Highlights
-
•
This study is the first ever report on the virus diversity among the hot springs of Sikkim, India.
-
•
The study revealed the dominance of Siphoviridae, Myoviridae and Phycodnaviridae in the two hot spring solfataric mud sediments.
-
•
The metavirome ecology of the two hot springs have dsDNA viromes in abundance.
-
•
Giant DNA viruses such as Pandoravirus and Pithovirus were found through metagenomic approach.
Keywords: Virus, Hot springs, Metavirome, Solfataric mud sediments, Sikkim himalayas, ICTV
Abstract
Viruses are the most prodigious repertory of the genetic material on the earth. They are elusive, breakneck, evolutionary life particles that constitute a riveting concealed world. Environmental viruses have been obscurely explored, and hence, such an intriguing world of viruses was studied in the Himalayan Geothermal Belt of Indian peninsula at Sikkim corridor through hot springs. The hot springs located at the North Sikkim district were selected for the current study. The solfataric mud sediment samples were pooled from both the hot springs. The virus community showed significant diversity among the two hot springs of Yume Samdung. Reads for viruses among the mud sediments at Old Yume Samdung hot springs (OYS) was observed to be 11% and in the case of New Yume Samdung hot springs (NYS) it was 6%. Both the hot springs were abundant in dsDNA viromes. The metavirome reads in both the OYS and NYS hot spring mud sediments showed the predominance of Caudovirales; Herpesvirales; Ortervirales among which viral reads from Siphoviridae, Myoviridae, Phycodnaviridae and Podoviridae were abundantly present. Other viral communities belonged to families like Baculoviridae, Mimiviridae, Parvoviridae, Marseilleviridae etc. Interestingly, in the case of NYS, the unassigned group reads belonged to some unclassified giant DNA viruses like genera Pandoravirus and Pithovirus. Other interesting findings were – reads for Badnavirus having ds (RT-DNA) was exclusively found in NYS whereas Rubulavirus having ss(-)RNA was exclusively found in OYS sample. This is the first ever report on viruses from any hot springs of Sikkim till date.
Graphical abstract
Introduction
Viruses are the most magnanimous repository of the genetic components on the earth and interestingly less than 0.1% of the environmental viruses have been unearthed (Zhang et al., 2018; Bin Jang et al., 2019; Roux et al., 2019). The focus on the virulent or pathogenic viruses or virus like particles have been significantly pursued by the researchers focused on their impact on human health as prima facie. The temporal phylogeny and neutral biodiversity models revealed that the environmental viral diversity is due to the higher immigration rates of globally distributive metaviromes rather than that of due to the spontaneous mutation (Zablocki et al., 2018). This implies that the viruses can selflessly get exchanged even in geographically distant locations via aerosols or droplets as proven by metacommunity sequencing projects (Bolduc et al., 2015). This unique and interesting phenomena has some serious fundamental insinuations to apprehend, as this viral dissemination or viral airing in the various ecosystem will help us to decode the role played by viruses in evolution of life and understand their genetic mobility or its purpose.
The bacteriophages (viruses infecting bacteria) or archeophages (viruses infecting archea) are ubiquitously present in the earth's biosphere (Rohwer, 2003; Kimura et al., 2008; Garrett et al., 2010; He et al., 2017; Sharma et al., 2018; Wang et al., 2018) and plays an undermining role which is yet to be completely understood. The geothermal habitats have been least explored for viruses in the Himalayan Geothermal Belt (HGB). Only few cases of archeal viruses, thermophages (thermophilic viruses), and eukaryotic viruses have been studied worldwide (Breitbart et al., 2004; Schoenfeld et al., 2008; Peng et al., 2012; Prangishvili, 2013; Dellas et al., 2014; Prangishvili et al., 2017). Very few thermophages from the various hot springs were reported till date with their genome sequences (Zablocki et al., 2018). Recently many researchers have explored their diversity from various environmental niches and are trying to decipher their ecological roles (Zhang et al., 2018; Bin Jang et al., 2019; Roux et al., 2019; Simmonds et al., 2017; Munson-McGee et al., 2018; Munson-McGee et al., 2018; Koonin and Dolja, 2018; Krupovic et al., 2018).
The Sikkim Himalayas are located at the Himalayan Geothermal Belt (HGB) and hence many thermal springs are found here. The hot springs of Sikkim (Tatopani or Tsha chu) are considered as social elixir (Das et al., 2012) and hosts many thermophilic and thermo-tolerant microbes and viruses in its ecosystem comprising of the thermal mineral water and solfataric mud. The deep solfataric mud sediments encompassing the thermal hot spring, hosts numerous enriched microbial housing multi-complexes, build through the microbial network signaling mechanisms which develops an intriguing relationship among each other. The microbial assemblages create their own biofilm suburbs coating the rocks, or any substratum available within their reach (Anderson et al., 2014). These dwelling viruses or virus-like-particles through their signaling modes, surrogate regular exchange of micronutrients, food, genes and energy within their niche through “host-cellular infection machinery” as proved through the bacterial whole genome sequence analysis (Munson-McGee et al., 2018; Beiko et al., 2005; Snyder et al., 2007; Ray et al., 2012). They can stay viable beneath the deep subsurface and move along the thermal gradient cutting across the soil depths. This elaborative metabolic adaptability has motivated the researchers to study these extreme hydrothermal niches where the fluid flux and ecological slants bares the microbial settlements to various biogeochemical constituents governed through abiotic factors like extreme temperature, pH, salinity and pressure. For example, it has been reported that for every centimeter scale depth in hydrothermal vents there is possibility of extensive variations in the various environmental parameters of the fluid fluxes (Anderson et al., 2014; Anderson et al., 2013).
Till date only bacteriological studies have been done in the hot springs of Sikkim where phylum Firmicutes is in majority. Geobacillus is the predominant culturable genus along with few representatives of Anoxybacillus and Bacillus (Najar et al., 2018). A novel culturable bacterium Geobacillus yumthangensis was discovered from the Yumthang hot spring (Najar et al., 2018). The predominant 16S rRNA metagenomes were clustered from various bacterial genus like Acinetobacter, Flavobacterium, Ignavibacterium, Sediminibacterium, Thermodesulfovibrio, Rhodococcus, Dietzia and Arthobacter respectively (Najar et al., 2020). So, it would be interesting to observe whether these bacteria acts as hosts or there are other bacterial hosts apart from these metagenomes.
However, no studies on the viral diversity have been reported till date on the hot springs of Sikkim Himalayas. Here, a comprehensive first ever metagenomic data on the virosphere at hot springs of Sikkim is being presented.
Materials and methods
Sampling site
The geographical position of the coordinates and the elevation (above mean sea level) of the hot springs were measured with the help of GPSMAP 78S (Garmin, India).
Sample collection
The in-situ hot spring conditions were recorded by HORIBA, Japan – an automated portable field instrument for analyzing water parameters. Sulphite, fluoride and chloride contents of the hot spring was tested using HiMedia AQUACheck Testing Kits, India (WT012-1NO; WT004-1NO and WT005-1NO) respectively in situ. The solfataric mud sediments (1000 gm) were aseptically collected from the hot springs of OYS and NYS in sterile sample containers (Wang et al., 2014). The mud samples were then carefully transported (temperature being maintained at 4 °C) to the laboratory for DNA extraction analysis.
DNA extraction and quality check
Total environmental DNA was extracted from the two soil samples by DNA extraction method using Illumina TruSeqⓇ DNA Library Prep Kits (Wang et al., 2014). The quality of eDNA of both the samples was analyzed using Agilent 2100 Bioanalyzer and the quantity was checked by using Nanodrop.
DNA sequencing and quality control
NYS and OYS eDNA were subjected to standard paired end DNASeq library preparation using Illumina recommended protocol. Samples were sequenced using 101 bp paired end module sequencing. The DNA library was checked for quality control using NGSQC tool kit (Patel and Jain, 2012) to get the high quality (HQ) filtered reads in fastq format from the two samples. Stringent criteria of 80:30 was used to obtain high quality filtered reads wherein more than 80% HQ bases each having phred scores >=30 were considered for building genome assembly.
Denovo metagenome assembly
HQ filtered paired-end libraries of the samples of NYS and OYS were assembled separately using metaSPADES assembler with default parameters (Nurk et al., 2017). The metagenomic data obtained from MetaSPAdes was run through MaxBin 2.0 (Wu et al., 2016) using default parameters.
Taxonomical classification and abundance
The putative microbial population reads from both the samples were classified using Kraken2 against the reference database containing all Ref-Seq bacterial, archaeal, fungi and virus genomes with 0.1 confidence threshold (Wood et al., 2019). After classification by Kraken2, Bracken was used to re-estimate the different microbial abundances at taxa levels from species to phylum using a read length parameter of 150 (Lu et al., 2017).
Gene prediction and functional gene annotation
The gene prediction for protein coding genes (ORFs) was predicted using Prodigal (Hyatt et al., 2010). ORFs shorter than 180 nucleotides were excluded from further analysis by default. tRNA genes were predicted with the ARAGORN program (Laslett and Canback, 2004) and Ribosomal RNA genes (5S, 5.8S, 16S, 18S, 23S, 28S) were identified and classified using rRNAFinder (Wheeler and Eddy, 2013). All ORFs were searched against KEGG Orthology (KO) terms (Kim et al., 2016) and Gene Ontology (GO) terms to predict the gene functional classification (Tatusov et al., 2003). ORFs without any search outcomes were annotated as “hypothetical protein”.
