is found at an altitude of about 2340 m above sea level (a.s.l.) (Wafula and Murunga, 2020). It records a pH of 7 and temp of 22°C annually. Samples were collected from five different locations at lake Olbolosat. 1: 0° 8′ 43.008′' S, 36° 26′ 26.664′' E and 2338 m a.s.l., 2: 0° 10′ 45.264′' S, 36° 26′ 46.392′' E and 2335 m a.s.l. 3: 0° 4′ 38.352′' S, 36° 24′ 58.068′' E and 2336 m a.s.l. 4: 0° 9′ 28.98′' S, 36° 25′ 56.712′' E and 2347 m a.s.l. 5: 0° 7′ 26.976′' S, 36° 25′ 49.224′' E and 2339 m a.s.l. Lake Oloiden a saline-alkaline water body is about 4-7.5 Km2. Lake Oloiden becomes fresh when the water overflows from L. Naivasha during the rain seasons (Luo et al., 2017). The lake is located at a latitude of 0° 48′S and 36° 16′E. Lake Oloiden is located at 1995 meters above sea level and it records a pH of 9 and a temperature of 25°C annually. Samples were collected from four different points; 1:0° 47′ 59.496′' S, 36° 16′ 45.444′' E and 1885 m a.s.l., 2: 0° 49′ 6.744′' S, 36° 15′ 49.392′' E and 1890 m a.s.l. 3: 0° 48′ 33.66′' S, 36° 16′ 36.624′' E and 1884 m a.s.l. 4: 0° 49′ 32.772′' S, 36° 16′ 38.748′' E and 1895 m a.s.l.
2.2. Measurement of physical parameters
The geographical position for the sampling sites in terms of longitude, latitude and elevation were taken using Global Positioning System (GARMIN eTrex 20, USA). The onsite metadata for total dissolved solids (TDS), dissolved oxygen (DO), electrical conductivity (EC) and temperature for each sampling point were measured using an Electrical Chemical Analyzer (Jenway – 3405, UK). The pH was measured with a portable pH-meter (Oakton pH 110, Eutech Instruments Pty. Ltd, USA) and confirmed with indicator strips (Merck, range 5-10) (Supplementary Table S1).
2.3. Sample collection
Water, microbial mats, wet, and dry sediments samples were randomly collected in triplicates. This was done by scooping wet and dry sediments with a hand shovel into sterile 1000ml plastic containers. Approximately upper 5mm microbial mats were scraped off using a sterile shovel into 1000ml containers. The sterile plastic containers (1000ml) were also used to fetch water from both lakes. All samples were transported at 4°C to Jomo Kenyatta University of Agriculture and Technology laboratories for analysis.
2.4. Chemical analysis of the water, microbial mats, dry and wet samples
The chemical properties (calcium, ammonium, sodium, magnesium, copper, iron, manganese, zinc, potassium, nitrate, nitrite, phosphate, sulfate, bicarbonate, chloride and fluoride) of the water, mats, dry and wet samples collected from lakes Olbolosat and Oloiden were analysed as described by Dawood and Sanad, (2014). Cations such as Cu+, Fe2+, NH4+, Mn2+, Mg2+, Na+, Ca2+, K+ were analyzed using Atomic Absorption Spectrophotometry (AAS) while anions such as F−, NO2− and PO43− SO43−, HCO3−, and Cl− were analyzed using Mass spectrometry.
2.5. Environmental nucleic acid extraction
The water samples (approximately 500ml) for nucleic acid extraction were filtered using a 0.22μM microfilter (Whatman) and filter papers. The obtained samples were centrifuged at 10,000 × g for 10 min. Pellets obtained were resuspended in 5ml of phosphate buffer saline solution. The pellets were placed in 2ml Eppendorf tubes and stored at -75°C ready for DNA extraction. DNA was extracted from 0.5g of the microbial mat, dry and wet sediment separately. DNA from all samples were differently extracted using DNeasy® Powersoil® Kit (Qiagen, USA) following the standard manufacturer's protocol. The extracted DNA samples were quantified using a Nano™Drop 2000/c Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and evaluated with 1.5% agarose gel.
2.6. Amplicon library preparation
The conserved sequences flanking the hypervariable V3-V4 region of the 16S rRNA gene served as primer sites to generate PCR amplicons. The sequencing libraries were generated according to the 16S Metagenomic Sequencing library preparation part # 15044223 Rev. B using a Herculase II Fusion DNA polymerase Nextera XT Index kit v2 according to the manufacturer's protocol (Illumina). The following primers were used; 341 F (5′- GTGCCAGCMGCCGCGGTAA-3′) that had barcode and 806R (5′- GGACTACHVGGGTWTCTAAT-3′) according to (Caporaso et al., 2012). The amplicon libraries were constructed under the following PCR conditions: initial denaturation at 94°C for 3 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 53°C for 40 seconds and extension at 72°C for 1 minute, and final elongation at 72°C for 5 minutes. The quality of PCR products was assessed on 1.2% agarose gel to determine whether the amplification was successful. Multiple samples tagged with different barcodes were pooled together in equimolar ratios based on their DNA concentrations from the gel results. Pooled samples were purified to remove adapters using calibrated AMPure XP magnetic beads. The prepared libraries were sequenced with the Illumina Miseq platform at Macrogen, Inc. Seoul, South Korea).
2.7. Sequence data analysis
The amplicon library was purified and sequenced on an Illumina Miseq sequencing-by-synthesis (SBS) technology, a proprietary reversible terminator-based platform (Macrogen, Inc. Seoul, South Korea). After sequencing, the barcodes and amplicon primer sequences were removed. Low-quality sequences were identified by denoising and filtered out of the dataset. Reads with <300 base pairs after phred20-based quality trimming, sequences with ambiguous base calls and those with homopolymer runs exceeding 5bp were removed (Reeder and Knight, 2010). The 16S rRNA high-throughput sequencing data was analyzed by following the workflow by Callahan et al., (2016a). The Divisive Amplicon Denoising Algorithm 2 (DADA2) pipeline that uses set of Illumina sequenced paired-end fastq files that have been demultiplexed was used. The end product was Amplicon sequence variant (ASV) table (Prodan et al., 2020). Quality checking, filtering and trimming of fastq files were performed using DADA2 v 1.16 package at 99.9 % similarity level and 1% divergence of ASVs in R v 4.02. High-resolution ASVs were inferred after filtering the reads using the DADA2 pipeline (Callahan et al., 2016b). Taxonomy was assigned to ASVs based on the naïve Bayesian classifier against the Silva v138 database. Demultiplexed high-quality sequence reads were deposited in the Sequence Read Archive, the National Centre for Biotechnology Information with study accession number PRJNA723886 (Accessions SAMN18836534-SAMN18836548) freely available for download.
2.8. Statistical analysis
Statistical data was done using phyloseq v 1.32.0, tidyverse v 1.3.1, compositions v 2.0.1, vegan v 2.5.7, ALDEx2 v 1.20.0, ggcorrplot v 0.1.3, ggrepel v 0.9.1, and superheat v 0.1.0. The resulting ASVs were used to calculate alpha diversity indices (Observed, Chao1, ACE, Shannon, Simpson, and inverted Simpson). The package venerable v 3.1.0.9000 was used to visualize the comparison of shared ASVs between the two lakes and sample types. Rarefaction curves were also visualized using the resulting ASVs to determine library sizes among the samples. IndVal function in the labdsv package (https://cran.r-project.org/web/packages/labdsv/labdsv.pdf) was used to identify taxon-habitat association patterns. IndVal function identifies taxa as an indicator species based on their independence abundance in the total data sets.
