Highlights
-
•
The established thinking is that host response, rather than M. leprae strain variation, is the reason for the broad variation of symptoms in the leprosy disease spectrum
-
•
Recent discoveries suggest that macrophage polarization also plays a significant role in the spectrum of leprosy disease but to what degree it contributes is not fully established.
-
•
Utilizing DNA from three different strains of M. leprae DNA to stimulate human macrophages under three polarization conditions we observed, that the transcriptome upon stimulation with DNA from M. leprae strain Thai-53 was larger compared to strains NHDP and Br4923, independent of the macrophage polarization condition.
-
•
We also found that macrophage polarization affects the responses to M. leprae DNA, with up-regulation of numerous interferon stimulated genes.
-
•
These findings provide a deeper understanding of the role of macrophage polarization in the recognition of M. leprae DNA, with the potential to improve leprosy treatment strategies.
Keywords: RNAseq, Mycobacterium leprae, Leprosy, Macrophage polarization
Abstract
Infection with Mycobacterium leprae, the causative organism of leprosy, is still endemic in numerous parts of the world including the southwestern United States. The broad variation of symptoms in the leprosy disease spectrum range from the milder tuberculoid leprosy (paucibacillary) to the more severe and disfiguring lepromatous leprosy (multibacillary). The established thinking in the health community is that host response, rather than M. leprae strain variation, is the reason for the range of disease severity. More recent discoveries suggest that macrophage polarization also plays a significant role in the spectrum of leprosy disease but to what degree it contributes is not fully established. In this study, we aimed to analyze if different strains of M. leprae elicit different transcription responses in human macrophages, and to examine the role of macrophage polarization in these responses.
Genomic DNA from three different strains of M. leprae DNA (Strains NHDP, Br4923, and Thai-53) were used to stimulate human macrophages under three polarization conditions (M1, M1-activated, and M2).
Transcriptome analysis revealed a large number of differentially expressed (DE) genes upon stimulation with DNA from M. leprae strain Thai-53 compared to strains NHDP and Br4923, independent of the macrophage polarization condition. We also found that macrophage polarization affects the responses to M. leprae DNA, with up-regulation of numerous interferon stimulated genes.
These findings provide a deeper understanding of the role of macrophage polarization in the recognition of M. leprae DNA, with the potential to improve leprosy treatment strategies.
Graphical Abstract
1. Introduction
Mycobacterium leprae is an acid-fast, obligate-intracellular bacterium and the causative organism of leprosy, a disease still endemic in numerous parts of the world (Programme, 2016, Fischer, 2017), including the southwestern United States (Sharma et al., 2015, Program, 2016). It causes various degrees of skin abnormalities, eye damage, respiratory damage, and peripheral neuropathy in humans leading to stigmatization of individuals and communities throughout human history, and still affecting millions of people worldwide to this day (Fisher 2017). The spectrum of disease ranges in severity from the milder tuberculoid leprosy (paucibacillary) to the more severe and disfiguring lepromatous leprosy (multibacillary) (Gaschignard et al., 2013) (Fig. 1).
Fig. 1.
Leprosy spectrum and mechanisms of pathogenesis. From least to most severe: tuberculoid (TT), intermediate borderline tuberculoid (BT), borderline borderline (BB), borderline lepromatous (BL), lepromatous leprosy (LL). Macrophage polarization: M1 showing granulomas and M2 displaying “foamy” macrophages. Boxes depict the cytokine profile in the clinical pole of disease.
The established thinking in the health community is that strain variations of M. leprae are of little consequence to disease symptoms and that differential expression of tuberculoid leprosy and lepromatous leprosy are due only to variations in host response, such as TH1 or TH2 and macrophage polarization (Kibbie et al., 2016, Mi et al., 2020). Differential macrophage polarization in skin lesions of leprosy has been described, with M1 phenotype predominantly present in granulomas of TT patients, whereas macrophages in LL granulomas exhibit an M2 phenotype (Mi et al., 2020, Fachin et al., 2017).
Although the genome of M.leprae has been highly conserved over the past ten centuries (Schuenemann et al., 2013), comparison of variable-number tandem repeats polymorphisms has shown to be useful in effectively discriminating M. leprae strains (Truman et al., 2004). Further genetic differences, which led to the establishment of a new species such as Mycobacterium lepromatosis, are associated with a more severe presentation, known as diffuse lepromatous leprosy (Sharma et al., 2019)(Sharma CID 2020).
Genetic analysis has also shown the geographic restriction of strains in humans (Matsuoka et al., 2006) and primates (Honap et al., 2018). Some evidence exists suggesting that specific M.leprae genotype may be associated with a particular clinical pole. Multibacillary form, LL, is more frequent in places like Saudi Arabia (Alotaibi et al., 2016). Analysis of LL skin samples from India has demonstrated that TTC repeats are abundant in these samples, and differ from other M.leprae strains from India (Chokkakula et al., 2014). Geographical patterning is also observed in the Pacific Islands (Blevins et al., 2020), where multibacillary disease predominates (Woodall et al., 2011). Paucibacillary disease, on the other hand, is predominant in other parts of the world like Brazil (Marciano et al., 2018).
