Highlights
-
•
Four bacterial strains with record resistance to Cu(II) have been isolated.
-
•
Genomic sequences are available for genome mining and studying the genetic determinants of bacterial resistance to Cu(II).
-
•
Genomic sequences provide the foundation necessary for transcriptional and functional studies of genes encoding for Cu(II) resistance mechanisms in Pseudomonas spp.
-
•
The data will be of interest for a broad community of biotechnologists and microbiologists and will aid in developing novel technologies for copper detoxification in contaminated soils and industrial wastewaters.
Keywords: Сopper-resistant microorganisms; Environmental biotechnologies; Soil isolates; Arctic; Antarctic, and Ukrainian soils
Abstract
Environmental copper pollution causes major destruction to ecological systems, which require the development of environmentally friendly biotechnological, in particular, microbial methods for copper removal. These methods rely on the availability of microorganisms resistant to high levels of copper. Here we isolated four bacterial strains with record resistance to up to 1.0 M Cu(II). The strains were isolated from ecologically diverse soil samples, and their genomes were sequenced. A 16S rRNA sequence-based phylogenetic analysis identified that all four isolates belong to the genus Pseudomonas. Particularly, strains UKR1 and UKR2 isolated from Kyiv region in Ukraine were identified as P. lactis and P. panacis, respectively, and strains UKR3 and UKR4 isolated from Svalbard Island in the Arctic Ocean and Galindez Island in Antarctica, respectively, were identified as P. veronii. Initial in-silico screening for genes encoding copper resistance mechanisms showed that all four strains encode copper resistance proteins CopA, CopB, CopD, CopA3, CopZ, as well as two-component regulatory system CusRS, all known to be associated with metal resistance in Pseudomonas genus. Further detailed studies will aim to characterize the full genomic potential of the isolates to enable their application for copper bioremediation in contaminated soils and industrial wastewaters.
Graphical abstract
1. Introduction
Environmental copper pollution causes major destruction to ecological systems (Abraham and Susan, 2017; Brennecke et al., 2016; Fashola et al., 2016). Copper-polluted sites are commonly associated with the urbanized areas in many countries, including Ukraine and the USA (Dovgalyuk, 2013; Schiff et al., 2007), and result from the heavy applications of copper-containing pesticides to control bacterial and fungal plant diseases (Husak, 2015). Other sources of copper contamination include industrial wastewaters (Al-Saydeh et al., 2017), metal mines, and tailing sites (Abraham and Susan, 2017; Fashola et al., 2016). Chemical and physical copper removal methods, such as adsorption, cementation, electrodialysis, electro-winning, photocatalysis, and membrane filtration have been developed (Al-Saydeh et al., 2017). However, these methods are expensive and environmentally hazardous. An environmentally friendly alternative is the development of biotechnological, in particular, microbial methods for copper removal (Cydzik-Kwiatkowska and Zielińska, 2016; Parungao et al., 2007; Rajbanshi, 2008; Yang et al., 2017). These methods require microorganisms that are resistant to high levels of copper. For bioremediation efforts, identifying such microorganisms and understanding their molecular mechanisms of resistance and detoxification of hazardous copper compounds are of utmost importance (Volentini et al., 2011; Andreazza et al., 2010; Ölmezoǧlu et al., 2012). In aqueous solutions, copper levels are affected by its solubility that can be altered during biochemical conversions, for example, reduction to insoluble and non-toxic copper(I) compounds (Volentini et al., 2011). Bioavailability of copper compounds is impacted by their complexation, precipitation, or adsorption (C.A.Flemming, 1989). The ability of microorganisms to withstand high levels of copper relies on adopting one or more of these mechanisms in natural ecosystems and is often exploited for bioremediation of heavy metal contaminated ecosystems (Andreazza et al., 2010; Ölmezoǧlu et al., 2012).
Recent studies of copper-resistant microorganisms using biochemical, bioinformatics and metalloproteomics approaches revealed several genetic determinants of copper resistance. They include cop genes encoding tightly regulated Cu+-sensing transcriptional regulators, chaperones, transporters, sequestering molecules that together constitute copper resistance mechanisms in various bacteria. Cop proteins can reside in the membrane, periplasm, or be secreted into the extracellular space, where they control the copper levels preventing its toxicity to bacterial cells (Argüello et al., 2013). For example, CopA and CopB belong to the family of P-type ATPases and couple the unidirectional Cu+ efflux to the hydrolysis of ATP (Argüello et al., 2013; Palmgren and Nissen, 2011; Samanovic et al., 2012). The Cu+-sensing transcriptional regulator CueR together with the two homologous metal chaperones CopZ1 and CopZ2 form a copper resistance system, whereby CopZ1 delivers Cu+ to the CueR sensor, while CopZ2 functions as a fast-response Cu+-sequestering storage protein (Novoa-Aponte et al., 2019). In eukaryotes, the common soil fungus Rhizophagus irregularis was shown to express two genes encoding copper transporters of the CTR family, RiCTR1 and RiCTR2 (Gómez-Gallego et al., 2019). Some of the genes are encoded on plasmids such as copA in Sphingomonas sp. and Stenotrophomonas sp. isolated from copper-polluted agricultural soils (Altimira et al., 2012), others are chromosomally encoded, such as cue and cus, identified in Escherichia coli (Outten et al., 2001).