Principal Component Analysis (PCA) was used to show the component ordinations between the sample type and environmental factors using the prcomp function. Similarly, PCA and Hierarchical clustering of both the environmental factors and taxa were used to show the relationship in microbial community composition between samples. Cluster analysis of environmental factors and taxa were based on the Euclidian and Bray-Curtis dissimilarity, respectively. Function aldex.plot in package ALDEx2 v 1.20.0 was used to determine the differentially abundant ASVs and the significance was tested at we.ep < 0.05. Packages phyloseqGraphTest, Intergraph, gridExtra, and ggnetwork were used to visualize the co-correlation network patterns of the indicator ASVs in the study. Edge-weighted positive values indicated the co-correlation between the ASVs.
One-way analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA) was used to explain the difference between taxa beta diversities and environmental factors. PERMANOVA was used to determine the effects of environmental factors and nutrients on the distribution of microbial communities based on standardized categories with 999 permutations. Pearson's correlation coefficient analysis between environmental factors and microbial community structure was based on the product-moment correlation on variance-stabilizing transformed counts. The significance of the predictor environmental factors was tested using Mantel test.
3. Results
3.1. Characteristics of sampling sites
The sampling sites were selected based on distinct physicochemical parameters and location. All the physicochemical concentrations varied between sampling sites and locations. Hierarchical clustering of physicochemical parameters of water samples using Euclidean distance matrix revealed two distinct clusters according to geographical location and chemical concentrations (Supplementary Figure 1). The pH (H2O) concentrations ranged between 9.1 to 10.6 in L. Oloiden and 7.3 to 8 in L. Olbolosat. Bicarbonates and chlorides were distinctly clustered due to their high concentrations across the sites. The concentration of bicarbonate ranged between 668ppm to 939.7ppm and 190ppm to 246.3ppm in Lakes Oloiden and Olbolosat, respectively. Concentrations of Chlorides ranged between 1062ppm to 1875.7ppm and 486ppm to 560ppm in Lakes Oloiden and Olbolosat, respectively. The concentration of Sulfate ions, Sodium ions, and Potassium ions also influenced the hierarchical cluster into distinct geographical locations. Levels of Sulfate ions ranged between 65.3ppm to 79.4ppm in L. Oloiden. This was averagely higher than the concentration of Sulfate ions (38.3ppm to 81.4ppm) in L. Olbolosat. The concentration of Sodium ions ranged between 71ppm to 111.3ppm and 20.5ppm to 84ppm in Lakes Oloiden and Olbolosat, respectively. Consequently, the concentration of Potassium ions ranged between 0.2ppm to 83.6ppm and 0.3ppm to 33ppm in Lakes Oloiden and Olbolosat, respectively. Zinc and Manganese were highly concentrated in L. Olbolosat (0.1 – 68.5ppm and 0.4 – 58.1ppm, respectively) than L. Oloiden (0 0.9ppm and 0.1 – 0.5ppm, respectively). Ammonium, Nitrate, TDS, and DO concentration were, however, homogenously distributed across the lakes and between sampling sites (Supplementary Figure 1)
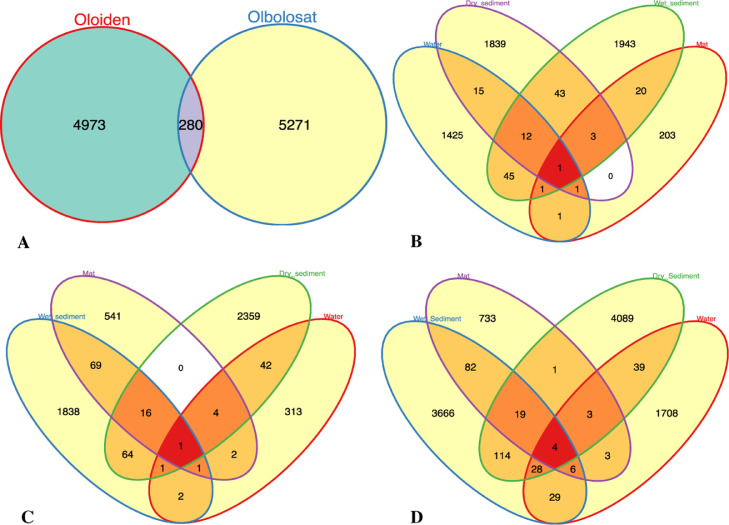
3.2. Diversity of prokaryotic communities
We obtained a total of 274,892 sequence reads after removing chimeras from 15 DNA amplicon data sets. Total ASV richness at 1% distance amounted to 10,523 ASVs distributed among 17 highly abundant phyla and 54 highly abundant genera. The ASV per data set ranged between 1 and 392 (Supporting Information S1). Out of the 10,523 ASVs, 280 (2.66%) ASVs were shared between lakes Oloiden and Olbolosat. Consequently, lakes Oloiden and Olbolosat constituted 4973 (47.27%) ASVs and 5271 (50.09%) ASVs, respectively (Fig. 1a). The distribution of shared ASVs between sample types revealed an overlap between water and wet sediments in L. Olbolosat. Dry sediments and the microbial mat had the lowest overlap in the same lake (Fig. 1b). Wet sediments and mat had the greatest overlap while dry sediment and mat had the lowest overlap in L. Oloiden (Fig. 1c). Overall, wet sediments and dry sediments shared a larger number of ASVs while dry sediment and mat had the lowest overlap across lakes Olbolosat and Oloiden (Figure 1d). Further, the high overlap between sample types was supported by the ANOSIM that revealed a significant difference in ASV composition between the two lakes (r = 0.191, p = 0.048), and between the sample types (r = 0.6667, p = 0.001).
Fig. 1.
a) The distribution of shared ASVs between lakes Olbolosat and Oloiden b) Distribution of ASVs between sample types in lake Olbolosat c) Distribution of ASVs between sample types in lake Oloiden d) Distribution of ASVs between sample types in both lakes.
3.3. Alpha-diversity
Alpha diversity indices were significantly (p < 0.05) different among samples (Supplementary Figure 1 and Table 1). The number of sequences per sample ranged between 2730 to 34827 with a mean value of 18326.13 and 12533.76 standard deviations (Table 1). Dry sediments from both lakes had the highest number of sequences ZC1 and EC1 (Table 1). All the indices except Simpson were highest in the dry sediment sample (EC1) followed by the dry sediment sample (EC2) both from lake Oloiden. The lowest alpha diversity was in the water sample (EA2) from lake Oloiden) (Supplementary Figure 2 and Table 1). The rarefaction curve also supported the difference in α-diversity since species diversity was highest in the dry sediment sample (EC1). The rarefaction curves were generated based on ASVs at 99.9% similarity and they supported the difference in α-diversity indices since species diversity was highest in the dry sediment (ZC1) and lowest in the water sample EA1 (Supplementary Figure 3). The rarefaction curves also indicated that all the sites were far from being exhaustively sampled.