In this study, we aim to analyze if bacterial genomic DNA from different strains of M. leprae (Singh and Cole, 2011, Truman et al., 2011) elicit different responses in human macrophages and the role of macrophage polarizations in these responses.
2. Material and methods
2.1. Cells and cell stimulation conditions
We utilized human monocytic cells (THP-1) under an established protocol for macrophage induction and polarization, creating M1, M1 activated (M1a), and M2 macrophages (Rey–Giraud et al., 2012). 5 × 105 cells/ml in 12-well tissue culture treated plates for 6 days in the presence of either 100 ng/ml rHuGM-CSF (M1) or 100 ng/ml rHuM-CSF (M2). For M1 activation, monocytes were first incubated with rHuM-CSF (Peprotech) for 3 days followed by stimulation with 10 ng/ml LPS (Sigma) and 50 ng/ml rHuIFN-γ (Roche) for 3 additional days (Rey–Giraud et al., 2012). Stimulation was performed with 500 ng of genomic DNA from three different strains of M. leprae, NHDP (NT-19350), Br4923 (NR-19351), and Thai-53 (NR-19352), acquired from BEI Resources (Manassa, VA). We utilized these strains as each one of them belongs to a different genetic subtype (Singh and Cole, 2011, Truman et al., 2011).
Genomic DNA was isolated from contaminating proteins and polysaccharides by organic extraction and precipitation with isopropanol respectively (Belisle and Sonnenberg, 1998). Polyethilenimine (PEI) (Polyplus) was used as an endo-lysosomal bacterial nucleic acid delivery system (Cervantes et al., 2013, Suh et al., 2012, Bieber et al., 2002), according to manufacturer's instructions.
2.2. RNA Seq and Transcriptome analysis
Total RNA was extracted after cell stimulation assays. Unstimulated cells were used as controls. A cDNA library was prepared and sequenced in an Illumina NextSeq 500 v2 High Output 150 cycle sequencing at the Center for Genomic Innovations (UConn, CT) to generate paired end (PE) 75bp reads. Sequence reads were trimmed and filtered based on read quality using Sickle (N et al., 2011) and Trimmomatic (Bolger et al., 2014). Reads were aligned with Hisat2 to the grch38 human reference genome (Pertea et al., 2016). Aligned reads were counted using htseq (Anders et al., 2015). Reproducibility of the sample replicates was checked by Principle Component Analysis (PCA) in R (RC, 2017). Computation of differentially expressed (DE) genes was done using DESeq2 (Love et al., 2014) comparing the stimulated to unstimulated cells. A comparison of the significant genes with an absolute fold change of greater than 2 was done between the different M. leprae strains for the different cell types (M1, M1a, and M2) compared to the unstimulated control, i.e. M1 stimulated with Thai-53, Br4923, or NHDP DNA. The number of DE genes that overlap was displayed in a venn diagram using the VennDiagram (v.1.6.20) package in R (v.3.5.2) Chen (2018). Association networks and GO term analysis were performed using the online STRING program (v.11) (Szklarczyk et al., 2019) for the identified overlapping DE genes.
Additionally, comparisons were also made across the three types of cells (M1 and M2 expression compared to M1a) for each the M. leprae strain Thai-53, NHDP, Br4283, i.e. M2 versus M1a where both were stimulated with Thai-53 DNA. Heatmaps were created using the log-fold change of the genes where there was at least one value had an absolute fold change value > 2 and adjusted p-value < 0.05 in the various comparisons. The values were normalized across the row and displayed within the R package gplots (v3.0.4) using heatmap.2 (Warnes et al. 2020).
Network analyses were performed based on NetworkAnalyst tool (Zhou et al., 2019). The overrepresentation analysis using hypergeometric test was performed to identify significant overlap with gene-sets or pathways. The enrichment analysis was performed against KEGG. Macrophage specific gene co-expression networks were constructed based on significant genes. Highly correlated genes in pairwise gene expression profile were measured and mapped to immuno-navigator database. Genes that were up-regulated together in a similar manner were shown as connected nodes in the network.
3. Theory
Deciphering the process of macrophage polarization to enhance anti‐microbial defense or to dampen detrimental inflammation is of great importance in the pathogenesis of leprosy, as it affects clinical disease (Shapouri-Moghaddam et al., 2018). Macrophage polarization appears to play a significant role in the spectrum of leprosy disease but to what degree it contributes to the clinical presentation is not fully established (Pinheiro et al., 2018, Fallows et al., 2016). We have recently shown that different strains of Mycobacterium tuberculosis elicit differential NF-kB and IRF responses in human macrophages (Cervantes et al., 2019). We aim to first determine if different strains of M. leprae (Singh et al., 2015) elicit different transcription responses in human macrophages and second to evaluate the role of macrophage polarization in the response to M. leprae DNA. Our study will provide a deeper understanding of the role of macrophage polarization and activation in the recognition of M. leprae which could lead to better future treatment strategies.
