Highlights
-
•
Ubiquitously present bacterial Toxin-Antitoxin (TA) modules consist of stable toxin associated with labile antitoxin.
-
•
Classification of TAs modules based on inhibition of toxin through antitoxin in 8 different classes.
-
•
Variety of specific toxin targets and the abundance of TA modules in various deadly pathogens.
-
•
Specific role of TAs modules in conservation of the resistant genes, emergence of persistence & biofilm formation.
-
•
Proposed antibacterial strategies involving TA modules for elimination of multi-drug resistance.
Keywords: Toxin-Antitoxin modules, Promoter, Translation, Persistence, Biofilms, PSK, Antibacterial, artificial activation
Abstract
Toxin-antitoxin (TA) modules are ubiquitous gene loci among bacteria and are comprised of a toxin part and its cognate antitoxin part. Under normal physiological conditions, antitoxin counteracts the toxicity of the toxin whereas, during stress conditions, TA modules play a crucial role in bacterial physiology through involvement in the post-segregational killing, abortive infection, biofilms, and persister cell formation. Most of the toxins are proteinaceous that affect translation or DNA replication, although some other intracellular molecular targets have also been described. While antitoxins may be a protein or RNA, that generally neutralizes its cognate toxin by direct interaction or with the help of other signaling elements and thus helps in the TA module regulation. In this review, we have discussed the current state of the multifaceted TA (type I–VIII) modules by highlighting their classification and specific targets. We have also discussed the presence of TA modules in the various pathogens and their role in antibiotic persistence development as well as biofilm formation, by influencing the different cellular processes. In the end, assembling knowledge about ubiquitous TA systems from pathogenic bacteria facilitated us to propose multiple novel antibacterial strategies involving artificial activation of TA modules.
Graphical Abstract
1. Introduction
Earlier, it was found that antibiotics, which have the potency to kill bacteria, are not successful to sterilize cultures (Bigger, 1944; Hobby et al., 1942). Later, Bigger observed a distinct subpopulation of bacteria that manage and survive in an intensive antibiotic environment and he called them persisters. Numerous bacterial infections like Staphylococcus aureus in prosthetic implant infections, Mycobacterium tuberculosis in pulmonary infections, etc. are major life-threatening health issues and are related to the antibiotic treatment defeat due to bacterial persistence (Fauvart et al., 2011). Persisters are not only resistant to antibiotics but also often protected from the immune defense of hosts. For example, they may hide in different niches like the stomach (Helicobacter pylori), central nervous system (Treponema pallidum), biofilms (Pseudomonas aeruginosa), macrophages or granulomas (Mycobacterium tuberculosis), and gallbladder (Salmonella typhi) (Jayaraman, 2008). A streak of significant investigations regarding bacterial persistence was done and it was deduced that the involvement was of intrinsic genetic factors like toxin-antitoxin (TA) modules (Dörr et al., 2010; Gutierrez et al., 2017; Moyed & Bertrand, 1983). Therefore, it becomes much important to investigate the functions of TA modules in various pathogenic bacterial strains.
Bacterial TA modules are primarily associated with various physiological activities like apoptosis, growth arrest, gene regulation, and survival (Buts et al., 2005; Gerdes et al., 2005; Hayes & Van, 2011; Kim et al., 2018). In 1983, TA modules were discovered on the Escherichia coli plasmid (Ogura & Hiraga, 1983). As an addiction module, these systems were involved in the maintenance of the genetic element. TA modules are formed with a toxin part associated with an antitoxin part and are encoded on the extrachromosomal unit or chromosomal unit. Extrachromosomal encoded TA modules belong to plasmid stabilization and cell viability (Magnuson, 2007; Monti et al., 2007), while chromosomal encoded TA modules are involved in biofilm formation, persister cell formation, growth arrest, and multidrug tolerance (Korch & Hill, 2006; Vazquez et al., 2006; Wang & Wood, 2011; Yamaguchi et al., 2011). A host cell is influenced by the toxin part which inhibits DNA replication, protein translation, and cell wall formation, while an antitoxin part neutralizes the toxic effect of its associated toxin part (Buts et al., 2005; Harms et al., 2018; Prysak et al., 2009).
The high prevalence of TA modules in bacteria makes them capable of slow growth and resulting in a dormant state. A total of 88 TA modules were carried by pathogenic strain Mycobacterium tuberculosis (Ramage et al., 2009; Yu et al., 2020), while only 5 TA modules were harbored by non-pathogenic strain Mycobacterium smegmatis (Robson et al., 2009; Yu et al., 2020), which is relatively fast-growing. Apart from human pathogens, an entomopathogen named Xenorhabdus nematophila has a total of 39 TA modules and helps it for surviving in insects by the formation of non-replicating persisters (Yadav & Rathore, 2020; Yadav & Rathore, 2018). Some specific TA systems also have been well characterized in non-human pathogens for example; Agrobacterium tumefasciens (Choi et al., 2021; Denkovskienė et al., 2020; McGillick et al., 2019), Erwinia amylovora (Fineran et al., 2009; Peng et al., 2021; Unterholzner et al., 2013), Xanthomonas sp. (Granato et al., 2019; Martins et al., 2016; Triplett et al., 2016), Xylella fastidiosa (Lee et al., 2014; Merfa et al., 2016; Santiago et al., 2016), and Acetobacter pasteurianus (Xia et al., 2019).
Thus, a high number of TA modules act as stumbling blocks for the treatment of bacterial diseases as they overcome antibiotic stresses. In this review, we have discussed the TA modules classification, functions, mode of toxin action, and their possible roles in bacterial physiology. We have also emphasized the future perspectives of the most abundantly found type II TA modules in bacterial genomes and followed the recent shreds of evidence to connect them with bacterial persistence.
2. The paradigm of TA modules
Some of TA modules are prevalent in the bacterial chromosomes as well as on plasmids and are generally found activated in different stress conditions (Fraikin et al., 2020; Hayes & Van, 2011; Page & Peti, 2016). TA modules consist of a toxin gene and another antitoxin gene, where antitoxin acts as an inhibitor to the toxin and antagonizes its toxic effect (Fernandez et al., 2016; Hayes & Van, 2011; Page & Peti, 2016). Antitoxin generally precedes the toxin gene and this upstream location has the benefit of its production over the toxin (Yang & Walsh, 2017). Toxin functions only inside the TA producer cell and is not secreted outside the cell. Thus, these are different from other exotoxins and endotoxins (Hayes & Van, 2011; Kedzierska & Hayes, 2016). However, there are exceptions to this, some toxins are secreted and can be involved in bacterial-host interactions (Santiago et al., 2016; Triplett et al., 2016). The toxin and antitoxin are also different from one another in terms of their stability and lifespan (Buts et al., 2005). Under stressful environments, antitoxin exhibits shortened lifespan as it can selectively be degraded. whereas, the toxin has a longer life span and is much more stable (Brzozowska & Zielenkiewicz, 2013).
Expression of toxins inhibits bacterial cell growth by acting on some essential cellular processes like membrane integrity, cell wall synthesis, transcription, translation, replication, and formation of the cytoskeleton (Unterholzner et al., 2013). On the other hand, toxic effects of toxin are normalized by interaction with its cognate antitoxin (Hayes & Van, 2011). The antitoxin can also compete and retrieve the toxin from its target site thus resuming the bacterial growth. Once the cell experiences normal environmental conditions after stress, it indicates a higher affinity towards toxin, as compared to its target (Buts et al., 2005; Maisonneuve & Gerdes, 2014).
TA modules were initially discovered to be present on plasmids exhibiting plasmid maintenance through post-segregational killing (PSK) and were described as “addiction modules” because, after cell division, they confirm the death of daughter cells that do not inherit such plasmids (Lobato et al., 2016; Van & De, 2009). Later, these modules were also found on the bacterial chromosome, where they encouraged programmed cell death of few cells out of the total population, in an altruistic manner thus releasing stress reliving components for survival in harsh conditions (Carmona & Xavier, 2012). The presence of TA operons in bacterial chromosomes suggests that they might have been acquired by horizontal gene transfer. TA modules were also involved in abortive infection. The mechanism which impairs the propagation of bacteriophage inside the bacterial host with altruistic suicide of the infected bacterial cell is known as abortive infection. TA modules are activated in infected cells before the phage replication, resulting in cell death, in a way supporting the survival of other cells in the bacterial population (Dy et al., 2014a; Dy et al., 2014b).
Some non-conventional tripartite TA modules were also identified in the different bacterial TA contents (Harms et al., 2018). These TA modules are also derived from type II TA modules. For example, in E. coli O157: H7, the paaRAE2 TA module contains a parE toxin, a paaA antitoxin, and an additional regulator protein paaR (Hallez et al., 2010). Likely, in Streptococcus pyogenes, the ω-ɛ-ζ TA module is composed of an ɛ-ζ TA module with ω as an additional transcriptional regulator (Volante et al., 2014). Some other examples of tripartite TA modules have been identified in Bacillus and M. tuberculosis, where a chaperone is included in the TA module (Bordes et al., 2016; Chan et al., 2016).
3. Classification of TA modules
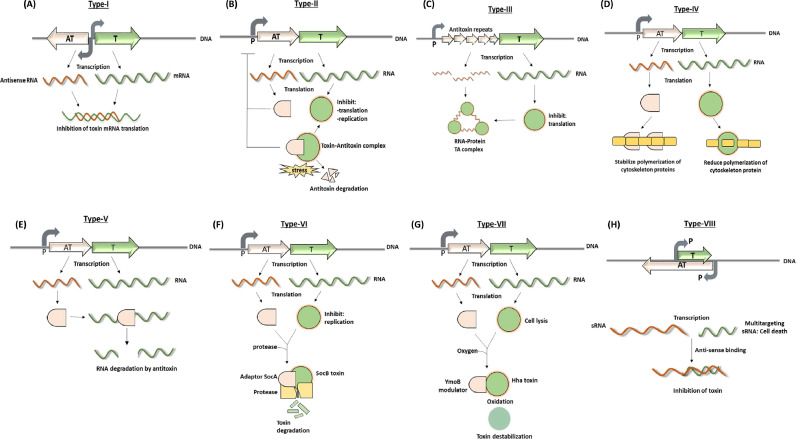
Till now, several reviews highlighting TA modules stated six primary classes of TA modules. The classification of TA modules is based on the type of interaction between antitoxin and toxin or mode of inhibition of toxin by antitoxin as illustrated in Fig. 1 (Song & Wood, 2020). Recent advances in this context categorize them into eight different classes including two newly described classes of TA systems. In type I to type VII TA modules, the toxins are generally proteins, whereas, in type VIII TA modules, it is a small RNA (Choi et al., 2018; Song & Wood, 2020). In the case of type I, type III and type VIII TA modules, antitoxins are small noncoding RNAs while in type II, type IV, type V, type VI and type VII TA modules are small proteins (Song & Wood, 2020).
Fig. 1.
The eight main types and regulatory mechanisms that control the activation of TA modules. (A–H) The illustrations schematically describe modes of action to neutralize toxins (green) by cognate antitoxins (orange) in type I–VIII TA modules (details are explained in the main text). TA genomic location and promoter positions are depicted with colored and grey arrows, respectively. RNAs are sketched with curly colored lines.
(A) Type I TA module
B) Type II TA module
(C) Type III TA module
(D) Type IV TA module
(E) Type V TA module
(F) Type VI TA module
(G) Type VII TA module
(H) Type VIII TA module
3.1. Type I
In the type I TA module, an antisense RNA is the antitoxin that inhibits the translation from mRNA of the toxin, as shown in Fig. 1A. The first known example in this class is the hok-sok TA module (Gerdes et al., 1986). Toxins in type I TA modules are mostly small hydrophobic peptides that target the integrity of bacterial membranes, causing obstruction in membrane potential as well as cell division. These proteins have predicted conserved domains of α-helical transmembrane proteins and enable them to form pores in the membrane-like phage holins, while their mechanism of action and cellular functions are found to be highly diverse (Brielle et al., 2016). Toxin and antitoxin can either be arranged in an overlapping manner, convergently transcribing the gene pairs as in the hok-sok TA system, or can also be located apart divergently, transcribing gene pairs as in the tisB-istR TA system. For the downregulation of toxin, the antitoxin is expressed as unstable small RNA which acts in different ways. In the particular case of E. coli, symER, the antisense RNA base pairs with the region of stable toxin mRNA, overlapping with Shine-Dalgarno sequence, inhibiting the translation of symE toxin, by preventing ribosomal binding to Shine-Dalgarno sequence (Kawano, 2012). In, hok-sok and ldrD-rdlD systems in E. coli, the antitoxin works by inhibiting the translation of leader peptides. In the case of hok-sok, sok (suppression of killing) antitoxin base pairs with the ribosomal binding site of mok (modulation of killing), which is otherwise an essential leader peptide and thus, indirectly prevent translation of hok toxin. Similarly, ldrD overlapping with ldrD toxin is a designated open reading frame (ORF) like mok. The rdlD antitoxin obstructs the translation of the ldrD leader peptide and inhibits the translation of the ldrD toxin (Brantl & Jahn, 2015). For tisB-istR TA module in E. coli, is present a hundred nucleotides upstream of the translation initiation site (TIR) of tisB toxin, and ribosomal standby, or loading site is located, which is required for highly structured TIR initiation. istR antitoxin binds to this ribosomal standby site location, competing with ribosomes in binding and inhibiting the translation of tisB toxin (Wagner & Unoson, 2012). In addition to this translational inhibition, the duplex formed by toxin-antitoxin RNA is the target for cellular RNases. Therefore, binding of the antitoxin ultimately results in the degradation of toxin RNA.
