ABSTRACT
Autophagy is thought to be involved in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. However, how SARS-CoV-2 interferes with the autophagic pathway and whether autophagy contributes to virus infection in vivo is unclear. In this study, we identified SARS-CoV-2-triggered autophagy in animal models, including the long-tailed or crab-eating macaque (Macaca fascicularis), human angiotensin-converting enzyme 2 (hACE2) transgenic mice, and xenografted human lung tissues. In Vero E6 and Huh-7 cells, SARS-CoV-2 induces autophagosome formation, accompanied by consistent autophagic events, including inhibition of the Akt-mTOR pathway and activation of the ULK-1-Atg13 and VPS34-VPS15-Beclin1 complexes, but it blocks autophagosome-lysosome fusion. Modulation of autophagic elements, including the VPS34 complex and Atg14, but not Atg5, inhibits SARS-CoV-2 replication. Moreover, this study represents the first to demonstrate that the mouse bearing xenografted human lung tissue is a suitable model for SARS-CoV-2 infection and that autophagy inhibition suppresses SARS-CoV-2 replication and ameliorates virus-associated pneumonia in human lung tissues. We also observed a critical role of autophagy in SARS-CoV-2 infection in an hACE2 transgenic mouse model. This study, therefore, gives insights into the mechanisms by which SARS-CoV-2 manipulates autophagosome formation, and we suggest that autophagy-inhibiting agents might be useful as therapeutic agents against SARS-CoV-2 infection.
IMPORTANCE Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic with limited therapeutics. Insights into the virus-host interactions contribute substantially to the development of anti-SARS-CoV-2 therapeutics. The novelty of this study is the use of a new animal model: mice xenografted with human lung tissues. Using a combination of in vitro and in vivo studies, we have obtained experimental evidence that induction of autophagy contributes to SARS-CoV-2 infection and improves our understanding of potential therapeutic targets for SARS-CoV-2.
KEYWORDS: autophagy, animal model, SARS-CoV-2, 3-MA, VPS34
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), and its rapid global spread resulted in a pandemic with more than 180 million laboratory-confirmed cases by the end of June 2021 (1, 2). The most common symptoms caused by SARS-CoV-2 are fever, fatigue, and a dry cough, while patients with underlying diseases may develop severe complications, including severe acute respiratory distress syndromes and shock (3, 4). Therefore, therapeutics capable of effectively treating this infection are urgently needed, and insights into the molecular mechanisms of SARS-CoV-2 infection would contribute to the development of treatments for infected individuals.
Autophagy has been implicated in virus-host interactions, and coronavirus has been reported to be capable of hijacking the endoplasmic reticulum-Golgi intermediate compartment to form viral proteins and complete virions, which are associated with autophagy (5). Autophagy is an evolutionarily conserved cellular process involving the formation of double-membrane-bound autophagosomes that enclose flawed or damaged cytoplasmic cargoes, including long-lived proteins and defective organelles. These autophagosomes eventually fuse with lysosomes to generate autolysosomes, where the phagocytosed material is degraded (6–8). Nonstructural protein 6 (NSP6) of SARS-CoV, which has been classified in the same genus as the SARS-CoV-2 and Middle East respiratory syndrome coronavirus (MERS-CoV), promotes the generation of autophagosomes but restricts their expansion and maturation into autolysosomes (9, 10). In addition, recent studies have reported that the accessary protein ORF3a of SARS-CoV-2 also blocked the fusion of autophagosomes and lysosomes in host cells via its binding with homotypic fusion and protein sorting (HOPS) complexes (11, 12). Evidence showed that SARS-CoV-2 increased the number of autophagosomes and the autophagy inhibitor 3-MA inhibited virus replication in vitro (13). Nevertheless, how SARS-CoV-2 acts on the signaling pathway involved in autophagosome formation remains unknown, and no animal study related to autophagy function in SARS-CoV-2 infection has been reported.
A high-throughput screening study showed that autophagy modulators might be potential drug candidates for SARS-CoV-2 therapy (14). Based on a WHO report from March 2020, 12 FDA-approved drugs have been repurposed and clinically trialed as potential COVID-19 treatments. (15, 16). Among them, six drugs have been reported as autophagy regulators, for example, chloroquine (CQ)/hydrochloroquine, highlighting the need for further research in this area (6). CQ and hydrochloroquine have previously been found to regulate autophagy at its late stage, by the inhibition of autolysosome formation, have been proven to be effective antivirals against SARS-CoV-2 in vitro, and have been previously used as first-line drugs for the treatment of SARS-CoV-2 in several clinical studies (17–21). However, a study in nonhuman primates has revealed that hydrochloroquine does not inhibit SARS-CoV-2 infection (22). Ultimately, it was found to have no beneficial effects on mortality rates in patients hospitalized with COVID-19 (23). This event emphasizes the need for adequate preclinical studies in different animal models to investigate how autophagy contributes to SARS-CoV-2 infection.
Here, we report results from SARS-CoV-2 infection conducted on established animal models, Macaca fascicularis and human angiotensin-converting enzyme 2 (hACE2)-transgenic mice, in addition to the original SARS-CoV-2 infection model based on human lung xenografts in mice (24–26). Mice engrafted with fetal human lung tissue that is capable of developing mature pulmonary structure represent a powerful tool for clinical translation of drug candidates and has been reported before, for example, in Nipah virus infection, which causes severe and often lethal respiratory damage in humans (27). Here, we report enhanced autophagy in SARS-CoV-2-infected human lung transplants, lung tissues of the Macaca fascicularis, and lung tissues of hACE2 transgenic mice. Moreover, we found that SARS-CoV-2 induces autophagosome formation, accompanied by consistent autophagic events, including inhibition of the Akt-mTOR pathway, and activation of the ULK-1-Atg13 and VPS34-VPS15-Beclin1 complexes, but blocks autophagosome-lysosome fusion. Finally, we provide evidence that autophagy initiation is critical for SARS-CoV-2 infection and inhibition of autophagy suppresses viral replication and improves lung injury in vivo.
RESULTS
SARS-CoV-2 activates autophagy in lesioned lung lobes of Macaca fascicularis.
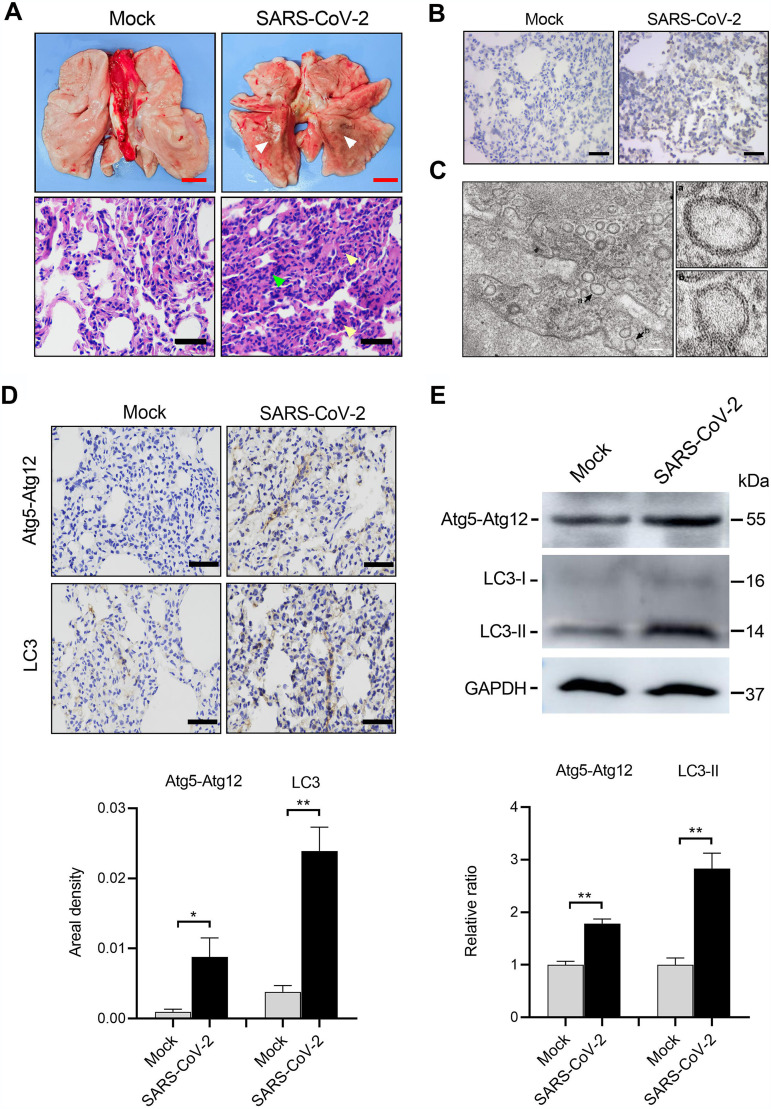
Recently, nonhuman primates have been established as effective animal models that reflect pathological changes caused by SARS-CoV-2 infection (24, 28). Macaca fascicularis macaques were intratracheally mock treated or infected with 1.3 × 106 median tissue culture infectious doses (TCID50) of SARS-CoV-2 and sacrificed at 7 days postinfection (dpi). Lung tissues from the SARS-CoV-2-infected monkeys displayed large areas of red dark lesions in the lower bilateral lobes, as well as patchy hemorrhaging (Fig. 1A). Studies looking at microscopic hematoxylin and eosin (H&E) staining revealed diffuse thickening of the alveolar septum with infiltration of segmented granulocyte cells and damaged alveolar structure, indicating interstitial pneumonia (Fig. 1A). In situ hybridization showed positive signaling in the virus-infected tissues (Fig. 1B). SARS-CoV-2 infection was further corroborated by electron microscopy observation of substantial viral replication structures of double-membrane vesicles (DMV) (Fig. 1C). The lung tissues with severe lesions from SARS-CoV-2-infected animals and corresponding areas from mock-infected lungs were collected and analyzed for autophagy. Atg5-Atg12 and LC3-II proteins are critical components of the phagophore or autophagosome membrane. Elevated expression levels of Atg5-Atg12 and LC3-I/LC3-II (indicative of autophagy activation) after SARS-CoV-2 infection were confirmed by immunohistochemistry (Fig. 1D) and Western blotting (Fig. 1E), compared to those in mock-infected lungs.