4. Results
4.1. DNA from different strains of M. leprae elicited different gene expression in human macrophages
To observe if DNA from the different strains would elicit different DE genes in stimulated macrophages, we first filtered out all expressed genes with at least one zero (missing) value sample and then used the fold change of each of NHDP, Br4923, and Thai-53 compared to unstimulated macrophages for each of the expressed genes. We observed each of the different M. leprae strains elicited different gene expression in human macrophages (Fig. 2) (A complete list of genes is shown in Supplemental file 1). Common genes found between M1 stimulated with Thai-53 and Br4923 DNA included: OASL, IFIT2, IFIT1, and CXCL10 (Fig. 2A and Table 1). These four overlapped genes from the M1 stimulated with Thai53 and Br4923 DNA had eight significant GO terms and formed one significant STRING network cluster: 4Fe-4S single cluster domain, and Interferon-induced protein with tetratricopeptide. Interestingly, 311 distinct DE genes were observed in M1 uniquely stimulated by Thai-53 strain. We found 204 significant GO terms for the M1 macrophages stimulated with Thai53 DNA with top terms including “defense response to virus” (GO:0051607), “defense response to other organism” (GO:0098542), and “innate immune response” (GO:0045087). Stimulation of M1 activated (M1a) macrophages with DNA from each of three different strains identified 0, 61, and 1 DE genes for the NHDP, Thai-53, and Br4923 DNA respectively (Fig. 2B). For M1a, Thai53 DNA-stimulated cells had 157 significant GO terms and 13 network clusters. Finally, M2 showed 14, 514, and 2 DE genes for the NHDP, Thai-53, and Br4923 DNA stimulations (Fig. 2C). M2 cells stimulated with Thai-53 and Br4923 DNA both contained IFIT1, while M2-stimulated with Thai-53 and NHDP DNA contained MDGA1, SLAMF7, IL4I1, and C3. Commonly expressed DE genes upon stimulation of different M. leprae strains are summarized in Table 1. Thai53 DNA-stimulated M2 cells showed 410 significant GO terms and 42 network clusters. All GO terms and network cluster lists can be found in Supplemental file 2 with the associated genes for each. In this aim, we challenged macrophage with DNA from different M.leprae strains and observed that there were both commonly and uniquely expressed genes by each M.leprae strains. These genes were identified to belong to various immune cellular pathways.
Fig. 2.
(A) Venn diagram of the number of DE genes in human macrophages across the DNA from strains NHDP (Yellow), Thai-53 (Blue), and Br4923 (Green) used for stimulation. (A) M1, (B) M1activated and (C) M2 macrophages. All DE genes comprise those whose fold change is greater than 2 and adjusted p-value less than 0.05 in each NHDP, Thai-53, and Br4923.
Table 1.
| M1 | M2 | M2 |
| Thai-53 and Br4923 | NHDP and Thai-53 | Thai-53 and Br4923 |
| OASL | MDGA1 | IFIT1 |
| IFIT1 | SLAMF7 | |
| IFIT2 | IL4I1 | |
| CXCL10 | C3 |
4.2. Macrophage polarization in the responses to M. leprae DNA exposure
4.2.1. Unique gene expression profile in M1a compared to M1 and M2 polarization
We then aimed to analyze the role of macrophage polarization in the response to M.leprae DNA. We were particularly interested in the activated M1 phenotype (M1a) that were generated by additional LPS and IFN-γ. DE genes occurring in M1a compared to M1 and M2 were identified for Thai-53, Br4923 and NHDP DNA stimulations. M1a phenotype expressed 4 unique DE genes compared to M1 when M.leprae strain Br4923 was applied (Fig. 3A). Using DNA from different M. leprae strains for stimulation showed different profiles based on macrophage polarization status. When DNA from M.leprae strain NHDP was used, M1a expressed 259 and 234 unique DE compared to M1 and M2 respectively, while expressing 416 common genes (Fig. 3B). Lastly, when M1a cells were stimulated with DNA from M.leprae strain Thai-53, we observed 387 and 240 unique DE compared to M1 and M2 respectively, while expressing 815 common genes (Fig. 3B). (Supplemental file 3 displays the list of these significant DE genes, and supplemental file 4 lists the significant GO terms and network clusters).
Fig. 3.
Venn diagrams comparing the DE genes after M. leprae DNA strain stimulation for compared to M1a against M1 (magenta) and M2 (cyan) (absolute fold change value >2 and an adjusted p-value <0.05). A:M. leprae Br4923, B:M. leprae NHDP and C:M. leprae Thai-53 DNA stimulation.
