Highlights
-
•
Antibiotics cause the development of antibiotic resistance genes (ARG).
-
•
Antibiotics entering the environment will create a significant health threat.
-
•
Animals and crops being given antibiotic are harmful to humans.
-
•
Bacteria and algae are ideal candidates for the bioremediation of environmental antibiotics.
Keywords: Pharmaceutical products, Emerging pollutants, Antibiotic resistance genes, Livestock production
Abstract
In developing countries, the use of antibiotics has helped to reduce the mortality rate by minimizing the deaths caused by pathogenic infections, but the costs of antibiotic contamination remain a major concern. Antibiotics are released into the environment, creating a complicated environmental problem. Antibiotics are used in human, livestock and agriculture, contributing to its escalation in the environment. Environmental antibiotics pose a range of risks and have significant effects on human and animal health. Nevertheless, this is the result of the development of antibiotic-resistant and multi-drug-resistant bacteria. In the area of health care, animal husbandry and crop processing, the imprudent use of antibiotic drugs produces antibiotic-resistant bacteria. This threat is the deepest in the developing world, with an estimated 700,000 people suffering from antibiotic-resistant infections each year. The study explores how bacteria use a wide variety of antibiotic resistance mechanism and how these approaches have an impact on the environment and on our health. The paper focuses on the processes by which antibiotics degrade, the health effects of these emerging contaminants, and the tolerance of bacteria to antibiotics.
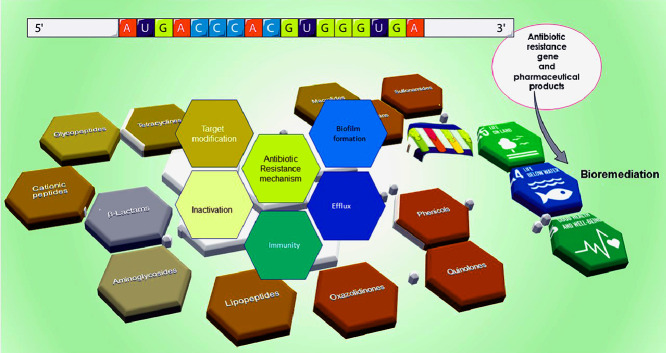
Graphic abstract
1. Introduction
A health care product such as a medicine is manufactured and/or used by a great number of people all over the world. In addition to human intake, pharmaceuticals are also widely used in ag agriculture and livestock production. The main source of environmental antibiotics is from its extensive use in animal production (Kinney and Heuvel, 2020). Many studies have shown that drugs are available in different environmental media, which give rise to concerns over potential adverse effects (Ebele et al., 2017). In recent times, there has been increasing knowledge of the unintentional presence of pharmaceutical products in various areas (e.g. ground water, soil and organisms) capable of adversely affecting - the biotic components . In almost every environmental matrix in all continents, pharmaceutical residues have been found (Aus der Beek et al., 2016). The database indicates that drugs or its metabolites have been identified in 71 countries throughout all continents. Such countries were then divided into five regions recognised by the United Nations. The UNSDGs (United Nations Sustainable Development Goals) have a strong emphasis on improving the environment (UN, 2015). The objective of the SDG is to conquer contaminated drinking water and soil with a view to providing a sustainable improvement in the health of millions of people. It includes surface water (waterways, lakes, rivers, estuaries, streams, and sea), groundwater and wastewater treatment plants (WWTP)(Zhou et al., 2020). Dugs or its metabolites are now widely detected in the geosphere (Riaz et al., 2018), and biosphere (Bartrons and Peñuelas, 2017), (Christou et al., 2017). Polar regions are now reported to have pharmaceutical contaminants, the most untouched environment on earth (González-Alonso et al., 2017).
Antibiotics are chemical therapeutic agents (pharmaceutical product) used in human, plant and animal for management of infectious diseases. A significant fraction of antibiotics produced annually are however used in non-therapeutic applications all over the world (Chattopadhyay, 2014) . A good part of it is used as growth promoters and not for infection-related therapy (Oliver et al., 2011). Approximately 24.6 million pounds of antibiotics are being used each year in livestock farming (Van et al., 2020). Low dose antibiotics were found to promote animal and bird growth (Kumar et al., 2018). It is at that point, antibiotics have become a global practise of adding to animal feeds to boost growth.
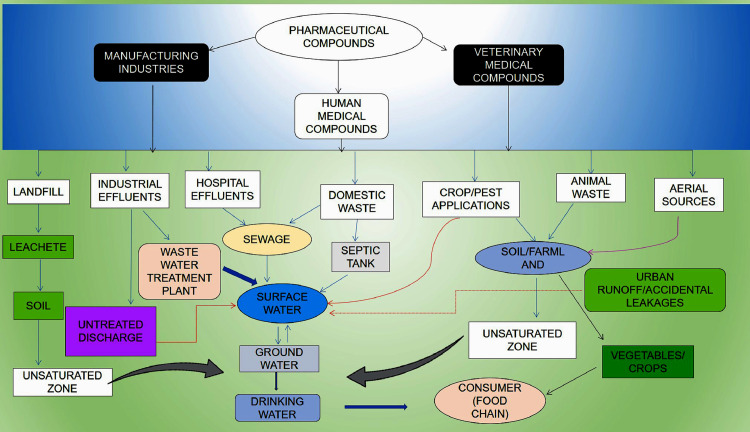
While the side effects of drugs have been well studied, the potential adverse effects of antibiotics on the environment are much less understood and have only recently become a matter of concern (Boxall, 2004). Pharmaceuticals have also been given more attention due to the rise of water pollution. In comparison, there is relatively less attention given to the release of pharmaceuticals into coastal environments and the impacts it has on the marine life. Researchers are now paying greater attention to preventing problems associated with environmental antibiotics (Gaw et al., 2014). The sources, impacts and trends of pharmaceuticals in the environment are defined in Fig 1. Sewage pollution can be a major concern of the ecosystem. In economically marginal countries, raw sewage may not be treated before being discharged into the environment. In many Asian cities, people still depend on septic tanks for disposal of sewage which pollutes the surface and groundwater with pharmaceutical products. Leachate generated through municipal solid waste landfill containing disposed antibiotics penetrate the surface and groundwater eliciting the development of antibiotic resistance genes (ARGs) and antimicrobial-resistant bacteria (ARB) (Wu et al., 2015).
Fig. 1.
Pathways of pharmaceuticals to the environment.
Once pharmaceuticals end up in the environment, they eventually penetrate the soil and water, and they are absorbed by the crops grown in those soils and water bodies. Another problem is the use of antibiotics in agriculture because they have been linked to the development of antibiotic-resistant bacterial strains in agricultural soil. Several drugs found to be used for human medicine are excreted and discharged at high-level into the drainage water. Pharmaceuticals will take years to get removed from the environment and be bioaccumulated in the general food supply. Current findings indicates that many drugs cannot be fully extracted from wastewater treatment plants. Drugs residues have been found within the soil, groundwater, and even marine ecosystems. Taking antibiotics can prevent deadly illnesses. However, in low income countries, antibiotics have been used improperly, thus resulting in the development of antibiotic-resistant bacteria. This article discusses how antibiotics entering the environment creates a health problem, mechanisms of antibiotic resistance genes and antimicrobial-resistant bacteria. The modes of action and resistance of widely used antibiotics, and potential effects of antibiotic pollution on both microbial communities and higher organisms are discussed in the article.
2. Animal antibiotics and its impact on humans
Antibiotics are used in animal feed and as a purpose of performance enhancement and improved production of livestock and animal husbandry. In 1959, tetracyclines, penicillins, streptomycins and bacitracin were used as growth promoters in cattle, pigs, chickens and ducks feed. Antibiotics that are being used are: sulphacetamide, oxytetracycline, tetracycline, tylosine, chloramphenicol, bacitracin, streptomycin, erythromycin, neomycin sulphate, lincomycin, oleandomycin, virginiamycin and bambermycin (Granados-Chinchilla and Rodríguez, 2017). In addition to the animal antibiotics, some chemical forms of antimicrobial agents are also applied, such as aresenate nitrofurans and sulfonamides (Vass et al., 2008; Ma et al., 2020). Arsenic compounds include arsanilic acid, 3-nitro-4-hydroxy-phenylarsonic acid and sodium arsenite, furazolidone- and nitrofurazone-containing nitro-furanic compounds (Yang et al., 2016). Various chemicals are added to prevent ducks and chickens from diarrhoea and malnourishment caused by coccidiosis and histomoniasis (Muthamilselvan et al., 2016). Meat consumption is on the rise due to an increase in animal products in our food. The number of antibiotics in some meat products is very high. Meat obtained from animals that receive antibiotics is of a higher quality than meat obtained from animals that have not received antibiotics (Chattopadhyay, 2014). This application of antibiotics in chicken feed has really improved productivity and egg lay (Mingmongkolchai and Panbangred, 2018). Cattle feed with mixed antibiotic have improved their health considerably. After dosing of antibiotics, the rate of bovine respiratory disease (BRD) cases decreased (Gallo and Berg, 1995). In all phases of pig development, antibiotic usage has been shown to promote animal health, increase growth rate, decrease mortality and morbidity, and reduce incidence of subclinical diseases (Cromwell, 2002).
Antibiotics lead to the development of ARGs, carried by the bacterial population in the environment, that provides selective pressures on the bacteria in the environment. The occurrences of ARG has proved to be very dangerous to human beings and other living species (Hinchliffe et al., 2018). When antimicrobial resistance keeps killing more and more people, scientists predict that it will become a major problem in the next 50 years; if the problem remains unattended , number of deaths would skyrocket to 300 million (Bunce and Hellyer, 2018).
