Highlights
-
•
The canonical P2 × 7-Caspase-1 pathway is necessary for secretion of IL-1β in oral tissues and macrophages infected with P. gingivalis.
-
•
P2 × 7 receptor controls bacterial load of F. nucleatum and P. gingivalis in macrophages and in mice.
-
•
Caspase-11 is essential for F. nucleatum-induced secretion of IL-1β in macrophages, limits F. nucleatum infection in macrophages and in mice, and is required for cell death induced by F. nucleatum infection.
-
•
The canonical inflammasome is activated preferentially in response to P. gingivalis infection, while the noncanonical inflammasome plays a predominant role during F. nucleatum infection.
Keywords: Porphyromonas gingivalis, Fusobacterium nucleatum, Oral cavity, Inflammasome, Purinergic receptor
Abstract
We examined the involvement of the P2 × 7 receptor and the canonical and noncanonical inflammasomes in the control of single-species or dual-species infection by the periodontal bacteria Porphyromonas gingivalis and Fusobacterium nucleatum in cells and mice. Stimulation of the P2 × 7 receptor leads to activation of the canonical NLRP3 inflammasome and activation of caspase-1, which leads to cleavage of pro-IL-1β to IL-1β, a key cytokine in the host inflammatory response in periodontal disease. The non-canonical inflammasome pathway involves caspase-11. Thus, wildtype (WT), P2 × 7−/−, caspase-11−/− and caspase-1/11−/− mice were co-infected with both bacterial species. In parallel, bone marrow-derived macrophages (BMDMs) from WT mice and the different knockout mice were infected with P. gingivalis and/or F. nucleatum, and treated or not with extracellular ATP, which is recognized by P2 × 7. F. nucleatum infection alone promoted secretion of IL-1β in BMDMs. Conversely, the canonical pathway involving P2 × 7 and caspase-1 was necessary for secretion of IL-1β in BMDMs infected with P. gingivalis and in the mandible of mice coinfected with P. gingivalis and F. nucleatum. The P2 × 7 pathway can limit bacterial load in single-species and dual-species infection with P. gingivalis and F. nucleatum in BMDMs and in mice. The non-canonical pathway involving caspase-11 was required for secretion of IL-1β induced by F. nucleatum infection in BMDMs, without treatment with ATP. Caspase-11 was also required for induction of cell death during infection with F. nucleatum and contributed to limiting bacterial load during F. nucleatum infection in BMDMs and in the gingival tissue of mice coinfected with P. gingivalis and F. nucleatum. Together, these data suggest that the P2 × 7-caspase-1 and caspase-11 pathways are involved in the immune response against infection by P. gingivalis and F. nucleatum, respectively.
Graphical abstract
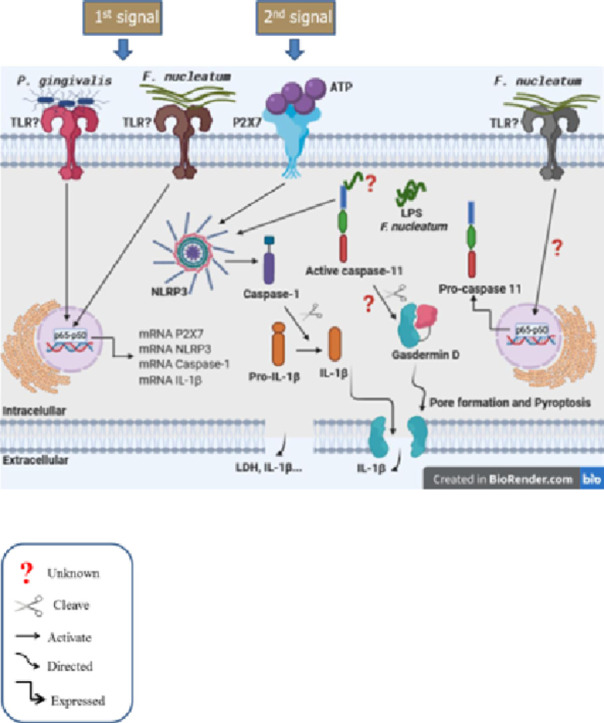
1. Introduction
Periodontal disease (PD) is a worldwide public health problem that affects mainly middle and low-income countries (Peres et al., 2019), but it is estimated that almost half of American adults also have mild, moderate, or severe periodontitis (Eke et al., 2012). According to the World Health Organization, about 20% of adults worldwide have some form of chronic periodontitis (Natto and Hameedaldain, 2019), and severe periodontitis is considered to be the sixth most prevalent disease in the world (Eke et al., 2015; Tonetti et al., 2015). Furthermore, PD can contribute to other inflammatory diseases as shown by clinical and experimental studies (Arimatsu et al., 2014; Bui et al., 2019).
Porphyromonas gingivalis is an anaerobic gram negative bacterium present in subgingival plaques and is one of the microorganisms associated with chronic periodontitis (Hajishengallis, 2015). P. gingivalis presents several virulence factors that allow the bacteria to colonize the host tissues under hostile environments, to obtain nutrients, to subvert immune responses, and to favor the formation of dysbiotic communities (Hajishengallis, 2015).
Other constituents of the dental biofilm participate in the development of oral inflammation and are associated with the disease, including the bacterium Fusobacterium nucleatum (Whitmore and Lamont, 2014). F. nucleatum can function as a bridge in the formation of dental plaque, aggregating other microorganisms, but can also invade several cells, including oral epithelial cells, macrophages, and keratinocytes (Gursoy et al., 2008; Han et al., 2000; Weiss et al., 2000; Kolenbrander et al., 2010).
Coinfection of P. gingivalis with F. nucleatum can facilitate adherence and invasive capacity in cells in the gingival tissue (Li et al., 2015; Saito et al., 2008; Saito et al., 2012). In addition, F. nucleatum produces a capnophilic environment that is conducive to the growth of P. gingivalis and that metabolizes molecular oxygen and hydrogen peroxide (Diaz et al., 2002).
Inflammation resulting from oral dysbiosis is mediated by neutrophils, macrophages, dendritic cells as well as acquired immunity cells, T and B lymphocytes (Lira-Junior and Figueredo, 2016). Macrophages correspond to about 5–7% of inflammatory infiltrates in injured periodontal tissue. Despite occupying a relatively small proportion, macrophages participate in the first line of defense, possessing phagocytic ability and regulatory functions, recruiting immune cells and activating immune and non-immune cells (Carcuac and Berglundh, 2014; Thorbert-Mros et al., 2015).
We and others (Chotjumlong et al., 2013; Kim et al., 2018) found that purinergic signaling is involved in the immune response to infection by periodontopathogenic bacteria in vivo and in vitro. Thus, P. gingivalis infection increases expression of the purinergic P2 × 7 receptor in infected murine macrophages (Morandini et al., 2014) and in the maxilla of infected mice (Ramos-Junior et al., 2015). This receptor was also required for IL-1β production, leukocyte recruitment to the site of infection, and control of bacterial load in an air pouch model of infection (Almeida-da-Silva et al., 2019).
Ligation of the ionotropic P2 × 7 receptor by extracellular ATP leads to activation of the NLRP3 inflammasome (NOD-, LRR- and pyrin domain-containing protein) via the canonical pathway. This in turn results in activation of caspase-1, which is responsible for cleaving the 33-kDa pro-IL-1β and 24-kDa pro-IL-18 into their biologically active forms, 17-kDa IL-1β and 18-kDa IL-18, respectively (Swanson et al., 2019). The NLRP3 inflammasome has been shown to play a role in the host's inflammatory response in PD (Pan et al., 2019; Yucel-Lindberg and BÃ¥ge, 2013), and the pro-inflammatory cytokine IL-1β contributes to induction of bone resorption (Chen et al., 2014; Huynh et al., 2017).
Caspase-11 (represented in humans by its orthologs, caspase-4 and caspase-5) is expressed broadly in immune and non-immune cells, and mediates inflammatory responses through the noncanonical pathway, in which the NLRP3 inflammasome is activated by gram negative bacteria such as Escherichia coli, Salmonella typhimurium and Burkholderia thailandensis (Kayagaki et al., 2011; Broz et al., 2012; Aachoui et al., 2013). The activation of caspase-11 occurs during both cytosolic and non-cytosolic bacterial infections, and it is induced by the detection of cytosolic LPS, which is translocated to the host cell's cytosol during infection with extracellular bacteria (Roberts and Yilmaz, 2015; Russo et al., 2018). Both caspase-1 and caspase-11 induce pyroptosis, an inflammatory cell death characterized by pore formation in the cell membrane, cell edema, osmotic lysis and release of cytosolic contents into the extracellular medium (Lamkanfi, 2011). These contribute to the elimination of infectious microorganisms within the cell (Uchiyama and Tsutsui, 2015).
Immune responses to single-species infections (Bui et al., 2016; Hajishengallis et al., 2009; Johnson et al., 2018) and dual-species infections (Saito et al., 2012 Nov; Taxman et al., 2012) by P. gingivalis and F. nucleatum have been investigated in animal models, such as rats and mice, and in cells. To date, there have been studies on the involvement of the purinergic P2 × 7 receptor and caspase-1 and caspase-11 in the control of single-species infection only by P. gingivalis, and no studies on co-infection by P. gingivalis and F. nucleatumin vivo and in vitro models. Therefore, we conducted the present study to investigate the canonical and non-canonical pathways of NLRP3 inflammasome activation during co-infection, which are closer to mimicking the conditions of polymicrobial infections in the oral cavity than the single-species infection reported by most studies to date.
2. Materials and methods
2.1. Reagents
All chemicals were purchased from Sigma-Aldrich (San Louis, Missouri, USA), except as noted below. Primer pairs were purchased from Exxtend Biotecnologia Ltda. Sybr Green PCR Master Mix and 96-well MicroAmp Optical reaction plates were purchased from Applied Biosystems (Foster City, California, USA). TRIzol reagent was purchased from Invitrogen Life Technologies (Carlsbad, California, USA). MultiScribecⓇ Reverse Transcriptase, dNTP Mix and oligo dT Primers were purchased from Invitrogen Life Technologies. Murine IL-1β ELISA reagents and Murine TNF-α ELISA reagents were purchased from R&D Systems, (Minneapolis, Minnesota, USA). The 96-well ELISA plates were obtained from CorningⓇ (Corning, New York, USA). AnaerocultⓇ A was purchased from Merck Millipore (Burlington, Massachusetts, USA). Sulfamethoxazole (0.87 mg/mL) and trimethoprim (0.17 mg/mL) (Bactrim 200/40 mg) were obtained from a pharmacy.
2.2. Bacterial strains and growth conditions
From frozen stocks at −80 °C, Porphyromonas gingivalis (strain ATCC 33277) cells were grown on brain and heart infusion (BHI) agar (1.85% BHI; 1.5% agar; 0.5% yeast extract; 0.5 g/L L-cysteine; 0.00005% vitamin K; 0.0005% hemin; 5% defibrinated sheep blood). The ATCC 33277 strain of P. gingivalis is more invasive in models of intracellular infection (Lamont et al., 1995). Fusobacterium nucleatum (strain ATCC 25586) cells were grown on BHI agar plate and Tryptic Soy Broth (TSB) (1.55% BHI; 1.48% TSB; 1.7% agar; 0.5% yeast extract; 1% menadione solution (0.5 mg/mL); 1% hemin solution (1 mg/mL); 5% defibrinated sheep blood). Both bacterial strains grew in an anaerobic jar system with AnaerocultⓇ A anaerobic generators for 7 days at 37 °C. For experiments, isolated colonies of P. gingivalis and F. nucleatum were selected and inoculated in supplemented specific broth for the growth of each bacterium and grown for 48 h in anaerobic jars under the same conditions as for previous growth[25]. Bacterial quantification was performed using spectrophotometry at 550 nm.
2.3. In vivo studies
2.3.1. Mice and in vivo study design
To prevent any effect of estrogens on oral tissue, male C57BL/6 (Wild-Type—WT), P2 × 7−/- (Pfizer, USA), Caspase11−/− (Casp11−/−) and Caspase1/11−/- (Casp1/11−/−) (Genentech, USA), at 6 weeks old (body mass average: 25 g), obtained from the Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro (IBCCF/UFRJ). The animals were weighed for random conditioning, together, per group, in microisolators for seven days for adaptation in an animal experimental vivarium at the IBCCF/UFRJ (24 ± 1 °C; 12-h light/dark cycle). The experimental phase was carried out after the approval of this study by the Animal Use Ethics Committee (CEUA) of UFRJ, under protocol number 080/19. During all stages of the study, the care proposed by the Guide for the Care and Use of Laboratory Animals (1996) was followed, and the number of animals was based on the minimum number needed to achieve the study's aims.