Despite the considerable scientific advances in the understanding of the mechanisms of microbial copper resistance, the number of cooper-resistant organisms and their resistance to copper are limited. To our knowledge, the maximum reported levels of Cu(II) enabling bacterial growth is 472 mM determined for Thiobacillus ferrooxidans ATCC 19859 (Brahmahrakash et al., 1998).
This study aimed to isolate bacterial strains with a greater resistance to copper and sequence their genomes to potentiate discoveries of new genetic determinants of copper detoxification. We report the isolation and genomic sequencing of four highly copper resistant Pseudomonas strains, and the preliminary analysis of their genomic determinants of copper resistance and detoxification.
2. Theory
Microbial growth and interaction with metals are theoretically permissible if the redox potential of the metal transformation reaction is within the zone of thermodynamic stability of water (the standard redox potential Eо′1 is from –414 to +814 mV) even at high metal concentration (Hovorukha et al., 2018). In this study, we used Pourbaix diagrams (Pourbaix, 1974) and Nernst equations (Lehninger et al., 1993) to determine the relationships between pH and solubility of Cu(II) compounds as well as the range of redox potential (Eh) values and the concentrations of Cu(II) compounds during their reduction to Cu(I). These calculations enabled the thermodynamic prognosis of theoretical possibility of microbial interactions with Cu(II) at the concentrations of up to 1.0 M. Guided by this novel approach, we isolated four strains with record level of resistance to Cu(II).
3. Materials and methods
3.1. Soil samples
Soil samples were collected from three geographic location: Kyiv region of Ukraine in April, 2018, Galindez Island in Antarctica in January, 2008 and Svalbard Island in the Arctic Ocean in August, 2010. Immediately upon collection, the samples were frozen and stored at −20 °C until further use. The concentration of copper in soil samples was determined by atomic absorption spectroscopy method (da Silva Medeiros et al., 2020).
3.2. Preparation of stock solution of Cu(II) and copper containing nutrient medium
To avoid precipitation of Cu(II), we optimized the preparation of growth media as follows. Stock solution of Cu(II) (1.333 M) in citrate was prepared by dissolving CuSO4∙5H2O in an aqueous solution of 1.94 M Na3C6H5O7 (pH 3.5). The pH of the solution was adjusted to 6.5 by adding NaHCO3. The obtained solution was transferred to a borosilicate glass bottle, hermetically sealed, and sterilized by boiling for 20 min in a water bath. This solution was then added to the previously autoclaved nutrient broth or nutrient agar (kept liquid by incubating at 45 °C) to adjust the final concentration of Cu(II) to 1.0 M. The sterility of the media was tested by incubating a control nutrient agar plate and 5 mL of nutrient broth in a tube at 30 °C for 7 days, and no growth was observed. The level of Cu(II) in the solutions was tested by colorimetric determination (Prekrasna and Tashyrev, 2015). The maximum levels of Cu(II) in nutrient agar (tested prior solidifying) and nutrient broth were 236 mM and 1.1 M, respectively. The concentration of Cu(II) in liquid media remained stable over time.
3.3. Isolation of highly copper resistant microorganisms
For strains isolation, 1.0 g of soil was inoculated into 10 mL of nutrient broth containing up to 1.0 M Cu(II) followed by incubation at 30 °C for 10 days. Adding soil to nutrient broth was used only as an initial step. To avoid any effect that soil may have on complexing copper and hence decreasing its effective concentration in solution, no soil was added during all the subsequent sub-culturing steps performed by transferring 0.5 ml collected from the top of the enriched culture into a fresh nutrient broth containing up to 1.0 M Cu(II). After at least three passages, individual isolates were obtained by streaking on nutrient agar containing increasing levels of Cu(II) up to 157 mM and incubation at 30 °C. The isolates were passaged on Cu(II)-containing nutrient agar until pure cultures were obtained. The isolated strains were tested for their ability to grow in nutrient broth containing 1.0 M Cu2+. Microbial cell growth was monitored by light microscopy and counting colony forming units (CFU) on nutrient agar.
3.4. Studying the ability of isolates to interact with Cu(II)
To test the ability of isolates to accumulate Cu(II) or reduce Cu(II) to Cu(I) in colonies, they were grown on nutrient agar plates amended with 80 mM Cu2+. Accumulation of Cu(II) in colonies was determined by hydrogen sulfide test (Prekrasna and Tashyrev, 2015) and observation of blue coloration of the colonies (Irawati et al., 2018). The ability of microorganisms to reduce Cu(II) was determined by the formation of insoluble brown Cu2O↓ that was visible within the colonies (Hovorukha et al., 2018).