2.1. Resistance arises from using antibiotics in animals
Many animals and fish have high levels of resistant bacteria in their intestines. Bacteria can be exchanged in many different manners, from animals to the environment (e.g. farms, animal markets and transport), and vice-versa (Kong et al., 2020). Bacteria capable of contaminating meat and other foodstuffs through slaughtering and feeding animals, exist in the environment (Pothakos et al., 2015). Animal waste also includes resistant, pathogenic bacteria. People are frequently infected through handling contaminated food, handling contaminated items and/or animals, and drinking or fishing in contaminated bodies of water. Improper use of antibiotics in animals can ultimately result in the production of antibiotic-resistant strains. The development of antibiotic resistance in bacteria is a major concern. According to reports by the World Health Organization, human deaths due to antibiotic resistant organisms have been higher than those due to diseases, including HIV, in recent years (Salgame et al., 2013). Although antibiotic resistance strains were mostly associated with antibiotic uses, resistance development is also detected in some bacterial population that are totally separated from human involvement (Bhullar et al., 2012). In the context of the crisis, the involvement of antibiotics in animal feed can be justified. The extensive use of antibiotics in large-scale animal farms raises the amount of antibiotic resistant bacteria that have been detected in these farms. The debates are encouraged by the studies regarding the isolation of animal bacteria that are genetically resistant to antibiotics. This shows that if antibiotics are not used properly, bacterial strains are developed that are resistant to those antibiotics. Understanding the susceptibility of different strains to multiple antibiotic treatments is essential in developing strategies to combat rising antibiotic resistance and to understand the advent of drug-resistant bacterial strains (Dawan and Ahn, 2020).
3. Antibiotic use in food crops a rising concern
Agricultural management activities, including increased use of agricultural pesticides and fertilizers, are often judged purely on the basis of what they contribute to the cost of production, such as lowering total production costs and increasing productivity, without taking into account possible environmental consequences (Udeigwe et al., 2015). The increase in the agricultural production is a result of the increased use of pesticides and fertilizers. Protection of crop diseases, insect pests and weed control in agriculture require the implementation of fertilization and pesticide applications (Prasad et al., 2017). However, pesticide incidences have been detected in food, microbes, human blood and adipose tissue.
In the 1950s, when antibiotics were being introduced for use in human medicine, opportunities for their use in stopping plant diseases were explored. It is known that nearly 40 antibiotics were screened in order to control plant diseases (McManus and Stockwell, 2000). The bactericides available in the 1950s and 1960s to farmers differ from those on which low dosage and insignificant toxicity for plants are found. Antibiotics are used to treat various types of bacterial diseases, such as fire blight of apples and pears. The amount of drugs used in the cultivation of plants is comparatively small in comparison to the amount used in human and veterinary medicine and in the production of livestock (McManus et al., 2002). Streptomycin was the main antibiotic used in crops (Plantomycin, Agrept, Fructocin) to protect against fire blight since it was introduced in 1955. Streptomycin is formulated in various forms as streptomycin sulfate and streptomycin nitrate as antibiotic. The application of antibiotics in United States in cash crops every year was estimated to be 19,550–65,227 kilograms (McManus et al., 2002). The Environmental Protection Agency (EPA) allowed growers to use drugs to protect against diseases which destroyed citrus trees. The decision was criticized by experts at the US Centers for Disease Control and Prevention and stated that the approach could harm human health by promoting antimicrobial resistance (Nelson, 2019). The agricultural use of antibiotics is already closely regulated by some countries. The European Union has prohibited both streptomycin and oxytetracycline from agricultural use.
3.1. The rise of antibiotic resistance in crops and consequences
Animal waste can contaminate fruit and vegetables. Vegetable plants may also contain bacteria believed to have the potential to cause injury to plants due to resistance to antibiotics. In addition, a significant portion of antibiotics remains unused after spraying. The root vegetables which grow in soil containing biosolids, may have more ARGs (Murray et al., 2019).
The overuse of antibiotics has been known to have negative effects on the gene pool and food chain (e.g., via fresh antibiotic producing bacteria). It would include all the resistance genes that could increase the risk (Schwaiger et al., 2011). Fertilization of the soil using raw animal waste, irrigation by agricultural waste water or release of farm waste into rivers are major factors for spread of ARGs (Hu et al., 2017).
Animal wastes may carry a majority of the germs that cause human disease. Many of these microbes, often resistant to antimicrobial drug, contaminate our food chain (Table 1). A number of different organisms have acquired the genes necessary to become resistant to aminoglycoside-based drugs, and are listed in Table 1. The mechanism incudes acetyltransferase (ACT); methyltransferase (MET); nucleotidyltransferase (NUT); and phosphotransferase (PHT).
Table 1.
A number of different bacteria acquired new genes capable of producing aminoglycoside drug resistance(Adapted from Van Hoek et al., 2011).
| Genera | Gene name | Length(nt) | Coding region | Accession number | Mechanism |
|---|---|---|---|---|---|
| Acinetobacter, Pseudomonas | aph (3)-Va | 780 | 103..882 | X07753 | PHT |
| Alcaligenes | aph (3)-VIb | 780 | 4934..5713 | AJ627643 | PHT |
| Bacillus, Campylobacter, Enterococcus, Staphylococcus, | aph (3)-III | 795 | 604..1398 | M26832 | PHT |
| Campylobacter | aph (3)-Via | 753 | 131..1036 | M29953 | PHT |
| Campylobacter | ant (6)-Ib | 858 | 27482..28339 | FN594949 | NUT |
| Campylobacter, Enterococcus, Staphylococcus, Streptococcus | sat4A | 543 | 38870..39412 | X92945 | ACT |
| Corynebacterium | aadA9 | 837 | 26773..27609 | AJ420072 | NUT |
| Enterococcus, Staphylococcus | spc | 783 | 331..1113 | X02588 | MET |
| Enterococcus, Staphylococcus | ant (6)-Ia | 909 | 22..930 | AF330699 | NUT |
| Enterococcus, Staphylococcus | ant (9)-Ia | 783 | 331..1113 | X02588 | NUT |
| Escherichia | sat3A | 543 | 221..763 | Z48231 | ACT |
| Escherichia | npmA | 660 | 3069..3728 | AB261016 | MET |
| Escherichia | aadA8b | 792 | 1174..1965 | AM040708 | NUT |
| Escherichia, Pseudomonas, Salmonella | sph | 801 | 6557..7354 | X64335 | NUT |
| Klebsiella, Vibrio | aadA8 | 792 | 1..792 | AF326210 | NUT |
| Legionella | aph (9)-Ia | 996 | 151..1146 | U94857 | PHT |
| Pseudomonas | rmtA | 756 | 6677..7432 | AB120321 | MET |
| Pseudomonas | aadA10 | 834 | 2807..3640 | U37105 | NUT |
| Pseudomonas | aac (6) | 441 | 1392..1832 | AY553333 | ACT |
| Pseudomonas | aac | 555 | 1985..2539 | AJ628983 | ACT |
| Pseudomonas | aac (6) | 402 | 81..482 | DQ302723 | ACT |
| Pseudomonas | aac (6) | 555 | 2092..2646 | EU912537 | ACT |
| Serratia, Pseudomonas | aac (3)-If | 465 | 61..525 | AY884051 | ACT |
| Stenotrophomonas | aph (3)-IIc | 813 | 2377498..2378310 | AM743169 | PHT |
| Streptomyces | aph (4)-Ib | 999 | 232..1230 | X03615 | PHT |
| Streptomyces | aph (6)-Ia | 924 | 1..924 | AY971801 | PHT |
| Streptomyces | aph (9)-Ib | 993 | 7526..8518 | U70376 | PHT |
Fruit and vegetable products can be contaminated with antibiotic-resistant bacteria before, during, and after harvesting due to cross contamination. Food poisoning is a serious problem, and it is understood that many people die because of food poisoning, including children under 5 years old. World Health Organization (WHO) estimates that bacteria are the most common microorganisms causing food poisoning in the world. This can also result in more illness as antibiotics do not affect the bacteria that is resistant to antibiotics (AL-Mamun et al., 2018). ARGs present in treated municipal waste water and water used for irrigation may have a potential to cause infection for fresh fruit and vegetables, especially after irrigation (Thanner et al., 2016). Irrigation water is one of the sources of bacterial contamination for vegetable crops grown before harvest. Due to scarcity of research on ARG, only limited data has been obtained on agriculture policies and on the use of irrigation water and manure (Hernando-amado et al., 2019).
The use of antimicrobial chemicals in the growth of crops is minimal compared to their use in protecting livestock or human health. The problem is linked to the application of the antimicrobial agents in plants and that infiltrate soil and water. The application of antimicrobial agents in crops is limited to streptomycin, which is utilized on a variety of crops for the treatment of plant diseases. The main problem with streptomycin application is that it gets strongly bound by the soil. The application of all antibiotics, however, must be studied in the context of complex farming characteristics in which adsorption of soil particles, precipitation or micro-organism degradationcan have a serious effect on antimicrobial agents stabilization and activity (Hernando-amado et al., 2019).
4. Antibiotic resistance in microbes
It is recognised that the rise of resistance to important bacterial diseases is one of the greatest problems encountered by most countries all over the world (Botelho et al., 2019). In studies of WHO, antibiotic resistance is on the list of three biggest public health threats to be addressed in 21st century (Prestinaci et al., 2015). According to a report published in 2006, the losses from multi-drug-resistant diseases in Europe alone were over USD 20 billion (Tillotson and Zinner, 2017).
The antibiotics used for years to treat bacterial diseases have been now acquired resistance, but today scientists continue to study new ways to synthesize and modify natural antibiotics in order to fight more stubborn types of microbes. There is evidence to say that bacteria can survive antibiotic stress by means of varied mechanism (Trinh et al., 2018). It is stated that after the antibiotics efflux from the body the antibiotic resistance can be develop in the microbes (Martínez and Baquero, 2002).
The functional groupings of the antibiotic modifying co-substrate enzymes, e.g., ATP, may be transferable via covalent modification of antibiotics via acetylation, phosphorylation, adenylation, nucleotidylation, ribosylation or glycosylation. Furthermore, changes in cell surface receptors, redox systems, and hydrolysis can lead to multi-drug resistance (Rajivgandhi et al., 2018). Various types of normal bacterial genes that become activated under severe antibiotic stress, produces resistant enzymes even under normal conditions (Cruz-Loya et al., 2019).
