Abstract
Second messenger (p)ppGpp mediated stress response plays a crucial role in bacterial persistence and multiple drug resistance. In E. coli, (p)ppGpp binds to RNA polymerase and upregulates the transcription of genes essential for stress response while concurrently downregulating the expression of genes critical for growth and metabolism. Recently, the family of alarmone molecules has expanded to pppGpp, ppGpp, pGpp & (pp)pApp as distinct members. These molecules may help in fine-tuning stress responses in different hostile conditions. Do all of these molecules bind to RNA polymerase? Do they compete with each other or complement each other's functions is still not clear. Earlier, others and we have synthesized artificial analogs of (p)ppGpp that inhibited (p)ppGpp synthesis and long-term survival in M. smegmatis and in B. subtilis suggesting that analogs could compete with each other. Understanding the interplay of these molecules will allow deciphering novel pathways that can be potentially subjected to the therapeutic intervention. In this article, we have reviewed newly characterized second messengers and discussed their mode of action. We have also documented the progress made to-date in understanding the molecular basis of regulation of transcription by second messenger ppGpp, pppGpp, and pGpp.
Keywords: Second messenger, ppGpp, Stress response, Transcription, pGpp, pppApp
Introduction
In 1969, Professor Mike Cashel reported the appearance of a magic spot (MS) on a thin layer chromatography sheet in the cell extract sample from E. coli cells subjected to amino acid starvation (Cashel and Gallant, 1969). This magic spot was identified as ppGpp, and it was observed to inhibit RNA synthesis (Cashel and Gallant, 1969). In the next few decades, (p)ppGpp was recognized for its critical role in heat stress response, tolerance to antibiotics, overall nutritional starvation response, and virulence (Kim et al., 2018; Kudrin et al., 2017; Li et al., 2015; Pulschen et al., 2017). Interestingly, most antibiotics target pathways linked to the exponential growth phase, and such selective targeting leads to the relative accumulation of cells in other phases like the stationary phase. As (p)ppGpp is an important second messenger in the stationary phase and crucial for associated phenotypes including biofilm formation, targeting of (p)ppGpp associated pathways, could constitute a vital non-conventional therapeutic approach (Diaz-Salazar et al., 2017; Syal et al., 2017; Syal et al., 2017). Second messenger (p)ppGpp do not exist in humans, so targeting its synthesis or associated pathways in bacteria will not have any side effects. Further, the recent discoveries highlight its direct role in virulence, and antibiotic tolerance which makes it an ideal drug target (Chatnaparat et al., 2015; Holley et al., 2015). In E. coli, (p)ppGpp binds to RNA polymerase to give a stress response, but the binding site of ppGpp on RNAP remained controversial for a very long (Vrentas et al., 2008). In the late 1980s and early 1990s, multiple studies validated that ppGpp binds to RNA polymerase (RNAP) in Escherichia coli and plays a direct role in the regulation of transcription (Glass et al., 1986). Chatterji et al. used photocrosslinking methodology, where azido labeled ppGpp was used for crosslinking with RNAP and determined the C-terminal domain of the beta-subunit as the binding site (Chatterji et al., 1998; Reddy et al., 1995). Hernandez et al. used a thio-derivative of ppGpp, and observed its binding to the N-terminal domain of beta’-subunit (Toulokhonov et al., 2001). Hernandez et al. explained the discrepancy with the former study based on the proximity of the N-terminal domain of beta’ to the C-terminal domain of the beta subunit. More importantly, the different positions of a crosslinking group are thio- and azido groups on guanine moiety of ppGpp molecule (Toulokhonov et al., 2001). In 2004, a 2.7 Angstrom crystal structure of ppGpp-RNAP from Thermus thermophilus revealed the active site of RNAP as the binding site. But, the latter study was contested due to the lack of omega subunit in the RNAP crystal structure, contradictory mutational analysis, and the inability of Thermus thermophilus to give a stringent response (Artsimovitch et al., 2004). More recently, Steitz et al. solved the crystal structure of RNAP from E. coli and soaked it with ppGpp (Zuo et al., 2013). The latter group reported the binding site at the interface of β’−ω subunit. This study was further confirmed by a concurrent mutational analysis study from the Richard Gourse group (Ross et al., 2013).
Did (p)ppGpp crystal structure with RNA polymerase solve the mystery of regulation of stringent response?