3.5. DNA extraction and whole genomic sequencing
Genomic DNA was extracted from each of the four isolates using standard phenol/chlorophorm method (Neumann et al., 1992) and sequenced on the Illumina MiSeq platform using 2 × 300 paired-end chemistry (Novogene). Generated reads were quality filtered using standard Illumina settings resulting in 10,025,497-13,114,132 reads. All quality-filtered reads were assembled using the short read de Brujin graph assembly (Compeau et al., 2011) program Velvet (Zerbino and Birney, 2008). Velvet assembly run-time settings were a k-mer value of 31 bp and a minimum contig coverage value of 10 × . In an attempt to identify plasmid sequences, the quality-filtered reads were assembled using the pipeline plasmidSpades (Antipov et al., 2016) using the default parameters. The phylogenetic position of the isolates was determined by 16S rRNA gene sequence analysis. The initial screening of the genomes for genes encoding copper resistance mechanisms was conducted through the IMG platform (Chen et al., 2019).
3.6. Data accession
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the Bioproject accession number PRJNA565195, Biosample accession numbers SAMN12736602, SAMN12736603, SAMN12736604, and SAMN12736605, and genome accession numbers VWXW00000000, VWXV00000000, VWXU00000000, VWXT00000000 for Pseudomonas lactis UKR1, Pseudomonas panacis UKR2, Pseudomonas veronii UKR3 and Pseudomonas veronii UKR4, respectively.
4. Results and discussion
4.1. Thermodynamic calculations of the redox potential of the reaction of Cu2+ reduction to Cu2O
We calculated the pH value below which Cu(II) compounds are highly soluble and stable in aqueous solutions using the Pourbaix diagrams (Pourbaix, 1974) that characterize the thermodynamically stable forms of chemical compounds in aqueous solutions at pH and Eh coordinates. The pH of the transformation of soluble Cu(II) compounds into insoluble Cu(OH)2↓ (according to Reaction 1) was calculated as follows: log[Cu2+] = 9.21 – 2pH; from which, pH = 4.6, indicating that Cu2+ precipitates at pH > 4.6.
The Eh of Cu2+ reduction (according to Reaction 2) was calculated at pH 4.6 and 1.0 M Cu2+ concentration applying the equation:
| (1) |
The Nernst equation was used to calculate the effect of the increased Cu2+ concentration on the value of Eh of Reaction 2 by substituting in the equation:
| (2) |
where Eо – the redox potential of the reaction, V; n – the number of electrons involved in the reaction.
The values of Eh for different copper concentrations was calculated and the results are shown in Fig. 1.
Fig. 1.
Redox potential of Cu(II) solutions as concentration increases from 0.0001 µM to 1.0 M at pH = 4.6 (the level of Eh was calculated at pH 4.6 since the Cu(II) compounds precipitate at higher pH values).
The calculated redox potential of Cu(II) reduction according to Reaction 2 increased from +180 mV to +475 mV as the concentration of Cu(II) increased from 0.0001 µM to 1.0 M (Fig. 1). These redox potential values are within the zone of water thermodynamic stability, and hence we expected the feasibility of microbial life at concentrations up to 1.0 M Cu(II), with the reduction of Cu(II) to Cu(I) (Hovorukha et al., 2018, Pourbaix, 1974) being the most probable outcome.
4.2. Isolated strains are resistant to and interact with Cu(II)
Guided by the calculations above, we attempted and successfully isolated four strains resistant to the presence of up to 1.0 M Cu(II). These highly copper resistant bacterial strains named UKR1, UKR2, UKR3, and UKR4 were isolated from diverse ecological niches including soils from central Ukraine, Galindez Island in Antarctica, and Svalbard Island in the Arctic Ocean (Table 1). The soil samples were tested for Cu(II) levels and showed concentrations ranging from 0.11 to 0.37 mM/dm3 of dry sample (Table 1). All four strains were able to grow in nutrient broth containing up to 1.0 M Cu(II) (Table 1), which, to our knowledge, is a record resistance to Cu(II) in bacteria that significantly exceeds the previously reported resistance levels to Cu(II) (up to 472 mM) (Rajbanshi, 2008, Brahmahrakash et al., 1998, Andrade et al., 2019). When grown on nutrient agar in the presence of high Cu2+ concentrations, the four strains accumulated Cu(II) within the colonies as indicated by the blue coloration of the colonies (Fig. 2A). While in the presence of lower Cu2+ concentrations (2.0 mM to 20 mM), the isolates were able to reduce Cu(II) to Cu(I) compounds that are insoluble and appear brown (Fig. 2B). Thus, the type interaction with Сu(II) depended on its concentration in the growth media.
Table 1.