2.3.2. F. nucleatum and P. gingivalis in vivo infection model
The animals (7 weeks old) were treated with sulfamethoxazole (0.87 mg/mL) and trimethoprim (0.17 mg/mL) in drinking water ad libitum for 10 days, followed by 3 days without antibiotics. Then the mice were anesthetized and infected in the oral cavity with 50 µL of 2% carboxymethylcellulose containing 109 CFU (colony forming units) of each bacterium applied at the surface of the mandibular and maxilla molar teeth using a micropipette and sterile tip (Ramos-Junior et al., 2015). For coinfection, the inoculum containing P. gingivalis was applied 4 h after the inoculum containing F. nucleatum, considering that Feuille et al. (1996) observed that this time led to significantly increased capacity of P. gingivalis to form large phlegmonous lesions. Three inoculations were applied at 2-day intervals (a total period of 7 days). Inocula containing bacteria were prepared immediately prior to administration. Bacterial inoculation induced an immune response in an animal model similar to periodontal disease in humans (Graves et al., 2012). The animals received the NuvitalⓇ Commercial Diet, CR1, Brazil, and had free access to food and water (filtered and autoclaved) throughout the experiment. At the end of the experimental period, on day 8, the animals were euthanized in a CO2 chamber and then the upper and lower dental arches were removed for separation into two hemiarcades per cut. Subsequently, gingival tissue and mandible fragment were collected for further analysis and were stored in a biofreezer at −80 °C.
2.3.3. Tissue homogenate preparations
Mandible homogenates were prepared in ice cold using radioimmunoprecipitation assay buffer (RIPA) buffer (pH 7.4) with protease inhibitor cocktail (for use with mammalian cell and tissue extracts; Sigma-Aldrich, San Louis, Missouri, USA). Subsequently, a 1:10 dilution of the homogenates was performed using PBS buffer (pH 7.4) for subsequent measurement of total proteins using a BCA Kit (bicinchoninic acid) (Bioagency, São Paulo, Brazil), following the manufacturer's instructions with absorbance reading at 562 nm.
2.4. In vitro studies
2.4.1. Cell culture
For bone marrow collection and macrophage differentiation, we used 8-week-old C57BL/6 (WT), P2 × 7−/−, caspase11−/− and caspase-1/11−/- mice. Bone marrow cells were isolated from the femurs and tibias and were used for macrophage differentiation using L929 cell conditioned media (LCCM) as a source of macrophage colony stimulating factor (M-CSF) (Morandini et al., 2014). As previously described (Morandini et al., 2014; Almeida-da-Silva et al., 2019), cells were resuspended in 10 mL RPMI 20/30 differentiation medium [RPMI1640 supplemented with 20% fetal bovine serum (FBS- GibcoⓇ), 30% LCCM, 100 U/mL penicillin, 100 U/mL streptomycin, 1 mM L-glutamine]. Cells were seeded in culture plates and incubated at 37 °C in a 5% CO2 atmosphere. Three days later, an additional 10 mL of 20/30 RPMI was added per incubation plate for another four days. On day 7, the adherent cells were collected with 10 mL of sterile ice-cold phosphate buffered saline (PBS, pH 7.4). The cells were centrifuged at 220 g for 5 min and resuspended in 10 ml RPMI culture medium with 10% FBS. The cells were then seeded at 106/mL on the day prior to the experiments. The macrophage purity of these preparations was above 90%, assessed by flow cytometry using Alexa Fluor 488 anti-F4/80 (eBioscience™, Waltham, Massachusetts, USA).
2.4.2. F. nucleatum and P. gingivalis infection in vitro model
A total of 106 bone marrow-derived macrophages (BMDMs) from C57BL/6 (WT), P2 × 7−/−, caspase11−/− and caspase1/11−/− mice were plated on 24-well plates in 500 µL RPMI 1640 medium (1% FBS). After 18 h of plating, macrophages were infected with P. gingivalis and/or F. nucleatum at multiplicities of infection (MOI) of 100 and 20, respectively. P. gingivalis were added to the cells along with F. nucleatum. After 2 h of the bacterium and macrophage interaction, 200 µg/ml of metronidazole and 300 µg/ml of gentamicin was added to each well and incubated for a further 1 h, 37 °C in a 5% CO2 atmosphere to eliminate non-phagocytic bacteria. The supernatants were then discarded and the cells were washed three times with PBS, then 500 µL of RPMI 1640 medium (1% SFB) was added. At 30 min before completing 6 h of infection, 3 mM ATP was added (or not) to cells. After this treatment, the supernatants were collected and stored at −80 °C. Subsequently, the cells were washed three times with PBS and then 500 µL of RPMI 1640 medium (1% SFB) were added. The cells were incubated at 37 °C in a 5% CO2 atmosphere for another 12 h. After 18 h of infection, the supernatants were collected and stored at −80 °C and cells were harvested with 500 µL TRIzol reagent. This infection protocol was used throughout the study. The choice of MOIs were based on our results of dose-response experiments (see supplementary data Figs. S1 and S2). MOIs of 100 for P. gingivalis and MOI of 20 for F. nucleatum appeared to be the ideal doses for the study.
2.4.3. Cytotoxicity assay
Cells were infected as described, and lactate dehydrogenase (LDH) levels were dosed in the supernatant at 6 and 18 h of infection. LDH release was quantified using an LDH Cytotoxicity Assay Kit (BioclinⓇ, Belo Horizonte, Brazil) according to the manufacturer's instructions.
2.4.4. Cytokine analysis
For in vivo tests, the mandible homogenates were initially diluted (1:10) with PBS buffer (pH 7.4). For in vitro tests, cell-free supernatants were used without prior dilution. The levels of cytokines IL-1β (cat.DY401, DuoSet ELISA - R&D Systems) and TNF-α (cat.DY410, DuoSet ELISA - R&D Systems) were assayed using ELISA with an R&D SystemsⓇ kit as per manufacturer's instructions. Cytokines were quantified as absorbance at 450 nm in an ELISA plate reader, and their concentrations were determined by interpolation of a standard curve. Results were expressed as pg/mL.
2.4.5. RNA and DNA isolation and real-time RT-PCR
RNA and DNA extraction from cells and gingival tissues was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. After the extraction step, samples were analyzed for RNA and DNA quantity and purity by absorbance, without prior dilution, using NanoVue Plus™ (Biochrom, Harvard Bioscience, US, Holliston, MA, USA). For bacterial load quantification, a calculation was performed to achieve DNA concentrations of 40 ng/µL and to standardize sample concentrations prior to DNA amplification. For the mRNA quantification, 1 µg RNA was used for cDNA (complementary DNA) synthesis in Eppendorf Mastercycler Gradient PCR Instrument using Reverse Transcriptase Enzyme. Homology of the selected primers was verified by a study carried out in the BLAST program of the National Center for Biotechnological Information (http://www.ncbi.nlm.nih.gov/BLAST/). The finished primer sequence (Table 1) was synthesized (Exxtend Solutions in Oligo, Campinas, SP, Brazil) and purified by desalination. Amplification was performed in a real-time quantitative PCR apparatus using the Sybr Green Reagent in StepOnePlus™ Real-Time PCR System programmed as follows: initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 1 min for the final elongation of the DNA strands. In all reactions, DNA from reference strains of the studied microorganisms was used as a positive control. To determine the quantification of relative bacterial levels was performed by interpolation with a bacterial standard curve obtained by multiple serial dilutions of 10 with DNA extracted from pure cultures (P. gingivalis strain ATCC 33,277 and F. nucleatum strain ATCC 25,586) through PureLinkⓇ. Genomic DNA Mini Kit (ThermoFisher Scientific, Alto de Pinheiros, SP, BR) according to manufacturer's instructions. To determine the quantification of the relative levels of gene expression, the delta Ct method was used after normalization with endogenous β-actin, which was chosen as the reference gene. Differences in gene expression levels were calculated using the 2−ΔΔCt comparative method using the reference gene, as well as the uninfected bone marrow-derived macrophages (BMDMs) and gingival tissue from C57BL/6 (WT) and knockout animals as controls.
Table 1.
Primers used for PCR amplifications.
| Gene | Protein | Primer sequence (5′ – 3′) | |
| P2RX7 | P2 × 7 | Forward: | TTCAGGCAGGCAGTATCACTC |
| Reverse: | CCACGGGAAAGACACAGGTAG | ||
| CASP11 | Casp-11 | Forward: | ATGGCTGAAAACAAACACCCT |
| Reverse: | TCAGTTGCCAGGAAAGAGGTAG | ||
| CASP1 | Casp-1 | Forward: | GGAAGCAATTTATCAACTCAGTG |
| Reverse: | GCCTTGTCCATAGCAGTAATG | ||
| IL1B | Il-1β | Forward: | TTCAGGCAGGCAGTATCACTC |
| Reverse: | CCACGGGAAAGACACAGGTAG | ||
| TNF | Tnf-α | Forward: | TTCAGGCAGGCAGTATCACTC |
| Reverse: | CCACGGGAAAGACACAGGTAG | ||
| NLRP3 | Nlrp-3 | Forward: | GCT CAG CTC TGA CCT CT |
| Reverse: | AGG TGA GGC TGC AGT TGT CT | ||
| TNFRSF11A | Rank | Forward: | TTAAGCCAGTGCTTCACGGG |
| Reverse: | ACGTAGACCACGATGATGTCGC | ||
| TNFSF11 | Rankl | Forward: | CAGAAGATGGCACTCACTGCA |
| Reverse: | CACCATCGCTTTCTCTGCTCT | ||
| Fusobacterium nucleatum (Target Species) | Foward: | CTT AGG AAT GAG ACA GAG ATG | |
| Reverse: | TGA TGG TAA CAT ACG AAA GG | ||
| Porphyromonas gingivalis (Target Species) | Foward: | CGCAGACGACAGAGAAGACA | |
| Reverse: | ACGGACAACCTGTTTTTGATAATCCT | ||
| ACT | β-actin | Forward: | TAT GCC AAC ACA GTG CTG TCT GG |
| Reverse: | TAC TCC TGC TTG CTG ATC CAC AT |
2.4.6. Flow cytometry immunophenotyping
To examine the profile of inflammatory cells, submandibular lymph nodes were removed. The organs were then macerated using a cell strainer (0.40 µm) in an ice-cold Petri dish using a 1-mL syringe plunger (rough part). Subsequently, the cells were centrifuged at 220 g, 4 °C for 10 min, discarding the supernatants and resuspending pellets in 1 mL RPMI 1640 (3% SFB). Then, the cells were counted in hemocytometer with 0.2% trypan blue. The volume corresponding to 5 × 105 cells, allowing adjustment of the amount of cells to be used for labeling, was pipetted into round-bottom 96-well plates and the cells were centrifuged again at 220 g, 4 °C for 4 min. The supernatants were discarded and the cells were resuspended in 10 µl Fc block (CD16/CD32; 0.5 mg/mL, Clone 93) diluted in FACS buffer (1: 100) and incubated for 15 min at 4 °C. Then, 10 µL/well of the conjugated antibody was added to the respective fluorochrome (diluted in FACS buffer (1: 100)), containing (0.1–10 µg/mL) and the isotype controls diluted (1: 100) and incubated for 30 min, 4 °C under light protection and then, added 200 µL/well of FACS buffer, centrifuged the cells (220 g, 4 °C for 4 min) and we discarded the supernatants. A repeat wash was performed with 200 µL FACS buffer, cells were centrifuged (220 g, 4 °C for 4 min) and the supernatants were discarded. Cells were resuspended in 250 µL/well PBS x 1. Then, the contents of the wells were transferred to FACS tubes and FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, USA) acquisition was performed. Data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR, USA). The percentage data were multiplied by the total amount of cells of the corresponding sample and then divided by 100. The results were then expressed as total number of cells.
2.5. Statistical analysis
For all variables, normality was assessed using the Kolmogorov–Smirnov test. Parametric variables were evaluated using one-way analysis of variance (ANOVA), followed by t-test or Tukey test for comparisons between groups. The Kruskal–Wallis test or Mann–Whitney test was used to evaluate non-parametric variables with corresponding post hoc analysis. Results were presented as mean±standard error (SEM). P values < 0.05 were considered statistically significant. GraphPadⓇPrism version 5.1 for Windows (San Diego, CA, USA) was used.