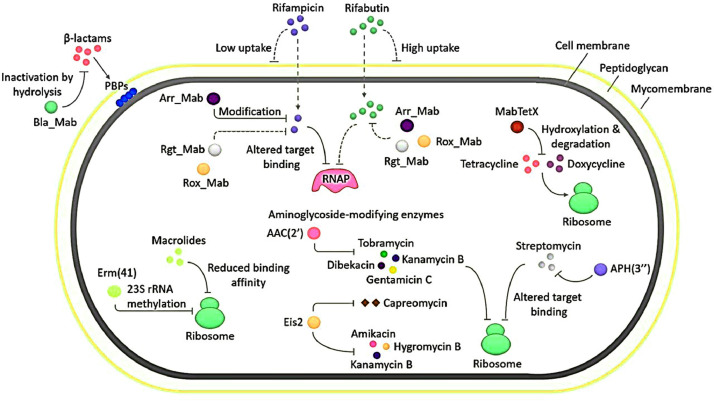
Different antibiotics work on different parts within bacteria that can be lethal, targeting a wide range of cellular components including the genetic material, proteins, the cytoplasmic membrane, and other cell wall components (Fig. 2). Antibiotics have high binding affinities for the targets that they aim to kill or inhibit, and so adversely affect the normal functioning of their targets (Methé et al., 2012; Fiegna and Velicer, 2005). Biomass is produced by high bacterial cell densities or is produced by a bacterial consortium using quorum sensing (Maurice et al., 2018). The negatively charged biofilm matrix prevents positively charged antibiotics (gentamicin and streptomycin, for example) from penetrating the bacterial matrix. Mycobacterium abscessus are resistant to a wide range of antibacterial drugs. The presence of some enzymes can alter and/or degrade antibiotics, and also enables to resist the action of several groups of antibiotics. This figure shows the mechanisms of resistance to antibiotics treatment in M. abscessus (Fig. 2).
Fig 2.
In Mycobacterium abscessus, aminoglycoside covalently modifying enzymes that modify the aminoglycoside groups from the antibiotic molecule via acetyltransferases - AAC (2׳), Eis2; and phosphotransferase - APH (3׳׳) (Luthra et al., 2018).
The large bacterial density within the biofilm consortium raises the likelihood of resistant bacteria being selected through antibiotic pressure by increase in horizontal gene transfer (HGT) ratios and mutation frequency (Li et al., 2018). One of the most well-known examples of antibiotic resistance has occurred from aminoglycoside covalently modifying enzymes (AMEs) that modify the hydroxyl or aminoglycoside groups from the antibiotic molecules. Many AMEs have been described so far and have become the world's dominant resistance mechanism for aminoglycosides. Another traditional example of enzymatic alteration of an antibiotic is the modification in chloramphenicol, an antibiotic which inhibits protein synthesis through its interaction with the peptidyl transfer centre of ribosomal unit 50S. CATs (chloramphenicol acetyltransferases) mainly drives the chemical modification of chloramphenicol. In both Gram-positive and Gram-negative bacteria several CAT genes have been described (Goodale et al., 2020).
The ecosystem may also be affected by resistant bacteria from farms. The use of manure in farmlands spread resistant bacteria and active antibiotic metabolites. A research was conducted in 2006, has shown that after treatment of farms with pig manure slurry, antibiotic-resistant bacteria were identified from the ground surface (Sengeløv et al., 2003).
The direct transmission between animals and farmworkers of resistant enterococci was shown in several studies (Da Costa et al., 2013). Most importantly, while the molecular enterococci subtypes of human and animals are highly heterogeneous, several studies on animals, food and humans have identified identical or closely related subtypes that support the importance of the route of food transmission (Argudín et al., 2017).
4.1. Commonly used antibiotics, mechanisms of action and resistance
Alexander Fleming's discovery of penicillin in pharmacology and medicine in 1928 marked a turning point, but the search for chemical compounds with inhibition of microbial growth began much earlier. In the year of 1871, J. B. Sanderson and his colleagues reported that the strain of Penicillium mould inhibited bacterial growth, but the actual effect was effectively demonstrated by E. Duchesne in the year 1897. Penicillin was first reported in 1938, but soon thereafter in 1939–1940, S. Waksman and R. Dubos has reported more than 20 potential antibiotics. Resistance has been discovered and recognised since then, along with the discovery of new antibiotics. The main failure of all antibiotics and other drug industries is that all organisms are resistant to antibiotics (Tacconelli et al., 2017). Horizontal Gene Transfer (HGT) is the transfer of genes from one bacterial cell to another, including ARGs, creating resistance to antibiotics. HGT is a serious threat and concern for both humans and animals including the bacterial population (Kivits et al., 2018). Microorganisms have been able to develop resistance by avoiding specific drug-target interactions such as DNA, RNA or protein, by rejecting antibiotics from the cells through the efflux pump and through biotic and abiotic processes through ARGs. The functioning of ARGs also extends to the fight against survival between bacterial strains that naturally produce antibiotics in order to reduce competition for the same resources. The structural characteristics of the cell walls such as extra membrane and exo-polysaccharide and extracellular DNA and the aggregation of bacteria on biofilms (Qi et al., 2016) prevent antibiotics from reaching their targets in order to increase their resistance (Galaon et al., 2019). The biofilm is suitable for the acquisition and distribution of resistance genes among bacterial populations (Fux et al., 2005). According to Acar and Moulin, the ability of the bacteria to establish antibiotic resistance dependent on the availability of the bacteria to respond to the selective pressure of the antibiotic (Acar and Moulin, 2006). One of the most efficient bacterial defense mechanisms is the efflux pump through which bacteria eliminate ingested chemical compounds, including antibiotics (Table 2). These bacterial efflux pumps may expel certain substrates or may have very broad specificities and may transport multiple substrates. In addition, there is a high mechanistic homology of the bacterial efflux pump with multi-drug resistance (MDRs) mechanisms for eukaryotic cells. Regarding this, a study showed that LmrA, an ATP-based ABC transporter in Lactococcus lactis, involves pumping amphiphilic compounds from the inside of the bacterium in antibiotic resistance systems and is a P-glycoprotein homologue, also an ATP-based ABC transporter involved in tumour cell resistance to chemotherapy (Endicott and Ling, 1989). Due to the high homology between human P-glycoprotein and bacterial LmrA, LmrA genes have been inserted into fibroblast cells that have produced resistance to P-glycoprotein. In addition, P-glycoprotein-specific inhibitors retain their inhibitory properties with LmrA (Veen and Callaghan, 1998). The interchange ability of these genes of different origins shows that the resistance mechanism caused by efflux-pumps is maintained from bacteria to humans.
Table 2.
Mechanism that causes bacteria to become resistant to antibiotics (Adapted from Peterson and Kaur, 2018).
| Mechanism of antibiotic resistance | Selected examples | Gene location | Reference |
|---|---|---|---|
| Antibiotic modification/degradation | Aminoglycoside modifying enzymes (AME):AAC; APH; ANT Streptomycin-6- phosphotransferase | Chromosome S. griseus (smk) | (Shinkawa et al., 1985; Mak et al., 2014) |
| Antibiotic efflux | ABC transporter DrrAB (Dox) OtrC (oxytetracycline) | Chromosome S. peucetius (drrAB), S. rimosus (otrC) | (Yu et al., 2012; Li et al., 2014) |
| Antibiotic sequestration by special proteins | Sequestration TlmA, BlmA, ZbmA (bleomycin) | Chromosome S. hindustanus (tlmA); S. verticillus (blmA); S. flavoviridis (zbmA) | (Gatignol et al., 1988; Rudolf et al., 2015; Sugiyama et al. 1994) |
| Antibiotic target modification | Low affinity penicillin-binding proteins (PBP) Class A Class B | Chromosome Streptomyces species | (Ogawara, 2016a, Ogawara, 2016b) |
| Antibiotic target bypass | DNA gyrase subunit B (novobiocin) | Chromosome S. sphaeroides (gyrBR) | (Schmutz et al., 2003) |
| Antibiotic target protection | Antibiotic removal DrrC (Dox) OtrA (oxytetracycline) | Chromosome S. peucetius (drrC), S. rimosus (otrA) | (Mak et al., 2014; Prija and Prasad, 2017) |
4.2. Superbugs and super-resistance bacteria
The term "superbug" refers to microbial strains with a higher amount, and or, exhibiting multiple mutations that provide antibiotic resistance, which are particularly important for research. A highly dangerous, multi drug-resistant pathogen called Mycobacterium tuberculosis is found in both developing countries and developed nations. Some life-threatening, multi drug-resistant pathogen include: Campylobacter jejuni, Acinetobacter baumannii, Citrobacter freundii, Enterococcus faecalis, Enterococcus faecium, Clostridium difficile, Klebsiella sp., Escherichia coli, Haemophilus influenzae, Proteus mirabilis, Pseudomonas aeruginosa, Staphylococcus epidermidis, Streptococcus pneumonia, and S. aureus. Some virulence cases, such as MRSA (Methicillin-resistant Staphylococcus aureus), may acquire super-resistance and transmissibility (Kaur, 2014). As a common pathogen, M. tuberculosis (tuberculosis causing bacteria) has developed along with humans and infects nearly one-third of the people in the world (Davies, 1996). A spectrum of infectious diseases in humans and animals are caused by E. coli, Salmonella enterica and K. pneumoniae. It has been observed that the class of antibiotics, β-lactams, and their associated enzymes has significantly contributed to the growth of antibiotic resistance. Several groups and classes of β-lactamases with unique genetic mutations have been identified, including novel genes and mutant irradiation (Alvarez-ord, 2020).
Infections and transmission of the β-lactam antibiotic resistance gene have been reported to occur due to horizontal gene transfer (HGT) processes in both the community and the hospital. P. aeruginosa has been a major nosocomial threat of burn wound infection in hospital acquired diseases. Again, antibiotic resistant mechanisms have coincided with the introduction of new antibiotic derivatives and have compromised more effective therapies (such as β-lactam and aminoglycosides). Staphylococcus aureus, also known as 'superbug', has long been associated with humans. Three out of four people carry it in their nose and it can cause skin infections such as boils. Multi-drug resistant bacteria, M. tuberculosis is one of the leading causes of hospital-acquired infections. In 1959, the main discovery and introduction of the first designer anti-resistance antibiotic, methicillin was assumed to be a safe defense against penicillinase-producing bacteria, but within 3 years, the appearance of methicillin-resistant S. aureus was shown to be a reality. MRSA was considered incurable, although other resistant strains were discovered (Kaur et al., 2013).
MRSA has been spreading rapidly throughout the community and is one of the most virulent of the community acquired pathogens. While there are various types of mec genes, CA-MRSA has most of its properties and has acquired a wide variety of mec gene clusters to acquire new or make additional carriers for more dangerous bacteria that cause major illness (Li et al., 2009)
Superbugs are common in the environment and their impact is greatly worsened in problematic and impoverished environment. The most dangerous ones often cause deadliest diseases for humans. At the top of the list of superbug Vibrio cholerae should be placed (Ahmad et al., 2018).
