Highlights
-
•
The most active archaea in the epimural community were different from that of the liquid and content-associated community, but the exact taxonomy requires further identification.
-
•
Rumen epithelial attached methanogens may not contribute to differences in CH4 production and variations in feed efficiency.
-
•
Families Campylobacteraceae and Neisseriaceae, which contain oxygen scavenging bacteria were significantly more active on the epithelium of efficient cattle.
Keywords: Beef cattle, Feed efficiency, Rumen epithelium, Mucosal associated microbiota, Total RNA sequencing, 16S rRNA transcripts
Abstract
To date, the role of ruminal epithelial attached microbiota in cattle feed efficiency is undefined. In this study, we aimed to characterize transcriptionally active bacteria and archaea attached to the rumen epithelial wall and to determine whether they differ in cattle with varied feed efficiency. RNA-sequencing was performed to obtain the rumen epithelial transcriptomes from 9 of the most efficient (low RFI) and 9 of the most inefficient (high RFI) animals. The bacteria and archaea 16S rRNA transcripts were identified using an in-house developed pipeline, enriched from filtered reads that did not map to the bovine genome. Archaea from unclassified genera belonging to the Euryarchaeota phylum showed the most activity on the rumen epithelium of low RFI (81.3 ± 1.9%) and high RFI (76.4 ± 3.0%) steers. Bacteria from the Succinivibrionaceae family showed the greatest activity of bacteria on the low RFI (28.7 ± 9.0%) and high RFI (33.9± 8.8%) epithelium. Of the bacterial families, Campylobacteraceae and Neisseriaceae had significantly greater activity on the low RFI epithelium (p < 0.05) and are known to play a role in oxygen scavenging. Greater activity of rumen epithelial attached oxygen scavenging bacteria may provide more optimal feed fermentation conditions, which contributes to high fermentation efficiency in the rumen.
1. Introduction
Ruminant animals, such as cattle, contain a large and diverse community of microorganisms within the rumen that consist of bacteria, archaea, protozoa, and fungi (Kamra, 2005). The rumen microbes are often classified into three populations based on their localization: the rumen fluid-associated microbes, the feed particle-associated microbes, and the rumen epithelial wall-associated microbes (McCowan et al., 1978; Olubobokun and Craig, 1990). Among these three fractions, the functions of the liquid-associated and feed particle-associated microbes have been extensively studied. The microbes of these two fractions play a major role in feed degradation and fermentation, which generates a large supply of substrates for the host to utilize (Bergman, 1990). The end products of fermentation, such as hydrogen and carbon dioxide, are converted to methane (CH4) by methanogenic archaea that belong to the phylum Euryarchaeota (Liu and Whitman, 2008; Morgavi et al., 2010).
Differences in the composition and activity of the rumen microorganisms may contribute to variation in host feed efficiency through their influence on feed digestion, fermentation, and CH4 production. The feed efficiency of an individual can be measured by residual feed intake (RFI), which is calculated as the difference between actual feed intake and expected feed requirements for growth and maintenance (Basarab et al., 2003). Animals with low RFI (L-RFI) are considered feed efficient while those with high RFI (H-RFI) are deemed feed inefficient. Feed efficient cattle produce approximately 25% less CH4 than inefficient cattle (Hegarty et al., 2007) and consume 3.77 kg less feed per day for similar growth and body weight (Basarab et al., 2003). Studies of methanogens within the rumen fluid of cattle differing in feed efficiency, have found that the abundance of total methanogens were not different between RFI groups (Zhou et al., 2009; Zhou et al., 2010). However, the feed inefficient individuals had a more diverse methanogenic community and greater abundance of Methanosphaera stadtmanae and Methanobrevibacter sp. Strain AbM4 compared to efficient animals (Zhou et al., 2009). When the ruminal fluid bacterial community was assessed, it was discovered that the abundance of particular bacterial phylotypes were associated with feed efficiency (Hernandez-Sanabria et al., 2012). In a latter study comparing the active feed-associated microbiota between L- and H-RFI steers, it was found that both the bacterial community and the archaeal community was more active in H-RFI steers compared to that of L-RFI steers (Li et al., 2017). Such variation was proposed to be one of the driving forces contributing to the variation of host feed efficiency.
To date, the previous findings have been solely focused on rumen digesta associated microbiota; and there have been no studies examining the rumen epithelial attached microbes (epimural community) in relation to feed efficiency. It has been found that the predominant bacteria identified on the epithelium belong to the phylum Firmicutes (Li et al., 2012). Characterization of epimural bacteria have found they have specific roles in the rumen such as; mucosal protection, urea hydrolysis, epithelial recycling, and oxygen scavenging (Holovska et al., 2002; Cheng and Wallace, 1979; McCowan et al., 1978; Cheng et al., 1979). The hydrolysis of urea from the rumen wall by ureolytic bacteria provides ammonia for microbial protein synthesis, which is a major source of protein for the host. Oxygen scavenging by facultative anaerobes on the epithelium removes oxygen, thereby protecting obligate anaerobes and maintaining optimal fermentation conditions inside the rumen.
