ABSTRACT
Cellular factors have important roles in all facets of the flavivirus replication cycle. Deciphering viral-host protein interactions is essential for understanding the flavivirus life cycle as well as development of effective antiviral strategies. To uncover novel host factors that are co-opted by multiple flaviviruses, a CRISPR/Cas9 genome wide knockout (KO) screen was employed to identify genes required for replication of Zika virus (ZIKV). Receptor for Activated Protein C Kinase 1 (RACK1) was identified as a novel host factor required for ZIKV replication, which was confirmed via complementary experiments. Depletion of RACK1 via siRNA demonstrated that RACK1 is important for replication of a wide range of mosquito- and tick-borne flaviviruses, including West Nile Virus (WNV), Dengue Virus (DENV), Powassan Virus (POWV) and Langat Virus (LGTV) as well as the coronavirus SARS-CoV-2, but not for YFV, EBOV, VSV or HSV. Notably, flavivirus replication was only abrogated when RACK1 expression was dampened prior to infection. Utilising a non-replicative flavivirus model, we show altered morphology of viral replication factories and reduced formation of vesicle packets (VPs) in cells lacking RACK1 expression. In addition, RACK1 interacted with NS1 protein from multiple flaviviruses; a key protein for replication complex formation. Overall, these findings reveal RACK1’s crucial role to the biogenesis of pan-flavivirus replication organelles.
IMPORTANCE Cellular factors are critical in all facets of viral lifecycles, where overlapping interactions between the virus and host can be exploited as possible avenues for the development of antiviral therapeutics. Using a genome-wide CRISPR knockout screening approach to identify novel cellular factors important for flavivirus replication we identified RACK1 as a pro-viral host factor for both mosquito- and tick-borne flaviviruses in addition to SARS-CoV-2. Using an innovative flavivirus protein expression system, we demonstrate for the first time the impact of the loss of RACK1 on the formation of viral replication factories known as 'vesicle packets' (VPs). In addition, we show that RACK1 can interact with numerous flavivirus NS1 proteins as a potential mechanism by which VP formation can be induced by the former.
KEYWORDS: ZIKV, crispr, flavivirus, host factor, organelle, screen, viral replication
INTRODUCTION
The Flavivirus genus includes dozens of virus species transmitted by arthropods and with high potential to inflict significant morbidity and mortality worldwide (1). Antiviral therapeutics are not available for the vast majority of these flaviviruses and vaccines remain limited in efficacy and not suitable for children and the elderly (2, 3). Thus, further investigation of host-viral interactions in the flavivirus replication-cycle may potentially expedite development of novel treatments and vaccines. Flaviviruses have single stranded positive sense RNA genomes, similar virion structure, and replicate exclusively in the cytoplasm. Binding and entry is mediated by numerous host receptors followed by endocytosis of the viral particle and release of viral RNA into the cytosol via endosomal acidification. The positive sense RNA genome is then translated to yield a large single polyprotein that is then cleaved by host and viral proteases to produce the structural proteins (capsid [C], pre-membrane [prM] and envelope [Env] and non-structural [NS] proteins [NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5]. Expression of the NS proteins induces invagination of the ER membrane, allowing formation of the replication complex; a viral specific membrane structure that harbors active viral replication (4). At all stages of the viral life cycle, interactions between host factors and viral proteins are crucial in the replication process and biogenesis of ER membrane rearrangement to allow formation and maintenance of the replication complex (5). Following budding into the ER, immature virions are transported via the Golgi pathway to enable release of viral particles via exocytosis. Thus, identification and characterization of virus-host interactions impacting replication will inform our understanding not only of flavivirus biology but possible targets for therapeutic intervention.
Recent advances in genome-wide screening technology provides a platform to further identify novel viral host factors (6). One such technology is clustered regularly interspaced short palindromic repeats (CRISPR) genome-wide knockout screens. In this context, the GeCKO (genome-wide CRISPR knockout) screening library has been used to investigate virus-host interactions across multiple viral families and mammalian cell types (7–11). Although identification of a number of host factors are reproducible from one CRISPR screen to the next, variations in experimental design and the screening process can influence the potential hits and thus host factors which may surface.
In this study, we sought to identify cellular proteins involved in ZIKV replication through the use of a genome-wide GeCKO screen. This screen identified Receptor for Activated C Kinase 1 (RACK1) as a critical host factor for not only ZIKV infection, but other mosquito and tick-borne flaviviruses. RACK1 is a highly conserved multifunctional protein and a member of the tryptophan-aspartate repeat (WD-repeat) family of proteins that shares significant homology to the ß-subunit of G-proteins. Through its interaction directly or in a complex with other cellular proteins, it functions in shuttling proteins throughout the cell, anchoring proteins at particular locations, stabilizing protein activity and has specific roles in modulation of signaling pathways (12). Here, we show that RACK1 interacts with flavivirus NS1 protein and localizes to replication complexes to specifically alter the latter’s cellular morphology. Depletion of RACK1 resulted in both altered morphology and a decrease in the number of VPs generated by DENV and ZIKV non-structural protein expression. These results indicate that RACK1 through its interaction with NS1, is instrumental for the formation of morphologically normal and functional flaviviral replication complexes.
RESULTS
CRISPR/Cas9 genome-wide KO screen reveals RACK1 as a critical host factor for ZIKV replication.