4.2.2. Gene differential expression on macrophage polarization
From the list of significant genes, we selected a list of 18 genes that were up-regulated in M1a compared to either M1 or M2 upon stimulation with either NHDP or Tha-53 DNA, but down-regulated by stimulation with the other strain (Fig. 4). These included genes SCUBE2, AMOTL2, GCNT4, SELL, PAX5, TMCC3, and HEPH, which were up-regulated in M1a upon Thai-53 DNA stimulation but down regulated with NHDP DNA. It also showed that SEMA3D, CCL4, PGTGER3, and MMP1 were in turn up-regulated with NHDP DNA stimulation but some of these genes (SEMA3D and CCL4) only showed down-regulation in M1a/M1 of Thai-53 DNA stimulated, while others (PTGER3, and MMP1) showed down-regulation in M1a/M2 of Thai-53 DNA stimulated. CCL3 was unique in showing down-regulation on M1a/M2 upon either, NHDP or Thai-53 DNA stimulation (Fig. 4A). When looking at expression switches within a strain treatment, we identified CCL13 as being the sole gene that had opposite DE in M1a compared to M1 vs. M1 compared to M2 (Fig. 4B). CCL13 showed co-regulation with MMP1, PTGER3, CCL4, and TMCC3 (Fig. 4C). GO term analysis of these genes showed three significant pathways: leukocyte migration (GO:0050900), negative regulation of viral transcription (GO:0032897), and cellular response to interferon-gamma (GO:0071346).
Fig. 4.
Heat Map indicating DE genes in polarized macrophages. A. M1a compared to either M1 or M2 macrophages upon stimulation with either NHDP or Tha-53 DNA but downregulated by stimulation with the other strain. B. Heat map showing DE expression switches within a strain treatment. Genes are listed on the right-hand size. Green represents up-regulation and red represents down-regulation. C. Network analysis of genes presented in A.
4.2.3. Network analysis for pathway identification
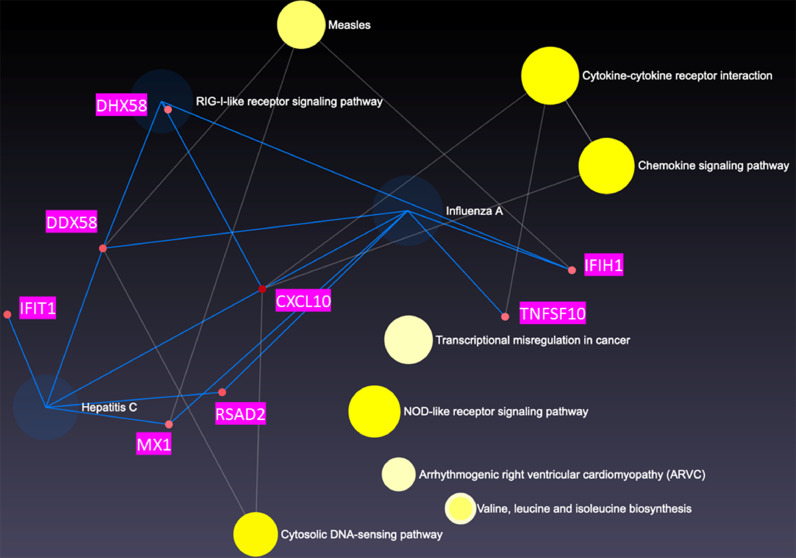
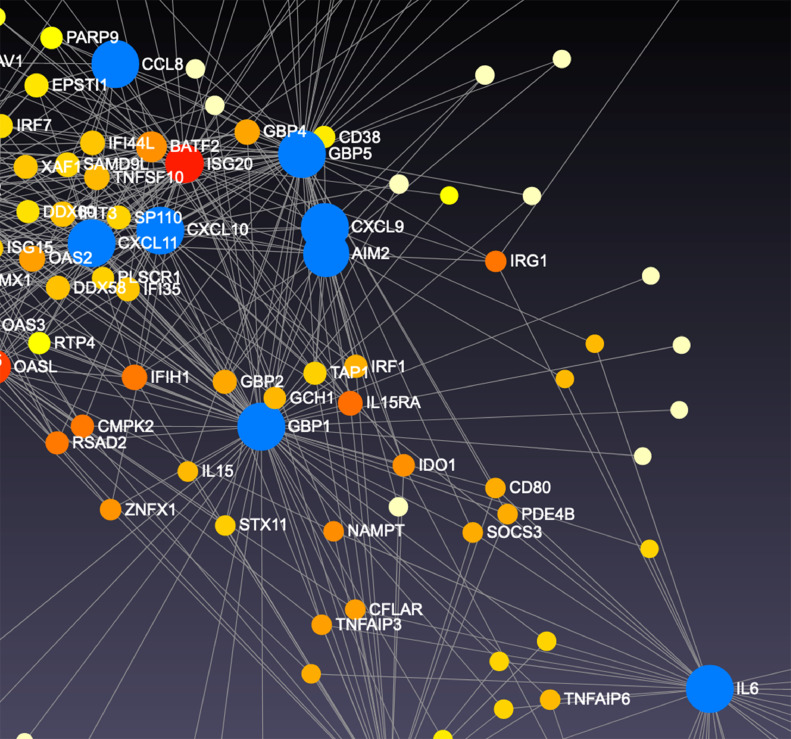
We then constructed a global enrichment network on cells stimulated with Thai-53 strain, given the large number of DE genes observed. We observed that DHX58, DDX58, IFIT1, CXCL10, MX1, RSAD2, IFIH1, and TNFS10 genes co-regulate influenza A (p = 0.0114), RIG-I-like receptor signal (p = 0.0224), and Hepatitis C pathways (p = 0.0305) (Fig. 5A) in M1 cells. When the network was constructed specifically for macrophage gene expression, CCL8, CXCL11, OASL, SP110, CXCL9, AIM2, GBP5, GBP1, and IL-6 were co-regulated in M1 cells (Fig. 5B). These genes were observed to significantly contribute to 5 macrophage gene pathways: toll-like receptor signal (p = 0.0044), cytokine-cytokine receptor signal (p= 0.0077), cytosolic DNA-sensing (p = 00001), NOD-like receptor signaling (p = 0.0092), and chemokine signaling (p = 0.0095). For M1a gene expression, the enrichment network analysis did not show any significant pathways based on our DE genes (Fig. 6A), after multiple comparison adjustments. However, the macrophage specific network analysis indicated 3 significantly distinct pathways: The toll-like receptor signal (p = 0.0134), chemokine signaling (p = 0.0246), and cytokine-cytokine receptor signaling (p = 0.038). CXCL11 along with HERC5 were major hubs controlling the expression of numerous interferon stimulated genes (ISGs) (Fig. 6B), and interferon regulatory factor (IRF)-7 (Fig. 6C). M2 cells showed cytokine-cytokine receptor signal (p = 0.0129) pathway as a distinct profile from enrichment network analysis. M2 macrophages showed numerous co-regulations of ISGs as well, including CXCL9, CXCL10, CXCL11, CCL7, CCL8, IFNB1, IFNL1, CCR4, OSM and CCL13 (Fig. 7). The macrophage specific network analysis didn't lead to any significant pathway based on our DE genes, after multiple comparison adjustment.
