Abstract
Background
The implementation of novel techniques as a complement to traditional disease surveillance systems represents an additional opportunity for rapid analysis.
Objective
The objective of this work is to describe a web-based participatory surveillance strategy among health care workers (HCWs) in two Swiss hospitals during the first wave of COVID-19.
Methods
A prospective cohort of HCWs was recruited in March 2020 at the Cantonal Hospital of St. Gallen and the Eastern Switzerland Children’s Hospital. For data analysis, we used a combination of the following techniques: locally estimated scatterplot smoothing (LOESS) regression, Spearman correlation, anomaly detection, and random forest.
Results
From March 23 to August 23, 2020, a total of 127,684 SMS text messages were sent, generating 90,414 valid reports among 1004 participants, achieving a weekly average of 4.5 (SD 1.9) reports per user. The symptom showing the strongest correlation with a positive polymerase chain reaction test result was loss of taste. Symptoms like red eyes or a runny nose were negatively associated with a positive test. The area under the receiver operating characteristic curve showed favorable performance of the classification tree, with an accuracy of 88% for the training data and 89% for the test data. Nevertheless, while the prediction matrix showed good specificity (80.0%), sensitivity was low (10.6%).
Conclusions
Loss of taste was the symptom that was most aligned with COVID-19 activity at the population level. At the individual level—using machine learning–based random forest classification—reporting loss of taste and limb/muscle pain as well as the absence of runny nose and red eyes were the best predictors of COVID-19.
Keywords: digital epidemiology, SARS-CoV-2, COVID-19, health care workers
Introduction
The COVID-19 pandemic is one of the greatest health challenges that societies around the globe have ever experienced. A range of instruments and ways to measure factors related to COVID-19 and the pandemic have been described [1-7]. COVID-19 presents a challenge for public health in general, while health care workers (HCWs) are at particular risk of acquiring COVID-19 [8]. Several studies using online forms have found they can be useful for tracking disease activity in different locations, including workplaces [9,10]. However, these technological platforms require timely, persistent, and ongoing engagement to generate valid and representative surveillance data [1]. In the context of collaboration and the collection of collective health information, digital epidemiology and participatory surveillance techniques have been demonstrated to be tools with great potential for helping to detect health threats [11-16]. Many strategies that involve daily reporting of symptoms through the voluntary participation of individuals have reported successful results [17,18]. Participatory surveillance by patients has been shown to have a complementary role in detecting syndromic clusters for several epidemiological challenges, such as COVID-19, seasonal influenza, or high-risk mass gatherings [17-22]. The implementation of novel techniques represents an additional opportunity for the rapid analysis of big data based on machine learning, thereby acting as a complement to traditional disease surveillance systems.
The objective of this work is to describe a web-based participatory surveillance strategy among HCWs in two Swiss hospitals during the first wave of the COVID-19 pandemic.
Methods
Study Design
A prospective cohort of HCWs was recruited in March 2020 at the Cantonal Hospital of St. Gallen and the Eastern Switzerland Children’s Hospital, Switzerland. Individuals aged 16 years and older were eligible. HCWs were enrolled in the study after accepting the electronic informed consent form. The anonymization of participants was carried out by using a management ID system with three levels; we anonymized the participants (user ID), surveys (survey ID), and their samples (order ID). No compensation was provided and participation was voluntary. A copy of the informed consent with all details about privacy and confidentiality is provided in Multimedia Appendix 1. The study was approved by the local ethics committee (Ethikkommission Ostschweiz; #2020-00502). All participants received a link via email to fill in a baseline questionnaire collecting data on pre-existing conditions at the start of the study. To improve the data quality and reduce reporting bias, mobile number validation was required; participants could only move forward if they input a token sent to their mobile phone. After completing the baseline form, participants became eligible to receive the daily SMS text message with an individualized link redirecting them to a secure web platform where they could fill in their symptom diary. To encourage participant engagement through the entire period, an SMS text message reminder was sent to those that did not fill in the symptom diary the day before. In the symptom diary, participants were asked about the type and severity of COVID-19 symptoms according to Table 1. Those that met SARS-CoV-2 testing criteria (ie, fever/feverishness, cough, shortness of breath, sore throat, or anosmia/ageusia) according to the Swiss Federal Office of Public Health (FOPH) were asked to schedule an appointment for a nasopharyngeal swab [23].
Table 1.
List of symptoms and consequences.
| Survey question topic | Type |
| Sore throat | Symptom |
| Cough | Symptom |
| Shortness of breath | Symptom |
| Runny nose | Symptom |
| Headache | Symptom |
| Diarrhea | Symptom |
| Anorexia/nausea | Symptom |
| Fever | Symptom |
| Chills | Symptom |
| Limb/muscle pain | Symptom |
| Loss of taste | Symptom |
| Itchy red eyes | Symptom |
| Feeling weak | Symptom |
| Fever-related muscle pain | Symptom |
| Took medicines | Consequence |
| Sought health care | Consequence |
| Missed work | Consequence |
| Hospitalized | Consequence |
For validation purposes, the positivity rate of the online survey was compared to the positivity rate of HCWs undergoing SARS-CoV-2 polymerase chain reaction (PCR) testing at the study institutions (independent of study participation). We tested both isolated symptoms and various combinations, including the FOPH testing criteria.
Data Analysis
For the analysis of time trends of symptoms, we used a locally weighted running line smoother (locally estimated scatterplot smoothing [LOESS]) [24], which is a nonparametric smoother with Gaussian noise added in the sine wave. This algorithm estimates the latent function in a pointwise fashion. This method is a supervised machine learning approach and was carried out to generate a moving average for scatterplot smoothing among the data points. Its function can be expressed as the following:
| ω (χ)=(1–|d|3)3 |
where d is the distance of the data point from the point on the fitter curve, scaled to lie in the range from 0-1. We then used a moving average with 7 days as the window size, aligned on the right.