Compared to the knowledge of liquid and feed-associated microbiota, that of the rumen epimural microbiota is limited. Chen et al. (2011) reported that the epimural bacteria Treponema sp., Ruminobacter sp. and Lachnospiraceae sp. were associated with subacute ruminal acidosis (SARA) in Holstein dairy cows. Petri et al. (2013) found that in beef cattle, epimural bacteria Atopobium, cc142, Lactobacillus, Olsenella, RC39, Sharpea, Solobacterium, Succiniclasticum and Syntrophococcus were associated with rumen acidosis. These studies suggest that both the epimural microbiota and the gene expression within host cells play important roles in maintaining rumen conditions that are more favorable for fermentation. Based on the functions of these microbiota, we speculate that the rumen epithelial attached bacteria could also influence cattle feed efficiency. The objective of this study was to investigate the differences in activity of rumen epithelial attached bacteria and archaea between low and high feed efficient beef cattle. While the previous studies mainly examined DNA samples, the activities of the epimural microbiota have not been examined. Therefore, the RNA level analysis was performed to assess the active epimural microbiota, where the relative abundance of bacterial and archaeal 16S rRNA transcripts obtained from RNA-sequencing (RNA-seq) was used to measure the potential activity of the epimural microbial community.
2. Materials and methods
2.1. Animals and rumen tissue collection
All experimental procedures were approved by the University of Alberta Animal Care and Use Committee for Livestock (Moore-2006-55) and animals were raised in accordance with the Canadian Council of Animal Care guidelines (CCAC, 1993). At 10 months old, 175 Hereford x Aberdeen Angus hybrid steers at the University of Alberta Roy Berg Kinsella Research Station (Alberta, Canada) were placed under feedlot conditions as described by Kong et al. (2016). Briefly, the growing diet was fed for 90 days and steers had a one-week adaptation period before starting a 90-day period on a finishing diet. Feed intake data was collected throughout the feeding trial using the GrowSafe automated feeding system (GrowSafe Systems Ltd., Airdrie, Alberta, Canada) and the RFI after the finishing diet was used to classify steers as L-RFI (feed efficient; RFI < -0.5), medium RFI (M-RFI; -0.5 ≤ RFI ≤ 0.5), or H-RFI (feed inefficient; RFI > 0.5) (Nkrumah et al., 2006). Steers were humanely euthanized after the feeding trial and rumen tissue (4-cm2) was collected from the central region of the ventral sac. The tissue samples were processed following Chen et al. (2011). Briefly, the tissue was rinsed with sterile phosphate-buffered saline (PBS, pH = 6.8) before being placed in RNAlater solution (Invitrogen, Carlsbad, CA) and stored at -80 °C until further processing. After thawing the rumen tissue, papillae (∼80 mg) were obtained using sterile scissors and scalpels. RNA extraction and sequencing were performed on the rumen papillae from nine extreme L-RFI (RFI = -1.40 to -2.33 kg/day) and nine extreme H-RFI (RFI = 1.32 to 3.23 kg/day) steers.
2.2. RNA extraction and sequencing
The mirVana kit (Ambion, Austin, TX) was used to isolate total RNA from rumen papillae (∼80 mg) according to the manufacturer's instructions. Then the RNA integrity, concentration, and purity of total RNA were measured using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and Nanodrop 2000 c spectrophotometer (Thermo Scientific, Wilmington, DE).
Rumen tissue samples of the top ten and bottom ten RFI animals were used for RNA extraction. And only 18 samples (N=9 for each RFI group) with an RNA integrity number greater than 7 were prepared for total RNA sequencing using the TruSeq RNA Sample Preparation v2 kit (Illumina, San Diego, CA) as previously reported (Kong et al., 2016). Briefly, the total RNA was directly subjected to cDNA synthesis without mRNA enrichment step, with the aim to identify host and microbial transcripts simultaneously. The resulting double-stranded cDNA was then subjected to end repair and 3’-end adenylation before ligation of index adapters. Polymerase chain reaction (PCR) (15 cycles) was used to enrich cDNA fragments that have an adapter attached and amplify the cDNA libraries. The cDNA libraries were then validated and quantified with the Agilent 2200 TapeStation (Agilent Technologies) and Qubit fluorometer (Invitrogen, Carlsbad, CA), respectively. Lastly, the libraries were sequenced on the Illumina HiSeq 2000 system at the McGill University and Génome Québec Innovation Centre (Québec, Canada) to obtain high quality paired-end reads (2 × 100 bp; Average quality score ≥ 33). The raw sequencing data have been deposited at publicly available NCBI's Gene Expression Omnibus Database with GEO Series accession number GSE76501 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76501).