4.3. Mobile genetic elements
Transposons also belong to the group of moving genetic elements called transposable elements, which includes tiny cryptic elements called insertion sequences, transposons and viral transpositions which help to move genetic material from one place to another (Aminov, 2011). Transposons are diverse, well-organized systems that are found in bacteria of different antibiotic resistance phenotypes. Many transposons are known to be associated with antibiotic resistance, like Tn5 and Tn10 which encode resistance to the drug kanamycin and the drug neomycin, but the Tn21 series is one of the largest and first known groups involved in antibiotic resistance accumulation and dissemination. The Tn21 is resistant to spectinomycin, streptomycin, and sulfonamides (Essa et al., 2003).
It is well known fact that mobile genetic elements can move between bacteria or move from bacteria to other species (Dimitriu et al., 2015). Mobile genetic elements are a portion of the DNA that contain enzymes and proteins that help to transfer the DNA around the cell. Recent research on mobile genetic elements plays a substantial role in the development and adaptation of bacteria and the resulting health risks caused by antibiotic resistant strains (Partridge et al., 2018). "Transposons" or "transposable elements" can facilitate "intracellular mobility" or "intracellular transfer" of genetic information (Munoz-Lopez and Garcia-Perez, 2010).
Bacterial plasmids are the tools used for gene transfer. Plasmids may be passed between bacterial cells, enabling transfer of new traits into other organisms. Plasmids contain a number of genes that make the cell immune to various kinds of antibiotics. One or more resistance genes may be carried on a plasmid. Many resistance plasmids encode their own virulence, and they promote their own transfer. Antibiotic resistance plasmids can harbour ARGs (Al-Riyami et al., 2018).
5. Transmission of antibiotic resistance gene reservoir, carrier and vector
Because of the presence of antimicrobial resistant bacteria, humans and animals are reservoirs for ARB, and the use of antimicrobial agents in livestock affects the general public health. The presence of vancomycin resistant and avoparcin resistant enterococci in foods reflects the widespread use of these glycopeptides’ antibiotics. Therefore, vancomycin and avoparcin resistant genes have already spread between livestock and humans. The first alarm was sounded in 1994. It has been proven that bacteria resistant to glycopeptide antibiotics can be isolated from various foods in the UK, Germany and Denmark. Epidemiological studies of poultry and swine populations have found that avoparcin exposure was positively associated with intestinal rot in broilers and pigs (Bager et al., 1997). The study demonstrated that avoparcin was often used as a growth promoter in breeding farms, which led to the emergence of vancomycin resistant Enterococcus faecium. Members of antimicrobial substances, such as macrolides (e.g., tylosin, spiramycin, evernimicin, avilamycin, streptogramin, virginiamycin and bacitracin) (Gambarotto et al., 2001) have also received most intense investigation (Manson et al., 2003). The rate at which antibiotic resistant bacteria are transmitted from livestock to the environment and in the food-production chain are crucial factors in the human resistance.
6. Detection and analysis of antibiotics
Chromatography techniques are most widely used for the automated system for detection and quality analysis of antibiotics (Detector, 2018). Other methods for determining antibiotics include electrophoresis (Dai et al., 2017), or enzyme-linked immunosorbent assay(ELISA) (Munteanu et al., 2018). In recent years, more attention has been paid to biologically based sensors, mainly due to the use of new nanomaterials for the construction of special nature-based sensors for the detection of antibiotics (Lan et al., 2017). The new bio-recognition sensors, including aptamers(oligonucleotide or peptide molecules), that can be more selective and specific for target proteins and vectors (Malhotra, 2016).
Analytical technologies (such as mass spectrometry and liquid chromatography/mass spectrometry) have helped to detect the presence of certain pharmaceuticals in environment at μg/L and ng/L concentrations (Azzouz et al., 2019). Determination and quantification of almost 3000 environmental compounds using these techniques has now been possible HPLC is a popular chromatography method widely used for extracting and detecting antibiotics for example virginiamycin (Wang et al., 2018). Because of the high sludge particle adsorption, analytical methods for the quantification of tetracyclines and sulfonamides in the wastewater, soil, sludge, and manure have created difficultly in extraction processes (Pamreddy et al., 2013. Results from studies suggested that in the influents and effluents of water purification plants sulfamethoxazole, trimethoprim, and ofloxacin are present (Barancheshme and Munir, 2018). Several antibiotics were extracted from solid samples using ultrasound extraction method and an extracting solvent via a microwave extraction method (Salman et al., 2018) and their concentration observed by liquid chromatography (Lehotay and Lightfield, 2018), mass spectrometry or tandem mass spectrometry (Bou, 2016). To determine how long antibiotics remain in the environment, researchers must examine their physical and chemical properties as well as their transformations (Yan et al., 2018).
7. Biodegradation of antibiotics
Biotic and abiotic processes lead to degradation of antibiotics. Biotic factor include microorganisms and abiotic factor includes hydrolysis, sorption, reactions of oxidation, photolysis, and reduction (Massé et al., 2014; Tiwari et al., 2016). The susceptibility to abiotic degradation depends on the chemical structure, and approximately 70% of the antibiotics administered may be discovered unchanged or degraded in the environment to different products. Some of these antibiotics pose a greater risk than the diseases they treat (Zhang et al., 2016). The structure of tetracyclines is smaller than cephalexin and degrades more quickly with UV and photolysis (Azimi and Nezamzadeh-ejhieh, 2015). Tetracyclines, sulphonamides, tylosin, nitrofuran or (fluoro) antibiotics are all UV sensitive but fluoroquinolones are hydrolysis resistant. Oxygen and ozone are widespread abiotic factors and are involved in oxidation-related degradation of antibiotic such as sulfamethoxazole and oxytetracycline (Sirés and Brillas, 2012). In the presence of metal ions such as mercury, copper, zinc, cadmium and cobalt ions, beta-lactam antibiotics are degraded by breakdown of β-lactam ring (Kumar et al., 2019).
Enzymes play an important role in forming and breaking down of antibiotics, which is important in the antibacterial resistance mechanism. Research has shown that ARG expression, the production of antibiotic degrading enzymes, and antibiotic resistant bacteria are correlated. In addition, some studies indicated that ARGs involving the expression of aminoglycoside modification and β-lactamase enzymes, had been present in the environment long before manmade antibiotics had been used for therapeutic purposes (Costa et al., 2011). Several possible pathways for biodegradation (LC-MS/MS identification) of ciprofloxacin by a mixed culture of Gamma-proteobacteria, Bacteroidia and Beta-proteobacteria has been elucidated (Liao et al., 2016). Several ciprofloxacin biodegradation products generated by deamination (C–N bond cleavage), hydroxylation, defluorination (C–F bond cleavage) and dealkylation (C–C-bond cleavage) were detected.
In a review, Chen et al. outline the recent progress towards understanding the biodegradation of sulfonamide antibiotics in natural sludge systems by microorganisms (Chen and Xie, 2018). The authors described the biodegradation pathway of the sulphonamides at aniline or amino heteroaromatic sulfonamide structure mainly through hydroxylation and acetylation reactions. Other chemical changes result from cleavage of C–N, C–S and C–C bond along with oxidation. Liao et al. reported bacterial strains, including Proteobacteria and Acidobacteria, Firmicutes and Bacteroidetes capable of degrading antibiotic compounds (for example sulfamethazine) (Liao et al., 2015). The sulfanilamide (SAD) biodegradation have been promoted by Firmicutes, Bacteroidetes, and Chryseobacterium . Rhodopirellula baltica, Pseudomonas sp., Micrococcus luteus, Methylibium petroleiphilum, Oligotropha carboxidovoran, Delftia acidvorans, and Acinetobacter, are some potential bacterial strains which are able to biodegrade many of sulfonamide antibiotics (Islas-espinoza et al., 2012) . Some of these micro-organisms have a quite high biodegradation rate which can even reach 100 % for sulfamethoxazole.
Beside antibiotics, microbial degradation is also used to rapidly and efficiently get rid of other pharmaceuticals (Rasmus et al., 2005). This biodegradation is supported by both bacteria and fungi. Numerous products are produced in microbial biodegradations. However, carbamazepine, acetaminophen, atenolol, ibuprofen, propranolol, indomethacin, mefenamic acid, and more have been found to resist degradation in wastewater treatment, and remain in the aquatic environment (Yamamoto et al., 2009). A combination of adsorption, photodegradation and biodegradation has been achieved to effectively remove ibuprofen and other resilient medicines (Santos et al., 2009). The success of bio-degradation of pharmaceuticals can be improved, but depends greatly on the amount of time that sludge is retained and on the hydraulic retention in WWTPs, the flow rate of the water and the microbial diversities in both sludge and water (Ingerslev and Halling-s, 2001).
Despite the fact that most antibiotics are persistent in the environment, .some of them are biodegradable or partially degradable (Thiele-bruhn, 2003), and are generally slow (Gartiser et al., 2007). Microbes in surface water, soil, as well as in sewage treatment systems, degrade antibiotics. The sediments with ampicillin, oxytetracycline, doxycycline, and thiamphenicol were significantly degraded, but josamycin remained largely undegraded (Maki et al., 2006). Studies suggests that josamycin resistant bacteria either cannot destroy drugs or inhibit other antibiotics in josamycin resistant sediments. Similarly, β-lactam antibiotics such as penicillin is biodegraded by bacterial β-lactamases (Kümmerer, 2009). However, there is very little degradation of fluoroquinolone antibiotics in aquatic systems and sediments (Lai and Lin, 2009).
8. Bioremediation of antibiotics for environmental safety
Antibiotics are important compounds used to treat and prevent infections in humans as well as animals (Bunce and Hellyer, 2018). Antibiotics are released into the environment via farm manure and through its use in medical treatment, often polluting the environment. Antibiotics persist in the environment (Ezzariai et al., 2018). Bioremediation refers to the practice of using living organisms to extract and/or detoxify chemicals. The various approaches of bioremediation are either in-situ or ex-situ. The in-situ bioremediation method would involve the removal of contamination on the premises while ex-situ refers to the removal of the contamination that must be treated elsewhere (Ezzariai et al., 2018). Bacteria are the main organisms used for bioremediation. Bacteria-mediated recovery is a valuable and environmentally friendly way to remove antibiotics from the environment. The bacteria to be used in the process of bioremediation should be able to survive under extreme conditions (Morikawa, 2006). A study involved researching the use of bacterial methods to remove antibiotics, including biodegradation, from public areas and drinking water (Al-Gheethi et al., 2015). Another report suggests that the treatment of β-lactamase effluent and wastewater treatment of cephalexin, ceftaroline, ampicillin, and amoxicillin has been shown to be successful when B. subtilis 1556WTNC is applied (Al-Gheethi et al., 2014).