Features of resistance and interaction of isolated Pseudomonas strains with Cu(II).
| Isolated strains | Colony accumulation of Cu(II)a | Reduction of Cu(II) to Cu(I)b | Growth in Nutrient Broth at 1000 mM Cu(II)c | Growth on Nutrient Agar with Cu(II), mMd | Cu(II) concentration, mM/dm3 of dry sample | Isolation source and location |
|---|---|---|---|---|---|---|
| P. lactis UKR1 | + | + | + | 157 | 0.13 | Chernozem soil, Kyiv region, Ukraine |
| P. panacis UKR2 | + | + | + | 110 | 0.11 | Chernozem soil, Kyiv region, Ukraine |
| P. veronii UKR3 | + | + | + | 157 | 0.36 | Arctic soil, Svalbard Isl., Norway, Arctic |
| P. veronii UKR4 | + | + | + | 157 | 0.37 | Antarctic soil, Galindez Isl., Antarctica |
Observed as blue colonies.
Observed as brown colonies.
Observed by light microscopy and sowing on nutrient agar followed by colony counting.
The maximum Cu(II) concentration supporting growth on solid media. The maximum solubility of Cu(II) in nutrient agar was 236 mM. Note that all four isolates were able to grow in liquid media in the presence of 1.0 M Cu(II).
Fig. 2.
(A) Growth of UKR1 strain on nutrient agar containing 80 mM of Cu(II). The blue coloration of the colonies is indicative of Cu(II) accumulation (Irawati et al., 2018). UKR2, UKR3, and UKR4 behaved similarly. (B) Reduction of Cu(II) to Cu(I) by the four isolates during growth in presence of 16 mM of Cu(II). The brown colour is indicative of the formation of insoluble brown Cu2O within the colonies (Hovorukha et al., 2018).
4.3. Phylogenetic analysis and general genomic features of the isolated strains
Small subunit rRNA sequence-based phylogenetic analysis identified that all four strains belonged to the genus Pseudomonas. BlastN against the GenBank 16S ribosomal DNA database (limited to species) identified Pseudomonas sp. strain UKR1 from Kyiv region as P. lactis (100% identity to P. lactis strain DSM 29167; NR_156986), Pseudomonas sp. strain UKR2 from Kyiv region as P. panacis (99.21% identity to P. panacis strain CG201106; NR_043195), and the two isolates from Svalbard and Galindez Islands (Pseudomonas sp. strain UKR3 and UKR4) as P. veronii (99.15%, and 99.61% identity with P. veronii strain CIP 104663; NR_028706.1, respectively). These phylogenies were confirmed by alignment and Maximum Likelihood tree construction (Fig. 3).
Fig. 3.
A maximum likelihood tree based on 16S rRNA genes alignment was constructed in Mega (Kumar et al., 2016) and showed the phylogenetic position of the isolated Pseudomonas strains (shown in red). GenBank or IMG gene accession numbers are shown for unwedged species. Bootstrap values (from 100 replicates) are shown for nodes with more than 50 bootstrap support. The tree was rooted (but the outgroup is not shown for better visualization) using the 16S rRNA sequence of Sphingomonas paucimobilis (GenBank accession number AM237364.1).
The genomes of the four strains were sequenced, and the assembly statistics and general genomic features are summarized in Table 2.
Table 2.
Genome assembly statistics and general genomic features of the four Pseudomonas isolates sequenced in this study.
| P. lactis strain UKR1 | P. panacis strain UKR2 | P. veronii strain UKR3a | P. veronii strain UKR4a | |
|---|---|---|---|---|
| Assembly statistics | ||||
| N50 (bp) | 7122 | 10124 | 11442 | 12356 |
| Largest assembled contig (bp) | 53476 | 68185 | 125256 | 148322 |
| Total number of contigs assembled | 1139 | 839 | 927 | 884 |
| General genomic features | ||||
| Genome Size (bp) | 5939518 | 6257067 | 7204846 | 7102407 |
| Gene Count | 5648 | 5936 | 6961 | 6765 |
| % GC | 60 | 60 | 60 | 61 |
| % Coding Bases | 89.6 | 89.51 | 89.54 | 89.72 |
| Number of 16S rRNA genes | 1 | 1 | 1 | 1 |
| Number of 23S rRNA genes | 1 | 1 | 1 | 1 |
| Number of tRNA genes | 25 | 29 | 30 | 28 |
| Number of genes | ||||
| with function prediction | 4696 | 4760 | 5673 | 5559 |
| without function prediction | 872 | 1089 | 1187 | 1107 |
| with COGs | 3922 | 3949 | 4628 | 4657 |
| GenBank WGS accession number | VWXW01 | VWXV01 | VWXU01 | VWXT01 |
The plasmid assembly algorithm plasmidSPAdes identified and assembled a few contigs in the two strains UKR3 and UKR4 as belonging to plasmids. These contigs ranged in length between 1.1-1.5 Kb and encoded 1-2 genes each. The IMG scaffold IDs for these potential plasmids are: 2806336179 and 2806336314 in strain UKR3, and 2806337104 and 2806337222 in strain UKR4.
