Abstract
Heterochromatin is characteristically the last portion of the genome to be replicated. In polytene cells, heterochromatic sequences are underreplicated because S phase ends before replication of heterochromatin is completed. Truncated heterochromatic DNAs have been identified in polytene cells of Drosophila and may be the discontinuous molecules that form between fully replicated euchromatic and underreplicated heterochromatic regions of the chromosome. In this report, we characterize the temporal pattern of heterochromatic DNA truncation during development of polytene cells. Underreplication occurred during the first polytene S phase, yet DNA truncation, which was found within heterochromatic sequences of all four Drosophila chromosomes, did not occur until the second polytene S phase. DNA truncation was correlated with underreplication, since increasing the replication of satellite sequences with the cycE1672 mutation caused decreased production of truncated DNAs. Finally, truncation of heterochromatic DNAs was neither quantitatively nor qualitatively affected by modifiers of position effect variegation including the Y chromosome, Su(var)2052, parental origin, or temperature. We propose that heterochromatic satellite sequences present a barrier to DNA replication and that replication forks that transiently stall at such barriers in late S phase of diploid cells are left unresolved in the shortened S phase of polytene cells. DNA truncation then occurs in the second polytene S phase, when new replication forks extend to the position of forks left unresolved in the first polytene S phase.
Eukaryotic chromosomes contain two distinct chromatin domains, euchromatin and heterochromatin. Euchromatin typically encompasses a majority of the chromosome and contains most genes and other unique sequences. Heterochromatin encompasses a smaller proportion of the chromosome, typically around the centromere and telomeres, and is enriched in noncoding, highly repetitive satellite sequences (22). The presence of heterochromatin and its association with repetitive sequences is a conserved feature of eukaryotic chromosomes and is found in most, if not all, organisms including yeast, flies, and humans. Heterochromatin is defined cytologically by its dark staining and condensed appearance and genetically by its ability to suppress gene expression. Numerous protein components of heterochromatin have now been identified, including the SIR proteins in yeast (18); HP1, Su(var)3-7, and ORC proteins in higher eukaryotes (58); specific acetylated isoforms of histone H4 (3, 56); and variant forms of histone H3 (34). Our understanding of how these and other proteins contribute to the structure of heterochromatin and how that structure relates to the dynamic functions of heterochromatin is beginning to be broadened (4, 38).
Another conserved characteristic of heterochromatin is its late replication during S phase; it is most often the last portion of the genome to be replicated (31). The mechanism for late replication is not known, although the condensed structure of heterochromatin and its ability to suppress transcription suggest that it might also suppress its own replication. If heterochromatin suppresses replication origins, for example, heterochromatic sequences might be late in replicating because initiation must occur at distant euchromatic origins or because suppressed origins fire only in late S (53). Alternatively, replication fork elongation might be substantially slower through heterochromatin than through euchromatin, resulting in longer and therefore later replication of heterochromatic sequences (17, 47).
A particularly dramatic consequence of the late replication of heterochromatic sequences is found in the polytene chromosomes of Drosophila melanogaster. The copy number of euchromatic sequences in the polytene chromosomes of the larval salivary gland exceeds 1,000c, while heterochromatic satellite sequences within the same chromosomes are severely underrepresented, remaining at or close to a copy number of 2c (11, 13, 43). Underrepresentation is likely to be a consequence of the failure of satellite sequences to be replicated during the polytene cell cycle because of a shortened S phase that ends before the satellite sequences are fully replicated (30). Consistent with this model, specific mutations in cyclin E, a regulator of S phase in both diploid and polytene cells of Drosophila (46), can increase the copy number of satellite sequences in polytene chromosomes by increasing the length of S phase and allowing late replication of satellite sequences to be completed (30).
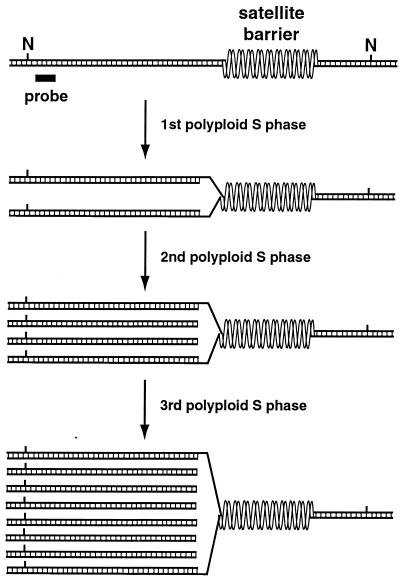
DNA molecules of discontinuous structure must occur between the fully replicated euchromatic and underreplicated heterochromatic regions of a polytene chromosome. DNA structures consisting of nested replication forks have been proposed to populate such regions (26); however, truncated linear DNAs rather than fork-containing molecules have been found in these regions, prompting reconsideration of the underreplication model of heterochromatic underrepresentation (14, 15, 51). In this study, we characterized the temporal pattern of heterochromatic DNA truncation during the development of polytene chromosomes. The data suggest that replication barriers within heterochromatin inhibit fork elongation through satellite sequences. Replication forks stalled at such barriers fail to be resolved during the shortened S phase of polytene cells, causing underreplication of satellite sequences. Truncated linear DNAs initially form and then accumulate as replication forks repeatedly extend to the same barriers during the multiple S phases that generate polytene chromosomes.
MATERIALS AND METHODS
Drosophila strains and crosses.
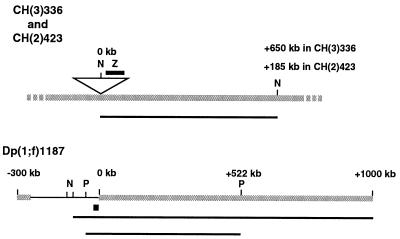
Information about Drosophila genes and nomenclature may be found at Flybase (http://flybase.bio.indiana.edu). Flies were maintained at 23°C and 55% relative humidity on cornmeal-brewer's yeast-glucose medium. Strains CH(3)336 and CH(2)423, which contain single heterochromatic P-element insertions, were described by Zhang and Spradling (63). Ovaries used for cyclin E analysis (see Fig. 3) were isolated from adult females of strain cycE1672/CyO; CH(3)336. Homozygous cycE1672/cycE1672 females are viable but sterile and can be recovered from this stock (30). Ovaries used for the analysis of Dp1187 (see Fig. 4) were isolated from adult females of strain In(1)sc8 Df(1)sc8, y; Dp(1;f)1187, y+. Salivary glands used for the Y chromosome modifier analysis (see Fig. 5A) were isolated from male larvae of genotype X/Y; CH(3)336/+, which were obtained from cross X/X; CH(3)336 × X/Y, and from male larvae of genotype X/O; CH(3)336/+, which were obtained from cross X/X; CH(3)336 × X∧Y. Salivary glands used for the Y chromosome modifier analysis shown (Fig. 5B) were isolated from male larvae of genotype X/Y, y; DP(1;f)1187, y+, which were obtained from cross X/X, y; × X/Y, y, Dp(1;f)1187, y+, and from male larvae of genotype X/O; Dp(1;f)1187, y+, which were obtained from cross X∧X, y × X/Y, y, Dp(1;f)1187, y+. Su(var)2052/+; CH(3)336/+ flies were obtained from cross Su(var)2052/CyO × CH(3)336. Female or male CH(3)336, P[ry+] flies and female or male Dp(1;f)1187, y+ flies were crossed to y1;ry506 animals to obtain CH(3)336 and Dp(1;f)1187 chromosomes, respectively, transmitted from either the female or male parent.