Fig. 5.
Network analysis of DE genes presented in M1 macrophages upon stimulation with genomic DNA M.leprae strain Thai-53. A. Highly correlated genes in pairwise gene expression. B Genes that were upregulated together in a similar manner shown as connected nodes in the network. C Detail of B.
Fig. 6.
Network analysis of DE genes presented in M1 activated macrophages upon stimulation with genomic DNA M.leprae strain Thai-53. A. Highly correlated genes in pairwise gene expression. B. Genes that were upregulated together in a similar manner shown as connected nodes in the network. C. Detail of B.
Fig. 7.
Network analysis of DE genes presented in M2 macrophages upon stimulation with genomic DNA M.leprae strain Thai-53. A. Highly correlated genes in pairwise gene expression. B Genes that were upregulated together in a similar manner shown as connected.
5. Discussion
When investigating an infectious disease the environment, host, and pathogen all play a role in disease progression. Variation in M. leprae genome has been shown in findings from current molecular epidemiology studies in China (Xing et al., 2009, Weng et al., 2011) and Brazil (Fontes et al., 2017), as well as in a study comparing strains and phylogeny from ancient and modern M. leprae strains (Schuenemann et al., 2018). Genetic analysis has shown that Thai-53 strain displays remarkable genetic variability (Truman et al., 2004). Despite the genetic variability within this pathogen (Singh et al., 2015, Benjak et al., 2018), the current consensus is that symptoms and disease course are dictated by host response (Fischer, 2017), with a focus on T-helper lymphocytes as the primary host mediators (de Sousa et al., 2017). However, recent research is now indicating that macrophage polarization also plays a central role in the clinical presentation of leprosy (Fachin et al., 2017, Pinheiro et al., 2018, Fallows et al., 2016). We herein showed that genomic DNA from various strains of M. leprae can induce different transcriptomes in human macrophages and that these responses are affected by macrophage polarization.
We utilized genomic DNA from three strains as each one of them belongs to a different genetic subtype (Singh and Cole, 2011, Truman et al., 2011).
We observed a large number of DE genes upon stimulation with DNA from M. leprae strain Thai-53 compared to strains NHDP and Br4923, independent of the macrophage polarization condition. A few genes were common in M1 macrophages after stimulation with DNA from different strains. These included OASL, which inhibits antimicrobial peptides expression and bacterial killing preventing M. leprae clearance (de Toledo-Pinto et al., 2016), IFIT1, and CXCL10). All these three genes have previously shown to be associated with type I IFN–activated pathway in THP1 derived macrophages (Zhang et al., 2019). Type I IFNs are associated with disseminated and progressive lepromatous lesions (Teles et al., 2013, P et al., 2019). Shared genes observed in M2 macrophages included MDGA1, SLAMF7, IL4I1, and C3. MDGA1 has been reported in integrated stress response (Kovaleva et al., 2016). SLAMF7 is observed in all forms of leprosy (Belone Ade et al., 2015) and in M1 macrophages as well (Schulz et al., 2019). IL4I1 has been seen up-regulated in DCs after NOD2 stimulation with correlation with leprosy patients with limited disease (Schenk et al., 2012). C3 plays a role in opsonization of M. leprae (Schorey et al., 1997).