Statistical analysis
Diversity indices, rarefaction curve and network analysis was calculated using PAST software (Najar et al., 2020). At taxonomic level species, alpha diversity was calculated using Shannon Diversity and significance was tested using T-test / ANOVA Statistical method. The beta diversity was calculated using PCoA ordination method and Bray-Curtis index distance method. Significance was tested using permutational MANOVA (PERMANOVA) statistical method.
Data availability and accession
Raw metagenomic reads were submitted to Sequence Read Archive (SRA), NCBI to obtain the Bio-Sample and Sequence Read Archive (SRA) accession numbers. The OYS data, Bio-Project accession obtained is PRJNA485728; Bio-Sample accession is SAMN09813281 and SRA is SRP158027 for the sample name OYSMUD4. The NYS data Bio-Project accession obtained is PRJNA485701; Bio-Sample accession is SAMN09813022 and SRA is SRP158029 for the sample name NYSMUD4.
Results
Study area
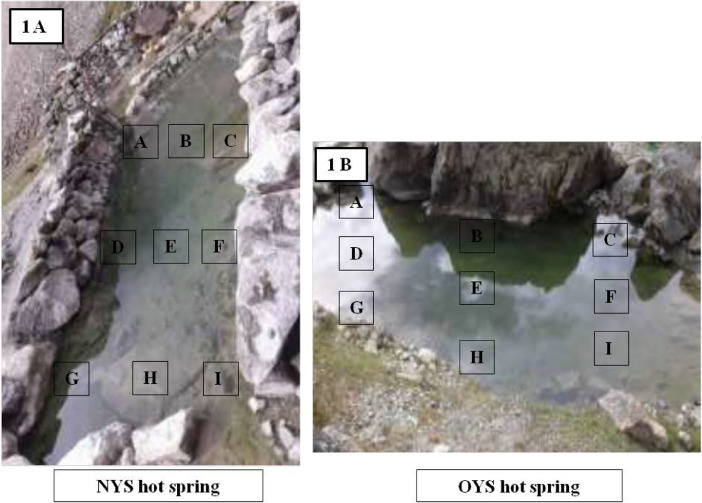
The hot springs located at the North Sikkim district were selected for the current study. NYS (New Yume Samdung) and OYS (Old Yume Samdung) hot springs are situated at the Yume Samdung valley (Fig. 1). In local dialect, hot springs are referred to as the “Tsha Chuu/Tatopani” (Das et al., 2012). Both the OYS and NYS hot springs are closely located in the same geographical vicinity with the latter being near the river Lachung chu. The temperature of the OYS hot spring during sample collection was 61 °C with alkaline conditions and the NYS was slightly cooler than it being 57 °C with near about same pH (Tables 1 and 2). The solfataric mud sediment samples were pooled from nine grid sections encompassing the whole hot spring (Fig. 1). The soil samples were preserved in-situ by storing in the thermally insulated sampling box packed with dry ice and ice gel bags.
Fig. 1.
Sampling site and study area.
Sampling procedure: Fig. 1A is the New Yume Samdung Tsha/Tatopani (NYS) and fig. 1B is that of the Old Yume Samdung Tsha Chu/Tatopani (OYS) located at the Yume Samdung Valley of North Sikkim district, Sikkim which falls under the Himalayan Geothermal belt (HGB). The solfataric mud sediments were collected from these two hot springs and studied. A, B, C, D, E, F, G, H and I being the individual sampling sections of a sampling site from where the soil samples were collected aseptically. From each site, approximately 100 gm of soil sediments was excavated and kept at the sampling container. Thus, finally a good representation of the complete hot spring site was obtained as the soil samples were pooled from the nine grid sections. Thus, for a single hot spring site we had 900 gm samples and the remaining 100 gm was obtained from the outlet of the hot spring making the final volume to 1000 gm.
Table 1.
Sampling site conditions during the sample collection.
| Hot Springs | Latitude | Longitude | Elevation | Temperature | pH | Conductivity (in mScm−1) | %DO | TDS |
|---|---|---|---|---|---|---|---|---|
| NYS | 27°55′085’’N | 88°41′595’’E | 4665m | 57 °C | 9.1 | 0.395 | 30.9 | 0.289 |
| OYS | 27°55′091’’N | 88°41′586’’E | 4668m | 61 °C | 8.77 | 0.25 | 48.6 | 0.158 |
*as measured on site through HORIBA, Japan.
Table 2.
On-site physicochemical analysis of the hot springs.
| Hot Springs | Fluoride (as F in gL−1) | Chloride (as Cl in gL−1) | Sulphite (as Na2SO3 in gL−1) |
|---|---|---|---|
| NYS | 0.1 | 0.5 | 0.7 |
| OYS | 0.2 | 1.0 | 0.8 |
*as measured on site through HiMedia Aqua Check Test Kit, India (colorometric test).
Microbiome data analysis
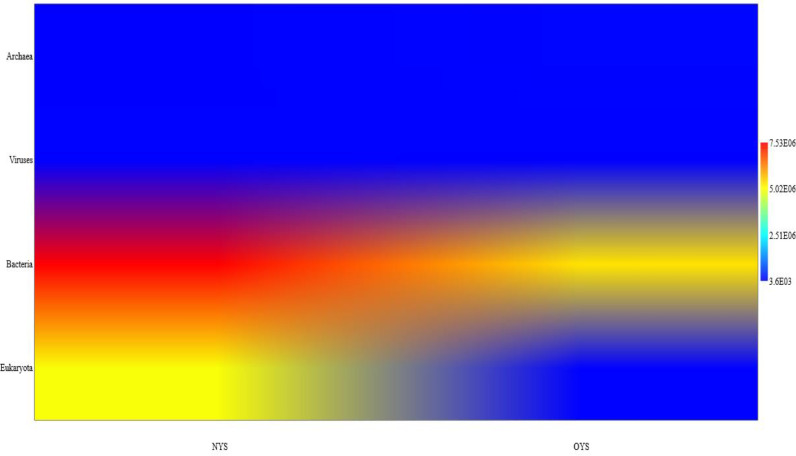
A total 49,570,898 (49.57 Million) paired end reads for NYS sample and 56,681,402 (56.68 Million) paired end for OYS sample was generated using the Illumina sequencing method. Out of 49,570,898 generated sequencing reads for NYS sample, 34,594,204 (34.59 Million) HQ reads with phred score >=30 was obtained after quality filtering. Similarly out of 56,681,402 generated sequencing reads for OYS, 40,319,936 (40.32 Million) HQ reads with phred score >=30 was obtained after quality filtering. These HQ reads were processed further for taxonomy classification and abundance analysis by Kracken2 and Bracken for understanding the microbiome data. The NYS and OYS hot spring solfataric mud sediments, had all the three domain of life and viruses in it. In the NYS hot spring, Archaea had 3789 reads (∼0.03%), Eukarya had 4,956,671 reads (∼39.8%), Bacteria had 7,528,447 reads (∼60.2%) and virus had 16,958 reads (∼0.17%) whereas from the OYS hot spring mud sediments, Archaea had 56,944 reads (∼1.06%), Eukarya had 24,508 reads (∼0.5%), Bacteria had 5,296,073 reads (∼98.42%) and virus had 3596 reads (∼0.07%) respectively. Thus, the hot spring microbiome had more bacteria and eukaryotic reads with less percentage of viruses (Fig. 2). Due to this limited detection of viromes in the thermal ecosystem caused the research impetus to further explore the viral diversity and their functional proteins present in the hot spring mud sediments of Sikkim.
Fig. 2.
The matrix plot here shows that in NYS sample there is higher amount of reads for bacterial species followed by the eukaryotic species as compared to that obtained for OYS microbiome reads. Blue color indicates lower percentage of abundance and increase in abundance is shown through red color.
Diversity indices and rarefaction curve
Alpha diversity indices such as Simpson Index, Shannon H, Fisher alpha and Chao 1 values and beta diversity (Table 3) showed great diversity among the viromes present in both the hot springs of Sikkim. The rarefaction curve for the metavirome showed that there is species richness in OYS mud sediments as compared to that of the NYS hot spring mud sediments (Supplementary Figure 1). Through alpha diversity it was found that there is also higher intra-species variation among the viral species in the OYS hot spring mud sediments (Supplementary Figure 2). Beta diversity among both the hot spring samples measured through Whittaker Index depicts that 70% metavirome reads were unique to each habitat i.e., they are found exclusively in the respective NYS or OYS hot springs only and 30% were commonly found in both the hot springs. Thus, we got more virus reads exclusively present either only in the NYS or OYS hot spring mud sediments (Figs. 3,4)
Table 3.
Diversity indices of the metaviromes in the OYS and NYS samples.
| Diversity indices | NYS | OYS |
|---|---|---|
| Viral reads | 1490 | 2329 |
| Simpson index (lci, hci)* | 0.9085 (0.89, 0.91) | 0.98 (0.98, 0.98) |
| Shannon H index (lci, hci)* | 3.411 (3.26, 3.44) | 5.13 (5.01, 5.12) |
| Fisher alpha index | 57.79 | 155 |
| Chao-1 value | 635.6 | 765.7 |
| Whittaker Index (Beta diversity) | 0.86965 | |
*where lci and hci indicates the lower CI and higher CI at 95% confidence.