3.2. Type II
The most widely studied class of all TA modules is type-II modules, where both toxin and antitoxin are small proteins, as illustrated in Fig. 1B. For neutralization of toxin, the antitoxin forms a protein-protein complex with the toxin and acts as a tight-binding inhibitor (Goeders & Van, 2013). The first known example of this class is ccdAB (Ogura & Hiraga, 1983). Unlike type I, both genes present here are transcribed under the same promoter. Generally, the antitoxin is located upstream of the toxin gene but in some cases for example higBA, rnlAB, hicAB, and mqsRA has reverse genetic organization where antitoxin is located downstream of the toxin gene (Tian et al., 2001). Under stress conditions such as nutrient deprivation, plasmid loss, high temperature, oxidative stress, bacteriophage infection, or antibiotic pressure, the cell ensures proteolytic degradation of antitoxin thus decreasing the antitoxin concentration and liberating the toxin (Chukwudi & Good, 2015; Coussens & Daines, 2016; Wang et al., 2011; Wang & Wood, 2011). Lon, ClpAP, and ClpXP are some ATP-dependent proteases involved in antitoxin degradation under these stress conditions. The toxin protein exhibits toxicity in several ways such as MqsR, MazF, HigB, and RelE toxins act as sequence-specific endoribonucleases (Christensen & Gerdes, 2003; Hurley & Woychik, 2009; Zhang et al., 2003), while CcdB and ParE toxins affect DNA replication through targeting the enzyme DNA gyrase. Some type II TA modules are categorized into superfamilies based on the functional and structural characteristics of the toxin, namely, relBE, mazEF, vapBC, ccdAB, parDE, higAB, hipBA, and Phd–Doc. Earlier each toxin family was thought to be associated with a specific family of antitoxin, however recent studies revealed the existence of some hybrid systems in which a TA locus may contain toxin and antitoxin both from different families. The active sites of the toxin are found to be sterically blocked by antitoxin in type II as well as type III TA systems. Thus, mutations in active sites of the toxin may alter the toxic effect of the toxin. The mechanism of regulation of TA operons by toxin-antitoxin ratio at the transcriptional level is termed as “Conditional Cooperativity”. Heteromers of different stoichiometric ratios of toxins and antitoxins are formed, whereas the best transcriptional repressor is the complex with the intermediate ratio of both the proteins. Conditional cooperativity was quantitatively analyzed in relBE TA loci of E. coli. When the antitoxin RelB is present in excess over the toxin RelE, it forms the dimer RelB2 which can inhibit the relBE promoter. While the 2:1 complex RelB2: RelE exhibits the strongest transcriptional inhibition of the relBE promoter, therefore RelE toxin itself acts as a transcriptional co-repressor. Although, the 2:2 complex (RelB2: RelE2) is unable to bind to the promoter and thus the transcription remains activated (Cataudella et al., 2012). Surprisingly, all the TA systems known so far are regulated by conditional cooperativity suggesting that it is a common characteristic feature of all the TA loci (Cataudella et al., 2013).
The first identified TA module carried on the F plasmid of E. coli was type II ccdAB and was described to play a vital role in plasmid maintenance through coupled host cell division and plasmid proliferation (Ogura & Hiraga, 1983). ccdA encodes for a toxin and ccdB encodes for an antitoxin protein, where CcdA antitoxin is specifically degraded by Lon protease. CcdA dimers can bind to the operator sequence with low affinity, CcdB toxin bridges the CcdA dimers and enhances the binding affinity of complex to the operator showing strong transcriptional inhibition (Vandervelde et al., 2017). ccdAB and parDE are well-studied modules in terms of gyrase poisoning. In comparison to plasmid encoding CcdB toxin, many non-conserved residues are present in the antitoxin and gyrase binding sites of the chromosomally encoded toxin. This suggests that these two CcdB toxins appear to have different affinities towards their cognate CcdA antitoxin and gyrase that possibly indicate distinct cellular roles of plasmid and chromosomally encoded TA modules (De et al., 2012). Many important and diverse roles are exhibited by different operons corresponding to type II modules, such as different virulence factors are influenced by higAB operon in Pseudomonas aeruginosa (Wood & Wood, 2016), whereas some other modules including yefM-yoeB assists in the survival of bacteria inside the host cell (Chan et al., 2011), and mqsRA shows involvement in biofilm formation (Yamaguchi et al., 2009). The toxin HipA contributes to the formation of persisters (Jayaraman, 2008).
3.3. Type III
These types of TA systems consist of RNA antitoxin that directly interacts with toxin protein, as depicted in Fig. 1C. There are mainly three known superfamilies of type III TA modules, which are toxIN, tenpIN, and cptIN (Blower et al., 2012). The toxIN TA system in the plasmid of plant pathogen Pectobacterium atrosepticum was the first discovered type III TA system (Fineran et al., 2009). The ToxN toxin exhibits endoribonuclease activity and can form a macromolecular complex by interacting with the RNA antitoxin (ToxI). Trimeric toxIN complex is formed through the interaction of three ToxN proteins and three ToxI monomers, resulting in ToxN toxin inhibition (Brantl & Jahn, 2015). An array of direct repeats and short inverted repeats precedes the toxN gene. Thus, toxI antitoxin is composed of direct repeats of 36 nucleotides (AGGTGATTTGCTACCTTTAAGTGCAGCTAGAAATTC). Such antitoxin repeats are described as the key features of type III TA systems (Goeders et al., 2016). toxIN TA system was primarily described as a system of protection against infecting bacteriophages through abortive infection, by promoting the altruistic suicide of the infected cell (Blower et al., 2009).
3.4. Type IV
In class type IV, the antitoxin and toxin both are proteins and do not directly interact with each other, as shown in Fig. 1D. The toxin target interaction is usually inhibited by the competitive binding of antitoxin to the toxin. The first reported type IV TA operon found in E. coli was cbeA-cbtA (Masuda et al., 2012). The functional analysis of cbeA-cbtA stated that the CbtA toxin is responsible for reducing the polymerization of MreB and FtsZ (cytoskeleton proteins), thus changing the bacterial morphology (Masuda et al., 2012). When cells were induced for expression of CbtA toxin, after several hours they formed swollen round lemon-shaped cells. These lemon-shaped cells eventually lysed with excess prolonged-expression (Heller et al., 2017). It was also observed that the CbeA antitoxin inhibits the toxin interaction with the targets by stabilizing polymers of MreB and FtsZ.
3.5. Type V
The enzyme antitoxin of the type V TA module does not directly bind to the toxin but is capable of degrading mRNAs of the corresponding toxin, as illustrated in Fig. 1E. ghoST was first studied as a type V TA system, which encodes for a small GhoT toxin protein that can damage the cell membrane, and an antitoxin GhoS exhibiting sequence-specific endoribonuclease activity towards mRNA of GhoT toxin (Wang et al., 2012).
3.6. Type VI
The socAB operon in gram-negative bacteria Caulobacter crescentus (it is now called Caulobacter vibroides) was first identified as a type VI TA module (Aakre et al., 2013). The protein toxin SocB strongly binds with the β-sliding clamp, thus repressing the elongation of replication. The antitoxin SocA acts as a proteolytic adaptor protein that binds to the SocB toxin and shows protease-mediated degradation of the SocB toxin as illustrated in Fig. 1F (Aakre et al., 2013; Markovski & Wickner, 2013).
3.7. Type VII
The newly classified type-VII modules involve antitoxins that are found to be enzymatically modifying the toxins. These enzymatic modifications are made through transient interactions, instead of primarily through binding as in the type-II TA system. There are few newly discovered and distinct TA systems where antitoxin is an enzyme directly targeting the cognate toxin. These TA modules are grouped in a separate class referred to as type-VII TA systems (Wang et al., 2020).
The first such TA module studied was the hha-tomB system in Yersinia enterocolitica (Marimon et al., 2016) and E. coli (Garcia et al., 2008). It is also a part of the first identified group of TA modules in biofilms (Barrios et al., 2006). Hha is a hemolysin expression modulating protein that results in cell lysis and reduced biofilm formation, but with an increased dispersal. While antitoxin TomB is a toxin overexpression modulator in biofilms that previously was known as YbaJ and inactivates Hha toxin in presence of oxygen. The antitoxin activity of TomB is oxygen-dependent as it promotes oxidation of Hha single conserved cysteine, Cys18 to SOxH species (sulfenic RSOH, sulfinic RSO2H, and sulfonic acid RSO3H). Thus, the oxidation destabilizes the Hha toxin protein as depicted in Fig. 1G.The Hha/TomB TA system acting as an oxygen sensor (Marimon et al., 2016) refers to its relevance for biofilms so that the bacterial cells under anoxic conditions would show reduced growth on account of Hha toxin activity.
HepT/MntA is another example of an enzyme antitoxin. A set of 2 genes encoding higher eukaryotes & prokaryotes nucleotide (HEPN) binding domain protein, and its associated minimal NTase (MNT) domain protein has been anticipated to represent one of the most abundant TA operons. The antitoxin MNT domain protein followed by its neighboring toxin HEPN domain protein has been first confirmed in a halophilic bacterium Halorhodospira halophila SL1 (Sberro et al., 2013), and then in a cryophilic bacterium Shewanella oneidensis (Yao et al., 2015). HEPN domain toxin protein with active RX4-6H motif act as endoribonuclease (Jia et al., 2018; Yao et al., 2015), whereas MntA antitoxin (MNT domain protein) functions as an NTase enzyme chemically modifying HepT toxin (HEPN domain protein) in archaea as well as bacteria. The in-vitro analysis through enzymatic assay revealed the transfer of 3 AMPs sequentially by MntA to HepT using ATP as a substrate (Yao et al., 2020). Furthermore, structural studies confirmed the transfer of 3 AMPs to Tyr104 in HepT, which is next to the active RNase domain RX4H. Thus, polyadenylation of HepT toxin through MntA reduces the HepT toxicity.
Rv1045/Rv1044 in Mycobacterium tuberculosis is also a TA system having antitoxin, which functions as an enzyme. Rv1045/Rv1044 further proposed to be renamed as Tgl/TakA is a specific module, which was predicted to be a type-IV TA system as no direct interaction could be confirmed between toxin and antitoxin (Dy et al., 2014a). Later, the antitoxin TakA was demonstrated to function as serine protein kinase neutralizing TglT toxin through phosphorylation of Ser78 (Yu et al., 2020). TakA antitoxin acting as an enzyme for toxin inactivation & the transient interaction of TakA and TglT makes TglT/TakA system distinct in comparison to type-IV TA systems. TglT toxin is a tRNA nucleotidyl transferase, which is responsible for the addition of pyrimidines (C, U) to the 3’-CCA acceptor arm of uncharged tRNA (Cai et al., 2020). Thus, the TglT toxin prevents the charging of tRNA. TglT toxin is homology to AbiEii, which belongs to the abortive infection (Abi) system of Streptococcus agalactiae (Dy et al., 2014a).
3.8. Type VIII
In the most recently described type VIII modules, the antitoxin masks the activity of the first time found small RNAs toxin by anti-sense binding as shown in Fig. 1H. The first identified example of this class is sdsR-ryeA (Choi et al., 2018). SdsR and RyeA are two small RNAs and located at opposite DNA strands with the same locus. Previously, SdsR was known as RyeB and classified as the RpoS regulon (Levi et al., 2014; Peano et al., 2015). The SdsR toxin regulates several mRNA targets, thus acting as multi-targeting sRNA through repressing mutS and tolC in Escherichia coli and different genes including crp, stpA, ompD & tolC in Salmonella (Frohlich et al., 2016; Gutierrez et al., 2013). The highest expression of SdsR toxin is in the stationary phase. The mutation in the promoter of antitoxin ryeA shifts sdsR expression to an earlier growth phase. The ectopic expression of SdsR caused cell death, whereas cell death was relieved by overexpression of RyeA thus it represented a novel type of TA module in which both the antitoxin and the toxin are small RNAs (Choi et al., 2018). The degradation of RyeA antitoxin by RNase BN is responsible for its lower abundance in the early exponential phase (Gupta et al., 2019). Furthermore, RyeA is found to be an acid stress-inducible RNA (Gupta et al., 2020). Homologous SdsR/RyeA elements are present in multiple enterobacterial species ((Fröhlich et al., 2012), (Fröhlich et al., 2016)). Thus, such TA modules may be widespread throughout the family Enterobacteriaceae.