2.3. Identification and quantification of bacterial and archaeal 16S rRNA transcripts
There was a total of 41,991,418 ± 6,835,817 (mean ± SD) high quality, paired reads (i.e. transcripts) generated per sample from RNA-seq that were first aligned to the bovine reference genome, UMD3.1 (Ensembl v83.31) using the software packages TopHat2 (v2.0.9; Kim et al., 2013) and Bowtie2 (v2.1.0; Langmead and Salzberg, 2012). The overall read alignment rate to the bovine reference genome was 85 ± 3.72 % and the transcripts were analyzed by Kong et al. (2016). Forward and reverse reads that could not be aligned to the bovine genome (12,905,961 ± 4,882,393 (mean ± SD)) were combined to increase the length and then input to SortMeRNA (v1.99-beta; Kopylova et al., 2012) to obtain bacterial and archaeal 16S rRNA reads. The joined 16S rRNA sequences (2,687,989 ± 1,350,507 (mean ± SD)) were then input into the Mothur program (Schloss et al., 2009) and analyzed using an in-house developed pipeline (Li et al., 2016). As the transcripts could be mapped to different regions of 16S rRNA genes which lead to the over-estimating of the microbes, we used an in-house built regional enriched references for taxonomic classification. Briefly, archaeal 16S rRNA sequences were mapped to an in-house reference based on the V6-V8 regions-enriched rumen-specific archaea database (Janssen and Kirs, 2008) with addition of newly identified phylotypes, and bacterial 16S rRNA sequences were mapped the V1-V3 region-enriched Greengenes database (version gg_13_5_99; DeSantis et al., 2006). De novo chimera detection was conducted using UCHIME (Edgar, 2010) with default settings, and the taxonomic assignment of chimera-removed sequences was performed using the naive Bayesian method with a minimum confidence of 0.8. Bacteria were only classified down to the family level, while archaea were classified to the genus level because the sequences (96-200 bp) were too short to be reliably classified at the lower taxonomic levels. Three animals from each of the RFI groups were removed from further analyses due to their low number of transcripts detected for bacteria and archaea. The proportion of 16S rRNA transcripts for every bacterial and archaeal phylotype was determined for every sample and the average proportion for each phylotype was calculated for each RFI group. Only phylotypes with an average abundance of 16S rRNA transcripts ≥ 0.5% in at least one RFI group are described in this study. After filtering low abundance phylotypes, Kruskal-Wallis Test was used to determine statistical significance (FDR adjusted p < 0.05) between RFI groups in the relative abundance of 16S rRNA transcripts for each remaining phylotype.
3. Results & Discussion
3.1. Identification of microbial transcriptional activity using RNA-seq method
The commonly used DNA-based methods for examining the epimural microbiota can only reveal the composition of the microbial community and not the microbial activity. Therefore, this study examined the epithelial attached microbes using RNA-seq, allowing for the measurement of the transcriptional activity of microbes in relation to feed efficiency. This adds more knowledge about the rumen microbiota in addition to previous studies mainly focused on the rumen liquid-associated and particle-associated microbiota.
Both the archaeal and bacterial community were identified through an RNA-seq approach. There were 59,166 ± 35,148 (mean ± SD) bacterial 16S rRNA transcripts (ranged from 17,293 to 96,379) and 31,999 ± 20,416 (mean ± SD) archaeal 16S rRNA transcripts (ranged from 10,245 to 84,639) identified per epithelial sample (n = 6 per group). The percentage of the microbial rRNA transcripts of each sample (15–25%) was comparable with Mann et al. (2018) (2.33–7.74%), where a similar approach was also applied to reveal the active microbial community of the rumen epithelium. The good's coverage of the obtained microbial transcripts of each sample was above 0.90, indicating the assigned transcripts were sufficient for downstream analyses. It should be noted that a very strict filtering criteria (good's coverage > 0.90) was applied to ensure the obtained data can fully represent the microbial community, therefore six samples (10,058 ± 4,681 bacterial transcripts with range 3,683 to 14,120; 2,460 ± 1,088 archaeal transcripts with range 1,255 to 4,334) were removed from downstream analyses due to their low good's coverage. As the samples with lower coverage may lead to biased taxonomy assignments and further influence downstream statistical analyses, they were therefore excluded.
3.2. Identification and quantification of bacterial 16S rRNA transcripts
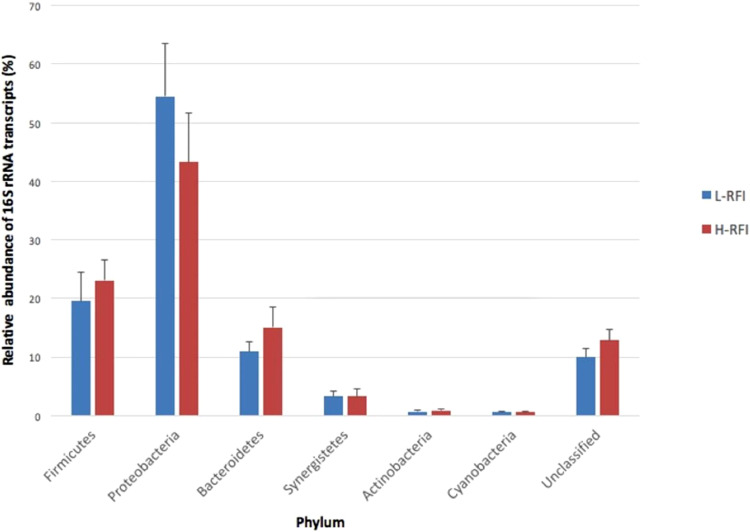