Bioremediation has recently received attention due to its promise in removing hazardous compounds from contaminated soil and water, thus being an effective and cost-effective method compared to conventional methods. Previously, Tasho and Cho (2016) reviewed how pharmaceutical compounds end up in the environment and are made use of by plants. Recently, algae are used in bioremediationofcontaminants such as antibiotics. Algae can be used in water treatment methods that are cost-effective and non-intrusive, and also safe and effective for antibiotic removal. In order to remove cephalosporins, a new treatment called alga-activated sludge syatem was implemented (Guo and Chen, 2015). Similarly, a green alga Chlorella pyrenoidosa e efficiently removes antibiotics such as cefradine, cephalexin, cefixime and ceftazidime (Yu et al., 2017). Using of microalgal strains (Chlamydomonas sp. Tai-03, Mychonastes sp. YL-02, and Chlorella sp. Cha-01) improves the elimination of the antibiotics 7-amino cephalosporin acid (7-ACA) (Waseem et al., 2017). C. pyrenoidosa has also been evaluated for the removal of ceftazidime (Yu et al., 2017). Mycoremediation, a type of bioremediation, involves using fungi to degrade contaminants. Over the past few years, very useful reviews of the role of fungi in biodegradation of pharmaceuticals and pesticides have been published (Naghdi et al., 2018).
9. Conclusion and future perspectives
Antibiotics have been present in the environment for a long time, but adverse effects have only been observed in the last few decades. Despite numerous publications on their acute and chronic effects on flora, fauna, including human being, their individual and combined health effects are unclear. A more comprehensive understanding of the environmental fate, impact and potential risks of of these antibiotics is required.
Therefore advance studies should be conducted to estimate the effectiveness of biodegradation of environmental antibiotices and the reducing the development of new antibiotic-resistant infections. In order to develop new prevention methods against the growing trend of antibiotic resistance, an in-depth understanding of the underlying mechanisms of resistance is crucial. Thus, efforts to develop antibiotics should be sustained, long-lasting and consistent. In order to implement more quickly, for more cost-efficient results, more affordable technology is needed. The potential risks of environmental exposure of pharmaceuticals are associated with eco-toxicological effects and impact on the ecosystem process. Given the threats to human health, it is necessary to remove environmental antibiotics
Credit Author Statement
Niharika Koch: Collection of data from published papers, review and assistance in the creation of Fig. 1 and the draft manuscript; Nazim F. Islam: Scientific advice and manuscript revision; Songita Sonowal: Data compilation and formatting support; Ram Prasad: Concept, advice on scientific research and proofreading; Hemen Sarma: The design concept, the content, original draft, as well as the visualization have been prepared and the manuscript revised.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgment
This study did not obtain any particular grant and was not funded by any sources.
Contributor Information
Ram Prasad, Email: ramprasad@mgcub.ac.in.
Hemen Sarma, Email: hemens02@yahoo.co.in.
References
- Acar J.F., Moulin G. Antimicrobial resistance at farm level Resistant bacterial clones on the farm. World. 2006;25:775–792. [PubMed] [Google Scholar]
- Ahmad S., Raza S., Uddin R., Azam S.S. AC SC. J. Mol. Graph. Model. 2018 doi: 10.1016/j.jmgm.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Al-Gheethi A.A.S., Lalung J., Noman E.A., Bala J.D., Norli I. Removal of heavy metals and antibiotics from treated sewage effluent by bacteria. Clean Technol. Environ. Policy. 2015;17:2101–2123. doi: 10.1007/s10098-015-0968-z. [DOI] [Google Scholar]
- Al-Gheethi A.A.S., Norli I., Lalung J., Azlan A.M., Farehah Z.A.N., Kadir M.O.A. Biosorption of heavy metals and cephalexin from secondary effluents by tolerant bacteria. Clean Technol. Environ. Policy. 2014;16:137–148. doi: 10.1007/s10098-013-0611-9. [DOI] [Google Scholar]
- AL-Mamun M., Chowdhury T., Biswas B., Absar N. Food poisoning and intoxication: a global leading concern for human health. Food Saf. Preserv. 2018:307–352. doi: 10.1016/b978-0-12-814956-0.00011-1. [DOI] [Google Scholar]
- Al-Riyami I.M., Ahmed M., Al-Busaidi A., Choudri B.S. Antibiotics in wastewaters: a review with focus on Oman. Appl. Water Sci. 2018;8:199. doi: 10.1007/s13201-018-0846-z. [DOI] [Google Scholar]
- Alvarez-ord A. 2020. Extended Spectrum β -Lactamase (ESBL) Producing Escherichia coli in Pigs and Pork Meat in the European Union. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R.I. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011 doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argudín M.A., Deplano A., Meghraoui A., Dodémont M., Heinrichs A., Denis O., Nonhoff C., Roisin S. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics. 2017 doi: 10.3390/antibiotics6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- aus der Beek T., Weber F.A., Bergmann A., Hickmann S., Ebert I., Hein A., Küster A. Pharmaceuticals in the environment-Global occurrences and perspectives. Environ. Toxicol. Chem. 2016;35:823–835. doi: 10.1002/etc.3339. [DOI] [PubMed] [Google Scholar]
- Azimi S., Nezamzadeh-ejhieh A. Elsevier B.V; 2015. Enhanced activity of clinoptilolite-supported hybridized PbS-CdS semiconductors for the photocatalytic degradation of a mixture of tetracycline and cephalexin. [DOI] [Google Scholar]
- Azzouz A., Palacios L., Souhail B., Ballesteros E. A multi-residue method for GC-MS determination of selected endocrine disrupting chemicals in fish and seafood from European and North African markets. Environ. Res. 2019;178 doi: 10.1016/j.envres.2019.108727. [DOI] [PubMed] [Google Scholar]
- Bager, F., Madsen, M., Christensen, J., Aarestrup, F.M., 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms 1, 95–112. [DOI] [PubMed]
- Barancheshme F., Munir M. 2018. Strategies to Combat Antibiotic Resistance in the Wastewater Treatment Plants 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrons M., Peñuelas J. Pharmaceuticals and personal-care products in plants. Trends Plant Sci. 2017;22:194–203. doi: 10.1016/j.tplants.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Bhullar K., Waglechner N., Pawlowski A., Koteva K., Banks E.D., Johnston M.D., Barton H.A., Wright G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012;7:1–11. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho J., Grosso F., Peixe L. Antibiotic resistance in Pseudomonas aeruginosa – Mechanisms, epidemiology and evolution. Drug Resist. Updates. 2019;44 doi: 10.1016/j.drup.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Bou G. Universal protocol for the rapid automated detection of carbapenem-resistant Gram-negative bacilli directly from blood cultures by matrix-assisted laser desorption /ionisation time-of- flight mass spectrometry (MALDI-TOF / MS) Servicio de Microbiolog. Int. J. Antimicrob. Agents. 2016 doi: 10.1016/j.ijantimicag.2016.08.024. [DOI] [PubMed] [Google Scholar]
- Boxall A.B.A. The environmental side effects of medication. EMBO Rep. 2004 doi: 10.1038/sj.embor.7400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce J.T., Hellyer P. Antibiotic resistance and antibiotic prescribing by dentists in England 2007–2016. Br. Dent. J. 2018;225:81–84. doi: 10.1038/sj.bdj.2018.525. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M.K. Use of antibiotics as feed additives: a burning question. Front. Microbiol. 2014;5:1–3. doi: 10.3389/fmicb.2014.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xie S. Science of the total environment overview of sulfonamide biodegradation and the relevant pathways and microorganisms. Sci. Total Environ. 2018;640–641:1465–1477. doi: 10.1016/j.scitotenv.2018.06.016. [DOI] [PubMed] [Google Scholar]
- Christou A., Karaolia P., Hapeshi E., Michael C., Fatta-Kassinos D. Long-term wastewater irrigation of vegetables in real agricultural systems: concentration of pharmaceuticals in soil, uptake and bioaccumulation in tomato fruits and human health risk assessment. Water Res. 2017;109:24–34. doi: 10.1016/j.watres.2016.11.033. [DOI] [PubMed] [Google Scholar]
- Costa V.M.D., King C.E., Kalan L., Morar M., Sung W.W.L., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R., Golding G.B., Poinar H.N., Wright G.D. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- Cromwell G.L. Why and how antibiotics are used in swine production. Animal Biotechnol. 2002;13:7–27. doi: 10.1081/ABIO-120005767. [DOI] [PubMed] [Google Scholar]
- Cruz-Loya M., Kang T.M., Lozano N.A., Watanabe R., Tekin E., Damoiseaux R., Savage V.M., Yeh P.J. Stressor interaction networks suggest antibiotic resistance co-opted from stress responses to temperature. ISME J. 2019;13:12–23. doi: 10.1038/s41396-018-0241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa P.M., Loureiro L., Matos A.J.F. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int. J. Environ. Res. Public Health. 2013;10:278–294. doi: 10.3390/ijerph10010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T., Duan J., Li X., Xu X., Shi H., Kang W. 2017. Determination of Sulfonamide Residues in Food by Capillary Zone Electrophoresis with On-Line Chemiluminescence Detection Based on an Ag (III) Complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. Microbiología; Madrid, Spain: 1996. Origins and Evolution of Antibiotic Resistance. [DOI] [PubMed] [Google Scholar]
- Dawan J., Ahn J. Assessment of cross-resistance potential to serial antibiotic treatments in antibiotic-resistant Salmonella Typhimurium. Microbial Pathog. 2020;148 doi: 10.1016/j.micpath.2020.104478. [DOI] [PubMed] [Google Scholar]
- Detector, F., 2018. Aqueous ultrasound-assisted extraction for the determination of fluoroquinolones in mangrove sediment by high-performance liquid chromatography and fluorescence detector 29, 24–32.