Table 1.
Alpha diversity indices computed on all ASVs-based microbial taxonomic units within 16S rRNA.
| Site | Number of Sequences | Observed | Chao1 | ACE | Shannon | Simpson | InverseSimpson |
|---|---|---|---|---|---|---|---|
| EA1 | 2730 | 197 | 224.029 | 240.110 | 4.627 | 0.986 | 71.759 |
| EA2 | 3344 | 212 | 222.150 | 236.312 | 4.832 | 0.990 | 96.863 |
| EB1 | 26691 | 1181 | 1205.265 | 1238.172 | 6.375 | 0.997 | 342.655 |
| EB2 | 16645 | 933 | 989.953 | 1053.067 | 6.181 | 0.997 | 351.963 |
| EC1 | 33878 | 1331 | 1357.750 | 1401.321 | 6.696 | 0.998 | 623.953 |
| EC2 | 28223 | 1259 | 1291.659 | 1343.116 | 6.624 | 0.998 | 582.833 |
| ED1 | 5223 | 293 | 300.065 | 307.122 | 5.136 | 0.991 | 117.433 |
| ED2 | 6495 | 343 | 352.220 | 360.925 | 5.492 | 0.995 | 201.261 |
| ZA1 | 5146 | 379 | 400.468 | 415.263 | 5.309 | 0.993 | 142.890 |
| ZA2 | 28819 | 1164 | 1189.084 | 1224.216 | 6.446 | 0.998 | 415.700 |
| ZB1 | 29579 | 1220 | 1237.243 | 1270.553 | 6.555 | 0.998 | 525.634 |
| ZB2 | 23268 | 1015 | 1023.797 | 1051.801 | 6.402 | 0.998 | 465.894 |
| ZC1 | 34827 | 1187 | 1219.591 | 1245.735 | 6.569 | 0.998 | 531.790 |
| ZC2 | 26807 | 886 | 897.938 | 916.577 | 6.271 | 0.998 | 406.068 |
| ZD2 | 3217 | 229 | 255.640 | 255.299 | 4.912 | 0.990 | 100.626 |
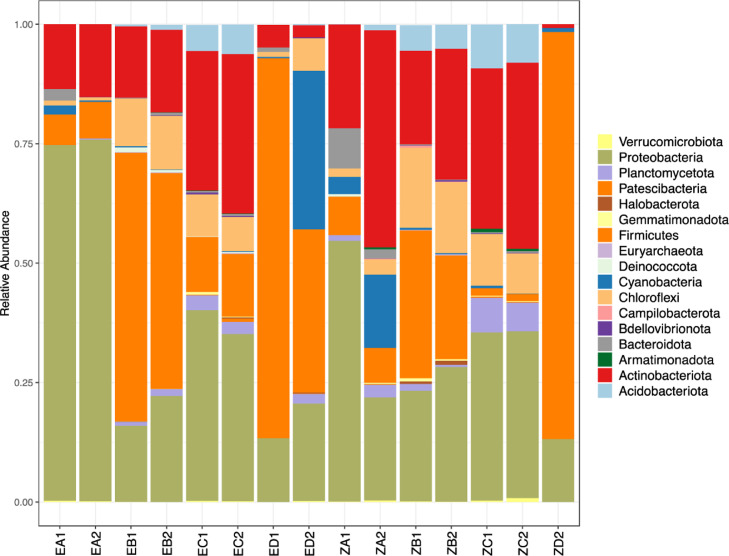
3.4. Abundance of the prokaryotic taxa
Composition analysis of the 16S rRNA ASVs comprised of the kingdom Bacteria (88.24%) and Archaea (11.76%). These results suggest that bacteria were the dominant taxa and the prominent phyla were Proteobacteria (33.8%), Firmicutes (27.3%), Actinobacteriota (21.2%), Chloroflexi (6.8%), Cyanobacteria (3.8%), Acidobacteriota (2.8%), Planctomycetota (1.9%), Bacteroidota (1.1%), Deinococcota (0.16%), Gemmatimonadota (0.16%), Verrucomicrobiota 61(0.12%), Bdellovibrionota (0.12%), Armatimonadota (0.12%), Patescibacteria (0.1%), and Campilobacterota (0.05%). The prominent Archaea phyla in the prokaryotic composition were Halobacterota (0.14%), and Euryarchaeota (0.06%). The remaining 0.93% were occupied by other less abundant phyla (<0.002%) phyla (Figure 2). Though Proteobacteria was the dominant phyla in the bacterial kingdom, the trend changed between sample types. For instance, the microbial mat samples were dominated by the Firmicutes (66.3%) against Proteobacteria (15.6%) (Supporting Information S2 and Fig. 2). The dominant phylum Proteobacteria was represented by two classes, α-Proteobacteria (75.3%) and γ-Proteobacteria (24.7%) that constituted 38 and 19 families, respectively (Supporting Information S3). Among the abundant families belonging to class α-Proteobacteria were Xanthobacteraceae (18.3%), Rhodobacteraceae (12%), Sphingomonadaceae (11.5%), and Berinckiaceae (9.3%). Among the abundant families that belonged to class γ-Proteobacteria were Pseudomonadaceae (35.4%), Moraxellaceae (26.6%), Comamonadaceae (15%), and Oxalobacteraceae (6.9%). The abundant archaea were represented by four classes, Methanobacteria (33.3%), Methanocellia (28.6%), Methanosacinia (23.8%), and Methanomicrobia (14.3%) (Supporting Information S3).
Fig. 2.
Relative abundance of the most predominant phyla in various sample types collected within Lakes Olbolosat and Oloiden.
Firmicutes was the second most abundant phylum that represented by class Clostridia and four families Peptostreptococcaceae (74%), Clostridia (20%), Lachnospiraceae (4%), and Peptostreptococcales-Tissierellales_fa (2%). Class Clostridia dominated wet sediments from Lake Olbolosat (wet sediment, 15.54-28.66%; Dry sediment, 0.65-13.3%; water, 0.14-5.66%) compared to Lake Oloiden wet sediments (wet sediment, 5.58-8.01%; Dry sediment, 0.68-7.96%; water, 0.49-3.16%). Clostridia were, however, highly abundant in Lake Oloiden microbial mat samples (4.08-5.96%) compared to the Lake Olbolosat microbial mat samples (0-0.13%). Dominant families, Peptostreptococcaceae dominated microbial mat samples (1-3.1%). Lachnospiraceae dominate wet sediment (1%), and microbial mat (1.8%), while Peptostreptococcales-Tissierellales_fa dominated the mat (1.1%).
The phylum Actinobacteriota was the third (Fig. 2) most abundant between the sample types and it was represented by five classes. Among the dominant classes were Actinobacteria (78.6%) and Acidimicrobiia (15.6%) (Supporting Information S3). Actinobacteria were highly abundant in L. Oloiden (Dry sediment, 9.38-16.61%; wet sediment, 0.44-14.57%; water, 9.5-14.56%; microbial mat, 0.05-0.23%) compared to L. Olbolosat (Dry sediment, 1.79-4.16%; wet sediment, 5.15-8.4%; water, 0.54-13.8%; mat, 0-0.82%). The abundance of class Acidimicrobiia varied between sample types across the two lakes. They were highly dominant in the L. Oloiden water samples (water, 3.18-54.49%; wet sediment, 0-5.75%; Dry sediment, 4.12-5.38%; mat, 0-0.31%) compared to L. Olbolosat water samples (water, 0.49-7.29%; wet sediment, 5.4-8.69%; Dry sediment, 1.04-3.61%; mat, 0-0.25%) (Supporting Information S3).