3. Results
3.1. P2 × 7 is required for expression and secretion of IL-1β, and control of infection by P. gingivalis and F. nucleatum in vivo
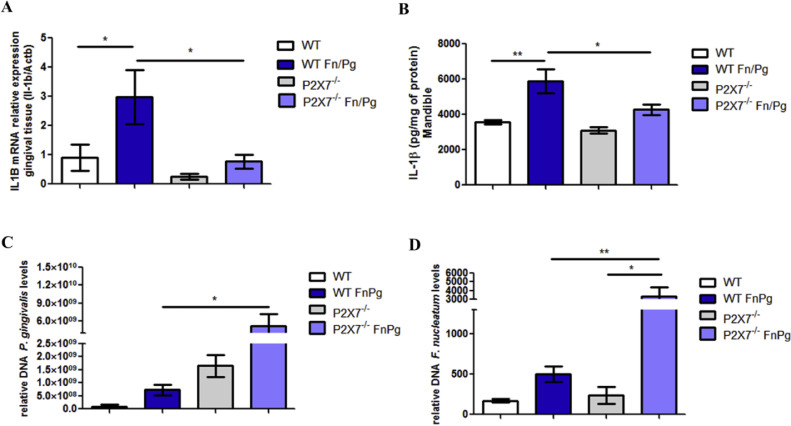
Considering the role of P2 × 7 receptor in NLRP3 inflammasome activation and subsequent secretion of IL-1β (Swanson et al., 2019), we examined the participation of P2 × 7 in the expression and secretion of IL-1β in the gingival tissue and mandible, respectively, of C57BL/6 and P2 × 7−/− mice co-infected by P. gingivalis and F. nucleatum. We observed that there were lower levels of IL-1β mRNA (Fig. 1A) and protein expression of this cytokine (Fig. 1B) in the gingival tissue and mandible, respectively, of P2 × 7−/− co-infected animals than in co-infected wildtype (WT) animals. Levels of P. gingivalis (Fig. 1C) and F. nucleatum (Fig. 1D), measured as the amount of specific DNA, in the gingival tissue of co-infected P2 × 7−/− animals were larger than in co-infected WT animals.
Fig. 1.
P2 × 7 is important for the control of infection by P. gingivalis and F. nucleatum in vivo. (A) Relative levels of IL-1β mRNA are shown in gingival tissue using real-time PCR. (B) Levels of IL-1β are shown in the mandible using ELISA. (C) Relative amount of P. gingivalis DNA in gingival tissue by real-time PCR. (D) Relative amount of F. nucleatum DNA are shown in the gingival tissue by real-time PCR of C57BL/6 (WT) and P2 × 7−/- animals infected or not infected orally with 109 CFU of each strain of P. gingivalis (ATCC 33,277) and F. nucleatum (ATCC 25,586). The data represent the mean ± SEM. (A) and (B)n = 4; (C) WT FnPg = 9, P2 × 7−/- FnPg = 6; (D) WT FnPg = 7, P2 × 7−/- FnPg = 6. The data were analyzed using one-way ANOVA, followed by the Tukey test. * p <0.05; **p <0.005. Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
These results suggest that P2 × 7 is important for the secretion of IL-1β in the mandible of coinfected mice and for the control of infection by P. gingivalis and F. nucleatum in gingival tissue of coinfected mice.
3.2. Caspase-1 contributes to the immune response against P. gingivalis while caspase-11 acts primarily against infection by F. nucleatum in vivo
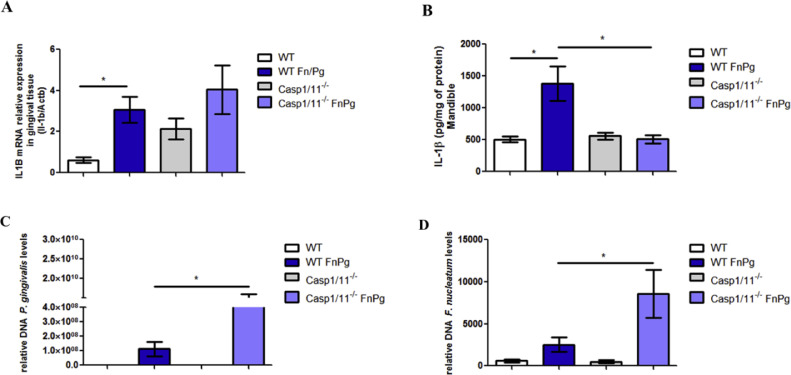
Next, we characterized the downstream pathway of canonical activation of the NLRP3 inflammasome by P2 × 7 in vivo. We found that neither caspase-1 nor caspase-11 were necessary for IL-1β expression increases induced by bacterial co-infection in the gingival tissue, because the levels of IL-1β mRNA in the gingival tissue of co-infected caspase-1/11−/− mice were similar to those of co-infected WT animals (Fig. 2A). However, both caspase-1 and caspase-11 were important for the secretion of this cytokine in the mandible of animals coinfected by the bacteria, because the levels of IL-1β in the mandible were lower in this tissue in co-infected caspase-1/11−/− animals than in the co-infected WT animals (Fig. 2B). When evaluating the bacterial load, we found more P. gingivalis and F. nucleatum DNA in the gingival tissue of the co-infected caspase-1/11−/− animals than in the co-infected WT animals (Fig. 2C and 2D).
Fig. 2.
Caspase-1/11−/- mice demonstrate greater susceptibility to infection by P. gingivalis and F. nucleatum in vivo. (A) Relative levels of IL-1β mRNA are shown in the gingival tissue using real-time PCR. (B) Levels of IL-1β are shown in the mandible by ELISA. (C) Relative amount of P. gingivalis DNA and (D)F. nucleatum DNA in gingival tissue using real-time PCR of C57BL/6 (WT) and caspase-1/11−/- animals infected or not infected orally with 109 CFU of each strain of P. gingivalis (ATCC 33,277) and F. nucleatum (ATCC 25,586). The relative amounts of DNA were calculated as described in the Methods. The data represent the mean ± SEM. (A) WT FnPg = 9, caspase-1/11−/- FnPg n = 11 and (B) WT FnPg n = 6, caspase-1/11−/- FnPg n = 6 were analyzed using the Kruskal–Wallis test followed by the Dunn's test. (C) WT FnPg = 8, caspase-1/11−/- FnPg = 9 and (D) WT FnPg = 10, caspase-1/11−/- FnPg = 7 were analyzed using the Mann–Whitney test. * p<0.05 Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
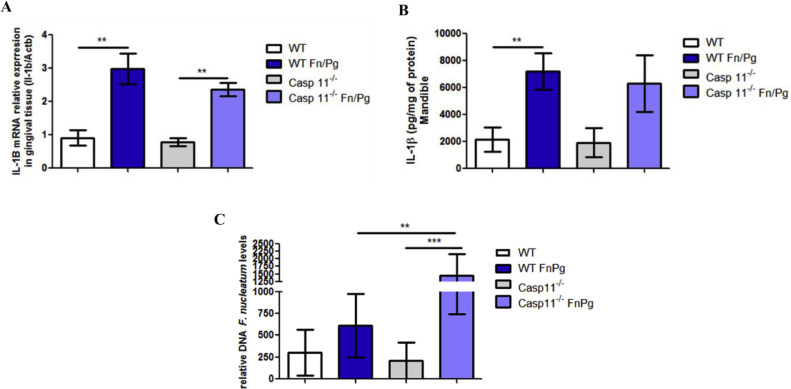
When we evaluated the participation of caspase-11 in co-infected animals, we found that caspase-11 was not necessary for the expression of IL-1β in gingival tissue (Fig. 3A) nor for secretion of this cytokine in the mandible (Fig. 3B), since no differences were found between co-infected WT and caspase-11−/− animals. However, we found greater amounts of F. nucleatum DNA in the gingival tissue of the caspase-11−/− animals co-infected than in co-infected WT animals (Fig. 3C).
Fig. 3.
Caspase-11 is critical for controlling F. nucleatum infection in vivo(A) Relative levels of IL-1β mRNA shown in the gingival tissue using real-time PCR. (B) Levels of IL-1β are shown in the mandible by ELISA. (C) Relative amount of F. nucleatum DNA in gingival tissue using real-time PCR of C57BL/6 (WT) and Caspase-11−/− animals infected or not infected orally with 109 CFU of each strain of P. gingivalis (ATCC 33,277) and F. nucleatum (ATCC 25,586). The relative amounts of DNA were calculated as described in the Methods. The data represent the mean ± SEM. (A) WT FnPg = 4, Caspase-11−/- FnPg = 4 and data were analyzed by one-way ANOVA, followed by the Tukey test. (B) WT FnPg = 7, Caspase-11−/- FnPg = 9 and data were analyzed using the Mann–Whitney test. (C) WT FnPg = 8, Caspase-11−/- FnPg = 6 and data were analyzed using one-way ANOVA followed by the Tukey test. **p <0.005; *** p <0.001 Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
Together, these results suggest that caspase-1 is required for the secretion of IL-1β in the mandible of co-infected mice and acts to control infection by P. gingivalis in gingival tissue, whereas caspase-11 is required to control infection by F. nucleatum in gingival tissue of coinfected mice.
3.3. Co-infection in vivo by P. gingivalis and F. nucleatum increased the expression of inflammatory markers in gingival tissue and altered the profile of inflammatory cells in submandibular lymph nodes
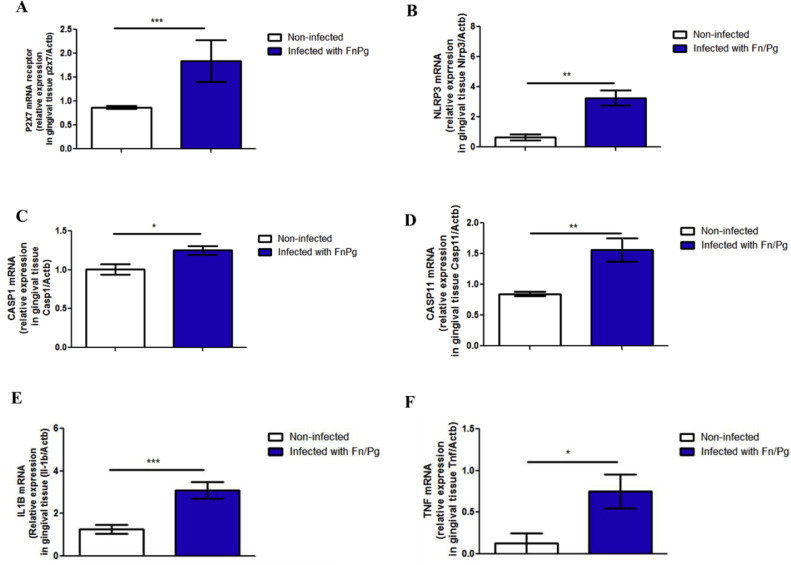
We verified the effect of oral co-infection by P. gingivalis and F. nucleatum on the targets of the canonical and non-canonical pathways of NLRP3 inflammasome activation in C57BL/6 mice. We found significantly higher amounts of P2 × 7 mRNA in the gingival tissue than in uninfected animals (Fig. 4A), as well as significantly higher levels of NLRP3 mRNA (Fig. 4B), caspase-1 mRNA (Fig. 4C), caspase-11 mRNA (Fig. 4D), IL-1β mRNA (Fig. 4E), and TNF-α mRNA (Fig. 4F).
Fig. 4.
Oral co-infection increases expression of pro-inflammatory markers. Relative mRNA levels of (A) P2 × 7 receptor, (B) NLRP3, (C) caspase-1, (D) caspase-11, (E) IL-1β, and (F) TNF-α in the gingival tissue using real time PCR of C57BL/6 (WT) mice co-infected orally or not with 109 CFU of each bacterial species, F. nucleatum strain (ATCC 25,586) and P. gingivalis (ATCC 33,277). The data represent the mean ± SEM. (A) WT = 10; WT FnPg = 9 and data were analyzed using the Mann–Whitney test. (B) WT= 4; WT FnPg = 7, (C) WT = 4; WT FnPg = 4, (D) WT = 6; WT FnPg = 7, (E) WT = 14; WT FnPg = 15 and were analyzed using two-tailed unpaired t-test. (F) WT = 5; WT FnPg = 7 and data were analyzed using the Mann–Whitney test. * p <0.05; **p <0.005; *** p <0.001 Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
Taken together, these data suggest that oral coinfection in mice for one week stimulates the expression of molecules of the canonical and non-canonical pathways of NLRP3 inflammasome activation.
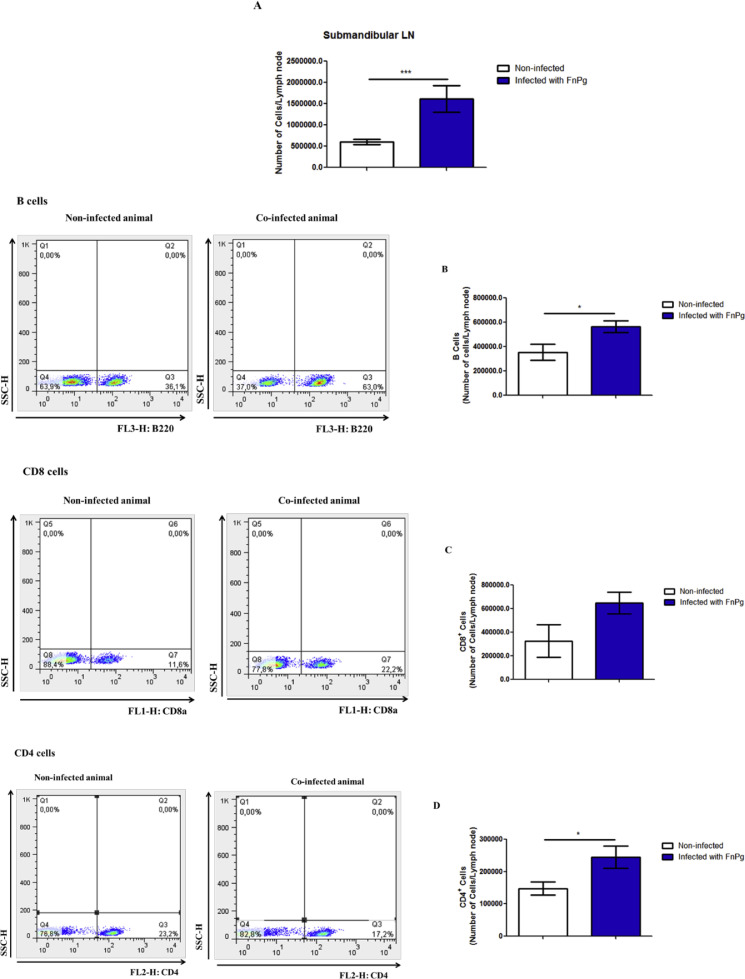
We then examined the effect of coinfection by P. gingivalis and F. nucleatum on the profile of inflammatory cells in the submandibular lymph nodes. We found that coinfection led to an increased number of cells in the submandibular lymph nodes (Fig. 5A). This increase was also due to the number of B cells (Fig. 5B), CD8+ cells (Fig. 5C) and CD4+ cells (Fig. 5D) that was increased in the infected animals compared with uninfected animals.