It is worth considering that crystallization was carried out by soaking and not co-crystallization. The RNA polymerase in crystal structure presents only one conformation and soaking of ppGpp over it would indicate the binding of ppGpp to that specific conformation. Since RNAP is a highly active molecule, tracing of binding of ppGpp to one conformation of RNAP cannot reveal the whole story. Also, what if RNAP binds (p)ppGpp in the presence of some other unknown factor? Evidently, Gourse et al. reported the DksA dependent binding site of ppGpp on RNA polymerase (Ross et al., 2016). So the possibility of more binding sites of (p)ppGpp on RNAP cannot be ruled out. In 2015, we used the DRaCALA assay to follow the binding of ppGpp and pppGpp to the RNA polymerase. DRaCALA assay was devised for c-di-GMP messenger and other small molecules (Roelofs et al., 2011). But, ppGpp and pppGpp were the exceptional cases as unlike c-di-GMP, which is a cyclic ring of two GMP molecules, ppGpp and pppGpp are different by a pyrophosphate from its precursor molecule GDP and GTP, respectively. So, we were unsure if we could precisely determine the binding of ppGpp or pppGpp by DRaCALA assay. We confirmed that ppGpp or pppGpp binding could also be followed by the DRaCALA assay (Syal and Chatterji, 2015). Soon, other groups adopted the DRaCALA assay too for following the binding of ppGpp. We observed multiple crosslinked peptides by mass spectrometry, where two of them pinpointed to the β’-ω subunit pocket that coincided with the studies from Gourse et al. and Steitz et al.. In contrast, the third peptide was located at the C-terminal domain of β- subunit (Syal and Chatterji, 2015). Interestingly, we observed that pppGpp binds more strongly to RNAP and has overlapped binding sites with ppGpp. With the discovery of potential functional differences in pppGpp, ppGpp, and pGpp, the investigations for deciphering the molecular basis of their binding, regulation, and function have become even more relevant. Also, how these nucleotide messengers cross-talk is worth pursuing. Previously, we have determined the biophysical parameters of binding of ppGpp to RNAP by isothermal titration calorimetry. We have shown that ppGpp and pppGpp compete to bind to RNA polymerase in in-vitro conditions. We have used 1:1 binding stoichiometry model for following the binding of ppGpp to RNAP from E. coli and observed the binding of ppGpp to RNAP in the order of micromolar range (Bhardwaj et al., 2018). However, the basal level of ppGpp is itself in micromolar range. It shoots to millimolar range in stress so we still do not understand the significance of micromolar binding affinity observed by us (Bhardwaj et al., 2018) especially in physiological conditions.
Earlier, Cashel group conclusively showed the differential regulation of transcription by ppGpp and pppGpp in E. coli. But, the mode of action was not clear. Then, we showed differential binding of ppGpp and pppGpp, explaining the different modes of regulation of transcription by ppGpp and pppGpp (Syal and Chatterji, 2015).
These discoveries opened a new series of questions like-are ppGpp and pppGpp different or same (as they were considered for so long)? Do ppGpp and pppGpp perform different functions? Do they bind at the same site on RNA polymerase? Can one compete against the other? Are their effects cumulative? Which one is more effective? (Fig. 1)
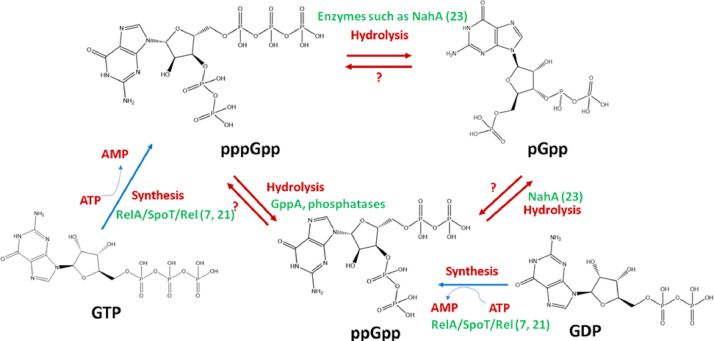
Fig. 1.
(p)ppGpp family, precursors and byproducts (Yang et al., 2020)- The RSH enzymes mediate the synthesis of pGpp, ppGpp and pppGpp by transfer of a pyrophosphate group to GMP/GDP/GTP. Further, interconversion of pppGpp to ppGpp is processed by guanosine pentaphosphatase (GppA) and translational GTPases (Hauryliuk et al., 2015).