There were no significant differences in the relative abundance of transcripts between the RFI groups (adjusted p > 0.1) for each phylum. Based on the previous studies using DNA-based methods, we hypothesized that the activity of rumen epithelial bacteria from the phylum Firmicutes would be the highest since Firmicutes was identified as the predominant bacterial phylum on the epithelium (Li et al., 2012). Surprisingly, the Proteobacteria phylum had the most abundant 16S rRNA transcripts in both RFI groups followed by Firmicutes and Bacteroidetes (Fig. 1). There was a higher relative abundance of transcripts from Proteobacteria on the L-RFI epithelium (54.5 ± 9.0% (mean ± SEM)) than on the H-RFI epithelium (43.3 ± 8.4%) (adjusted p < 0.1). Other phyla detected were Firmicutes, Bacteroidetes, Synergistetes, Actinobacteria, and Cyanobacteria (Fig. 1). This suggests that although Firmicutes had the highest abundance on the epithelium, it is not necessary that they have the highest activity (at RNA level). In the study by Kang et al. (2013), it was also reported that the activity of Proteobacteria in the rumen was higher than Firmicutes even though Proteobacteria had lower abundance. Although a harsh cell lysis technique was included in the current study, the predominance of Proteobacteria of the epimural tissue may be because they are more easily be lysed compared to Firmicutes (Yuan et al., 2012). The currently available databases contain far more versions of proteobacteria reads compared to other bacterial phyla. As there were large numbers of unidentified reads, they may belong to Firmicutes or Bacteroidetes or other phyla, yet there is no supporting evidence due to the lack of more in-depth reference databases. However, in our previous study where three microbial profiling methods were compared for the same samples, RNA-seq and RNA amplicon-seq did reflected a Proteobacteria predominant community while DNA amplicon-seq showed a Bateroidetes predominant community (Li et al., 2016), which may further support our results that although Proteobateria may not be numerically predominant, they are the most active microbial group within the rumen epithelium. Petri et al. (2020) reported that the rumen epimural microbiota may be affected by the different feed additives offered in animal feed. A recent study by Anderson et al. (2021) claimed that the variation in rumen epimural microbiota can be attributed to multiple factors such as different host species, geographic region, diet, age, farm management practice, time of year, and hypervariable region sequenced, yet a core community was found across studies. Therefore, we speculated that the variation in epimural bacteria between the current study and the other studies may also be due to these multiple factors.
Fig. 1.
The relative abundance of 16S rRNA transcripts belonging to each bacterial phylum on the rumen epithelium of L-RFI (n = 6; blue) and H-RFI (n = 6; red) beef steers. Relative abundance is given as a percentage and data are presented as mean ± SEM (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
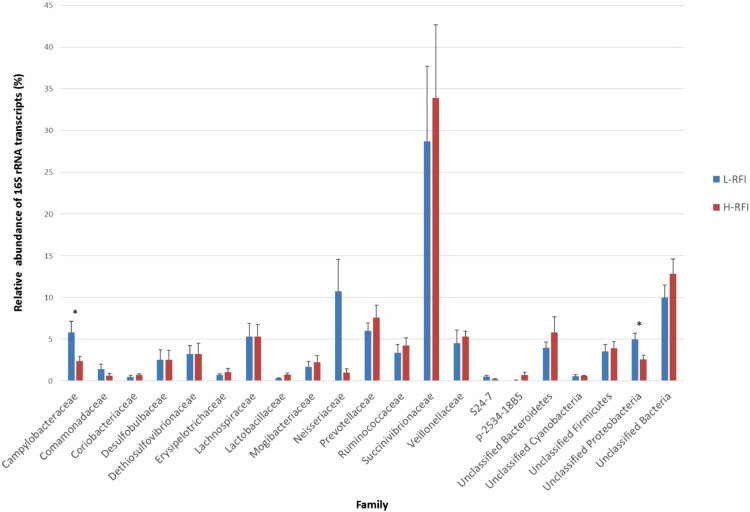
When the bacterial 16S rRNA transcripts were classified to the family level, the identified families included Campylobacteraceae, Comamonadaceae, Coriobacteriaceae, Desulfobulbaceae, Dethiosulfovibrionaceae, Erysipelotrichaceae, Lachnospiraceae, Lactobacillaceae, Mogibacteriaceae, Neisseriaceae, Prevotellaceae, Ruminococcaceae, Succinivibrionaceae, Veillonellaceae, S24-7, p-2534-18B5, unclassified Bacteroidetes, unclassified Cyanobacteria, unclassified Firmicutes, unclassified Proteobacteria, and families that could not be classified below kingdom bacteria (Fig. 2). Succinivibrionaceae, which belongs to the Proteobacteria phylum, had the highest relative abundance of bacterial transcripts on both the L-RFI (28.7 ± 9.0%) and H-RFI (33.9 ± 8.8%) epithelium. Although Succinivibrionaceae transcripts were numerically higher in the H-RFI group, it was not statistically significant (adjusted p > 0.1). The Succinivibrionaceae family contains the genera Anerobiospirillum, Ruminobacter, Succinimonas, and Succinivibrio (Stackebrant and Hespell, 2006). It is known that Succinivibrio dextrinosolvens has urease and other nitrogen assimilation enzymes such as glutamine synthetase and glutamate (Patterson and Hespell, 1985). Although the Succinivibrionaceae family contains ureolytic species to convert urea to ammonia, the activity of this family does not significantly differ between RFI groups. However, analysis at the genus or species level is needed to examine whether there are differences in the activity of urease producers between L- and H- RFI cattle.
Fig. 2.