FIG 1.
SARS-CoV-2 activates autophagy in lesioned lung lobes of the Macaca fascicularis. Macaca fascicularis macaques were intratracheally mock infected (n = 3) or infected (n = 3) with 1.3 × 106 TCID50 of SARS-CoV-2 and then sacrificed at 7 dpi. (A) Gross pathology and histopathology of the lungs from mock- or SARS-CoV-2-infected monkeys. A large area of dark red lesions was observed on the virus-infected lung surface (white arrowheads). H&E staining showed inflammatory cell infiltration (yellow arrowheads) and thickened alveolar walls (green arrowheads) after SARS-CoV-2 infection. (B) Representative images of in situ hybridization for SARS-CoV-2 RNA in the lung tissues. (C) Electron microscope images of virus replication complexes (black arrows). (D and E) Autophagy activation was examined by immunohistochemistry staining (D) and Western blotting (E), and increased Atg5-Atg12 and LC3-I/LC3-II expressions were observed in the lung tissues from SARS-CoV-2-infected Macaca fascicularis. Data were expressed as means ± SEM from three experiments. *, P < 0.05; **, P < 0.01. Red scale bars = 1 cm, black scale bars = 50 μm, and white scale bars = 500 nm.
SARS-CoV-2 increases autophagosome formation but blocks autophagosome-lysosome fusion.
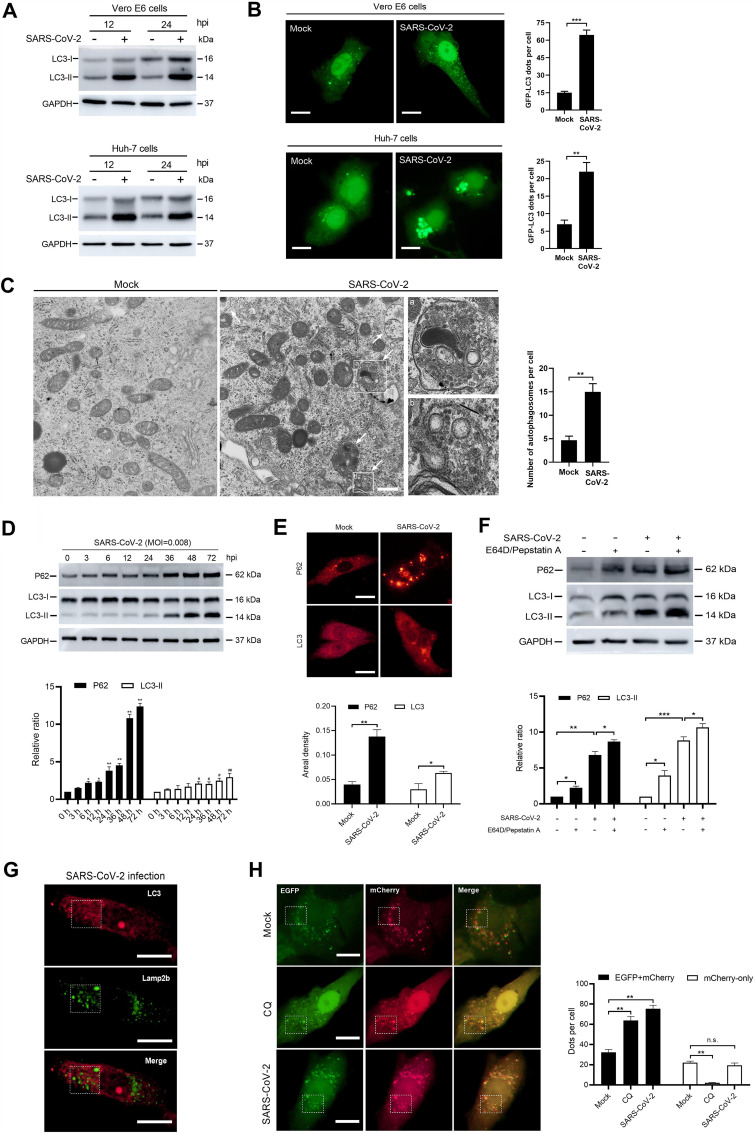
LC3-I is predominantly located in the cytoplasm and is converted into an active phosphatidylethanolamine-conjugated form, LC3-II, upon activation of autophagy. LC3-II incorporates into the extending phagophore membrane and controls its elongation into complete autophagosomes, thereby making LC3-II a reliable autophagosomal marker (29, 30). Western blotting assays displayed a marked increase in the expression levels of LC3-II at 12 and 24 h after SARS-CoV-2 infection in both Vero E6 and Huh-7 cells (Fig. 2A), suggesting an increased number of autophagosomes caused by SARS-CoV-2. Autophagosomes represent the intermediate vesicular compartment during autophagy flux, which includes cargoes for transportation into lysosomes, and degradation (29). To directly observe the accumulation of autophagosomes, Vero E6 and Huh-7 cells were transfected with green fluorescent protein (GFP)-LC3, which caused them to become decorated with GFP-LC3. We found that SARS-CoV-2 increased GFP-LC3 puncta in both Vero E6 and Huh-7 cells at 24 h postinfection (hpi) (Fig. 2B). Using electron microscopy, we further examined Vero E6 cells treated with vehicle or SARS-CoV-2 at 6 hpi and found an increased number of double-membrane autophagosomes in the Vero E6 cells infected with live virus compared to that in noninfected cells (Fig. 2C).
FIG 2.
SARS-CoV-2 increases autophagosome formation but blocks autophagosome-lysosome fusion. Vero E6 and Huh-7 cells were treated with a vehicle or SARS-CoV-2 at an MOI of 0.008. (A) LC3 expression at 12 and 24 hpi in Vero E6 and Huh-7 cells. Samples were analyzed by Western blotting. (B) GFP-LC3 accumulation in mock- and SARS-CoV-2-infected Vero E6 and Huh-7 cells at 24 hpi. Cells were transfected with GFP-LC3 plasmids and analyzed for autophagosome accumulation. Theamount of GFP-LC3 per cell was calculated from at least 30 cells from each group. Scale bar = 5 μm. (C) Electron microscope images of autophagosomes (white arrows) in Vero E6 cells at 6 hpi. The number of autophagosomes per cell was calculated from at least 10 cells. Scale bar = 500 nm. (D) p62 and LC3 expression in Vero E6 cells at indicated time points after SARS-CoV-2 infection. Samples were analyzed by Western blotting. (E) Immunofluorescence analysis of p62 and LC3 in mock- or SARS-CoV-2-infected Vero E6 cells at 24 hpi. The areal density of p62 and LC3 was calculated from at least 30 cells for each group. Scale bar = 5 μm. (F) Western blotting of p62 and LC3 expression in Vero E6 cells that had been treated or not for 2 h with E64D (10 μg/ml) and pepstatin A (10 μg/ml) before mock or SARS-CoV-2 infection. Samples were collected at 24 hpi. (G) Immunofluorescence analysis of LC3 and lamp2b colocalization in SARS-CoV-2-infected Vero E6 cells at 24 hpi. Scale bar = 5 μm. (H) Colocalization analysis of EGFP and mCherry in mock-infected, CQ-treated, and SARS-CoV-2-infected Vero E6 cells, pretransfected with EGFP-mCherry-LC3 plasmids. In the CQ group, 40 μM drug was added to the cells 2 h before infection. Samples were collected at 24 hpi. The numbers of EFGP-mCherry (yellow) and mCherry-only dots were calculated from at least 30 cells from each group. Scale bar = 5 μm. Data were expressed as means ± SEM from three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant.
The accumulation of autophagosomes can result from the following two events during autophagic flux: (i) increased biogenesis of complete autophagosomes and (ii) reduced fusion of autophagosomes and lysosomes (29, 31). Recent studies have shown that SARS-CoV-2 blocks autophagosome maturation into autolysosomes (11, 12). In accordance, our results displayed an increased abundance of both p62 and LC3 proteins in SARS-CoV-2-infected Vero E6 cells over time (Fig. 2D), suggesting impaired autophagic degradation of p62 substrates, which serve as a readout for clearance of autophagic cargo (29). Immunofluorescence analysis showed a similar rise in p62 and LC3 expression at 24 hpi (Fig. 2E). In addition, no colocalization of LC3 and lamp2b, a biomarker for lysosomes, was observed in the SARS-CoV-2-infected Vero E6 cells at 24 hpi (Fig. 2G), indicating that SARS-CoV-2 impedes the fusion of autophagosomes with lysosomes.