Fig. 5.
Oral co-infection with periodontopathogens increases the number of B cells, CD4+and CD8+ cells in the submandibular lymph node. (A) Number of submandibular lymph node cells, (B) number of B cells, (C) number of CD8+ cells, and (D) number of CD4+ cells from C57BL/6 mice infected or not infected orally with 109 CFU of each bacterial species, P. gingivalis (ATCC 33,277) and F. nucleatum (ATCC 25,586). The data represent the mean ± SEM. (A) WT = 13; WT FnPg = 17 and were analyzed using the Mann–Whitney test. (B) WT = 9; WT FnPg = 10, (C) WT = 3; WT FnPg = 3 and (D) WT = 12; WT FnPg = 16 and were analyzed using the two-tailed unpaired t-test. * p <0.05; **p <0.005; *** p <0.001 Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum; LN: lymph node.
3.4. Pathways involved in F. nucleatum and P. gingivalis induction of IL-1β secretion in murine macrophages
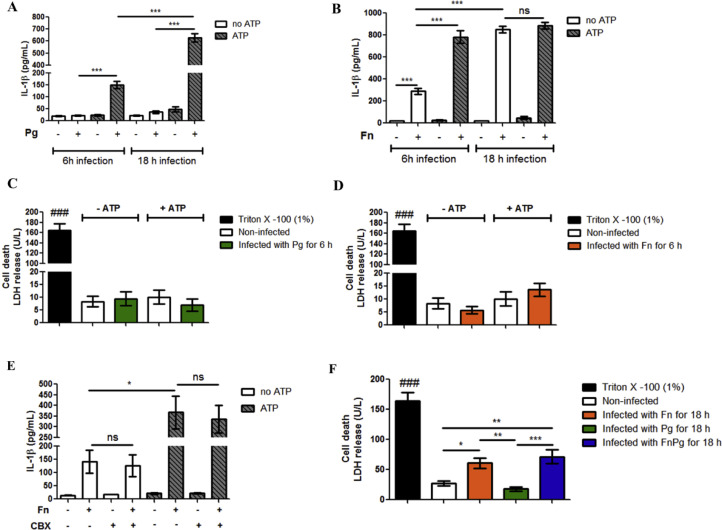
To explore the mechanisms involved in the canonical and non-canonical pathways of NLRP3 inflammasome activation induced by infection with P. gingivalis and F. nucleatum, we characterized the pathways during infection in vitro, using bone-marrow derived macrophages (BMDMs). Thus, we investigated IL-1β secretion induced by P. gingivalis and F. nucleatum infection in BMDMs from C57BL/6 mice. We found that P. gingivalis after 6 h and 18 h of infection induced IL-1β secretion in BMDMs only following treatment with 3 mM ATP, which acts as a second signal for NLRP3 inflammasome activation. After 18 h, the levels of IL-1β secretion were significantly higher than after 6 h of infection (Fig. 6A).
Fig. 6.
F. nucleatum infection by itself promotes IL-1β secretion. (A) and (B) Levels of IL-1β in the supernatants of BMDMs of C57BL/6 (WT) animals after 6 h or 18 h of in vitro infection by P. gingivalis (ATCC 33,277) (Pg) (MOI 100) or F. nucleatum (ATCC 25,586) (Fn) (MOI 20) treated or not with 3 mM ATP (30 min before completing 6 h or 18 h of infection). (C) and (D) Lactate dehydrogenase (LDH) levels in the supernatant of BMDMs of C57BL/6 (WT) animals after 6 h in vitro infection for Pg (MOI 100) or Fn (MOI 20). (E) IL-1β levels in the supernatants of BMDMs of C57BL/6 (WT) animals after 6 h of in vitro infection with Fn (MOI 20) treated or not with 3 mM ATP (30 min before completing 6 h of infection) and 50 µM carbenoxolone (CBX) (1 h and 10 min before completing 6 h of infection). (F) Lactate dehydrogenase (LDH) levels in the supernatants of BMDMs of C57BL/6 (WT) animals after 18 h in vitro infection for Pg (MOI 100) and/or Fn (MOI 20). The data represent the mean ± SEM and were analyzed using one-way ANOVA, followed by the Tukey test. (A)n = 3 per group; (B)n = 6 per group at 6 h of infection and n = 3 per group at 18 h of infection; (C)n = 4 per group; (D)n = 5 per group; (E)n = 3 per group; (F)n = 6 per group. * p<0.05; **p<0.005; *** p<0.001. ###p<0.001 in relation to the other groups. ns: not significant. Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
When the BMDMs were infected for 6 h and 18 h with F. nucleatum, infection by itself (without ATP treatment) was sufficient to induce IL-1β secretion, with the IL-1β release being much higher after 18 h than after 6 h (Fig. 6B). Potentiation of IL-1β secretion was observed with ATP treatment after 6 h of infection, but not after 18 h (Fig. 6B).
Infection with some gram-negative bacteria can induce pyroptosis (Broz et al., 2012; Case et al., 2013). Thus, we measured cytotoxicity during infection by measuring LDH levels released from host cells into the extracellular medium. We observed that, after 6 h of infection, neither P. gingivalis nor F. nucleatum bacteria induced cytotoxicity (Fig. 6C and D).
Since the pannexin-1 channel induces ATP release in macrophages (Pelegrin and Surprenant, 2006) and activates P2 × 7 purinergic receptors, culminating in IL-1β release (Iglesias et al., 2008), we evaluated the participation of pannexin-1 in IL-1β secretion induced by F. nucleatum infection. We used carbenoxolone (CBX), widely used as a pannexin-1 inhibitor (Connors, 2012), in BMDMs infected with F. nucleatum for 6 h with and without ATP treatment. No changes in IL-1β levels were observed (Fig. 6E).
Since ATP did not increase the secretion of IL-1β in BMDMs after 18 h of infection with F. nucleatum (Fig. 6B), we examined whether bacterial infection was cytotoxic after 18 h of infection. We found that F. nucleatum infection caused a significant increase in LDH release from infected cells, compared with uninfected BMDMs (Fig. 6F).
These results show that, unlike P. gingivalis infection, F. nucleatum infection by itself (without ATP treatment) promotes IL-1β secretion in murine macrophages, it does not require the pannexin-1 channel, and it induces cell death after 18 h of infection.
3.5. Contribution of P2 × 7 to the immune response against P. gingivalis and F. nucleatum
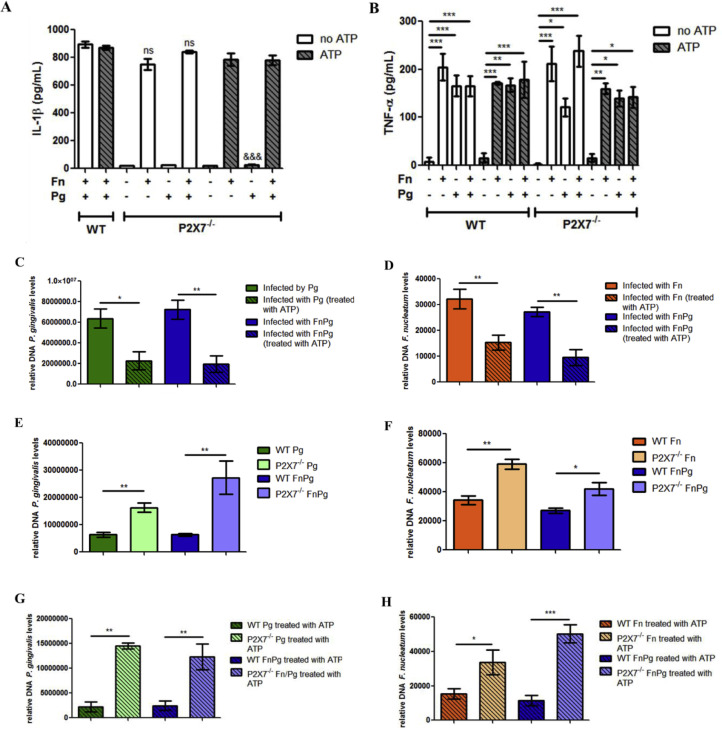
We next evaluated the participation of P2 × 7 in IL-1β secretion induced by bacterial infections in BMDMs derived from P2 × 7−/− and WT mice. In the absence of P2 × 7 in BMDMs, there was a significant decrease in IL-1β levels induced by P. gingivalis for 18 h following treatment with 3 mM ATP, compared with WT BMDMs infected with P. gingivalis and treated with ATP (Figs. 7A and 6A). Conversely, infection with F. nucleatum by itself was sufficient to induce IL-1β secretion in the supernatants of P2 × 7−/- BMDMs and in the supernatant of WT BMDMs (Figs. 7A and 6B). However, F. nucleatum was cytotoxic after 18 h of infection (Fig. 6F).
Fig. 7.
P2 × 7 controls the infection by P. gingivalis and F. nucleatum in murine macrophages. (A) Levels of IL-1β and (B) levels of TNF-α in the supernatants of BMDMs of C57Bl/6 (WT) and P2 × 7−/− animals after 18 h of in vitro infection with P. gingivalis (ATCC 33,277) (Pg) (MOI 100) and/or F. nucleatum (ATCC 25,586) (Fn) (MOI 20) treated or not with 3 mM ATP (30 min before completing 18 h of infection). (C) and (D) Relative amount of Pg DNA and Fn DNA, respectively, using real-time PCR in BMDMs from C57Bl/6 (WT) animals after 18 h of in vitro infection by Pg (MOI 100) and/or Fn (MOI 20). (E) and (F) Amount of Pg DNA and Fn DNA, respectively, using real-time PCR in BMDMs from C57Bl/6 (WT) and P2 × 7−/− animals after 18 h of in vitro infection by Pg (MOI 100) and/or Fn (MOI 20). (G) and (H) Amount of Pg DNA and Fn DNA, respectively, using real-time PCR in BMDMs from C57Bl/6 (WT) and P2 × 7−/− animals after 18 h of in vitro infection by Pg (MOI 100) and/or Fn (MOI 20) treated or not with 3 mM ATP (in the last 30 min of the total 6 h of infection). The data represent the mean ± SEM and were analyzed using one-way ANOVA, followed by the Tukey test. (A) and (B)n = 3 per group; (C), (D), (E) and (F)n = 4 per group; (G)n = 3 per group; (H)n = 4 per group. * p <0.05; **p <0.005; *** p <0.001. (A) *** p<0.001;&&&p<0.001 compared to the WT cells infected with Pg and treated with ATP (Fig. 6A). ns: not significant compared with their respective (WT) mono and co-infected controls, without ATP treatment (Fig. 6B). Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
As a control, we measured the effect of infection on secretion of a cytokine that does not depend on activation of the inflammasome. We observed that TNF-α levels were similar in the BMDMs infected with one or both bacterial species in the cells derived from the knockouts with or without treatment with 3 mM ATP (Fig. 7B).
Since P2 × 7 was required for secretion of IL-1β during P. gingivalis infection, we checked the bacterial load in P2 × 7−/− BMDMs and WT BMDMs infected with F. nucleatum or P. gingivalis alone, or both bacterial species, to determine whether the P2 × 7 pathway had an effect on the bacterial load in murine macrophages. We observed that treatment with 3 mM ATP reduced the bacterial infection in both monoinfected and coinfected WT BMDMs, leading to a significant decrease in the levels of P. gingivalis (Fig. 7C) and F. nucleatum (Fig. 7D) after 18 h of infection, as measured by their DNA content. In the absence of P2 × 7 in BMDMs, there was an increase in the levels of P. gingivalis and F. nucleatum in infected cells when compared with WT BMDMs for both mono- and co-infection for 18 h (Fig. 7E and F). Similar results were observed when WT BMDMs and P2 × 7−/− BMDMs were infected separately with single bacterial species or co-infected with both bacterial species for 18 h and treated with 3 mM ATP (Fig. 7G and H).