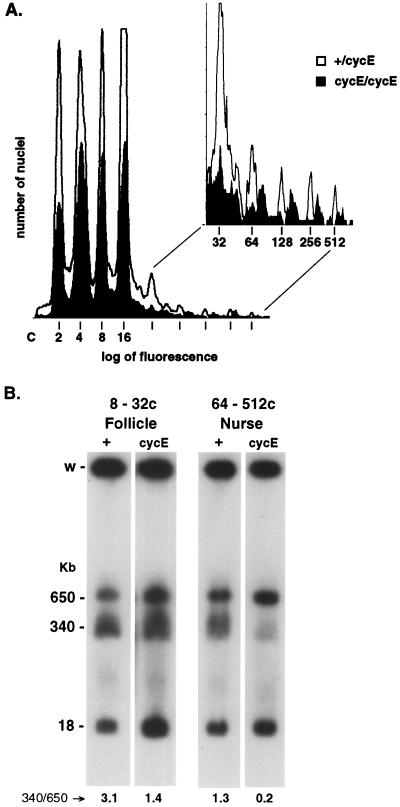
FIG. 3.
Truncation of heterochromatic DNAs is correlated with the extent of underreplication. (A) Nuclei from flies carrying the CH(3)336 chromosome that were either heterozygous (open) or homozygous (solid) for the cycE1672 mutation were isolated from ovaries and sorted by flow cytometry into discrete ploidy classes. The increased fluorescence observed for nuclei from the homozygous mutant flies is due to increased replication of heterochromatic sequences (30). (B) Nuclei from follicle cells (8c to 32c) and from nurse cells (64c to 512c) were collected and pooled. DNA was isolated and analyzed as described for Fig. 2. The 18-kb DNA is the NotI fragment flanking the euchromatic P element inserted into the cycE gene that creates the cycE1672 mutation.
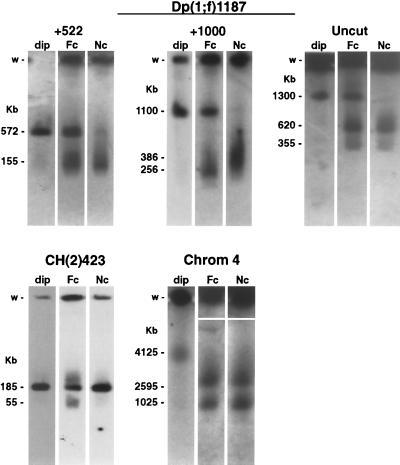
FIG. 4.
Truncation of heterochromatic DNAs occurs on all chromosomes. Nuclei from flies carrying the Dp(1;f)1187, CH(2)423, or wild-type fourth chromosome (Chrom 4) were isolated from ovaries and sorted by flow cytometry into discrete ploidy classes. Nuclei from 2c to 4c follicle cells (dip), from 8c to 32c follicle cells (Fc), and from 64c to 512c nurse cells (Nc) were collected and pooled. DNA was isolated, restriction endonuclease digested, fractionated by pulsed-field gel electrophoresis, and analyzed by in-gel hybridization. Dp(1;f)1187 DNA was digested with PmeI (+522) or NotI (+1000) or left uncut. CH(2)423 DNA was digested with NotI, and fourth-chromosome DNA was uncut. DNAs detected in 2c to 4c follicle cells (dip) are full-length restriction fragments or chromosomes, and the smaller DNAs detected in follicle (Fc) and nurse cells (Nc) are truncated molecules. Autoradiograms of the follicle and nurse cell samples for the fourth chromosome had to be manually aligned due to severe skewing of the lanes during electrophoresis in the region of the wells.
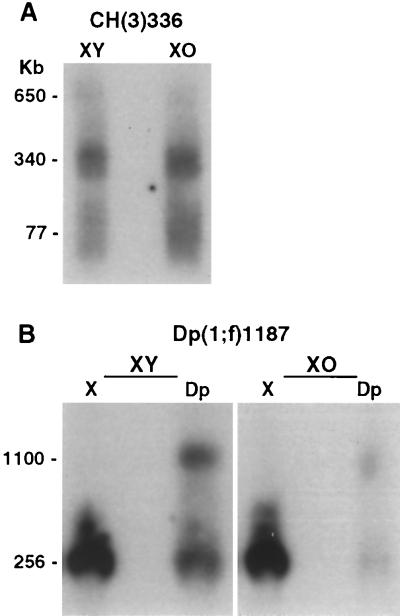
FIG. 5.
Truncation of heterochromatic DNA is not sensitive to modifiers of PEV. (A) Salivary glands were dissected from X/Y or X/O third-instar larvae carrying the CH(3)336 chromosome. Total salivary gland DNA was isolated and analyzed as described for Fig. 2. (B) Salivary glands were dissected from X/Y or X/O third-instar larvae carrying the Dp(1;f)1187 chromosome. Total salivary gland DNA was isolated, digested with NotI, and analyzed using a two-dimensional electrophoresis technique that physically separates DNA molecules derived from the normal X chromosome and Dp(1;f)1187 chromosome, both of which are present in these animals. Without separation, hybridization to the X chromosome NotI fragment would obstruct the hybridization signal from truncated Dp(1;f)1187 DNAs.
DNA isolation and analysis.
Fluorescence-activated cell sorter analysis of nuclei was done essentially as described by Lilly and Spradling (30). Briefly, ovaries dissected from adult females were homogenized, and nuclei were isolated on sucrose gradients. After the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), they were sorted on a Becton Dickinson FACS Vantage using a Nova Enterprise UV laser at 15 mW. Nuclei of specific ploidy classes were collected and prepared in agarose inserts. Approximately 400 pairs of ovaries were dissected, and 106 nuclei were collected for each ploidy class (see Fig. 2B). DNAs used for the Y chromosome modifier analysis (see Fig. 5) were isolated from whole salivary glands dissected from roaming third-instar larvae.