4. Specific Targets of TA modules
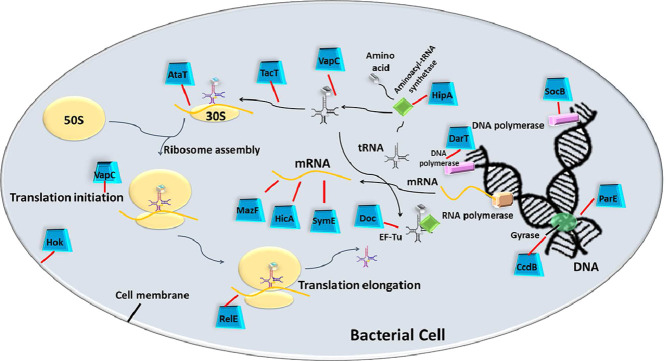
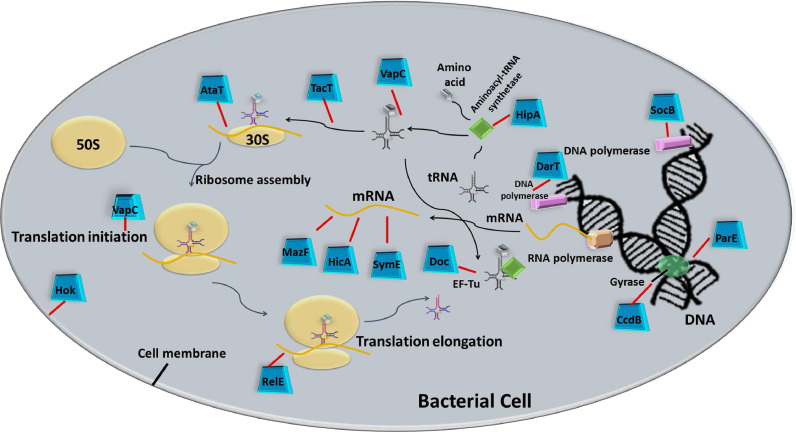
Several years of extensive research have reported diverse cellular targets of the TA modules. Many vital processes of the bacterial cell, including DNA replication, transcription, protein translation, biosynthesis of the cell wall as well as the formation of the cytoskeleton are altered by active TA modules, as illustrated in Fig. 2 and listed in Table 1. Protein translation is found to be the most popular target for the majority of TA modules (Harms et al., 2018). The translation is a complex process involving stepwise ribosomal assembly and harmonized motion of translational machinery, with the inclusion of each amino acid to the nascent polypeptide chain. The overall complexity of protein synthesis comes up with multiple possibilities of intervention, which are explored by different antibiotics, secreted toxins, and bacteriocins. The translation is thus targeted by type II TAs in several ways, starting from cleavage of RNA transcripts before as well as during the process of translation, to interrupting the tRNA charging or amino acid delivery to the elongating polypeptide chain. Some toxins may also affect ribosomal biogenesis.
Fig. 2.
Intracellular molecular targets for TA encoded Toxins. The illustration depicts how the molecular targets of typical TA encoded toxins influence the vital processes of bacterial cells. Active free toxins interfere with cellular processes such as cell-wall synthesis, DNA replication, transcription, and translation that ultimately may result in the formation of bacterial persisters.
Table 1.
Intracellular molecular activities of TA modules
| S.No. | TA types | Toxin | Antitoxin | Toxin targets |
Affected cellular processes | Examples | References |
| 1. | Type –I | Protein | RNA | Bacterial membrane | Biosynthesis of cell membrane |
hoh-sok tisB-istR |
(K. Gerdes et al., 1986) (Vogel et al., 2004) |
| 2. | Type-II | Protein | Protein | DNA gyrase Sequence-specific mRNA Initiator tRNA EF-Tu elongation factor glu-tRNA synthetase |
DNA replication Translation Translation Translation Translation |
ccdAB parDE relBE mazEF vapBC phd-doc hipBA |
(Bernard & Couturier, 1992) (Jiang et al., 2002) (Christensen & Gerdes, 2003) (Yonglong Zhang et al., 2003) (K. S. Winther & Gerdes, 2011) (Cruz et al., 2014) (Germain et al., 2013) |
| 3. | Type-III | Protein | RNA | mRNA | Translation | toxIN | (Fineran et al., 2009) |
| 4. | Type-IV | Protein | Protein | MreB and FtsZ (cytoskeleton proteins) | Cell morphology | cbtA-cbeA | (Masuda et al., 2012) |
| 5. | Type-V | RNA | Protein | Cell membrane | Biosynthesis of cell membrane | ghoST | (Wang et al., 2012) |
| 6. | Type-VI | Protein | Protein | β-sliding clamp | DNA replication | socAB | (Markovski & Wickner, 2013) |
| 7. | Type-VII | Protein | Protein | Biofilm | Biofilm | hha-tomB | (Marimon et al., 2016) |
| 8. | Type-VIII | RNA | RNA | Repression of YhcB (inner membrane protein) |
Cell morphology | sdsR-ryeA | (J. S. Choi et al., 2018) |
There are few toxins responsible for mRNA hydrolysis and are either solvent-exposed or ribosome-linked mRNA. mazEF is one of the well-studied type II TA operons in E. coli. The majority of MazF toxins specifically cleaves upstream of the 5’-ACA-3’nucleobases in mRNA transcripts. It behaves as a sequence-specific endoribonuclease, completely inhibiting the translation (Munoz et al., 2004; Zhang et al., 2003) like VapC and RelE toxins. Some MazF toxins are manifested to target 23S rRNA, 16S rRNA, and tRNA (Schifano et al., 2016), as recently described that MazF-mt9 toxin present in Mycobacterium tuberculosis specifically targets tRNA substrate (Schifano et al., 2016). Specific RNA pools are targeted or cleaved selectively by these endoribonuclease toxins and can be an added advantage to respond against diverse metabolic and environmental stimuli (Moll & Engelberg, 2012). The gene names of hicAB operon owe to their genetic locus linked to hif contiguous (pilus gene cluster) in Haemophilus influenza. HicA toxin holds a specific double-stranded RNA binding domain (~50 amino acids) and in addition to binding, all HicA toxins can also hydrolyze RNA. The mRNA degradation is observed through the HicA toxin in E. coli, whereas no consensus has been reported for the cleavage (Jørgensen et al., 2009). HicA from Sinorhizobium meliloti was experimentally shown to degenerate purified rRNA (Thomet et al., 2019). HicA3 from Yersinia pestis cleaves the in-vitro transcribed mRNA (Bibi et al., 2014). However, RelE toxins are responsible for cleavage between the 2nd and 3rd position of the codon of mRNA in the ribosomal A site. Toxins of the RelE family describe low sequence identity (11-20%) but retains conserved folds similar to T1, SA2, and U2 (ribosome independent endoribonuclease) (Neubauer et al., 2009).
Although all toxins corresponding to type II systems do not cleave mRNA, some toxins without degrading mRNA can halt the synthesis of proteins by interfering with the translational machinery. For instance, glutamate transfer RNA synthetase is phosphorylated by HipA toxin, causing inhibition of protein synthesis (Germain et al., 2013). Doc toxin from the phd-doc system is responsible for the inactivation of an important translation elongation factor EF-Tu through its phosphorylation (Castro et al., 2013; Cruz et al., 2014). So far, VapC toxin of VapBC module is characterized for targeting initiation tRNA (tRNAfMet), distinct elongation tRNA, and SRL (Sarcin-Rich loop) of 23S rRNA (Winther et al., 2016; Winther et al., 2013; Winther & Gerdes, 2011). Two GNAT toxins (AtaT and TacT) are newly identified type-II toxins with acetyltransferase activity (Jurėnas et al., 2017). Translation initiation is inhibited by AtaT toxin through acetylation of methionine charged on initiator tRNA. Whereas, TacT toxin is responsible for the acetylation of elongator tRNAs.
Classes of TA modules other than type II are not well investigated yet for their cellular targets. However, the type I Fst toxin disturbs the membrane integrity (Brinkman et al., 2013), Hok toxin depolarizes the cytoplasmic membrane inducing cellular damage, and biosynthesis of cell envelope is disrupted by type I BsrG toxin (Jahn et al., 2015). While the type III toxIN complex is well defined but the specific intracellular target of endoribonuclease toxin ToxN is still unknown. The type IV CbtA toxin reduces the polymerization of cytoskeleton proteins (MreB and FtsZ), which play an important role in the maintenance of cell morphology (Masuda et al., 2012). Whereas, the type V GhoT toxin is found responsible for disrupting the cell membrane thus forming lysed cells (ghost cells) (Wang et al., 2012), and type VI SocB toxin shows a strong affinity towards β sliding clamp, which promotes the processivity of the polymerase during DNA replication (Aakre et al., 2013). In type VII, Hha toxin is a hemolysin expression modulating protein that results in cell lysis (Marimon et al., 2016) while in type VIII, SdsR toxin is a RpoS regulon (Levi et al., 2014; Peano et al., 2015).
Surprisingly, TA modules and some antibiotics share common cellular targets. For example, both CcdB (Bernard & Couturier, 1992) and ParE (Jiang et al., 2002) toxin target the DNA gyrase. The enzyme relaxing the supercoil of DNA has two GyrA subunits and two GyrB subunits. When a transient break is created in one of the segments of DNA by two GyrA subunits, another segment passes through it to release the supercoil (Dao et al., 2005). The toxin binds in between, forming a dead-end complex and thus blocking DNA polymerase passage, therefore halting the replication. Such inhibition of DNA replication is also observed by the binding of quinolone antibiotics to the DNA gyrase complex (Kohanski et al., 2010). Another example is the inhibition of cell wall synthesis by phosphorylation of essential nucleotide sugar UDP-N-acetyl glucosamine by the action of zeta toxin of non-conventional tripartite ω-ε-ζ TA module (Moreno-Del Álamo et al., 2019). This is very much similar to the mechanism of penicillin antibiotic, which inhibits peptidoglycan synthesis (Kohanski et al., 2010). These remarkably similar targets of antibiotics and TA modules may indulge in novel insights for developing new antimicrobial drugs.
5. The abundance of TA modules in pathogens
The bacterial TA modules are important factors involved in bacterial persistence, abortive infection, maintenance of plasmids, and biofilm formation (Jayaraman, 2008; (Renbarger and Baker, 2017)). All these biological roles of TA modules support the pathogenicity of bacteria, neglecting the effects of antibiotics, and giving rise to multidrug-resistant strains. Multidrug resistance is one of the highest threats to public health globally. TA modules, particularly type II, are highly abundant in pathogenic bacteria as compared to non-pathogenic bacteria (Kang et al., 2018; Lobato et al., 2016). Recent advances in bioinformatics also revealed that intestinal microbiota shows a high abundance of type III TA modules (Kang et al., 2018).
In the past few years, a group of bacteria has been highlighted by the infectious disease society of America and has been named as “ESKAPE pathogens” (Pendleton et al., 2013). These pathogens are efficient in escaping the bactericidal action of antibiotics. This specific group of pathogens acronymically constitutes Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (Pendleton et al., 2013). These pathogens were extensively studied for the presence of multiple TA operons. Gram-positive bacteria S.aureus is a virulent pathogen causing endocarditis, skin infections, pneumonia, and food poisoning. Here, the worldwide issue was the emergence of methicillin-resistant S. aureus (MRSA). Mainly three type II TA is present in the genome of S. aureus, including mazEF, yefM-yoeB, and ω-ε-ζ operon (Schuster & Bertram, 2016). A member of the Enterobacteriaceae family, K. pneumoniae is responsible for causing severe damage to human lungs, urinary tract infection, and intra-abdominal infections. This Gram-negative opportunistic pathogen normally resides in the human intestine. K. pneumoniae isolate exhibits resistance to carbapenem antibiotics. Ten completely sequenced genomes of K. pneumoniae were analyzed for distribution of TA operons and a total of 212 putative type II TA locus were found in K. pneumoniae strains (Wei et al., 2016). A. baumannii is another Gram-negative opportunistic pathogen, primarily associated with hospital-acquired infections. This shows a high mortality rate of up to 75%. In the genomes of A. baumannii, a total of 15 TA operons were found, out of which five type II TAs were shown to be functional, namely hicAB, higAB, relBE, splAT, and cheAT (Jurenaite et al., 2013). A Gram-negative bacteria P. aeruginosa is a leading cause of nosocomial infections. It affects immune-compromised individuals causing chronic infections including burn wound infections, bacterial keratitis, and cystic fibrosis. This pathogen possesses relBE, yefM-yoeB, parDE, mazEF, higBA, parDE, vapBC-type TA module (Bonnin et al., 2013; Fernandez et al., 2016; Savari et al., 2016).