These results suggest that P2 × 7 was necessary for the secretion of IL-1β induced by P. gingivalis and that P2 × 7 controls the infection by P. gingivalis and F. nucleatum in murine macrophages.
3.6. Role of P2 × 7, caspase-1, and caspase-11 in the immune response against P. gingivalis and F. nucleatum infection
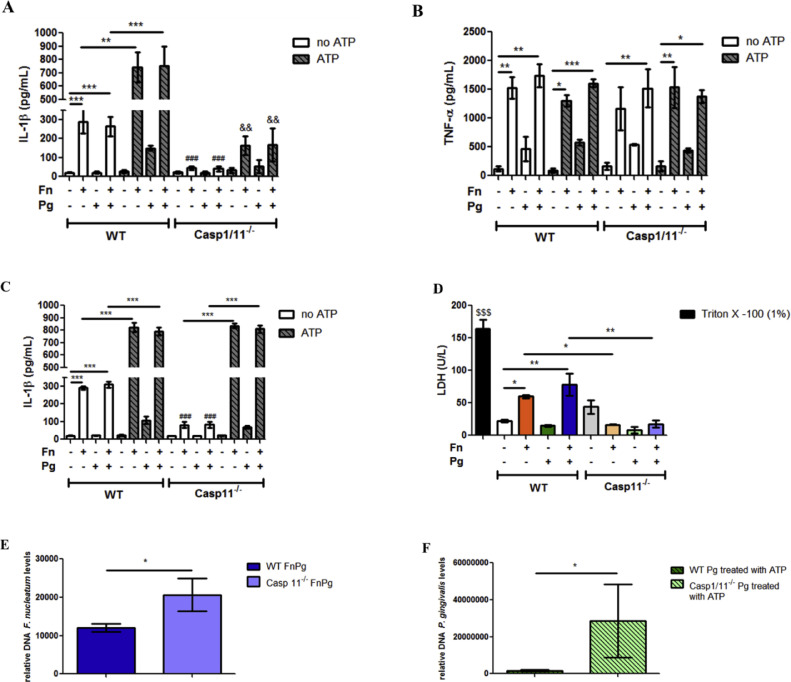
We next characterized the involvement of caspase-1 in inflammasome activation during infection. For this, we used BMDMs derived from caspase-1/11−/− and WT mice. After infection of BMDMs with F. nucleatum for 6 h, we found a significant decrease in IL-1β levels in the supernatants of infected caspase-1/11−/− BMDMs compared with infected WT BMDMs (Fig. 8A). The same significant decrease was observed in the supernatant of caspase-1/11−/− BMDMs infected for 6 h with P. gingivalis and treated with ATP, compared with monoinfected WT BMDMs (Fig. 8A and see supplementary data Fig. S3A). We did not detect any effect on TNF-α secretion in BMDMs of caspase-1/11−/− that were infected singly or dually with the two bacterial species, treated or not with ATP, compared with infection or treatment of WT cells (Fig. 8B).
Fig. 8.
Caspase-11 is required during F. nucleatum infection to IL-1β secretion and cell death independently of caspase-1. (A) Levels of IL-1β and (B) TNF-α in the supernatants of BMDMs of C57Bl/6 (WT) and caspase-1/11−/- animals after 6 h of in vitro infection by P. gingivalis (ATCC 33,277) (Pg) (MOI 100) and/or F. nucleatum (ATCC 25,586) (Fn) (MOI 20) treated or not with ATP (3 mM) (30 min before completing 6 h of infection). (C) Levels of IL-1β in the supernatants of BMDMs of C57BL/6 (WT) and caspase-11−/- animals after 6 h of in vitro infection by Pg (MOI 100) and/or Fn (MOI 20) treated or not with 3 mM ATP (30 min before completing 6 h of infection). (D) Lactate dehydrogenase (LDH) levels in the supernatants of WT BMDM after 18 h in vitro infection by Pg (MOI 100) and/or Fn (MOI 20). (E) Relative amounts of Fn DNA using real-time PCR in BMDMs from WT and caspase-11−/- animals after 18 h of in vitro infection by Pg (MOI 100) and Fn (MOI 20). (F) Amounts of Pg DNA using real-time PCR in BMDMs from WT and caspase-1/11−/- animals after 18 h of in vitro infection by Pg (MOI 100) treated with 3 mM ATP (30 min before completing 6 h of infection). The data represent the mean ± SEM. (A), (B), (C) and (D)n = 3 per group and data were analyzed by one-way ANOVA, followed by the Tukey test. (E) WT FnPg (n = 7), caspase-11−/- FnPg (n = 5) data were analyzed by unpaired t-test and (F) WT Pg ATP (n = 6), caspase-1/11−/- Pg ATP (n = 5) data were analyzed using the Mann–Whitney test. * p <0.05; **p <0.005; *** p <0.001. ### p <0.001 compared with their respective (WT) mono and co-infected controls not treated with ATP. && compared with their respective (WT) mono and co-infected controls treated with ATP. $$$ p<0.001 compared to the other groups. Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
Since caspase-11 participates in the non-canonical pathway of NLRP3 inflammasome activation and it is involved in clearance of some infections (Roberts and Yilmaz, 2015; Russo et al., 2018; Huang et al., 2019), we examined the effect of the absence of caspase-11 in BMDMs derived from caspase-11−/− mice on secretion of IL-1β. We found that monoinfection with F. nucleatum and coinfection with F. nucleatum and P. gingivalis in caspase-11−/- BMDMs resulted in significant decreases in the levels of IL-1β secreted, compared with infections in WT BMDMs, with and without ATP treatment (Fig. 8C). However, when BMDMs from caspase-11−/− that had been monoinfected with F. nucleatum or coinfected with both bacterial species were treated with ATP, there was a significant increase in IL-1β secretion compared with secretion from caspase-11−/- BMDMs monoinfected with F. nucleatum and coinfected without treatment with ATP, suggesting that F. nucleatum infection induces two IL-1β secretion pathways: one is caspase-11-dependent and ATP-independent, and other is caspase-1-, ATP- and P2 × 7-dependent. When examining caspase-11−/− BMDMs monoinfected with P. gingivalis treated or not with ATP, there was no difference in IL-1β levels compared with WT BMDMs monoinfected with P. gingivalis (see supplementary data Fig. S3B).
Since caspase-11 activation can lead to pyroptosis (Huang et al., 2019), we evaluated the cytotoxicity in caspase-11−/- BMDMs infected with F. nucleatum, as we already observed that F. nucleatum infection induces cytotoxicity after 18 h of infection in BMDMs (Fig. 6F). We found that, in the absence of caspase-11 in BMDMs monoinfected with F. nucleatum or co-infected with both species for 18 h, LDH levels were lower compared with F. nucleatum-monoinfected or coinfected WT cells (Fig. 8D). We then examined whether caspase-11 was involved in control of infection by P. gingivalis and F. nucleatum in BMDMs. We observed that the absence of caspase-11 led to an increase in F. nucleatum levels in co-infected caspase-11−/- BMDMs at 18 h, compared with the co-infected WT cells (Fig. 8E), suggesting that caspase-11 is involved in the immune response during F. nucleatum infection. In caspase-11−/− BMDMs infected with P. gingivalis for 18 h, there was no difference in the levels of P. gingivalis, compared with monoinfected WT BMDMs (see supplementary data Fig. S3C). Caspase-1/11−/− BMDMs monoinfected with P. gingivalis for 18 h and treated with ATP showed significantly higher levels of P. gingivalis than monoinfected WT BMDMs treated with ATP (Fig. 8F).
Taken together, these results suggest that the immune response through the canonical pathway of NLRP3 inflammasome activation involving P2 × 7 is predominant in the response against P. gingivalis. The activation of caspase-11 is required during F. nucleatum infection to induce NLRP3 inflammasome activation followed of IL-1β secretion and cell death independently of caspase-1, and appears to be the predominant immune response against F. nucleatum.
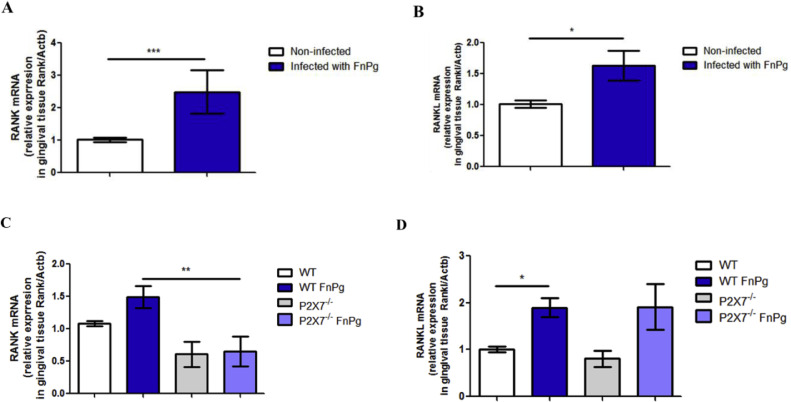
To determine whether oral co-infection affected the expression of bone demineralization markers during one week of infection, we measured RANK and RANK-L expression in the gingival tissue of C57BL/6 (WT) mice. Interestingly, we found greater expression of bone demineralization markers, RANK and RANKL mRNA in infected (Fig. 9A and B), than in uninfected animals. Since the cytokine IL-1β contributes to alveolar bone demineralization (Chen et al., 2014; Huynh et al., 2017), we then examined the effect of the P2 × 7 receptor on bone demineralization markers. Interestingly, we observed that, in the gingival tissue of P2 × 7−/− animals, there were lower levels of RANK mRNA than in co-infected WT animals (Fig. 9C), although the same result was not found for RANKL mRNA, which was similar in the gingival tissue of co-infected P2 × 7−/− and WT animals (Fig. 9D).
Fig. 9.
Oral co-infection increases expression of osteoclastogenic markers. (A) RANK mRNA and (B) RANKL mRNA, in the gingival tissue using real time PCR of C57BL/6 (WT) mice co-infected orally or not with 109 CFU of each bacterial species, F. nucleatum strain (ATCC 25,586) and P. gingivalis (ATCC 33,277). (C) Relative levels of RANK mRNA and (D) of RANKL are shown in the gingival tissue using real-time PCR from C57BL/6 (WT) and P2 × 7−/− animals infected or not infected orally with 109 CFU of each strain P. gingivalis (ATCC 33,277) and F. nucleatum (ATCC 25,586). The data represent the mean ± SEM. (A) WT = 8; WT FnPg = 7, (B) WT = 8; WT FnPg = 10, and were analyzed using the two-tailed unpaired t-test. (C) WT FnPg = 10, P2 × 7−/- FnPg = 8; (D) WT FnPg = 8, P2 × 7−/- FnPg = 5. The data were analyzed using one-way ANOVA, followed by the Tukey test. * p <0.05; **p <0.005; *** p <0.001 Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum.
These data show that, during this period of coinfection, there was an increase in the expression of bone demineralization markers stimulated by coinfection.
4. Discussion
Many studies have shown the importance of IL-1, particularly IL-1β, in the development of periodontal disease (Aral et al., 2020; Cheng et al., 2020). In an experimental model of periodontitis in the primate Macaca fascicularis, IL-1 and TNF antagonists reduced the recruitment of inflammatory cells and osteoclasts in the vicinity of the bone, and reduced bone loss, thereby demonstrating that these pro-inflammatory cytokines are important for the development of periodontitis (Assuma et al., 1998). In our study, we found that coinfection with P. gingivalis and F. nucleatum in mice increased expression of pro-inflammatory cytokines IL-1β and TNF-α in gingival tissue and increased protein levels of IL-1β in the mandibles of infected mice. Our study is the first to combine both animal and cell culture to describe the molecules involved in inflammasome activation during co-infection of P. gingivalis and F. nucleatum. This model more closely represents the polymicrobial etiology of periodontitis than most previous studies performed using mono-infections.
Our group has investigated the role of the P2 × 7 receptor during host immune responses against intracellular pathogens (Morandini et al., 2014; Ramos-Junior et al., 2015; Almeida-da-Silva et al., 2019; Moreira-Souza et al., 2017). The P2 × 7 receptor induces intracellular microbicidal responses, including reactive oxygen species (ROS) production, lysosome-phagosome fusion and IL-1β production (Moreira-Souza et al., 2017; Savio and Coutinho-Silva, 2019). Considering that extracellular ATP is a damage-associated molecular pattern (DAMP) that activates the purinergic P2 × 7 receptor, we treated mono- and co-infected macrophages with ATP, and observed a reduction in both bacterial loads. In contrast, macrophages derived from P2 × 7−/− mice that were mono- or co-infected and treated with ATP showed no reduction in bacterial loads, thereby demonstrating that the P2 × 7 receptor was involved in controlling both P. gingivalis and F. nucleatum load in murine BMDMs. During a mouse model of oral co-infection, we observed increased expression of P2 × 7 in gingival tissues. This receptor was required for controlling the bacterial load of both P. gingivalis and F. nucleatum in gingival tissue. It was shown by our group that infection with P. gingivalis increased the expression of P2 × 7 in vitro (Morandini et al., 2014) and in vivo (Ramos-Junior et al., 2015), and contributed to control of P. gingivalis infection in vivo (Almeida-da-Silva et al., 2019).