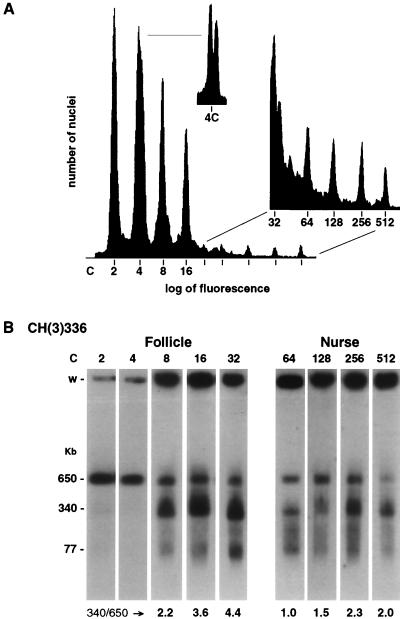
FIG. 2.
Truncation of heterochromatic DNAs occurs after the second polytene S phase. (A) Nuclei from flies carrying the CH(3)336 chromosome were isolated from ovaries and sorted by flow cytometry into discrete ploidy classes. (B) Nuclei from each ploidy class were collected, and their DNA was isolated, restriction digested, fractionated by pulsed-field gel electrophoresis, and analyzed by Southern blot hybridization. The ploidy of nuclei analyzed is indicated above each autoradiogram. Nuclei of 2c to 32c ploidy are predominantly from follicle cells, and nuclei of 64c to 512c ploidy are predominantly from nurse cells. The sizes of the DNA molecules are indicated in kilobase pairs, and the ratio of abundance of the 340-kb molecules to the 650-kb full-length restriction fragment is indicated below each autoradiogram. The well of each lane is indicated (w).
DNA was prepared in agarose inserts by standard procedures as described previously (16). The DNA was fractionated by contour-clamped homogeneous electric field pulsed-field gel electrophoresis on a Bio-Rad DRII or CHEF Mapper apparatus. The two-dimensional agarose gel electrophoresis technique used to separate the X and Dp1187 hybridization signals in samples analyzed in the experiment in Fig. 5B has been described previously (16). Briefly, DNA was first digested with NotI and fractionated by pulsed-field gel electrophoresis. A lane of the first-dimension gel was excised, and the DNA in the gel was digested in situ with a second enzyme. The DNA was then fractionated by conventional electrophoresis in a direction perpendicular to the direction of the first electrophoresis. The restriction sites for the second enzyme are polymorphic on the X and Dp1187 chromosomes; therefore, the X and Dp1187-derived restriction fragments differ in size and therefore are separated in the second dimension. After pulsed-field gel or two-dimensional electrophoresis, DNA was analyzed by Southern blot hybridization (45; also see reference 16). In-gel hybridizations, which provide a more sensitive and accurate method for detecting DNAs separated by pulsed-field gel electrophoresis (28), were used for the experiments in Fig. 4. Both Southern blot and in-gel hybridizations were done using the procedures of Church and Gilbert (6). Probes were labeled with 32P to specific activities of approximately 109 cpm/μg by random priming (12). Hybridization signals were analyzed qualitatively using Kodak XAR5 film and quantitatively using stored PhosphorImager screens and ImageQuant software (Molecular Dynamics).
Clones of bacterial lacZ sequences were used as hybridization probes for analysis of CH(3)336 and CH(2)423. Subclones of X chromosome genomic DNA used as hybridization probes for analysis of Dp1187 have been described previously (15) and included a 2.6-kb AseI-HindIII fragment used for the experiments in Fig. 4 and a 0.8-kb BglII-PstI fragment used for the experiments in Fig. 5, both from the sc8 region of Dp1187. A 3.6-kb EcoRI fragment containing the zfh-2 gene was used for the analysis of the fourth chromosome (see Fig. 4) (32).
RESULTS
Truncation of heterochromatic DNAs occurs after the second polytene S phase.
Defined heterochromatic restriction fragments were studied in Drosophila by using chromosomes CH(3)336 and Dp(1;f)1187 (Dp1187) (Fig. 1). These chromosomes have been used extensively to study the structure and function of Drosophila heterochromatin (15, 24, 25, 37, 62, 63). CH(3)336 has a P element inserted into pericentromeric heterochromatin in region h54-57 of chromosome 3. The P element is inserted into an “island” of complex sequences consisting primarily of clustered transposable elements (62). Interspersion of such islands among large tracks of satellite repeats is the normal sequence structure of Drosophila heterochromatin (8, 27). Dp1187 was derived from the X chromosome first by a large pericentric inversion, In(1)sc8, which directly juxtaposed centromere-proximal heterochromatin and distal euchromatic sequences, followed by a larger internal deletion that created the 1300-kb Dp1187 minichromosome (27). The euchromatic sequences of Dp1187 directly adjoin 1.688 satellite repeats (16), while on the normal X chromosome, these euchromatic sequences are located approximately 15 Mb from pericentromeric heterochromatin (27).
FIG. 1.
Structure of CH(3)336, CH(2)423, and Dp(1;f)1187 chromosomes. A portion of the CH(3)336 or CH(2)423 chromosome is shown containing a P-element transposon (triangle) inserted into heterochromatic sequences (gray line) on the third [CH(3)336] or second [CH(2)423] chromosome. lacZ sequences within the P element were used as a probe (dark line above the P element) to detect the 650-kb [CH(3)336] or 185-kb [CH(2)423] full-length NotI restriction fragment (dark line below the chromosome). The entire Dp(1;f)1187 chromosome is shown, with heterochromatic (gray line) and euchromatic (black line) sequences indicated. The coordinate system used defines 0 kb as the position of the sc8 euchromatic-heterochromatic breakpoint. Euchromatic sequences used as a probe (fat black line) and full-length restriction fragments (thin black lines) are indicated below the chromosome. N, NotI; P, PmeI; Z, lacZ.
It has been demonstrated previously that restriction fragments that extend from euchromatic into heterochromatic sequences of a chromosome are truncated in size in polytene cells (15). These truncated molecules are likely to be the discontinuous DNAs that were predicted to form between regions of fully replicated euchromatic and underreplicated heterochromatic sequences of a polytene chromosome. In this earlier study, however, the presence of truncated heterochromatic DNAs was correlated only with polytene tissues and not with the specific developmental transition from a diploid to a polytene cell cycle (15). To determine when DNA truncation occurs relative to the underreplication of heterochromatic sequences, chromosomes were analyzed during the transition from a diploid to a polytene structure.