Other important pathogens such as Shigella flexneri capable of causing chronic diarrhea in humans also harbors vapBC, parDE, mazEF, and relBE type II TA pairs (Dienemann et al., 2011; Hosseini et al., 2019; Sengupta & Austin, 2011). The pathogenic strain of E. coli, for example, E. coli O157 is characterized as a human enterohemorrhagic pathogen responsible for hemorrhagic diarrhea and renal failure. This particular pathogenic strain of E. coli retains ccdAB, parDE, higAB, hicAB, mazEF, relBE and many more TA modules that are not listed here. An acid-fast bacterium Mycobacterium tuberculosis, which can persist longer in macrophages of the human respiratory system in a non-replicating and drug-tolerant state also has a total of 88 chromosomally encoded TA modules, out of which 81 are well known and seven are putative TA modules (Ramage et al., 2009; Yu et al., 2020). Thus, these pathogens are highly notable for their overabundance of TA modules.
TA systems have different demonstrated roles; still, there is relatively less knowledge about the functional significance of these modules in plant pathogens. A recent survey revealed that Erwinia amylovora which causes fire blight disease in apple and pear comprises six conserved type–II and type–IV TA systems. Out of six, only three TA systems (Doc/PhD, ParE/RHH, and CbtA/CbeA) were validated to be functional (Shidore et al., 2019). The hok-sok type-I TA module is also well characterized in E. amylovora (Peng et al., 2019). Multiple putative toxins or homologs of toxins from TA modules are identified from the genome of widely studied tumor-inducing bacterium Agrobacterium tumefasciens. The rod-shaped, Gram-negative bacteria causes crown gall disease in plants. The pemIK, yoeB-yefM, and mazEF are functionally characterized TA systems of A.tumefasciens (Choi et al., 2021; Denkovskienė et al., 2020; McGillick et al., 2019). Xylella fastidiosa is a major pathogenic bacteria responsible for many plant diseases including alfalfa dwarf, pony peach, coffee leaf scorch, and economically important pierce's a disease in grapes. DinJ/RelE and MqsRA are the two functional TA systems having different roles in the regulation of X. fastidosa growth and virulence (Lee et al., 2014; Burbank & Stenger, 2017; Merfa et al., 2016).
To attain a much better understanding for purpose of such modules in bacterial cell biology, highly comprehensive strategies for spotting novel TA loci are essential. A discovery-oriented database is recently created and named TASmania, in response to this specific need. Researchers annotated over 41000 assemblies out of the Ensembl Bacteria database, bringing about recognition of >2*106 candidate TA loci (Akarsu et al., 2019). TASmania allows the identification of a larger number of putative TA loci (type I-IV) because it has greater flexibility in comparison to the initially used TAfinder search tool in TADB 2.0. Therefore, providing initiation for future experimental analysis of TAs in many other pathogenic bacteria.
6. TA modules conserving resistant genes
TA modules actively participate in the maintenance of resistant genes of plasmids as well as genomic islands ((Díaz-Orejas et al., 2017); Yang & Walsh, 2017). Conjugative plasmids are the potential reservoirs of resistant genes capable of disseminating antibiotic resistance among bacteria (Carattoli, 2013; Mathers et al., 2015). These resistance genes are responsible for the emergence of persistence in harsh environments, i.e. antibiotic pressure linking plasmids to clinical failure of treatment through antibiotics (Andersson & Hughes, 2011; Li & Webster, 2018). Plasmids are known to be extrachromosomal or mobile genetic elements of the host cell imposing a metabolic burden (Norman et al., 2009; Velmurugan et al., 2003). Thus, plasmids can easily be eliminated from the genome of host bacteria when it does not experience any selective pressure, resulting in loss of multiple resistant genes. TA modules like ccdAB and hok-sok are involved in the stabilization of plasmids (Harms et al., 2018; Kroll et al., 2010). These TAs are responsible for the selective elimination of cells that fail to acquire plasmid during cell division, thus indirectly promoting the maintenance of resistant genes. The genes in a bacterial genome, acquired through horizontal gene transfer thus forming genomic islands are also stabilized by TA modules (Van & De, 2009). A Gram-negative bacterium Vibrio cholerae includes an integrative and conjugative constituent (SXT) mediating tolerance against several antibiotics (Das et al., 2020). A specific TA operon mosAT within SXT is recognized for promoting the stability of SXT (Wozniak & Waldor, 2009). When SXT is at risk of loss, the active MosT toxin minimizes the number of cells lacking SXT and the expression of active mosAT operon participates in SXT maintenance among the bacterial population (Wozniak & Waldor, 2009). This provides evidence for the participation of TA modules in maintaining the resistant genes.
7. The emergence of persistence through TA modules
In studies involving antibiotics, it was observed that a small population of cells survived as dormant cells and had a different mechanism from traditional antibiotic resistance (Fisher et al., 2017; Wood et al., 2013). This ability of few genetically homogeneous cells to survive in stress conditions by entering the dormant state temporarily is termed as “persistence”. The reduced metabolic activity and growth rate of cells are the main characteristics of persistence (Orman & Brynildsen, 2013; Wood et al., 2013). These persister cells are phenotypic variants but not genetic mutants (Wen et al., 2014). It has been unambiguously confirmed that TA modules have influential roles in the formation of persister cells (Kedzierska & Hayes, 2016; Maisonneuve & Gerdes, 2014).
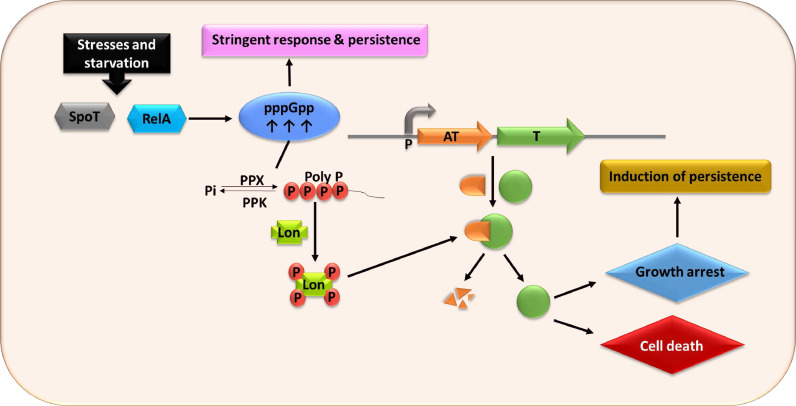
It was noticed that, as compared to wild-type strain under different antibiotic stress, the persistence was reduced more than 100 folds in mutant formed by successive deletion of 10 mRNAs, encoding TA pairs from the E. coli chromosome (Maisonneuve et al., 2011). TA modules are well studied as stress-responsive modules that are activated under certain stress conditions and thus form persisters (Harms et al., 2016; Wang & Wood, 2011). The hipBA was the first identified and elaborated TA module associated with persister cell formation in E. coli (Lou et al., 2008). Two major stress responses involved in TA activation are stringent response and SOS response (Maslowska et al., 2019; Strugeon et al., 2016). In stringent response as illustrated in Fig. 3, persister cell formation is mediated by a signaling nucleotide (p)ppGpp, which modulates the activation of TA modules through the cascade involving the protease enzyme (Lon) and inorganic phosphate (poly P). The higher level of (p)ppGpp serving as alarmone in exponentially growing cells activates the TA modules resulting in growth arrest. In response to nutrient starvation, RelA or SpoT is activated in bacteria promoting (p)ppGpp accumulation resulting in exopolyphosphatase (PPX) inhibition, which is a cellular enzyme degrading poly P. Active polyphosphate kinase (PPK) catalyzes the polyP stimulating expression of Lon protease to degrade antitoxin, liberating toxin and thus leading to growth arrest (Paul et al., 2019). When the degradation rate of antitoxin through Lon protease is reduced to a normal level, the antitoxin accumulation quenches the toxin reviving the normal growth of cells. This specific mechanism of TA activation is very well supported through mathematical modeling (Gelens et al., 2013). For example, lack of amino acid isoleucine leads to the increment in (p)ppGpp level and up-regulates multiple TA operons like dinJ/yafQ, mazEF, hicAB, relBE, mqsRA, and yafNO (Gutierrez et al., 2017). TAs are activated to reduce bacterial growth in response to stress and the growth rate of cells is proposed to be inversely related to the emergence of persistence. Thus, it seems that persistence depends on the growth rate, rather than exclusively depending on (p)ppGpp.
Fig. 3.
TA modules and pppGpp mediated persister pathway. Exposure to stress, such as nutrient deprivation, activated SpoT, and RelA that synthesize the alarmone (p)ppGpp. Further increment in the level of the alarmone and by inhibition of exopolyphosphatase (PPX), inorganic polyphosphate (PolyP) is accumulated. This leads to the activation of bacterial proteases, such as Lon, that preferentially cleave antitoxins, leaving an excess of the toxin. Thus, exposure to stress may result in the rapid cell death of the most actively growing cells and also activates a very small fraction of cells into the persister state by cellular growth arrest.
SOS response is another stress response supporting oxidative stress and antibiotic stress tolerance through induction of some type I and types II TA modules (Fraikin et al., 2020; Yang & Walsh, 2017). This is a bacterial cell response triggered by an obstruction in DNA replication, resulting in the mass of single-stranded DNA (Yamaguchi & Inouye, 2011). Two important proteins RecA and LexA act as regulators of SOS response (Cohen et al., 2008). Activation of RecA proteins initiates the mechanism of SOS response and in turn, the activated RecA inactivates the LexA repressor (Cohen et al., 2008). The LexA repressor is responsible for inhibiting the expression of SOS genes, which exhibit a role in cell growth, mutagenesis, and DNA repair (Kreuzer, 2013). Thus, it is a very crucial mechanism for the emergence of persisters in response to DNA damaging antibiotics, such as ciprofloxacin and fluoroquinolone (Dorr et al., 2009). tisB-istR type I TA locus in E. coli is the first identified TA module contributing to persistence through SOS induction (Yang & Walsh, 2017). Use of antibiotic ciprofloxacin which causing DNA damage resulted in a sharp downfall in the number of tolerant persister cells when a specific TA locus type-I tisAB/istR1 was knocked out from E. coli cells, indicating TA dependent formation of antibiotics tolerant cells (Dorr et al., 2010).
For TisB expression, the SOS response must be induced first as this toxin is regulated by LexA (Weel et al., 2008). While the IstR antidote shows an affinity towards LexA independent promoter and downregulates the expression of TisB toxin thus exhibiting RNase III dependent cleavage of the toxin mRNA (Vogel et al., 2004). Therefore, under normal conditions when SOS response is not triggered, constitutive transcription of antitoxin IstR inactivates TisB toxicity. In response to antibiotics targeting DNA replication, RecA is activated resulting in cleavage of LexA repressor and induction of SOS response (Cohen et al., 2008). The IstR antitoxin regulating the LexA promoter is degraded (Vogel et al., 2004). Whereas, TisB toxin shows gradual accumulation and binding towards cytoplasmic membrane leading to membrane damage, reduction in ATP level as well as proton motive force (Yang & Walsh, 2017). All this results in a reduced rate of DNA, RNA, and protein synthesis, and blockage of drug intake (Dorr et al., 2010; Unoson & Wagner, 2008). Finally, the bacterial growth slows down and antibiotic tolerant persisters are formed.
Similarly, the SOS response can also activate symER, dinQ-agrB, and hok-sok type-I TA modules as well as yafNO and dinJ-yafQ type-II TA modules (Baharoglu & Mazel, 2014; Berghoff & Wagner, 2017; Weel et al., 2013). In the case of dinJ-yafQ, the persister formation increases with YafQ toxin production (Hu et al., 2015). YafQ toxin significantly reduces the levels of tryptophanase (TnaA) by degrading its mRNA, which has 16 putative cleavage sites for YafQ toxin (Hu et al., 2015). In the stationary phase, RpoS activates tnaA, while YafQ inhibits the rpoS expression (Kwan et al., 2015). The reduced levels of RpoS and TnaA protein force limited the production of indole (tryptophanase activity product), which in turn inhibits the persistence (Hu et al., 2015).