4.4. Initial screening of the genomes revealed genes encoding copper resistance mechanisms
Initial screening of the four genomes for genes encoding copper resistance proteins was done through the IMG platform (Chen et al., 2019). All four strains encode for the copper resistance proteins A, B, C, and D (Chan et al., 2016; Chong et al., 2016; Cooksey, 1993), copper exporting ATPase copA3 (Chong et al., 2016), copper chaperone copZ (Quintana et al., 2017), as well as the two-component regulatory system cusRS (Adams, 2016), all known to determine copper resistance in Pseudomonas genus (Fig. 4). Compared to previously studied copper-resistant or copper-tolerant microorganisms (reference sequences shown in Fig. 5), percentage similarities of CopA, CopB, CopC, and CopD encoded by the four Pseudomonas strains studied here ranged within 49.1–100%, 34.7–100%, 47.5–98.4%, and 34–99.6%, respectively. Maximum likelihood phylogenetic trees comparing proteins encoded by the four Pseudomonas strains studied here to other copper resistant species are shown in Fig. 5.
Fig. 4.
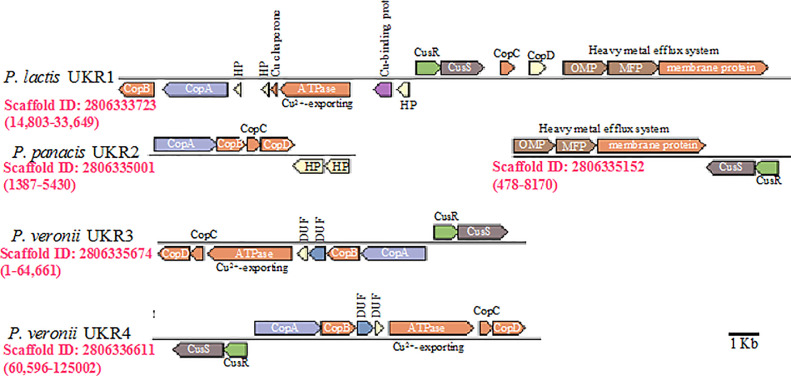
Genes predicted to encode copper resistance in the genomes of the four Pseudomonas species studied. Shown are regions of the chromosome encoding the copper resistance Cop system (copA, copB, copC, and copD), the Cu2+−exporting ATPase, the two-component signal transduction system Cus (cusRS), and the type I secretion system heavy metal efflux system (OMP, outer membrane protein; MFP, membrane fusion protein; and efflux system membrane protein). HP, hypothetical protein; DUF, domain of unknown function. IMG scaffold IDs are shown below each scaffold in red. Copper resistance genes in strain UKR2 were encoded on two scaffolds and are shown as separate regions. The scaffolds shown are most probably chromosomal as they were not identified by the plasmidSPades as belonging to plasmids. A scale bar is shown in the lower right corner.
Fig. 5.
Maximum likelihood phylogenetic trees based on the alignment of (A) CopA, (B) CopB, (C) CopC, and (D) CopD proteins encoded in the four studied Pseudomonas genomes (shown in red text) and other copper-resistant species. GenBank accession numbers are shown for reference sequences, while IMG gene IDs are shown for the Pseudomonas genes from this study. Alignment and tree construction were conducted in Mega (Kumar et al., 2016), and bootstrap values (from 100 replicates) are shown for nodes with more than 50 bootstrap support. Trees are mid-point rooted. Clades A and B refer to the phylogenetically distinct copies of Cop proteins encoded by strains UKR3 and UKR4 as explained in text.
Interestingly, the copper resistance genes of P. lactis strain UKR1 (encoded on scaffold ID 2806333723), P. veronii strain UKR3 (encoded on scaffold ID 2806335674), and P. veronii strain UKR4 (encoded on scaffold ID 2806336611) are all flanked by integrative conjugative mobile elements (including the genes encoding for excisionase, integrase, and conjugative transfer ATPase necessary for the excision and subsequent integration into the chromosome) suggestive of their transfer by conjugation.
Notably, several copies of the copper resistance genes copA, copB, copC, and copD were encoded in three Pseudomonas genomes belonging to strains UKR1, UKR3, and UKR4 (only one such copy is depicted in Fig. 4 for each of these genomes). Intra-strain copies of the same gene were not identical, and ranged in similarity between 42 and 75.6% at the protein level (Table 3). The different copies of the copper resistance genes copA, copB, copC, and copD encoded by Pseudomonas lactis strain UKR1 were phylogenetically similar (Fig. 5) and belonged to a clade that included other Gamma- and Alpha-Proteobacteria species (Clade A in Fig. 5). This clade also included the genes from Pseudomonas panacis strain UKR2. Interestingly, Pseudomonas veronii strains UKR3, and UKR4 encoded phylogenetically distinct copies of each of the copper resistance genes (Fig. 5), one of which belonged to the Gamma/Alpha-Proteobacteria clade (Clade A in Fig. 5) that also included the Pseudomonas lactis strain UKR1, and Pseudomonas panacis strain UKR2 genes, while the other was more closely related to Beta-Proteobacteria species (Clade B in Figure 5). Whether these multiple copies were all acquired by horizontal gene transfer from other copper resistant microorganisms, or originated by gene birth via gene duplication remains to be investigated. Regardless of their origin, the presence of multiple copies of copper-resistance genes in these strains strongly suggest their potential importance in the presence of high copper concentration.