Phylum Chloroflexi was the fourth most dominant represented by class Anaerolineae (45.7%), Chloroflexia (44.6%), and Ktedonobacteria (9.5%) (Supporting Information S3). Class Anaerolineae was abundant in L. Olbolosat (2.2-34.18%) compared to L. Oloiden (0-0.84%). They were associated with dry sediments (34.18%), water samples (23.5%), wet sediments (13.14%), lower abundance was associated with microbial mat samples (0-0.84%). Class Chloroflexia was highly abundant in L. Oloiden (Dry sediment, 6.95-15.23%; wet sediment, 0.05-14.51%; water, 7.01-8.36%; mat, 0.00-3.46%) compared to L. Olbolosat (Dry sediment, 0.89-9.83%; wet sediment, 10.63-12.26%; water, 0.08-10.71%; mat, 0.05%). Similarly, class Ktedonobacteria was highly abundant in Lake Oloiden (Dry sediment, 1.03-52.82%; wet sediment, 0.0-9.44%; water, 0.0-35.09%; microbial mat, 0%) compared to L. Olbolosat (Dry sediment, 0%; wet sediment, 0.34-0.51%; water, 0.0-0.77%; mat, 0.0%).
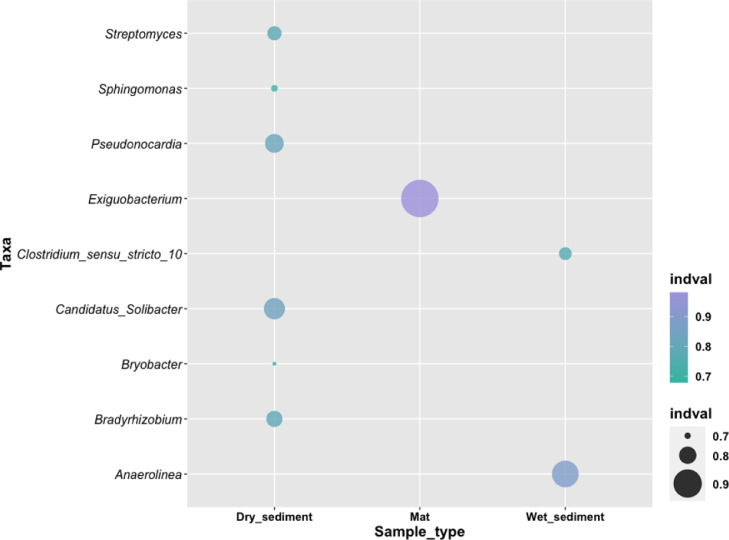
Indicator species analysis using lambdsv package revealed most genera affiliated to major phyla were associated with wet sediments, dry sediments, and microbial mat samples across lakes Olbolosat and Oloiden (Supplementary Table 1). The representatives of the genera associated with the wet sediments were distributed among Proteobacteria (Bradirhizobium, Sphingomonas, p <0.05), Firmicutes (Clostridium_sensu_stricto_10, Exiguobacterium, p <0.05), Actinobacteriota (Pseudonocardia, Streptomyces, p <0.05), Chloroflexi (Anaerolinea, p <0.05), and Acidobacteriota (Candidatus_Solibacter, Bryobacter, p <0.05) (Supplementary Table 1). Genus Anaerolinea was the indicator species in the wet sediments with an indicator value of 0.88 and it was closely followed by genus Clostridium_sensu_stricto_10 at a 0.76 indicator value. Genus Candidatus_Solibacter was the indicator species in dry sediments with an indicator value of 0.84, while Exiguobacterium was the indicator species in the microbial mat with an indicator value of 0.98 (Supplementary Table 1 and Fig. 3). The frequency of indicator species ranged between 6 to 10. Genera belonging to phylum Firmicutes (Clostridium_sensu_stricto_10 and Exiguobacterium) were represented with the highest (10) and lowest (6) frequency in wet sediments and microbial mat, respectively (Supplementary Table 1).
Fig. 3.
Indicator species within lake Olbolosat and Oloiden at the genus level.
3.5. Bacterial/Archaeal community composition and correlation with environmental factors
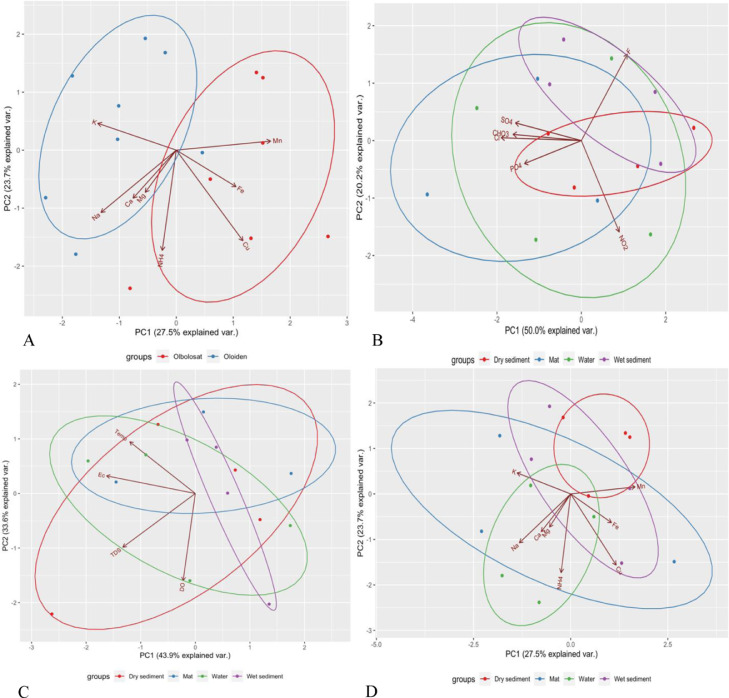
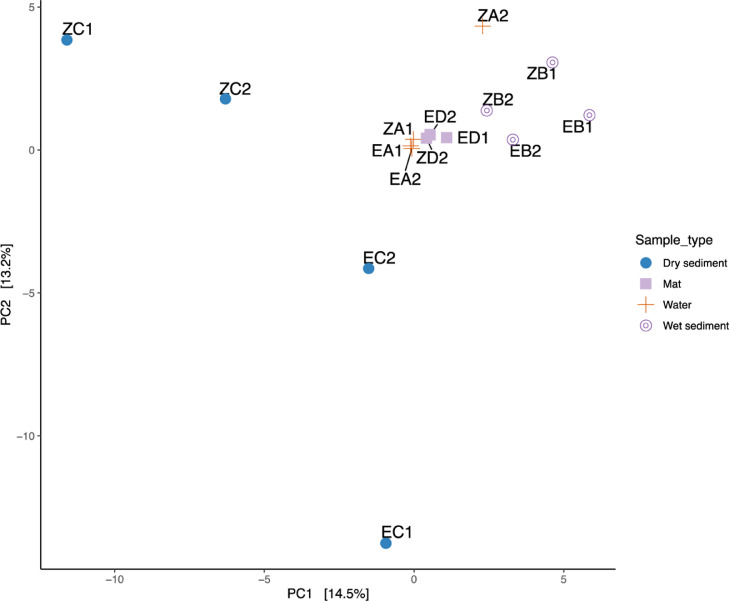
Principal Component Analysis indicated that community compositions were clustered according to the locations (Lakes) of the sampling sites (Fig. 4a). Microbial community compositions were also clustered based on the sample types across the lakes (Fig. 4b-4d). The distribution of the ASVs in lake Olbolosat was associated with Cu+, Fe2+, and Mn2+, while the distribution of ASVs in lake Oloiden was associated with Mg2+, Na+, and K+ (Fig. 4a). The concentration of anions F− were associated with the distribution of ASVs in the wet sediments while the distribution of ASVs in the water samples and dry sediments were associated with NO2− and PO43−, respectively. The concentration of SO43 −, HCO3−, and Cl− were positively associated with water samples (Fig. 4b). The concentration of TDS and DO were associated with the distribution of ASVs in water samples, while the distribution of ASVs in dry sediments samples were associated with the electrical conductivity and temperature (Fig. 4c). Similarly, the distribution of the ASVs between water and microbial mat samples was associated with NH4+, Na+, and Mg2+, while Fe2+ concentrations were associated with the water and wet sediments. Mn2+ and Cu+ were distinctly associated with dry sediments and microbial mat, respectively. Potassium ions (K+) were, however, associated with the microbial mat samples (Fig. 4d).