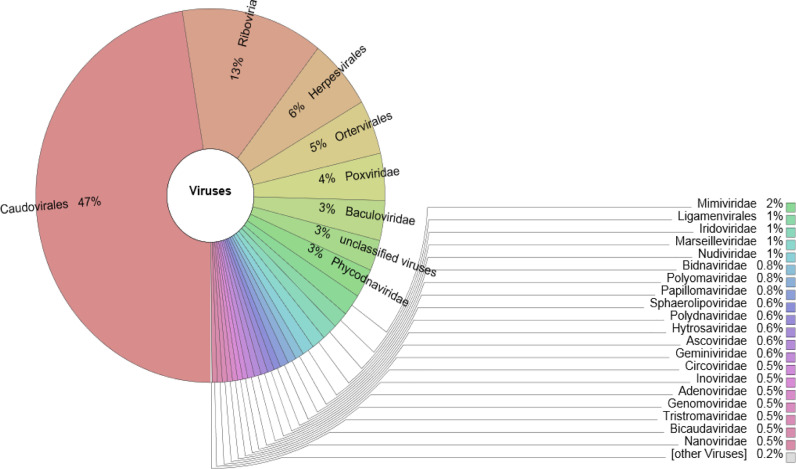
Fig. 3.
Krona chart analysis showing the predominant viral community structure present in NYS sample.
Fig. 4.
Krona chart analysis showing the predominant viral community structure present in OYS sample.
Community analysis of the metavirome reads
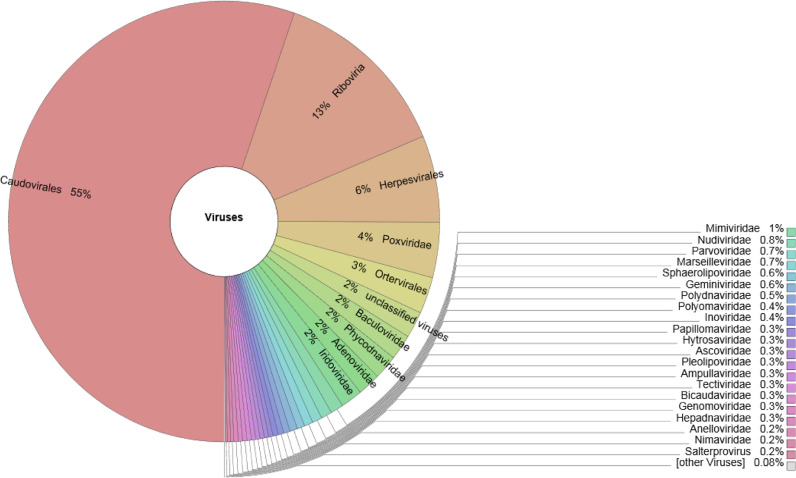
The metavirome reads in both the hot spring mud sediments showed the predominance of order Caudovirales (NYS=47%; OYS=55%); Herpesvirales (NYS=5%; OYS=6%); Ortervirales (NYS=4%; OYS=4%) (Figs. 6,7). The unassigned order (3%) found in NYS metavirome interestingly, belonged to some giant unclassified DNA viruses like Pandoravirus and Pithovirus genus. Whereas the unassigned order (2%) in case of OYS metavirome, belonged to various families like Baculoviridae (2%), Phycodnaviridae (2%), Adenoviridae (2%), Iridoviridae (2%), Mimiviridae (2%) and Nudiviridae (1%) respectively.
Fig. 6.
Genus level classification of the metavirome reads of OYS sample presented in percentage.
Fig. 7.
Species level classification of the metavirome reads of NYS sample presented in percentage.
Caudovirales showed the predominance of the viral families belonging to Siphoviridae, Myoviridae, Podoviridae, Herelleviridae and Ackermannviridae respectively. Among them, Siphoviridae and Myoviridae had higher number of viral members found through the metavirome reads present in the samples. Siphoviridae of the NYS sample had representation from various sub-families such as Bclasvirinae, Pclasvirinae and Guernseyvirinae majorly whereas in case of OYS it also had representation from Nymbaxtervirinae, Tunavirinae, Arquatrovirinae apart from the above found families in NYS sample. Myoviridae metavirome reads found in the hot spring muds of NYS showed the candidates from various sub-families like Tevenvirinae, Ounavirinae, Eucampyvirinae etc. whereas sub-families Peduovirinae and Vequintavirinae was found only in the hot spring muds of OYS. Herpesvirales was found to possess two families - Herpesviridae (NYS=77%; OYS=82%), and Alloherpesviridae (NYS=21%; OYS=13%) were the predominant ones in both the samples but in the case of OYS metavirome, another family Malacoherpesviridae (4%) representatives was also found. Family Herpesviridae had metavirome reads from all its three sub-families – Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae. From the Order Ortervirales community, in case of NYS metavirome, Retroviridae had metavirome reads from Gammaretrovirus and Betaretrovirus respectively whereas in the OYS sample it had viral members from the genera Alpharetrovirus, Betaretrovirus, Gammaretrovirus and Simiispumavirus respectively.
Thus, it was observed that although the virus reads were less in abundance in the microbiome yet they showed highest diversity among themselves. Exploring them and classifying into their respective genus and species classification level helped us to have better idea of their diversity.
Genus level classification of the metavirome
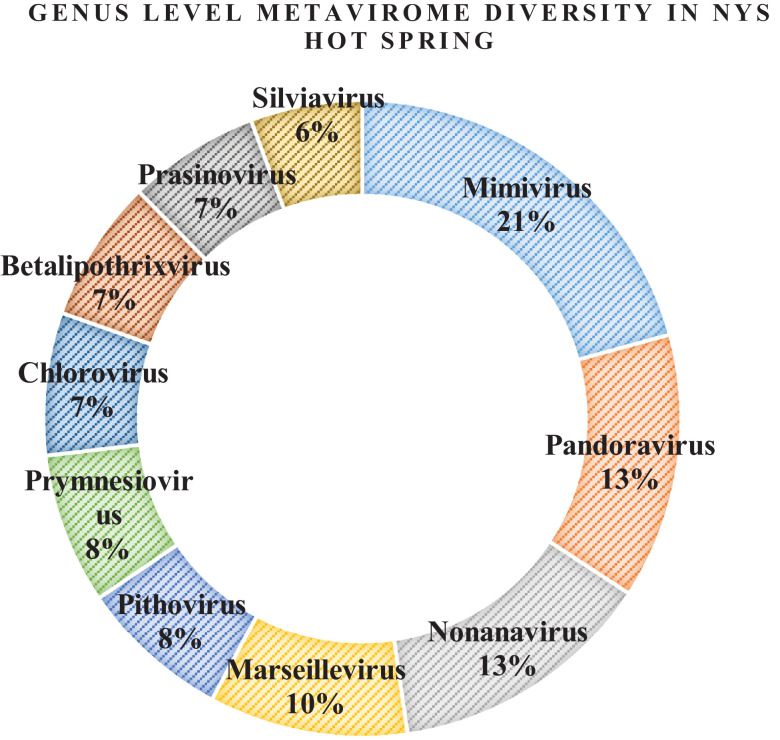
At the NYS hot spring solfataric mud sediments, the genus level classification among the top ten predominant viral genera present were - Mimivirus (21%), Pandoravirus (13%), Nonanavirus (13%), Marseillevirus (10%), Pithovirus (8%), Prymnesiovirus (8%), Chlorovirus (7%), Betalipothrixvirus (7%), Prasinovirus (7%) and Silviavirus (6%) respectively. All of them had dsDNA as their genetic material (Fig. 5). However, the exclusive genera present only at this hot spring were – Badnavirus, Cornellvirus, Babuvirus, Biquartavirus, Alphamesonivirus, Ampelovirus, Mosigvirus, Soupsvirus etc. whose metavirome reads were found (Table 4). As per the Baltimore Classification (BC), Badnavirus viral members having ds (RT-DNA) belonged to group VII, Babuvirus viral members having ssDNA belonged to group II; Alphamesonivirus and Ampelovirus viral members having ss(+)RNA (positive sense) belonged to group IV and the rest others belonged to group I having dsDNA as their genetic material. This was an interesting discovery of positive sense RNA viruses in the hot springs of Sikkim which seems to be dominated by dsDNA viruses.
Fig. 5.
Genus level classification of the metavirome reads of NYS sample presented in percentage.
Table 4.
Predominant viral genera exclusively present only in the NYS metavirome.
| Genus | Family | Strandness (ss/ds) | DNA/RNA | Reads |
|---|---|---|---|---|
| Betalipothrixvirus | Lipothrixviridae | – | – | 51 |
| Silviavirus | Herelleviridae | ds | DNA | 40 |
| Badnavirus | Caulimoviridae | ds | DNA (RT) | 32 |
| Cornellvirus | Siphoviridae | ds | DNA | 11 |
| Babuvirus | Nanoviridae | ss | DNA | 03 |
| Biquartavirus | Myoviridae | ds | DNA | 02 |
| Alphamesonivirus | Mesoniviridae | ss (+) | RNA | 02 |
| Ampelovirus | Closteroviridae | ss (+) | RNA | 02 |
| Mosigvirus | Myoviridae | ds | DNA | 02 |
| Soupsvirus | Siphoviridae | ds | DNA | 02 |
* where, ds = double strandness; ss = single strandness; (+) = positive sense.