- Dimitriu T., Misevic D., Lindner A.B., Taddei F. Mobile genetic elements are involved in bacterial sociality. Mobile Genet. Elem. 2015;5:7–11. doi: 10.1080/2159256x.2015.1006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebele A.J., Abou-Elwafa Abdallah M., Harrad S. Pharmaceuticals and Personal Care Products (PPCPs) in the freshwater aquatic environment. Emerg. Contam. 2017 doi: 10.1016/j.emcon.2016.12.004. [DOI] [Google Scholar]
- Endicott J.A., Ling V. 1989. The Biochemistry of p-Glycoprotein-Mediated Multidrug Resistance! [DOI] [PubMed] [Google Scholar]
- Essa A.M.M., Julian D.J., Kidd S.P., Brown N.L., Hobman J.L. Mercury resistance determinants related to Tn21, Tn1696, and Tn5053 in enterobacteria from the preantibiotic era. Antimicrob. Agents Chemother. 2003;47:1115–1119. doi: 10.1128/AAC.47.3.1115-1119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzariai A., Hafidi M., Khadra A., Aemig Q., El Fels L., Barret M., Merlina G., Patureau D., Pinelli E. Human and veterinary antibiotics during composting of sludge or manure: global perspectives on persistence, degradation, and resistance genes. J. Hazard. Mater. 2018;359:465–481. doi: 10.1016/j.jhazmat.2018.07.092. [DOI] [PubMed] [Google Scholar]
- Fiegna F., Velicer G.J. 2005. Exploitative and Hierarchical Antagonism in a Cooperative Bacterium 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux C.A., Costerton J.W., Stewart P.S., Stoodley P. 2005. Survival Strategies of Infectious Biofilms 13. [DOI] [PubMed] [Google Scholar]
- Galaon T., Banciu A., Chiriac F.L., Nita-Lazar M. Biodegradation of antibiotics: the balance between good and bad. Roman. J. Ecol. Environ. Chem. 2019;1:16–25. doi: 10.21698/rjeec.2019.103. [DOI] [Google Scholar]
- Gallo G.F., Berg J.L. Efficacy of a feed-additive antibacterial combination for improving feedlot cattle performance and health. Can. Vet. J. 1995;36:223–229. [PMC free article] [PubMed] [Google Scholar]
- Gambarotto, K., Ploy, M., Dupron, F., Giangiobbe, M., 2001. Occurrence of vancomycin-resistant enterococci in pork and poultry products from a cattle-rearing area of France 39, 2354–2355. https://doi.org/10.1128/JCM.39.6.2354 [DOI] [PMC free article] [PubMed]
- Gartiser, S., Urich, E., Alexy, R., Ku, K., 2007. Ultimate biodegradation and elimination of antibiotics in inherent tests 67, 604–613. https://doi.org/10.1016/j.chemosphere.2006.08.038 [DOI] [PubMed]
- Gatignol A., Durand H., Tiraby G. Bleomycin resistance conferred by a drug-binding protein. FEBS Lett. 1988;230:171–175. doi: 10.1016/0014-5793(88)80665-3. [DOI] [PubMed] [Google Scholar]
- Gaw S., Thomas K.V., Hutchinson T.H. Sources, impacts and trends of pharmaceuticals in the marine and coastal environment. Philos. Trans. R. Soc. B: Biol. Sci. 2014 doi: 10.1098/rstb.2013.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Alonso S., Merino L.M., Esteban S., López de Alda M., Barceló D., Durán J.J., López-Martínez J., Aceña J., Pérez S., Mastroianni N., Silva A., Catalá M., Valcárcel Y. Occurrence of pharmaceutical, recreational and psychotropic drug residues in surface water on the northern Antarctic Peninsula region. Environ. Pollut. 2017;229:241–254. doi: 10.1016/j.envpol.2017.05.060. [DOI] [PubMed] [Google Scholar]
- Goodale A., Michailidis F., Watts R., Chok S.C., Hayes F. Characterization of permissive and non-permissive peptide insertion sites in chloramphenicol acetyltransferase. Microb. Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104395. [DOI] [PubMed] [Google Scholar]
- Granados-Chinchilla F., Rodríguez C. Tetracyclines in food and feedingstuffs: from regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017 doi: 10.1155/2017/1315497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Chen J. Application of alga-activated sludge combined system (AASCS) as a novel treatment to remove cephalosporins. Chem. Eng. J. 2015;260:550–556. doi: 10.1016/j.cej.2014.09.053. [DOI] [Google Scholar]
- Hernando-amado S., Coque T.M., Baquero F., Martínez J.L. One health and global health perspectives. Nat. Microbiol. 2019;4 doi: 10.1038/s41564-019-0503-9. [DOI] [PubMed] [Google Scholar]
- Hinchliffe S., Butcher A., Rahman M.M. The AMR problem: demanding economies, biological margins, and co-producing alternative strategies. Palgrave Commun. 2018;4 doi: 10.1057/s41599-018-0195-4. [DOI] [Google Scholar]
- Hu Y., Cheng H., Tao S. Environmental and human health challenges of industrial livestock and poultry farming in China and their mitigation. Environ. Int. 2017;107:111–130. doi: 10.1016/j.envint.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Ingerslev, F., Halling-s, B., 2001. Biodegradability of metronidazole, olaquindox, and tylosin and formation of tylosin degradation products in aerobic soil i manure slurries 320, 311–320. https://doi.org/10.1006/eesa.2000.2026 [DOI] [PubMed]
- Islas-espinoza, M., Reid, B.J., Wexler, M., Bond, P.L., 2012. Soil bacterial consortia and previous exposure enhance the biodegradation of sulfonamides from pig manure 140–151. https://doi.org/10.1007/s00248-012-0010-5 [DOI] [PubMed]
- Kaur N. Prevalence and antibiotic susceptibility pattern of methicillin resistant staphylococcus aureus in tertiary care hospitals. Br. Biotechnol. J. 2014;4:228–235. doi: 10.9734/BBJ/2014/4245. [DOI] [Google Scholar]
- Kaur N., Prasad R., Varma A. Antibiotic resistance among clinical isolates of Staphylococcus aureus and usefulness of antibiogram. Int. J. Pharma Bio Sci. 2013;4:B957–B964. [Google Scholar]
- Kinney C.A., Heuvel B.Vanden. Current Opinion in Environmental Science and Health; 2020. Translocation of Pharmaceuticals and Personal Care Products after Land Application of Biosolids. [DOI] [Google Scholar]
- Kivits T., Broers H.P., Beeltje H., van Vliet M., Griffioen J. Presence and fate of veterinary antibiotics in age-dated groundwater in areas with intensive livestock farming. Environ. Pollut. 2018;241:988–998. doi: 10.1016/j.envpol.2018.05.085. [DOI] [PubMed] [Google Scholar]
- Kong Y., Li M., Li R., Shan X., Wang G. Evaluation of cholesterol lowering property and antibacterial activity of two potential lactic acid bacteria isolated from the intestine of snakehead fish (Channa argus) Aquac. Rep. 2020;17 doi: 10.1016/j.aqrep.2020.100342. [DOI] [Google Scholar]
- Kumar, A., Patyal, A., Panda, A.K., 2018. Sub-therapeutic use of antibiotics in animal feed and their potential impact on environmental and human health : a comprehensive review Sub-therapeutic use of antibiotics in animal feed and their potential impact on environmental and human health : a comp. https://doi.org/10.21088/jafst.2321.1628.6118.3
- Kumar M., Jaiswal S., Sodhi K.K., Shree P., Singh D.K., Agrawal P.K., Shukla P. Antibiotics bioremediation: perspectives on its ecotoxicity and resistance. Environ. Int. 2019;124:448–461. doi: 10.1016/j.envint.2018.12.065. [DOI] [PubMed] [Google Scholar]
- Kümmerer K. Chemosphere antibiotics in the aquatic environment – a review – part I. Chemosphere. 2009;75:417–434. doi: 10.1016/j.chemosphere.2008.11.086. [DOI] [PubMed] [Google Scholar]
- Lai H., Lin J. Chemosphere Degradation of oxolinic acid and flumequine in aquaculture pond waters and sediments. Chemosphere. 2009;75:462–468. doi: 10.1016/j.chemosphere.2008.12.060. [DOI] [PubMed] [Google Scholar]
- Lan L., Yao Y., Ping J., Ying Y. Biosensors and Bioelectronics recent advances in nanomaterial-based biosensors for antibiotics detection. Biosens. Bioelectron. 2017;91:504–514. doi: 10.1016/j.bios.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Lehotay, S.J., Lightfield, A.R., 2018. Simultaneous analysis of aminoglycosides with many other classes of drug residues in bovine tissues by ultrahigh-performance liquid chromatography – tandem mass spectrometry using an ion-pairing reagent added to final extracts 1095–1109. [DOI] [PubMed]
- Li L., Guo C., Fan S., Lv J., Zhang Y., Xu Y., Xu J. 2018. Dynamic Transport of Antibiotics and Antibiotic Resistance Genes Under Different Treatment Processes in a Typical Pharmaceutical Wastewater Treatment Plant. [DOI] [PubMed] [Google Scholar]
- Li, M., An, B., Villaruz, A.E., Braughton, K.R., Jiang, X., Deleo, F.R., Chambers, H.F., Lu, Y., Otto, M., 2009. Staphylococcus aureus 106, 5883–5888. [DOI] [PMC free article] [PubMed]
- Li W., Sharma M., Kaur P. The DrrAB efflux system of streptomyces peucetius is a multidrug transporter of broad substrate specificity. J. Biol. Chem. 2014;289:12633–12646. doi: 10.1074/jbc.M113.536136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, X., Li, B., Zou, R., Dai, Y., Xie, S., Yuan, B., 2016. Biodegradation of antibiotic ciprofloxacin : pathways, influential factors, and bacterial community structure. https://doi.org/10.1007/s11356-016-6054-1 [DOI] [PubMed]
- Liao, X., Li, B., Zou, R., Xie, S., Yuan, B., 2015. Antibiotic sulfanilamide biodegradation by acclimated microbial populations. https://doi.org/10.1007/s00253-015-7133-9 [DOI] [PubMed]