Next, we examined whether the formation of autophagosomes was also triggered by SARS-CoV-2 infection. CQ and E64D/pepstatin A can block autophagic flux at the terminal stages and prevent lysosomal degradation by inhibiting lysosomal acidification and protease function, respectively. These compounds lead to increased accumulation of p62 and LC3 aggregates which are dependent upon extant, complete autophagic events (29, 32). Western blotting showed higher levels of both p62 and LC3-II in SARS-CoV-2-infected Vero E6 cells pretreated with E64D/pepstatin A compared to those in cells treated with phosphate-buffered saline (PBS) (Fig. 2F), suggesting that SARS-CoV-2 infection also triggered autophagosome formation in addition to blocking its maturation. Finally, we further monitored the movement of LC3 from autophagosomes to lysosomes using enhanced GFP (EGFP)-mCherry-LC3-transfected Vero E6 cells. The EGFP signal from the mCherry-EGFP-LC3 fusion protein is sensitive to acidic pH compared to the mCherry fluorophore and is quenched under the low-pH conditions within the autolysosomes (32). We found that SARS-CoV-2 infection markedly increased yellow (EGFP-mCherry) puncta in infected Vero E6 cells at 24 hpi compared to those in mock-infected cells (Fig. 2H), indicating an increased autophagosome accumulation caused by SARS-CoV-2 infection. In addition, despite a rise in yellow puncta, no significant decrease in mCherry-only signal was observed in SARS-CoV-2-infected cells compared to mock-infected ones, while such mCherry-only puncta were hardly seen in the CQ-treated cells (Fig. 2H), suggesting that SARS-CoV-2 induced formation of autophagosomes.
Autophagosome formation induced by SARS-CoV-2 promotes virus replication via the VPS34 complex.
To further elucidate the mechanism by which SARS-CoV-2 can induce autophagosome formation, we analyzed a series of signaling cascades involved in autophagosome formation, notably, the mTORC1 signaling pathway, representing the most common upstream autophagy suppression regulator (29). The activity of mTOR can be monitored by the phosphorylation of its downstream substrates, including 4E-BP1, p70S6 kinase, and the ribosomal protein S6 (33, 34). In Vero E6 cells infected with SARS-CoV-2, the relative ratio of p-mTOR, p-4E-BP1 and pS6 to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were reduced over time (Fig. 3A), suggesting attenuated mTOR signaling. Adenosine 5′-monophosphate-activated protein kinase (AMPK) and Akt are two of the upstream control points of mTOR signaling. While AMPK suppresses mTOR activation, Akt produces the opposite effect (30). Western blotting showed a corresponding decrease in p-Akt abundance in Vero E6 cells after SARS-CoV-2 infection (Fig. 3A), given the inhibited mTOR activity, while the expressions of p-AMPK and its downstream substrates, such as p-TSC-2 and p-Raptor, were downregulated after SARS-CoV-2 infection (Fig. 3B), suggesting the inhibition of Akt-mTOR pathway by SARS-CoV-2.
FIG 3.
Autophagy induced by SARS-CoV-2 promotes virus replication in Vero E6 cells via the VPS34 complex. SARS-CoV-2-infected (MOI, 0.008) Vero E6 cells were harvested at different time points (0 h, 3 h, 6 h, 12 h, 24 h, 36 h, 48 h, and 72 h) after infection. (A to F) Alterations in the signaling pathway involved in the cellular autophagy machinery were analyzed by Western blotting, and this included the Akt-mTOR pathway (A), AMPK-TSC2/Raptor pathway (B), p-ULK1 (Ser 757), p-Atg13 (Ser 355), Atg13 proteins (C), VPS34-VPS15-Beclin1 complex (D), Atg14 protein (E), and phagophore and autophagosome membrane-associated proteins (F). (G) Viral load in SARS-CoV-2-infected Vero E6 cells pretreated with SAR405 (1 μM) or 3-MA (5 mM). Cell samples were collected at 0 h, 24 h, 48 h, and 72 h after infection and analyzed by RT-qPCR. (H) Viability of Vero E6 cells treated with 3-MA after SARS-CoV-2 infection (MOI, 0.008) for 72 h. (I) Western blotting of Atg14 knockdown efficiency in Vero E6 cells. (J) Viral load in SARS-CoV-2-infected Vero E6 cells transfected with control or Atg14 siRNA. Samples were harvested at 24 hpi. (K) Western blotting of alterations in Atg5-related proteins in Atg5−/− Vero E6 cells. (L) Viral load in SARS-CoV-2-infected Atg5+/+ and Atg5−/− Vero E6 cells at 24 hpi. Data were expressed as means ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we examined the downstream events associated with mTOR inhibition. The dephosphorylation of Atg1/ULK1 at Ser 757 caused by mTOR inactivation can initiate autophagy (29). Activated ULK1 phosphorates multiple downstream targets that function in autophagy flux, such as Atg13 at Ser 355 and Beclin-1 at Ser 15. As shown in Fig. 3C and D, dephosphorylation of ULK1 at Ser 757, as well as phosphorylation of Atg13 at Ser 355 and Beclin-1 at Ser15, was observed in SARS-CoV-2-infected Vero E6 cells. These results suggest phosphorylation events caused by the active form of ULK1 and mTOR inhibition.
In mammalian cells, VPS34 is thought to exert its effect downstream of mTOR and the ULK1 complex, and it appears to be the only class III phosphatidylinositol-3-phosphate (PI3P) kinase to generate the majority of PI3P, which is necessary for phagophore elongation (29, 35). VPS34 also facilitates the formation of the Atg5-Atg12-Atg16L1 complex, which represents components of the phagophore membrane (36). To determine whether VPS34 functions in SARS-CoV-2-induced autophagy, we examined the core VPS34 complex (Vps34-Vps15-Beclin1) and found that the expression of VPS34, VPS15, p-Beclin1 (Ser 15), and Beclin1, as well as Atg5-Atg12 and Atg16L1, was upregulated over time up to 72 hpi in SARS-CoV-2-infected Vero E6 cells (Fig. 3D and F), indicating activation of the VPS34-VPS15-Beclin1 complex by SARS-CoV-2 infection. We further measured the expression of Atg14, which can enhance the stability of Beclin1 and act as a regulatory subunit in the VPS34-VPS15-Beclin1-Atg14 complex, promoting phagophore formation (36, 37). We found that in SARS-CoV-2-infected Vero E6 cells, the abundance of Atg14 first increased up to 36 hpi but then decreased thereafter, which is inconsistent with the VPS34 complex, indicating a temporary autophagy-inducing function of the Atg14-containing VPS34 complex upon SARS-CoV-2 infection.
After an overall detection of SARS-CoV-2-induced signaling cascades related to autophagy induction, we sought to determine whether activation of autophagy contributes to virus infection. We first inhibited initiation of autophagy by a widely used autophagy inhibitor, 3-MA, which persistently inhibits class I PI3P kinase and temporarily inhibits VPS34 activity. 3-MA (5 mM) was used to treat Vero E6 cells for 2 h prior to SARS-CoV-2 infection, and reverse transcription-PCR (RT-PCR) analysis showed that the viral load was significantly reduced at 24, 48, and 72 hpi compared to that in the mock-treated group (Fig. 3G), indicating that autophagy activation was involved in SARS-CoV-2 replication. In addition, 3-MA (0.3125 mM, 0.625 mM, and 1.25 mM) improved viability of Vero E6 cells after SARS-CoV-2 infection for 72 h (Fig. 3H), suggesting that autophagy-associated cell death was caused by SARS-CoV-2.
To further determine the key elements involved in virus replication, SAR405 (1 μM), a specific VPS34 inhibitor, was used to treat cells for 2 h prior to SARS-CoV-2 infection, and RT-PCR analysis showed that the viral load was significantly reduced at 24, 48, and 72 hpi in Vero E6 cells compared to that in the mock-treated group (Fig. 3G), indicating that autophagy and VPS34 activation were required for SARS-CoV-2 replication. We also consistently found that knockdown of Atg14 by siRNA reduced virus copy number in SARS-CoV-2-infected Vero E6 cells at 24 hpi (Fig. 3I and J), suggesting a positive role for the Atg14-containing VPS34 complex in SARS-CoV-2 infection. However, in Atg5−/− Vero E6 cells, the viral load stayed at the same level as that seen with the Atg5+/+ cells (Fig. 3K and L), suggesting that the SARS-CoV-2-induced formation of Atg5-containing complexes was not associated with virus replication.
Taken together, these findings suggest that SARS-CoV-2 infection inhibited the Akt-mTOR pathway and further dephosphorylated ULK1 to an active form which may be able to initiate autophagy. In addition, the VPS34-VPS15-Beclin1 complex increased in functions which may facilitate elongation of phagophore membranes and autophagosome formation, and VPS34 complex is closely involved in SARS-CoV-2 replication.
Autophagy inhibition suppresses SARS-CoV-2 replication and ameliorates pneumonia in hACE2 transgenic mice.