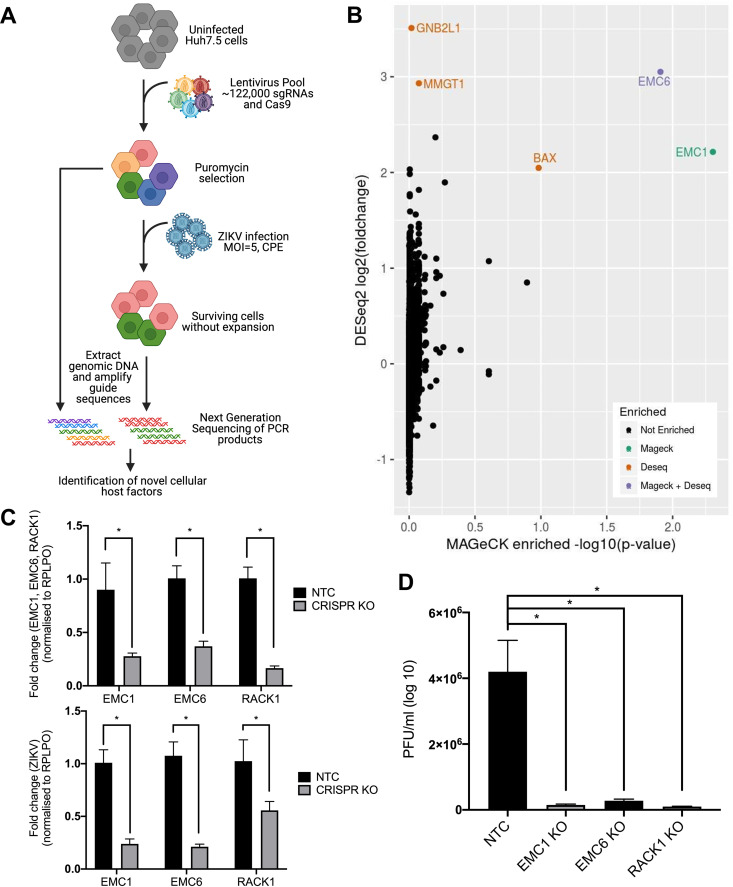
To identify host genes required for the ZIKV life cycle, we performed a lentiviral-mediated whole genome CRISPR screen to uncover sgRNAs that rendered cells resistant to ZIKV-induced cytopathic effect (CPE) (Fig. 1A) (Data Set S1). The LentiCRISPRv2-GecKO library was stably transduced into Huh7.5 cells, selected with puromycin, and infected with ZIKV (MR766, MOI = 5). In contrast to other screening approaches that expanded surviving cells following viral infection (8, 10), we harvested genomic DNA from unamplified surviving cells to identify host factors that may otherwise be under-represented following expansion as a consequence of altered cell proliferation. sgRNAs sequences from surviving cells were PCR amplified and sequenced (Illumina NextSeq) and analyzed using the CaRpools software package (Fig. 1B) (13). We identified several genes which were statistically enriched using DESeq2 and/or MAGeCK, some of which were previously identified as playing a role in viral replication. These include the ER membrane protein complex proteins (EMC), EMC1, MMGT1 (EMC5) and EMC6, as well as BAX. (Fig. 1B). However, our most significant hit identified was for sgRNAs targeting GNB2L1, otherwise known as RACK1. Interestingly, RACK1 has previously been shown to be an important host factor in virus infections including HCV (14, 15). In addition, a proteomics screen involving DENV NS1 also identified RACK1 as an interacting partner and thus was pursued further (16). To confirm our screen results, we generated independent EMC1, EMC6 and RACK1 CRISPR polyclonal knockout Huh7.5 cell lines and infected with ZIKV (MR766) and assessed viral replication and spread by qRT-PCR and plaque assay (Fig. 1C and D). As expected, both ZIKV RNA accumulation and the production of infectious viral particles were significantly reduced in cells depleted for EMC1 and EMC6 expression, and similar replication defects were observed in the absence of RACK1. The identification of EMC1 and EMC6 validate our screening approach and thereby identify RACK1 as a potential host factor involved in ZIKV replication.
FIG 1.
Identification of RACK1 as a critical host factor for the ZIKV life cycle. (A) Schematics of the genome-wide CRISPR/Cas9 screen with ZIKV strategy. (B) Enriched genes after CaRpools analysis isolated several genes of interest including proteins from the EMC complex (EMC1, MMGT1, EMC6) and GNB2L1 (RACK1). (C) Independent polyclonal CRISPR knockouts were performed for EMC1, EMC6 and RACK1, infected with ZIKV (MOI = 2) and qRT-PCR utilised to determine levels of both the respective knockout gene transcripts as well as ZIKV RNA. (D) Plaque assays were also performed concurrently with EMC1, EMC6 and RACK1 knockout cell lines to determine levels of ZIKV virions within the supernatant. All data are *P < 0.05 and n = 3 biological triplicates, representative of similar repeat experiments.
Confirmation of RACK1 as a critical host factor for ZIKV infection.
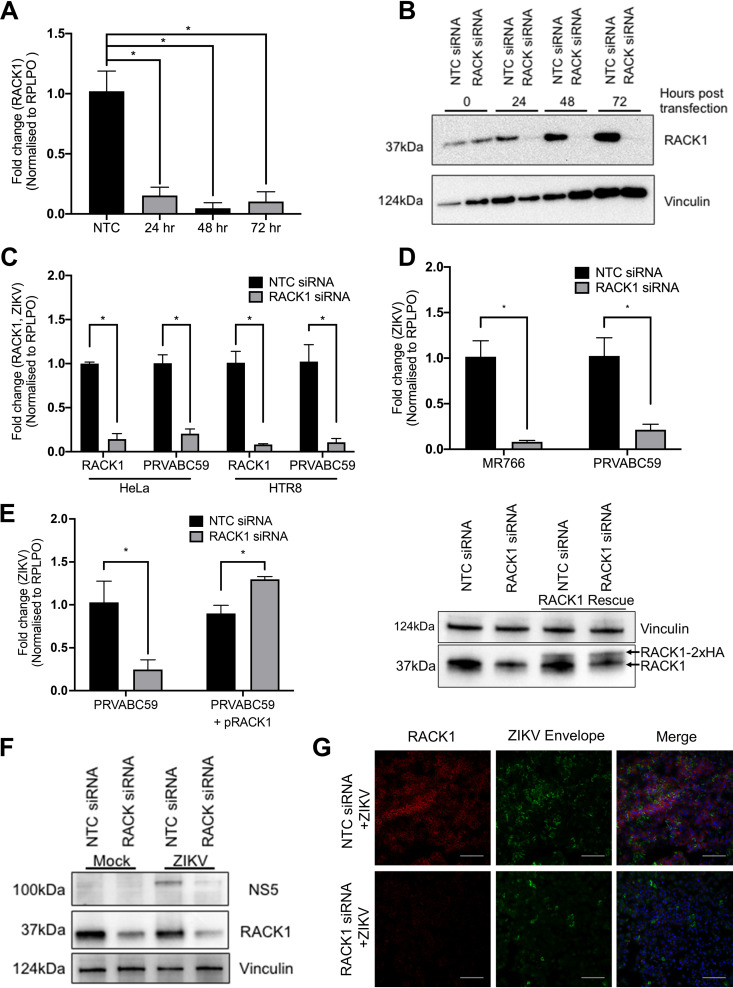