Ovaries from adult flies contain two abundant polytene cell types, follicular epithelial cells and nurse cells. Isolated nuclei from whole adult ovaries can be separated into specific ploidy classes by flow cytometry (Fig. 2A). The 2c to 16c nuclei are predominantly from the abundant follicular epithelial cells, and the 32c to 512c nuclei are from the less abundant but higher-ploidy nurse cells (50). The 4c peak is a double peak that consists of nuclei from diploid follicle cells that have replicated their DNA completely and nuclei from follicle cells that have committed to polytene development and underreplicated their heterochromatic sequences during their first polytene S phase (Fig. 2A). Nuclei larger than 4c have only a single fluorescence maximum since all polytene cells underreplicate their DNA comparably with each S phase.
Nuclei from ovaries of CH(3)336 flies that contain a P element inserted into third-chromosome heterochromatin were separated by flow cytometry, and nuclei of each ploidy class were collected. The heterochromatic restriction fragment flanking the P element was analyzed by Southern blot hybridization from nuclei of each ploidy class (Fig. 2B). DNA from the 2c and 4c nuclei contained only the full-length restriction fragment of 650 kb. Truncated DNAs appeared abruptly in 8c nuclei and were present in all nuclei of 8c and higher ploidy. The truncated DNAs were heterogeneous in size, ranging from 77 kb up to the size of the 650-kb full-length restriction fragment, although molecules of 340 kb and, to a lesser extent, 77 kb were prominent. Quantitative differences between follicle cells and nurse cells in the abundance of truncated DNAs were observed: truncated DNAs were more abundant in follicle cells than in nurse cells. For example, in follicle cells, DNAs truncated to 340 kb were were always more abundant than the full-length restriction fragment and the 77-kb truncated DNA was readily detected, while in nurse cells, DNAs truncated to 340 kb were not always more abundant than the full-length restriction fragment and the 77-kb DNA was not always detected (Fig. 2B). In both cell types, the abundance of truncated DNAs appeared to increase with increasing ploidy, and this increase may occur more rapidly in follicle cells than in nurse cells.
Unexpectedly, initial truncation of heterochromatic DNAs did not correlate temporally with the start of heterochromatic underreplication. Recall that half of the 4c follicle cell nuclei had begun polytene development and underreplicated their DNA (Fig. 2A), yet no truncated DNAs were detected in the 4c sample (Fig. 2B). Even if the truncated DNAs are the product of stalled replication forks that subsequently brake in vivo or in vitro, they should be present in the underreplicated 4c nuclei. Lack of temporal correlation between the appearance of the truncated DNAs and the start of heterochromatic underreplication raised the possibility that truncation is not a consequence of underreplication but instead is caused by some other cellular process that occurs during polytene development.
Truncation of heterochromatic DNAs is correlated with the extent of underreplication.
If truncation of heterochromatic DNA is a consequence of underreplication, the abundance of truncated DNAs should correlate with the extent of underreplication. This hypothesis was tested by characterizing truncated DNAs in flies containing a mutation in cyclin E that increases the extent of heterochromatic replication. Cyclin E is a regulator of S phase in both diploid and polytene cells of Drosophila (46). CycE1672 is a viable hypomorphic P-element mutation of cyclin E that increases the replication of satellite sequences, principally in the polytene chromosomes of ovarian nurse cells, leading to a decrease in heterochromatic underrepresentation (30). CycE1672 is proposed to lengthen S phase, allowing more time for late replication of satellite sequences (30). Flow cytometry analysis showed that the DNA content in nurse cell nuclei of cycE1672 flies increased significantly, as expected (Fig. 3A). The DNA content in follicle cell nuclei also increased a slight but detectable amount (Fig. 3A). The CH(3)336 chromosome was crossed into a cycE1672 background, and the heterochromatic restriction fragment flanking the P element was analyzed by Southern blot hybridization in samples of pooled follicle cell nuclei (8c to 32c) and pooled nurse cell nuclei (64c to 512c) from wild-type and cycE1672 mutant flies (Fig. 3B). Truncated DNAs in mutant cycE1672 flies were significantly less abundant in nurse cell nuclei and slightly less abundant in follicle cell nuclei than were truncated DNAs in the wild-type flies (Fig. 3B). Thus, truncation of heterochromatic DNAs does correlate with the extent of heterochromatic underreplication. When more replication of satellite sequences was allowed, fewer truncated DNAs were produced (Fig. 3). We would predict that if replication of satellite sequences were complete, no truncated DNAs would be formed. These results strongly support the hypothesis that DNA truncation is a consequence of heterochromatic underreplication (Fig. 3B), even though truncated DNAs do not appear until the second cycle of underreplication (Fig. 2B).
Truncation of heterochromatic DNAs occurs on all chromosomes.
Truncation of heterochromatic DNA was also analyzed on the Dp1187, second, and fourth chromosomes. Ovary nuclei were isolated from flies carrying the Dp1187 minichromosome and sorted by flow cytometry into three classes: diploid nuclei (2c to 4c), follicle cell nuclei (8c to 32c), and nurse cell nuclei (64c to 512c). Truncated DNAs were analyzed in the three samples by in-gel hybridization. Analysis was done on both undigested and restriction enzyme-digested DNA. Consistent with what was observed for CH(3)336, only full-length Dp1187 molecules were detected in diploid nuclei while truncated DNAs were detected in the polytene nuclei of both follicle cells and nurse cells (Fig. 4). Significant quantitative differences between follicle cells and nurse cells were observed in the abundance of truncated Dp1187 DNAs. Full-length Dp1187 chromosomes were abundant in follicle cell nuclei but were barely detectable in nurse cell nuclei, suggesting that replication of heterochromatic sequences of Dp1187 is more efficient in follicle cells (Fig. 4). Qualitative differences were also observed. The population of truncated Dp1187 DNAs formed in nurse cell nuclei and revealed by NotI restriction digestion was 130 kb larger, on average, than was the population of truncated DNAs formed in follicle cell nuclei (Fig. 4, +1000 samples). This observation is consistent with earlier experiments in which truncated DNAs of two different sizes were observed in NotI-digested Dp1187 DNA isolated from whole ovaries (15). Finally, two distinct species of truncated Dp1187 molecules were detected in uncut DNA from both follicle cell and nurse cell nuclei (Fig. 4).