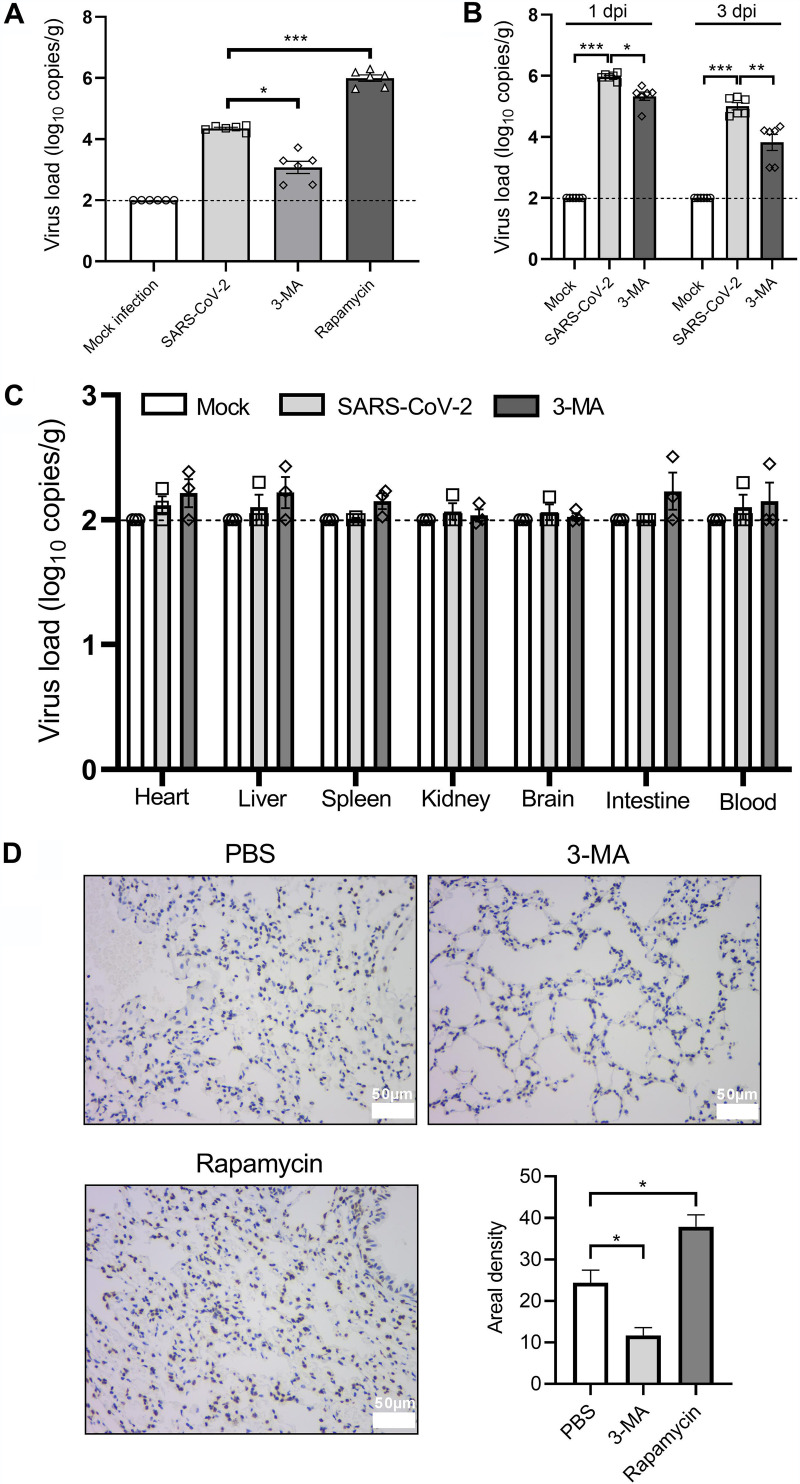
Transgenic mice expressing the hACE2 receptor driven by the mouse ACE2 promoter are a well-established animal model for SARS-CoV-2 infection (26). Transgenic mice expressing hACE2 were intratracheally treated with a vehicle or infected with 104 TCID50 of SARS-CoV-2 and sacrificed at 1, 3, and 5 dpi. Immunohistochemistry analysis showed increased abundance of LC3 in lung tissues from the SARS-CoV-2-infected hACE2 transgenic mice on 5 dpi (Fig. 4A), indicating an induction of autophagy. To further determine the role of autophagy in SARS-CoV-2 infection in vivo, we intraperitoneally treated SARS-CoV-2-infected mice with 3-MA (30 mg/kg of body weight, once daily for 6 consecutive days) to block autophagy and rapamycin (4 mg/kg, once daily for 6 consecutive days) to trigger autophagy. The infected mice displayed severe dark red lesions in large parts of the inferior lobe of the left lung and local hemorrhaging in the inferior lobe of the right lung at 5 dpi (Fig. 4B). 3-MA alleviated these pneumonia-like symptoms, as opposed to the autophagy-inducing agent rapamycin, which further aggravated lung lesions (Fig. 4B). Similar effects were observed at the microscopic level where 3-MA ameliorated the pneumonia-associated pathological changes in SARS-CoV-2-infected lung tissues, including necrosis and abscission of the bronchiolar endothelial cells, peribronchiolar and perivascular infiltration of segmented granulocytes, and thickened alveolar septum with inflammatory exudation and proliferation of fibroblasts, as well as severe destruction of alveolar structure (Fig. 4D to F and G). In contrast, rapamycin increased the pathological changes in the lung tissue associated with SARS-CoV-2 infection (Fig. 4H). Neither SARS-CoV-2 infection nor drug treatment had any effect on body weight in the hACE2 transgenic mice (Fig. 4C). It is worth noting that among these mice, all infected animals remained alive. The mild disease progression may be due to the relatively low levels of hACE2 expression driven by the mouse ACE2 promoter compared to other types of promoters that control hACE2 expression in transgenic mice, including human cytokeratin 18 (K18) and hepatocyte nuclear factor-3/forkhead homologue 4 (HFH4) promoters (38–41). Since the death rate is relatively low in COVID-19 patients, this more resistant hACE2 transgenic mouse, still with significant pathological symptoms, is useful to understand the pathogenesis of SARS-CoV-2 (42).
FIG 4.
Autophagy inhibition ameliorates pneumonia in hACE2 transgenic mice. hACE2 transgenic mice (n = 6) were intratracheally inoculated with 104 TCID50 of SARS-CoV-2 and intraperitoneally administered either PBS, 3-MA (30 mg/kg), or rapamycin (4 mg/kg) for 6 consecutive days, with the first dose given 2 h before infection. Mice were sacrificed at 5 dpi and lung tissues were collected for immunohistochemistry analysis and H&E staining. (A) Immunohistochemistry analysis of LC3 expression. Scale bar = 50 μm. (B) Gross pathology of the lung. Scale bar = 2 mm. (C) Body weight changes from the different groups. (D) Histological score based on H&E staining indicating pathological lesions in lung tissues from mock-infected and SARS-CoV-2-infected mice treated with PBS (indicated as Mock infection, SARS-CoV-2 group), and SARS-CoV-2-infected mice treated with 3-MA or rapamycin (indicated as 3-MA, rapamycin group). The score used for the measurement of damage is detailed in Materials and Methods. (E to H) Representative H&E-stained images from each group. The numbered panels display enlarged images from the indicated black circular areas on the left and illustrate necrosis and exfoliation of the bronchiolar endothelial cells (black arrowheads), and infiltration of segmented granulocytes around the bronchiolar area (yellow arrowheads) (1), exudation of inflammatory cells into the perivascular tissues (yellow arrowheads) (2), and destruction of alveolar structure and thickened alveolar septum, with inflammatory exudation (black arrowheads) (3). Scale bar = 200 μm. Data were expressed as means ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we examined viral replication in the infected animals by real-time quantitative PCR (RT-qPCR) analysis and found that SARS-CoV-2 N gene copy numbers increased significantly in lung tissues at 1, 3, and 5 dpi compared to those in mock-infected tissues (Fig. 5A and B). The highest viral load was observed at 1 dpi and decreased as described previously with time (26). Apparently, 3-MA reduced the viral yield in lung tissues at 1, 3, and 5 dpi (Fig. 5A and B). Compared to the mock infection group, no significant viral yield was detected in other primary organs caused by SARS-CoV-2 infection or 3-MA treatment, suggesting that SARS-CoV-2 replicates specifically in the lung tissues of hACE2 transgenic mice, and such tissue tropism was not affected by 3-MA administration (Fig. 5C). In contrast to 3-MA, rapamycin substantially promoted the viral yields in SARS-CoV-2-infected lung tissues (Fig. 5A). In situ hybridization analysis consistently displayed contradictory findings caused by 3-MA and rapamycin treatment in SARS-CoV-2 RNA levels in lung tissues at 5 dpi (Fig. 5D). Collectively, enhanced autophagy induction promotes SARS-CoV-2 infection, while autophagy inhibition suppresses SARS-CoV-2 replication and ameliorates virus-associated pneumonia in this model.
FIG 5.