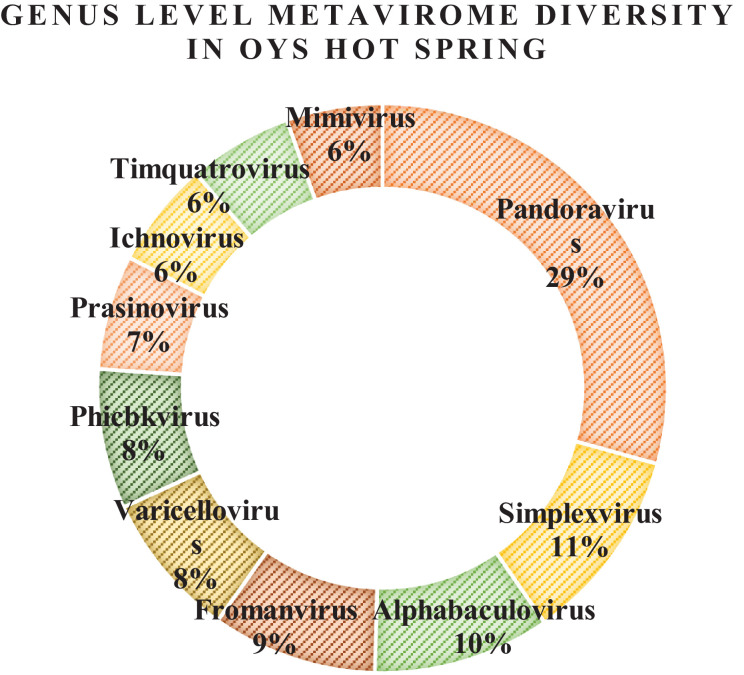
On the other hand, at the OYS hot spring solfataric mud sediments, the genus level classification among the top ten predominant viral genera present were - Pandoravirus (29%), Simplexvirus (11%), Alphabaculovirus (10%), Fromanvirus (9%), Varicellovirus (8%), Phicbkvirus (8%), Prasinovirus (7%), Ichnovirus (6%), Timquatrovirus (6%) and Mimivirus (6%) respectively. Here also, all the virions had dsDNA as their genetic material (Fig. 6). Again, the exclusive genera present only at this hot spring were – Badnavirus, Cornellvirus, Babuvirus, Biquartavirus, Alphamesonivirus, Ampelovirus, Mosigvirus, Soupsvirus etc. whose metavirome reads were found (Table 5). As per the Baltimore Classification (BC), Rubulavirus viral members having ss(-)RNA (negative sense) belonged to group V and the rest others belonged to group I having dsDNA as their genetic material. This is a first ever report and an interesting discovery of negative sense RNA virus metavirome reads in the hot springs of Sikkim dominated by dsDNA viruses.
Table 5.
Predominant viral genera exclusively present only in the OYS metavirome.
| Genus | Family | Strandness(ss/ds) | DNA/RNA | Reads |
|---|---|---|---|---|
| Simplexvirus | Herpesviridae | ds | DNA | 116 |
| Aviadenovirus | Adenoviridae | ds | DNA | 33 |
| Anatolevirus | Siphoviridae | ds | DNA | 31 |
| Redivirus | Siphoviridae | ds | DNA | 31 |
| Rubulavirus | Paramyxoviridae | ss (-) | RNA | 30 |
| Twortvirus | Herelleviridae | ds | DNA | 29 |
| Yuavirus | Siphoviridae | ds | DNA | 28 |
| Ripduovirus | Myoviridae | ds | DNA | 26 |
| Hpunavirus | Myoviridae | ds | DNA | 24 |
| Kochitakasuvirus | Podoviridae | ds | DNA | 23 |
*where, ds = double strandness; ss = single strandness; (-) = negative sense.
The commonly genera shared by both the hot spring mud sediments were abundant in Pandoravirus, Mimivirus, Nonanavirus, Alphabaculovirus, Prasinovirus, Fromanvirus, Varicellovirus, Phicbkvirus, Marseillevirus, Timquatrovirus which formed the major population of their virosphere.
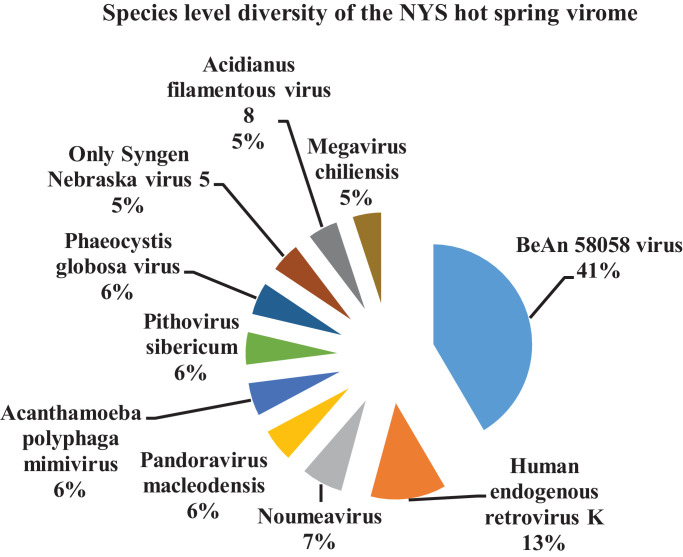
Species level classification of the metavirome
At the NYS hot spring solfataric mud sediments, the species level classification among the top ten predominant viral species present were - BeAn 58,058 virus (41%); Human endogenous retrovirus K (13%); Noumeavirus (7%); Pandoravirus macleodensis (6%); Pithovirus sibericum (6%); Acanthamoeba polyphaga mimivirus (6%); Phaeocystis globosa virus (6%); Only Syngen Nebraska virus 5 (5%); Acidianus filamentous virus 8 (5%) and Megavirus chiliensis (5%) respectively (Fig. 7). The species exclusively present only in this hot spring found were - Dishui lake phycodnavirus 1, Cacao swollen shoot Ghana L virus, Equid alphaherpesvirus 1, Klebsiella virus K64-1, Mycobacterium virus Goose, Hubei sobemo-like virus 28, Banana bunchy top virus, Orgyia leucostigma nucleopolyhedrovirus, Shamonda orthobunyavirus etc. respectively as detected through metavirome reads (Table 6).
Table 6.
Predominant viral species exclusively present only in the NYS metavirome.
| Species | Host | Reads |
|---|---|---|
| Acidianus filamentous virus 8 | Archaea | 50 |
| Dishui lake phycodnavirus 1 | Eukarya | 47 |
| Cacao swollen shoot Ghana L virus | Eukarya | 31 |
| Equid alphaherpesvirus 1 | Eukarya | 18 |
| Klebsiella virus K64-1 | Bacteria | 14 |
| Mycobacterium virus Goose | Bacteria | 12 |
| Hubei sobemo-like virus 28 | Eukarya | 8 |
| Banana bunchy top virus | Eukarya | 3 |
| Orgyia leucostigma nucleopolyhedrovirus | Eukarya | 3 |
| Shamonda orthobunyavirus | Eukarya | 2 |
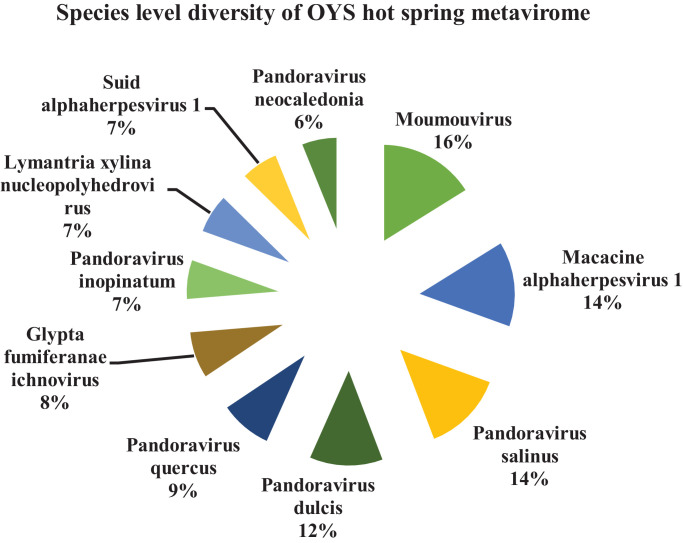
Similarly, at the OYS hot spring solfataric mud sediments, the species level classification among the top ten predominant viral species present were - Moumouvirus (16%); Macacine alphaherpesvirus 1 (14%); Pandoravirus salinus (14%); Pandoravirus dulcis (12%); Pandoravirus quercus (9%); Glypta fumiferanae ichnovirus (8%); Pandoravirus inopinatum (7%); Lymantria xylina nucleopolyhedrovirus (7%); Suid alphaherpesvirus 1 (7%) and Pandoravirus neocaledonia (6%) respectively on the basis of their reads (Fig. 8). The species exclusively present only in this hot spring found were - Bovine alphaherpesvirus 5; Micromonas sp. RCC1109 virus MpV1; Propionibacterium virus B3; Mammalian rubulavirus 5; Staphylococcus virus Twort; Pigeon aviadenovirus A etc. as per the metavirome reads (Table 7).
Fig. 8.
Species level classification of the metavirome reads of OYS sample presented in percentage.
Table 7.