Second-chromosome heterochromatin was analyzed using chromosome CH(2)423, which has a P element inserted into heterochromatic sequences in pericentromeric region h42-43 (Fig. 1) (63). Ovary nuclei were isolated from flies carrying the CH(2)423 chromosome and sorted by flow cytometry as described above. Truncation of the flanking heterochromatic restriction fragment was analyzed by Southern blot hybridization. Only the full-length restriction fragment was detected in diploid nuclei; truncated DNAs were detected only in the polytene nuclei of follicle cells (Fig. 4). It is interesting that truncated DNAs were most abundant in follicle cell nuclei for CH(2)423 and CH(3)336 (Fig. 2 and 4) but were most abundant in nurse cell nuclei for Dp1187 (Fig. 4). Since the abundance of truncated DNAs is inversely correlated with the extent of heterochromatic replication (Fig. 3), these observations suggest that the efficiency of replicating heterochromatic sequences varies between different regions of heterochromatin in the same cell and is not a genomewide property of all heterochromatic sequences in any single cell type.
Truncation of heterochromatic DNAs was also analyzed on the wild-type fourth chromosome, which is small enough to be directly resolved by pulsed-field gel electrophoresis (32). Ovary nuclei isolated from flies carrying a wild-type fourth chromosome were sorted by flow cytometry, and their DNA was analyzed by in-gel hybridization. Full-length 4,125-kb chromosomes were detected only in diploid nuclei whereas truncated chromosomes of 2,595 and 1,025 kb were detected in polytene nuclei of both follicle cells and nurse cells (Fig. 4). No full-length fourth chromosomes were detected in either follicle or nurse cell nuclei (Fig. 4). Except for the difference in size, the relative pattern of truncated DNAs observed for the fourth chromosome was strikingly similar to the pattern observed for uncut Dp1187 (Fig. 4). This suggests that underreplication of satellite sequences and the resulting truncation of heterochromatic DNAs occurs by the same mechanism on both the Dp1187 and fourth chromosomes. A quantitative difference between the Dp1187 and fourth chromosomes was observed in follicle cell nuclei, where full-length Dp1187 chromosomes were detected but full-length fourth chromosomes were not (Fig. 4).
Truncation of heterochromatic DNA is not sensitive to modifiers of PEV.
Since packaging of DNA into heterochromatin suppresses transcription, we considered whether heterochromatin might play a similar role in suppressing replication and thereby contribute to heterochromatic underreplication in polytene cells. If the heterochromatin structure contributes to underreplication, genetic modifiers of that structure might alter the truncated DNAs that form as a consequence of underreplication. Heterochromatin structure can be altered using modifiers of position effect variegation (PEV), which occurs when gene transcription becomes variably suppressed due to the proximity of a gene to heterochromatin (21, 60). Several modifiers of PEV, including the Y chromosome, Su(var)2052, parental source, and temperature, were tested, but none significantly influenced the formation of truncated heterochromatic DNAs. The Y chromosome is a strong suppressor of PEV (49) and suppresses PEV of the rosy gene on CH(3)336 (63) and the yellow gene on Dp1187 (25). Truncation of heterochromatic DNA on the CH(3)336, Dp1187, and fourth chromosomes was analyzed in salivary glands from X/O and X/Y animals. The Y chromosome had no effect on the truncation of CH(3)336 heterochromatin (Fig. 5A) or fourth-chromosome heterochromatin (data not shown), but it did have a quantitative effect on the overall replication of the Dp1187 chromosome (Fig. 5B). Total Dp1187 DNA was less abundant in salivary glands from X/O animals than in those from X/Y animals (Fig. 5B). This effect appeared to be a general influence on total Dp1187 copy number, consistent with earlier analysis of PEV on Dp1187 (25), and not a specific effect on truncation of heterochromatic DNAs. The truncated DNAs detected in X/O animals, although less abundant, were the same size and of the same abundance relative to the full-length restriction fragment as those observed in X/Y animals (Fig. 5B). This suggests that Dp1187 replication that occurred in X/O salivary glands was subject to the same relative amount of underreplication producing the same truncated DNAs. Su(var)2052 is a suppressor of PEV and creates a missense mutation in the HP1 protein, a known component of heterochromatin (41). The Su(var)2052 mutation had no effect on the truncation of CH(3)336 heterochromatin in follicle cell, nurse cell, or salivary gland chromosomes (data not shown). Transmission of a chromosome through the female or male germ line influences the extent of PEV on that chromosome in the subsequent generation, with passage through the male germ line enhancing PEV (49). Parental source effects have been demonstrated for the Dp1187 chromosome (25). The parental source, however, had no effect on the truncation of heterochromatic DNAs on the CH(3)336 or Dp1187 chromosomes analyzed in the follicle and nurse cells of the ovary (data not shown). Finally, temperature influences PEV, with low temperatures during development enhancing PEV and high temperatures suppressing PEV (49). High temperatures suppress PEV of genes on heterochromatic P elements like the one on CH(3)336 (63). Flies grown at 18 and 27°C showed no difference in the truncation of heterochromatic DNA on the CH(3)336, DP1187, or fourth chromosomes in follicle and nurse cells of the ovary (data not shown).
DISCUSSION
Model for heterochromatic underreplication and the structure of polytene chromosomes.
Heterochromatic underreplication in polytene cells occurs when S phase ends before late replication of heterochromatic DNA has been completed. This is probably due to changes in cyclin E regulation (30). Premature termination of S phase would produce unresolved replication forks between fully replicated euchromatic and underreplicated heterochromatic sequences (26). The truncated heterochromatic DNAs described in this report, however, were not detected in the initial 4c nuclei that underreplicate their DNA, suggesting that truncation is not caused by simple breakage of unresolved replication forks (Fig. 2). Instead, truncated DNAs were detected after the second underreplication S phase. How could the abundance of truncated DNAs be dependent on the extent of underreplication yet require two underreplication S phases to form (Fig. 2 and 3)? We propose that truncated DNAs are produced when replication forks in the second polytene S phase extend to the position where unresolved replication forks formed in the first polytene S phase (Fig. 6). DNA synthesized during the first S phase provides the template strands for DNA synthesis in the second S phase, and since these template strands simply end at unresolved replication forks, replication in the second S phase would produce truncated linear DNAs (Fig. 6). This “replication runoff” model not only explains why truncated DNAs would appear specifically in the second S phase but also explains why their abundance would be dependent on the degree of underreplication; specifically, the fraction of replication forks left unresolved in the first S phase would determine the abundance of truncated DNAs produced in the second S phase. Furthermore, the gradual increase in the abundance of truncated DNAs that is observed as nuclei increase in ploidy (Fig. 2) would be caused by forks that complete replication in early S phases but are left unresolved in later S phases, leading to the production of additional truncated DNAs. While the replication runoff model explains the origin of truncated linear DNAs, it does not preclude the possibility that underreplication also creates DNA molecules with structures of sufficient complexity that they are unable to migrate out of the sample wells during electrophoresis (Fig. 2). Such molecules, if they exist, would probably have structures more complex than simple replication forks (14).
FIG. 6.