Fig. 4.
Principal component analysis based on a Bray-Curtis similarities matrix between sample types and environmental factors. (a) Ordination of Lakes and cations. (b) Ordination of anions and sample types. (c) Ordination of physical parameters and sample types. (d) Ordination of cations and sample types.
The effects of environmental factors on the prokaryotic diversity and community composition were supported by the Permutation Multivariate Analysis of Variance (PERMANOVA). Nine out of the seventeen factors strongly influenced the variation between samples and distribution of ASVs with R2 ranging from 0.38 to 0.71 and p < 0.05 (Table 2). The effect of temperature on the ASV distribution in water sample was about two times (R2 = 0.71, p = 0.002) the impact of nitrite (R2 = 0.38, p = 0.045). The distribution of ASVs in the microbial mat and water samples were strongly influenced by the Magnesium (R2 = 0.70, p = 0.005), while the distribution of ASVs and sample variations in the wet sediment were influenced by Manganese (R2 = 0.58, p = 0.008). A subset analysis of a correlation between the ions and the ASVs revealed Magnesium and Manganese as the greatest predictors of microbial community structures (r = 0.39). Further, analysis of environmental factors also revealed dissolved oxygen (DO) and Temperature as the best predictors (r = 0.17) of microbial community structures in dry sediments and water samples, respectively. Consequently, the subset analysis of a correlation between the cations and ASVs revealed sulfate as the predictor of the distribution of microbial community structure in the microbial mat and water samples (r = 0.17). However, Mantle statistical test Pearson's product-moment correlation did not show any significant correlation between community similarity and spatial proximity to each of the environmental factors. Ordination cluster analysis showed that sample types clustered according to the composition of microbial community structure. Sample types with similar microbial community structures clustered together, while those with unique community structures clustered independently or further away (Fig. 5).
Table 2.
Results from individual PERMANOVA analyses on selected environmental factors, using Pearson's product-moment correlation on variance-stabilizing transformed counts.
| Factor | R2 | p |
|---|---|---|
| Temperature (Temp) | 0.71 | 0.002** |
| Magnesium (Mg2+) | 0.70 | 0.005** |
| Manganese (Mn2+) | 0.58 | 0.008** |
| Bicarbonate (HCO3−) | 0.56 | 0.005** |
| Sulphate (SO43 −) | 0.54 | 0.007** |
| Chloride (Cl−) | 0.46 | 0.023* |
| Total Dissolve Substances (TDS) | 0.44 | 0.031* |
| Dissolve Organic (DO) | 0.42 | 0.042* |
| Nitrite (NO2−) | 0.38 | 0.045* |
| Copper (Cu+) | 0.31 | 0.123 |
| Electron Conductivity (Ec) | 0.20 | 0.249 |
| Fluoride (F−) | 0.09 | 0.564 |
| Potassium (K+) | 0.06 | 0.713 |
| Sodium (Na+) | 0.04 | 0.778 |
| Ammonium (NH4+) | 0.04 | 0.786 |
| Iron (Fe2+) | 0.00 | 0.782 |
| Phosphate (PO43−) | 0.00 | 0.993 |
P values with an asterisk *are significant.
Fig. 5.
PCA ordination plot for all sample types, using Bray-Curtis dissimilarity distance, colored by sample type.
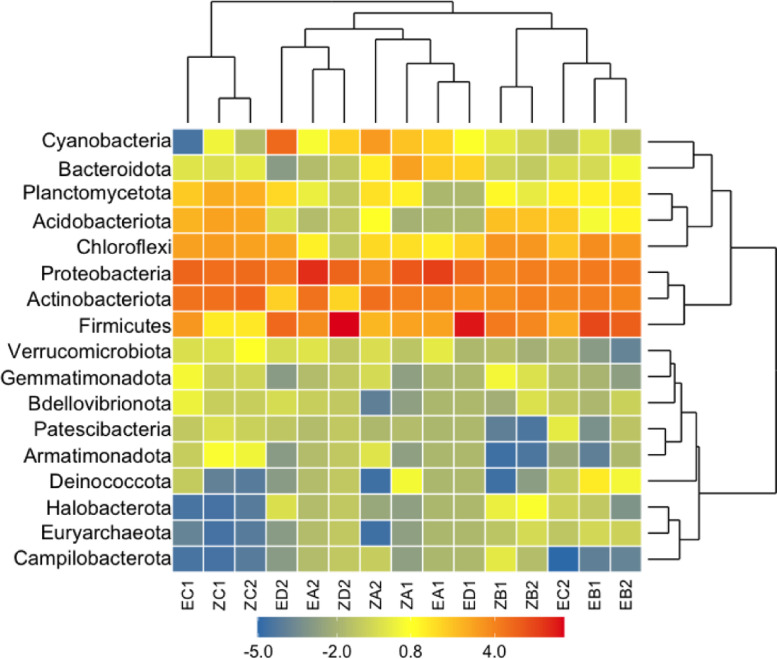
The clustering and distribution of ASVs were also supported by the hierarchical clustering in which samples were clustered according to the sample types. The hierarchical cluster of the highly abundant ASVs also indicated that major phyla and minor phyla clustered separately (Fig. 6). The ordination and hierarchical cluster results were significantly (r = 0.07143, p = 0.041) supported by the ANOSIM. Further, aldex.ttest and aldex.effect functions revealed a total of 5 differentially abundant (we.ep < 0.05) ASVs from 70 indicators ASVs between the sample types (Supplementary Table 2). Most ASVs were, however, non-differentially abundant across the samples. The differentially abundant ASVs formed 7.1% of the total indicator ASVs from the sampling sites. Similarly, an evaluation of the ASV correlation at the phylum level revealed a positive co-correlation network pattern of the indicator ASVs (Supplementary Figure 4). Among the 5 differentially abundant ASVs, 80% belonged to phylum Actinobacteriota and 20% belonged to phylum Proteobacteria. Further, the differentially abundant ASVs were classified down to genus level representing phylum Actinobacteriota; Pseudarthrobacter (40%), Paracoccus (20%), Micromonospora (20%), and phylum Proteobacteria; Methylosistis (20%) (Supporting Information S3).