To confirm that RACK1 is an important host factor for ZIKV replication, siRNAs targeting RACK1 were utilised as a complementary method of abrogating RACK1 expression. Time-course knockdown experiments with siRNAs targeting RACK1 showed that mRNA (Fig. 2A) and subsequently protein levels (Fig. 2B) are reduced significantly after 24 h siRNA transfection and remain low for at least 72 h posttransfection. Since the genome-wide CRISPR KO screen was performed with the liver derived Huh7.5 cell-line, we also thought it was imperative to determine whether perturbation of ZIKV replication was evident in cells of non-hepatic origin. siRNA knockdown in both HeLa and the placental cell line HTR8, followed by infection with ZIKV (MR766), revealed a reduction in RACK1 mRNA expression that was associated with a significant decrease in ZIKV RNA (Fig. 2C). Our original genome-wide CRISPR KO screen utilised the MR766 ZIKV strain of African lineage, which through continued passage in mouse brains may have resulted in adaptations and changes in host factor reliance (17). Thus, HeLa cells with RACK1 siRNA knockdown were infected with ZIKV MR766 or PRVABC59, the latter being of Asian lineage, and revealed a similar impact on ZIKV replication for both strains, indicating that RACK1 is important for replication of both African and Asian ZIKV lineages (Fig. 2D). Rescue experiments were performed involving transfection of an expression plasmid encoding siRNA-resistant RACK1 into cells which have depleted RACK1 expression and infection with ZIKV (PRVABC59). These cells showed restoration of ZIKV RNA levels (Fig. 2E). To ensure that the loss of ZIKV RNA by RACK1 knockdown resulted in perturbation in active viral replication and NS protein expression, immunoblot detection was performed and revealed a significant decrease in ZIKV NS5 protein levels in the absence of RACK1 expression (Fig. 2F). In a complementary approach, immunofluorescence microscopy was employed that revealed a reduction in ZIKV envelope protein staining following siRNA knockdown of RACK1 (Fig. 2G). Together, these findings indicate that RACK1 is an important proviral host factor for different ZIKV strains and in multiple cell types.
FIG 2.
RACK1 is a critical host factor for ZIKV infection. Huh7 cells were transfected with either NTC or RACK1 siRNA and cells harvested for RNA extraction and qRT-PCR (A) or Western blot (B) at indicated time points. (C) HeLa and HTR8 cells were transfected with either NTC or RACK1 siRNA prior to infection with ZIKV (PRVABC59, MOI = 2). 48 hpi, RNA was harvested and qRT-PCR performed. (D) HeLa cells were infected with either ZIKV MR766 or PRVABC59 (MOI = 2) 24 h post NTC/RACK1 siRNA transfection and RNA harvested for qRT-PCR 48 h postinfection. (E) HeLa cells were co-transfected with NTC/RACK1 siRNA with or without plasmid encoding siRNA resistant HA-tagged RACK1. 48 h posttransfection, cells were infected with ZIKV PRVABC59 (MOI = 1) and RNA was harvested for qRT-PCR 24 h postinfection. Prior to infection, WCL was also harvested for immunoblot analysis with mouse RACK1 antibodies. Vinculin served as sample loading control. (F) HeLa cells were infected with PRVABC59 (MOI = 1) 24 h post NTC/RACK1 siRNA transfection and 48 h postinfection, WCL was harvested for immunoblot analysis with chicken flavivirus NS5 and mouse RACK1 antibodies. Vinculin served as sample loading control (G) Immunofluorescence was performed on endogenous RACK1 and ZIKV envelope by transfecting NTC/RACK1 siRNA into HeLa cells and infection with ZIKV (PRVABC59, MOI = 1) 24 h posttransfection. Cells were fixed 48 hpi and stained as indicated (scale bar: 250 μm). All data are *P < 0.05 and n = 3 biological triplicates (except for B and F which are n = 1), representative of similar repeat experiments.
RACK1 is required for flavivirus replication.