The cytokine IL-1β is a key mediator of the host's response to infections (Eskan et al., 2008). In a study by Almeida-da-Silva et al. (2019), it was shown that IL-1β leads to P. gingivalis elimination by autocrine IL-1 receptor activation in vivo and in THP-1 cells (Almeida-da-Silva et al., 2019). Secretion of the active form of IL-1β is due to the activation of inflammatory caspases and this activation is regulated by the inflammasome (Groslambert and Py, 2018). The ATP-P2 × 7 pathway is one of the most potent stimuli for NLRP3 inflammasome activation (Munoz-Planillo et al., 2013), which is followed by secretion of IL-1β. We observed that P2 × 7 was required for the expression and secretion of IL-1β in the mandible of co-infected animals, suggesting that IL-1β is involved in controlling the intracellular bacterial load. Our data support the notion that the P2 × 7 receptor is an important modulator of intracellular oral pathogens (De Andrade et al., 2019; Almeida-da-Silva et al., 2016; Coutinho-Silva and Eduardo Baggio, 2021) such as P. gingivalis and F. nucleatum. Future studies will further evaluate the possible mechanism involved in P2 × 7-mediated elimination of oral bacteria.
Evaluation of the downstream pathways of NLRP3 activation via P2 × 7 showed an increase in the mRNA expression of NLRP3, caspase-1 and caspase-11 in the gingival tissues of co-infected animals compared to controls. Bostanci et al. (2009) found increased expression of NLRP3 in the gingival tissue of patients with gingivitis compared with healthy individuals (Bostanci et al., 2009). Park et al. (2014) observed that the expression of NLRP3 and caspase-1 was increased in the gingival tissue of patients with periodontitis compared with healthy individuals (Park et al., 2014). In our study, we observed in vivo that the absence of both caspase-1 and −11 led to increased bacterial loads of both P. gingivalis and F. nucleatum in the gingival tissue. When we verified the production of IL-1β in the mandibles of caspase-1/11−/− animals, IL-1β levels were lower than those of the co-infected WT animals. However, caspase-1 and caspase-11 were dispensable for IL-1β mRNA expression in the gingival tissue of mice infected with both bacterial species. Similarly, Almeida-da-Silva et al. (2019) found higher loads of P. gingivalis in exudates of caspase-1/11−/− mice infected with P. gingivalis in an air pouch model, compared with infected WT animals (Almeida-da-Silva et al., 2019). Our findings and previous studies support the idea that inflammasome activation leading to IL-1β secretion is an important host immune response against oral bacterial infections.
Infection by some Gram negative bacteria induces the expression of pro-caspase-11 mRNA, and caspase-11 detects and is activated intracellularly by LPS from these bacteria (Huang et al., 2019). Activated caspase-11 promotes the fusion between the phagosomes containing the pathogen and lysosomes, and inhibits replication of the pathogen in the intracellular environment (Akhter et al., 2012). We observed that caspase-11 controlled the load of F. nucleatum in the gingival tissue of co-infected animals and in co-infected murine macrophages.
Aachoui et al. (2013) demonstrated that caspase-11 protected animals from infection by the intracellular pathogens Burkholderia thailandensis and B. pseudomallei (Aachoui et al., 2013). Perlee et al. (2020) also noted that caspase-11 was important for the control of Klebsiella pneumoniae in the lungs, and resulted in the presence of IL-1β in pulmonary homogenates (Perlee et al., 2020). Unlike our results for F. nucleatum infection, we found that caspase-11 did not affect the bacterial load of P. gingivalis in murine BMDMs (see supplementary data Fig. S3B and C). This is thus the first study to report the involvement of caspase-11 in a mono- and co-infection model in vivo and in vitro by P. gingivalis and F. nucleatum.
As previously observed, P. gingivalis infection of BMDMs or gingival epithelial cells (GECs) (Almeida-da-Silva et al., 2019; Yilmaz et al., 2010) on its own does not activate the NLRP3 inflammasome, but requires an exogenous danger signal such as extracellular ATP to activate the inflammasome and induce IL-1β secretion (Olsen and Yilmaz, 2016). Our data corroborate those of previous studies showing that murine BMDMs infected with P. gingivalis do not secrete IL-1β unless these infected cells are also treated with ATP. In contrast, we found that, in murine BMDMs, F. nucleatum infection by itself was sufficient to induce secretion of IL-1β, suggesting the non-canonical pathway of NLRP3 activation by caspase-11 for the secretion of IL-1β. We previously showed that F. nucleatum infection by itself also induced IL-1β secretion in GECs (Bui et al., 2016). When murine macrophages were infected with F. nucleatum and treated with ATP, the levels of IL-1β increased further, suggesting that ATP had an additional effect on the activation of NLRP3 via the canonical inflammasome in F. nucleatum-infected cells. Corroborating our findings, Taxman et al. (2012) reported high levels of IL-1β induced by F. nucleatum infection of murine BMDMs in the absence of ATP, and that ATP treatment potentiated IL-1β secretion (Taxman et al., 2012). For IL-1β secretion to occur, cell lysis is not necessary (Evavold et al., 2018). Together, our data and those of previous studies support the view that P. gingivalis and F. nucleatum induce inflammasome activation and IL-1β secretion through different pathways.
Because we showed that there was no further increase in IL-1β levels after 18 h of infection by F. nucleatum and ATP treatment, we then analyzed possible bacterial cytotoxic effects. We found that infection with either F. nucleatum or P. gingivalis was not cytotoxic after 6 h of infection; however, infection with only F. nucleatum was cytotoxic after 18 h of infection. Consistent with our results, Fleetwood et al. (2017) found that there was no LDH release from BMDMs infected with P. gingivalis for 24 h (Fleetwood et al., 2017); and similar results were obtained by Jung et al. (2015), who studied P. gingivalis infection in THP-1 cells (Jung et al., 2015). Our data are consistent with the view that P. gingivalis can modulate host biology in order to promote its own survival and establish persistent infections (Yao et al., 2010; Roberts et al., 2017).
In our study, we found that IL-1β secretion induced by F. nucleatum in murine BMDMs did not depend on pannexin-1, a non-selective cell plasma membrane channel (Adamson and Leitinger, 2014; Pelegrin and Surprenant, 2007). This channel is highly expressed in human and murine macrophages, is associated with the ligation of P2 × 7, and is involved in activation of caspase-1 with subsequent release of IL-1β (Pelegrin and Surprenant, 2006). However, pannexin-1 mediates some IL-1β release after non-canonical activation of NLRP3 via caspase-11, independently of P2 × 7 (Lamkanfi and Dixit, 2014; De And Martinon, 2015).
We observed that P2 × 7 was necessary for ATP-induced IL-1β secretion in P. gingivalis-infected BMDMs for 18 h, as we previously reported (Morandini et al., 2014; Ramos-Junior et al., 2015; Almeida-da-Silva et al., 2019). However, F. nucleatum infection resulted in IL-1β secretion independently of ATP treatment in 18 h of infection. Moreover, we observed that, at 18 h of infection, F. nucleatum is cytotoxic. When evaluating the downstream pathway of NLRP3 activation via P2 × 7 in vitro, we found that IL1-β levels were lower in caspase-1/11−/− BMDMs after single-species and dual-species infection by F. nucleatum and P. gingivalis, regardless of ATP treatment. Ramos-Junior et al. (2015) found that the absence of both caspases 1 and 11 did not affect the expression of IL-β in BMDMs infected with P. gingivalis (Ramos-Junior et al., 2015).
Caspase-11 promotes IL-1β secretion via the NLRP3 inflammasome (Kayagaki et al., 2011; Rathinam et al., 2012). In our study, we found that caspase-11 was important for the secretion of IL-1β in murine BMDMs induced by F. nucleatum infection, without treatment with ATP, while caspase-11 did not affect the levels of IL-1β induced by P. gingivalis. Similar to our findings, Kayagaki et al. (2011) found that LPS-primed caspase-11−/− BMDMs, when infected with C. difficile toxin B (CTB) or E. coli or C. rodentium or V. cholerae, failed to secrete IL-1β (Kayagaki et al., 2011). In our mouse co-infection model, the absence of caspase-11 did not influence the secretion of IL-1β in the animals' mandibles, suggesting activation of the inflammasome NLRP3 via the canonical inflammasome. Similar results were found using co-infected murine macrophages treated with ATP. Casson et al. (2013) found that Yersinia pseudotuberculosis also depends on caspase-11 for the secretion of IL-1β and induction of cell death (Casson et al., 2013). Caspase-11 limits the intracellular replication of bacteria such as S. typhimurium and Burkholderia species in epithelial cells and macrophages (Stowe et al., 2015).
Caspase-11 directly induces pyroptosis independently of NLRP3 or caspase-1 (Kayagaki et al., 2011; Huang et al., 2019; Man et al., 2017). In our study, caspase-11 was also involved in the cytotoxicity induced by F. nucleatum. Caspase-11-dependent cell death induction by F. nucleatum also represents a way to control the bacterial load in infected cells. However, caspase-11 was not involved in P. gingivalis load control in murine BMDMs (see supplementary data Fig. S3B and C). Gram-negative bacteria such as P. gingivalis (Coats et al., 2009) and Yersinia pestis (Brodsky et al., 2010) have the ability to change the composition of lipid A from their LPS, thereby preventing the activation of caspase-11 and manipulating the host's immune response. This hypothesis may explain the inability of P. gingivalis to activate the inflammasome via caspase-11 as we previously discussed (De Andrade et al., 2019). Our study is the first to describe the role of the non-canonical inflammasome during F. nucleatum-induced IL-1β secretion and control of bacterial load.
The inflammasome plays a critical role in periodontal disease, as NLRP3-deficient mice fail to develop the disease in mouse models of study (Yamaguchi et al., 2017), and polymorphisms leading to increased expression of NLRP3 and IL1B are linked to periodontitis susceptibility in humans (De Alencar et al., 2020). Characterization of the mechanisms of inflammasome activation during periodontitis should contribute to a better understanding of the pathophysiology of the disease. In fact, a number of studies suggested the use of NLRP3 pharmacological inhibitors for the treatment of various diseases including diabetes, arthritis and cardiovascular diseases (Marchetti, 2019; Van Hout et al., 2017; Zahid et al., 2019). Our study supports the notion that the NLRP3 inflammasome should be considered as a potential target for treatment and/or prevention of periodontitis.
T cells and B cells appear to be important sources of RANKL in periodontal disease (Kawai et al., 2006). When we examined the profile of inflammatory cells in the submandibular lymph nodes of animals coinfected with P. gingivalis and F. nucleatum, we found an increase in the total number of cells in the submandibular lymph nodes. This enhancement was partially due to the increase in the number of B cells, CD8+ T cells and CD4+ T cells compared with uninfected animals.
The Th1 subset appears to be involved in stable injuries in periodontal disease. B cells were observed predominantly in the progression of periodontal lesions, along with profiles of Th2 cells (Gemmell and Seymour, 2004; Okui et al., 2014). However, further studies are needed to verify the involvement of each subset of T cells and also the presence of B cells in different periods of periodontal disease, since there is some disagreement among recent studies (Okui et al., 2014). Our results showed that, after one week of infection, it was already possible to see an increase in the number CD4+ T cells, responsible for stimulating the production of pro-inflammatory cytokines. Future studies should track the activation and migration of lymphocytes from regional lymph nodes to gingival tissues and correlate the presence of cells to specific immune responses.
Zitzmann et al. (2005) reported an increase of B cells instead of T cells in gingivitis lesions in an experimental model of gingivitis (Zitzmann et al., 2005). Mahanonda et al. (2016) found memory B cells in the gums of patients with gingivitis, suggesting previous activation by periodontal microorganisms (Mahanonda et al., 2016). In another study, B cells in response to a bacterium associated with periodontal disease in vivo led to upregulation of RANKL expression (Han et al., 2009). Gemmell et al. (2002) found that BALB/c mice co-infected with F. nucleatum and P. gingivalis displayed a higher percentage of cytokines from CD8+ T cells than from CD4+ T cells in the spleen (Gemmell et al., 2002).
The process of alveolar bone resorption in humans and animal models involves RANKL (receptor-activator of nuclear factor-κB ligand), which binds to RANK (receptor activator of nuclear factor κ B) in osteoclast precursors, leading to differentiation into active cells that produce proteolytic enzymes that degrade bone (Chen et al., 2014; Hajishengallis and Korostoff, 2017). We found a slight significant increase in the expression of RANK and RANKL in the gingival tissue of co-infected animals. Molon et al. (2014) reported increased RANKL expression in C57BL/6 mice monoinfected with P. gingivalis and coinfected with F. nucleatum in a gavage infection model (De Molon et al., 2014), while Lin et al. (2017) found that oral infection with P. gingivalis induced an increase in the area of bone resorption; they also observed increased levels of secreted RANKL (Lin et al., 2017).