Fig. 6.
Hierarchical clustering of phyla assessing the relationship between sample types and prokaryotic taxa based on Bray-Curtis dissimilarity of the highly abundant taxa at the phylum.
4. Discussion
4.1. Alpha diversity
The study revealed that prokaryotic diversity in lake Oloiden was lower than that of lake Olbolosat. These lakes are distinguished by their salinity and freshness as in lake Oloiden and lake Olbolosat, respectively. Prokaryotic biodiversity results were in accordance with studies by (Luo et al., 2017) who reported that microbial diversity of soda lakes is normally lower than that of freshwater ecosystems. The number of ASVs recovered was lower in water samples as compared to other samples. This is in line with studies by Cleary and Polónia (2018) who reported OTU richness being lowest in water biotope against sediments and microbial mats. We found that α-diversity indices across lake Olbolosat and lake Oloiden were similar to indices reported from other tropical lakes in the same latitude (Inceoʇlu et al., 2015).
4.2. The abundance of the prokaryotic taxa
Our study is additional study to the known recent extensive studies in Africa for microbial communities in extreme environments (Mwirichia et al., 2011; Kambura et al., 2016; Ghilamicael et al., 2017; Josiah et al., 2018) . Though there are still limited studies on the prokaryotic diversity of the fast disappearing fresh and saline-alkaline ecosystems along the equator, the current study revealed the abundance of prokaryotic community structures similar to those determined along the equatorial ecosystems (Mwirichia et al., 2011; Inceoʇlu et al., 2015; Kambura et al., 2016; Ghilamicael et al., 2017; Cleary and Polónia, 2018). The higher relative abundance of phyla Proteobacteria, Firmicutes, Actinobacteriota, Chloroflexi, Cyanobacteria, Acidobacteriota, Planctomycetota and Bacteroidota comprised most of the observed taxa, perhaps indicating either that inland water ecosystems are similar regardless of seasons and geographical locations, or that high-level taxonomic assessment harbors important species-level variation (Mhuireach et al., 2019).
The phylum Proteobacteria was the predominant phylum observed among the samples across the two lakes, but was highly abundant in water samples. Proteobacteria are essential in the biodegradation and biogeochemical processes within the marine ecosystems (Huang et al., 2017). Only α-Proteobacteria and γ-Proteobacteria are the abundant classes in the two lakes with α-Proteobacteria being the predominant class within the phylum Proteobacteria. The abundant α-Proteobacteria within lake Oloiden were recovered from the dry sediment samples as compared to wet sediments, water and microbial mats. Families (Xanthobacteraceae, Rhodobacteraceae, Sphingomonadaceae, and Berinckiaceae) that belong to the abundant α- and γ-Proteobacteria are beneficial partners in plant-microbe interactions, especially nitrogen fixation, legume nodulation and methanotrophic activities (Grube and Berg, 2015). Pseudomonadaceae, Moraxellaceae, Comamonadaceae, and Oxalobacteraceae that belong to γ-Proteobacteria are important in the biodegradation of low and high molecular weight organic matter in the ecosystem with low levels of oxygen concentration (Mahmoudi et al., 2015).
Similarly, α-Proteobacteria and γ-Proteobacteria are regarded as primary colonizers and they prepare the ecosystem for subsequent colonization according to (Abed et al. 2019). They mutually coexist and establish a heterotrophic bacterial film in the mats. They sense environmental signals such as NA+ flux in the surrounding water thereby shifting from motile to sessile lifestyle (Belas, 2014). Actinobacterota and Firmicutes are secondary colonizers depending on the nutrients, microbial, or decay products provided by the primary colonizers (Abed et al., 2019). The interaction between the primary and secondary colonizers such as competition, cooperation, mutual exclusion, limited dispersal or availability can result in a segregated pattern of biofilm formation by such microbes over time (Abed et al., 2019).
The phylum Firmicutes was the second most abundant phylum recovered from lakes Olbolosat and Oloiden. They are ubiquitous in aquatic ecosystems probably due to their spores that quickly germinate during favorable conditions (Kiama et al., 2021). Adherence of Firmicutes to the surfaces could be a strategy used for survival and evolution as a community in the lake ecosystem (Prieto-Barajas et al., 2018). Firmicutes are also known to be good bio-degraders of organic pollutants such as petroleum hydrocarbons polychlorinated biphenyl and hexahydro-1,3,5-trinitro-1,3,5-triazine (Zhang et al., 2015). Clostridia was the predominant class among Firmicutes retrieved from wet sediments within L. Olbolosat as compared to L. Oloiden. The trend, however, changed in lake Oloiden in which Clostridia were highly abundant in the microbial mats compared to lake Olbolosat. Dominant families in the class Clostridia were Peptostreptococcaceae in the microbial mat samples, Lachnospiraceae in the wet sediment, and mat, while Peptostreptococcales-Tissierellales_fa dominated the mat samples. Earlier studies have reported Clostridia species as precursors for sulfate reduction. They are also important in the anaerobic decomposition of organic matter in aquatic ecosystems (Wang et al., 2016).
Actinobacteria and Acidiimicrobiia (Actinobacteriota) were also retrieved from the current study. Most members of this group are associated with high mol G+C content (Sivalingam et al., 2019). The high mol G+C content is important in the adaptation to unfavorable environmental conditions, tolerant to antagonism factors, and susceptibility to low mutation rates (Sivalingam et al., 2019). Other abundant phyla in the study included Cyanobacteria, Acidobacteriota, Planctomycetota, Bacteroidota, Deinococcota, Gemmatimonadota, Verrucomicrobiota, Bdellovibrionota, Armatimonadota, Patescibacteria, and Campilobacterota, which were present in low percentages. Sequences affiliated with Halobacterota and Euryarchaeota were detected from the archaeal kingdom and have been reported to be methanogens within the lake ecosystems (He et al., 2020).
Currently, archaea are known to be ubiquitous (Li et al., 2019) unlike in the earlier studies that they were believed to inhabit only the extreme ecosystems (Cavicchioli et al., 2011). Our study revealed four archaeal classes; Methanobacteria, Methanocellia, Methanosacinia, and Methanomicrobia. Methanogens are highly distributed in stringently anaerobic and natural ecosystems such as freshwater and marine sediments, rice fields, deep-sea hydrothermal vents, marine mud volcanoes, mangroves and hot springs (Laskar et al., 2018; Li et al., 2019). Their activities in the environment are linked to the emission of methane gas that significant contributes to a great increase in global warming (Laskar et al., 2018).