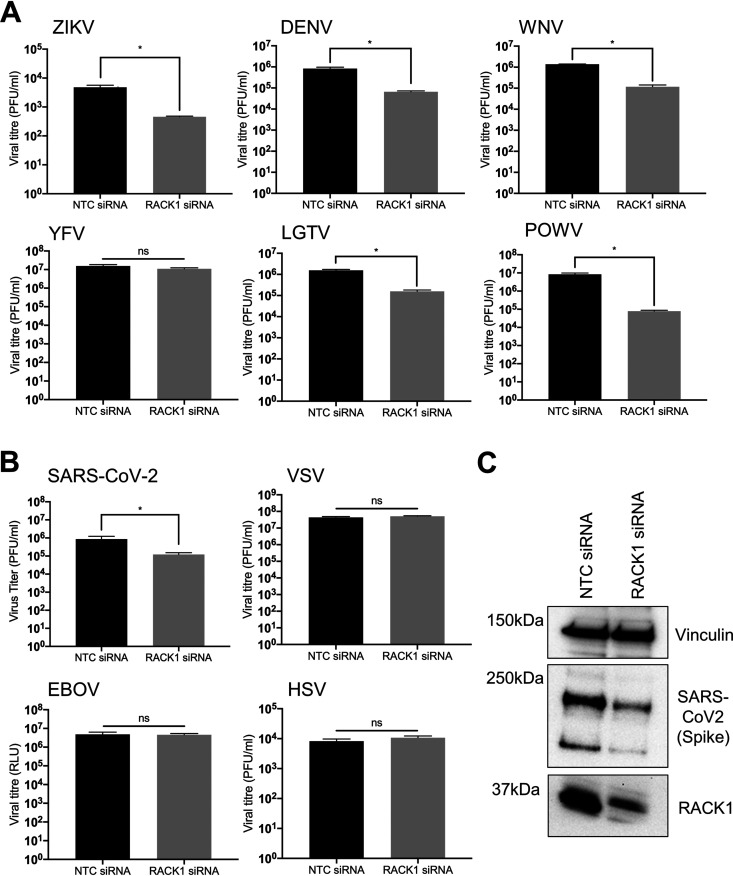
To determine whether RACK1 is also a proviral host factor for other flaviviruses, we explored whether other mosquito or tick-borne flavivirus species require RACK1 for virus replication. Compared to siRNA non-targeting control (NTC) cells, knockdown of RACK1 had a significant impact of at least a 1 log10 decrease in infectious virus release of the mosquito-borne flaviviruses including ZIKV (strain 2013 French Polynesia), WNVNY99 and DENV (strain NGC, serotype 2). Interestingly, there was no effect on YFV (strain 17D) replication in the absence of RACK1 expression (Fig. 3A). Demonstrating that this effect is not specific to mosquito-borne flaviruses, we also noted that the tick-borne flaviviruses, Langat virus (LGTV; an attenuated member of the TBEV serocomplex) and Powassan virus (POWV; strain LB) also had a dependency for expression of RACK1. In fact, of all the flaviviruses investigated, POWV revealed the greatest dependency on RACK1 with a consistent 2 log10 decrease in viral RNA following RACK1 depletion (Fig. 3A). We next investigated if other RNA viruses required a dependency for RACK1. Using an EBOV luciferase reporter virus, we noted no significant impact of RACK1 on viral replication in comparison to the NTC siRNA (Fig. 3B). Interestingly, RACK1 was required for SARS-CoV-2 replication, as shown by reduced infectious virus release (approximately 1 log10) and intracellular spike protein expression (Fig. 3B and Supp. 1). No requirement for RACK1 was observed for either VSV (negative strand RNA virus) or HSV (DNA virus) replication. Collectively, this suggests that positive strand RNA viruses of the flavivirus genus exhibit dependency for RACK1 expression.
FIG 3.
RACK1 is required for flavivirus replication. Huh7 cells (or A549-ACE2 for SARS-CoV-2) were transfected with either NTC/RACK1 siRNA and infected with the following viruses 24 h posttransfection: (A) ZIKV (MOI = 0.5), DENV (MOI = 0.5), WNVNY99 (MOI = 0.5), YFV (MOI = 0.5), LGTV (MOI = 0.5), POWV (MOI = 0.5), (B) SARS-CoV-2 (MOI = 0.1), VSV (MOI = 0.5), EBOV/MAY (103 TCID50) and HSV (MOI = 0.05). Plaque assays were performed to determine viral titers within the supernatant. (C) A549 cells stably expressing the ACE2 receptor was transfected with NTC/RACK1 siRNA prior to infection with SARS-CoV-2 (MOI = 0.1). WCL was harvested and immunoblot analysis performed with primary (anti-vinculin, anti-SARS-CoV-2 spike and anti-RACK1) and secondary (anti-mouse conjugated HRP) antibodies. Vinculin served as sample loading control (n = 3). Data are n = 3 biological triplicates (except for C which is n = 1), representative of similar repeat experiments (*P < 0.05, ns=non-significant).
RACK1 expression is important prior to establishment of viral RNA replication.
To gain insight into the mechanism of action of RACK1, we next determined at which stage of the viral life cycle RACK1 was functioning. To achieve this, we used a DENV sub-genomic replicon expressing Renilla Luciferase (Rluc). Following transfection of in vitro transcribed DENV sub-genomic (SGR) replicon RNA, replication is established that recapitulates the complete RNA replication cycle, although infectious virions are not produced due to the lack of structural proteins required for virion morphogenesis. Using this approach, Rluc is expressed at low levels following immediate translation of the input RNA and increases thereafter during genome amplification. Huh7.5 cells were pre-treated with NTC or RACK1 siRNA prior to transfection with RNA for DENV SGR or DENV SGR containing a GND mutation in the NS5 protein encoding the viral RNA-dependent RNA polymerase to control for input RNA mediated luciferase expression (Fig. 4A). In the absence of RACK1, DENV SGR RNA replication was supressed at late but not early time points following transfection of RNA suggesting that primary viral translation was not impacted (Fig. 4B). Furthermore, the pattern of Rluc expression in the absence of RACK1 from DENV SGR GND was similar to that of the replication competent DENV SGR confirming no impact on viral translation. In addition, we also infected siRNA treated cells with DENV (16681) and as expected, DENV RNA levels were similarly significantly reduced when RACK1 expression was decreased prior to infection with DENV (16681) infectious virus (Fig. 4C). We also investigated the impact of RACK1 depletion following establishment of viral replication (Fig. 4D). siRNA treatment of cells with NTC or RACK1 postinfection with DENV or transfection of DENV SGR RNA, revealed no impact on viral replication as determined by qRT-PCR or luciferase assays respectively (Fig. 4E and F). Collectively, these results suggest that virus entry and translation of viral RNA are not impacted by RACK1 depletion, whereas RACK1 is important for viral RNA genome replication.
FIG 4.
RACK1 is important prior to flavivirus replication complex formation. (A) Huh7.5 cells were transfected with NTC/RACK1 siRNA for 24 h prior to (B) transfection of DENV2 SGR/SGR GND RNA or (C) infection with DENV2. (B) Cells transfected with DENV-SGR RNA were lysed at the appropriate time points and luciferase assay performed to determine Renilla Luciferase levels. (C) RNA was harvested and extracted from DENV-infected cells 24 hpi and qRT-PCR performed. (D) In contrast, Huh7.5 cells were transfected with DENV2 SGR/SGR GND RNA (E) or infected with DENV2 (F) for 24 h prior to transfection of NTC/RACK1 siRNA for 24 h. (E) Cells transfected with the DENV SGR RNA were lysed at the appropriate time points and luciferase assay performed immediately to determine Renilla Luciferase levels. (F) RNA from DENV infected cells was extracted and qRT-PCR performed (*P < 0.05). Data are n = 3 biological triplicates (except for C which is n = 1), representative of similar repeat experiments (*P < 0.05, ns=non-significant). Data are n = 3 biological triplicates, representative of similar repeat experiments.