The relative abundance of 16S rRNA transcripts belonging to each bacterial family on the rumen epithelium of L-RFI (n = 6; blue) and H-RFI (n = 6; red) beef steers. Relative abundance is given as a percentage and data are presented as mean ± SEM (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Of the bacterial families, Campylobacteraceae, Neisseriaceae, and unclassified Proteobacteria had significantly greater transcripts on the L-RFI epithelium (adjusted p < 0.05) than on the H-RFI epithelium. The L-RFI group had 5.8 ± 1.3% of transcripts from Campylobacteraceae, while the H-RFI group had 2.4 ± 0.5%. Neisseriaceae contributed 10.8 ± 3.8% and 1.0 ± 0.4% of total transcripts on the L- and H- RFI epithelium, respectively. Unclassified Proteobacteria had 5.0 ± 0.8% transcript abundance in the L-RFI group and 2.6 ± 0.5% in the H-RFI group. Both Campylobacteraceae and Neisseriaceae are Proteobacteria. Campylobacteraceae contains the genera Campylobacter and Arcobacter, which are known to colonize mucosal surfaces (Lastovica et al., 2014). Most Campylobacter species have oxidase activity that catalyzes the reduction of oxygen to water (Lastovica et al., 2014). Neisseriaceae, which contains the genus Neisseria, are also known to colonize mucosal surfaces and have oxidase activity (Jurtshuk and Milligan, 1974). Because the Campylobacteraceae and Neisseriaceae families have oxygen scavenging functions and have significantly higher activity in L-RFI steers, our data suggests that L-RFI animals have better oxygen removal from the rumen and maintenance of the anaerobic condition for fermentation. Optimal fermentation conditions result in effective feed fermentation and may contribute to high feed efficiency by providing an increased supply of energetic substrates for the host to utilize as energy. It is known that the rumen digesta and liquid-associated microbes are obligate anaerobic organisms, any trace amount of oxygen may influence their fermentation capacity within the rumen. The higher activity of oxygen scavenging microbes in the rumen epithelium of L-RFI steers may therefore ensure the strict anaerobic condition in the rumen, thus the content and liquid-associated microbes can be more effective in digesting the feed.
Furthermore, there was significantly lower transcript abundance from Lactobacillaceae in the L-RFI group (0.3% ± 0.0%) compared to the H-RFI group (0.8 ± 0.2%) (adjusted p < 0.05). Lactobacillaceae belongs to the Firmicutes phylum, and it includes species that metabolize sugars to lactic acid, which lowers the pH of their environment. This suggests that the H-RFI epithelium may have a greater localized production of lactic acid, which may damage the rumen epithelium and have a negative effect on feed efficiency by decreasing absorptive capacity. In this study, however, we did not check the whole rumen epithelium for damage.
3.3. Identification and quantification of archaeal 16S rRNA transcripts
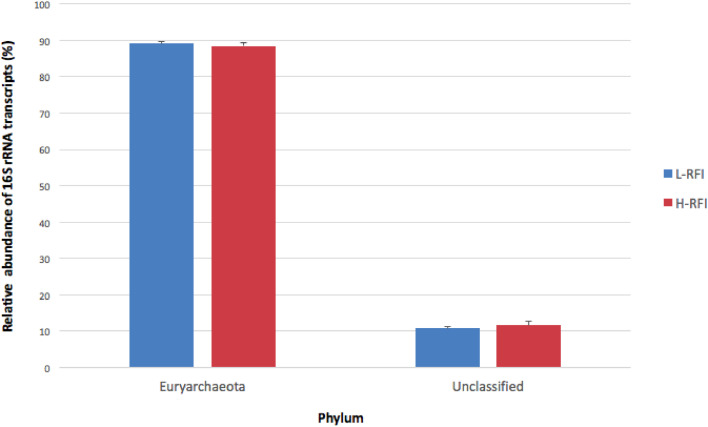
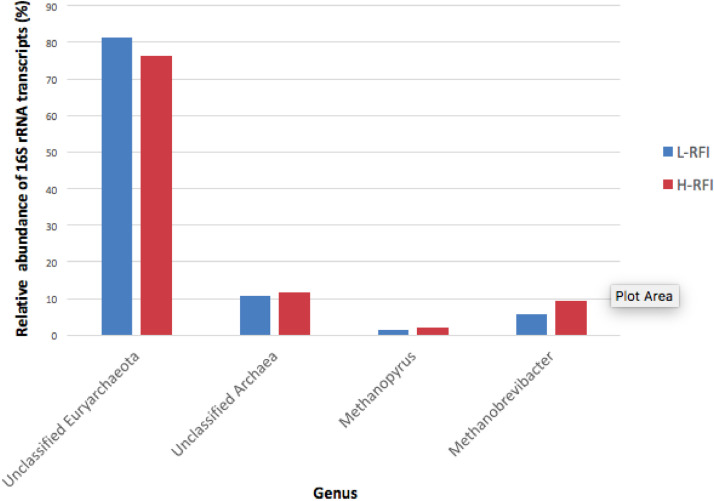
At the phylum level, Euryarchaeota and an unclassified phylum were detected from the transcripts (Fig. 3). Euryarchaeota contributed to 89.2 ± 0.6% (mean ± SEM) and 88.4 ± 1.0% of the relative abundance of archaeal 16S rRNA transcripts from the epithelial tissue of the L- and H- RFI groups, respectively. The archaeal genera consisted of Methanopyrus, Methanobrevibacter, unclassified genera from the phylum Euryarchaeota, and genera that could not be classified at any taxonomic level below kingdom archaea (Fig. 4). The majority of the archaeal 16S rRNA transcripts were from genera that were unclassified Euryarchaeota. They contributed 81.3 ± 1.9% and 76.4% ± 3.0% of total archaeal transcripts from the L- and H- RFI epitheliums, respectively. There was also a high relative abundance of transcripts from the archaeal genera that could not be classified below kingdom and from Methanobrevibacter. The unclassified archaea had 10.8 ± 0.6% relative transcript abundance on the L-RFI epithelium and 11.6 ± 1.0% on the H-RFI epithelium. Methanobrevibacter relative transcript abundance was lower on the L-RFI epithelium (5.8 ± 1.5%) compared to on the H-RFI epithelium (9.3 ± 2.8%). However, there was no significant difference found between the RFI groups in the relative abundance of 16S rRNA transcripts for any of the archaeal phylotypes (adjusted p > 0.1).