We also found that macrophage polarization affects the responses to M. leprae DNA. Genes DE in M1 macrophages upon stimulation with DNA from strain Thai-53 included pro-inflammatory receptor TARM1 (Radjabova et al., 2015), M2-associated SELL (Mould et al., 2019), and IFN-γ and IFN-β associated TMCC3 (Zhang et al., 2010). On the other hand, stimulation of NHDP DNA on M1 macrophages induced CCL4, an innate immunity cytokine associated with leprosy occurring in households (van Hooij et al., 2020), M2-associated MMP-1 (Jager et al., 2016), and PGE2 receptor PTGER3, which is associated with immune evasion of mycobacteria (Behar et al., 2010). CCL13 was clearly up-regulated in M1 activated cells compared to M1 regardless of the strain, as well in M2 macrophages upon Thai-53 DNA stimulation. This cytokine is involved in many chronic inflammatory diseases (Mendez-Enriquez and Garcia-Zepeda, 2013). No common genes were found upon stimulation with Br4923 DNA.
As stimulation with DNA from strain Thai-53 yielded a large amount of DE genes, we were able to construct a comprehensive network analysis of co-regulated genes in each of the macrophage polarization conditions. We observed that in M1 and M1a cells there were numerous up-regulated ISGs. In M1a specifically, CXCL-11 and IFNB1 were up-regulated, underscoring Type I IFN pathway activation. CXCL10 and CXCL11 are induced by IFN-γ and by IFN-β, whereas CXCL9 induction is restricted to IFN-γ Groom and Luster (2011). The inflammatory chemokines CXCL9, CXCL10, and CXCL11 share an exclusive chemokine receptor named CXC chemokine receptor 3 (CXCR3). The CXCL11-CXCR3 axis is crucial for macrophage resistance to mycobacterial infection (Torraca et al., 2015). Increased CXCL10 without correlation with IFN-γ, is characteristic of Type 1 reaction, a systemic inflammatory syndrome seen in BL (Scollard et al., 2011). IFNL1 (IL-29), reported to be greatly down-regulated in LL and BL forms of leprosy, was also an important node in M1a (Berrington et al., 2014). CXCL11 along with HERC5 were major hubs controlling the expression of numerous (ISGs). HERC5 is up-regulated by M. leprae-induced Type I IFN signature, and is a major regulator of this pathway (P et al., 2019).
IRF-7 was revealed from the network in M1a cells, which is in line with knowledge of THP-1 derived macrophages utilizing this transcription factor after recognition of Mycobacterium tuberculosis (Mtb), and hypothesized to help patients resist Mtb infection (Zhang et al., 2019). TLR-9 has been shown to sense M. leprae DNA (Dias et al., 2016), and may be using IRF-7 as a transcription factor for Type I IFN induction.
In summary, despite the accepted thinking that genomic variation of M. leprae may not substantially contribute to the clinical manifestations of the disease, we have shown here that DNA from different M. leprae strains are able to induce differential gene expression in human macrophages. M1 activated macrophages presented a marked differential expression of genes involved in Type I IFN regulation, macrophage activation, pathogen DNA recognition, and recruitment of effector cells to site of inflammation upon stimulation with M. leprae genomic DNA.
6. Conclusions
While the current belief in the science and health community indicate that only host response effects gene expression and clinical exhibitions of leprosy, our data suggests M. leprae strain genomic variations may be able to induce differential gene expression in human macrophages. Activated M1 polarized macrophages presented a marked differential expression of genes involved in Type I IFN regulation, macrophage activation, pathogen recognition, pathogen DNA recognition, and recruitment of effector cells to site of inflammation upon stimulation with M. leprae genomic DNA.
Author contributions
JC conceptualized the study. AM, KVH, and JC performed the experiments. BYH, JM and SK performed the bioinformatics analysis. JC, BYH, AM, KVH, and JM prepared the manuscript.
Declaration of Competing Interest
None.
Acknowledgments
Acknowledgments
None.
Funding
This work was supported in part by Grant 5U54MD007592 from the National Institute on Minority Health and Health Disparities (NIMHD), a component of the National Institutes of Health (NIH).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2020.100015.
Appendix. Supplementary materials
References
- Alotaibi M.H., et al. The demographic and clinical characteristics of leprosy in Saudi Arabia. J. Infect. Public Health. 2016;9(5):611–617. doi: 10.1016/j.jiph.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar S.M., Divangahi M., Remold H.G. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 2010;8(9):668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle J.T., Sonnenberg M.G. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 1998;101:31–44. doi: 10.1385/0-89603-471-2:31. [DOI] [PubMed] [Google Scholar]
- Belone Ade F., et al. Genome-wide screening of mRNA expression in leprosy patients. Front Genet. 2015;6:334. doi: 10.3389/fgene.2015.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjak A., et al. Phylogenomics and antimicrobial resistance of the leprosy bacillus mycobacterium leprae. Nat. Commun. 2018;9(1):352. doi: 10.1038/s41467-017-02576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrington W.R., et al. Differential dermal expression of CCL17 and CCL18 in tuberculoid and lepromatous leprosy. PLoS Negl Trop. Dis. 2014;8(11):e3263. doi: 10.1371/journal.pntd.0003263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber T., et al. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. J. Control Release. 2002;82(2-3):441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- Blevins K.E., et al. Evolutionary history of mycobacterium leprae in the pacific islands. Philos. Trans. R Soc. Lond B Biol. Sci. 2020;375(1812) doi: 10.1098/rstb.2019.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes J.L., et al. Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes. J. Leukoc Biol. 2013;94(6):1231–1241. doi: 10.1189/jlb.0413206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes J.L., et al. Vitamin D modulates human macrophage response to mycobacterium tuberculosis DNA. Tuberculosis (Edinb) 2019 doi: 10.1016/j.tube.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. VennDiagram: Generate High-Resolution Venn and Euler Plots. R package version 1.6.20. 2018; Available from: https://CRAN.R-project.org/package=VennDiagram.