Model for heterochromatic underreplication and the structure of polytene chromosomes. Double-stranded DNA in a hypothetical region of Drosophila pericentromeric heterochromatin is shown, with NotI restriction sites (N) and a region of probe homology indicated. Satellite repeats inhibit their own replication by blocking fork elongation. The barrier formed by satellite repeats could reflect an intrinsic property of highly repetitious sequences or could result from the compaction of satellite repeats into a chromatin structure distinct from other heterochromatic sequences. The first polytene S phase ends before replication forks stalled at satellite barriers are resolved, causing heterochromatic underreplication. Truncated linear DNAs are generated in the second polytene S phase, when replication forks extend to the same barriers where forks were left unresolved in the first polytene S phase. DNAs are shown with blunt ends for clarity, but they would probably have staggered ends, particularly on lagging strands. Once produced, truncated DNAs would be replicated in each subsequent S phase.
In the replication runoff model, the sizes of truncated DNAs are determined by the locations of unresolved replication forks produced in the first underreplication S phase (Fig. 6). Although they formed a heterogeneous population, most truncated DNAs were of preferred sizes (Fig. 2 and 4), suggesting that unresolved forks were located at preferred positions when S phase ended. While those positions could be where actively elongating replication forks happen to be “caught” when S phase ends, the data suggest that the size of truncated DNAs is not directly determined by the length of the S phase. For example, the cycE1672 mutation reduced the abundance of truncated DNAs by increasing the length of S phase (Fig. 3) (30), yet the truncated DNAs detected in cycE1672 flies were not larger, as would be expected if the length of the S phase were determining the size of truncated DNAs (Fig. 3). How does extending S phase allow more heterochromatic replication without changing the size of truncated DNAs? If, during S phase, replication forks stall for extended periods at barriers to fork elongation, the locations of such barriers would be the preferred positions at which replication forks would most often be caught when the S phase ends. By extending the length of the S phase, the cycE1672 mutation would increase the probability that forks are able to traverse these barriers and complete replication before S phase ends, but the forks still left unresolved would most often be located at the same barriers, producing the same sized truncated DNAs. Transient stalling of replication forks at fixed barriers would also explain the pattern of truncated DNAs produced in CH(3)336 heterochromatin. The same 340- and 77-kb truncated DNAs were detected in all tissues analyzed, but their abundances relative to the full-length restriction fragment differed significantly. In 64c nurse cell nuclei, truncated DNAs were approximately equal in abundance to the full-length molecules (Fig. 2); in 8c follicle cell nuclei, truncated DNAs were about twofold more abundant than full-length molecules (Fig. 2); and in the 1,024c nuclei of salivary glands, only truncated DNAs, including a significant amount of 77-kb DNAs, and no detectable full-length molecules were detected (Fig. 5). These observations are consistent with stalling of replication forks at the same 340- and 77-kb barriers in all three tissues but with the probability of forks traversing these barriers before S phase ends differing, with forks being caught least often in nurse cells and most often in salivary glands.
Tissue-specific differences in the abundance of truncated DNAs, as discussed above for CH(3)336, appear to be a property of a given region of heterochromatin and not a consequence of tissue-specific differences in overall S-phase length, which would affect all regions of heterochromatin in the same way. For example, truncated DNAs were more abundant in follicle cells than nurse cells for heterochromatin assayed on the second and third chromosomes (Fig. 2 and 4) but the opposite was true for heterochromatin assayed on the Dp1187 chromosome, where truncated DNAs were more abundant in nurse cells than in follicle cells (Fig. 4). Therefore, whether a replication fork is able to complete replication or is left unresolved when S phase ends must be determined, at least in part, by more regional parameters, such as the location of origins used to replicate specific segments of heterochromatin or when those origins fire during S phase.
Underreplication in polytene chromosomes occurs primarily to satellite sequences (13, 19, 20). Complex sequences present in heterochromatin, such as islands of transposable elements, can replicate efficiently despite their heterochromatic location (1, 62). If replication barriers exist in heterochromatin, satellite sequences themselves could be the barriers that inhibit their own replication. At least some satellite sequences can have strong intrinsic curvature (33, 42), and some microsatellite repeats can cause stalling of DNA polymerases (44). Such properties multiplied over hundreds of contiguous kilobases of repetitious DNA might present a barrier to fork elongation. It is also possible that compaction of satellite sequences into heterochromatin could inhibit fork elongation. While the inability of PEV modifiers to influence underreplication suggests that heterochromatin structure does not play a role (Fig. 5), PEV modifiers have been identified based on their effects on transcription, not replication, and the components of heterochromatin that suppress transcription may not be the same as those that suppress replication. Furthermore, PEV modifiers have usually been selected as mutations that influence the ability of heterochromatin to act at a distance, typically across a euchromatic-heterochromatic breakpoint. The heterochromatin into which satellite sequences are packaged may not be as sensitive to PEV modifiers as the heterochromatin that “spreads” into normally euchromatic sequences. Finally, the size difference that was observed between truncated Dp1187 DNAs detected in follicle cells versus nurse cells (Fig. 4) is difficult to explain if the sizes of truncated DNAs were determined solely by the primary sequence, which is the same in both cell types. Such size differences are more easily explained if the barrier were a chromatin structure whose position relative to underlying sequence could differ between cells. Thus, satellite sequence, either directly or through their compaction into heterochromatin, may inhibit their own replication by blocking fork elongation.
The replication runoff model of polytene chromosome structure predicts that DNA molecules with double-stranded free ends are produced at sites of stalled replication forks (Fig. 6). Since polytene cells undergo repeated S phases despite experiencing incomplete DNA replication, as well as skipping mitosis, they clearly lack normal checkpoint controls (29, 52, 61) and therefore might accumulate DNA containing double-stranded free ends without triggering the cell cycle arrest that such structures would induce in diploid cells. In fact, using ligation-mediated PCR, truncated Dp1187 molecules have been cloned from ovarian nurse and follicle cells, and they have structures consistent with the presence in these polytene tissues of Dp1187 DNAs containing double-stranded free ends (B. Calvi and A. Spradling, personal communication). On occasion, polytene cells might still attempt to repair double-stranded free ends and, in so doing, might create ectopic fibers, a characteristic feature of polytene chromosomes. Nonhomologous end-joining reactions between free-ended DNAs at different loci would produce ectopic ligations between regions of the genome that were underreplicated but not necessarily homologous by sequences, both of which are characteristic features of ectopic fibers (64). Ectopic ligations would also create chimeric molecules more complex in structure than the simple truncated DNAs proposed in Fig. 6 and would complicate attempts to localize replication barriers, since the size of truncated DNAs would be determined not only by the position of fork arrest but also by structural changes introduced by ectopic ligation. Mapping of a series of truncated Dp1187 DNAs onto the known heterochromatic sequence structure of Dp1187 and assuming a simple truncation of DNA resulted in differing positions of putative replication barriers depending on the restriction enzyme used for the analysis (15). Perhaps the source of this ambiguity is, in part, a consequence of ectopic ligations.