Predominant viral species exclusively present only in the OYS metavirome.
| Species | Host | Reads |
|---|---|---|
| Macacine alphaherpesvirus 1 | Eukarya | 82 |
| Glypta fumiferanae ichnovirus | Eukarya | 46 |
| Lymantria xylina nucleopolyhedrovirus | Eukarya | 39 |
| Suid alphaherpesvirus 1 | Eukarya | 37 |
| Bovine alphaherpesvirus 5 | Eukarya | 35 |
| Micromonas sp. RCC1109 virus MpV1 | Bacteria | 33 |
| Propionibacterium virus B3 | Bacteria | 31 |
| Mammalian rubulavirus 5 | Eukarya | 30 |
| Staphylococcus virus Twort | Bacteria | 29 |
| Pigeon aviadenovirus A | Eukarya | 29 |
The commonly shared viral species among these two hot springs were - BeAn 58,058 virus; Human endogenous retrovirus K; Moumouvirus; Pandoravirus salinus; Pandoravirus macleodensis; Megavirus chiliensis; Pandoravirus dulcis; Noumeavirus; Acanthamoeba polyphaga mimivirus; Pandoravirus neocaledonia as per the available reads observed in the metavirome.
Functional metavirome diversity
On the basis of the virions functional genes, it was found that in the NYS hot spring mud sediments it was dominated by the bacteriophages (six major species) as compared to that of the eukaryotic phages (four major species). Top ten diversity profiling among the functional gene reads showed the following diversity among the NYS metavirome - Pneumococcus phage Dp-1, Acanthamoeba polyphaga mimivirus (APMV), Staphylococcus phage L54a, Fowlpox virus (FPV), Microplitis demolitor bracovirus (MdBV), Escherichia phage Mu, Bacillus phage phi105, Heliothis armigera granulosis virus (HaGV), Escherichia phage lambda and Escherichia phage N15 respectively (Table 8).
Table 8.
NYS metavirome species diversity on the basis of their functional proteins.
| Phage | Host | Genus | Family | Order | NYS reads |
|---|---|---|---|---|---|
| Pneumococcus phage Dp-1 | Bacteria | – | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 3865 |
| Acanthamoeba polyphaga mimivirus (APMV) | Eukarya | Mimivirus | Mimiviridae | – | 1737 |
| Staphylococcus phage L54a | Bacteria | – | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 1369 |
| Fowlpox virus (FPV) | Eukarya | Avipoxvirus | Poxviridae | – | 955 |
| Microplitis demolitor bracovirus (MdBV) | Eukarya | Bracovirus | Polydnaviridae | – | 300 |
| Escherichia phage Mu | Bacteria | Muvirus | Myoviridae (phages with contractile tails) | Caudovirales | 292 |
| Bacillus phage phi105 | Bacteria | – | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 237 |
| Heliothis armigera granulosis virus (HaGV) | Eukarya | Betabaculovirus | Baculoviridae | – | 191 |
| Escherichia phage lambda | Bacteria | Lambdavirus | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 137 |
| Escherichia phage N15 | Bacteria | Ravinvirus | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 107 |
Similarly, in the OYS hot springs mud sediments it was also dominated by the bacteriophages (eight major species) followed by the eukaryotic phages (one major species). Interestingly, one major archaeal phage species was found ATV – in the OYS hot spring metavirome functional reads. Top ten diversity profiling among the functional gene reads showed the following diversity among the OYS metavirome - Bacillus phage SPbeta, Escherichia phage P1, Pneumococcus phage Dp-1, Acanthamoeba polyphaga mimivirus (APMV), Escherichia phage lambda, Acidianus two-tailed virus (ATV), Salmonella phage P22, Salmonella virus Ike, Escherichia phage lambda and Escherichia phage Mu respectively (Table 9).
Table 9.
Species level diversity of the OYS metavirome on the basis of their functional proteins.
| Phage | Host | Genus | Family | Order | NYS reads |
|---|---|---|---|---|---|
| Bacillus phage SPbeta | Bacteria | Spbetavirus | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 3147 |
| Escherichia phage P1 | Bacteria | Punavirus | Myoviridae (phages with contractile tails) | Caudovirales | 1885 |
| Pneumococcus phage Dp-1 | Bacteria | – | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 1863 |
| Acanthamoeba polyphaga mimivirus (APMV) | Eukarya | Mimivirus | Mimiviridae | – | 1813 |
| Escherichia phage lambda | Bacteria | Lambdavirus | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 1699 |
| Acidianus two-tailed virus (ATV) | Archeae | Bicaudavirus | Bicaudaviridae | – | 917 |
| Salmonella phage P22 | Bacteria | Lederbergvirus | Podoviridae (phages with short tails) | Caudovirales | 869 |
| Salmonella virus IKe | Bacteria | Lineavirus | Inoviridae | – | 744 |
| Escherichia phage lambda | Bacteria | Lambdavirus | Siphoviridae (phages with long non-contractile tails) | Caudovirales | 720 |
| Escherichia phage Mu | Bacteria | Muvirus | Myoviridae (phages with contractile tails) | Caudovirales | 700 |
Thus, the functional genes depicted that the bacteriophage diversity is predominant in the both of the hot springs.
Functional characterization of the metavirome
On the basis of the functional proteins such as :- (a) viral capsid associated proteins (b) viral tail associated proteins (c) viral host cell processing associated proteins (d) viral genetic information processing associated proteins and (e) viral metabolic process associated proteins, metaviromes were characterized. Depending on these proteins, the functional diversity of the metavirome was analysed in both the hot spring of Sikkim.
The capsid of the virion plays a crucial role in host cell pathogenesis and attachment. Six contigs were found aligning to various major capsid proteins [GO:0,039,620; GO:0,019,028; GO:0,030,430; GO:0,019,028; GO:0,030,430; GO:0,019,028; GO:0,019,028; GO:0,019,028; GO:0,030,430; GO:0,039,620]. The highest reads obtained for capsid protein was for Escherichia phage Mu gene F Mup30 responsible for putative capsid assembly protein F [GO:0,030,430 and GO:0,019,012]. The other associated viral capsid proteins found mostly belonged to bacteriophages (Supplementary Table 1). The viral tail associated proteins identified through annotation belonged to various proteins like Pre-neck appendage protein [GO:0,098,024; GO:0,098,671; GO:0,098,994]; Head-tail preconnector protein; Tail completion protein [GO:0,019,012]; Tail sheath protein (TSP) [GO:0,019,012; GO:0,098,027; GO:0,030,430; GO:0,098,027; GO:0,030,430; GO:0,098,027]; Baseplate wedge protein [GO:0,019,012; GO:0,098,025]; Tail fiber protein [GO:0,098,024]; Tape measure protein (TMP) [GO:0,098,015]; Tip attachment protein J etc. (Supplementary Table 2). Here, also most of the tail associated proteins belonged to various bacteriophages. The highest reads among the tail associated proteins was found to be Pneumococcus phage Dp-1 gene TMP ORF52. Similarly, various other viral host cellular processing associated proteins were found like endolysin [GO:0,030,430] and other cell defence proteins (Supplementary Table 3). Among the viral genetic information processing associated proteins were found responsible for replication, transcription and translation process like polymerases, topoisomerases, integrases etc. (Supplementary Table 4). Few metabolic process associated proteins were also annotated having various activities such as hydrolase activity [GO:0,016,787]; metal ion binding [GO:0,046,872]; metallopeptidase activity [GO:0,008,237] etc. (Supplementary Table 5).
Discussion
Currently, the diversity of hot spring phages, remains under-studied in the Himalayan Geothermal Belt (HGB). Sikkim lies in this belt and has experienced many earthquakes and other natural hazards. So, these unique ecosystems always are at ecological risk of being extinct. The Sikkim geothermal areas have both artificial thermal pools and natural thermal pools for bathing and tourism purposes. Our study focusses on the natural thermal pools where the solfataric mud sediments can be found. The bathers frequently come in contact with these mud sediments and they also apply these solfataric muds on their skin due to its balneotherapeutic properties (Das et al., 2012). The solfataric mud sediments have a characteristic sulfurous odor which is due to the dissolved sulfur compounds present in the hot springs. The characteristics of the hot spring water (NYS and OYS) as tested through the colorimetric analysis suggests that it is rich in sulphite content and that might be due to the dissolved minerals such as pyrites which are known for emitting sulphide gas. Thus, in this pretext our study is of very prime significance as it concerns the human health for the spring goers and bathers. Hence, we have studied the culture independent virobiome present in the hot solfataric mud sediments as detected through the Next Generation Sequencing. It is the first ever study and holds a sheer importance to understand the environment sustainability and ecological vulnerability associated with it.