Autophagy inhibition suppresses SARS-CoV-2 replication in lung tissues from hACE2 transgenic mice. hACE2 transgenic mice (n = 6) were intratracheally inoculated with 104 TCID50 of SARS-CoV-2 and intraperitoneally administered either PBS, 3-MA (30 mg/kg), or rapamycin (4 mg/kg) for 6 consecutive days, with the first dose given 2 h before infection. Mice were sacrificed at 1, 3, and 5 dpi and lung tissues and primary organs were collected for RT-qPCR and in situ hybridization analysis. (A) RT-qPCR analysis of SARS-CoV-2 N gene copies in lung tissues at 5 dpi. (B and C) RT-qPCR analysis of SARS-CoV-2 N gene copies in lung tissues at 1 dpi and 3 dpi (B) and primary organs at 5 dpi (C) from mock- or SARS-CoV-2-infected mice with or without 3-MA treatment. (D) Representative images and analysis of in situ hybridization for SARS-CoV-2 RNA in lung tissues at 5 dpi. Scale bar = 50 μm. Data were expressed as means ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Autophagy inhibition suppresses SARS-CoV-2 replication and ameliorates pneumonia in xenografted human lung tissues.
The xenografted human lung mouse model previously used to study Nipah virus infection gives clinical readouts for studying virus infections in pulmonary disease (27). To further investigate the role of autophagy initiation during SARS-CoV-2 infection, we studied this new mouse model of SARS-CoV-2 infection using fetal human lung tissues engrafted into the dorsal subcutaneous space of NCG mice. SARS-CoV-2 (105.5 TCID50) was injected into the fetal human lung, and mice were sacrificed at 12 hpi, 24 hpi, 48 hpi, 72 hpi, and 5 dpi for tissue harvesting (Fig. 6A). RT-qPCR analysis showed that SARS-CoV-2 N and E gene copies continued to increase with time (Fig. 6B and C). The viral titer of live SARS-CoV-2 consistently increased significantly after infection and reached a peak on 48 hpi (Fig. 6D), indicating SARS-CoV-2 replication in human lung tissues. At the microscopic level, we observed a mature pulmonary structure of the distal respiratory tract containing primitive alveolar spaces, as well as bronchioles, in the mock-infected human lungs. SARS-CoV-2 infection caused severe damage to this alveolar structure with diffuse inflammatory infiltration of granulocytes and macrophages, extensive fibrosis, and connective tissue hyperplasia, as well as alveolar endothelial cell death (Fig. 6J), indicating that SARS-CoV-2 induced pneumonia-like pathological changes.
FIG 6.
Autophagy inhibition suppresses SARS-CoV-2 replication and ameliorates pneumonia in xenografted human lung tissues. NCG mice engrafted with human lung tissues were inoculated with 105.5 TCID50 of SARS-CoV-2 or a vehicle. PBS or 3-MA (30 mg/kg) was intraperitoneally injected for 6 consecutive days, with the first dose given 2 h before infection. Mice were sacrificed at 12 hpi, 24 hpi, 48 hpi, 72 hpi, and 5 dpi and xenografted human lung tissues were isolated for RT-qPCR, virus titration, immunohistochemistry, and H&E staining. (A) Schematic diagram of the SARS-CoV-2 challenge, drug treatment, and sample collection schedule. (B and C) RT-qPCR analysis of SARS-CoV-2 N (B) and E (C) gene copies in the mock- or SARS-CoV-2-infected human lung tissues, along the indicated timeline. (D) Viral titers of the SARS-CoV-2-infected human lung tissues collected at the indicated times. (E and F) RT-qPCR analysis of SARS-CoV-2 N (E) and E (F) gene copies at 3 and 5 dpi in the mock- or SARS-CoV-2-infected human lungs treated with PBS or 3-MA. (G) Viral titers of SARS-CoV-2-infected human lung tissues from PBS- or 3-MA-treated mice at 3 and 5 dpi. (H) In situ hybridization analysis of SARS-CoV-2 RNA in PBS or 3-MA-treated human lung tissues after SARS-CoV-2 infection at 5 dpi. (I) Immunohistochemistry of LC3 expression in mock- or SARS-CoV-2-infected human lung tissues from PBS or 3-MA-treated mice at 5 dpi. (J) Representative H&E-stained images of mock- or SARS-CoV-2-infected human lung tissues from PBS- or 3-MA-treated mice at 5 dpi. Scale bar = 50 μm. Data were expressed as means ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we determined whether autophagy inhibition conferred protection on human lung after SARS-CoV-2 infection by treating with 3-MA intraperitoneally once daily for 6 consecutive days with the first dose given 2 h prior to infection (Fig. 6A). The induction of autophagy caused by SARS-CoV-2 was confirmed by the presence of elevated LC3 expression in the lungs which was blocked by 3-MA (Fig. 6I). RT-qPCR and in situ hybridization analysis indicated a suppression of viral yield caused by 3-MA at 3 dpi and 5 dpi (Fig. 6E and F and H). Consistent suppression of live virus particles was also observed in 3-MA-treated human lungs (Fig. 6G). Histopathology examination exhibited improved morphology of the alveolar space and bronchiolar structure by a reduction in inflammatory exudation in 3-MA-treated human lung tissues at 5 dpi (Fig. 6J). The body weights of the mice were not affected by either SARS-CoV-2 infection or drug treatment (data not shown). Collectively, NCD mice xenografted human lung appear to be a suitable model for SARS-CoV-2 infection, and autophagy inhibition suppresses SARS-CoV-2 replication and ameliorates pneumonia-like symptoms.
DISCUSSION
Autophagy is thought to be involved in SARS-CoV-2 infection (6, 8, 43, 44). In the present study, using animal infection-based models such as Macaca fascicularis, hACE2 transgenic mice, and human lung xenografts in mice, we identified autophagy induced by SARS-CoV-2 infection (Fig. 1, 4A, and 6I). We also showed increased autophagosome accumulation in SARS-CoV-2-infected Vero E6 and Huh-7 cells (Fig. 2), which is consistent with a recent study demonstrating that SARS-CoV-2 ORF3a protein triggered GFP-LC3 accumulation in HeLa-hACE2 cells (45). From our findings, we have concluded that the accumulation of autophagosomes in SARS-CoV-2-infected Vero E6 cells resulted from both suppression of autophagosome-lysosome fusion and activation of autophagosome formation (Fig. 2). Consistent with our findings, the maturation of autophagosomes into autolysosomes was blocked by the SARS-CoV-2 ORF3a protein, SARS-CoV NSP6, and MERS-CoV infection (9, 11, 12, 46).
By use of Western blotting, we screened the signaling cascade involved in autophagosome formation, and our results showed an inhibition in the Akt-mTOR pathway, which is thought to be involved in upstream activation signals for the induction of autophagy in SARS-CoV-2-infected Vero E6 cells (Fig. 3A). In addition, SARS-CoV-2 failed to activate the AMPK-Raptor/TSC-2 pathway, another pathway thought to inhibit mTOR activity (Fig. 3B). Consistent with this mTOR inhibition, dephosphorylation of ULK1 at Ser 757 and phosphorylation of Atg13 at Ser 355 were also observed (Fig. 3C). Furthermore, we observed an activation of the VPS34-VPS15-Beclin1 complex and increased abundance of its substrates Atg5-Atg12 and Atg16L1, which serve as components of the phagophore membrane and are essential for membrane elongation into autophagosomes (Fig. 3D and F) (29).
To determine the relationship between these autophagic events and SARS-CoV-2 replication, we assessed virus yields in Vero E6 cells and looked at the inhibition of virus replication using 3-MA and VPS34 inhibitor SAR405, as well as the knockdown of Atg14 protein, which reached a peak at 24 h after SARS-CoV-2 infection and may act as a subunit of the VPS34 complex (Fig. 3G, I, and J). Consistently, recent studies reported that attenuation of VPS34 kinase activity suppressed SARS-CoV-2 replication in a human airway epithelial cell line and human lung tissues (47, 48). We further demonstrated a role of autophagy induction in SARS-CoV-2-induced cell death, as observed by increased cell proliferation in 3-MA-treated Vero E6 cells after virus infection (Fig. 3H). Moreover, we found that SARS-CoV-2 replication was independent of Atg5, since the genetic ablation of Atg5 had no effect on virus yields in SARS-CoV-2-infected Vero E6 cells (Fig. 3K and L). A previous study using Atg5−/− mice also demonstrated a dispensable role for Atg5 in the replication of coronavirus murine hepatitis virus (49). However, in a different study, Atg5 knockout reduced SARS-CoV-2 replication efficiency in MEF-hACE2 cells (45). This finding, which differs from our results, may be attributed to the use of different cell lines for SARS-CoV-2 infection, and therefore, further studies using different infection models are needed to determine the function of Atg5 during SARS-CoV-2 replication.
We further applied two different animal models to simulate SARS-CoV-2 infection in vivo. In our hACE2 transgenic mouse model, we demonstrated that inhibition of autophagy suppressed SARS-CoV-2 replication and alleviated virus-associated pneumonia in the lungs. In contrast, autophagy induction by rapamycin promoted virus replication and aggregated the lung lesions (Fig. 4 and 5). It is worth noting that all infected mice were free of brain infection, as indicated by the unaltered brain SARS-CoV-2 N gene copies in virus-infected mice compared to those in controls (Fig. 5C). In contrast, brain infection was reported for some of the hACE2 transgenic mice (38–41). This may be due to different infection methods of SARS-CoV-2. Since there were few early publications involving a SARS-CoV-2 infection model when we began this study, to ensure effective lung infection with SARS-CoV-2, we infected hACE2 transgenic mice by intratracheal inoculation, which is a common infection route for SARS-CoV-2 in nonhuman primates (22, 25, 28), rather than the intranasal route. Intranasal inoculation of SARS-CoV-2 may contribute to brain infection and corresponding fatality, since nerve terminals in the oral and nasal mucosa, as well as the olfactory nerves, might be potential entry sites for SARS-CoV-2 neurotropic infection (50).