Heterochromatic replication origins.
Sequences flanking the CH(3)336 P element were fully replicated in polytene chromosomes despite their heterochromatic location, and the level of replication was unaffected by the Y chromosome (Fig. 5) (62). Even though replication is not suppressed, sequences flanking the P element are likely to be compacted into heterochromatin, since expression of the rosy gene present on the CH(3)336 P element is subject to strong heterochromatin-induced suppression (63). This contrasts to the Dp1187 chromosome, whose replication is both suppressed by heterochromatin and sensitive to modulation by the Y chromosome (Fig. 5) (25). Why should replication in these two chromosomes differ with respect to their sensitivity to heterochromatin? The answer may be that sequences flanking the CH(3)336 P element occur normally within heterochromatin (63), while sequences of Dp1187 contain an abnormal euchromatic-heterochromatic junction (27). Origins that replicate the heterochromatic sequences flanking the CH(3)336 P element may be specifically adapted to function in heterochromatin, analogous to heterochromatic genes, which must be located in heterochromatin for proper expression (60). On the other hand, Dp1187 may be replicated by euchromatic origins that are suppressed by their proximity to heterochromatin on the rearranged chromosome, analogous to the suppression of euchromatic genes by heterochromatin-induced PEV. The existence of replication origins specifically adapted to a heterochromatic environment is supported by the replication behavior of the light gene. light is fully replicated in polytene chromosomes despite its location within pericentromeric heterochromatin, but when it is placed into a euchromatic environment by chromosome inversion, its replication is suppressed consistent with the presence near light of a replication origin requiring a heterochromatic environment for function (21a).
Implications for diploid cells.
If replication barriers exist in diploid cells such that stalled replication forks occur normally when cells replicate heterochromatic sequences, this would have interesting implications for the biology of heterochromatin. For example, heterochromatic sequences are characteristically the last portion of the genome to be replicated (31). The mechanism of late replication is not known, but origins responsible for replicating heterochromatin might be intrinsically late firing or might be located so far into euchromatin that passive replication of heterochromatic sequences does not occur until late S phase (53). If replication barriers exist in heterochromatin, however, inhibited fork elongation could also cause late replication. Elongating replication forks might arrive at heterochromatin in early S or mid-S phase but might be unable to elongate through heterochromatic barriers until late S phase, when a stochastic process or a specific cellular event finally allows replication to be completed. In this scenario, such barriers would have a significant impact on the replication timing of specific regions of heterochromatin, and replication timing may play an important role in centromere formation, particularly in relation to the temporal expression pattern of the centromere-specific histone variant CENP-A/Cse4 (9, 23, 48).
By their nature, replication forks cause dramatic changes in the structure of both DNA and chromatin. Stalled forks may even create transient double-stranded free ends via isomerization of the replication fork and annealing of newly synthesized DNA strands (2, 10, 35). If stalled forks are common within heterochromatin, the structural perturbations they create could make heterochromatin prone to DNA rearrangements. For example, stalled forks could create preferred insertion sites for transposable elements (5, 36, 55, 59) and could explain, in part, why heterochromatic domains characteristically contain a high density of transposons (7, 39, 40, 54). Structural polymorphisms are also characteristic of heterochromatic domains (57) and might arise by unequal exchange within satellite repeats when a cell misinterprets a fork stalled at a heterochromatic barrier as evidence of DNA damage and attempts to repair the “lesion” by homologous recombination.
Given the unique characteristics of heterochromatin, it is perhaps not unexpected that the way in which cells replicate heterochromatin might differ in important aspects from the way in which they replicate euchromatin. Studying those differences will provide important insights into the biology of heterochromatin.
ACKNOWLEDGMENTS
We are grateful to A. Spradling, P. Zhang, and G. Karpen for fly strains and to J. Lock for zfh-2 DNA. We acknowledge the Wadsworth Center Cellular Immunology and Molecular Genetics Core Facilities for assistance with flow cytometry and contour-clamped homogeneous electric field electrophoresis, respectively.
This work was supported by grant GM53476 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Berghella L, Dimitri P. The heterochromatic rolled gene of Drosophila melanogster is extensively polytenized and transcriptionally active in the salivary gland chromocenter. Genetics. 1996;144:117–125. doi: 10.1093/genetics/144.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidnenko V, Seigneur M, Penel-Colin M, Bouton M-F, Ehrlich S D, Michel B. sbcB sbcC null mutations allow RecF-mediated repair of arrested replication forks in rep recBC mutants. Mol Microbiol. 1999;33:846–857. doi: 10.1046/j.1365-2958.1999.01532.x. [DOI] [PubMed] [Google Scholar]
- 3.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown K E, Guest S S, Smales S T, Hahm K, Merkenschlager M, Fisher A G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Greenblatt I M, Dellaporta S L. Molecular analysis of Ac transposition and DNA replication. Genetics. 1992;130:665–676. doi: 10.1093/genetics/130.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copenhaver G P, Nickel K, Kuromori T, Benito M-I, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell L D, McCombie W R, Martienssen R A, Marra M, Preuss D. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 8.Cryderman D E, Cuaycong M H, Elgin S C, Wallrath L L. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma. 1998;107:277–285. doi: 10.1007/s004120050309. [DOI] [PubMed] [Google Scholar]
- 9.Csink A K, Henikoff S. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- 10.Defossez P-A, Prusty R, Kaeberlein M, Lin S-J, Ferrigno P, Silver P A, Keil R L, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 11.Dickson E, Boyd J B, Laird C D. Sequence diversity of polytene chromosome DNA from Drosophila hydei. J Mol Biol. 1971;61:615–627. doi: 10.1016/0022-2836(71)90067-2. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg A P, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 13.Gall J G, Cohen E H, Polan M L. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33:319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- 14.Glaser R L, Karpen G H, Spradling A C. Replication forks are not found in a Drosophila minichromosome demonstrating a gradient of polytenization. Chromosoma. 1992;102:15–19. doi: 10.1007/BF00352285. [DOI] [PubMed] [Google Scholar]