The Spearman rank correlation coefficient was used to verify the statistical dependence between symptoms and test positivity, using a monotonic function described by the following formula [25]:
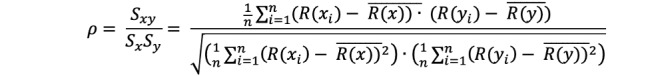 |
It is critical to identify significant temporal deviations throughout the period including the impact of seasonality in such high frequency data inputs. Therefore, we applied the Seasonal-Hybrid Extreme Studentized Deviate (S-H-ESD) algorithm [26], which uses a modified Seasonal-Trend decomposition procedure based on LOESS [27]. This technique allows for the identification of change points over time, recognizing when the signal frequency (FOPH classification) was positive (increasing) or negative (decreasing). The missing value was handled by spline interpolation, the maximal anomaly ratio was 0.1, and a piecewise median time window of 2 weeks was chosen.
Finally, to classify participants according to the probability of having symptoms compatible with COVID-19, we used the random forest algorithm. This is an ensemble learning method based on decision trees, which increases the accuracy of classification for both training and test data [28]. Specifically, this algorithm is a predictor consisting of an assembly of randomized base regression trees {rn(x,Θm,Dn),m ≥ 1}, where Θ1,Θ2,... are independent and identically distributed (IID) outputs of a randomizing variable Θ. These random trees are pooled to form the following aggregated regression estimate [29]:
where 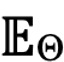 denotes expectation with respect to the random parameter, conditionally on X and the data set Dn.
denotes expectation with respect to the random parameter, conditionally on X and the data set Dn.
To explain how the random forest technique was used in this study, a summary of its parameters along with a prediction matrix for the model was generated. In addition, a receiver operating characteristic (ROC) curve was created to evaluate the binary classification of the model. We split the data, using 70% of entries for model training and 30% of entries for the test set. To determine which variables were more or less important for predicting the outcome, we used a boxplot chart.
Algorithms and techniques were programmed and deployed in R language, using the Exploratory [30] framework. The data collection system was developed using JotForm [31] as well as a proprietary solution and was hosted at Amazon Web Services, using EC2 and S3 instances. The SMS text messaging system used Twilio’s [32] application programming interface to send out the messages.
Results
From March 23, 2020, to August 23, 2020, a total of 127,684 SMS text messages were sent, generating 90,414 valid reports among 1004 participants, achieving a weekly average of 4.5 (SD 1.9) reports per user. Female gender (n=755, 75.2%) was more prevalent than male (n=249, 24.8%) among participants, reflecting the general HCW population in these hospitals. The median age was 39 years, with a mean of 40.2 (SD 11.3) years. Figure 1 shows the temporal distribution of symptoms of respiratory infection over the study period, using LOESS regression. In total, 1.49% (n=15) of participants reported a positive PCR result during the study period. The first peak of the bimodal curves clearly parallels the reference curve of individuals in the hospital who tested positive, representing the first COVID-19 wave in the region. The second peak appears between July 2020 and August 2020, with a much lower signal in the reference curve of individuals who tested positive.
Figure 1.
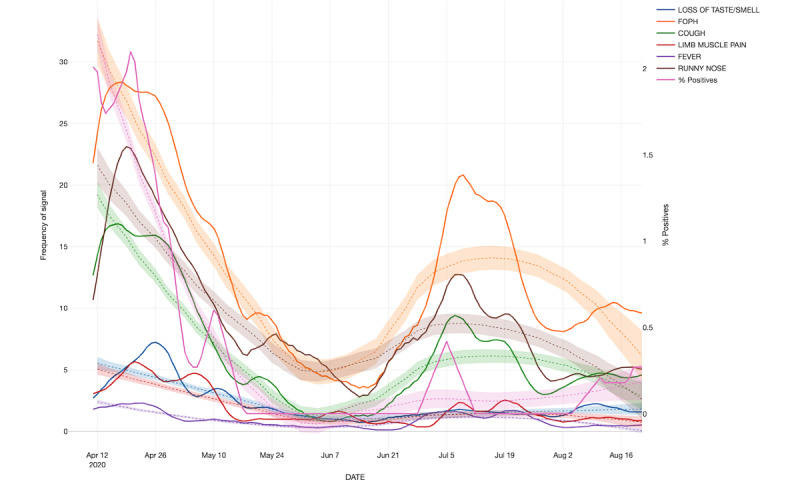
Temporal distribution and LOESS regression of symptoms related to acute respiratory infection in health care workers at two hospitals in Switzerland. FOPH: cases documented by the Federal Office of Public Health; LOESS: locally estimated scatterplot smoothing.
Regarding anomaly detection over time, Figure 2 shows whether a signal of symptoms was expected (based in the past trends) or if it represented a positive or negative anomaly, meaning a significant increase or decrease in the frequency of recorded symptoms. Table 2 indicates the change points that were statistically significant, including the difference observed when compared with the expected amount. The positive anomalies happened in three different periods; two of them occurred during the highest activity of the first wave and the third occurred between July and August, representing a possible second wave. However, as mentioned above, no second (or third) wave was seen in the reference curve.
Figure 2.