Moreover, we evaluated a new mouse model involving xenografted human lung during SARS-CoV-2 infection and confirmed that the virus was able to replicate in human lung tissues and the human lung went on to mimic SARS-CoV-2-associated pneumonia (Fig. 6B to D, H, and J). We consistently found that autophagy inhibition restrained virus infection and ameliorated the pathological changes seen in human lung tissues, demonstrating the need for autophagy initiation in SARS-CoV-2 infection (Fig. 6E to H and J). However, despite the presence of macrophages and granulocytes, the leukomonocytes and NK cells were deficient in NCG mice, thereby avoiding transplantation rejection (27). This model may be more suitable for studies of innate immunity, and therefore, future studies are needed to look at the inflammatory cytokines released by the xenografted human lung tissue after SARS-CoV-2 infection to further explore its application.
In summary, by combination of in vitro and in vivo studies, we have confirmed that autophagy induction contributes to SARS-CoV-2 infection, and we suggest that autophagy-inhibiting agents might be useful as therapeutic agents against SARS-CoV-2 infection. Our study provides a preliminary foundation for further research into virus-cell interactions and the development of autophagy-related antiviral therapeutics.
MATERIALS AND METHODS
Cell lines, viruses, and chemicals.
African green monkey kidney Vero E6 cells (ATCC; CRL-1586) and human hepatocarcinoma Huh-7 cells (JCRB, 0403) were cultured in Dulbecco’s modified Eagle medium (DMEM; Sigma-Aldrich; D5796) containing 10% fetal bovine serum (FBS; Invitrogen; 10270), 50 U ml−1 of penicillin, and 50 μg ml−1 of streptomycin. The Atg5−/− Vero E6 cell line was purchased from Ubigene Biosciences (Guangzhou, China) and maintained in the same growth medium as described above.
The SARS-CoV-2 strain BetaCoV/Beijing/IME-BJ01/2020 was originally isolated by C. F. Qin’s laboratory (Beijing, China) as described previously (24). Virus stocks were propagated and titers were determined on Vero E6 cells. All experiments with infectious SARS-CoV-2 virus were conducted in a biosafety level 3 laboratory in the Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
3-MA (HY-19312), rapamycin (HY-10219), E64D (HY-100229), pepstatin A (HY-P0018), SAR405 (HY-12481), and VPS34-IN-2 (HY-12473) were purchased from MedChemExpress (Monmouth Junction, NJ). VPS34-IN-1 (A15874) was purchased from Adooq Bioscience (Irvine, CA). CQ (C6628) was purchased from Sigma-Aldrich (St. Louis, MO).
Antibodies.
Antibodies against Atg5 (2630), p-AKT (4060), AKT (4685), p-mTOR (5536), mTOR (2983), p-S6 (4858), S6 (2317), p-4E-BP1 (2855), 4E-BP1 (9644), p-AMPK (2535), AMPK (5831), p-TSC-2 (3614), TSC-2 (4308), p-Raptor (2083), Raptor (2280), p-Atg13 (26839), Atg13 (13273), ULK1 (8054), p-ULK1 (Ser757) (14202), p-ULK1 (Ser555) (5869), VPS15 (14580), VPS34 (4263), Beclin1 (3495), p-Beclin1 (84966), Atg14 (96752), Atg16L1 (8089), p62 (16177), and GAPDH (5174) were obtained from Cell Signaling Technology (Danvers, MA). Anti-LC3 antibody (L8918) was obtained from Sigma-Aldrich. Anti-Lamp2b antibody (66301-1-Ig) was purchased from Proteintech (Chicago, IL). Horseradish peroxidase (HRP)-conjugated secondary antibodies (7074 or 7076), Alexa Fluor 647-conjugated secondary antibodies (4414), and Alexa Fluor 488-conjugated secondary antibodies (4408) were purchased from Cell Signaling Technology.
Plasmid and small interfering RNA (siRNA) transfection.
The GFP-LC3 plasmid (psetz-gfplc3) was purchased from InvivoGen (San Diego, CA). The pmCherry-EGFP-LC3 plasmid (PPL70001-2a) was purchased from Bioworld Technology (New York, NY). At 24 h after seeding, the Vero E6 and Huh-7 cells were transfected with 1.5 μg of plasmid using Lipofectamine 3000 reagent (Thermo Fisher Scientific; L3000015), and experiments were conducted 48 h posttransfection.
The control siRNA (sc-37007) and siRNAs against Atg14 (sc-92229) were purchased from Santa Cruz Biotechnology (CA). At 24 h after seeding, the Vero E6 cells were transfected with 50 nM siRNA using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific; 13778150), and experiments were conducted 48 h posttransfection.
Western blotting.
Cells were seeded into 6-well plates and infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.008 1 day later. In the E64D (10 μg/ml)-treated and pepstatin A (10 μg/ml)-treated groups, drugs were used for treatment 2 h before SARS-CoV-2 infection. At various time points as indicated, cells were collected and lysed and protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Beyotime, Shanghai, China; P0010). The proteins were mixed with loading buffer and denatured by boiling, and then equal amounts (30 μg) were electrophoresed, transferred onto a polyvinylidene difluoride (PVDF) membrane, and then incubated overnight at 4°C with primary antibodies as described previously. After a further incubation with HRP-conjugated secondary antibodies, specific protein bands were visualized using Pierce ECL Western blotting substrate (Thermo Fisher Scientific; 32106) and an Amersham Imaging 600 system. In the siRNA gene silencing and Atg5−/− cell-associated experiments, cells were collected without SARS-CoV-2 infection and subjected to Western blotting as described above. For the lung tissues isolated from Macaca fascicularis, samples were collected and homogenized in lysis buffer and the tissue lysate was denatured and processed as described above.
TEM.
Cells were mock or SARS-CoV-2 infected for 6 h and then fixed with 2.5% glutaraldehyde. Areas containing cells were block mounted and sliced. Ultrathin sections were stained with uranyl acetate and lead citrate before transmission electron microscopy (TEM) analysis. The number of autophagosomes per cell was calculated, and at least 30 cells were included. For the lung tissues isolated from Macaca fascicularis, samples were fixed with 2.5% glutaraldehyde and processed as described above.
RT-qPCR analysis.
Vero E6 cells were seeded into 6-well plates at a density of 2 × 105/well, and after culture for 24 h, SARS-CoV-2 was infected at an MOI of 0.008 and incubated for the specified time. Viral RNA in the supernatant was isolated with the QIAamp viral RNA kit (Qiagen; 52906) following the manufacturer’s instructions. Viral copy numbers were then detected by absolute quantitative real-time PCR (RT-qPCR) methodology using the HiScript II one-step RT-qPCR SYBR green kit (Vazyme Biotech, Nanjing, China) and an ABI 7500 real-time PCR system (Applied Biosystems, CA). The protocol for the RT-qPCR was as follows: 50°C for 15 min and 95°C for 30 s, followed by 45 cycles at 95°C for 10 s and 63°C for 35 s. The specific primers used to detect the SARS-CoV-2 N and E genes were as follows: for the N gene, 5′-GGGGAACTTCTCCTGCTAGAAT-3′ (forward) and 5′-CAGACATTTTGCTCTCAAGCT-3′ (reverse), and for the E gene, 5′-CGATCTCTTGTAGATCTGTTCTC-3′ (forward) and 5′-ATATTGCATTGCAGCAGTACGCACA-3′ (reverse). The sequences of the TaqMan probe were as follows: for the N gene, 5′-FAM-TTGCTGCTGCTTGACAGATT-6-carboxytetramethylrhodamine (TAMRA)-3′, and for the E gene, 5′-6-carboxyfluorescein (FAM)-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1-3′. For lung tissues or xenografted human lung tissues isolated from mice, samples were collected and homogenized in PBS. Viral RNA in the homogenate was isolated and processed as described above.
Analysis of LC3 aggregates.
Vero E6 and Huh-7 cells were transfected with GFP-LC3 plasmid or pmCherry-EGFP-LC3 plasmid as described previously (51). At 48 h after transfection, SARS-CoV-2 was infected at an MOI of 0.008. In the CQ-treated groups, CQ (40 μM) was used for treatment 2 h before SARS-CoV-2 infection. Images were captured 12 and 24 h postinfection using a fluorescence microscope (Olympus, Tokyo, Japan; BX43) and the average numbers of GFP/EGFP- or mCherry-containing dots in 30 cells were counted at a magnification of ×40.
Immunofluorescence analysis.