RACK1 is instrumental to the formation of the flavivirus replication complex.
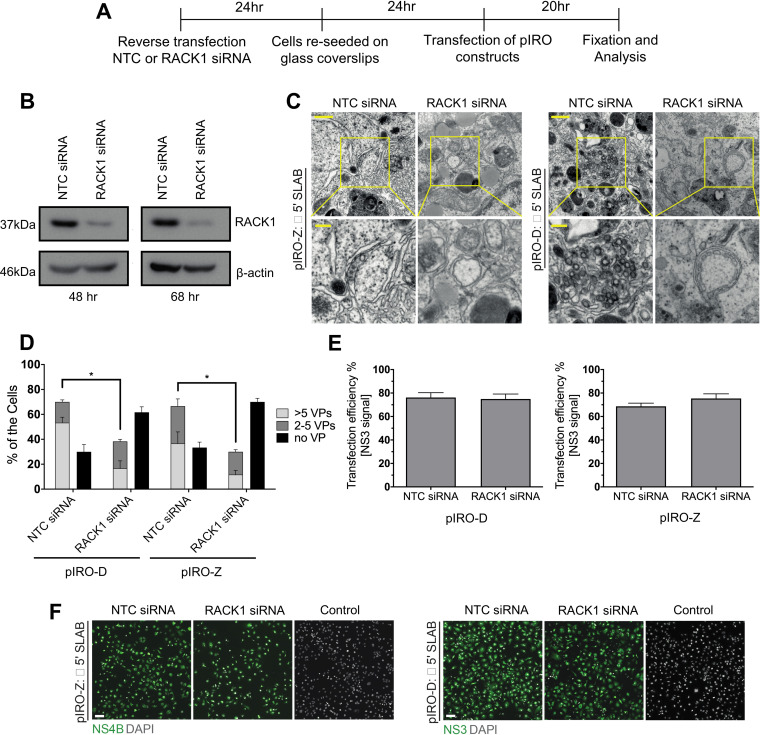
Following translation and processing of the viral polyprotein, a hallmark of flavivirus replication is rearrangement of ER membranes to form single membrane vesicle packets (VPs) that house sites of viral RNA replication in a replication complex (RC) (18). However, the requirement of RACK1 for viral replication will inevitably impact RC formation and thus uncoupling polyprotein synthesis from viral replication is necessary to determine the role of RACK1 in flavivirus RNA replication. To achieve this, we used a replication-independent expression system, termed pIRO (plasmid induced replication organelle formation) that induces DENV and ZIKV vesicle packets that are morphologically indistinguishable from those of infected cells (19). Huh7/Lunet-T7 cells were reverse transfected with either NTC or RACK1 siRNA, following which cells were transfected with either ZIKV or DENV pIRO constructs and 20 h later cells were fixed and processed for immunofluorescence and TEM (Fig. 5A). Transfection efficiencies determined for both ZIKV NS4B and DENV NS3 were comparable between siRNA treatments (Fig. 5E and F). Cells transfected with siRNAs targeting RACK1 revealed a significant decrease in RACK1 expression at 48 and 68 h post treatment that correlated with significant impairment of VP formation for both DENV and ZIKV pIRO systems (Fig. 5B and C). In cells treated with NTC siRNA and expressing pIRO-DENV or -ZIKV constructs, distinct ER membrane alterations were visible representing VPs, however this was not the case for cells lacking RACK1 expression (Fig. 5C). Furthermore, VP quantification by TEM were classified into three categories; sections with no VPs, sections with 2–5 VPs and sections with > 5 VPs. For cells transfected with either pIRO-DENV or -ZIKV, there was a significant increase in the numbers of sections lacking VPs when RACK1 expression was depleted (Fig. 5D). Conversely, under the same conditions, we also noted a decrease in the number of sections harboring VPs. Collectively, our results show that RACK1 is an essential host factor for mediating the biogenesis of convoluted ER membrane structures and VPs essential for RNA viral replication.
FIG 5.
RACK1 is critical for the formation of the flavivirus replication complex. (A) Huh7/Lunet-T7 cells were reverse transfected with NTC/RACK1 siRNA for 24 h prior to re-seeding onto glass coverslips. Cells were transfected with pIRO-DENV or -ZIKV constructs for 20 h before fixation and electron microscopy. (B) Immunoblot analysis performed to evaluate RACK1 levels 48 and 72 h posttransfection of siRNA. ß-actin served as sample loading control. (C) Thin-section TEM images of altered morphology of VPs upon transfection of pIRO-DENV or -ZIKV constructs when RACK1 expression is reduced. Upper panel scale bar: 500 nm. Lower panels are magnifications of yellow squared areas in the upper panel images. Lower panel scale bar: 100 nm. (C) For each condition, VPs present within whole-cell sections from 100 cells were counted (E&F) Transfection efficiency of pIRO-DENV and -ZIKV constructs were analyzed via immunofluorescence with anti-NS3 and NS4B primary antibodies and appropriate secondary antibodies and multiple fields of view quantified by counting positive staining cells and normalization to total number of cells. Data are n = 3 biological triplicates (except for C which is n = 1), representative of similar repeat experiments (*P < 0.05).
RACK1 is required for the positioning of flavivirus molecular components during virus replication.