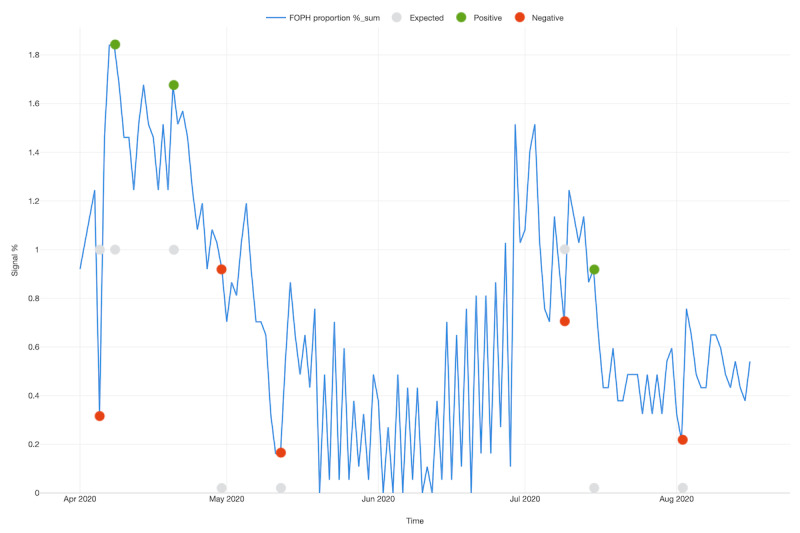
Temporal distribution of the FOPH proportion of positives, indicating which types of anomalies occurred in health care workers in two hospitals in Switzerland. FOPH: Federal Office of Public Health.
Table 2.
Significant (P<.05) timepoints for anomaly detection in health care workers, Switzerland.
| Date | Federal Office of Public Health proportion of positives | Expected | Difference from expected | Anomaly type |
| 05/04/2020 | .324675325 | 1 | –.675324675 | Negative |
| 08/04/2020 | 1.83982684 | 1 | .83982684 | Positive |
| 20/04/2020 | 1.677489177 | 1 | .677489177 | Positive |
| 30/04/2020 | .91991342 | 0 | .91991342 | Negative |
| 12/05/2020 | .162337662 | 0 | .162337662 | Negative |
| 09/07/2020 | .703463203 | 1 | –.296536797 | Negative |
| 15/07/2020 | .91991342 | 0 | .91991342 | Positive |
| 02/08/2020 | .216450216 | 0 | .216450216 | Negative |
A correlation matrix between symptoms and a positive PCR test result for SARS-CoV-2 is shown in Figure 3, while in Figure 4, the significance matrix shows the positive and negative correlations, as well as the nonsignificant ones. The symptom with the strongest correlation with a positive PCR result was loss of taste. Conversely, symptoms such as red eyes or runny nose were negatively associated with a positive test (Table 3).
Figure 3.
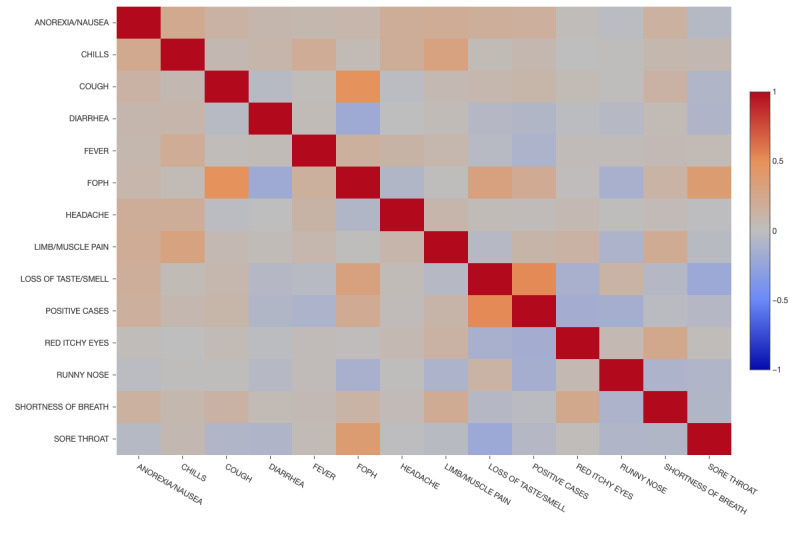
Correlation matrix using the Spearman method for symptoms and positive results in health care workers in two hospitals in Switzerland during the study period. FOPH: Federal Office of Public Health.
Figure 4.
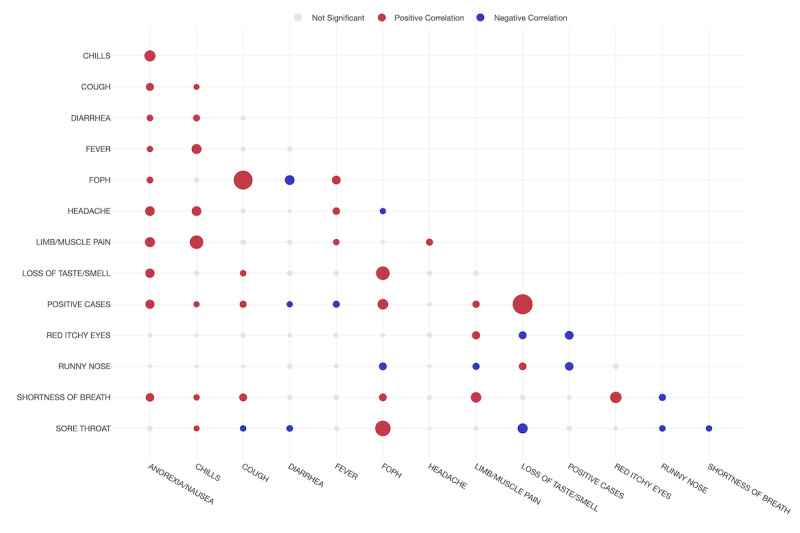
Significance matrix showcasing the positive and negative correlations between variables in health care workers in two hospitals in Switzerland during the study period. A larger dot represents a higher correlation.