Vero E6 cells were seeded into 12-well plates at a density of 1 × 105/well and incubated for 24 h. Then, cells were infected with SARS-CoV-2 at an MOI of 0.008 and cultured for a further 24 h. The cells were then fixed with ice-cold 4% (wt/vol) paraformaldehyde for 1 h and washed three times with PBS. Next, cells were blocked with 0.3% Triton X-100 and 5% bovine serum albumin (BSA) for 1 h. After washing a further three times with PBS, specific primary antibodies were added and incubated overnight at 4°C. Cells were then washed and incubated with Alexa Fluor 647-conjugated anti-mouse antibody and/or Alexa Fluor 488-conjugated anti-rabbit antibody for 1 h, and images were captured using a fluorescence microscope and analyzed with Image-pro plus 6.0 (Media Cybernetics, Inc., MD). Images were transformed into black and white, and a unified standard was set up to identify positive signals. The integrated optical density (IOD) and pixel area of the cells were measured, and areal density was calculated as IOD/corresponding pixel area of the target cell.
Cell viability assays.
Cell viability was assessed with Cell Counting Kit-8 (CCK-8) reagent (Dojindo Molecular Technologies, Rockville, MD). Vero E6 cells were seeded into 96-well plates at a density of 7 × 103/well and incubated for 24 h. Then, 3-MA (0, 0.3125, 0.625, or 1.25 mM) was given 2 h prior to virus treatment. After incubation for 72 h, the CCK-8 reagent was added (10 μl per well), and values for OD at 450 nm were determined with a multifunction microplate reader after 2 h of incubation.
Animal experiments.
Male Macaca fascicularis macaques, weighing 4 to 6 kg, were randomly distributed into experimental groups (mock infection, n = 3; SARS-CoV-2, n = 3). Each monkey was intratracheally mock infected or infected with 1.3 × 106 median tissue culture infectious doses (TCID50) of SARS-CoV-2 in 4 ml of DMEM under anesthesia with tiletamine-zolazepam (Zoletil). On day 7 postinfection (dpi), animals were sacrificed by exsanguination and lung tissues were collected for Western blotting, histopathological examination, TEM analysis, immunohistochemistry staining, and in situ hybridization analysis.
Female hACE2 transgenic mice (C57BL/B6 background), weighing 30 to 32 g, were obtained from the National Institute for Food and Drug Control (Beijing, China). The transgenic mice express hACE2, which was driven by the mouse ACE2 promoter as described previously (26). Mice were randomly allocated into experimental groups (n = 6). 3-MA (30 mg/kg) or rapamycin (4 mg/kg) was given intraperitoneally 2 h prior to SARS-CoV-2 injection and then once daily for a total of 6 days. Each mouse was intratracheally administered DMEM (mock infection) or 104 TCID50 of SARS-CoV-2 in 30 μl of DMEM under anesthesia (intraperitoneally administered barbiturates). The body weights were measured daily. At 1, 3, and 5 dpi, mice were sacrificed by cervical dislocation and the lung tissues, as well as primary organs, were isolated for RT-qPCR analysis, histopathological examination, immunohistochemistry staining, and in situ hybridization analysis.
Male NCG (NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22) mice, weighing 22 to 29 g and bearing a fragment of human fetal lung in the dorsal subcutaneous space, were kindly provided by Yong-guang Yang from the First Hospital of Jilin University, Changchun, China. The xenografted fetal human lungs develop mature pulmonary structure as described previously (27). Mice were randomly allocated into experimental groups (n = 3). The 3-MA (30 mg/kg) was added intraperitoneally 2 h prior to SARS-CoV-2 infection and once daily for a total of 6 days. Human lung tissues were subjected to intragraft inoculation with 105.5 TCID50 of SARS-CoV-2 in 100 μl of DMEM or a vehicle, using the same volume of DMEM. The body weights were measured daily. At 12 hpi, 24 hpi, 48 hpi, 72 hpi, and 5 dpi, mice were sacrificed by cervical dislocation and human lung tissues were isolated for RT-qPCR analysis, viral titration, immunohistochemistry staining, and histopathological analysis.
All animals were housed at biosafety level 3 and given free access to standard pellet feed and water. All experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (52). Discarded tissue from deceased human fetuses was obtained from the first hospital of Jilin University with written informed consent.
Histopathological examination.
Lung tissues were fixed in 4% paraformaldehyde solutions for 7 days, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E) according to standard protocols. Images were captured using light microscopy and analyzed as described previously (53). To distinguish lung histopathology, the severity of damage in bronchioles, alveoli, and blood vessels was graded, and the total score was accumulated. The degree of damage was scored as follows: 0, normal structure; 1, mild infiltration, epithelial cell thickening, and congestion; 2, moderate epithelial cell death, infiltration, focal exudation, or widened alveolar septum; and 3, severe epithelial cell death, diffuse inflammatory cell infiltration, or collapse of alveolar structure.
Immunohistochemistry analysis.
Lung tissues were fixed in 4% paraformaldehyde solution for 7 days, then paraffin embedded, and cut into 3- to 4-μm sections. Sections were deparaffined and rehydrated and antigens were retrieved using a citric acid (pH 6.0) antigen retrieval buffer. Next, the endogenous peroxidase activity was blocked using 3% hydrogen peroxide at room temperature for 25 min. After washing three times with PBS, 3% BSA was added to tissue sections and the tissues were sealed for 30 min. Sections were then incubated overnight with primary antibodies against LC3 and Atg5 at 4°C. The next day after washing, the sections were incubated with HRP-conjugated secondary antibodies for 50 min and 3,3′-diaminobenzidine (DAB) color-developing solution was added, followed by counterstaining with hematoxylin stain solution, for nuclear staining. The final DAB-labeled images were captured by microscopy. The nucleus became blue after hematoxylin staining, and DAB-positive sections were stained brownish yellow. Images were analyzed with Image-pro plus 6.0, and a unified standard was set up for positive signaling of brownish yellow. The positive IOD and pixel area from three visual fields in each slice were measured, and areal density was calculated as IOD/corresponding tissue pixel area.
RNA in situ hybridization assay.
Lung tissues were fixed in 4% paraformaldehyde solution containing 0.1% diethyl pyrocarbonate (DEPC) for 72 h. Tissues were then dehydrated, paraffin embedded, and sectioned into thin slices. Endogenous peroxidase was blocked, and the sections were hybridized with specific probes, the sequence of which was as follows: 5′-digoxigenin (DIG)-ACTACAGCCATAACCTTTCCACATACCGCAGAC-DIG-3′. The DIG label was detected using an anti-DIG-HRP antibody. After incubation with DAB, the images were captured by light microscopy and the areal density was analyzed as described previously using Image-Pro Plus 6.0.
Measurement of viral titers.
Lung tissues were homogenized and serially diluted in serum-free DMEM growth medium. Tenfold dilutions were added to 96-well plates in a volume of 100 μl/well. Vero E6 cells were seeded in the plates 1 day before at a density of 7 × 103/well. Next, 100 μl of DMEM with 4% FBS, 100 IU/ml of penicillin, and 100 μg/ml of streptomycin were added 2 h postinfection to reach a final volume of 200 μl per well. Cells were then cultured at 37°C with 5% CO2 for 4 days. Viral titers were calculated using the Reed-Muench method in the form of TCID50.
Statistics.
Statistical significance between groups was determined using GraphPad Prism, version 8.0. Data were presented as means ± standard errors of the means (SEM) in all experiments and analyzed using a t test or analysis of variance followed by a two-tailed t test, and a P value of <0.05 was considered to be statistically significant.
Data availability.
Source data in this work are available in the GitHub repository at https://github.com/13382029577/SARS-CoV-2-autophagy.
ACKNOWLEDGMENTS
We express our gratitude to EditSprings for the expert linguistic services provided.
This study was funded by the Research and Development of New Coronavirus Pneumonia Recombinant Protein Vaccine (grant no. 20YF003) as part of The New Coronavirus Pneumonia (NCP) Epidemic Prevention and Control Emergency Scientific Research Project in Changchun City. This study was also supported by the Key Projects of Science and Technology Boosting Economy in 2020 (grant no. SQ2020YFF0417940).
Xiao Li, Mingyao Tian, and Ningyi Jin conceived and designed the animal experiments. Chao Shang performed the experiments and wrote the paper. Xinyu Zhuang, He Zhang, Yiquan Li, Yilong Zhu, Jing Lu, Chenchen Ge, Jianan Cong, Tingyu Li, and Nan Li performed the experiments. Chao Shang analyzed the data. Xiao Li revised the paper. All authors have read and approved the final paper.
We declare no competing interests.
Contributor Information
Mingyao Tian, Email: linjiaxiaoya@163.com.
Ningyi Jin, Email: lixiao06@mails.jlu.edu.cn.
Xiao Li, Email: skylee6226@163.com.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 30 June 2021. Coronavirus disease (COVID-19) situation report. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---29-june-2021.