8. TA modules associated with the biofilm formation
Bacterial cells can aggregate on both biotic and abiotic solid surfaces forming a multicellular complex enveloped by an exopolysaccharide matrix known as biofilms (Garnett & Matthews, 2013; Limoli et al., 2015). These biofilms help bacteria to survive in harsh environmental conditions and antibiotic tolerance is a hallmark for mature biofilms (Singh et al., 2017). Many bacteria capable of biofilm formation are associated with chronic infections in humans. Pathogenic bacteria such as Pseudomonas aeruginosa uses the strategy of biofilm formation to persist against the host defense mechanism (Mulcahy et al., 2014). The slimy layer over the bacterial cells in a biofilm does not allow antibiotics to penetrate (Roy et al., 2018). Apart from failure in the diffusion of antibiotics, some modifications in lipopolysaccharide, slow growth, and degradation of antibiotics have been observed in biofilm-associated drug-resistant cells of P. aeruginosa (Hall & Mah, 2017). A representative of type II TA system mqsRA has shown to be directly associated with biofilm formation (Merfa et al., 2016). The antitoxin MqsA antagonizes the toxin and is linked with default stress responses. The protein MqsA acts as a repressor of stress regulator RpoS. Low concentrations of RpoS reduces GMP levels, ultimately limiting the biofilm formation (Soo & Wood, 2013). Upon stress, Lon and ClpXP proteases rapidly degrade the unstable antitoxin MqsA, leading to RpoS accumulation. This finally switches the highly motile state of planktonic cells to a sessile state thus forming biofilms (Soo & Wood, 2013). Other TA modules like mazEF, relBE, yefM-yoeB, and dinJ-yafQ are also reported to influence biofilm formation in E. coli (Kim et al., 2009).
9. Targeting TA modules for eliminating antibiotic persistence
For eliminating persisters without any failure, some possible molecular targets must be examined to develop antipersister strategies. Since bacterial growth reduction can be achieved by targeting TA modules, they are expected to be involved in eliminating persisters (Paul et al., 2019; Ying Zhang et al., 2012). No homologs of bacterial TA genes and pre-existing resistance against toxins of TA systems are found in humans, thus TAs can be served as the ideal targets for the production of novel antibacterial drugs (Paul et al., 2019).
There are several proposed antibacterial strategies involving TA modules, yet they are not many supported with experimental data in most cases (Kang et al., 2018; Williams & Hergenrother, 2012). These multiple strategies involve the artificial activation of TA systems either directly or indirectly. The direct activation involves the use of specific molecules that can interfere with TA complexes and disrupt stoichiometry of active toxin & antitoxin. Whereas, in indirect activation, other cellular targets are triggered having interconnected functions with TA systems (Rownicki et al., 2020). The most convenient way that can be studied is to use some peptides or small compounds as a strong inhibitor of TA modules, preventing the formation of TA complex as shown in Fig. 4A. Designing peptides to inhibit TA and using protein-protein interaction to artificially activate the toxins is the common plan of action (Fernandez et al., 2016; Bienstock, 2012; Williams & Hergenrother, 2012). This strategy was used to activate the toxin of the pemIK TA module in B. anthracis. C-terminal fragment of protein antitoxin PemI was found to be responsible for interaction with PemK toxin (Agarwal et al., 2010; Chopra et al., 2011).
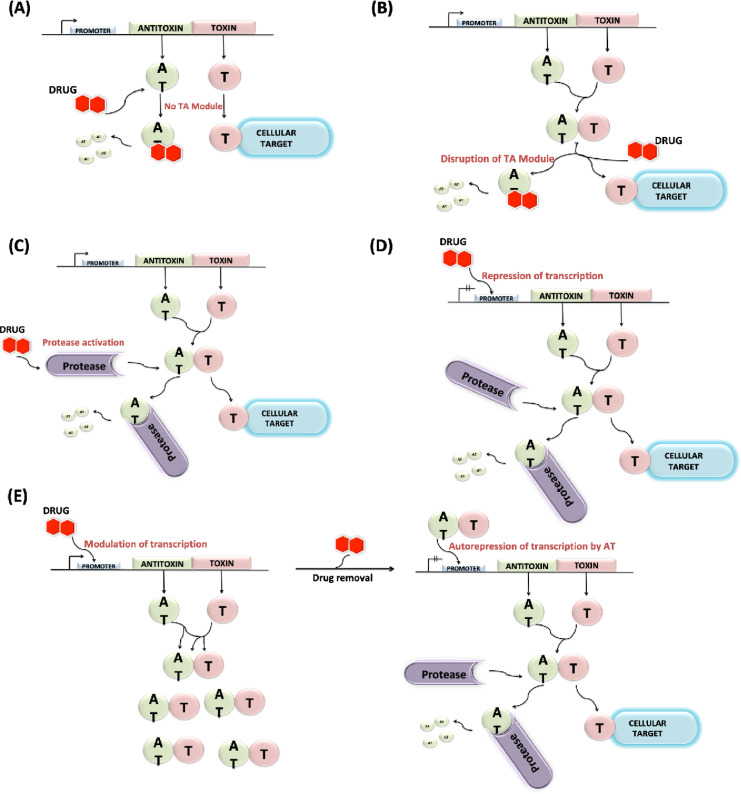
Fig. 4.
Different antibacterial strategies involving artificial activation of TA modules; (A) Prevention of TA complex formation; (B) Disruption of TA complexes; (C) Activation of cellular proteases degrading antitoxins; (D) Inhibition of TA transcription; (E) Overexpression of the TA system and subsequent removal of the activating drug (see text for details).
We can also use toxin mimicking peptides rather than antitoxin mimicking peptides for stronger inhibition of TA complex formation. For example, three peptides were designed to mimic the TA interface of the vapBC TA module (Lee et al., 2015). Disrupting the TA complex by the use of high-affinity peptide inhibitors displaces the toxin from its associated antitoxin. This forcibly liberates the toxin causing it to act onto its cellular target resulting in lethal effects to the TA producer cells, as illustrated in Fig. 4B. Although, not a single drug is designed till now to target TA interfaces, however, computational drug designing methods such as virtual screening and molecular docking may assist in the identification of such drug candidates (Bienstock, 2012).
Initially, in 2010, Lioy and colleagues through oligopeptide library screening using high throughput methods identified 17 amino acid long oligopeptide interfering ε-ζ TA system in Streptococcus pyogenes (Lioy et al., 2010). However, their results for disruption of ε-ζ TA assembly were not further replicable which may be due to weak binding of oligopeptide. But at the same time, their research proved the concept for using the strategy providing antimicrobial action.
The accelerated antitoxin degradation from its non-toxic TA complex through overexpression of cellular proteases is envisioned to be another fruitful antibiotic strategy. Therefore, increment in protease levels or designing molecules responsible for specifically activating these proteases may be an effective strategy indirectly activating the toxins as depicted in Fig. 4C. For achieving such a condition plasmid harboring cloned gene for protease can be introduced into the host cell. For example, overproduction of Lon protease-activated YoeB, leading to toxin-dependent mRNA degradation, translation inhibition, and eventually cell death (Christensen et al., 2004). Inhibition of bacterial cell division and cell death are the outcomes of such uncontrolled proteolysis, possibly involving toxin participation (Brotz et al., 2005).
A different antimicrobial strategy involving quorum sensing factors triggering TA systems was developed by Engelberg-Kulka and Kumar, in which they tried an approach to target the MazEF system. Their strategy involves the use of extracellular death factors or EDFs (group of pentapeptides) secreted by bacteria (Engelberg et al., 2005; Kumar & Engelberg, 2014). Another conceivable approach is to repress the transcription of TA genes, interrupting the constant availability of the less stable antitoxin (Brown et al., 2013; Hayes & Kedzierska, 2014). This strategy may involve drug candidates that can efficiently bind to the promoter sequence of the TA operon thus silencing the gene transcription, as illustrated in Fig. 4D. To avoid complex formation with a toxin, some biomolecules enhancing promoter binding can be designed based on structural information of the antitoxin (Lee & Lee, 2016). One therapeutic approach can be utilizing agents responsible for modulating transcription of two genes. So, designing activator protein-like drug molecules ultimately attract few proteins to the promoter region, activating RNA polymerase and resulting in overexpression of the TA system. This overexpression of genes provides excessive stable TA complexes. While subsequent removal of activating drug would show autorepression of TA system, excess free toxin after antitoxin degradation may cause bacterial cell death, as illustrated in Fig. 4E.
A typical approach utilizes TA systems, which involve inhibition of antitoxin production without modifying the translation of toxin (Lee & Lee, 2016; Unterholzner et al., 2013). This strategy is based on the fact that both genes of a type-II TA system are co-transcribed to having distinct Shine-Dalgarno sequences. Therefore, inhibiting antitoxin translation through artificially designed sequence-specific antisense PNA (peptide nucleic acid) would not disturb the toxin protein translation (Harms et al., 2018; Saberi et al., 2016). An experiment using antisense PNA for targeting MazE and HipB antitoxins in E. coli resulted in effectual cell growth inhibition. Whereas, no changes in relative levels of toxin mRNAs were reported, presenting a proof of concept (Rownicki et al., 2018).
Some other approach involves stringent response induction for indirect activation of toxins. Naturally, starvation and some stress signals activate the stringent response mediated by guanosine 3, 5 bispyrophosphate (ppGpp) alarmone (Jimmy et al., 2020). For example, in E. coli an antisense PNA conjugated with (KFF)3K carrier peptide, targeting thyA gene encoding for thymidylate synthase induced thymine starvation. As a consequence, MazF toxin production was triggered via ppGpp accumulation in bacterial cells. The significant drop in mRNA levels of thyA with complementary anti-thyA PNA treatment and resultant inhibition of growth certified the effectiveness of strategy involving silencing (Rownicki et al., 2018).
Specifically, type-II TA modules can easily be targeted using any of the above-mentioned strategies. The direct use of toxin as an antibiotic drug can also be an effective approach, while this has several therapeutic limitations such as expensive bioprocessing and poor oral availability (Lee & Lee, 2016). The need for an effective delivery system for a toxin to reach the pathogenic habitat in the host body without affecting the normal microflora and human cells is another issue. Using recombinant bacteriophages for delivering the desired toxin gene would be a more sophisticated procedure so that the high expression of toxin inside the pathogen can lead to cell death (Huys et al., 2013).
Although TA systems are being potential targets for eliminating antibiotic resistance, we cannot be in a hurry to assess their therapeutic potential in the clinical system. Fishing out the most appropriate strategy should be the foremost priority for developing new antimicrobial-based TA activators as drug discovery and development demands a huge amount of time and cost. Selection of correct TA target is also a crucial step, which must be contingent upon their clinical relevance involving their prevalence in antibiotic-resistant clinical isolates, human cell cytotoxicity, and the required drug delivery mode. It might be a highly advantageous approach to use TA activators combined with conventional antibiotics for targeting a broad range of antibiotic-resistant bacterial strains.
10. Concluding remarks
In summary, we have the staggering molecular classification of TA modules with diverse biological functions in bacteria. With various examples, we linked persister cell formation, biofilm formation, and bacterial resistance to TA modules. Further, we argue about the abundance of TA modules in bacterial pathogens and toxin's specific targets. We also discussed some novel antibacterial strategies involving TA modules to get rid of multiple drug tolerance. However, the molecular phenomena of many TA modules, their regulation, and their association with different biological functions in bacteria are still ambiguous. Therefore, the future aspects of bacterial TA biology should include TAome analysis, TA role in bacterial physiology, the molecular structure of TA modules, association with bacterial pathogenicity and virulence, evolutionary origin, and mechanism of TA modules. Moreover, a comparison of orthologous and paralogous TA modules is also an attractive field of study. An intense knowledge and novel approaches of TA modules are much required to facilitate the development in biotechnology and fight against bacterial resistance.
Author contributions
Mohit Yadav: Writing - Original draft preparation, Writing - Reviewing and Editing
Garima Singh: Writing - Original draft preparation, Writing - Reviewing and Editing
Chaitali Ghosh: Writing - Reviewing and Editing
Jitendra Singh Rathore: Writing - Reviewing and Editing, Funding acquisition
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this review article.
Acknowledgments
Work in Jitendra Singh Rathore's laboratory is currently funded by Project No. 37(1658)/15-EMR-II from the Council of Scientific & Industrial Research (CSIR), India. Mohit Yadav received a senior research fellowship (SRF) from the Council of Scientific & Industrial Research (CSIR), India.