- 15.Glaser R L, Leach T J, Ostrowski S E. The structure of heterochromatic DNA is altered in polyploid cells of Drosophila melanogaster. Mol Cell Biol. 1997;17:1254–1263. doi: 10.1128/mcb.17.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser R L, Spradling A C. Unusual properties of genomic DNA molecules spanning the euchromatic-heterochromatic junction of a Drosophila minichromosome. Nucleic Acids Res. 1994;22:5068–5075. doi: 10.1093/nar/22.23.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenfeder S A, Newlon C S. Replication forks pause at yeast centromeres. Mol Cell Biol. 1992;12:4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 19.Hammond M P, Laird C D. Chromosome structure and DNA replication in nurse and follicle cells of Drosophila melanogaster. Chromosoma. 1985;91:267–278. doi: 10.1007/BF00328222. [DOI] [PubMed] [Google Scholar]
- 20.Hammond M P, Laird C D. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma. 1985;91:279–286. doi: 10.1007/BF00328223. [DOI] [PubMed] [Google Scholar]
- 21.Henlkoff S. Position-effect variegation in Drosophila: recent progress. In: Russo V E A, Martienssen R A, Riggs A D, editors. Epigenetic mechanisms of gene regulation. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 319–334. [Google Scholar]
- 21a.Howe M. The cis-effects of heterochromatin on gene activity and polyteny in Drosophila melanogaster. Ph.D. thesis. Seattle: University of Washington; 1999. [Google Scholar]
- 22.John B. The biology of heterochromatin. In: Verma R S, editor. Heterochromatin: molecular and structural aspects. Cambridge, United Kingdom: Cambridge University Press; 1988. pp. 1–147. [Google Scholar]
- 23.Karpen G H, Allshire R C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 24.Karpen G H, Spradling A C. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpen G H, Spradling A C. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990;63:97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laird C D. DNA of Drosophila chromosomes. Annu Rev Genet. 1973;7:177–204. doi: 10.1146/annurev.ge.07.120173.001141. [DOI] [PubMed] [Google Scholar]
- 27.Le M-H, Duricka D, Karpen G H. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics. 1995;141:283–303. doi: 10.1093/genetics/141.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leach T J, Glaser R L. Quantitative hybridization to genomic DNA fractionated by pulsed-field gel electrophoresis. Nucleic Acids Res. 1998;26:4787–4789. doi: 10.1093/nar/26.20.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehner C F, O'Farrell P H. The roles of Drosophila cyclins A and B in mitotic control. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilly M A, Spradling A C. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- 31.Lima de Faria A, Jaworska H. Late DNA synthesis in heterochromatin. Nature. 1968;217:138–142. doi: 10.1038/217138a0. [DOI] [PubMed] [Google Scholar]
- 32.Locke J, McDermid H E. Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma. 1993;102:718–723. doi: 10.1007/BF00650898. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Balbas A, Rodriguez-Campos A, Garcia-Ramirez M, Sainz J, Carrera P, Aymami J, Azorin F. Satellite DNAs contain sequences that induce curvature. Biochemistry. 1990;29:2342–2348. doi: 10.1021/bi00461a019. [DOI] [PubMed] [Google Scholar]
- 34.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;44:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 35.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore J K, Haber J E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 37.Murphy T D, Karpen G H. Localization of centromere function in a Drosophila minichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 39.Neitzel H, Kalscheuer V, Henschel S, Digweed M, Sperling K. Beta-heterochromatin in mammals: evidence from studies in Microtus agrestis based on the extensive accumulation of L1 and non-L1 retroposons in the heterochromatin. Cytogenet Cell Genet. 1998;80:165–172. doi: 10.1159/000014974. [DOI] [PubMed] [Google Scholar]
- 40.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Transposable elements are stable structural components of Drosophila melanogaster. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platero J S, Hartnett T, Eissenberg J C. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radic M Z, Lundgren K, Hamkalo B A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987;50:1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- 43.Rudkin G T. Non-replicating DNA in Drosophila. Genetics. 1969;61:227–238. [PubMed] [Google Scholar]
- 44.Samadashwily G M, Raca G, Mirkin S M. Trinucleotide repeats affect DNA replication in vivo. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sauer K, Knoblich J A, Richardson H, Lehner C F. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 1995;9:1327–1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- 47.Scott R S, Truong K Y, Bos J-M H. Replication initiation and elongation fork rates within a differentially expressed human multicopy locus in early S phase. Nucleic Acids Res. 1997;25:4505–4512. doi: 10.1093/nar/25.22.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelby R D, Vafa O, Sullivan K F. Assembly of CENP-A into centromreic chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spofford J B. Position-effect variegation in Drosophila. In: Ashburner M, Novitski E, editors. Genetics and biology of Drosophila. 1C. London, United Kingdom: Academic Press, Ltd.; 1976. pp. 955–1018. [Google Scholar]
- 50.Spradling A C. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. I. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- 51.Spradling A C, Karpen G, Glaser R, Zhang P. Evolutionary conservation of developmental mechanisms: DNA elimination in Drosophila. In: Spradling A, editor. 50th Symposium of the Society of Developmental Biology. New York, N.Y: Wiley-Liss; 1993. pp. 39–53. [Google Scholar]
- 52.Stern B, Ried G, Clegg N J, Grigliatti T A, Lehner C F. Genetic analysis of the Drosophila cdc2 homolog. Development. 1993;117:219–232. doi: 10.1242/dev.117.1.219. [DOI] [PubMed] [Google Scholar]
- 53.Stevenson J B, Gottschling D E. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun X, Wahlstrom J, Karpen G. Molecular Structure of a functional Drosophila centromere. Cell. 1997;91:1007–1019. doi: 10.1016/s0092-8674(00)80491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teng S-C, Kim B, Gabriel A. Retrotransposon reverse transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 56.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 57.Verma R S. Heteromorphisms of heterochromatin. In: Verma R S, editor. Heterochromatin: molecular and structural aspects. Cambridge, United Kingdom: Cambridge University Press; 1988. pp. 276–292. [Google Scholar]
- 58.Wallrath L L. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 59.Warbrick E, Heatherington W, Lane D P, Glover D M. PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res. 1998;26:3925–3932. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiler K, Wakimoto B. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 61.Whitfield W, Gonzalez C, MacDonald-Codina G, Glover D. The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition. EMBO J. 1990;9:2563–2572. doi: 10.1002/j.1460-2075.1990.tb07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang P, Spradling A C. The Drosophila salivary gland chromocenter contains highly polytenized subdomains of mitotic heterochromatin. Genetics. 1995;139:659–670. doi: 10.1093/genetics/139.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang P, Spradling A C. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc Natl Acad Sci USA. 1994;91:3539–3543. doi: 10.1073/pnas.91.9.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhimulev I F. Polytene chromosomes, heterochromatin, and position effect variegation. Adv Genet. 1998;37:1–566. doi: 10.1016/s0065-2660(08)60341-7. [DOI] [PubMed] [Google Scholar]