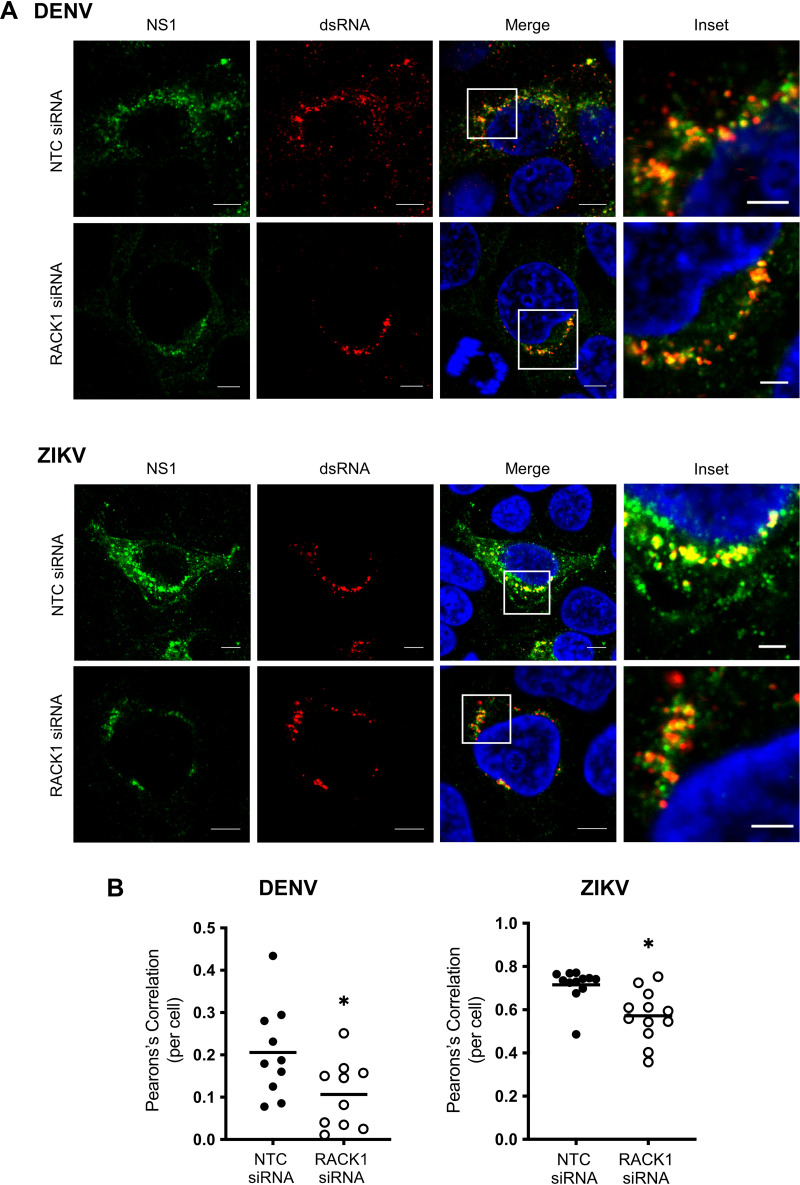
The NS1 protein across the flaviviruses has a major role in the virus life cycle, particularly in VP formation (20). Given our results above and based on a previous study identifying NS1 interaction with RACK1 (16), we next investigated the impact of RACK1 depletion on the subcellular localization of NS1 and dsRNA as a marker of replication complex formation following either DENV or ZIKV infection. Under normal infection conditions, confocal microscopy revealed NS1 localization to a punctate and reticular subcellular foci which significantly colocalised with dsRNA, representing modified ER-associated replication complexes as previously demonstrated (Fig. 6). However, following RACK1 depletion, there was a significant decrease in NS1 expression as expected, and in particular, altered ZIKV NS1 localization to a predominantly peri-nuclear localization juxtaposed to dsRNA punctate foci. This altered NS1 and dsRNA localization is consistent with our EM studies in which we showed a decrease in the frequency of VPs and their altered morphology (Fig. 5D).
FIG 6.
RACK1 is required for the positioning of flavivirus molecular components during virus replication. Huh7 cells were transfected with NTC/RACK1 siRNA for 24 h prior to infection of DENV2 or ZIKV PRVABC59 (MOI = 1). (A) Cells were subjected to indirect immunofluorescence labeling with primary (anti-NS1 and anti-dsRNA) and secondary (mouse anti-IgG 488 and anti-IgM-555) antibodies. DAPI was used to stain nuclear DNA in the merged images. (B) The signal of both channels was thresholded to remove background noise with green channel designated as region of interest (ROI) and Pearson’s correlation coefficient (PCC) was then determined and plotted as a scatterplot (scale bar: 5 μm, inset scale bar: 2 μm, images representative of n = 3 biological replicates, colocalization was calculated for 10 NTC and 10 RACK1 siRNA treated Huh7.5 cells transfected with either DENV2 or ZIKV PRVABC59).
RACK1 interacts with multiple flavivirus NS1 within the ER lumen.
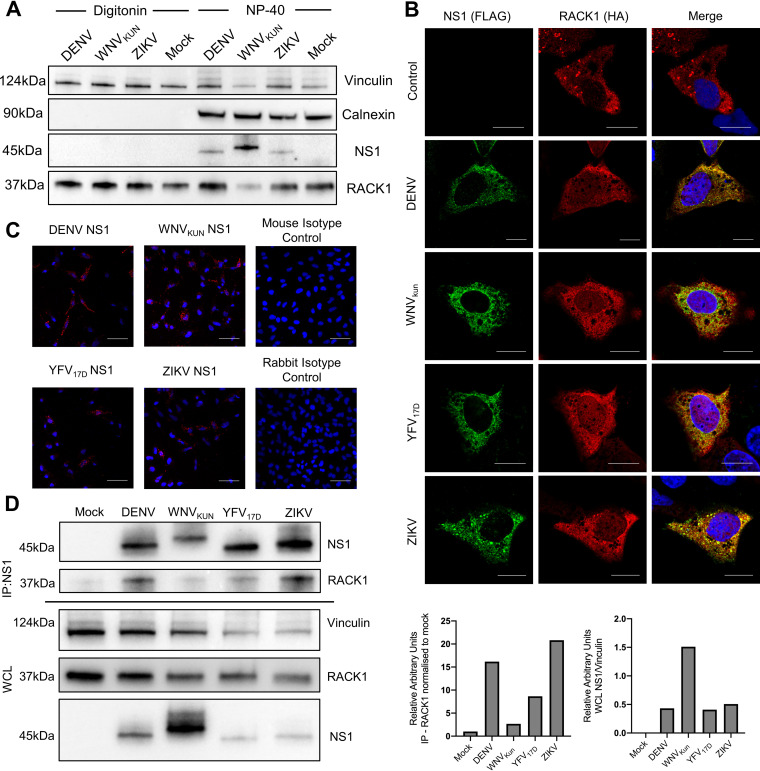
Re-modeling of ER membranes is required for flavivirus replication complex formation, driven by interactions between viral and ER-associated host factors (21). One of the viral proteins critical for VP biogenesis is NS1, which localizes to the ER lumen (22). In addition, a recent DENV NS1 interactome map has identified RACK1 as an interaction partner (16). Therefore, to investigate whether RACK1 was associated with the ER and in a position to potentially interact with NS1, HeLa cells were infected with DENV, West Nile Virus (Kunjin strain) (WNVKun) or ZIKV. Immunoblotting was utilised to determine levels of both flavivirus NS1 and RACK1 within fractional whole cell lysates (WCL) containing cytosolic (digitonin-sensitive) and membranous organelles (NP-40-sensitive) which include the ER (23). Flavivirus NS1 (NS1’ for WNVKUN) was only present within the NP-40 fraction, indicating its predominant localization within the membranous organelles, such as the ER lumen as expected (Fig. 7A). However, RACK1 was present in both the digitonin and NP-40 fraction, indicating RACK1 localization within the cytosol and membranous organelles and thus potentially capable of interacting spatially with RACK1 within the replication complex.