On top of it, do pGpp also have a role in stress response, and how it is connected to the function of (p)ppGpp is not clear. In Enterococcus faecalis, regulatory effects of pGpp synthesized by the Small Alarmone Synthetase have been investigated (Gaca et al., 2015). It would be worth investigating its binding kinetics to RNA polymerase too.
Most have explored only the possibility of differential regulation of transcription by ppGpp and pppGpp (Fig. 2). What determines their proportion in the cell remains an open question. Is it a simple percentage of their substrates/precursors or some other factor? Even more interesting would be the determinants of the stability of each of them inside the cell under different conditions? Are they completely interconvertible? What triggers the interconversion? Can pGpp form ppGpp and pppGpp? pppGpp can be hydrolyzed to ppGpp and pGpp or ppGpp to pGpp. If they have the same or similar function, then why interconversion? These are open questions that require extensive research (Syal et al., 2015a; Syal and Chatterji, 2018; Yang et al., 2020).
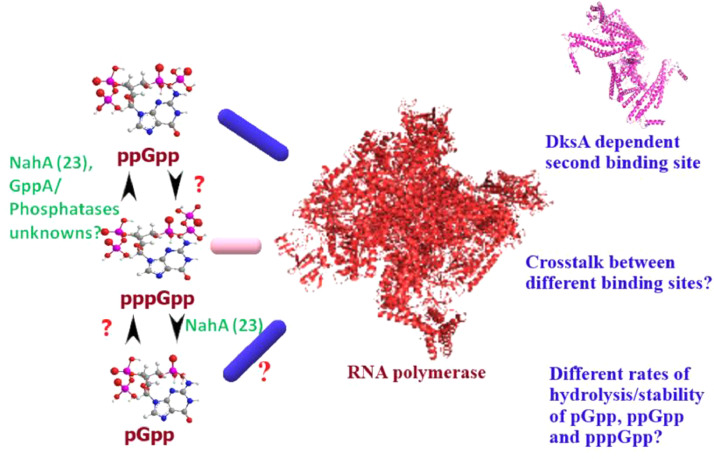
Fig. 2.
This schematic highlights cross-talk between ppGpp, pppGpp, and pGpp, and their corresponding binding sites with each other. How variable rates of hydrolysis and synthesis of pGpp, ppGpp and pppGpp regulates transcription is not well understood. Also, the dynamics of different binding sites and coordination with each other is incompletely understood (Yang et al., 2020).
(p)ppApp vs. (p)ppGpp
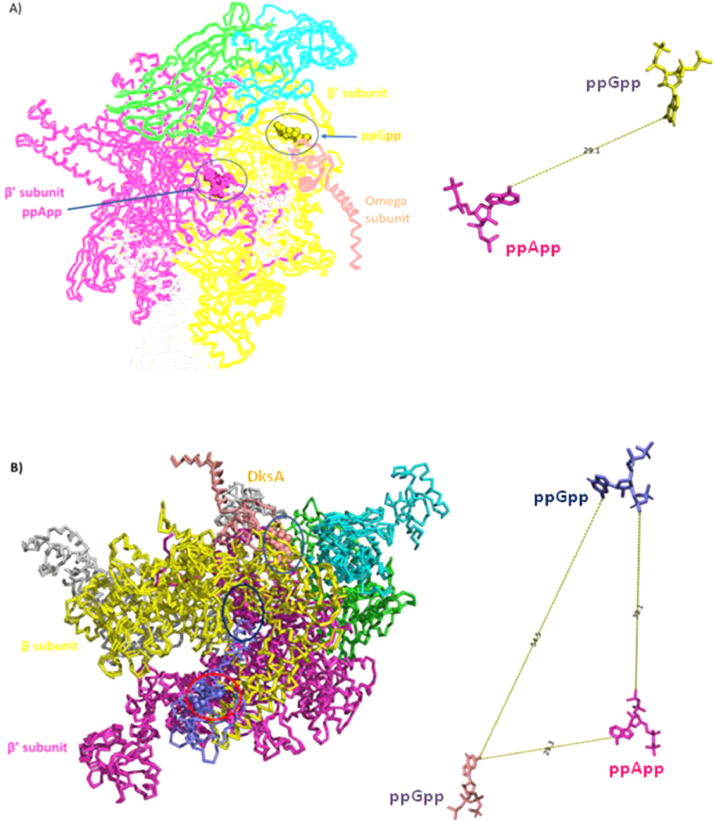
Earlier, (p)ppApp was observed in actinobacteria and B. subtilis (Oki et al., 1976), and it was showed to increase the transcription of rRNA gene in vitro (Travers, 1978). Recently, the crystal structure of Escherichia coli RNAP-(p)ppApp complex revealed the binding of ppApp near the active site. We have used PyMOL to overlay the structures of ppGpp-RNAP and ppApp-RNAP to have a visual understanding of the binding site of ppGpp and ppApp (Fig. 3). Bruhn-Olszewska et al. followed the regulatory effects of (p)ppApp on E. coli rrnB promoter. As reported earlier, unlike (p)ppGpp, (p)ppApp activated the transcription at rrnB promoter, and interestingly DksA opposed such activation. The authors observed that ppGpp and pppApp when present together, the resulting effects are conditional and depend on the sequence of incubation. However, the molecular mechanism of pppApp action on RNAP is still unknown (Bruhn-Olszewska et al., 2018).