- Chokkakula S., et al. Strain tryping and strain differentiation of mycobacterium leprae by TTC repeats. Int. J. Pharmacol. 2014;10(3):168–174. [Google Scholar]
- de Sousa J.R., Sotto M.N., Simoes Quaresma J.A. Leprosy as a complex infection: breakdown of the Th1 and Th2 immune paradigm in the immunopathogenesis of the disease. Front Immunol. 2017;8:1635. doi: 10.3389/fimmu.2017.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Toledo-Pinto T.G., et al. STING-Dependent 2′-5′ oligoadenylate synthetase-like production is required for intracellular mycobacterium leprae survival. J. Infect. Dis. 2016;214(2):311–320. doi: 10.1093/infdis/jiw144. [DOI] [PubMed] [Google Scholar]
- Dias A.A., et al. DNA sensing via TLR-9 constitutes a major innate immunity pathway activated during erythema nodosum leprosum. J. Immunol. 2016;197(5):1905–1913. doi: 10.4049/jimmunol.1600042. [DOI] [PubMed] [Google Scholar]
- Fachin L.R., et al. Immunohistochemical assessment of cell populations in leprosy-spectrum lesions and reactional forms. Histol. Histopathol. 2017;32(4):385–396. doi: 10.14670/HH-11-804. [DOI] [PubMed] [Google Scholar]
- Fallows D., et al. Mycobacterium leprae alters classical activation of human monocytes in vitro. J. Inflamm. (Lond) 2016;13:8. doi: 10.1186/s12950-016-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. Leprosy - an overview of clinical features, diagnosis, and treatment. J. Dtsch Dermatol. Ges. 2017;15(8):801–827. doi: 10.1111/ddg.13301. [DOI] [PubMed] [Google Scholar]
- Fontes A.N.B., et al. Genotyping of mycobacterium leprae for better understanding of leprosy transmission in Fortaleza, Northeastern Brazil. PLoS Negl Trop. Dis. 2017;11(12) doi: 10.1371/journal.pntd.0006117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschignard J., Scurr E., Alcais A. [Leprosy, a pillar of human genetics of infectious diseases] Pathol. Biol. (Paris) 2013;61(3):120–128. doi: 10.1016/j.patbio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Groom J.R., Luster A.D. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 2011;89(2):207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honap T.P., et al. Mycobacterium leprae genomes from naturally infected nonhuman primates. PLoS Negl Trop. Dis. 2018;12(1) doi: 10.1371/journal.pntd.0006190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager N.A., et al. Distribution of matrix metalloproteinases in human atherosclerotic carotid plaques and their production by smooth muscle cells and macrophage subsets. Mol. Imaging Biol. 2016;18(2):283–291. doi: 10.1007/s11307-015-0882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbie J., et al. Jagged1 instructs macrophage differentiation in leprosy. PLoS Pathog. 2016;12(8) doi: 10.1371/journal.ppat.1005808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva I.E., et al. Intermedin/adrenomedullin 2 is a stress-inducible gene controlled by activating transcription factor 4. Gene. 2016;590(1):177–185. doi: 10.1016/j.gene.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano L., et al. Epidemiological and geographical characterization of leprosy in a Brazilian hyperendemic municipality. Cad Saude Publica. 2018;34(8) doi: 10.1590/0102-311X00197216. [DOI] [PubMed] [Google Scholar]
- Matsuoka M., et al. Genotypic analysis of mycobacterium leprae isolates from Japan and other Asian countries reveals a global transmission pattern of leprosy. FEMS Microbiol. Lett. 2006;261(1):150–154. doi: 10.1111/j.1574-6968.2006.00346.x. [DOI] [PubMed] [Google Scholar]
- Mendez-Enriquez E., Garcia-Zepeda E.A. The multiple faces of CCL13 in immunity and inflammation. Inflammopharmacology. 2013;21(6):397–406. doi: 10.1007/s10787-013-0177-5. [DOI] [PubMed] [Google Scholar]
- Mi Z., Liu H., Zhang F. Advances in the immunology and genetics of leprosy. Front. Immunol. 2020;11:567. doi: 10.3389/fimmu.2020.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould K.J., et al. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight. 2019;4(5) doi: 10.1172/jci.insight.126556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N J., F. J, Sickle . 2011. A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ files. [Google Scholar]
- P R.A., et al. The cell fate regulator NUPR1 is induced by mycobacterium leprae via type I interferon in human leprosy. PLoS Negl Trop Dis. 2019;13(7) doi: 10.1371/journal.pntd.0007589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro R.O., et al. Innate immune responses in leprosy. Front. Immunol. 2018;9:518. doi: 10.3389/fimmu.2018.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Program N.H.s.D. Department of Health and Human Services; 2016. A Summary of Hansen's Disease in the United States-2015. [Google Scholar]
- Programme W.-G.L. 2016. Global Leprosy Strategy 2016-2020: Accelerating towards a leprosy-free world. [Google Scholar]
- Radjabova V., et al. TARM1 is a novel leukocyte receptor complex-encoded ITAM receptor that costimulates proinflammatory cytokine secretion by macrophages and neutrophils. J. Immunol. 2015;195(7):3149–3159. doi: 10.4049/jimmunol.1401847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RC, T., R: A language environment for statistical computing., R.F.f.S. Computing, Editor. 2017: Vienna, Austria.