Fig. 3.
The relative abundance of 16S rRNA transcripts belonging to each archaeal phylum on the rumen epithelium of L-RFI (n = 6; blue) and H-RFI (n = 6; red) beef steers. Relative abundance is given as a percentage and data are presented as mean ± SEM (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Fig. 4.
The relative abundance of 16S rRNA transcripts belonging to each archaeal genus on the rumen epithelium of L-RFI (n = 6; blue) and H-RFI (n = 6; red) beef steers. Relative abundance is given as a percentage and data are presented as mean ± SEM (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Based on the previous studies, Archaea belonging to Methanobrevibacter was the most predominant in the liquid and solid phase of rumen microbiota, we hypothesized that this genus of archaea is also the most active in the rumen epimural archaeal community.
It has been found that the most abundant rumen epimural methanogens, as characterized by 16S rRNA gene sequencing (DNA based method), are from the order Methanobacteriales and the genus Methanobrevibacter (Pei et al., 2010; Seedorf et al., 2015). The active archaeal community represented by the transcripts obtained in the current study differed significantly as compared to the previous studies. Such variation between DNA-level and RNA-level assessment suggests that while Methanobrevibacter are highly abundant at the DNA level, they are not the most transcriptionally active archaeal genus on the epithelium. Methanogens from unclassified genera of Euryarchaeota might be highly active on the epithelium, indicating that they may be responsible for a majority of the CH4 production on the epithelium and that methanogenesis may be a major role of epimural archaea. However, their abundance is low compared to their activity. It has been reported that only 25.5% of epithelial associated methanogens are unidentified Euryarchaeota based on a clone library method (Pei et al., 2010). Other than methanogens, the unclassified genera belonging to Euryarchaeota may also possibly include halobacteria and thermophilic archaea (Gao and Gupta, 2007). The high proportion of unclassified reads in RNA-seq datasets signify the advanced capability of RNA-seq to detect the archaeal community on the epithelium. However, a rumen-specific archaea reference with the most updated data included is necessary for better taxonomically describe the unclassified reads. Further pure culture-based studies need to be conducted to identify the unclassified Euryarchaeota to determine which methanogenic genus is the most active on the epithelium and to study its functions. Additionally, we found that archaea that could not be classified below kingdom level (unclassified archaea) also seem to be quite active on the epithelium (8.2% - 14.2% relative activity), which may have roles other than in methanogenesis. This is the first study detecting a phylum other than Euryarchaeota and Crenarcheota in the rumen (Shin et al., 2004; Janssen and Kirs, 2008). One of the limitations of the current study is that owing to the lack of well-annotated rumen-specific archaeal databases, many of the archaeal reads were not able to be assigned to known taxa, making it difficult to clearly address how epimural archaeal community contribute to host RFI. Although the Rumen and Intestinal Methanogen-DB (RIM-DB) developed by Seedorf et al. (2014) may allow better taxa assignment, it has not been updated recently. Therefore, we have adopted the V6-V8 regions-enriched database published by Janssen and Kris (2008) and added newly identified phylotypes as the reference used in this study, aiming to allow a wider coverage of the reads. However, as shown by the data, such amendment is still far from sufficient to provide a clear overview of the archaeal community as our reads were very short and may not overlap or only with a few bp overlap between each other even if they were all from the same regions (V6-V8 regions for archaea). As such, it is necessary to include newly identified rumen methanogen phylotypes into this reference (e.g. an updated version of RIM-DB) so that to allow more precise taxa assignment. Using other marker genes (e.g. mcrA gene) for archaeal taxa assignment may be an alternate method to increase the resolution of the archaeal community. However, as archaea/bacteria ratio of 16S rRNA gene was proposed to be associated with methane emissions (Tapio et al., 2017), examining 16S rRNA gene allows further analyses with different purposes. Future work to improve the coverage of the rumen archaea reference database and to enhance sequencing depth to achieve higher archaeal reads may be helpful to identify the unclassified archaeal phylum to determine how they contribute to the functions of rumen epithelium.
Moreover, we had predicted there would be lower methanogen activity on the L-RFI epithelium since L-RFI steers produce less CH4 than H-RFI steers, however, we found no significant difference between the L- and H- RFI epithelium in the relative activity of any of the archaeal phylotypes. This suggests that the difference in CH4 production between the RFI groups may be predominantly due to differences in the activity of the methanogens in the rumen content. Therefore, studying the activity of methanogens associated with rumen content may be a superior indicator of methanogenesis.