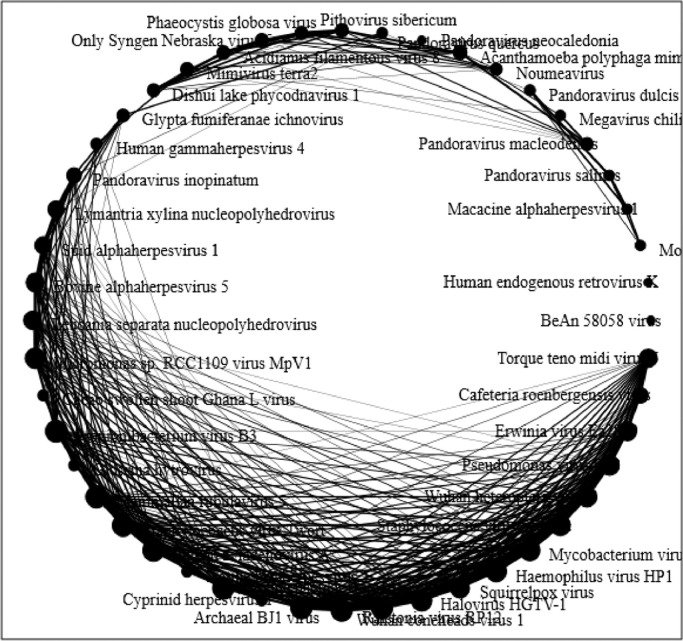
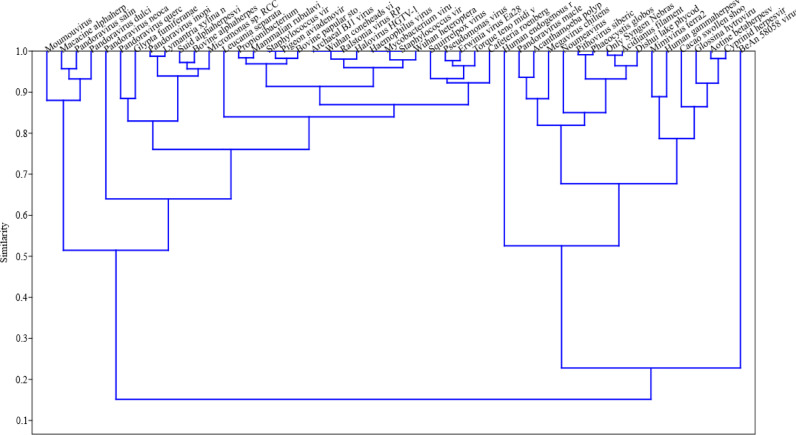
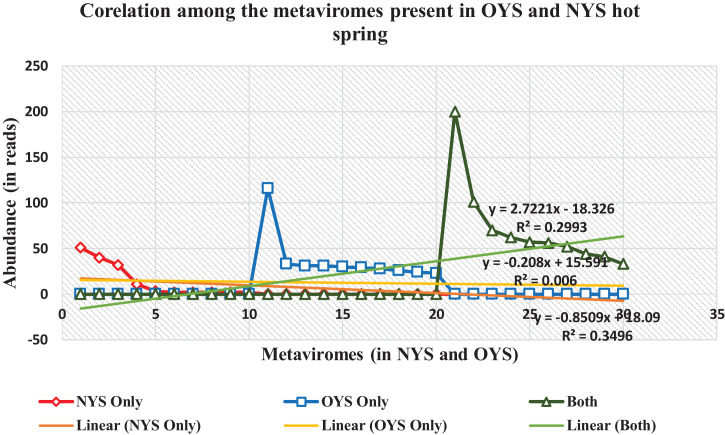
The selected hot springs are located at such a pristine conditions that their virome study plays a crucial role in understanding the microbial biodiversity. As, it is the first ever study of its kind on the hot spring solfataric mud sediments, it was more of an exploration study rather than establishing the ecological roles and significances of the viromes found. Thus, through the higher Shannon Index (Supplementary Figure 3) it was understood that OYS was rich in viral species and had more dominance over the NYS reads. But, there was common viromes shared between both the hot springs as shown through the network analysis (Fig. 9) which states their probabilistic inter-dependence on their common hosts of eukaryotes, bacteria and archaea. Clustering the viromes showed that they grouped among each other phylogenetically which suggests that they might have evolved around their hosts (Fig. 10). There was also a positive correlation among the viromes present exclusively in the NYS and OYS hot spring mud sediments and the common viromes present in both the samples (Fig. 11).
Fig. 9.
Network Analysis: Using average distance matrix based on the tetranucleotide frequencies a networking circular model for the metaviromes was calculated. Each viral metagenomic reads (edge cut off at 80%) are represented as singular nodal points and the node size is governed by the weight of the edges connecting the node. The edges of the network shows the correlation between the viral genomes and their nucleotide frequency abundances. The weight of the edges correlates the frequency between the nodes.
Fig. 10.
Cluster analysis for the predominant species of NYS and OYS metaviormes through UPGMA paired ends constructed through Bray-Curtis method.
Fig. 11.
Correlation plot for the predomiannt species of NYS and OYS metaviromes.
Geothermal spring habitats present on the land surface of the earth are the most desired ecosystems to study the isolation, characterization and discovery of the viruses and majority of these habitats exhibited archeaphages only (phages/viruses infecting arechea) (Zablocki et al., 2018; Peng et al., 2012; Prangishvili, 2013; Dellas et al., 2014; Prangishvili et al., 2017). However, the hot spring virus community also comprises of thermophages and eukaryotic phages apart from their well discovered counterparts i.e., the archeaphages. Geothermal springs from the Himalayan Geothermal Belt have been least explored in terms of viruses. However, one such hot spring to report for the viruses from this geothermal belt was from the Manikaran hot springs. It is located in Uttarakhand state of Indian Himalayan Geothermal Belt, whose virosphere in sediments and microbial mats have been unearthed through metagenomics. It was found that they were abundant in dsDNA phages which is trend similar to found at Sikkim. They had huge abundance of archaeophages from Fuselloviridae family and bacteriophages from Myoviridae, Podoviridae and Siphoviridae (Sharma et al., 2018). They also characterized and reported novel phages for Enterobacter host. Thus, due to the very limited study on the viral diversity and habitats, in these ecosystem; their evolution and patterns are yet to be deciphered. Most of the hot spring thermophages till date have been generally found from the five major viral taxonomical families of – Inoviridae, Myoviridae, Siphoviridae, Sphaerolipoviridae and Tectiviridae (Zablocki et al., 2018). Similarly, from the metavirome reads suggests that the NYS hot spring mud sediments had major viral taxonomical families comprising of Mimiviridae, Siphoviridae and Phycodnaviridae and some exclusively families such as Marseilleviridae, Herelleviridae, Caulimoviridae, Nanoviridae, Myoviridae, Mesoniviridae, Closteroviridae etc. were also present.
Thermus infecting phages have been majorly reported from the various hot springs worldwide and the first discovered virus from hot spring - Thermus myovirus ϕYS40 was the also the first to be isolated from Japan almost four decades earlier (Sakaki and Oshima, 1976). Soon, after this great discovery, slowly many studies followed, leading to virus or VLPs discovery from worldwide (Rachel et al., 2002). But archeal phages then stole the limelight being frequently discovered in the hot springs. In the studied hot springs, very few archaeal phage reads were found in Sikkim. Only in the case of NYS hot spring mud sediment sample, some genes corresponding to Acidianus filamentous virus 8 were found which suggests it to be the lone archael representative in our studied ecological niche.
Very few reports on the thermophages have made it more intriguing to understand both about their host and conditions in the ecosystem relating to this mysterious scarcity of their population. However, the NYS metavirome reads corresponded with many thermophages like Pneumococcus phage Dp-1, and Staphylococcus phage L54a which were predominant in it and also few other bacteriophages such as Escherichia phage Mu, Bacillus phage phi105, Escherichia phage lambda, Escherichia phage N15 were also found. But in the case of, OYS sample it had majority of Bacillus phage SPbeta, Escherichia phage P1, Escherichia phage lambda, Salmonella phage P22 and Salmonella virus Ike respectively. Both the hot spring mud samples had moderate reads for Pneumococcus phage Dp-1.
Zablocki categorized the thermophages on the basis of their ecosystem adaptability to produce optimal concentrations at their optimum infective temperature and pH to infect their suitable host (Zablocki et al., 2018): (i) Thermophage (40 °C–71 °C, pH 6.5–7.5), (ii) Hyperthermophage (72 °C –98 °C, pH 6.5–7.5), (iii) Acidothermophage (40 °C –71 °C, pH 1–5) and (iv) Acidohyperthermophage (72 °C–98 °C, pH 1–5). Thus, the phages present in the NYS (57 °C, pH 9.1) and OYS (61 °C, pH 8.77) hot springs can be newly classified as “Alkalothermophages” as they can survive at slightly alkaline conditions. But, these criterion should also hold for the host growth conditions as well because for viruses or VLPs host specificity plays a very crucial role. Without hosts, they can virtually remain suspended as particles in the sediments (Bäckström et al., 2019) waiting for their fate “to multiply”. Although there are so many suitable hosts for the phages to infect but somehow they went undetected and also the strictness of isolating and culturing them as per ICTV made it more constraints in their discovery. But recent studies like MIUViG (Roux et al., 2019) are substantial metagenomic initiatives and suggestions which should be implemented and accepted by ICTV so as to introspect and accumulate the inventory of environmental viruses.
The environmental virobiomes usually contains three predominant viruses or virus-like-particles – (i) RNA viruses (ii) DNA viruses and (iii) retroid viruses (RT-DNA based genetic segments). But, Diemer and Stedman, discovered a new form of viruses - “RNA-DNA hybrid virus – RDHV” from their viral diversity study in Boiling Springs Lake (BSL) of Lassen Volcanic National Park, USA. The novel putative BSLV genome showed homology to both ssRNA and ssDNA viruses, giving an important dimension to viral evolution (Diemer and Stedman, 2012). Similarly, Bolduc reported the first ever Archea infecting RNA viral genomes from the Yellowstone National Park, USA hot springs (Bolduc et al., 2012). This unique contig assemblage shed light on the unique viral RNA metagenomes which contained genes encoding for RNA-dependent RNA-polymerase (RdRp) – a characteristic feature of positive strand RNA viruses. They showed homology to the capsid protein of birnaviruses, nodaviruses and tetraviruses. Thus, they concluded that these positively sense RNA strands are the primordial originators to eukaryotic RNA viruses (Bolduc et al., 2012). From the OYS metavirome the reads were obtained for negative sense strand RNA, VLPs belonged to Rubulavirus which was completely absent in the NYS metavirome reads. Similarly, in the case of NYS metavirome, only positive sense strand RNA VLPs were found for Alphamesonivirus belonging to Mesoniviridae and Ampelovirus belonging to Closteroviridae which was completely absent in OYS reads. And through the functional metagenomics characterization, genes encoding for the RdRp protein was also found in the reads, supporting the evidence of positively strand RNA viruses to be present in the hot spring.