References
- Aakre C.D., Phung T.N., Huang D., Laub M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol Cell. 2013;52(5):617–628. doi: 10.1016/j.molcel.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Mishra N.K., Bhatnagar S., Bhatnagar R. PemK toxin of Bacillus anthracis is a ribonuclease: An insight into its active site, structure, and function. J Biol Chem. 2010;285(10):7254–7270. doi: 10.1074/jbc.M109.073387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akarsu H., Bordes P., Mansour M., Bigot D.J., Genevaux P., Falquet L. TASmania: A bacterial toxin-antitoxin systems database. PLoS Comput Biol. 2019;15(4) doi: 10.1371/journal.pcbi.1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D.I., Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev. 2011;35(5):901–911. doi: 10.1111/j.1574-6976.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- Baharoglu Z., Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev. 2014;38(6):1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- Berghoff B.A., Wagner E.G.H. RNA ‑ based regulation in type I toxin – antitoxin systems and its implication for bacterial persistence. Curr Genet. 2017;63(6):1011–1016. doi: 10.1007/s00294-017-0710-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226(3):735–745. doi: 10.1016/0022-2836(92)90629-X. [DOI] [PubMed] [Google Scholar]
- Bibi-Triki S., de la Sierra-Gallay I.L., Lazar N., Leroy A., Van Tilbeurgh H., Sebbane F., Pradel E. Functional and structural analysis of HicA3-HicB3, a novel toxin-antitoxin system of yersinia pestis. J Bacteriol. 2014;196(21):3712–3723. doi: 10.1128/JB.01932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienstock R.J. Computational Drug Design Targeting Protein-Protein Interactions. Curr Pharm Des. 2012;18(9):1240–1254. doi: 10.2174/138161212799436449. [DOI] [PubMed] [Google Scholar]
- Bigger, J. W. (1944). TREATMENT OF STAPHYLOCOCCAL INFECTIONS WITH PENICILLIN BY INTERMITTENT STERILISATION. The Lancet., 244(6320), 497–500. https://doi.org/10.1016/S0140-6736(00)74210-3.
- Blower T.R., Fineran P.C., Johnson M.J., Toth I.K., Humphreys D.P., Salmond G.P.C. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol. 2009;191(19):6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower Tim R., Short F.L., Rao F., Mizuguchi K., Pei X.Y., Fineran P.C., Luisi B.F., Salmond G.P.C. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012;40(13):6158–6173. doi: 10.1093/nar/gks231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin R.A., Poirel L., Nordmann P., Eikmeyer F.G., Wibberg D., Pühler A., Schlüter A. Complete sequence of broad-host-range plasmid pNOR-2000 harbouring the metallo-β-lactamase gene blavim-2 from Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68(5):1060–1065. doi: 10.1093/jac/dks526. [DOI] [PubMed] [Google Scholar]
- Bordes P., Sala A.J., Ayala S., Texier P., Slama N., Cirinesi A.M., Guillet V., Mourey L., Genevaux P. Chaperone addiction of toxin-antitoxin systems. Nat Commun. 2016;7:13339. doi: 10.1038/ncomms13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Jahn N. SRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol Rev. 2015;39(3):413–427. doi: 10.1093/femsre/fuv003. [DOI] [PubMed] [Google Scholar]
- Brielle R., Pinel-Marie M.L., Felden B. Linking bacterial type I toxins with their actions. Curr Opin Microbiol. 2016;30:114–121. doi: 10.1016/j.mib.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Brinkman C.L., Bumgarner R., Kittichotirat W., Dunman P.M., Kuechenmeister L.J., Weaver K.E. Characterization of the effects of an rpoC mutation that confers resistance to the fst peptide toxin-antitoxin system toxin. J Bacteriol. 2013;195(1):156–166. doi: 10.1128/JB.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brötz-Oesterhelt H., Beyer D., Kroll H.P., Endermann R., Ladel C., Schroeder W., Hinzen B., Raddatz S., Paulsen H., Henninger K., Bandow J.E., Sahl H.G., Labischinski H. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med. 2005;11(10):1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- Brown B.L., Lord D.M., Grigorius S., Peti W., Pages R. The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J Biol Chem. 2013;288(2):1286–1294. doi: 10.1074/jbc.M112.421008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowska I., Zielenkiewicz U. Plasmid Regulation of toxin – antitoxin systems by proteolysis. Plasmid. 2013;70(1):33–41. doi: 10.1016/j.plasmid.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Buts L., Lah J., Dao-Thi M.H., Wyns L., Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30(12):672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Cai Y., Usher B., Gutierrez C., Tolcan A., Mansour M., Fineran P.C., Condon Ciarán, Neyrolles O., Genevaux P., Blower T.R. A nucleotidyltransferase toxin inhibits growth of Mycobacterium tuberculosis through inactivation of tRNA acceptor stems. Sci Adv. 2020;6(31) doi: 10.1126/sciadv.abb6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. International Journal of Medical Microbiology Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303(6–7):298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Xavier J.B. Altruistic cell death and collective drug resistance. Mol Syst Biol. 2012;8:627. doi: 10.1038/msb.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Roa D., Garcia-Pino A., De Gieter S., Van Nuland N.A.J., Loris R., Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol. 2013;9(12):811–817. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataudella I., Sneppen K., Gerdes K., Mitarai N. Conditional Cooperativity of Toxin - Antitoxin Regulation Can Mediate Bistability between Growth and Dormancy. PLoS Computational Biology. 2013;9(8) doi: 10.1371/journal.pcbi.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataudella I., Trusina A., Sneppen K., Gerdes K., Mitarai N. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res. 2012;40(14):6424–6434. doi: 10.1093/nar/gks297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.T., Espinosa M., Yeo C.C. Keeping the wolves at bay: Antitoxins of prokaryotic type II toxin-antitoxin systems. Front Mol Biosci. 2016;3 doi: 10.3389/fmolb.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.T., Nieto C., Harikrishna J.A., Khoo S.K., Othman R.Y., Espinosa M., Yeo C.C. Genetic regulation of the yefM-yoeB toxin-antitoxin locus of Streptococcus pneumoniae. J Bacteriol. 2011;193(18):4612–4625. doi: 10.1128/JB.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.S., Kim W., Suk S., Park H., Bak G., Yoon J., Lee Y. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 2018;15(10):1319–1335. doi: 10.1080/15476286.2018.1532252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W., Yamaguchi Y., Park J.-Y., Park S.-H., Lee H.-W., Lim B.-K., Otto M., Inouye M., Yoon M.-H., Park J.-H. Functional Characterization of the mazEF Toxin-Antitoxin System in the Pathogenic Bacterium Agrobacterium tumefaciens. Microorganisms. 2021;9(5):1107. doi: 10.3390/microorganisms9051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra N., Agarwal S., Verma S., Bhatnagar S., Bhatnagar R. Modeling of the structure and interactions of the B. anthracis antitoxin, MoxX: Deletion mutant studies highlight its modular structure and repressor function. J Comput Aided Mol Des. 2011;25(3):275–291. doi: 10.1007/s10822-011-9419-z. [DOI] [PubMed] [Google Scholar]
- Christensen S.K., Gerdes K. RelE toxins from Bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48(5):1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- Christensen S.K., Maenhaut-Michel G., Mine N., Gottesman S., Gerdes K., Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: Involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- Chukwudi C.U., Good L. The role of the hok /sok locus in bacterial response to stressful growth conditions. Microb Pathog. 2015;79:70–79. doi: 10.1016/j.micpath.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Cohen S.E., Foti J.J., Simmons L.A., Walker G.C. The SOS Regulatory Network. EcoSal Plus. 2008;3(1) doi: 10.1128/ecosalplus.5.4.3. [DOI] [PubMed] [Google Scholar]
- Coussens N.P., Daines D.A. Wake me when it's over – Bacterial toxin–antitoxin proteins and induced dormancy. Exp Biol Med. 2016;241(12):1332–1342. doi: 10.1177/1535370216651938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J.W., Rothenbacher F.P., Maehigashi T., Lane W.S., Dunham C.M., Woychik N.A. Doc toxin is a kinase that inactivates elongation factor Tu. J Biol Chem. 2014;289(11):7788–7798. doi: 10.1074/jbc.M113.544429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao-Thi M.H., Van Melderen L., De Genst E., Afif H., Buts L., Wyns L., Loris R. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol. 2005;348(5):1091–1102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
- Das B., Verma J., Kumar P., Ghosh A., Ramamurthy T. Antibiotic resistance in Vibrio cholerae: Understanding the ecology of resistance genes and mechanisms. Vaccine. 2020;38:A83–A92. doi: 10.1016/j.vaccine.2019.06.031. [DOI] [PubMed] [Google Scholar]
- De Jonge N., Simic M., Buts L., Haesaerts S., Roelants K., Garcia-Pino A., Sterckx Y., De Greve H., Lah J., Loris R. Alternative interactions define gyrase specificity in the CcdB family. Mol Microbiol. 2012;84(5):965–978. doi: 10.1111/j.1365-2958.2012.08069.x. [DOI] [PubMed] [Google Scholar]
- Denkovskienė E., Paškevičius Š., Stankevičiūtė J., Gleba Y., Ražanskienė A. Control of T-DNA transfer from Agrobacterium tumefaciens to plants based on an inducible bacterial toxin-antitoxin system. Mol Plant Microbe Interact. 2020;33(9):1142–1149. doi: 10.1094/MPMI-03-20-0067-R. [DOI] [PubMed] [Google Scholar]
- Díaz-Orejas R., Espinosa M., Yeo C.C. The importance of the expendable: Toxin-antitoxin genes in plasmids and chromosomes. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienemann C., Bøggild A., Winther K.S., Gerdes K., Brodersen D.E. Crystal Structure of the VapBC Toxin – Antitoxin Complex from Shigella flexneri reveals a Hetero-Octameric DNA-Binding Assembly. J Mol Biol. 2011;414(5):713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Lewis K., Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5(12) doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Vulić M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8(2) doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy R.L., Przybilski R., Semeijn K., Salmond G.P.C., Fineran P.C. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014;42(7):4590–4605. doi: 10.1093/nar/gkt1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy R.L., Richter C., Salmond G.P.C., Fineran P.C. Remarkable mechanisms in microbes to resist phage infections. Annu Rev Virol. 2014;1(1):307–331. doi: 10.1146/annurev-virology-031413-085500. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Hazan R., Amitai S. mazEF: A chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118(19):4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- Fauvart M., De Groote V.N., Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60(6):699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- Fernández-Bachiller M., Brzozowska I., Odolczyk N., Zielenkiewicz U., Zielenkiewicz P., Rademann J. Mapping protein–protein interactions of the resistance-related bacterial zeta toxin–epsilon antitoxin complex (ε2ζ2) with high affinity peptide ligands using fluorescence polarization. Toxins (Basel) 2016;8(7):222. doi: 10.3390/toxins8070222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-García L., Blasco L., Lopez M., Bou G., García-Contreras R., Wood T., Tomas M. Toxin-antitoxin systems in clinical pathogens. Toxins (Basel) 2016;8(7):227. doi: 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran P.C., Blower T.R., Foulds I.J., Humphreys D.P., Lilley K.S., Salmond G.P.C. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106(3):894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A., Gollan B., Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol. 2017;15(8):453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- Fraikin N., Goormaghtigh F., Van Melderen L. Type II Toxin-Antitoxin Systems: Evolution and Revolutions. J Bacteriol. 2020;202(7) doi: 10.1128/jb.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich K.S., Haneke K., Papenfort K., Vogel J. The target spectrum of sdsr small RNA in Salmonella. Nucleic Acids Res. 2016;44(21):10406–10422. doi: 10.1093/nar/gkw632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich K.S., Papenfort K., Berger A.A., Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40(8):3623–3640. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Contreras R., Zhang X.S., Kim Y., Wood T.K. Protein translation and cell death: The role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One. 2008;3(6) doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett J.A., Matthews S. Interactions in Bacterial Biofilm Development: A Structural Perspective. Curr Protein Pept Sci. 2013;13(8):739–755. doi: 10.2174/138920312804871166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelens L., Hill L., Vandervelde A., Danckaert J., Loris R. A General Model for Toxin-Antitoxin Module Dynamics Can Explain Persister Cell Formation in E. coli. PLoS Comput Biol. 2013;9(8) doi: 10.1371/journal.pcbi.1003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Bech F.W., Jørgensen S.T., Løbner-Olesen A., Rasmussen P.B., Atlung T., Boe L., Karlstrom O., Molin S., von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E.coli relB operon. EMBO J. 1986;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes Kenn, Christensen S.K., Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Germain E., Castro-roa D., Zenkin N., Gerdes K. Molecular Mechanism of Bacterial Persistence by HipA. Mol Cell. 2013:1–7. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- Goeders N., Chai R., Chen B., Day A., Salmond G.P.C. Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins (Basel) 2016;8(10) doi: 10.3390/toxins8100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders N., Van Melderen L. Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 2013;6(1):304–324. doi: 10.3390/toxins6010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios González, F. A., Zuo R., Ren D., Wood T.K. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol Bioeng. 2006;93(1):188–200. doi: 10.1002/bit.20681. [DOI] [PubMed] [Google Scholar]