3.4. Advantage of utilizing RNA-seq for microbial profiling
RNA-seq has been recently adopted for a wide range of studies, such as identifying the microbial community of the rumen epithelial microbiota (Mann et al., 2018), assessing the microbiota in human blood (Loohuis et al., 2018) and measuring the active microbes in soil (Bang-Andreasen et al., 2020). As shown in these studies, RNA-seq can be utilized as an alternative tool for microbial profiling. Since both the active microbiota and all other transcripts of the sample are assessed simultaneously, the obtained microbial data can be directly compared with transcripts of the host species. In the current study, although 100bp PE sequencing platform was used when the experiment was performed, and the reads were shorter compared to the recent sequencing platforms (250bp/300bp PE), all the obtained reads were assembled to contigs prior to processing. Therefore, the taxa assignment was based on the contigs which were long enough to cover multiple variable regions of the 16S rRNA gene of bacteria and archaea. The microbial data obtained in this study, together with our previous findings on host transcriptome of the same tissue samples (Kong et al. 2016) will allow us to perform downstream analyses identifying the host-microbial interactions in the rumen epithelium.
4. Conclusions
Approximately 80% of the archaeal activity on the rumen epithelium was from unclassified genera from phylum Euryarchaeota, which emphasizes the need for further studies to classify unknown rumen methanogens. Our study showed that the relative activity of archaeal phylotypes on the epithelium was not significantly different between L- and H- RFI steers, which suggests that rumen epithelial attached methanogens may not contribute to differences in CH4 production and variations in feed efficiency. The most transcriptionally active epithelial attached bacteria were from the phylum Proteobacteria, which was unexpected as Proteobacteria DNA abundance is not as high as Firmicutes on the epithelium (Li et al., 2012). There was no significant difference between RFI groups in the activity of ureolytic bacterial phylotypes; however, we found that the families Campylobacteraceae and Neisseriaceae, which contain oxygen scavenging bacteria were significantly more active on the L-RFI epithelium. Therefore, L-RFI steers may have more optimal rumen fermentation conditions for feed fermentation and generation of energetic substrates for energy production by the host.
CRediT authorship contribution statement
Rebecca S.G. Tan: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Mi Zhou: Data curation, Writing – original draft, Methodology, Writing – review & editing. Fuyong Li: Formal analysis, Methodology, Writing – review & editing. Le Luo Guan: Conceptualization, Resources, Software, Supervision, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Alberta Livestock and Meat Agency Ltd. (Edmonton, Canada; Project No.: 2015P008R), Alberta Agriculture and Forestry (Edmonton, Canada; Project No.: 2018F095R), and NSERC discovery grant funded this research. RK was also partly supported by the Queen Elizabeth II Graduate Scholarship.
Reference
- Anderson C.J., Koester L.R., Schmitz-Esser S. Rumen epithelial communities share a core bacterial microbiota: a meta-analysis of 16S rRNA gene Illumina MiSeq sequencing datasets. Front. Microbiol. 2021 doi: 10.3389/fmicb.2021.625400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang-Andreasen T., Anwar M.Z., Lanzen A., Kjoller R., Ronn R., Ekelund F., et al. Total RNA sequencing reveals multilevel microbial community changes and functional responses to wood ash application in agricultural and forest soil. FEMS Microbiol. Ecol. 2020;96:fiaa016. doi: 10.1093/femsec/fiaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basarab J.A., Price M.A., Aalhus J.L., Okine E.K., Snelling W.M., Lyle K.L. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 2003;83:189–204. [Google Scholar]
- Bergman E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Canadian Council on Animal Care (CCAC) In: Guide to the Care and Use of Experimental Animals. Olfert E.D., Cross B.M., McWilliams A.A., editors. CCAC; 1993. [Google Scholar]
- Chen Y., Penner G.B., Li M., Oba M., Guan L.L. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 2011;77:5770–5781. doi: 10.1128/AEM.00375-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.J., Wallace R.J. The mechanism of passage of endogenous urea through the rumen wall and the role of ureolytic epithelial bacteria in the urea flux. Brit. J. Nutr. 1979;42:553–557. doi: 10.1079/bjn19790147. [DOI] [PubMed] [Google Scholar]
- Cheng K.J., McCowan R.P., Costerton J.W. Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am. J. Clin. Nutr. 1979;32:139–148. doi: 10.1093/ajcn/32.1.139. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., et al. Greengenes, a chimera-checked 16s rrna gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Guo B., Gupta R.S. Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genom. 2007;8:86. doi: 10.1186/1471-2164-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty R.S., Goopy J.P., Herd R.M., McCorkell B. Cattle selected for lower residual feed intake have reduced daily methane production. J. Anim. Sci. 2007;85:1479–1486. doi: 10.2527/jas.2006-236. [DOI] [PubMed] [Google Scholar]
- Hernandez-Sanabria E., Goonewardene L.A., Wang Z., Durunna O.N., Moore S.S., Guan L.L. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 2012;78:1203–1214. doi: 10.1128/AEM.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holovska K., Lenartova V., Holovska K., Javorsky P. Characterization of superoxide dismutase in the rumen bacterium Streptococcus bovis. Vet. Med. 2002;47:38–44. [Google Scholar]