Table 3.
Number of genomic copies and intra- and inter-strain percentage similarities of CopA, CopB, CopC, and CopD copper-resistance proteins encoded by the four Pseudomonas genomes in the current study.
| Copper resistance Protein | Genome | # of copies per genome | Percentage similarity |
|||
|---|---|---|---|---|---|---|
| Pseudomonas sp. UKR1 | Pseudomonas sp. UKR2 | Pseudomonas sp. UKR3 | Pseudomonas sp. UKR4 | |||
| CopA | Pseudomonas sp. UKR1 | 2 | 71.5 | |||
| Pseudomonas sp. UKR2 | 1 | 74–84 | 100 | |||
| Pseudomonas sp. UKR3 | 4 | 52.7–89.8 | 55.3–82.6 | 51.9–75.6 | ||
| Pseudomonas sp. UKR4 | 4 | 52.7–90 | 55.3–82.6 | 51.9–99.8 | 51.9–75.6 | |
| CopB | Pseudomonas sp. UKR1 | 2 | 69.6 | |||
| Pseudomonas sp. UKR2 | 1 | 67.4–73.1 | 100 | |||
| Pseudomonas sp. UKR3 | 2 | 42.4–83.4 | 51–70.7 | 50.5 | ||
| Pseudomonas sp. UKR4 | 2 | 42.4–83.4 | 51–70.7 | 50.5–99.8 | 50.52 | |
| CopC | Pseudomonas sp. UKR1 | 2 | 68.5 | |||
| Pseudomonas sp. UKR2 | 1 | 67.7–77.3 | 100 | |||
| Pseudomonas sp. UKR3 | 3 | 61.3–100 | 57.6–73.4 | 58.8–68.6 | ||
| Pseudomonas sp. UKR4 | 3 | 61.3–100 | 57.6–73.4 | 58.8–100 | 58.7–68.5 | |
| CopD | Pseudomonas sp. UKR1 | 2 | 56.8 | |||
| Pseudomonas sp. UKR2 | 1 | 61.9–62.7 | 100 | |||
| Pseudomonas sp. UKR3 | 2 | 39.79–74 | 43.3–63.6 | 41.96 | ||
| Pseudomonas sp. UKR4 | 2 | 39.79–74 | 43.3–63.6 | 41.96–100 | 41.96 | |
Conclusions
Based on thermodynamic calculations, we hypothesized the theoretical possibility of microbial growth in the presence of 1.0 M Cu(II). This was experimentally verified, and here we report the isolation of four strains resistant to 1.0 M Cu(II) from diverse non-polluted soils. The genomes of the strains were sequenced, and the initial screening for genes encoding copper resistance mechanisms in Pseudomonas spp. revealed several genomic determinants of Cu(II) resistance. The isolates may serve as potential candidates for copper-containing wastewaters purification and bioremediation of copper-contaminated ecosystems.
CRediT authorship contribution statement
Olesia Havryliuk: Investigation, Writing - original draft. Vira Hovorukha: Investigation, Supervision. Marianna Patrauchan: Resources, Conceptualization, Writing - review & editing. Noha H. Youssef: Resources, Data curation, Investigation, Writing - review & editing. Oleksandr Tashyrev: Conceptualization, Methodology, Resources, Supervision.
Declaration of Competing Interest
The authors have no competing interests.
Acknowledgments
Acknowledgments
The authors are thankful to Anatoliy Shakhovsky (Institute of Cell Biology and Genetic Engineering of the National Academy of Sciences of Ukraine, Kyiv) for help in genomic DNA extraction and to Anastasiya Sachko (Yuriy Fedkovych Chernivtsi National University, Chernivtsi, Ukraine) for help in atomic absorption spectroscopy analysis of soil samples.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
A standard redox potential (Eо′, mV) – the redox potential of the reaction at 1M concentration of both the oxidized and reduced forms of the reacting compounds at the pH = 7.0
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2020.06.002.
Contributor Information
Olesia Havryliuk, Email: gav_olesya@ukr.net.
Vira Hovorukha, Email: vira-govorukha@ukr.net.
Marianna Patrauchan, Email: m.patrauchan@okstate.edu.
Noha H. Youssef, Email: noha@okstate.edu.
Oleksandr Tashyrev, Email: tach2007@ukr.net.