Fig. 3.
PyMOL was used for overlaying of the following crystal structures available in PDB database. A) ppGpp-RNAP (Steitz et al., Mol Cell, 2013; PDB ID: 4JKR) and ppApp-RNAP (Potrykus et al., Biochim Biophys Acta, 2018) B) ppApp-RNAP (Potrykus et al., Biochim Biophys Acta, 2018; PDB ID: 6BYU) and ppGpp/DksA-RNAP (Murakami et al., Mol Cell, 2018; PDB ID: 5VSW).
In 2019, Ahmad et al. characterized Tas1 enzyme that codes for an antibacterial toxin in Pseudomonas aeruginosa. Though it is structurally similar to RSH enzymes but it makes (p)ppApp and not (p)ppGpp (Ahmad et al., 2019). Interestingly, type VI secretion system injects it into competing neighboring cells, where it rapidly utilizes ATP and converts it into (pp)pApp leading to the quenching of metabolic processes. (pp)pApp has been shown to directly bind to PurF thereby inhibiting purine biosynthesis (Ahmad et al., 2019). Like (p)ppGpp, too much of (pp)App is also toxic in hydrolase mutants, and it can be countered by corresponding hydrolase (Geiger et al., 2014). Do non-toxic accumulation of (p)ppApp helps in sporulation of B. subtilis cells is not understood and requires investigation.
Many RSH enzymes, including 30 small alarmone synthetase (SAS), have been shown to produce (pp)pApp. Five of these SASs have been shown to be an integral part of the bicistronic toxin–antitoxin-like operons (toxSAS) systems (Irving et al., 2021; Jimmy et al., 2020). (pp)pApp and (pp)pGpp have been realized as the vital constituent of genetic modules that involve toxin (that inhibits cell growth) and a cognate antitoxin (neutralizes the toxin or its effects). In E. coli, the RelP/Q group of SAS have been observed to be non-toxic in the presence of their cognate antitoxin. Upon expression of the SAS and in the absence of its cognate antitoxin, accumulation of ppApp and ppGpp lead to the inhibition of cell growth. The toxin activity can be controlled by an antitoxins that can either bind and inactivate the toxin (Type II TA) or possess hydrolase activity thus breaking the accumulated alarmone molecules (Irving et al., 2021).
pGpp- A new second messenger or a potential stress response switch
In 1976s, the pGpp molecule was observed in the B. subtilis and actinobacteria but its physiological relevance was not clear for very long (Nishino et al., 1979; Oki et al., 1976). Jade Wang group from UW-Madison reported pGpp as a third alarmone molecule in Bacillus subtilis. It is worth mentioning here that stress response in B. subtilis does not involve (p)ppGpp mediated regulation of transcription (Yang et al., 2020). Wang et al. performed a screening of potential binding partners of pGpp, ppGpp, and pppGpp. They observed that ppGpp and pppGpp interact with similar proteins involved in the same or overlapping pathways like purine biosynthesis pathways and GTPase. In contrast, pGpp interacted with the inhibitory cascades of purine nucleotide biosynthesis alone and not with GTPase responsible for the assembly of ribosomes. A NahA hydrolase was identified that could convert (p)ppGpp to pGpp and may have a potential role in fine-tuning the stringent response. Upon deletion of NahA, authors reported slow growth recovery phenotype and survival (Yang et al., 2020). pGpp synthesis has also been confirmed by RSH enzymes in S. aureus (Yang et al., 2019), M. smegmatis (Petchiappan et al., 2020) and E. coli (Sajish et al., 2009). In gram-negative bacteria, the mode of action of pGpp remains largely unclear. Earlier, Gaca et al. showed that pGpp downregulated the function of GTP biosynthesis in E. faecalis (Gaca et al., 2015). They also reported inhibition (though lesser than (p)ppGpp) of transcription of rrnB in E. coli. Interestingly, both pGpp and ppGpp activated the RelA synthetase activity. Unlike Wang group, Gaca et al. concluded that pGpp function like (p)ppGpp and has similar regulatory functions in stress response (Gaca et al., 2015).