- Granato L.M., Picchi S.C., De Oliveira Andrade M., Martins P.M.M., Takita M.A., Machado M.A., De Souza A.A. The ECNA antitoxin is important not only for human pathogens: Evidence of Its Role in the Plant Pathogen Xanthomonas citri subsp. Citri. J Bacteriol. 2019;201(20) doi: 10.1128/JB.00796-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A.K., Siddiqui N., Yadav D., Arora L., Dutta T. Regulation of RyeA/SraC expression in Escherichia coli. Biochem Biophys Res Commun. 2019;516(3):661–665. doi: 10.1016/J.BBRC.2019.06.110. [DOI] [PubMed] [Google Scholar]
- Gupta A.K., Siddiqui N., Dutta T. A novel mechanism of RyeA/SraC induction under acid stress. Biochem Biophys Res Commun. 2020;525(2):298–302. doi: 10.1016/j.bbrc.2020.02.085. [DOI] [PubMed] [Google Scholar]
- Gutierrez A., Laureti L., Crussard S., Abida H., Rodríguez-Rojas A., Blázquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J., Matic I. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun. 2013;4 doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez Arnaud, Jain S., Bhargava P., Hamblin M., Lobritz M.A., Collins J.J. Understanding and Sensitizing Density-Dependent Persistence to Quinolone Antibiotics. Mol Cell. 2017;68(6):1147–1154. doi: 10.1016/j.molcel.2017.11.012. e3. [DOI] [PubMed] [Google Scholar]
- Hall C.W., Mah T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;010:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- Hallez R., Geeraerts D., Sterckx Y., Mine N., Loris R., Van Melderen L. New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol Microbiol. 2010;76(3):719–732. doi: 10.1111/j.1365-2958.2010.07129.x. [DOI] [PubMed] [Google Scholar]
- Harms A., Brodersen D.E., Mitarai N., Gerdes K. Toxins, Targets, and Triggers : An Overview of Toxin-Antitoxin Biology. Mol Cell. 2018;70(5):768–784. doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Harms A., Maisonneuve E., Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354(6318) doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- Hayes F., Kedzierska B. Regulating toxin-antitoxin expression: Controlled detonation of intracellular molecular timebombs. Toxins (Basel) 2014;6(1):337–358. doi: 10.3390/toxins6010337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Van Melderen L. Toxins-antitoxins: Diversity, evolution and function. Crit Rev Biochem Mol Biol. 2011;46(5):386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- Heller D.M., Tavag M., Hochschild A. CbtA toxin of Escherichia coli inhibits cell division and cell elongation via direct and independent interactions with FtsZ and MreB. PLoS Genet. 2017;13(9) doi: 10.1371/journal.pgen.1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobby G.L., Meyer K., Chaffee E. Observations on the Mechanism of Action of Penicillin. Exp. Biol. Med. (Maywood) 1942;50(2):281–285. doi: 10.3181/00379727-50-13773. [DOI] [Google Scholar]
- Hosseini N., Pourhajibagher M., Chiniforush N., Hosseinkhan N., Rezaie P., Bahador A. Modulation of Toxin-Antitoxin System Rnl AB Type II in Phage-Resistant Gammaproteobacteria Surviving Photodynamic Treatment. J Lasers Med Sci. 2019;10(1):21–28. doi: 10.15171/jlms.2019.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Kwan B.W., Osbourne D.O., Benedik M.J., Wood T.K. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ Microbiol. 2015;17(4):1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- Hurley J.M., Woychik N.A. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem. 2009;284(28):18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huys I., Pirnay J., Lavigne R., Jennes S., Vos D.De, Casteels M., Verbeken G. Paving a regulatory pathway for phage. Nature Publishing Group. 2013;14(11):951–954. doi: 10.1038/embor.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienstock R.J. Computational Drug Design Targeting Protein-Protein Interactions. Curr Pharm Des. 2012;18(9):1240–1254. doi: 10.2174/138161212799436449. [DOI] [PubMed] [Google Scholar]
- Jahn N., Brantl S., Strahl H. Against the mainstream: The membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol Microbiol. 2015;98(4):651–666. doi: 10.1111/mmi.13146. [DOI] [PubMed] [Google Scholar]
- Jayaraman R. Bacterial persistence: Some new insights into an old phenomenon. J Biosci. 2008;33(5):795–805. doi: 10.1007/s12038-008-0099-3. [DOI] [PubMed] [Google Scholar]
- Jia X., Yao J., Gao Z., Liu G., Dong Y.H., Wang X., Zhang H. Structure–function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN–MNT toxin–antitoxin system. J Biol Chem. 2018;293(18):6812–6823. doi: 10.1074/jbc.RA118.002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Pogliano J., Helinski D.R., Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol. 2002;44(4):971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- Jimmy S., Saha C.K., Kurata T., Stavropoulos C., Oliveira S.R.A., Koh A., Cepauskas A., Takada H., Rejman D., Tenson T., Strahl H., Garcia-Pino A., Hauryliuk V., Atkinson G.C. A widespread toxin−antitoxin system exploiting growth control via alarmone signaling. Mol Microbiol. 2020;117(19):10500–10510. doi: 10.1073/pnas.1916617117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen M.G., Pandey D.P., Jaskolska M., Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol. 2009;191(4):1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurenaite M., Markuckas A., Sužiedeliene E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J Bacteriol. 2013;195(14):3165–3172. doi: 10.1128/JB.00237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurėnas D., Garcia-Pino A., Van Melderen L. Novel toxins from type II toxin-antitoxin systems with acetyltransferase activity. Plasmid. 2017;93:30–35. doi: 10.1016/j.plasmid.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Kang S.M., Kim D.H., Jin C., Lee B.J. A systematic overview of type II and III toxin-antitoxin systems with a focus on druggability. Toxins (Basel) 2018;10(12) doi: 10.3390/toxins10120515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E.coli: Hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012;9(12):1520–1527. doi: 10.4161/rna.22757. [DOI] [PubMed] [Google Scholar]
- Kedzierska B., Hayes F. Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules. 2016;21(6) doi: 10.3390/molecules21060790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Kang S.M., Park S.J., Jin C., Yoon H.J., Lee B.J. Functional insights into the Streptococcus pneumoniae HicBA toxin-antitoxin system based on a structural study. Nucleic Acids Research. 2018;46(12):6371–6386. doi: 10.1093/nar/gky469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Wang X., Ma Q., Zhang X.S., Wood T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. Nucleic Acids Res. 2009;191(4):1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M.A., Dwyer D.J., Collins J.J. How antibiotics kill bacteria : from targets to networks. Nat Rev Microbiol. 2010;8(6):423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch S.B., Hill T.M. Ectopic Overexpression of Wild-Type and Mutant hipA Genes in Escherichia coli: Effects on Macromolecular Synthesis and Persister Formation. J Bacteriol. 2006;188(11):3826–3836. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K.N. DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb Perspect Biol. 2013;5(11) doi: 10.1101/cshperspect.a012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J., Klinter S., Schneider C., Voß I., Steinbüchel A. Plasmid addiction systems: Perspectives and applications in biotechnology. Microb Biotechnol. 2010;3(6):634–657. doi: 10.1111/j.1751-7915.2010.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Engelberg-Kulka H. Quorum sensing peptides mediating interspecies bacterial cell death as a novel class of antimicrobial agents. Curr Opin Microbiol. 2014;21:22–27. doi: 10.1016/j.mib.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Kwan B.W., Osbourne D.O., Hu Y., Benedik M.J., Wood T.K. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol Bioeng. 2015;112(3):588–600. doi: 10.1002/bit.25456. [DOI] [PubMed] [Google Scholar]
- Lee I.G., Lee S.J., Chae S., Lee K.Y., Kim J.H., Lee B.J. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: Implications for the design of novel antimicrobial peptides. Nucleic Acids Res. 2015;43(15):7624–7637. doi: 10.1093/nar/gkv689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Lee B.J. Structure, biology, and therapeutic application of toxin-antitoxin systems in pathogenic bacteria. Toxins (Basel) 2016;8(10) doi: 10.3390/toxins8100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.W., Tan C.C., Rogers E.E., Stenger D.C. Toxin-antitoxin systems mqsR/ygiT and dinJ/relE of Xylella fastidiosa. Phytopathology. 2014;87:59–68. doi: 10.1016/j.pmpp.2014.07.001. [DOI] [Google Scholar]
- Lévi-Meyrueis C., Monteil V., Sismeiro O., Dillies M.A., Monot M., Jagla B., Coppée J.Y., Dupuy B., Norel F. Expanding the RpoS/σS-network by RNA sequencing and identification of σS-controlled small RNAs in Salmonella. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0096918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Webster T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J Orthop Res. 2018;36(1):22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli D.H., Jones C.J., Wozniak D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol Spectr. 2015;3(3) doi: 10.1128/microbiolspec.mb-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy V.S., Rey O., Balsa D., Pellicer T., Alonso J.C. A toxin-antitoxin module as a target for antimicrobial development. Plasmid. 2010;63(1):31–39. doi: 10.1016/j.plasmid.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Lobato-Márquez D., Díaz-Orejas R., García-del Portillo F. Toxin-antitoxins and bacterial virulencea. FEMS Microbiol Rev. 2016;40(5):592–609. doi: 10.1093/femsre/fuw022. [DOI] [PubMed] [Google Scholar]
- Lou C., Li Z., Ouyang Q. A molecular model for persister in E.coli. J Theor Biol. 2008;255(2):205–209. doi: 10.1016/j.jtbi.2008.07.035. [DOI] [PubMed] [Google Scholar]
- Magnuson R.D. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189(17):6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E., Gerdes K. Molecular mechanisms underlying bacterial persisters. In Cell. 2014;157(3):539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E., Shakespeare L.J., Jørgensen M.G., Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A. 2011;108(32):13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marimon O., Teixeira J.M.C., Cordeiro T.N., Soo V.W.C., Wood T.L., Mayzel M., Amata I., García J., Morera A., Gay M., Vilaseca M., Orekhov V.Y., Wood T.K., Pons M. An oxygen-sensitive toxin-antitoxin system. Nat Commun. 2016;7 doi: 10.1038/ncomms13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovski M., Wickner S. Preventing bacterial suicide: A novel toxin-antitoxin strategy. Mol Cell. 2013;52(5):611–612. doi: 10.1016/j.molcel.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins P.M.M., Machado M.A., Silva N.V., Takita M.A., De Souza A.A. Type II toxin-antitoxin distribution and adaptive aspects on Xanthomonas genomes: Focus on Xanthomonas citri. Nat Commun. 2016;7(MAY):652. doi: 10.3389/fmicb.2016.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowska K.H., Makiela-Dzbenska K., Fijalkowska I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ Mol Mutagen. 2019;60(4):368–384. doi: 10.1002/em.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Tan Q., Awano N., Wu K.P., Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol. 2012;84(5):979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- Mathers A.J., Peirano G., Pitout J.D.D. The role of epidemic resistance plasmids and international high- risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillick J., Ames J.R., Murphy T., Reddem E., Bourne C.R. A YoeB toxin from A. tumefaciens has metal-dependent DNA cleaving activity. BioRxiv. 2019;795211 doi: 10.1101/795211. [DOI] [Google Scholar]
- Merfa M.V., Niza B., Takita M.A., De Souza A.A. The MqsRA toxin-antitoxin system from Xylella fastidiosa plays a key role in bacterial fitness, pathogenicity, and persister cell formation. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I., Engelberg-Kulka H. Selective translation during stress in Escherichia coli. Trends Biochem Sci. 2012;37(11):493–498. doi: 10.1016/j.tibs.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti M.C., Hernández-Arriaga A.M., Kamphuis M.B., López-Villarejo J., Heck A.J.R., Boelens R., Díaz-Orejas R., van den Heuvel R.H.H. Interactions of Kid-Kis toxin-antitoxin complexes with the parD operator-promoter region of plasmid R1 are piloted by the Kis antitoxin and tuned by the stoichiometry of Kid-Kis oligomers. Nucleic Acids Res. 2007;35(5):1737–1749. doi: 10.1093/nar/gkm073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Del Álamo M., Tabone M., Muñoz-Martínez J., Valverde J.R., Alonso J.C. Toxin ζ reduces the ATP and modulates the uridine diphosphate-n-acetylglucosamine pool. Toxins (Basel) 2019;11(1) doi: 10.3390/toxins11010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed H.S., Bertrand K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155(2):768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy L.R., Isabella V.M., Lewis K. Pseudomonas aeruginosa Biofilms in Disease. Microb Ecol. 2014;68(1):1–12. doi: 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez A.J., Santos-Sierra S., Berzal-Herranz A., Lemonnier M., Díaz-Orejas R. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 2004;567(2–3):316–320. doi: 10.1016/j.febslet.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Neubauer C., Gao Y.G., Andersen K.R., Dunham C.M., Kelley A.C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D.E. The Structural Basis for mRNA Recognition and Cleavage by the Ribosome-Dependent Endonuclease RelE. Cell. 2009;139(6):1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman A., Hansen L.H., Sørensen S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364(1527):2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A., 1983;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman M.A., Brynildsen M.P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother. 2013;57(7):3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R., Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12(4):208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- Paul P., Sahu B.R., Suar M. Plausible role of bacterial toxin–antitoxin system in persister cell formation and elimination. Mol Oral Microbiol. 2019;34(3):97–107. doi: 10.1111/omi.12258. [DOI] [PubMed] [Google Scholar]