Appendix. Supplementary materials
References
- Abraham M.R., Susan T.B. Water contamination with heavy metals and trace elements from Kilembe copper mine and tailing sites in Western Uganda; implications for domestic water quality. Chemosphere. 2017;169:281–287. doi: 10.1016/j.chemosphere.2016.11.077. [DOI] [PubMed] [Google Scholar]
- Adams M.W.W. Novel metal cation resistance systems from mutant fitness analysis of denitrifying Pseudomonas stutzeri. Appl. Environ. Microbiol. 2016;82:6046–6056. doi: 10.1128/AEM.01845-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saydeh S.A., El-Naas M.H., Zaidi S.J. Copper removal from industrial wastewater: a comprehensive review. J. Ind. Eng. Chem. 2017;56:35–44. doi: 10.1016/j.jiec.2017.07.026. [DOI] [Google Scholar]
- Altimira F., Yáñez C., Bravo G., González M., Rojas L.A., Seeger M. Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol. 2012;12:193. doi: 10.1186/1471-2180-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade C.J., Andrade L.M., Dias M., Mendes M.A., Nascimento C.A.O. Isolated Pseudomonas aeruginosa from mining site: proteome changes due to presence of copper. Current Trends Anal. Bioanal. Chem. 2019;1(1):42–49. doi: 10.36959/525/438. [DOI] [Google Scholar]
- Andreazza R., Pieniz S., Wolf L., Lee M.K., Camargo F.A.O., Okeke B.C. Characterization of copper bioreduction and biosorption by a highly copper resistant bacterium isolated from copper-contaminated vineyard soil. Sci. Total Environ. 2010;408:1501–1507. doi: 10.1016/j.scitotenv.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Antipov D., Hartwick N., Shen M., Raiko M., Lapidus A., Pevzner P.A. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32(22):3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- Argüello J.M., Raimunda D., Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 2013;4:1–14. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmahrakash G.P., Devasia P., Jagadish K.S., Natarajan K.A., Ramananda Rao G. Development of Thiobacillus ferrooxidans ATCC 19859 strain tolerant to copper and zinc. Bull. Mater. Sci. 1998;10(5):461–465. [Google Scholar]
- Brennecke D., Duarte B., Paiva F., Caçador I., Canning-Clode J. Microplastics as vector for heavy metal contamination from the marine environment. Estuar. Coast. Shelf. Sci. 2016;178:189–195. doi: 10.1016/j.ecss.2015.12.003. 2016. [DOI] [Google Scholar]
- C.A.Flemming J.T.Trevors. Copper toxicity and chemistry in the environment: a review. Water Air Soil Pollut. 1989;44:143–158. doi: 10.1007/BF00228784. [DOI] [Google Scholar]
- Chan K.-G., Chong T.-M., Adrian T.-G.-S., Kher H.L., Grandclément C., Faure D., Yin W.-F., Dessaux Y., Hong K.-W. Pseudomonas lini strain ZBG1 revealed carboxylic acid utilization and copper resistance features required for adaptation to vineyard soil environment, a draft genome analysis. J. Genom. 2016;4:26–28. doi: 10.7150/jgen.16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J.R., Seshadri R., Smirnova T., Kirton E., Jungbluth S.P., Woyke T., Eloe-Fadrosh E.A., Ivanova N.N., Kyrpides N.C. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucl. Acids Res. 2019;47(D1):D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong T.M., Yin W.-F., Chen J.-W., Mondy S., Grandclément C., Faure D., Dessaux Y., Chan K.-G. Comprehensive genomic and phenotypic metal resistance profile of Pseudomonas putida strain S13.1.2 isolated from a vineyard soil. AMB Express. 2016;6(1):1–7. doi: 10.1186/s13568-016-0269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeau P.E.C., Pevzner P.A., Tesler G. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 2011;29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D.A. Copper uptake and resistance in bacteria. Mol. Microbiol. 1993;7(1):1–5. doi: 10.1111/j.1365-2958.1993.tb01091.x. [DOI] [PubMed] [Google Scholar]
- Cydzik-Kwiatkowska A., Zielińska M. Bacterial communities in full-scale wastewater treatment systems. World J. Microbiol. Biotechnol. 2016;32:66. doi: 10.1007/s11274-016-2012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Medeiros D.C.C., Piechontcoski F., da Rocha Watanabe E.R.L., Chaves E.S., Inglez S.D. Fast and effective simultaneous determination of metals in soil samples by ultrasound-assisted extraction and flame atomic absorption spectrometry: assessment of trace elements contamination in agricultural and native forest soils from Paraná – Brazil. Environ. Monit. Assess. 2020;192(111):1–15. doi: 10.1007/s10661-020-8065-0. [DOI] [PubMed] [Google Scholar]