- Janssen P.H., Kirs M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008;74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtshuk P., Milligan T.W. Quantitation of the tetramethyl-p-phenylenediamine oxidase reaction in Neisseria species. Appl. Microbiol. 1974;28:1079–1081. doi: 10.1128/am.28.6.1079-1081.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamra D.N. Rumen microbial ecosystem. Curr. Sci. 2005;89:124–135. [Google Scholar]
- Kang S.H., Evans P., Morrison M., McSweeney C. Identification of metabolically active proteobacterial and archaeal communities in the rumen by DNA- and RNA- derived 16S rRNA gene. J. Appl. Microbiol. 2013;115:644–653. doi: 10.1111/jam.12270. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriotomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R.S.G., Liang G., Chen Y., Stothard P., Guan L.L. Transcriptome profiling of the rumen epithelium of beef cattle differing in residual feed intake. BMC Genom. 2016;17:592. doi: 10.1186/s12864-016-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E., Noe L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinform. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastovica A.J., On S.L.W., Zhang L. In: The Prokaryotes. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. Springer; 2014. The family Campylobacteraceae; pp. 307–335. [Google Scholar]
- Li F, Guan L.L. Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle. Applied and Environmental Microbiology. 2017;83(9) doi: 10.1128/AEM.00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Henderson G., Sun X., Cox F., Janssen P.H., Guan L.L. Taxonomic assessment of rumen microbiota using total RNA and targeted amplicon sequencing approaches. Front. Microbiol. 2016;7:987. doi: 10.3389/fmicb.2016.00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhou M., Adamowicz E., Basarab J.A., Guan L.L. Characterization of bovine ruminal epithelial bacterial communities using 16S rRNA sequencing, PCR-DGGE, and qRT-PCR analysis. Vet. Microbiol. 2012;155:72–80. doi: 10.1016/j.vetmic.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Liu Y., Whitman W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- Loohuis L.M.O., Mangul S., Ori A.P.S., Jospin G., Koslicki D., Yang H.T., Wu T., et al. Transcriptome analysis in whole blood reveals increased microbial diversity in schizophrenia. Trans. Psychiatry. 2018;8:96. doi: 10.1038/s41398-018-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E., Wetzels S.U., Wagner M., Zebeli Q., Schmitz-Esser S. Metatranscriptome sequencing reveals insights into the gene expression and functional potential of rumen wall bacteria. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCowan R.P., Cheng K.J., Bailey C.B.M., Costerton J.W. Adhesion of bacteria to epithelial cell surfaces within the reticulo-rumen of cattle. Appl. Environ. Microbiol. 1978;35:149–155. doi: 10.1128/aem.35.1.149-155.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgavi D.P., Forano E., Martin C., Newbold C.J. Microbial ecosystem and methanogenesis in ruminants. Animal. 2010;4:1024–1036. doi: 10.1017/S1751731110000546. [DOI] [PubMed] [Google Scholar]
- Nkrumah J.D., Okine E.K., Mathison G.W., Schmid K., Li C., Basarab A., Price M.A., et al. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 2006;84:145–153. doi: 10.2527/2006.841145x. [DOI] [PubMed] [Google Scholar]
- Olubobokun J.A., Craig W.M. Quantity and characteristics of microorganisms associated with ruminal fluid or particles. J. Anim. Sci. 1990;68:3360–3370. doi: 10.2527/1990.68103360x. [DOI] [PubMed] [Google Scholar]
- Patterson J.A., Hespell R.B. Glutamine synthetase activity in the ruminal bacterium Succinivibrio dextrinosolvens. Appl. Environ. Microbiol. 1985;50:1014–1020. doi: 10.1128/aem.50.4.1014-1020.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei C.X., Mao S.Y., Cheng Y.F., Zhu W.Y. Diversity, abundance and novel 16S rRNA gene sequences of methanogens in rumen liquid, solid and epithelium fractions of Jinnan cattle. Animal. 2010;4:20–29. doi: 10.1017/S1751731109990681. [DOI] [PubMed] [Google Scholar]
- Petri R.M., Neubauer V., Humer E., Kroger I., Reisinger N., Zebeli Q. Feed additives differentially impact the epimural microbiota and host epithelial gene expression of the bovine rumen fed diets rich in concentrates. Front. Microbiol. 2020 doi: 10.3389/fmicb.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R.M., Schwaiger T., Penner G.B., Beauchemin K.A., Forster R.J., McKinnon J.J., McAllister T.A. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Applied and Environmental Microbiology. 2013;79(12):3744–3755. doi: 10.1128/AEM.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf, Kittelmann S., Henderson G., Jassen P.H. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J. 2014;2:e494. doi: 10.7717/peerj.494. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf H., Kittelmann S., Janssen P.H. Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle. Appl. Environ. Microb. 2015;81:986–995. doi: 10.1128/AEM.03018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E.C., Choi B.R., Lim W.J., Hong S.Y., An C.L., Cho K.M., et al. Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16S rDNA sequence. Anaerobe. 2004;10:313–319. doi: 10.1016/j.anaerobe.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Hespell R.B. In: The Prokaryotes. Dworkin M., Falkow S., Rosenberg E., Schleifer K., Stackebrandt E., editors. Springer; 2006. The family Succinivibrionaceae; pp. 419–429. [Google Scholar]
- Tapio I., Snelling T.J., Strozzi F., Wallace R.J. The ruminal microbiome associated with methane emissions from ruminant livestock. Journal of Animal Science and Biotechnology. 2017;8 doi: 10.1186/s40104-017-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Cohen D.B., Ravel J., Abdo Z., Forney L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Hernandez-Sanabria E., Guan L.L. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 2009;75:6524–6533. doi: 10.1128/AEM.02815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Hernandez-Sanabria E., Guan L.L. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 2010;76:3776–3786. doi: 10.1128/AEM.00010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]