- Peano C., Wolf J., Demol J., Rossi E., Petiti L., De Bellis G., Geiselmann J., Egli T., Lacour S., Landini P. Characterization of the Escherichia coli δS core regulon by chromatin immunoprecipitation-sequencing (ChIP-seq) analysis. Sci Rep., 2015;5 doi: 10.1038/srep10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton J.N., Gorman S.P., Gilmore B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11(3):297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- Peng J., Triplett L.R., Schachterle J.K., Sundin G.W. Chromosomally encoded hok-sok toxin-antitoxin system in the fire blight pathogen Erwinia amylovora: Identification and functional characterization. Appl Environ Microbiol. 2019;85(15) doi: 10.1128/AEM.00724-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Triplett L.R., Sundin G.W. Activation of metabolic and stress responses during subtoxic expression of the type I toxin hok in Erwinia amylovora. BMC Genomics. 2021;22(1) doi: 10.1186/s12864-021-07376-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prysak M.H., Mozdzierz C.J., Cook A.M., Zhu L., Zhang Y., Inouye M., Woychik N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- Ramage H.R., Connolly L.E., Cox J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5(12) doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbarger T.L., Baker J.M., Matthew Sattley W. Slow and steady wins the race: an examination of bacterial persistence. AIMS Microbiol. 2017;3(2):171–185. doi: 10.3934/microbiol.2017.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson J., McKenzie J.L., Cursons R., Cook G.M., Arcus V.L. The vapBC Operon from Mycobacterium smegmatis Is An Autoregulated Toxin-Antitoxin Module That Controls Growth via Inhibition of Translation. J Mol Biol. 2009;390(3):353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Równicki M., Lasek R., Trylska J., Bartosik D. Targeting type II toxin–antitoxin systems as antibacterial strategies. Toxins (Basel) 2020;12(9) doi: 10.3390/toxins12090568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Równicki M., Pieńko T., Czarnecki J., Kolanowska M., Bartosik D., Trylska J. Artificial activation of Escherichia coli mazEF and hipBA toxin–antitoxin systems by antisense peptide nucleic acids as an antibacterial strategy. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, R., Tiwari, M., Donelli, G., & Tiwari, V. (2018). Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence., 9,(1), 522–554. https://doi.org/10.1080/21505594.2017.1313372. [DOI] [PMC free article] [PubMed]
- Saberi F., Kamali M., Najafi A., Yazdanparast A., Moghaddam M.M. Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell Mol Biol Lett. 2016;21(1) doi: 10.1186/s11658-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago A., da S., Mendes J.S., dos Santos C.A., de Toledo M.A.S., Beloti L.L., Crucello A., Horta M.A.C., Favaro M.T.d.P., Munar D.M.M., de Souza A.A., Cotta M.A., de Souza A.P. The antitoxin protein of a toxin-antitoxin system from Xylella fastidiosa is secreted via outer membrane vesicles. Front Microbiol. 2016;7:2030. doi: 10.3389/fmicb.2016.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savari M., Rostami S., Ekrami A., Bahador A. Characterization of toxin-antitoxin (TA) systems in Pseudomonas aeruginosa clinical isolates in Iran. Jundishapur J Microbiol. 2016;9(1) doi: 10.5812/jjm.26627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sberro H., Leavitt A., Kiro R., Koh E., Peleg Y., Qimron U., Sorek R. Discovery of Functional Toxin/Antitoxin Systems in Bacteria by Shotgun Cloning. Mol Cell. 2013;50(1):136–148. doi: 10.1016/j.molcel.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifano J.M., Cruz J.W., Vvedenskaya I.O., Edifor R., Ouyang M., Husson R.N., Nickels B.E., Woychik N.A. TRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2016;44(3):1256–1270. doi: 10.1093/nar/gkv1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C.F., Bertram R. Toxin-antitoxin systems of Staphylococcus aureus. Toxins (Basel) 2016;8(5) doi: 10.3390/toxins8050140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M., Austin S. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect Immun. 2011;79(7):2502–2509. doi: 10.1128/IAI.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidore T., Zeng Q., Triplett L.R. Survey of toxin-antitoxin systems in Erwinia amylovora reveals insights into diversity and functional specificity. Toxins (Basel) 2019;11(4) doi: 10.3390/toxins11040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Singh S.K., Chowdhury I., Singh R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol J. 2017;11(1):53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wood T.K. Toxin/Antitoxin System Paradigms: Toxins Bound to Antitoxins Are Not Likely Activated by Preferential Antitoxin Degradation. Adv Biosyst. 2020;4(3) doi: 10.1002/adbi.201900290. [DOI] [PubMed] [Google Scholar]
- Soo V.W.C., Wood T.K. Antitoxin MqsA represses curli formation through the master biofilm regulator. CsgD. Sci Rep. 2013;3 doi: 10.1038/srep03186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strugeon E., Tilloy V., Ploy M.C., Da Re S. The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. MBio. 2016;7(4) doi: 10.1128/mBio.00868-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomet M., Trautwetter A., Ermel G., Blanco C. Characterization of HicAB toxin-antitoxin module of Sinorhizobium meliloti. BMC Microbiol. 2019;19(1):1–12. doi: 10.1186/s12866-018-1382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q.B., Ohnishi M., Murata T., Nakayama K., Terawaki Y., Hayashi T. Specific protein-DNA and protein-protein interaction in the hig gene system, a plasmid-borne proteic killer gene system of plasmid Rts1. Plasmid. 2001;45(2):63–74. doi: 10.1006/plas.2000.1506. [DOI] [PubMed] [Google Scholar]
- Triplett L.R., Shidore T., Long J., Miao J., Wu S., Han Q., Zhou C., Ishihara H., Li J., Zhao B., Leach J.E. AvrRxo1 Is a Bifunctional type III Secreted effector and toxin-antitoxin system component with homologs in diverse environmental contexts. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0158856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoson C., Wagner E.G.H. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol Microbiol. 2008;70(1):258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- Unterholzner S.J., Hailer B., Poppenberger B., Rozhon W. Plasmid Characterisation of the stbD /E toxin – antitoxin system of pEP36, a plasmid of the plant pathogen Erwinia pyrifoliae. Plasmid. 2013;70(2):216–225. doi: 10.1016/j.plasmid.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Unterholzner S.J., Poppenberger B., Rozhon W. Toxin-antitoxin systems: Biology, identification, and application. Mob Genet Elements. 2013;3(5):e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L., De Bast M.S. Bacterial toxin-Antitoxin systems: More than selfish entities? PLoS Genet. 2009;5(3) doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandervelde A., Drobnak I., Hadži S., Sterckx Y.G.J., Welte T., De Greve H., Charlier D., Efremov R., Loris R., Lah J. Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon. Nucleic Acids Res. 2017;45(6):2937–2950. doi: 10.1093/nar/gkx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N., Lee H., Neyfakh A.A. Increased Persistence in Escherichia coli Caused by Controlled Expression of Toxins or Other Unrelated Proteins. J Bacteriol. 2006;188(10):3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan S., Mehta S., Uzri D., Jayaram M. Stable propagation of “selfish” genetic elements. J Biosci. 2003;28(5):623–636. doi: 10.1007/BF02703338. [DOI] [PubMed] [Google Scholar]
- Vogel J., Argaman L., Wagner E.G.H., Altuvia S. The small RNA istR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14(24):2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Volante A., Soberón N.E., Ayora S., Alonso J.C. The Interplay between Different Stability Systems Contributes to Faithful Segregation: Streptococcus pyogenes pSM19035 as a Model. Microbiol Spectr. 2014;2(4) doi: 10.1128/microbiolspec.plas-0007-2013. [DOI] [PubMed] [Google Scholar]
- Wagner E.G., Unoson C. The toxin-antitoxin system tisB-istR1: Expression, regulation and biological role in persister phenotypes. RNA Biol. 2012;9(12):1513–1519. doi: 10.4161/rna.22578. [DOI] [PubMed] [Google Scholar]
- Wang X., Kim Y., Hong S.H., Ma Q., Brown B.L., Pu M., Tarone A.M., Benedik M.J., Peti W., Page R., Wood T.K. general stress response. Nat. Chem. Biol. 2011;7(6):359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lord D.M., Cheng H.Y., Osbourne D.O., Hong S.H., Sanchez-Torres V., Quiroga C., Zheng K., Herrmann T., Peti W., Benedik M.J., Page R., Wood T.K. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012;8(10):855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wood T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yao J., Sun Y.C., Wood T.K. Type VII Toxin/Antitoxin Classification System for Antitoxins that Enzymatically Neutralize Toxins. Trends Microbiol. 2020;29(5):388–393. doi: 10.1016/j.tim.2020.12.001. [DOI] [PubMed] [Google Scholar]
- Weel-Sneve R., Bjørå M., Kristiansen K.I. Overexpression of the LexA-regulated tisAB RNA in E.coli inhibits SOS functions; implications for regulation of the SOS response. Nucleic Acids Res. 2008;36(19):6249–6259. doi: 10.1093/nar/gkn633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weel-Sneve R., Kristiansen K.I., Odsbu I., Dalhus B., Booth J., Rognes T., Skarstad K., Bjørås M. Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities. PLoS Genet. 2013;9(2) doi: 10.1371/journal.pgen.1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.Q., Bi D.X., Wei D.Q., Ou H.Y. Prediction of Type II Toxin-Antitoxin Loci in Klebsiella pneumoniae Genome Sequences. Interdiscip Sci. 2016;8(2):143–149. doi: 10.1007/s12539-015-0135-6. [DOI] [PubMed] [Google Scholar]
- Wen, Y., Behiels, E., & Devreese, B. (2014). Toxin-Antitoxin systems: Their role in persistence, biofilm formation, and pathogenicity. Pathog Dis., 70(3), 240–249. https://doi.org/10.1111/2049-632X.12145. [DOI] [PubMed]
- Williams J.J., Hergenrother P.J. Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol. 2012;20(6):291–298. doi: 10.1016/j.tim.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther K.S., Brodersen D.E., Brown A.K., Gerdes K. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat Commun. 2013;4 doi: 10.1038/ncomms3796. [DOI] [PubMed] [Google Scholar]
- Winther K.S., Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108(18):7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther K., Tree J.J., Tollervey D., Gerdes K. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 2016;44(20):9860–9871. doi: 10.1093/nar/gkw781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.K., Knabel S.J., Kwan B.W. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79(23):7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.L., Wood T.K. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen. 2016;5(3):499–511. doi: 10.1002/mbo3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.A.F., Waldor M.K. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5(3) doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K., Bao H., Zhang • Fuming, Linhardt R.J., Liang • Xinle. Characterization and comparative analysis of toxin-antitoxin systems in Acetobacter pasteurianus. J Ind Microbiol Biotechnol. 2019;46(3):869–882. doi: 10.1007/s10295-019-02144-y. [DOI] [PubMed] [Google Scholar]
- Yadav M., Rathore J.S. TAome analysis of type-II toxin-antitoxin system from Xenorhabdus nematophila. Comput Biol Chem. 2018;76:293–301. doi: 10.1016/j.compbiolchem.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Yadav M., Rathore J.S. The hipBAXn operon from Xenorhabdus nematophila functions as a bonafide toxin-antitoxin module. Appl Microbiol Biotechnol. 2020;104(7):3081–3095. doi: 10.1007/s00253-020-10441-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Inouye M. Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nat Rev Microbiol. 2011;9(11):779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Park J.H., Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284(42):28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Park J.H., Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- Yang Q.E., Walsh T.R. Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol Rev. 2017;41(3):343–353. doi: 10.1093/femsre/fux006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Guo Y., Zeng Z., Liu X., Shi F., Wang X. Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis. Microb Biotechnol. 2015;8(6):961–973. doi: 10.1111/1751-7915.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Zhen X., Tang K., Liu T., Xu X., Chen Z., Guo Y., Liu X., Wood T.K., Ouyang S., Wang X. Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system. Nucleic Acids Res. 2020;48(19):11054–11067. doi: 10.1093/nar/gkaa855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Gao X., Zhu K., Yin H., Mao X., Wojdyla J.A., Qin B., Huang H., Wang M., Sun Y.C., Cui S. Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism. Commun Biol. 2020;3(1) doi: 10.1038/s42003-020-0941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Ying, Yew W.W., Barer M.R. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother. 2012;56(5):2223–2230. doi: 10.1128/AAC.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Yonglong, Zhang J., Hoeflich K.P., Ikura M., Qing G., Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12(4):913–923. doi: 10.1016/S1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]