FIG 7.
RACK1 interacts with multiple flavivirus NS1 proteins within the ER lumen. (A) HeLa cells were infected with DENV, WNVKUN or ZIKV (MOI = 1) and 48 hpi, WCL were harvested via sequential incubation with digitonin and NP-40 lysis buffers. Immunoblot analysis was performed with primary (mouse anti-flavivirus NS1 4G4, mouse anti-RACK1 and rabbit anti-Calnexin) and secondary (anti-mouse and rabbit conjugated HRP) antibodies. Vinculin served as sample loading control. (B) Huh7.5 cells were transfected with DENV, WNVKUN, YFV17D or ZIKV NS1-FLAG and RACK1-HA tagged constructs. 24 h posttransfection, cells were fixed and stained with primary antibodies - anti-mouse FLAG/anti-rabbit HA, secondary antibodies – anti-mouse 488/anti rabbit 555. DAPI was used to stain for nuclear DNA (scale bar: 70 μm). (C) HeLa cells were transfected with DENV, WNVKUN, YFV17D or ZIKV NS1-FLAG and RACK1-HA tagged constructs. 24 h posttransfection, cells were fixed and proximity ligation assay performed. DAPI was used to stain for nuclear DNA (scale bar: 15 μm) (D) HeLa cells were infected with DENV2, WNVKUN, YFV17D or ZIKV (MOI = 2) for 24 h and WCL extracted for immunoblot analysis with primary (anti-vinculin, anti-NS1 and anti-RACK1) and secondary (anti-mouse conjugated HRP) antibodies. Vinculin served as sample loading control. Lysates were also used for immunoprecipitation using human anti-NS1 4G4 antibody and subjected to immunoblot analysis with primary (mouse anti-NS1 4G4 and -RACK1) and secondary (anti-mouse conjugated HRP) antibodies. Immunoblot densitometry quantification analysis was performed using Image J for RACK1 levels in flavivirus infected samples relative to mock infection after immunoprecipitation and levels of NS1 relative to loading control (vinculin) prior to immunoprecipitation. Data are n = 3 biological triplicates (except for A and D which is n = 1), representative of similar repeat experiments.
Given that both NS1 and RACK1 are integral to flavivirus VP formation and reside in the same cellular compartment, we next assessed if they interact. This was achieved by transfection of Huh7 cells with flavivirus NS1 expression constructs bearing an N-terminal FLAG-tag and RACK1 bearing an N-terminal HA-tag. Indeed, the co-expression of NS1 (DENV, WNVKun, YFV17D and ZIKV) and RACK1 revealed a reticular cytoplasmic staining pattern consistent with co-localization and robust interaction between NS1 and RACK1 also observed using proximity ligation assays (Fig. 7B and C). As overexpression can result in mis-localized proteins, we next sought to investigate the interaction of NS1 with endogenous RACK1 in HeLa cells infected with DENV, WNVKUN, YFV17D or ZIKV. Co-immunoprecipitation experiments were conducted to pull down NS1 (humanized NS1 4G4 antibody) from virus-infected HeLa cells, followed by immunoblotting to detect endogenous RACK1. Comparative analysis of NS1 expression revealed a strong interaction for DENV and ZIKV NS1 with endogenous RACK1 which was not as pronounced for WNVKun (NS1’) and YFD17D (Fig. 7D).
DISCUSSION
Host factors play critical roles in all facets of the flavivirus life cycle and identification of such factors is essential to not only our understanding of viral replication but also to exploit these host-virus interactions in the development of novel antiviral therapeutics. Although numerous host factors important for ZIKV and other flaviviruses have been identified, it is likely that additional host factors remain to be discovered. Therefore, to expand our understanding of virus-host interactions, we used a genome-scale CRISPR KO approach to screen for single guide RNAs (sgRNAs) that upon gene knockout conferred cell survival following infection with ZIKV. Similar approaches using genome-wide CRISPR KO screens have identified novel host factors essential for a range of viruses including the flaviviruses and include the OST, EMC and TRAP complexes (24). Although CRISPR technology has significant advantages to siRNA mediated screens in the form of improved consistency as a result of a reduction in false positives, differences in experimental parameters can result in bias during the recovery of sgRNAs and therefore influence novel host factors identified. In contrast to previous CRISPR screens in which cells are expanded (allowing for sgRNA enrichment) following virus induced CPE, we harvested cells immediately following virus induced cell death in the control cells to allow for identification of sgRNAs that may target genes that confer inherent defects in cell proliferation and that would otherwise be diluted and under-represented during the expansion process. Interestingly, using this approach we identified a limited number of sgRNAs compared to previous screens, indicating that the expansion stage may be critical for the enrichment of the abundance of gRNA sequences required to surpass the threshold of significance utilising the DESeq2 or MAGeCK algorithims. Nevertheless, among our top hits, in addition to RACK1 (GNBL21), our screen identified sgRNAs targeting the EMC complex (EMC1, EMC5 and EMC6) that have been previously identified in supporting expression of flaviviral NS4A and NS4B and thus confirms the validity and reproducibility of our approach (Fig. 1) (9, 11, 25). In addition, we identified sgRNAs targeting BAX, a member of the BCL2 family regulating apoptosis. This is not surprising, given that DENV and ZIKV both rely on BCLx to suppress activation of BAX to prolong cell survival (26). However, sgRNAs targeting RACK1 were the most significantly enriched in the current screen and thus we focused on the role of RACK1 in flavivirus biology.