Discussion
The first and second binding sites of (p)ppGpp on RNAP have been reported to be at the interface of beta’-omega and RNAP/DksA, respectively (Ross et al., 2016). With a thoughtful experiment, Gourse et al. followed the effects of (p)ppGpp on RNAP without nutrient starvation by RNA seq. They reported the list of 750 genes that are positively and negatively regulated by (p)ppGpp. However the promoter sequence preference of (p)ppGpp is still not well-understood (Sanchez-Vazquez et al., 2019). The targets of (p)ppGpp have expanded beyond RNA polymerase in E. coli (Zhang et al., 2018). As discussed before, in B. subtilis, purine nucleotide biosynthesis enzymes have been reported to be the primary target and not RNA polymerase (Kriel et al., 2012). p)ppGpp is a highly charged species with structural similarity to its precursor (GTP), which makes it capable of binding to many binding partners. The identification and characterization of the relevant interactions remain a challenge. Interestingly, DNA replication enzymes like primase have also been shown to be regulated by (p)ppGpp (Gourse and Keck, 2007). Evidently, (p)ppGpp regulates its own synthesis, and its analogs have been shown to inhibit ppGpp synthesis (Syal et al., 2017; Wexselblatt et al., 2012; Syal et al., 2015b). Though the required concentration of ppGpp analogues to inhibit (p)ppGpp synthesis is high, but these studies are the proof of concept that ppGpp analogues can be of therapeutic value. Interestingly, excess of ppGpp has also been shown to be toxic for the cell, making the (p)ppGpp hydrolase an attractive target (Kriel et al., 2012). In the light of the rapid emergence of antibiotic resistance in the last few decades, targeting the ppGpp mediated stress response, and associated pathways has become more appealing.
Additionally, attempts have been made to follow the cross-talk between c-di-GMP and ppGpp, if any. It should be noted that the reported concentrations of the two are extremely different, that is ppGpp shoots to the mM range in stress, but c-di-GMP never crosses the nano to micromolar range. We observed c-di-GMP binding affinity for Rel WT from M. smegmatis in the micromolar range (Syal et al., 2015a). Such high concentrations of c-di-GMP are difficult to achieve inside the cell. Still, it is possible that in invivo conditions, in a localized environment, or in the presence of some other factors, it may become significant. Also, the possibility of binding of ppGpp to the diguanylate cyclase is worth evaluating.
More research into domains of the interconnection of (pp)pApp and (pp)pGpp-mediated regulation is necessary for deciphering the network of nucleotide signaling cascades and their role in bacteria.
Declaration of Competing Interest
None to declare.
Acknowledgments
Acknowledgement
KS acknowledges Department of Biotechnology, Government of India for DBT-Ramalingaswamy Fellowship for funding.
Credit author statement
Conceptualization: KS, Resources: KS, Writing- Original Draft: KS Writing-Review and Editing: KS, NRS and MVNJR; Supervision: NRS and MVNJR are PhD students of KS.