- Luthra S., Rominski A., Sander P. The role of antibiotic-target-modifying and antibiotic-modifying enzymes in mycobacterium abscessusdrug resistance. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.02179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Xu S., Tang Z., Li Z., Zhang L. Biosafety and Health; 2020. Use of Antimicrobials in Food Animals and Impact of Transmission of Antimicrobial Resistance on Humans. [DOI] [Google Scholar]
- Mak S., Xu Y., Nodwell J.R. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 2014 doi: 10.1111/mmi.12689. [DOI] [PubMed] [Google Scholar]
- Maki T., Hasegawa H., Kitami H., Fumoto K. 2006. Bacterial Degradation of Antibiotic Residues in Marine Fish Farm Sediments of Uranouchi Bay and Phylogenetic Analysis of Antibiotic-Degrading Bacteria using 16S rDNA Sequences. [Google Scholar]
- Malhotra B.D. Author ’ s accepted manuscript. Biosens. Bioelectron. 2016 doi: 10.1016/j.bios.2016.03.004. [DOI] [Google Scholar]
- Manson, J.M., Keis, S., Smith, J.M.B., Cook, G.M., 2003. A clonal lineage of VanA-type enterococcus faecalis predominates in vancomycin-resistant enterococci isolated in New Zealand 47, 204–210. https://doi.org/10.1128/AAC.47.1.204 [DOI] [PMC free article] [PubMed]
- Martínez, L., Baquero, F., 2002. Interactions among strategies associated with bacterial infection : pathogenicity, epidemicity, and antibiotic resistance † 15, 647–679. https://doi.org/10.1128/CMR.15.4.647 [DOI] [PMC free article] [PubMed]
- Massé, D.I., Saady, N.M.C., Gilbert, Y., 2014. Potential of biological processes to eliminate antibiotics in livestock manure: an overview 146–163. https://doi.org/10.3390/ani4020146 [DOI] [PMC free article] [PubMed]
- Maurice N.M., Bedi B., Sadikot R.T. Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respiratory Cell Mol. Biol. 2018;58:428–439. doi: 10.1165/rcmb.2017-0321TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus P., Stockwell V. Antibiotics for plant diseases control: silver bullets or rusty sabers. APSnet Feature Articles. 2000 doi: 10.1094/apsnetfeature-2000-0600. [DOI] [Google Scholar]
- McManus P.S., Stockwell V.O., Sundin G.W., Jones A.L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002 doi: 10.1146/annurev.phyto.40.120301.093927. [DOI] [PubMed] [Google Scholar]
- Methé B.A., Nelson K.E., Pop M., Creasy H.H., Giglio M.G., Huttenhower C., Gevers D., Petrosino J.F., Abubucker S., Badger J.H., Chinwalla A.T., Earl A.M., Fitzgerald M.G., Fulton R.S., Hallsworth-Pepin K., Lobos E.A., Madupu R., Magrini V., Martin J.C., Mitreva M., Muzny D.M., Sodergren E.J., Versalovic J., Wollam A.M., Worley K.C., Wortman J.R., Young S.K., Zeng Q., Aagaard K.M., Abolude O.O., Allen-Vercoe E., Alm E.J., Alvarado L., Andersen G.L., Anderson S., Appelbaum E., Arachchi H.M., Armitage G., Arze C.A., Ayvaz T., Baker C.C., Begg L., Belachew T., Bhonagiri V., Bihan M., Blaser M.J., Bloom T., Bonazzi V.R., Brooks P., Buck G.A., Buhay C.J., Busam D.A., Campbell J.L., Canon S.R., Cantarel B.L., Chain P.S., Chen I.M.A., Chen L., Chhibba S., Chu K., Ciulla D.M., Clemente J.C., Clifton S.W., Conlan S., Crabtree J., Cutting M.A., Davidovics N.J., Davis C.C., Desantis T.Z., Deal C., Delehaunty K.D., Dewhirst F.E., Deych E., Ding Y., Dooling D.J., Dugan S.P., Michael Dunne W., Scott Durkin A., Edgar R.C., Erlich R.L., Farmer C.N., Farrell R.M., Faust K., Feldgarden M., Felix V.M., Fisher S., Fodor A.A., Forney L., Foster L., Di Francesco V., Friedman J., Friedrich D.C., Fronick C.C., Fulton L.L., Gao H., Garcia N., Giannoukos G., Giblin C., Giovanni M.Y., Goldberg J.M., Goll J., Gonzalez A., Griggs A., Gujja S., Haas B.J., Hamilton H.A., Harris E.L., Hepburn T.A., Herter B., Hoffmann D.E., Holder M.E., Howarth C., Huang K.H., Huse S.M., Izard J., Jansson J.K., Jiang H., Jordan C., Joshi V., Katancik J.A., Keitel W.A., Kelley S.T., Kells C., Kinder-Haake S., King N.B., Knight R., Knights D., Kong H.H., Koren O., Koren S., Kota K.C., Kovar C.L., Kyrpides N.C., La Rosa P.S., Lee S.L., Lemon K.P., Lennon N., Lewis C.M., Lewis L., Ley R.E., Li K., Liolios K., Liu B., Liu Y., Lo C.C., Lozupone C.A., Dwayne Lunsford R., Madden T., Mahurkar A.A., Mannon P.J., Mardis E.R., Markowitz V.M., Mavrommatis K., McCorrison J.M., McDonald D., McEwen J., McGuire A.L., McInnes P., Mehta T., Mihindukulasuriya K.A., Miller J.R., Minx P.J., Newsham I., Nusbaum C., O'Laughlin M., Orvis J., Pagani I., Palaniappan K., Patel S.M., Pearson M., Peterson J., Podar M., Pohl C., Pollard K.S., Priest M.E., Proctor L.M., Qin X., Raes J., Ravel J., Reid J.G., Rho M., Rhodes R., Riehle K.P., Rivera M.C., Rodriguez-Mueller B., Rogers Y.H., Ross M.C., Russ C., Sanka R.K., Sankar P., Fah Sathirapongsasuti J., Schloss J.A., Schloss P.D., Schmidt T.M., Scholz M., Schriml L., Schubert A.M., Segata N., Segre J.A., Shannon W.D., Sharp R.R., Sharpton T.J., Shenoy N., Sheth N.U., Simone G.A., Singh I., Smillie C.S., Sobel J.D., Sommer D.D., Spicer P., Sutton G.G., Sykes S.M., Tabbaa D.G., Thiagarajan M., Tomlinson C.M., Torralba M., Treangen T.J., Truty R.M., Vishnivetskaya T.A., Walker J., Wang L., Wang Z., Ward D.V., Warren W., Watson M.A., Wellington C., Wetterstrand K.A., White J.R., Wilczek-Boney K., Qing Wu Y., Wylie K.M., Wylie T., Yandava C., Ye L., Ye Y., Yooseph S., Youmans B.P., Zhang L., Zhou Y., Zhu Y., Zoloth L., Zucker J.D., Birren B.W., Gibbs R.A., Highlander S.K., Weinstock G.M., Wilson R.K., White O. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingmongkolchai S., Panbangred W. 2018. Bacillus Probiotics : an Alternative to Antibiotics for Livestock Production. [DOI] [PubMed] [Google Scholar]
- Morikawa M. Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J. Biosci. Bioeng. 2006;101:1–8. doi: 10.1263/jbb.101.1. [DOI] [PubMed] [Google Scholar]
- Munoz-Lopez M., Garcia-Perez J. DNA transposons: nature and applications in genomics. Curr. Genom. 2010;11:115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munteanu F.D., Titoiu A.M., Marty J.L., Vasilescu A. Detection of antibiotics and evaluation of antibacterial activity with screen-printed electrodes. Sensors (Switzerland) 2018;18 doi: 10.3390/s18030901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray R., Tien Y.C., Scott A., Topp E. The impact of municipal sewage sludge stabilization processes on the abundance, field persistence, and transmission of antibiotic resistant bacteria and antibiotic resistance genes to vegetables at harvest. Sci. Total Environ. 2019;651:1680–1687. doi: 10.1016/j.scitotenv.2018.10.030. [DOI] [PubMed] [Google Scholar]
- Muthamilselvan T., Kuo T.F., Wu Y.C., Yang W.C. Evidence-based Complementary and Alternative Medicine; 2016. Herbal Remedies for Coccidiosis Control: A Review of Plants, Compounds, and Anticoccidial Actions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghdi M., Taheran M., Kaur S., Kermanshahi-pour A. Removal of pharmaceutical compounds in water and wastewater using fungal oxidoreductase enzymes *. Environ. Pollut. 2018;234:190–213. doi: 10.1016/j.envpol.2017.11.060. [DOI] [PubMed] [Google Scholar]
- Nelson R. Controversial antibiotic use for crops approved in Florida. Lancet Infect. Dis. 2019;19:697–698. doi: 10.1016/S1473-3099(19)30318-4. [DOI] [PubMed] [Google Scholar]
- Ogawara H. Self-resistance in streptomyces, with special reference to β-lactam antibiotics. Molecules. 2016 doi: 10.3390/molecules21050605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawara H. Distribution of PASTA domains in penicillin-binding proteins and serine/threonine kinases of actinobacteria. J. Antibiot. 2016 doi: 10.1038/ja.2015.138. [DOI] [PubMed] [Google Scholar]
- Oliver S.P., Murinda S.E., Jayarao B.M. Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: a comprehensive review. Foodborne Pathog. Dis. 2011 doi: 10.1089/fpd.2010.0730. [DOI] [PubMed] [Google Scholar]
- Pamreddy A., Hidalgo M., Havel J., Salvadó V. Determination of antibiotics (tetracyclines and sulfonamides) in biosolids by pressurized liquid extraction and liquid chromatography–tandem mass spectrometry. J. Chromatogr. A. 2013 doi: 10.1016/j.chroma.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018 doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E., Kaur P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Frontiers in Microbiology. 2018;9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothakos V., Devlieghere F., Villani F., Björkroth J., Ercolini D. Lactic acid bacteria and their controversial role in fresh meat spoilage. Meat Sci. 2015;109:66–74. doi: 10.1016/j.meatsci.2015.04.014. [DOI] [PubMed] [Google Scholar]
- Prasad R., Bhattacharyya A., Nguyen Q.D. Nanotechnology in sustainable agriculture: recent developments, challenges, and perspectives. Front. Microbiol. 2017 doi: 10.3389/fmicb.2017.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestinaci F., Pezzotti P., Pantosti A. 1–10. Pathogens and Global Health; 2015. (Antimicrobial Resistance : A Global Multifaceted Phenomenon). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prija F., Prasad R. DrrC protein of Streptomyces peucetius removes daunorubicin from intercalated dnrI promoter. Microbiol. Res. 2017;202:30–35. doi: 10.1016/j.micres.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Qi, L., Li, H., Zhang, C., Liang, B., Li, J., Wang, L., Du, X., Liu, X., 2016. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in acinetobacter baumannii bacterial strains and growth conditions 7, 1–10. https://doi.org/10.3389/fmicb.2016.00483 [DOI] [PMC free article] [PubMed]