Table 3.
Correlation between symptoms and positive cases in health care workers in Switzerland for the period of the study.
| Symptoms | Correlation | Pairs | P value |
| Loss of taste | 0.5274 | Positive | <.001 |
| Federal Office of Public Health definition | 0.2189 | Positive | <.001 |
| Anorexia/nausea | 0.1698 | Positive | <.001 |
| Limb/muscle pain | 0.1103 | Positive | <.001 |
| Cough | 0.1032 | Positive | <.001 |
| Chills | 0.0731 | Positive | .002 |
| Headache | 0.0279 | Positive | .37 |
| Red itchy eyes | –0.1560 | Negative | .01 |
| Runny nose | –0.1508 | Negative | .001 |
| Fever | –0.1025 | Negative | .10 |
| Diarrhea | –0.0770 | Negative | .001 |
Finally, Table 4 shows the summary results from a random forest algorithm that was used to classify participants into SARS-CoV-2 positive and negative cases based on their indicated symptoms. The area under the ROC curve shows reasonable performance of the classification tree, with an accuracy of 88% for the training data and 89% for the test data (Figure 5). Nevertheless, while the prediction matrix showed good specificity (80.0%), sensitivity was low (10.6%; Table 5). Figure 6 shows the importance of symptoms and their capacity to predict the expected outcome based on the random forest algorithm, considering a P value of <.05. Loss of taste and limb/muscle pain were the most important variables for prediction of a positive result, while runny nose and red eyes were negatively correlated with the same outcome. Fever was a very weak predictor of a positive result.
Table 4.
Summary of the parameters of the random forest model.
| Data set | Area under the curve | F1 score | Accuracy rate | Misclassification rate | Precision | Recall |
| Training | .90375 | .68027 | .8839 | .11604 | .87719 | .5555 |
| Test | .87576 | .66331 | .89438 | .10561 | .91304 | .5206 |
Figure 5.
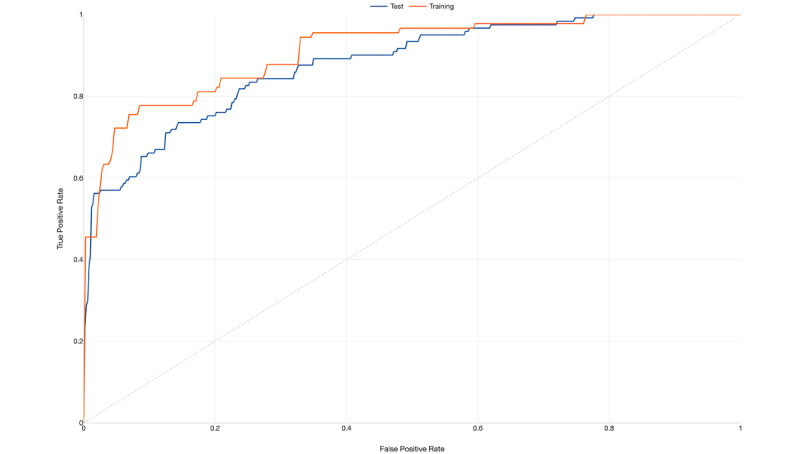
Receiver operating characteristic curve for the random forest model.
Table 5.
Prediction matrix for the random forest model.
| Data set and type (actual) | Data type (predicted) | ||||
|
|
TRUE, % | FALSE, % | |||
| Test | |||||
|
|
TRUE | 10.4 | 9.57 | ||
|
|
FALSE | .99 | 79.04 | ||
| Training | |||||
|
|
TRUE | 12.35 | 9.88 | ||
|
|
FALSE | 1.73 | 76.05 | ||
Figure 6.
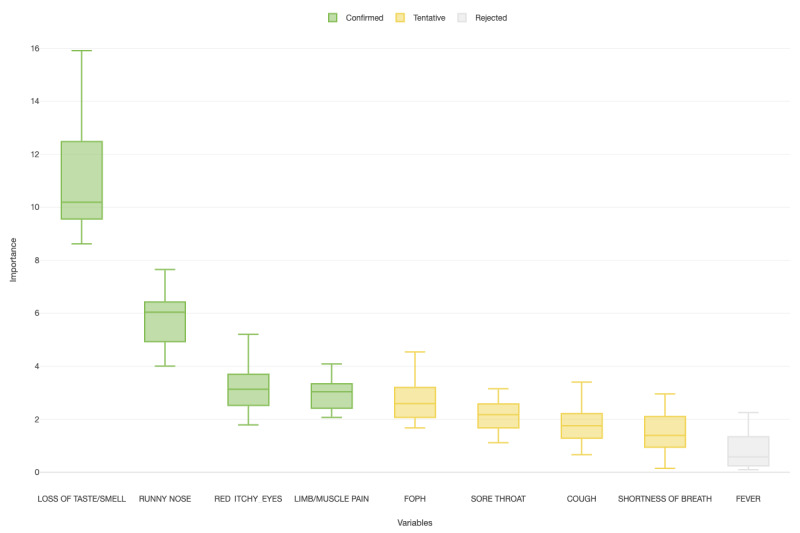
Boxplot of the importance of symptoms and their capacity to predict the expected outcome based on the random forest algorithm (P<.05). Loss of taste, limb/muscle pain, FOPH (Federal Office of Public Health), sore throat, cough, and shortness of breath were positively associated with the outcome. Runny nose and red itchy eyes were negatively associated with the outcome. Fever was neither positively nor negatively associated with the outcome.