The crenarchaeal VLPs metagenomes predominant in the hot springs of Italy, Iceland and YNP, USA usually belonged to Ampullaviridae, Bicaudaviridae, Lipothrixviridae and Rudiviridae (Gudbergsdóttir et al., 2016). Interestingly, the NYS metavirome reads showed the presence of crenarchaeal VLPs metagenomes from the Lipothrixviridae family. Although, it was not found in the OYS sample suggesting the uniqueness of the habitat. Many interesting morphologically and genetically unique viruses were discovered from the study of Pozzuoli hot springs in Italy, from which novel families Fuselloviridae, Globuloviridae, Guttaviridae, Lipothrixviridae and Rudiviridae was established (Zablocki et al., 2018; Häring et al., 2005; Ortmann et al., 2006). Similarly, from the Porcelana hot spring of Chile, viruses composing of thermophage Caudovirus like particles was found from the families of Myoviridae, Podoviridae and Siphoviridae; archeaphage (VLPs) from Clavaviridae and Lipothrixviridae families (Guajardo-Leiva et al., 2018). Interestingly, through metatrancriptomic studies, with an increase in temperature from 48 °C to 66 °C, the Caudovirales population decreased whereas Megavirales population increased. The most abundant family was Podoviridae (8%) in this hot spring but its population decreased with increase in temperature to 66 °C whereas Siphoviridae population increased six times at that temperature. Marseilleviridae, Mimiviridae and Phycodnaviridae families’ population was constant at all the temperature gradients of the metatranscriptomes (Guajardo-Leiva et al., 2018). Similarly, at the Manikaran hot springs, it was rich in Caudovirales and mostly were phylogenetically related to Myoviridae and others aligned with Podoviridae or Siphoviridae suggesting that only these three families VLPs were in majority. In the hot sediments they found that Siphoviridae was highly abundant whereas the microbial mats (comparatively cooler) had the dominance of Myoviridae (Sharma et al., 2018). Thus, it was in accordance with the Guajardo-Leiva et al. (2018) findings. As, our NYS hot springs had these similar VLPs it would be quite interesting to study such kind of population dynamics with respect to temperature from our hot spring viromes.
Bäckström and their team conducted extravagant viral diversity researches on NCLDV (Nucleocytoplasmic large DNA viruses) of eukaryotes reported from the Loki's Castle hydrothermal vents (Bäckström et al., 2019). They predominantly found the viral members belonging to various families like Ascoviridae, Asfarviridae, Iridoviridae, Marseilleviridae, Mimiviridae, Phycodnaviridae and Poxviridae and others were unclassified faustoviruses, molliviruses, pandoraviruses and pithoviruses respectively. Similarly, in our hot spring samples giant viruses like various species of Pandovirus and Pithovirus was found. Bäckström gave an interesting hypothesis regarding the virus evolution that NCLDV has characteristic reproductive features in infected cytoplasmic cells along with some gene encoding proteins responsible for viral replication and morphogenesis, hence they most probably evolved from a common primordial virus (Bäckström et al., 2019). They also concluded that as there was no host for LCV, hence, the viruses could not multiply in the vent sediments and remained as particles in water gradients precipice and are viable fossils.
Many limitations, such as not being to isolate these viruses in enrichments or obtain their complete genome is a major shortcoming of this study. However, on comparing the viral metagenomes from various other ecological geothermal areas like hot springs and hydrothermal vents, Sikkim hot springs promises and suggests to have unique virosphere diversity among all the other ecosystems.
Conclusion
The apparent shortcoming of this type of work - as in the case of most of the metagenomic studies occurs is that the viruses discussed here are culture independent reads only. They have not been cultivated in any host or enrichment medium. Hence, it is noteworthy to declare that our basic inference on the viruses of the solfataric mud sediments present in the hot spring ecosystem of Sikkim is very rudimentary in nature without any substantial cultural proofs. And all the conclusion drawn is merely from the genomic sequences constructed. The metagenomic studies rather discusses the probability of the presence of the biota and it equilibrates the culture dependent studies. Similarly, our data complements and shows the gap of research required to study the viruses of the hot springs and other geothermal areas. It should act as an adjunct to the culturable virology study dynamics. Being the first study of its kind in this geography, there is severe limitations even in literature comparisons as these ecosystem are still virgin in terms of researches.
The domain of life i.e. archea, bacteria and eukarya are being more easily cultivable and easy to work with as compared to viruses, virions or prions. The omics study has enhanced the ability to understand the microbial world better than the culture methodologies. These three domains of life has immense biomass and they forms the majority of the ecosystem whereas the viruses are the parasites infecting on them and deriving their energy out of their respective hosts ultimately killing in order to survive. This whole dynamics of predatory nature plays a very crucial role and thus knowing viruses have become more important for human mankind keeping the pandemic crisis situation faced recently as a grave human health hazards. It is due to the sheer lack of researches in the environmental virology and its impact that we do not have adept knowledge about their habitats and ecological roles. So, there is no universal tools or methodologies formulized to isolate them independently or study their morphology or taxonomy. Viruses do not have unique conserved consensus sequences among them which can be easily targeted to construct OTUs to create their phylogenies and study them as in the case of bacteria, archea, fungi and yeasts. Recent studies on metagenomic sequences and metaviromes by many groups, have started to shed light on the evolution of viruses and theories and hypotheses are being channeled to create and expand the virus genome databases. These database will help and support ICTV for more enhanced viral taxonomy classifications. Undoubtedly, the pace of viruses and their evolution has grown at such a pace that there is greater demand for knowing viruses and their relatives in correct taxonomical order. The databases submitted on viral diversity discussed through omics is far more outnumbered than that of complete isolated whole genome sequences on viruses. Polyphasic metagenomic approaches should be done to understand about the viral diversity present in the ecosystem. Thus, the various metavirome reads discussed here provides the kaleidoscopical overview of the viruses present in the hot springs of Sikkim.
Funding
The work was funded by the Department of Biotechnology, Government of India (DBT-NER/Health/45/2015 & BT/PR25092/NER/95/1009/2017).
CRediT author statement
Sayak Das: Conceptualization, Investigation, Methodology, Writing- Original draft preparation, Data Curation, Formal Analysis. Ankita Kumari: Software, Validation, Formal Analysis, Methodology. Mingma Thundu Sherpa: Investigation. Ishfaq Nabi Najar: Investigation. Nagendra Thakur: Writing - Review & Editing, Visualization, Supervision.
Declaration of Competing Interest
None to be declared.
Acknowledgement
SD would like to thank DST INPIRE, Department of Science and Technology, Govt. of India, for providing the INSPIRE FELLOWSHIP (IF130091) for the research work. The authors heartily thank and express their most sincere gratitude to Department of Forest, Govt. of Sikkim for giving the permission and kind cooperation and support during the field research work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2020.05.002.