- 3.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323:1061–1069. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. 2020. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8:475–481. 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ujike M, Taguchi F. 2015. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses 7:1700–1725. 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shojaei S, Suresh M, Klionsky DJ, Labouta HI, Ghavami S. 2020. Autophagy and SARS-CoV-2 infection: a possible smart targeting of the autophagy pathway. Virulence 11:805–810. 10.1080/21505594.2020.1780088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonam SR, Muller S, Bayry J, Klionsky DJ. 2020. Autophagy as an emerging target for COVID-19: lessons from an old friend, chloroquine. Autophagy 16:2260–2266. 10.1080/15548627.2020.1779467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mijaljica D, Klionsky DJ. 2020. Autophagy/virophagy: a “disposal strategy” to combat COVID-19. Autophagy 16:2271–2272. 10.1080/15548627.2020.1782022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottam EM, Whelband MC, Wileman T. 2014. Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10:1426–1441. 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, Britton P, Ktistakis NT, Wileman T. 2011. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7:1335–1347. 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao G, Zhao H, Li Y, Ji M, Chen Y, Shi Y, Bi Y, Wang P, Zhang H. 2021. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell 56:427–442.e5. 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Sun H, Pei R, Mao B, Zhao Z, Li H, Lin Y, Lu K. 2021. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov 7:31. 10.1038/s41421-021-00268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui X, Zhang L, Cao L, Huang K, Zhao Y, Zhang Y, Chen X, Lin X, Chen M, Jin M. 2021. SARS-CoV-2 promote autophagy to suppress type I interferon response. Signal Transduct Target Ther 6:180–183. 10.1038/s41392-021-00574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang R, Xu M, Zhu H, Chen CZ, Zhu W, Lee EM, He S, Zhang L, Zhao J, Shamim K, Bougie D, Huang W, Xia M, Hall MD, Lo D, Simeonov A, Austin CP, Qiu X, Tang H, Zheng W. 2021. Biological activity-based modeling identifies antiviral leads against SARS-CoV-2. Nat Biotechnol 39:747. 10.1038/s41587-021-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. 2020. Table of therapeutics against COVID-19 as [sic] 21st March 2020. https://www.who.int/blueprint/priority-diseases/key-action/Table_of_therapeutics_Appendix_17022020.pdf?ua=1.
- 16.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583:459–468. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, Li Y, Hu Z, Zhong W, Wang M. 2020. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 6:16. 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S, Lu R, Li H, Tan W, Liu D. 2020. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 71:732–739. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreani J, Le Bideau M, Duflot I, Jardot P, Rolland C, Boxberger M, Wurtz N, Rolain JM, Colson P, La Scola B, Raoult D. 2020. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb Pathog 145:104228. 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Tian Z, Yang X. 2020. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14:72–73. 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 22.Maisonnasse P, Guedj J, Contreras V, Behillil S, Solas C, Marlin R, Naninck T, Pizzorno A, Lemaitre J, Goncalves A, Kahlaoui N, Terrier O, Fang RHT, Enouf V, Dereuddre-Bosquet N, Brisebarre A, Touret F, Chapon C, Hoen B, Lina B, Calatrava MR, van der Werf S, de Lamballerie X, Le Grand R. 2020. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 585:584–587. 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- 23.Fiolet T, Guihur A, Rebeaud ME, Mulot M, Peiffer-Smadja N, Mahamat-Saleh Y. 2021. Effect of hydroxychloroquine with or without azithromycin on the mortality of COVID-19 patients: a systematic review and meta-analysis. Clin Microbiol Infect 27:920–921. 10.1016/j.cmi.2020.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang NN, Li XF, Deng YQ, Zhao H, Huang YJ, Yang G, Huang WJ, Gao P, Zhou C, Zhang RR, Guo Y, Sun SH, Fan H, Zu SL, Chen Q, He Q, Cao TS, Huang XY, Qiu HY, Nie JH, Jiang Y, Yan HY, Ye Q, Zhong X, Xue XL, Zha ZY, Zhou D, Yang X, Wang YC, Ying B, Qin CF. 2020. A thermostable mRNA vaccine against COVID-19. Cell 182:1271–1283.e16. 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, Xu W, Zhao Y, Li N, Zhang J, Liang H, Bao L, Xu Y, Ding L, Zhou W, Gao H, Liu J, Niu P, Zhao L, Zhen W, Fu H, Yu S, Zhang Z, Xu G, Li C, Lou Z, Xu M, Qin C, Wu G, Gao GF, Tan W, Yang X. 2020. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 182:713–721.e9. 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 27.Valbuena G, Halliday H, Borisevich V, Goez Y, Rockx B. 2014. A human lung xenograft mouse model of Nipah virus infection. PLoS Pathog 10:e1004063. 10.1371/journal.ppat.1004063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y, Yang L, Hu X, Xiong J, Jiang RD, Zhang HJ, Gao XX, Peng C, Min J, Chen Y, Si HR, Wu J, Zhou P, Wang YY, Wei HP, Pang W, Hu ZF, Lv LB, Zheng YT, Shi ZL, Yuan ZM. 2020. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in rhesus macaques. Cell Res 30:670–677. 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NYO, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai X-Y, Bailly Y, Baiocchi R, Baldini G, et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8:445–544. 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glick D, Barth S, Macleod KF. 2010. Autophagy: cellular and molecular mechanisms. J Pathol 221:3–12. 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weidberg H, Shvets E, Elazar Z. 2011. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80:125–156. 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 32.Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326. 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. 2006. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 281:8518–8527. 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 34.Erlich S, Alexandrovich A, Shohami E, Pinkas-Kramarski R. 2007. Rapamycin is a neuroprotective treatment for traumatic brain injury. Neurobiol Dis 26:86–93. 10.1016/j.nbd.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Burman C, Ktistakis NT. 2010. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett 584:1302–1312. 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Itakura E, Mizushima N. 2009. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy 5:534–536. 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 37.Itakura E, Kishi C, Inoue K, Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19:5360–5372. 10.1091/mbc.e08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yinda CK, Port JR, Bushmaker T, Offei Owusu I, Purushotham JN, Avanzato VA, Fischer RJ, Schulz JE, Holbrook MG, Hebner MJ, Rosenke R, Thomas T, Marzi A, Best SM, de Wit E, Shaia C, van Doremalen N, Munster VJ. 2021. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog 17:e1009195. 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang R-D, Liu M-Q, Chen Y, Shan C, Zhou Y-W, Shen X-R, Li Q, Zhang L, Zhu Y, Si H-R, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang H-J, Baric RS, Zhou P, Yang X-L, Shi Z-L. 2020. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182:50–58.e8. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oladunni FS, Park J-G, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, Olmo-Fontánez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Dick EJ, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11:6122. 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumari P, Rothan HA, Natekar JP, Stone S, Pathak H, Strate PG, Arora K, Brinton MA, Kumar M. 2021. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses 13:132. 10.3390/v13010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutz C, Maher L, Lee C, Kang W. 2020. COVID-19 preclinical models: human angiotensin-converting enzyme 2 transgenic mice. Hum Genomics 14:20. 10.1186/s40246-020-00272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calender A, Israel-Biet D, Valeyre D, Pacheco Y. 2020. Modeling potential autophagy pathways in COVID-19 and sarcoidosis. Trends Immunol 41:856–859. 10.1016/j.it.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brest P, Benzaquen J, Klionsky DJ, Hofman P, Mograbi B. 2020. Open questions for harnessing autophagy-modulating drugs in the SARS-CoV-2 war: hope or hype? Autophagy 16:2267–2270. 10.1080/15548627.2020.1779531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu Y, Wang X, Zhu Y, Wang Y, Yang X, Hu G, Liu C, Li J, Ren S, Xiao Z, Liu Z, Wang W, Li P, Zhang R, Liang Q. 2020. ORF3a mediated-incomplete autophagy facilitates SARS-CoV-2 replication. bioRxiv 10.1101/2020.11.12.380709. [DOI] [PMC free article] [PubMed]
- 46.Gassen NC, Niemeyer D, Muth D, Corman VM, Martinelli S, Gassen A, Hafner K, Papies J, Mosbauer K, Zellner A, Zannas AS, Herrmann A, Holsboer F, Brack-Werner R, Boshart M, Muller-Myhsok B, Drosten C, Muller MA, Rein T. 2019. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat Commun 10:5770. 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvas JA, Jureka AS, Nicolini AM, Chvatal SA, Basler CF. 2020. Inhibitors of VPS34 and lipid metabolism suppress SARS-CoV-2 replication. bioRxiv 10.1101/2020.07.18.210211. [DOI] [PMC free article] [PubMed]
- 48.Yuen CK, Wong WM, Mak LF, Wang X, Chu H, Yuen KY, Kok KH. 2021. Suppression of SARS‐CoV‐2 infection in ex‐vivo human lung tissues by targeting class III phosphoinositide 3‐kinase. J Med Virol 93:2076–2083. 10.1002/jmv.26583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, Mizushima NN, Denison MR, Virgin HW, IV.. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3:581–585. 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 50.Fenrich M, Mrdenovic S, Balog M, Tomic S, Zjalic M, Roncevic A, Mandic D, Debeljak Z, Heffer M. 2020. SARS-CoV-2 dissemination through peripheral nerves explains multiple organ injury. Front Cell Neurosci 14:229. 10.3389/fncel.2020.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura S, Noda T, Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460. 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 52.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 53.Yuan S, Wang R, Chan JF, Zhang AJ, Cheng T, Chik KK, Ye ZW, Wang S, Lee AC, Jin L, Li H, Jin DY, Yuen KY, Sun H. 2020. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat Microbiol 5:1439–1448. 10.1038/s41564-020-00802-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source data in this work are available in the GitHub repository at https://github.com/13382029577/SARS-CoV-2-autophagy.