Discussion
This study demonstrates the use of digital surveillance to monitor COVID-19 activity among HCWs. Loss of taste was the symptom that was most aligned with COVID-19 activity at the population level. At the individual level, using machine learning–based random forest classification, reporting loss of taste and limb/muscle pain as well as absence of runny nose and red eyes were the best predictors of COVID-19. The main strengths of the study are its high response rate and the comparison to a reference curve, which was based on documented PCR results in the same population.
Syndromic surveillance through participatory surveillance has been shown to be a feasible strategy to monitor COVID-19 activity [33], and is considered an important measure to inform the public health response to this pandemic [34]. Considering that engagement is a key element of a successful platform, our study—with an average response of 4.5 answers per week—has an excellent basis to produce valid and representative results. This high rate of engagement and participation is extraordinary when compared to other platforms [13,17,18,33], especially over a period of 5 months [35]. The easy-to-use survey, the defined population of HCWs from two different hospitals, and the regular interaction with study participants are potential reasons for this high response rate. It remains to be seen if these engagement indexes can be maintained when the study is scaled up to larger communities.
The temporal distribution of symptoms followed the trends represented in the first wave of COVID-19 in Switzerland [36,37]. However, the signals detected in July were not due to COVID-19, as shown by the reference curve. Interestingly, several HCWs tested positive for rhinovirus during this time period, suggesting that this was the reason for this wave. Of note, loss of taste, the most specific symptom of COVID-19, did not increase during this second wave.
Several other studies have shown that loss of taste is a good proxy for COVID-19 [38-41]. Although the specificity of this symptom is excellent, only about 20% of patients report loss of taste [42]. We conclude that the detection of loss of taste is very helpful to interpret findings at the population level, but less so at the individual patient level because of its low prevalence. The second most important positively associated symptom in our analysis was limb/muscle pain, which has also been noted by others [43]. Remarkably, runny nose and red eyes were very important negative predictors of COVID-19; this finding is particularly useful for when surveillance is performed during allergy season. However, both the sensitivity and specificity of a symptom depend on the background activity of other infections and allergies and might therefore be subject to change. The validity of a symptom may also change due to genetic adaptations in the dominant SARS-CoV-2 strain. During the study period, none of the new variants of SARS-CoV-2 (eg, B.1.1.7/Alpha) were circulating in Switzerland. Therefore, the symptoms described here cannot necessarily be extrapolated to a different circulating SARS-CoV-2 variant. However, syndromic surveillance through participatory surveillance may allow for the detection or validation of a different clinical presentation emerging from a new circulating strain. Indeed, a recent study describes small differences in COVID-19 symptoms in the general population in the United Kingdom depending on the variant [44].
Our study has a number of limitations. First, it was performed outside influenza season. Because influenza more often presents with constitutional symptoms than other respiratory viruses, distinguishing influenza from COVID-19 by analysis of symptoms is difficult. Second, we relied on participants self-reporting their symptoms, a method that is prone to bias. Third, generalizability of our data is limited because only one-fifth of the HCWs from our hospitals participated in the study; in addition, the spatial component could not be explored due to these same reasons. At the same time, this would be a very important parameter for evaluating whether SARS-CoV-2 is being regionally distributed, which would be useful to form a complete picture for disease surveillance purposes. The application of classification techniques based on machine learning, such as random forest classification, has its own limitations, as a large number of trees can make the algorithm too slow and ineffective for real-time predictions. In general, these algorithms are fast to train, but quite slow to create predictions once they are trained. A more accurate prediction requires more trees, which results in a slower model.
Nevertheless, we deem the presented surveillance tool highly useful in monitoring and predicting COVID-19 activity among our HCWs. Currently, we have expanded our HCW cohort to include over 5000 participants from over 20 institutions [45]. The analysis of data from different institutions will allow us to detect the clustering of cases in certain institutions, which might trigger targeted intervention measures in affected health care institutions. Additionally, these data allow for the detection of symptomatic HCWs who were either not tested or had a false-negative PCR result, and also for the discrimination of symptoms caused by SARS-CoV-2 from symptoms caused by other viruses, such as influenza. Further questions, which we aim to answer with the surveillance data generated in this larger cohort, include how long HCWs with documented SARS-CoV-2 infection (or vaccination) are protected against reinfection or how the emergence of viral variants might change the symptomatology of COVID-19.
Acknowledgments
This work was supported by the Swiss National Sciences Foundation (grants 31CA30_196544 and PZ00P3_179919 to PK), the Federal Office of Public Health (grant 20.008218/421-28/1), and the research fund of the Cantonal Hospital of St. Gallen. OLN acknowledges support from Rodrigo Paiva in the development of the technological platform.
Abbreviations
- FOPH
Federal Office of Public Health
- HCW
health care worker
- LOESS
locally estimated scatterplot smoothing
- PCR
polymerase chain reaction
- ROC
receiver operating characteristic
- S-H-ESD
Seasonal-Hybrid Extreme Studentized Deviate
Informed consent (in German).
Footnotes
Authors' Contributions: OLN and PK conceived of the presented idea and wrote the manuscript with support from TE, CK, MS, DF, WA, and PV. OLN carried out the analysis. All authors revised the final version of the manuscript. PK supervised the project.
Conflicts of Interest: None declared.