References
- Ahmad S., Wang B., Walker M.D., Tran H.R., Stogios P.J., Savchenko A., Grant R.A., McArthur A.G., Laub M.T., Whitney J.C. An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature. 2019;575:674–678. doi: 10.1038/s41586-019-1735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Bhardwaj N., Syal K., Chatterji D. The role of omega-subunit of Escherichia coli RNA polymerase in stress response. Genes Cells. 2018;23:357–369. doi: 10.1111/gtc.12577. [DOI] [PubMed] [Google Scholar]
- Bruhn-Olszewska B., Molodtsov V., Sobala M., Dylewski M., Murakami K.S., Cashel M., Potrykus K. Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro. Biochim. Biophys. Acta Gene Regul. Mech. 2018;1861:731–742. doi: 10.1016/j.bbagrm.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M., Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Chatnaparat T., Li Z., Korban S.S., Zhao Y. The bacterial alarmone (p)ppGpp is required for virulence and controls cell size and survival of Pseudomonas syringae on plants. Environ. Microbiol. 2015;17:4253–4270. doi: 10.1111/1462-2920.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji D., Fujita N., Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Salazar C., Calero P., Espinosa-Portero R., Jimenez-Fernandez A., Wirebrand L., Velasco-Dominguez M.G., Lopez-Sanchez A., Shingler V., Govantes F. The stringent response promotes biofilm dispersal in Pseudomonas putida. Sci. Rep. 2017;7:18055. doi: 10.1038/s41598-017-18518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A.O., Kudrin P., Colomer-Winter C., Beljantseva J., Liu K., Anderson B., Wang J.D., Rejman D., Potrykus K., Cashel M., et al. From (p)ppGpp to (pp)pGpp: characterization of regulatory effects of pGpp synthesized by the small alarmone synthetase of Enterococcus faecalis. J. Bacteriol. 2015;197:2908–2919. doi: 10.1128/JB.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Kastle B., Gratani F.L., Goerke C., Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J. Bacteriol. 2014;196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R.E., Jones S.T., Ishihama A. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VII. RNA polymerase is a target for ppGpp. Mol. Gen. Genet. 1986;203:265–268. doi: 10.1007/BF00333964. [DOI] [PubMed] [Google Scholar]
- Gourse R.L., Keck J.L. Magic spots cast a spell on DNA primase. Cell. 2007;128:823–824. doi: 10.1016/j.cell.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V., Atkinson G.C., Murakami K.S., Tenson T., Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley C.L., Zhang X., Fortney K.R., Ellinger S., Johnson P., Baker B., Liu Y., Janowicz D.M., Katz B.P., Munson R.S., Jr., et al. DksA and (p)ppGpp have unique and overlapping contributions to Haemophilus ducreyi pathogenesis in humans. Infect. Immun. 2015;83:3281–3292. doi: 10.1128/IAI.00692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving SE, Choudhury NR, Corrigan RM. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 2021;19:256–271. doi: 10.1038/s41579-020-00470-y. [DOI] [PubMed] [Google Scholar]
- Jimmy S., Saha C.K., Kurata T., Stavropoulos C., Oliveira S.R.A., Koh A., Cepauskas A., Takada H., Rejman D., Tenson T., et al. A widespread toxin-antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10500–10510. doi: 10.1073/pnas.1916617117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., Go J., Lee K.M., Oh Y.T., Yoon S.S. Guanosine tetra- and pentaphosphate increase antibiotic tolerance by reducing reactive oxygen species production in Vibrio cholerae. J. Biol. Chem. 2018;293:5679–5694. doi: 10.1074/jbc.RA117.000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell. 2012;48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudrin P., Varik V., Oliveira S.R., Beljantseva J., Del Peso Santos T., Dzhygyr I., Rejman D., Cava F., Tenson T., Hauryliuk V. Subinhibitory concentrations of bacteriostatic antibiotics induce relA-dependent and relA-independent tolerance to beta-lactams. Antimicrob. Agents Chemother. 2017:61. doi: 10.1128/AAC.02173-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Xie F., Zhang Y., Bosse J.T., Langford P.R. Wang C: Role of (p)ppGpp in viability and biofilm formation of actinobacillus pleuropneumoniae S8. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Gallant J., Shalit P., Palmer L., Wehr T. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J. Bacteriol. 1979;140:671–679. doi: 10.1128/jb.140.2.671-679.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki T., Yoshimoto A., Ogasawara T., Sato S., Takamatsu A. Occurrence of pppApp-synthesizing activity in actinomycetes and isolation of purine nucleotide pyrophosphotransferase. Arch. Microbiol. 1976;107:183–187. doi: 10.1007/BF00446837. [DOI] [PubMed] [Google Scholar]