4.3. Ecological roles of the retrieved indicator Taxa
Indicator organisms are useful in the study of the functional role of specific microbes within an ecosystem (Hartmann et al., 2012; Rime et al., 2015). Microbial mats comprise millions of microorganisms that belong to different taxa, that interact and exchange signals embedded in a matrix of exopolysaccharide and nutrients to enable the flow of energy, and resources for the survival of the community (Prieto-Barajas et al., 2018). Sediments within lakes are the main medium for migration and nutrients transformation that influence the nutrients contents within the lakes (Huang et al., 2017). The indicator species were associated with the wet sediment, dry sediment, and microbial mat across the two lakes. Genera associated with the wet sediments belonged to phylum Proteobacteria (Bradirhizobium and Sphingomonas), Firmicutes (Clostridium_sensu_stricto_10 and Exiguobacterium), Actinobacteriota (Pseudonocardia and Streptomyces), Chloroflexi (Anaerolinea), and Acidobacteriota (Candidatus_Solibacter and Bryobacter). Sphingomonas and Bradyrhizobium (Proteobacteria) influence the biodegradation of environmental pollutants Huang et al., (2017), while Exiguobacterium (Firmicutes), the indicator species in the microbial mat is a secondary colonizer that withstands harsh environmental conditions by the formation of spores (Kiama et al., 2021). Streptomyces and Pseudonocardia (Actinobacteriota) are important in the biodegradation of complex organic molecules in the lake ecosystem. They are also able to survive in harsh environmental conditions due to their high G+C content that is important in the adaptation to unfavorable environmental conditions, tolerance to antagonism factors, and susceptibility to low mutation rates (Sivalingam et al., 2019). Candidatus_Solibacter and Bryobacter (Acidobacteriota) are oligotrophic microbes adapted to low nutritional concentrations (Liao et al., 2019).
4.4. Community composition and correlation to environmental factors
Despite intensive anthropogenic activities that can influence changes in biogeochemical cycles and microbial ecology within inland water bodies such as lakes Olbolosat and Oloiden, these ecosystems are still rich in microbial communities that are dependent on environmental factors. Prokaryotic communities within lakes Olbolosat are highly dependent on Copper, Iron, Manganese and Ammonium while those from lake Oloiden are dependent on Magnesium, Sodium, Calcium and potassium. Copper, Iron, Manganese are micronutrients that are trace elements used as enzyme cofactors by microorganisms. Magnesium, Sodium and Potassium are macro elements that have been reported to contribute to the species richness and composition of prokaryotic communities (Liu et al., 2018).
5. Conclusion
The next-generation DNA sequencing data of 15 samples from lakes Olbolosat and Oloiden provides the first results characterizing water, microbial mats, wet and dry sediments from the two ecosystems. The majority of sequences (99.9%) were observed to belong to the kingdom bacteria. These results suggest that bacteria are the most dominant taxa within the two lakes. The bacterial phylum Proteobacteria was dominant across all samples. Moreover, the interaction network of prokaryotic communities within lakes Olbolosat and Oloiden displayed Proteobacteria to be highly positively connected with other microbes. This could be due to diverse metabolic mechanisms thus promoting the growth of the microbial community (Liao et al., 2019). The composition of ASVs in lake Olbolosat was highly dependent on Cu+, Fe2+, NH4+, and Mn2+, while L. Oloiden was dependent on Mg2+, Na+, Ca2+, and K+. Exiguobacterium was the indicator species in the microbial mat with the highest indicator value while Clostridium_sensu_stricto_10 recorded the highest indicator frequency from wet sediments. Differentially abundant ASVs belonged to phylum Actinobacteriota; Pseudarthrobacter, Paracoccus, Micromonospa and phylum Proteobacteria; Methylosistis.
Declaration of Competing Interest
The authors have not declared any conflict of interest.
Acknowledgments
Author's contribution
Conceived and designed the experiment: CWK. Contributed to the reagents/materials/sequencing cost/analysis tools: CWK. Conducted the experiments: CWK. Complied results: CWK. Analyzed the data: CWK ENW JOK. Write up and final editing of the manuscript: CWK ENW JOK. All authors CWK MMN AKK JNM VNM ENW RNK JOK read and approved the final manuscript.
Funding
The authors appreciate the International Foundation for Science (IFS) for funding the work (Grant No.1I1_W_036264).
Acknowledgments
We thank International Foundation for Science (IFS) for funding this work. Prof Elijah Ateka and Dr. Maina Mathara for providing laboratory space.
Availability of data and materials
The data is available on NCBI Sequence Read Archive with accession number PRJNA723886 (Accessions SAMN18836534-SAMN18836548).
Ethics approval and consent to participate
Not applicable.
New clinical tools and procedure
Not applicable.
Research authorization
The research was authorized by National Commission for Science, Technology and Innovation (NACOSTI) and Kenya Wildlife Service (KWS).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2021.100066.
Appendix. Supplementary materials
References
- Abed R.M.M., Al Fahdi D., Muthukrishnan T. Short-term succession of marine microbial fouling communities and the identification of primary and secondary colonizers. Biofouling. 2019;35:526–540. doi: 10.1080/08927014.2019.1622004. [DOI] [PubMed] [Google Scholar]
- Belas R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014;22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Holmes S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., Sankaran K., Fukuyama J.A., McMurdie P.J., Holmes S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research. 2016;5:1–48. doi: 10.12688/F1000RESEARCH.8986.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J.A., Smith G., Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R., Amils R., Wagner D., Mcgenity T. Life and applications of extremophiles. Environ. Microbiol. 2011;13:1903–1907. doi: 10.1111/j.1462-2920.2011.02512.x. [DOI] [PubMed] [Google Scholar]
- Cleary D.F.R., Polónia A.R.M. Bacterial and archaeal communities inhabiting mussels, sediment and water in Indonesian anchialine lakes. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2018;111:237–257. doi: 10.1007/s10482-017-0944-1. [DOI] [PubMed] [Google Scholar]
- Dawood D.H., Sanad M.I. Determination of Ions (Anion and Cation) By Ion Chromatography in Drinking Water From Talkha Territory and Some Its Villages, Dakahlia, Egypt. J. Agric. Chem. Biotechnol. 2014;5:215–226. doi: 10.21608/jacb.2014.49898. [DOI] [Google Scholar]
- Ghilamicael A.M., Budambula N.L.M., Anami S.E., Mehari T., Boga H.I. Evaluation of prokaryotic diversity of five hot springs in Eritrea. BMC Microbiol. 2017;17:1–13. doi: 10.1186/s12866-017-1113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube M., Berg G. Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Front. Microbiol. 2015;6:1–9. doi: 10.3389/fmicb.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M., Howes C.G., Vaninsberghe D., Yu H., Bachar D., Christen R., Henrik Nilsson R., Hallam S.J., Mohn W.W. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J. 2012;6:2199–2218. doi: 10.1038/ismej.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Li M., Zhen Y., Mi T., Yu Z. Bacterial and Archaeal Communities in Sediments from the Adjacent Waters of Rushan Bay (China) Revealed by Illumina Sequencing. Geomicrobiol. J. 2020;37:86–100. doi: 10.1080/01490451.2019.1666193. [DOI] [Google Scholar]