References
- 1.Wirth FN, Johns M, Meurers T, Prasser F. Citizen-Centered Mobile Health Apps Collecting Individual-Level Spatial Data for Infectious Disease Management: Scoping Review. JMIR mHealth uHealth. 2020 Nov 10;8(11):e22594. doi: 10.2196/22594. https://mhealth.jmir.org/2020/11/e22594/ v8i11e22594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaxman S, Mishra S, Gandy A, Unwin HJT, Mellan TA, Coupland H, Whittaker C, Zhu H, Berah T, Eaton JW, Monod M, Imperial College COVID-19 Response Team. Ghani AC, Donnelly CA, Riley S, Vollmer MAC, Ferguson NM, Okell LC, Bhatt S. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020 Aug;584(7820):257–261. doi: 10.1038/s41586-020-2405-7. doi: 10.1038/s41586-020-2405-7.10.1038/s41586-020-2405-7 [DOI] [PubMed] [Google Scholar]
- 3.Howell O'Neill P, Ryan-Mosley T, Johnson B. A flood of coronavirus apps are tracking us. Now it's time to keep track of them. MIT Technology Review. 2020. [2021-01-15]. https://www.technologyreview.com/2020/05/07/1000961/launching-mittr-covid-tracing-tracker/
- 4.Altmann S, Milsom L, Zillessen H, Blasone R, Gerdon F, Bach R, Kreuter F, Nosenzo D, Toussaert S, Abeler J. Acceptability of app-based contact tracing for COVID-19: Cross-country survey evidence. JMIR mHealth uHealth. 2020 Jul 24;:1–6. doi: 10.2196/19857. doi: 10.2196/19857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye Q, Zhou J, Wu H. Using Information Technology to Manage the COVID-19 Pandemic: Development of a Technical Framework Based on Practical Experience in China. JMIR Med Inform. 2020 Jun 08;8(6):e19515. doi: 10.2196/19515. http://www.hxkqyxzz.net/fileup/PDF/20120401.pdf .v8i6e19515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abeler J, Bäcker M, Buermeyer U, Zillessen H. COVID-19 Contact Tracing and Data Protection Can Go Together. JMIR mHealth uHealth. 2020 Apr 20;8(4):e19359. doi: 10.2196/19359. https://mhealth.jmir.org/2020/4/e19359/ v8i4e19359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker MJ, Fraser C, Abeler-Dörner L, Bonsall D. Ethics of instantaneous contact tracing using mobile phone apps in the control of the COVID-19 pandemic. J Med Ethics. 2020 Jul 04;46(7):427–431. doi: 10.1136/medethics-2020-106314. http://jme.bmj.com/lookup/pmidlookup?view=long&pmid=32366705 .medethics-2020-106314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams JG, Walls RM. Supporting the Health Care Workforce During the COVID-19 Global Epidemic. JAMA. 2020 Apr 21;323(15):1439–1440. doi: 10.1001/jama.2020.3972.2763136 [DOI] [PubMed] [Google Scholar]
- 9.Sim JXY, Conceicao EP, Wee LE, Aung MK, Wei Seow SY, Yang Teo RC, Goh JQ, Ting Yeo DW, Jyhhan Kuo B, Lim JW, Gan WH, Ling ML, Venkatachalam I. Utilizing the electronic health records to create a syndromic staff surveillance system during the COVID-19 outbreak. Am J Infect Control. 2021 Jun;49(6):685–689. doi: 10.1016/j.ajic.2020.11.003. http://europepmc.org/abstract/MED/33159997 .S0196-6553(20)30971-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler PP, Kahlert CR, Sumer J, Flury D, Güsewell S, Leal-Neto OB, Notter J, Albrich WC, Babouee Flury B, McGeer A, Kuster S, Risch L, Schlegel M, Vernazza P. Prevalence of SARS-CoV-2 antibodies among Swiss hospital workers: Results of a prospective cohort study. Infect Control Hosp Epidemiol. 2021 May;42(5):604–608. doi: 10.1017/ice.2020.1244. http://europepmc.org/abstract/MED/33028454 .S0899823X20012441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bach M, Jordan S, Hartung S, Santos-Hövener C, Wright MT. Participatory epidemiology: the contribution of participatory research to epidemiology. Emerg Themes Epidemiol. 2017 Feb 10;14(1):2–15. doi: 10.1186/s12982-017-0056-4. https://ete-online.biomedcentral.com/articles/10.1186/s12982-017-0056-4 .56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koppeschaar CE, Colizza V, Guerrisi C, Turbelin C, Duggan J, Edmunds WJ, Kjelsø C, Mexia R, Moreno Y, Meloni S, Paolotti D, Perrotta D, van Straten E, Franco AO. Influenzanet: Citizens Among 10 Countries Collaborating to Monitor Influenza in Europe. JMIR Public Health Surveill. 2017 Sep 19;3(3):e66. doi: 10.2196/publichealth.7429. https://publichealth.jmir.org/2017/3/e66/ v3i3e66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smolinski MS, Crawley AW, Olsen JM, Jayaraman T, Libel M. Participatory Disease Surveillance: Engaging Communities Directly in Reporting, Monitoring, and Responding to Health Threats. JMIR Public Health Surveill. 2017 Oct 11;3(4):e62. doi: 10.2196/publichealth.7540. https://publichealth.jmir.org/2017/4/e62/ v3i4e62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leal-Neto OB, Dimech GS, Libel M, Oliveira W, Ferreira JP. Digital disease detection and participatory surveillance: overview and perspectives for Brazil. Rev Saude Publica. 2016;50:17. doi: 10.1590/S1518-8787.2016050006201. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0034-89102016000100702&lng=en&nrm=iso&tlng=en .S0034-89102016000100702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salathé M. Digital epidemiology: what is it, and where is it going? Life Sci Soc Policy. 2018 Jan 04;14(1):1–5. doi: 10.1186/s40504-017-0065-7. http://europepmc.org/abstract/MED/29302758 .10.1186/s40504-017-0065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wójcik OP, Brownstein JS, Chunara R, Johansson MA. Public health for the people: participatory infectious disease surveillance in the digital age. Emerg Themes Epidemiol. 