- Rajivgandhi G., Maruthupandy M., Muneeswaran T., Anand M., Manoharan N. SC. Process Biochem. 2018;1 doi: 10.1016/j.procbio.2018.01.015. [DOI] [Google Scholar]
- Rasmus, H., Hansen, M., Kjølholt, J., Stuer-lauridsen, F., Ternes, T., Halling-sørensen, B., 2005. Assessment of the importance of sorption for steroid estrogens removal during activated sludge treatment 61, 139–146. https://doi.org/10.1016/j.chemosphere.2005.02.088 [DOI] [PubMed]
- Riaz L., Mahmood T., Khalid A., Rashid A., Ahmed Siddique M.B., Kamal A., Coyne M.S. Fluoroquinolones (FQs) in the environment: a review on their abundance, sorption and toxicity in soil. Chemosphere. 2018;191:704–720. doi: 10.1016/j.chemosphere.2017.10.092. [DOI] [PubMed] [Google Scholar]
- Rudolf J.D., Bigelow L., Chang C., Cuff M.E., Lohman J.R., Chang C.Y., Ma M., Yang D., Clancy S., Babnigg G., Joachimiak A., Phillips G.N., Shen B. Crystal structure of the zorbamycin-binding protein ZbmA, the primary self-resistance element in streptomyces flavoviridis ATCC21892. Biochemistry. 2015;54:6842–6851. doi: 10.1021/acs.biochem.5b01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgame, P., Yap, G.S., Gause, W.C., 2013. review Effect of helminth-induced immunity on infections with microbial pathogens 14. https://doi.org/10.1038/ni.2736 [DOI] [PMC free article] [PubMed]
- Salman M.S., Fadl M.A., Elkhateeb A., El-awady M.A. Kingdom of Saudi Arabia, and their Anticancer and Antimicrobial Activity; 2018. Chemical Composition of Hydrodistillation and Solvent Free Microwave Extraction of Essential Oils from Mentha piperita L . Growing in Taif. [Google Scholar]
- Santos, J.L., Aparicio, I., Callejón, M., Alonso, E., 2009. Occurrence of pharmaceutically active compounds during 1-year period in wastewaters from four wastewater treatment plants in Seville (Spain) 164, 1509–1516. https://doi.org/10.1016/j.jhazmat.2008.09.073 [DOI] [PubMed]
- Schmutz E., Mühlenweg A., Li S.M., Heide L. Resistance genes of aminocoumarin producers: two type II topoisomerase genes confer resistance against coumermycin A1 and clorobiocin. Antimicrob. Agents Chemother. 2003;47:869–877. doi: 10.1128/AAC.47.3.869-877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger K., Helmke K., Hölzel C.S., Bauer J. Antibiotic resistance in bacteria isolated from vegetables with regards to the marketing stage (farm vs. supermarket) Int. J. Food Microbiol. 2011;148:191–196. doi: 10.1016/j.ijfoodmicro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Sengeløv, G., Agersø, Y., Halling-sørensen, B., Baloda, S.B., Andersen, J.S., Jensen, L.B., 2003. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry 28, 587–595. [DOI] [PubMed]
- Shinkawa H., Sugiyama M., Nimi O., Nomi R. Molecular cloning and expression in Streptomyces lividans of a streptomycin 6-phosphotransferase gene from a streptomycin-producing microorganism. FEBS Lett. 1985;181:385–389. doi: 10.1016/0014-5793(85)80298-2. [DOI] [PubMed] [Google Scholar]
- Sirés I., Brillas E. Remediation of water pollution caused by pharmaceutical residues based on electrochemical separation and degradation technologies : a review. Environ. Int. 2012;40:212–229. doi: 10.1016/j.envint.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Thompson C.J., Kumagai T., Suzuki K., Deblaere R., Villarroel R., et al. Characterisation by molecular cloning of two genes from Streptomyces verticillus encoding resistance to bleomycin. Gene. 1994;151:11–16. doi: 10.1016/0378-1119(94)90626-2. [DOI] [PubMed] [Google Scholar]
- Tacconelli, E., Sifakis, F., Harbarth, S., Schrijver, R., Mourik, M. Van, Voss, A., Sharland, M., 2017. Personal view surveillance for control of antimicrobial resistance 3099, 1–8. https://doi.org/10.1016/S1473-3099(17)30485-1
- Tasho R.P., Cho J.Y. Science of the total environment veterinary antibiotics in animal waste, its distribution in soil and uptake by plants : a review. Sci. Total Environ. 2016;563–564:366–376. doi: 10.1016/j.scitotenv.2016.04.140. [DOI] [PubMed] [Google Scholar]
- Thanner S., Drissner D., Walsh F. Antimicrobial resistance in agriculture. mBio. 2016;7:1–7. doi: 10.1128/mBio.02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele-bruhn S. 2003. Pharmaceutical Antibiotic Compounds in Soils ± A Review. [Google Scholar]
- Tillotson G.S., Zinner S.H. Expert review of anti-infective therapy burden of antimicrobial resistance in an era of decreasing susceptibility. Exp. Rev. Anti-Infect. Therapy. 2017;15:663–676. doi: 10.1080/14787210.2017.1337508. [DOI] [PubMed] [Google Scholar]
- Tiwari B., Sellamuthu B., Ouarda Y., Drogui P., Tyagi R.D., Buelna G. Review on fate and mechanism of removal of pharmaceutical pollutants from wastewater using biological approach. Bioresour. Technol. 2016 doi: 10.1016/j.biortech.2016.11.042. [DOI] [PubMed] [Google Scholar]
- Trinh P., Zaneveld J.R., Safranek S., Rabinowitz P.M. One health relationships between human. Animal, Environ. Microbiomes : Mini-Rev. 2018;6:1–9. doi: 10.3389/fpubh.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeigwe T.K., Teboh J.M., Eze P.N., Stietiya M.H., Kumar V., Hendrix J., Mascagni H.J., Ying T., Kandakji T. Implications of leading crop production practices on environmental quality and human health. J. Environ. Manag. 2015;151:267–279. doi: 10.1016/j.jenvman.2014.11.024. [DOI] [PubMed] [Google Scholar]
- Van Hoek A.H.A.M., Mevius D., Guerra B., Mullany P., Roberts A.P., Aarts H.J.M. Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2011 doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van T.T.H., Yidana Z., Smooker P.M., Coloe P.J. Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J. Global Antimicrob. Resist. 2020;20:170–177. doi: 10.1016/j.jgar.2019.07.031. [DOI] [PubMed] [Google Scholar]
- Vass M., Hruska K., Franek M. Veterinarni Medicina; 2008. Nitrofuran Antibiotics: A Review on the Application, Prohibition and Residual Analysis. [Google Scholar]
- Veen, H.W. Van, Callaghan, R., 1998. letters to nature 291–295.
- Wang X., Wang M., Zhang K., Hou T., Zhang L., Fei C. Determination of virginiamycin M1 residue in tissues of swine and chicken by ultra-performance liquid chromatography tandem mass spectrometry. Food Chem. 2018;250:127–133. doi: 10.1016/j.foodchem.2018.01.024. [DOI] [PubMed] [Google Scholar]
- Waseem H., Williams M.R., Stedtfeld R.D., Hashsham S.A. Antimicrobial resistance in the environment. Water Environ. Res. 2017;89:921–941. doi: 10.2175/106143017X15023776270179. [DOI] [PubMed] [Google Scholar]
- Wu D., Huang Z., Yang K., Graham D., Xie B. Relationships between antibiotics and antibiotic resistance gene levels in municipal solid waste leachates in Shanghai, China. Environ. Sci. Technol. 2015;49:4122–4128. doi: 10.1021/es506081z. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Nakamura Yudai, Moriguchi S., Nakamura Yuki, Honda Y., Tamura I., Hirata Y., Hayashi A., Sekizawa J. Persistence and partitioning of eight selected pharmaceuticals in the aquatic environment : laboratory photolysis, biodegradation, and sorption experiments. Water Res. 2009;43:351–362. doi: 10.1016/j.watres.2008.10.039. [DOI] [PubMed] [Google Scholar]
- Yan Weifu, Xiao Y., Yan Weida, Ding R., Wang S., Zhao F. The effect of bioelectrochemical systems on antibiotics removal and antibiotic resistance genes : a review. Chem. Eng. J. 2018 doi: 10.1016/j.cej.2018.10.128. [DOI] [Google Scholar]
- Yang Z., Peng H., Lu X., Liu Q., Huang R., Hu B., Kachanoski G., Zuidhof M.J., Le X.C. Vol. 50. Environmental Science and Technology; 2016. Arsenic metabolites, including N-acetyl-4-hydroxy-m-arsanilic acid; pp. 6737–6743. (Chicken Litter from a Roxarsone-Feeding Study Involving 1600 Chickens). [DOI] [PubMed] [Google Scholar]
- Yu L., Yan X., Wang L., Chu J., Zhuang Y., Zhang S., Guo M. Molecular cloning and functional characterization of an ATP-binding cassette transporter OtrC from Streptomyces rimosus. BMC Biotechnol. 2012;12:1–12. doi: 10.1186/1472-6750-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhou Y., Wang Z., Torres O.L., Guo R., Chen J. Investigation of the removal mechanism of antibiotic ceftazidime by green algae and subsequent microbic impact assessment. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-04128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Yang Y., Huang C., Zhao L., Sun P. Kinetics and modeling of sulfonamide antibiotic degradation in wastewater and human urine by UV/H2 O2 and UV/PDS. Water Res. 2016;103:283–292. doi: 10.1016/j.watres.2016.07.037. [DOI] [PubMed] [Google Scholar]
- Zhou C.shuang, Wu J.wen, Dong L.li, Liu B.feng, Xing D.feng, Yang S.shan, Wu X.kun, Wang Q., Fan J.ning, Feng L.ping, Cao G.li. Removal of antibiotic resistant bacteria and antibiotic resistance genes in wastewater effluent by UV-activated persulfate. J. Hazard. Mater. 2020;388 doi: 10.1016/j.jhazmat.2020.122070. [DOI] [PubMed] [Google Scholar]