- Huang W., Chen X., Jiang X., Zheng B. Characterization of sediment bacterial communities in plain lakes with different trophic statuses. Microbiologyopen. 2017;6:1–14. doi: 10.1002/mbo3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inceoʇlu Ö., Llirós M., García-Armisen T., Crowe S.A., Michiels C., Darchambeau F., Descy J.P., Servais P. Distribution of Bacteria and Archaea in meromictic tropical Lake Kivu (Africa) Aquat. Microb. Ecol. 2015;74:215–233. doi: 10.3354/ame01737. [DOI] [PubMed] [Google Scholar]
- Josiah O.K., Huxley M.M., Hamadi I.B., Anne T.W.M., Jun U. Phylogenetic diversity of prokaryotes on the snow-cover of Lewis glacier in Mount Kenya. African J. Microbiol. Res. 2018;12:574–579. doi: 10.5897/ajmr2017.8750. [DOI] [Google Scholar]
- Kambura A.K., Mwirichia R.K., Kasili R.W., Karanja E.N., Makonde H.M., Boga H.I. Bacteria and Archaea diversity within the hot springs of Lake Magadi and Little Magadi in Kenya. BMC Microbiol. 2016;16:1–12. doi: 10.1186/s12866-016-0748-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiama C.W., Njire M.M., Kambura A.K., Mugweru J.N., Matiru V.N., Wafula E.N. Isolation and Characterization of Bacteria from Lakes Olbolosat and Oloiden. Kenya. African J. Microbiol. Res. 2021;15:1–19. doi: 10.5897/AJMR2020.9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskar F., Das Purkayastha S., Sen A., Bhattacharya M.K., Misra B.B. Diversity of methanogenic archaea in freshwater sediments of lacustrine ecosystems. J. Basic Microbiol. 2018;58:101–119. doi: 10.1002/jobm.201700341. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhu X., Zhang W., Zhu D., Zhou X., Zhang L. Archaeal communities in the deep-sea sediments of the South China Sea revealed by Illumina high-throughput sequencing. Ann. Microbiol. 2019;69:839–848. doi: 10.1007/s13213-019-01477-4. [DOI] [Google Scholar]
- Liao B., Yan X., Zhang J., Chen M., Li Y., Huang J., Lei M., He H., Wang J. Microbial community composition in alpine lake sediments from the Hengduan Mountains. Microbiologyopen. 2019;8:1–16. doi: 10.1002/mbo3.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Ding X., Tang X., Wang J., Li W., Yan Q., Liu Z. Macro and microelements drive diversity and composition of prokaryotic and fungal communities in hypersaline sediments and saline-alkaline soils. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Li H., Kotut K., Krienitz L. Molecular diversity of plankton in a tropical crater lake switching from hyposaline to subsaline conditions: Lake Oloidien. Kenya. Hydrobiologia. 2017;788:205–229. doi: 10.1007/s10750-016-2998-x. [DOI] [Google Scholar]
- Mahmoudi N., Robeson M.S., Castro H.F., Fortney J.L., Techtmann S.M., Joyner D.C., Paradis C.J., Pfiffner S.M., Hazen T.C. Microbial community composition and diversity in Caspian Sea sediments. FEMS Microbiol. Ecol. 2015;91:1–11. doi: 10.1093/femsec/fiu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina C.W., Sang J.K., Mutua B.M., Raude J.M. Bathymetric survey of Lake Naivasha and its satellite Lake Oloiden in Kenya; using acoustic profiling system. Lakes Reserv. Res. Manag. 2018;23:324–332. doi: 10.1111/lre.12247. [DOI] [Google Scholar]
- Mhuireach G., Betancourt-Román C.M., Green J.L., Johnson B.R. Spatiotemporal controls on the urban aerobiome. Front. Ecol. Evol. 2019;7:1–15. doi: 10.3389/fevo.2019.00043. [DOI] [Google Scholar]
- Mwirichia R., Cousin S., Muigai A.W., Boga H.I., Stackebrandt E. Bacterial diversity in the haloalkaline Lake Elmenteita. Kenya. Curr. Microbiol. 2011;62:209–221. doi: 10.1007/s00284-010-9692-4. [DOI] [PubMed] [Google Scholar]
- Oulas A., Pavloudi C., Polymenakou P., Pavlopoulos G.A., Papanikolaou N., Kotoulas G., Arvanitidis C., Iliopoulos I. Metagenomics: Tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform. Biol. Insights. 2015;9:75–88. doi: 10.4137/BBI.S12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Kumbhare S.V., Mhatre S.S., Chowdhury S.P., Shetty S.A., Marathe N.P., Bhute S., Shouche Y.S. Exploration of microbial diversity and community structure of Lonar Lake: The only hypersaline meteorite crater lake within basalt rock. Front. Microbiol. 2016;6:1–12. doi: 10.3389/fmicb.2015.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Barajas C.M., Valencia-Cantero E., Santoyo G. Microbial mat ecosystems: Structure types, functional diversity, and biotechnological application. Electron. J. Biotechnol. 2018;31:48–56. doi: 10.1016/j.ejbt.2017.11.001. [DOI] [Google Scholar]
- Prodan A., Tremaroli V., Brolin H., Zwinderman A.H., Nieuwdorp M., Levin E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS One. 2020;15:1–19. doi: 10.1371/journal.pone.0227434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J., Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods. 2010:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rime T., Hartmann M., Brunner I., Widmer F., Zeyer J., Frey B. Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Mol. Ecol. 2015;24:1091–1108. doi: 10.1111/mec.13051. [DOI] [PubMed] [Google Scholar]
- Silveira R., Silva M.R.S.S., de Roure Bandeira de Mello T., Alvim E.A.C.C., Marques N.C.S., Kruger R.H., da Cunha Bustamante M.M. Bacteria and Archaea Communities in Cerrado Natural Pond Sediments. Microb. Ecol. 2020;81:563–578. doi: 10.1007/s00248-020-01574-x. [DOI] [PubMed] [Google Scholar]
- Sivalingam P., Hong K., Pote J., Prabakar K. Extreme Environment Streptomyces : Potential Sources for New Antibacterial and Anticancer Drug Leads? Int. J. Microbiol. 2019;2019:1–20. doi: 10.1155/2019/5283948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wafula E.N., Murunga S.I. Isolation and Identification of Phosphate Solubilizing and Nitrogen-Fixing Bacteria from Lake Ol'Bolossat Sediments. Kenya. Mod. Appl. Sci. 2020;14:1–37. doi: 10.5539/mas.v14n10p37. [DOI] [Google Scholar]
- Wang N.F., Zhang T., Yang X., Wang S., Yu Y., Dong L.L., Guo Y.D., Ma Y.X., Zang J.Y. Diversity and Composition of Bacterial Community in Soils and Lake Sediments from an Arctic Lake Area. Front. Microbiol. 2016;7:1–9. doi: 10.3389/fmicb.2016.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Ma L., Jiang H., Wu G., Dong H. Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan Lakes. Sci. Reports- Nat. Brief. 2016;6:6–11. doi: 10.1038/srep25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang Y., Zhao L., Li Y., Xie S., Liu Y. Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl. Microbiol. Biotechnol. 2015;99:3291–3302. doi: 10.1007/s00253-014-6262-x. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhao T., Wang Q., Li L., Shen T., Gao G. Bacterial community composition in aquatic and sediment samples with spatiotemporal dynamics in large, shallow, eutrophic Lake Chaohu. China. J. Freshw. Ecol. 2019;34:575–589. doi: 10.1080/02705060.2019.1635536. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available on NCBI Sequence Read Archive with accession number PRJNA723886 (Accessions SAMN18836534-SAMN18836548).