2014 Jun 20;11(1):7–7. doi: 10.1186/1742-7622-11-7. https://ete-online.biomedcentral.com/articles/10.1186/1742-7622-11-7 .1742-7622-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leal Neto O, Dimech GS, Libel M, de Souza WV, Cesse E, Smolinski M, Oliveira W, Albuquerque J. Saúde na Copa: The World's First Application of Participatory Surveillance for a Mass Gathering at FIFA World Cup 2014, Brazil. JMIR Public Health Surveill. 2017 May 04;3(2):e26. doi: 10.2196/publichealth.7313. https://publichealth.jmir.org/2017/2/e26/ v3i2e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leal Neto O, Cruz O, Albuquerque J, Nacarato de Sousa M, Smolinski M, Pessoa Cesse EÂ, Libel M, Vieira de Souza W. Participatory Surveillance Based on Crowdsourcing During the Rio 2016 Olympic Games Using the Guardians of Health Platform: Descriptive Study. JMIR Public Health Surveill. 2020 Apr 07;6(2):e16119. doi: 10.2196/16119. https://publichealth.jmir.org/2020/2/e16119/ v6i2e16119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drew D, Nguyen LH, Steves CJ, Menni C, Freydin M, Varsavsky T, Sudre CH, Cardoso MJ, Ourselin S, Wolf J, Spector TD, Chan AT, COPE Consortium Rapid implementation of mobile technology for real-time epidemiology of COVID-19. Science. 2020 Jun 19;368(6497):1362–1367. doi: 10.1126/science.abc0473. http://europepmc.org/abstract/MED/32371477 .science.abc0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garg S, Bhatnagar N, Gangadharan N. A Case for Participatory Disease Surveillance of the COVID-19 Pandemic in India. JMIR Public Health Surveill. 2020 Apr 16;6(2):e18795. doi: 10.2196/18795. https://publichealth.jmir.org/2020/2/e18795/ v6i2e18795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo H, Lie Y, Prinzen FW. Surveillance of COVID-19 in the General Population Using an Online Questionnaire: Report From 18,161 Respondents in China. JMIR Public Health Surveill. 2020 Apr 27;6(2):e18576. doi: 10.2196/18576. https://publichealth.jmir.org/2020/2/e18576/ v6i2e18576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leal-Neto O, Santos F, Lee J, Albuquerque J, Souza W. Prioritizing COVID-19 tests based on participatory surveillance and spatial scanning. Int J Med Inform. 2020 Nov;143:104263. doi: 10.1016/j.ijmedinf.2020.104263. https://linkinghub.elsevier.com/retrieve/pii/S1386-5056(20)30853-4 .S1386-5056(20)30853-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Infektionskrankheiten melden. [2021-11-16]. https://www.bag.admin.ch/bag/de/home/krankheiten/infektionskrankheiten-bekaempfen/meldesysteme-infektionskrankheiten/meldepflichtige-ik/meldeformulare.html .
- 24.Garimella R. A Simple Introduction to Moving Least Squares and Local Regression Estimation. Los Alamos National Lab. 2017. [2021-11-17]. https://www.osti.gov/biblio/1367799-simple-introduction-moving-least-squares-local-regression-estimation .
- 25.Cleff T. Exploratory Data Analysis in Business and Economics: An Introduction Using SPSS, Stata, and Excel. Cham: Springer International Publishing; 2014. [Google Scholar]
- 26.Hochenbaum J, Vallis OS, Kejariwal A. Automatic anomaly detection in the cloud via statistical learning. arXiv. Preprint posted online on April 24, 2017 https://arxiv.org/abs/1704.07706 . [Google Scholar]
- 27.Cleveland RB, Cleveland WS, McRae JE, Terpenning I. STL: A Seasonal-Trend Decomposition Procedure Based on Loess. Journal of Official Statistics. 1990;6(1):3–73. https://www.wessa.net/download/stl.pdf . [Google Scholar]
- 28.Ho TK. Random decision forests. Proceedings of 3rd International Conference on Document Analysis and Recognition; 3rd International Conference on Document Analysis and Recognition; August 14-16, 1995; Montreal, QC. 1995. pp. 278–282. [DOI] [Google Scholar]
- 29.Biau G. Analysis of a random forests model. The Journal of Machine Learning Research. 2012;13(1):1063–1095. https://www.jmlr.org/papers/v13/biau12a.html . [Google Scholar]
- 30.Nishida K. Exploratory. 2020. [2021-04-14]. https://exploratory.io .
- 31.Jotform. 2020. [2021-04-10]. https://jotform.com .
- 32.Twilio - Communication APIs for SMS, Voice, Video and Authentication. [2021-01-01]. https://www.twilio.com/
- 33.Lapointe-Shaw L, Rader B, Astley CM, Hawkins JB, Bhatia D, Schatten WJ, Lee TC, Liu JJ, Ivers NM, Stall NM, Gournis E, Tuite AR, Fisman DN, Bogoch II, Brownstein JS. Web and phone-based COVID-19 syndromic surveillance in Canada: A cross-sectional study. PLoS One. 2020 Oct 2;15(10):e0239886. doi: 10.1371/journal.pone.0239886. https://dx.plos.org/10.1371/journal.pone.0239886 .PONE-D-20-18024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budd J, Miller BS, Manning EM, Lampos V, Zhuang M, Edelstein M, Rees G, Emery VC, Stevens MM, Keegan N, Short MJ, Pillay D, Manley E, Cox IJ, Heymann D, Johnson AM, McKendry RA. Digital technologies in the public-health response to COVID-19. Nat Med. 2020 Aug;26(8):1183–1192. doi: 10.1038/s41591-020-1011-4. doi: 10.1038/s41591-020-1011-4.10.1038/s41591-020-1011-4 [DOI] [PubMed] [Google Scholar]
- 35.Brownstein JS, Chu S, Marathe A, Marathe MV, Nguyen AT, Paolotti D, Perra N, Perrotta D, Santillana M, Swarup S, Tizzoni M, Vespignani A, Vullikanti AKS, Wilson ML, Zhang Q. Combining Participatory Influenza Surveillance with Modeling and Forecasting: Three Alternative Approaches. JMIR Public Health Surveill. 2017 Nov 01;3(4):e83. doi: 10.2196/publichealth.7344. https://publichealth.jmir.org/2017/4/e83/ v3i4e83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.COVID-19 Situation Updates. European Centre for Disease Prevention and Control. 2020. [2020-04-24]. https://www.ecdc.europa.eu/en/COVID-19-pandemic .