- Petchiappan A., Naik S.Y., Chatterji D. RelZ-mediated stress response in mycobacterium smegmatis: PGpp synthesis and its regulation. J. Bacteriol. 2020:202. doi: 10.1128/JB.00444-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulschen A.A., Sastre D.E., Machinandiarena F., Crotta Asis A., Albanesi D., de Mendoza D., Gueiros-Filho F.J. The stringent response plays a key role in Bacillus subtilis survival of fatty acid starvation. Mol. Microbiol. 2017;103:698–712. doi: 10.1111/mmi.13582. [DOI] [PubMed] [Google Scholar]
- Reddy P.S., Raghavan A., Chatterji D. Evidence for a ppGpp-binding site on Escherichia coli RNA polymerase: Proximity relationship with the rifampicin-binding domain. Mol. Microbiol. 1995;15:255–265. doi: 10.1111/j.1365-2958.1995.tb02240.x. [DOI] [PubMed] [Google Scholar]
- Roelofs K.G., Wang J., Sintim H.O., Lee V.T. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Sanchez-Vazquez P., Chen A.Y., Lee J.H., Burgos H.L., Gourse R.L. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell. 2016;62:811–823. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajish M., Kalayil S., Verma S.K., Nandicoori V.K., Prakash B. The significance of EXDD and RXKD motif conservation in Rel proteins. J. Biol. Chem. 2009;284:9115–9123. doi: 10.1074/jbc.M807187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vazquez P., Dewey C.N., Kitten N., Ross W., Gourse R.L. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8310–8319. doi: 10.1073/pnas.1819682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K., Bhardwaj N., Chatterji D. Vitamin C targets (p)ppGpp synthesis leading to stalling of long-term survival and biofilm formation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 2017:364. doi: 10.1093/femsle/fnw282. [DOI] [PubMed] [Google Scholar]
- Syal K., Chatterji D. Differential binding of ppGpp and pppGpp to E. coli RNA polymerase: Photo-labeling and mass spectral studies. Genes Cells. 2015;20:1006–1016. doi: 10.1111/gtc.12304. [DOI] [PubMed] [Google Scholar]
- Syal K., Chatterji D. Vitamin C: A natural inhibitor of cell wall functions and stress response in mycobacteria. Adv. Exp. Med. Biol. 2018;1112:321–332. doi: 10.1007/978-981-13-3065-0_22. [DOI] [PubMed] [Google Scholar]
- Syal K., Flentie K., Bhardwaj N., Maiti K., Jayaraman N., Stallings C.L., Chatterji D. Synthetic (p)ppGpp Analogue Is an Inhibitor of Stringent Response in Mycobacteria. Antimicrob. Agents Chemother. 2017:61. doi: 10.1128/AAC.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K., Maiti K., Naresh K., Chatterji D., Jayaraman N. Synthetic glycolipids and (p)ppGpp analogs: development of inhibitors for mycobacterial growth, biofilm and stringent response. Adv. Exp. Med. Biol. 2015;842:309–327. doi: 10.1007/978-3-319-11280-0_20. [DOI] [PubMed] [Google Scholar]
- Syal K., Joshi H., Chatterji D., Jain V. Novel pppGpp binding site at the C-terminal region of the Rel enzyme from Mycobacterium smegmatis. FEBS J. 2015;282:3773–3785. doi: 10.1111/febs.13373. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I.I., Shulgina I., Hernandez V.J. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta'-subunit. J. Biol. Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- Travers A.A. ppApp alters transcriptional selectivity of Escherichia coli RNA polymerase. FEBS Lett. 1978;94:345–348. doi: 10.1016/0014-5793(78)80973-9. [DOI] [PubMed] [Google Scholar]
- Vrentas C.E., Gaal T., Berkmen M.B., Rutherford S.T., Haugen S.P., Vassylyev D.G., Ross W., Gourse R.L. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O, Yavin E, Glaser G, Katzhendler J, Ben-Yehuda S. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Anderson B.W., Turdiev A., Turdiev H., Stevenson D.M., Amador-Noguez D., Lee V.T., Wang J.D. The nucleotide pGpp acts as a third alarmone in Bacillus, with functions distinct from those of (p)ppGpp. Nature Communications. 2020;11:5388. doi: 10.1038/s41467-020-19166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Xie S, Tang NY, Choi MY, Wang Y, Watt RM. The Ps and Qs of alarmone synthesis in Staphylococcus aureus. PLoS One. 2019;14 doi: 10.1371/journal.pone.0213630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zbornikova E., Rejman D., Gerdes K. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio. 2018:9. doi: 10.1128/mBio.02188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Wang Y., Steitz T.A. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]