- Rey-Giraud F., Hafner M., Ries C.H. In vitro generation of monocyte-derived macrophages under serum-free conditions improves their tumor promoting functions. PLoS One. 2012;7(8):e42656. doi: 10.1371/journal.pone.0042656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk M., et al. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat. Med. 2012;18(4):555–563. doi: 10.1038/nm.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey J.S., Carroll M.C., Brown E.J. A macrophage invasion mechanism of pathogenic mycobacteria. Science. 1997;277(5329):1091–1093. doi: 10.1126/science.277.5329.1091. [DOI] [PubMed] [Google Scholar]
- Schuenemann V.J., et al. Genome-wide comparison of medieval and modern mycobacterium leprae. Science. 2013;341(6142):179–183. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- Schuenemann V.J., et al. Ancient genomes reveal a high diversity of Mycobacterium leprae in medieval Europe. PLoS Pathog. 2018;14(5) doi: 10.1371/journal.ppat.1006997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D., et al. In-depth characterization of monocyte-derived macrophages using a mass cytometry-based phagocytosis assay. Sci. Rep. 2019;9(1):1925. doi: 10.1038/s41598-018-38127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scollard D.M., et al. Increased CXC ligand 10 levels and gene expression in type 1 leprosy reactions. Clin. Vaccine Immunol. 2011;18(6):947–953. doi: 10.1128/CVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A., et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018;233(9):6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- Sharma R., et al. Zoonotic leprosy in the Southeastern United States. Emerg. Infect. Dis. 2015;21(12):2127–2134. doi: 10.3201/eid2112.150501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., et al. Isolation of mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish M. leprae and M. lepromatosis. Clin. Infect. Dis. 2019 doi: 10.1093/cid/ciz1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., et al. Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis. Proc. Natl. Acad. Sci. USA. 2015;112(14):4459–4464. doi: 10.1073/pnas.1421504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Cole S.T. Mycobacterium leprae: genes, pseudogenes and genetic diversity. Future Microbiol. 2011;6(1):57–71. doi: 10.2217/fmb.10.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J., et al. Real-time gene delivery vector tracking in the endo-lysosomal pathway of live cells. Microsc. Res. Tech. 2012;75(5):691–697. doi: 10.1002/jemt.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles R.M., et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339(6126):1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraca V., et al. The CXCR3-CXCL11 signaling axis mediates macrophage recruitment and dissemination of mycobacterial infection. Dis. Model Mech. 2015;8(3):253–269. doi: 10.1242/dmm.017756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman R., et al. Genotypic variation and stability of four variable-number tandem repeats and their suitability for discriminating strains of Mycobacterium leprae. J. Clin. Microbiol. 2004;42(6):2558–2565. doi: 10.1128/JCM.42.6.2558-2565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman R.W., et al. Probable zoonotic leprosy in the southern United States. N. Eng. J. Med. 2011;364(17):1626–1633. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooij A., et al. Household contacts of leprosy patients in endemic areas display a specific innate immunity profile. Front. Immunol. 2020;11:1811. doi: 10.3389/fimmu.2020.01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes, G.R., et al. gplots: various R Programming Tools for Plotting Data. R package. 2020; Available from: https://CRAN.R-project.org/package=gplots.
- Weng X., et al. Transmission of leprosy in Qiubei County, Yunnan, China: insights from an 8-year molecular epidemiology investigation. Infect. Genet. Evol. 2011;11(2):363–374. doi: 10.1016/j.meegid.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodall P., Scollard D., Rajan L. Hansen disease among Micronesian and Marshallese persons living in the United States. Emerg. Infect. Dis. 2011;17(7):1202–1208. doi: 10.3201/eid1707.102036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., et al. VNTR typing studies of mycobacterium leprae in China: assessment of methods and stability of markers during treatment. Lepr Rev. 2009;80(3):261–271. [PubMed] [Google Scholar]
- Zhang S., et al. Delineation of diverse macrophage activation programs in response to intracellular parasites and cytokines. PLoS Negl Trop Dis. 2010;4(3):e648. doi: 10.1371/journal.pntd.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.W., et al. Characteristic genes in THP1 derived macrophages infected with mycobacterium tuberculosis H37Rv strain identified by integrating bioinformatics methods. Int. J. Mol. Med. 2019;44(4):1243–1254. doi: 10.3892/ijmm.2019.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.