Clinical data show that RANKL is enhanced in patients with chronic periodontitis and aggressive periodontitis, compared with healthy and gingivitis groups (Bostanci et al., 2007). Although we did not evaluate the secreted levels of RANK and RANKL, we found an inducing signal for the expression of these molecules during early stages of coinfection. The expression of these bone demineralization markers could increase even more during chronic stages of the co-infection. Whether the expression of these markers are further modulated or whether they are more proeminent during co-infection, compared to mono-infection models (Almeida-da-Silva et al., 2019; Yilmaz et al., 2010), remains to be elucidated.
5. Conclusion
Our study provides evidence that the canonical P2 × 7-caspase-1 pathway is necessary for the secretion of IL-1β induced by P. gingivalis in BMDMs and in mice, while P2 × 7 ligation leads to control of bacterial load of F. nucleatum in BMDMs and in mice, possibly through other mechanisms. Caspase-11 is an essential effector for the non-canonical activation of the NLRP3 inflammassome induced by F. nucleatum infection, resulting in the secretion of IL-1β in murine macrophages. Caspase-11 is also necessary for induction of cell death by F. nucleatum infection, independently of caspase-1. Thus, our study contributes to a better understanding of the host's immune response to coinfection with these periodontopathogenic bacteria. The immune responses and molecules evaluated in this study (P2 × 7 receptor, caspase-1, caspase-11, NLRP3) may be important targets for future research into therapeutic strategies for treatment and prophylaxis of periodontal disease.
Authors’ contributions
Kívia Queiroz de Andrade, Robson Coutinho-Silva and David M. Ojcius conceived the project, Kívia Queiroz de Andrade performed the experiments and made the analyses, reviewed the literature, wrote the first draft of the manuscript, and prepared the figures. Robson Coutinho-Silva and David M. Ojcius and Cássio Luiz Coutinho Almeida-da-Silva critically reviewed the paper and Robson Coutinho-Silva and David M. Ojcius supervised the project.
Ethical approval
Experimental protocol was approved by the Animal Use Ethics Committee (CEUA) of UFRJ, under protocol number 080/19.
Funding
The Brazilian funding agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant number 306,839/2019–9), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)–Finance Code 001, and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (grant number E-26/202.774/2018) are greatly appreciated for their research grants and fellowships. This study was partially supported by funds provided by The Regents of the University of California, Tobacco-Related Diseases Research Program, Grant Number T29FT0540 to CLCAS. The opinions, findings, and conclusions herein are those of the authors and not necessarily represent those of The Regents of the University of California or any of its programs.
Declaration of Competing Interest
Robson Coutinho-Silva and David M. Ojcius serve on the editorial board of Current Research in Microbial Sciences.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2021.100023.
Appendix. Supplementary materials
References
- Aachoui Y., Leaf I.A., Hagar J.A., Fontana M.F., Campos C.G., Zak D.E., et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339(6122):975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson S.E., Leitinger N. The role of pannexin1 in the induction and resolution of inflammation. FEBS Lett. 2014;588(8):1416–1422. doi: 10.1016/j.febslet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter A., Caution K., Abu K.A., Tazi M., Abdulrahman B.A., Abdelaziz D.H., et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity. 2012;37(1):35–47. doi: 10.1016/j.immuni.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-da-Silva C.L.C., Morandini A.C., Ulrich H., Ojcius D.M., Coutinho-Silva R. Purinergic signaling during Porphyromonas gingivalis infection. Biomed. J. 2016;39(4):251–260. doi: 10.1016/j.bj.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-da-Silva C.L.C., Ramos-Junior E.S., Morandini A.C., Rocha G.D.C., Marinho Y., Tamura A.S., et al. P2X7 receptor-mediated leukocyte recruitment and Porphyromonas gingivalis clearance requires IL-1β production and autocrine IL-1 receptor activation. Immunobiology. 2019;224(1):50–59. doi: 10.1016/j.imbio.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Aral K., Milward M.R., Kapila Y., Berdeli A., Cooper P.R. Inflammasomes and their regulation in periodontal disease: a review. J. Periodontal Res. 2020;55(4):473–487. doi: 10.1111/jre.12733. [DOI] [PubMed] [Google Scholar]
- Arimatsu K., Yamada H., Miyazawa H., Minagawa T., Nakajima M., Ryder M.I., et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep. 2014;4:4828. doi: 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assuma R., Oates T., Cochran D., Amar S., Graves D.T. IL-1 and TNF antagonists inhibit the inflammatory response and bone loss in experimental periodontitis. J. Immunol. 1998;160(1):403–409. [PubMed] [Google Scholar]
- Bostanci N., Ilgenli T., Emingil G., Afacan B., Han B., Töz H., et al. Gingival crevicular fluid levels of RANKL and OPG in periodontal diseases: implications of their relative ratio. J. Clin. Periodontol. 2007;34(5):370–376. doi: 10.1111/j.1600-051X.2007.01061.x. [DOI] [PubMed] [Google Scholar]
- Bostanci N., Emingil G., Saygan B., Turkoglu O., Atilla G., Curtis M.A., et al. Expression and regulation of the NALP3 inflammasome complex in periodontal diseases. Clin. Exp. Immunol. 2009;157(3):415–422. doi: 10.1111/j.1365-2249.2009.03972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky I.E., Palm N.W., Sadanand S., Ryndak M.B., Sutterwala F.S., Flavell R.A., et al. A yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 2010;7(5):376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., Ruby T., Belhocine K., Bouley D.M., Kayagaki N., Dixit V.M., et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490(7419):288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui F.Q., Johnson L., Roberts J., Hung S.C., Lee J., Atanasova K.R., et al. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1β and the danger signals ASC and HMGB1. Cell Microbiol. 2016;18(7):970–981. doi: 10.1111/cmi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui F.Q., Almeida-da-Silva C.L.C., Huynh B., Trinh A., Liu J., Woodward J., et al. Association between periodontal pathogens and systemic disease. Biomed. J. 2019;42(1):27–35. doi: 10.1016/j.bj.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcuac O., Berglundh T. Composition of human peri-implantitis and periodontitis lesions. J. Dent. Res. 2014;93(11):1083–1088. doi: 10.1177/0022034514551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case C.L., Kohler L.J., Lima J.B., Strowig T., de Zoete M.R., Flavell R.A., et al. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc. Natl. Acad. Sci. U.S.A. 2013;110(5):1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson C.N., Copenhaver A.M., Zwack E.E., Nguyen H.T., Strowig T., Javdan B., et al. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013;9(6) doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Wu W., Sun W., Zhang Q., Yan F., Xiao Y. RANKL expression in periodontal disease: where does RANKL come from? Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/731039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Wu Z., Li M., Shao M., Hu T. Interleukin-1β is a potential therapeutic target for periodontitis: a narrative review. Int. J. Oral Sci. 2020;12(1):2. doi: 10.1038/s41368-019-0068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotjumlong P., Bolscher J.G., Nazmi K., Reutrakul V., Supanchart C., Buranaphatthana W., et al. Involvement of the P2X7 purinergic receptor and c-Jun N-terminal and extracellular signal-regulated kinases in cyclooxygenase-2 and prostaglandin E2 induction by LL-37. J. Innate Immun. 2013;5(1):72–83. doi: 10.1159/000342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats S.R., Jones J.W., Do C.T., Braham P.H., Bainbridge B.W., To T.T., et al. Human toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell. Microbiol. 2009;11(11):1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors B.W. Tales of a dirty drug: carbenoxolone, gap junctions, and seizures. Epilepsy Curr. 2012;12(2):66–68. doi: 10.5698/1535-7511-12.2.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Silva R., Eduardo Baggio S.L. Purinergic signalling in host innate immune defence against intracellular pathogens. Biochem. Pharmacol. 2021 doi: 10.1016/j.bcp.2021.114405. [DOI] [PubMed] [Google Scholar]
- de Alencar J.B., Zacarias J.M.V., Tsuneto P.Y., Souza V.H., Silva C.O.E., Visentainer J.E.L., et al. Influence of inflammasome NLRP3, and IL1B and IL2 gene polymorphisms in periodontitis susceptibility. PLoS ONE. 2020;15(1) doi: 10.1371/journal.pone.0227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de G.A., Martinon F. Pyroptosis: caspase-11Â unlocks the gates of death. Immunity. 2015;43(5):835–837. doi: 10.1016/j.immuni.2015.10.024. [DOI] [PubMed] [Google Scholar]
- De Andrade K.Q., Almeida-da-Silva C.L.C., Coutinho-Silva R. Immunological pathways triggered by Porphyromonas gingivalis and Fusobacterium nucleatum: therapeutic possibilities? Mediators Inflamm. 2019;2019 doi: 10.1155/2019/7241312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Molon R.S., de Avila E.D., Boas Nogueira A.V., Chaves de Souza J.A., Avila-Campos M.J., de Andrade C.R., et al. Evaluation of the host response in various models of induced periodontal disease in mice. J. Periodontol. 2014;85(3):465–477. doi: 10.1902/jop.2013.130225. [DOI] [PubMed] [Google Scholar]
- Diaz P.I., Zilm P.S., Rogers A.H. Vol. 148. 2002. pp. 467–472. (Fusobacterium Nucleatum Supports the Growth of Porphyromonas Gingivalis in Oxygenated and Carbon-Dioxide-Depleted Environments Microbiol. (Read.)). [DOI] [PubMed] [Google Scholar]
- Eke P.I., Dye B.A., Wei L., Thornton-Evans G.O., Genco R.J. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J. Dent. Res. 2012;91(10):914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- Eke P.I., Dye B.A., Wei L., Slade G.D., Thornton-Evans G.O., Borgnakke W.S., et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J. Periodontol. 2015;86(5):611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan M.A., Benakanakere M.R., Rose B.G., Zhang P., Zhao J., Stathopoulou P., et al. Interleukin-1beta modulates proinflammatory cytokine production in human epithelial cells. Infect. Immun. 2008;76(5):2080–2089. doi: 10.1128/IAI.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold C.L., Ruan J., Tan Y., Xia S., Wu H., Kagan J.C. The pore-forming protein gasdermin d regulates interleukin-1 secretion from living macrophages. Immunity. 2018;48(1):35–44. doi: 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuille F., Ebersole J.L., Kesavalu L., Stepfen M.J., Holt S.C. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect. Immun. 1996;64(6):2094–2100. doi: 10.1128/iai.64.6.2094-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleetwood A.J., Lee M.K.S., Singleton W., Achuthan A., Lee M.C., O'Brien-Simpson N.M., et al. Metabolic remodeling, inflammasome activation, and pyroptosis in macrophages stimulated by Porphyromonas gingivalis and its outer membrane vesicles. Front. Cell. Infect. Microbiol. 2017;7:351. doi: 10.3389/fcimb.2017.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E., Seymour G.J. Immunoregulatory control of Th1/Th2 cytokine profiles in periodontal disease. Periodontol 2000. 2004;35:21–41. doi: 10.1111/j.0906-6713.2004.003557.x. [DOI] [PubMed] [Google Scholar]