- 19.Dovgalyuk A. Environmental contamination by toxic metals and its indication by plant test systems. Biol. Stud. 2013;7(1):197–204. doi: 10.30970/sbi.0701.269. [DOI] [Google Scholar]
- Fashola M., Ngole-Jeme V., Babalola O. Heavy metal pollution from gold mines: environmental effects and bacterial strategies for resistance. Int. J. Environ. Res. Public Health. 2016;13(11):1047. doi: 10.3390/ijerph13111047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gallego T., Benabdellah K., Merlos M.A., Jiménez-Jiménez A.M., Alcon C., Berthomieu P., Ferrol N. The Rhizophagus irregularis genome encodes two ctr copper transporters that mediate cu import into the cytosol and a ctr-like protein likely involved in copper tolerance. Front. Plant Sci. 2019;10:1–16. doi: 10.3389/fpls.2019.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovorukha V., Havryliuk O., Tashyreva H., Tashyrev O., Sioma I. Thermodynamic substantiation of integral mechanisms of microbial interaction with metals. Ecol. Eng. Environ. Protect. 2018;2:55–63. doi: 10.32006/eeep.2018.2.5563. [DOI] [Google Scholar]
- Husak V. Copper and copper-containing pesticides: metabolism, toxicity and oxidative stress. J. Vasyl Stefanyk Precarpathian Natl. Univ. 2015;2(1):38–50. doi: 10.15330/jpnu.2.1.38-50. [DOI] [Google Scholar]
- Irawati W., Ompusunggu N.P., Yuwono T. Influence of bacterial consortium for copper biosorption and accumulation. AIP Conf. Proc. 2018;2002 doi: 10.1063/1.5050168. 020072-1–020072-8. [DOI] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger A.L., Nelson D.L., Cox M.M. second ed. Worth Publishers; New York: 1993. Principles of Biochemistry. [Google Scholar]
- Neumann B., Pospiech A., Schairrer H.U. Rapid isolation of genomic DNA from Gram-negative bacteria. Trends Genet. 1992;8:332–333. doi: 10.1016/0168-9525(92)90269-a. [DOI] [PubMed] [Google Scholar]
- Novoa-Aponte L., Ramírez D., Argüello J.M. The interplay of the metallosensor CueR with two distinct CopZ chaperones defines copper homeostasis in Pseudomonas aeruginosa. J. Biol. Chem. 2019;294:4934–4945. doi: 10.1074/jbc.RA118.006316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ölmezoǧlu E., Kiratli Herand B., Öncel M.S., Tunç K., Özkan M. Copper bioremoval by novel bacterial isolates and their identification by 16S rRNA gene sequence analysis. Turkish J. Biol. 2012;36:469–476. doi: 10.3906/biy-1104-15. [DOI] [Google Scholar]
- Outten F.W., Huffman D.L., Hale J.A., O'Halloran T.V. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- Palmgren M., Nissen P. P-type ATPases. Annu. Rev. Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- Parungao M.M., Tacata P.S., Tanayan C.R.G., Trinidad L.C. Biosorption of copper, cadmium and lead by copper-resistant bacteria isolated from Mogpog river, Marinduque. Philippine. J. Sci. 2007;136(2):155–165. [Google Scholar]
- Pourbaix M. Atlas of electrochemical equilibria in aqueous solutions. Houston:NACE International. Mater. Sci. Forum. 1974:43–54. doi: 10.4028/www.scientific.net/msf.251-254.143. [DOI] [Google Scholar]
- Prekrasna I.P., Tashyrev O.B. Copper resistant strain Candida tropicalis RomCu5 interaction with soluble and insoluble copper compounds. Biotechnol. Acta. 2015;8:93–102. doi: 10.15407/biotech8.05.093. [DOI] [Google Scholar]
- Quintana J., Novoa-Aponte L., Argüello J.M. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J. Biol. Chem. 2017;292(38):15691–15704. doi: 10.1074/jbc.m117.804492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A. Rajbanshi, Study on heavy metal resistant bacteria in guheswori sewage treatment plant, Our nature6 (2008) 52–57. 10.3126/on.v6i1.1655. [DOI]
- Samanovic M.I., Ding C., Thiele D.J., Darwin K.H. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff K., Brown J., Diehl D., Greenstein D. Extent and magnitude of copper contamination in marinas of the San Diego region, California, USA. Mar. Pollut. Bull. 2007;54(3):322–328. doi: 10.1016/j.marpolbul.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Volentini S.I., Farías R.N., Rodríguez-Montelongo L., Rapisarda V.A. Cu(II)-reduction by Escherichia coli cells is dependent on respiratory chain components. BioMetals. 2011;24:827–835. doi: 10.1007/s10534-011-9436-3. [DOI] [PubMed] [Google Scholar]
- Yang Y., Hu M., Zhou D., Fan W., Wang X., Huo M. Bioremoval of Cu2+ from CMP wastewater by a novel copper-resistant bacterium Cupriavidus gilardii CR3: characteristics and mechanisms. RSC Adv. 2017;7:18793–18802. doi: 10.1039/c7ra01163f. [DOI] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.