Appendix. Supplementary materials
References
- Anderson R.E., Brazelton W.J., Baross J.A. The deep viriosphere: assessing the viral impact on microbial community dynamics in the deep subsurface. Rev. Mineral. Geochem. 2013;75:649–675. doi: 10.2138/rmg.2013.75.20. [DOI] [Google Scholar]
- Anderson R.E., Sogin M.L., Baross J.A. Evolutionary strategies of viruses, bacteria and archaea in hydrothermal vent ecosystems revealed through metagenomics. PLoS ONE. 2014;9(10) doi: 10.1371/journal.pone.0109696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström D., Yutin N., Jørgensen S.L., Dharamshi J., Homa F., Zaremba-Niedwiedzka K., et al. Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism. MBio. 2019;10 doi: 10.1128/mBio.02497-18. e02497-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko R.G., Harlow T.J., Ragan M.A. Highways of gene sharing in prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Jang H., Bolduc B., Zablocki O., Kuhn J.H., Roux S., Adriaenssens E.M., et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019;37:632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- Bolduc B., Shaughnessy D.P., Wolf Y.I., Koonin E.V., Roberto F.F., Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012;86(10):5562–5573. doi: 10.1128/JVI.0719611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc B., Wirth J.F., Mazurie A., Young M.J. Viral assemblage composition in Yellowstone acidic hot springs assessed by network analysis. ISME J. 2015;9(10):2162–2177. doi: 10.1038/ismej.2015.28. 10.10-38/ismej.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M., Wegley L., Leeds S., Schoenfield T., Rohwer F. Phage community dynamics in hot springs. Appl. Environ. Microbiol. 2004;70:1633–1640. doi: 10.1128/ae-m.70.3.1633-1640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Sherpa M.T., Sachdeva S., Thakur N. Hot springs of Sikkim (Tatopani): a Socio medical conjuncture which amalgamates religion, faith, traditional belief and tourism. Asian Acad Res. J. Soc. Sci. Human. 2012;1(4):80–93. [Google Scholar]
- Das S., Sherpa M.T., Thakur N. Sikkim's Tatopani – A balneotherapeutic prospect for community health in North East India. Int J Agric Food Sci Technol. 2012;3(2):149–152. [Google Scholar]
- Dellas N., Snyder J.C., Bolduc B., Young M.J. Archaeal viruses: diversity, replication, and structure. Annu. Rev. Virol. 2014;1:399–426. doi: 10.1146/annurev-virology-031413-085357. [DOI] [PubMed] [Google Scholar]
- Diemer G.S., Stedman K.M. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett R.A., Prangishvili D., Shah S.A., Reuter M., Stetter K.O., Peng X. Metagenomic analyses of novel viruses and plasmids from a cultured environmental sample of hyperthermophilic neutrophiles. Environ. Microbiol. 2010;12(11):2918–2930. doi: 10.1111/j.1462292-0.2010.022-66.x. [DOI] [PubMed] [Google Scholar]
- Guajardo-Leiva S., Pedrós-Alió C., Salgado O., Pinto F., Díez B. Active crossfire between Cyanobacteria and Cyanophages in phototrophic mat communities within hot springs. Front. Microbiol. 2018;9:2039. doi: 10.3389/fmicb.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbergsdóttir S.R., Menzel P., Krogh A., Young M., Peng X. Novel viral genomes identified from six metagenomes reveal wide distribution of archaeal viruses and high viral diversity in terrestrial hot springs. Environ. Microbiol. 2016;18(3):863–874. doi: 10.1111/1462-2920.13079. [DOI] [PubMed] [Google Scholar]
- Häring M., Rachel R., Peng X., Garrett R.A., Prangishvili D. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J. Virol. 2005;79(15):9904–9911. doi: 10.1128/JVI.79.15.9904-9911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T., Li H., Zhang X. Deep-sea hydrothermal vent viruses compensate for microbial metabolism in virus-host interactions. MBio. 2017;8 doi: 10.1128/mBio.0-089317. e00893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. 10.11-86/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim M.S., Koh A.Y., Xie Y., Zhan X. FMAP: functional mapping and analysis pipeline for metagenomics and metatranscriptomics studies. BMC Bioinform. 2016;17:1–8. doi: 10.1186/s12859-016-1278-0. 10.11-86/s12859-016-1278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Jia Z.J., Nakayama N., Asakawa S. Ecology of viruses in soils: past, present and future perspectives. Soil Sci. Plant Nutr. 2008;54:1–32. doi: 10.1111/j.1747-0765.2007.00197.x. [DOI] [Google Scholar]
- Koonin E.V., Dolja V.V. Metaviromics: a tectonic shift in understanding virus evolution. Virus Res. 2018;246:A1–A3. doi: 10.1016/j.virusres.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Cvirkaite-Krupovic V., Iranzo J., Prangishvili D., Koonin E.V. Viruses of archaea: structural, functional, environmental and evolutionary genomics. Virus Res. 2018;244:181–193. doi: 10.1016/j.virusres.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic. Acids. Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Breitwieser F.P., Thielen P., Salzberg S.L. Bracken: estimating species abundance in metagenomics data. PeerJ. Comput. Sci. 2017;3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- Munson-McGee J.H., Snyder J.C., Young M.J. Archaeal viruses from high-temperature environments. Genes (Basel) 2018;9(3):128. doi: 10.3390/genes9030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson-McGee J.H., Peng S., Dewerff S., Stepanauskas R., Whitaker R.J., Weitz J.S., et al . A virus or more in (nearly) every cell: ubiquitous networks of virus-host interactions in extreme environments. ISME J. 2018;12(7):1706–1714. doi: 10.1038/s41396-018-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najar I.N., Sherpa M.T., Das S., Das S., Thakur N. Microbial ecology of two hot springs of Sikkim: predominate population and geochemistry. Sci. Total Environ. 2018;637-638:730–745. doi: 10.1016/j.scitotenv.2018.05.037. 10.10-16/j.scitotenv.2018.05.037. [DOI] [PubMed] [Google Scholar]
- Najar I.N., Sherpa M.T., Das S., Verma K., Dubey V.K., Thakur N. Geobacillus yumthangensis sp. nov., a thermophilic bacterium isolated from a north-east Indian hot spring. Int. J. Syst. Evol. Micr. 2018;68(11):3430–3434. doi: 10.1099/ijsem.0.00-3002. [DOI] [PubMed] [Google Scholar]
- Najar I.N., Sherpa M.T., Das S., Thakur N. Bacterial diversity and functional metagenomics expounding the diversity of xenobiotics, stress, defense and CRISPR gene ontology providing eco-efficiency to Himalayan Hot Springs. Funct. Integr. Genomic. 2020 doi: 10.1007/s10142-019-00723-x. [DOI] [PubMed] [Google Scholar]
- Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann A., Wiedenheft B., Douglas T., Young M. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat. Rev. Microbiol. 2006;4:520–528. doi: 10.1038/nrmicro1444. [DOI] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Garrett R.A., She Q. Archaeal viruses—Novel, diverse and enigmatic. Sci. China Life Sci. 2012;55:422–433. doi: 10.1007/s11427-012-4325-8. [DOI] [PubMed] [Google Scholar]
- Prangishvili D., Bamford D.H., Forterre P., Iranzo J., Koonin E.V., Krupovic M. The enigmatic archaeal virosphere. Nat. Rev. Microbiol. 2017;15(12):724–739. doi: 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- Prangishvili D. The wonderful world of archaeal viruses. Ann. Rev. Microbiol. 2013;67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- Rachel R., Bettstetter M., Hedlund B.P., Haring M., Kessler A., Stetter K.O. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch. Virol. 2002;147(12):2419–2429. doi: 10.1007/s00705-002-0895-2. [DOI] [PubMed] [Google Scholar]
- Ray J., Dondrup M., Modha S., Steen I.H., Sandaa R.-.A., Clokie M. Finding a needle in the virus metagenome haystack – micro-metagenome analysis captures a snapshot of the diversity of a bacteriophage armoire. PLoS ONE. 2012;7(4):e34238. doi: 10.1371/journal.pone.0034238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F. Global phage diversity. Cell. 2003;113(2):141. doi: 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- Roux S., Adriaenssens E.M., Dutilh B.E., Koonin E.V., Kropinski A.M., Krupovic M., et al . Minimum Information about an Uncultivated Virus Genome (MIUViG) Nat. Biotechnol. 2019;37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki Y., Oshima T. A new lipid-containing phage infecting acidophilic thermophilic bacteria. Virology. 1976;75:256–259. doi: 10.1016/00426822(76)900246. [DOI] [PubMed] [Google Scholar]
- Schoenfeld T., Patterson M., Richardson P.M., Wommack E., Young M., Mead D. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 2008;74:4164–4174. doi: 10.1128/aem.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Schmidt M., Kiesel B., Mahato N.K., Cralle L., Singh Y., et al . Bacterial and archaeal viruses of Himalayan hot springs at Manikaran modulate host genomes. Front. Microbiol. 2018;9:3095. doi: 10.3389/fmicb.2018.03095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Adams M.J., Benko M., Breitbart M., Brister J.R., Carstens E.B., et al . Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- Snyder J.C., Wiedenheft B., Lavin M., Roberto F.F., Spuhler J., Ortmann A.C., et al . Virus movement maintains local virus population diversity. Proc. Natl. Acad. Sci. USA 27. 2007;104(48):19102–19107. doi: 10.1073/pnas.0709445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Dong H., Hou W., Jiang H., Huang Q., Briggs B.R., et al. Greater temporal changes of sediment microbial community than its waterborne counterpart in Tengchong hot springs, Yunnan Province. China. Sci. Rep. 2014;4:7479. doi: 10.1038/srep07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Guo Z., Feng H., Chen Y., Chen X., Li Z., et al. Novel Sulfolobus virus with an exceptional capsid architecture. J. Virol. 2018;92 doi: 10.1128/JVI.01727-17. e01727-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T.J., Eddy S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.W., Simmons B.A., Singer S.W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32(4):605–607. doi: 10.1093/bioinformatics/btv638. 10.10-93/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- Zablocki O., van Zyl L., Trindade M. Biogeography and taxonomic overview of terrestrial hot spring thermophilic phages. Extremophiles. 2018;22:827–837. doi: 10.1007/s00792-018-1052-5. [DOI] [PubMed] [Google Scholar]
- Zablocki O., van Zyl L.J., Kirby B., Trindade M. Diversity of dsDNA viruses in a South African hot spring assessed by metagenomics and microscopy. Viruses. 2018;9(11):348. doi: 10.3390/v9110348. 1810.33-90/v9110348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Shi M., Holmes E.C. Using metagenomics to characterize an expanding virosphere. Cell. 2018;172:1168–1172. doi: 10.1016/j.cell.2018.02.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw metagenomic reads were submitted to Sequence Read Archive (SRA), NCBI to obtain the Bio-Sample and Sequence Read Archive (SRA) accession numbers. The OYS data, Bio-Project accession obtained is PRJNA485728; Bio-Sample accession is SAMN09813281 and SRA is SRP158027 for the sample name OYSMUD4. The NYS data Bio-Project accession obtained is PRJNA485701; Bio-Sample accession is SAMN09813022 and SRA is SRP158029 for the sample name NYSMUD4.