- Gemmell E., Bird P.S., Carter C.L., Drysdale K.E., Seymour G.J. Effect of Fusobacterium nucleatum on the T and B cell responses to Porphyromonas gingivalis in a mouse model. Clin. Exp. Immunol. 2002;128(2):238–244. doi: 10.1046/j.1365-2249.2002.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves D.T., Kang J., Andriankaja O., Wada K., Rossa C., Jr Animal models to study host-bacteria interactions involved in periodontitis. Front. Oral Biol. 2012;15:117–132. doi: 10.1159/000329675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groslambert M., Py B.F. Spotlight on the NLRP3 inflammasome pathway. J. Inflamm. Res. 2018;11:359–374. doi: 10.2147/JIR.S141220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursoy U.K., Könönen E., Uitto V.J. Intracellular replication of fusobacteria requires new actin filament formation of epithelial cells. APMIS. 2008;116(12):1063–1070. doi: 10.1111/j.1600-0463.2008.00868.x. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G., Korostoff J.M. Revisiting the page & schroeder model: the good, the bad and the unknowns in the periodontal host response 40â years later. Periodontol 2000. 2017;75(1):116–151. doi: 10.1111/prd.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Wang M., Liang S. Induction of distinct TLR2-mediated proinflammatory and proadhesive signaling pathways in response to Porphyromonas gingivalis fimbriae. J. Immunol. 2009;182(11):6690–6696. doi: 10.4049/jimmunol.0900524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015;15(1):30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.W., Shi W., Huang G.T., Kinder H.S., Park N.H., Kuramitsu H., et al. Interactions between Periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 2000;68(6):3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Lin X., Seliger A.R., Eastcott J., Kawai T., Taubman M.A. Expression of receptor activator of nuclear factor-kappaB ligand by B cells in response to oral bacteria. Oral Microbiol. Immunol. 2009;24(3):190–196. doi: 10.1111/j.1399-302X.2008.00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Feng Y., Xiong G., Whyte S., Duan J., Yang Y., et al. Caspase-11, a specific sensor for intracellular lipopolysaccharide recognition, mediates the non-canonical inflammatory pathway of pyroptosis. Cell Biosci. 2019;9:31. doi: 10.1186/s13578-019-0292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh N.C., Everts V., Pavasant P., Ampornaramveth R.S. Interleukin-1β induces human cementoblasts to support osteoclastogenesis. Int. J. Oral Sci. 2017;9(12):e5. doi: 10.1038/ijos.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias R., Locovei S., Roque A., Alberto A.P., Dahl G., Spray D.C., et al. P2X7 receptor-pannexin1 complex: pharmacology and signaling. Am. J. Physiol. Cell Physiol. 2008;295(3):C752–C760. doi: 10.1152/ajpcell.00228.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L., Almeida-da-Silva C.L.C., Takiya C.M., Figliuolo V., Rocha G.M., Weissmüller G., et al. Oral infection of mice with Fusobacterium nucleatum results in macrophage recruitment to the dental pulp and bone resorption. Biomed. J. 2018;41(3):184–193. doi: 10.1016/j.bj.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.J., Jun H.K., Choi B.K. Contradictory roles of Porphyromonas gingivalis gingipains in caspase-1 activation. Cell. Microbiol. 2015;17(9):1304–1319. doi: 10.1111/cmi.12435. [DOI] [PubMed] [Google Scholar]
- Kawai T., Matsuyama T., Hosokawa Y., Makihira S., Seki M., Karimbux N.Y., et al. B and T lymphocytes are the primary sources of RANKL in the bone resorptive lesion of periodontal disease. Am. J. Pathol. 2006;169(3):987–998. doi: 10.2353/ajpath.2006.060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande W.L., Louie S., Dong J., et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kim H., Kajikawa T., Walsh M.C., Takegahara N., Jeong Y.H., Hajishengallis G., et al. The purinergic receptor P2X5 contributes to bone loss in experimental periodontitis. BMB Rep. 2018;51(9):468–473. doi: 10.5483/BMBRep.2018.51.9.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P.E., Palmer R.J., Jr., Periasamy S., Jakubovics N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010;8(7):471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M., Dixit V.M. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M. Emerging inflammasome effector mechanisms. Nat. Rev. Immunol. 2011;11(3):213–220. doi: 10.1038/nri2936. [DOI] [PubMed] [Google Scholar]
- Lamont R.J., Chan A., Belton C.M., Izutsu K.T., Vasel D., Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 1995;63(10):3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guo H., Wang X., Lu Y., Yang C., Yang P. Coinfection with Fusobacterium nucleatum can enhance the attachment and invasion of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans to human gingival epithelial cells. Arch. Oral Biol. 2015;60(9):1387–1393. doi: 10.1016/j.archoralbio.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Lin M., Hu Y., Wang Y., Kawai T., Wang Z., Han X. Different engagement of TLR2 and TLR4 in Porphyromonas gingivalis vs. ligature-induced periodontal bone loss. Braz. Oral Res. 2017;31:e63. doi: 10.1590/1807-3107BOR-2017.vol31.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Junior R., Figueredo C.M. Periodontal and inflammatory bowel diseases: is there evidence of complex pathogenic interactions? World J. Gastroenterol. 2016;22(35):7963–7972. doi: 10.3748/wjg.v22.i35.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanonda R., Champaiboon C., Subbalekha K., Sa-Ard-Iam N., Rattanathammatada W., Thawanaphong S., et al. Human memory B cells in healthy gingiva, gingivitis, and periodontitis. J. Immunol. 2016;197(3):715–725. doi: 10.4049/jimmunol.1600540. [DOI] [PubMed] [Google Scholar]
- Man S.M., Karki R., Briard B., Burton A., Gingras S., Pelletier S., et al. Differential roles of caspase-1 and caspase-11 in infection and inflammation. Sci. Rep. 2017;7:45126. doi: 10.1038/srep45126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti C. The NLRP3 Inflammasome as a pharmacological target. J. Cardiovasc. Pharmacol. 2019;74(4):285–296. doi: 10.1097/FJC.0000000000000718. [DOI] [PubMed] [Google Scholar]
- Morandini A.C., Ramos-Junior E.S., Potempa J., Nguyen K.A., Oliveira A.C., Bellio M., et al. Porphyromonas gingivalis fimbriae dampen P2X7-dependent interleukin-1β secretion. J. Innate Immun. 2014;6(6):831–845. doi: 10.1159/000363338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Souza A.C.A., Almeida-da-Silva C.L.C., Rangel T.P., Rocha G.D.C., Bellio M., Zamboni D.S., et al. The P2X7 receptor mediates toxoplasma gondii control in macrophages through canonical NLRP3 Inflammasome activation and reactive oxygen species production. Front. Immunol. 2017;8:1257. doi: 10.3389/fimmu.2017.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Planillo R., Kuffa P., Martínez-Colon G., Smith B.L., Rajendiran T.M. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38(6):1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natto Z.S., Hameedaldain A. Methodological quality assessment of meta-analyses and systematic reviews of the relationship between periodontal and systemic diseases. J. Evid. Based Dent. Pract. 2019;19(2):131–139. doi: 10.1016/j.jebdp.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Okui T., Aoki-Nonaka Y., Nakajima T., Yamazaki K. The role of distinct t cell subsets in periodontitisTÇöStudies from humans and rodent models. Curr Oral Health Rep. 2014;1(2):114–123. [Google Scholar]
- Olsen I., Yilmaz Ã. Modulation of inflammasome activity by Porphyromonas gingivalis in periodontitis and associated systemic diseases. J. Oral Microbiol. 2016;8:30385. doi: 10.3402/jom.v8.30385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Wang Q., Chen Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019;11(3):30. doi: 10.1038/s41368-019-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Na H.S., Song Y.R., Shin S.Y., Kim Y.M., Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect. Immun. 2014;82(1):112–123. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrin P., Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006;25(21):5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrin P., Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1beta release through a dye uptake-independent pathway. J. Biol. Chem. 2007;282(4):2386–2394. doi: 10.1074/jbc.M610351200. [DOI] [PubMed] [Google Scholar]
- Peres M.A., Macpherson L.M.D., Weyant R.J., Daly B., Venturelli R., Mathur M.R., et al. Oral diseases: a global public health challenge. Lancet. 2019;394(10194):249–260. doi: 10.1016/S0140-6736(19)31146-8. [DOI] [PubMed] [Google Scholar]
- Perlee D., de B.R., Florquin S., van der Poll T., van 't Veer C., de Vos A.F. Caspase-11 contributes to pulmonary host defense against Klebsiella pneumoniae and local activation of coagulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020;319(1):L105–L114. doi: 10.1152/ajplung.00422.2019. [DOI] [PubMed] [Google Scholar]
- Ramos-Junior E.S., Morandini A.C., Almeida-da-Silva C.L., Franco E.J., Potempa J., Nguyen K.A., et al. A Dual role for p2x7 receptor during Porphyromonas gingivalis infection. J. Dent. Res. 2015;94(9):1233–1242. doi: 10.1177/0022034515593465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Vanaja S.K., Waggoner L., Sokolovska A., Becker C., Stuart L.M., et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150(3):606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.S., Yilmaz Ó. Dangerous liaisons: caspase-11 and reactive oxygen species crosstalk in pathogen elimination. Int. J. Mol. Sci. 2015;16(10):23337–23354. doi: 10.3390/ijms161023337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.S., Atanasova K.R., Lee J., Diamond G., Deguzman J., Hee C.C., et al. Opportunistic pathogen Porphyromonas gingivalis modulates danger signal atp-mediated antibacterial NOX2 pathways in primary epithelial cells. Front. Cell. Infect. Microbiol. 2017;7:291. doi: 10.3389/fcimb.2017.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A.J., Behl B., Banerjee I., Rathinam V.A.K. Emerging insights into noncanonical inflammasome recognition of microbes. J. Mol. Biol. 2018;430(2):207–216. doi: 10.1016/j.jmb.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Inagaki S., Kimizuka R., Okuda K., Hosaka Y., Nakagawa T., et al. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 2008;54(3):349–355. doi: 10.1111/j.1574-695X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- Saito A., Kokubu E., Inagaki S., Imamura K., Kita D., Lamont R.J., et al. Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts. Microb. Pathog. 2012;53(5–6):234–242. doi: 10.1016/j.micpath.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio L.E.B., Coutinho-Silva R. Immunomodulatory effects of P2X7 receptor in intracellular parasite infections. Curr. Opin. Pharmacol. 2019;47:53–58. doi: 10.1016/j.coph.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Stowe I., Lee B., Kayagaki N. Caspase-11: arming the guards against bacterial infection. Immunol. Rev. 2015;265(1):75–84. doi: 10.1111/imr.12292. [DOI] [PubMed] [Google Scholar]
- Swanson K.V., Deng M., Ting J.P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19(8):477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxman D.J., Swanson K.V., Broglie P.M., Wen H., Holley-Guthrie E., Huang M.T., et al. Porphyromonas gingivalis mediates inflammasome repression in polymicrobial cultures through a novel mechanism involving reduced endocytosis. J. Biol. Chem. 2012;287(39):32791–32799. doi: 10.1074/jbc.M112.401737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbert-Mros S., Larsson L., Berglundh T. Cellular composition of long-standing gingivitis and periodontitis lesions. J. Periodontal Res. 2015;50(4):535–543. doi: 10.1111/jre.12236. [DOI] [PubMed] [Google Scholar]
- Tonetti M.S., Chapple I.L., Jepsen S., Sanz M. Primary and secondary prevention of periodontal and peri-implant diseases: introduction to, and objectives of the 11th European workshop on periodontology consensus conference. J. Clin. Periodontol. 2015;42(Suppl 16):S1–S4. doi: 10.1111/jcpe.12382. [DOI] [PubMed] [Google Scholar]
- Uchiyama R., Tsutsui H. Caspases as the key effectors of inflammatory responses against bacterial infection. Arch. Immunol. Ther. Exp. (Warsz) 2015;63(1):1–13. doi: 10.1007/s00005-014-0301-2. [DOI] [PubMed] [Google Scholar]
- van Hout G.P., Bosch L., Ellenbroek G.H., de Haan J.J., van Solinge W.W., Cooper M.A., et al. The selective NLRP3-inflammasome inhibitor MCC950 reduces infarct size and preserves cardiac function in a pig model of myocardial infarction. Eur. Heart J. 2017;38(11):828–836. doi: 10.1093/eurheartj/ehw247. [DOI] [PubMed] [Google Scholar]
- Weiss E.I., Shaniztki B., Dotan M., Ganeshkumar N., Kolenbrander P.E., Metzger Z. Attachment of Fusobacterium nucleatum pk1594 to mammalian cells and its coaggregation with Periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol. Immunol. 2000;15(6):371–377. doi: 10.1034/j.1399-302x.2000.150606.x. [DOI] [PubMed] [Google Scholar]
- Whitmore S.E., Lamont R.J. Oral bacteria and cancer. PLoS Pathog. 2014;10(3) doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Kurita-Ochiai T., Kobayashi R., Suzuki T., Ando T. Regulation of the NLRP3 inflammasome in Porphyromonas gingivalis-accelerated periodontal disease. Inflamm. Res. 2017;66(1):59–65. doi: 10.1007/s00011-016-0992-4. [DOI] [PubMed] [Google Scholar]
- Yao L., Jermanus C., Barbetta B., Choi C., Verbeke P., Ojcius D.M., et al. Porphyromonas gingivalis infection sequesters pro-apoptotic bad through Akt in primary gingival epithelial cells. Mol. Oral Microbiol. 2010;25(2):89–101. doi: 10.1111/j.2041-1014.2010.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O., Sater A.A., Yao L., Koutouzis T., Pettengill M., Ojcius D.M. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell. Microbiol. 2010;12(2):188–198. doi: 10.1111/j.1462-5822.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel-Lindberg T., BÃ¥ge T. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev. Mol. Med. 2013;15:e7. doi: 10.1017/erm.2013.8. [DOI] [PubMed] [Google Scholar]
- Zahid A., Li B., Kombe A.J.K., Jin T., Tao J. Pharmacological inhibitors of the NLRP3 inflammasome. Front. Immunol. 2019;10:2538. doi: 10.3389/fimmu.2019.02538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzmann N.U., Lindhe J., Berglundh T. Host response to microbial challenge following resective/non-resective periodontal therapy. J. Clin. Periodontol. 2005;32(11):1175–1180. doi: 10.1111/j.1600-051X.2005.00746.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.