- 37.Micallef S, Piscopo TV, Casha R, Borg D, Vella C, Zammit M, Borg J, Mallia D, Farrugia J, Vella SM, Xerri T, Portelli A, Fenech M, Fsadni C, Mallia Azzopardi C. The first wave of COVID-19 in Malta; a national cross-sectional study. PLoS One. 2020;15(10):e0239389. doi: 10.1371/journal.pone.0239389. https://dx.plos.org/10.1371/journal.pone.0239389 .PONE-D-20-20316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spinato G, Fabbris C, Polesel J, Cazzador D, Borsetto D, Hopkins C, Boscolo-Rizzo P. Alterations in Smell or Taste in Mildly Symptomatic Outpatients With SARS-CoV-2 Infection. JAMA. 2020 May 26;323(20):2089–2090. doi: 10.1001/jama.2020.6771. http://europepmc.org/abstract/MED/32320008 .2765183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sudre C, Lee K, Lochlainn M, Varsavsky T, Murray B, Graham MS, Menni C, Modat M, Bowyer RCE, Nguyen LH, Drew DA, Joshi AD, Ma W, Guo CG, Lo CH, Ganesh S, Buwe A, Pujol JC, du Cadet JL, Visconti A, Freidin MB, El-Sayed Moustafa JS, Falchi M, Davies R, Gomez MF, Fall T, Cardoso MJ, Wolf J, Franks PW, Chan AT, Spector TD, Steves CJ, Ourselin S. Symptom clusters in COVID-19: A potential clinical prediction tool from the COVID Symptom Study app. Sci Adv. 2021 Mar;7(12):eabd4177. doi: 10.1126/sciadv.abd4177. http://europepmc.org/abstract/MED/33741586 .7/12/eabd4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eliezer M, Hautefort C, Hamel A, Verillaud B, Herman P, Houdart E, Eloit C. Sudden and Complete Olfactory Loss of Function as a Possible Symptom of COVID-19. JAMA Otolaryngol Head Neck Surg. 2020 Jul 01;146(7):674–675. doi: 10.1001/jamaoto.2020.0832.2764417 [DOI] [PubMed] [Google Scholar]
- 41.Menni C, Valdes AM, Freidin MB, Sudre CH, Nguyen LH, Drew DA, Ganesh S, Varsavsky T, Cardoso MJ, El-Sayed Moustafa JS, Visconti A, Hysi P, Bowyer RCE, Mangino M, Falchi M, Wolf J, Ourselin S, Chan AT, Steves CJ, Spector TD. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020 Jul 11;26(7):1037–1040. doi: 10.1038/s41591-020-0916-2. http://europepmc.org/abstract/MED/32393804 .10.1038/s41591-020-0916-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bénézit F, Le Turnier P, Declerck C, Paillé C, Revest M, Dubée V, Tattevin P, RAN COVID Study Group Utility of hyposmia and hypogeusia for the diagnosis of COVID-19. Lancet Infect Dis. 2020 Sep;20(9):1014–1015. doi: 10.1016/S1473-3099(20)30297-8. http://europepmc.org/abstract/MED/32304632 .S1473-3099(20)30297-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nepal G, Rehrig JH, Shrestha GS, Shing YK, Yadav JK, Ojha R, Pokhrel G, Tu ZL, Huang DY. Neurological manifestations of COVID-19: a systematic review. Crit Care. 2020 Jul 13;24(1):421–11. doi: 10.1186/s13054-020-03121-z. https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-03121-z .10.1186/s13054-020-03121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vihta K, Pouwels K, Peto T, Pritchard E, Eyre DW, House T, Gethings O, Studley R, Rourke E, Cook D, Diamond I, Crook D, Matthews PC, Stoesser N, Walker AS, COVID-19 Infection Survey team Symptoms and SARS-CoV-2 positivity in the general population in the UK. Clin Infect Dis. 2021 Nov 08;:ciab945. doi: 10.1093/cid/ciab945.6423489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahlert CR, Persi R, Güsewell S, Egger T, Leal-Neto OB, Sumer J, Flury D, Brucher A, Lemmenmeier E, Möller JC, Rieder P, Stocker R, Vuichard-Gysin D, Wiggli B, Albrich WC, Babouee Flury B, Besold U, Fehr J, Kuster SP, McGeer A, Risch L, Schlegel M, Friedl A, Vernazza P, Kohler P. Non-occupational and occupational factors associated with specific SARS-CoV-2 antibodies among hospital workers - A multicentre cross-sectional study. Clin Microbiol Infect. 2021 Sep;27(9):1336–1344. doi: 10.1016/j.cmi.2021.05.014. https://linkinghub.elsevier.com/retrieve/pii/S1198-743X(21)00236-6 .S1198-743X(21)00236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Informed consent (in German).