Previous studies have identified a role for RACK1 in the life cycle of a number of unrelated viruses, the mechanism of which falls into two main categories; (i) RACK1 support of IRES mediated translation by virtue of RACK1 being localized to the 40s ribosomal subunit for viruses such as HCV, polio virus, cricket paralysis virus (CrPV) and picorna-like Drosophila C Virus (DCV), and (ii) RACK1-mediated intracellular signaling or as a scaffolding protein which assists the virus life cycle of viruses such as porcine reproductive and respiratory syndrome virus (PRRSV), IAV and HIV (14, 27–30). Our results indicate that the flaviviruses can now be added to the list of viruses for which RACK1 is a host dependency factor. Whether this is true for viral replication in the mosquito is unknown, however considering the high degree of homology (74% at AA level) between mammalian and mosquito RACK1, it is possible and further investigation is required (31). Using an siRNA depletion approach, we report that RACK1 is an essential host factor for multiple strains of ZIKV in different cell types and as expected given the similarities in their viral lifecycles, RACK1 was important for other mosquito (DENV, WNV) and tick-borne flaviviruses (LGTV and POWV). Interestingly, the vaccine strain YFV17D used in this study, is evolutionally distinct from other flaviviruses such as DENV and ZIKV and was not affected by RACK1 knockdown. While DENV and ZIKV have likely streamlined their lifecycles to require minimal host factors to improve efficiency, YFV evolves at a slower evolutionary rate compared to the former and therefore may utilize multiple redundant proteins to fulfil the same role as RACK1 (32). Another factor to consider is that although the vaccine strain of YFV results in active viral replication, this strain is both evolutionarily and replication divergent from circulating YFV, which may behave differently when RACK1 expression is lost (33, 34). It will be interesting to determine in future studies if WT strains of YFV are dependent on RACK1. We extended our RACK1 studies to determine if viruses outside the flavivirus genus also had a dependency for RACK1. Depletion of RACK1 had no impact on VSV or EBOV (negative strand RNA genome), or the DNA virus HSV. While not an exhaustive repertoire of viruses, our results suggest a level of selectivity for the dependency for RACK1 in viral replication-cycles.
From our initial studies, it was not apparent at what stage of the flavivirus life cycle RACK1 was playing a role. To investigate this further, we utilised a DENV SGR expressing Rluc to separate viral entry and infectious virus production from viral RNA replication. Following transfection of in vitro transcribed DENV SGR RNA into cells devoid of RACK1, we noted a significant reduction of DENV RNA replication suggesting that viral genome replication was inhibited. Given that RACK1 is a constituent protein of the eukaryotic ribosome and has been reported to play a role in HCV and CrPV IRES-dependent translation, it is possible that RACK1 may affect DENV 5’UTR cap dependent translation (14). This was improbable as input in vitro transcribed DENV SGR RNA (including that of the GND replication mutant) was translated with equal efficiency at early time points following transfection, ruling out RACK1 as playing a role at the level of translation. Moreover, the depletion of RACK1 post initiation of viral replication has no apparent impact on flavivirus replication. This indicates that RACK1 is a proviral host factor that must be present prior to the establishment of replication and supports the finding that it may target virus-induced ER membrane alteration and generation of vesicle packets and the virus replication complex.
Following flavivirus infection, expression of the viral NS proteins results in the reorganization of host ER membranes to generate viral replication organelles (ROs). These ROs are comprised of arrays of vesicle-like invaginations into rough ER membranes that have been designated as vesicle packets (VPs) (4). Evidence that VPs are the site of viral RNA replication stems from the detection of dsRNA (a flavivirus RNA genome replication intermediate) and viral replication components including viral NS proteins and essential host factors (35). However, while we have a good understanding of the architecture of flavivirus VPs, the host factors and mechanisms of biogenesis are not well understood. Evidence to suggest that RACK1 may play a role in flavivirus VP formation comes from the recent observation that it plays a role in the biogenesis of HCV ROs through an interaction with the HCV NS5A protein and ATG14L to induce autophagasome formation (15). Although this may be true, the mechanism by which HCV and DENV/ZIKV induce RO formation in addition to the differences observed in double membrane vesicle (DMV) morphology are fundamentally different and suggests that specific mechanisms may be at play (4, 15). For example, the HCV protein NS5A recruits and interacts with phosphatidylinositol 4-kinase-α (PI4Kα) resulting in increased local levels of PI4P that induces high amounts of localized phosphatidylinositol-4-phosphate that is required to modulate membrane lipid composition and drive the formation of double membrane vesicles and generation of HCV replication ROs (36). However, PI4P is dispensable for DENV RO formation (36). Moreover, the significant perturbation of NS1 and dsRNA localization following depletion of RACK1, and the requirement of RACK1 prior to infection combined with its interaction with NS1 (a key NS protein involved in VP formation) suggests that RACK1 may significantly influence VP formation.
A number of viral proteins and host factors have been implicated in flavivirus VP biogenesis, however evaluation of their role in this process has been problematic as deletion of these factors can impact on viral replication in the context of virus infection or using autonomous SGR based cell lines (37–39). To overcome this limitation, we adopted the recently described pIRO transcription-translation system that relies on T7 RNA polymerase and a heterologous IRES to achieve robust production and cleavage of DENV and ZIKV polyproteins with subsequent VP formation that is indistinguishable to VPs in infected cells (19). Thus, the production of VPs in this system is independent of replication and allows the study of host cells factors involved in RO production. Using this system coupled with siRNA knockdown of RACK1, we determined that RACK1 is important in the formation of VPs for both DENV and ZIKV. TEM revealed that in the absence of RACK1, the morphology of VPs for both DENV and ZIKV was significantly altered while quantification of VPs revealed that the percentage of cells with 2–5 or >5 VPs was also decreased with reduced RACK1 expression. These results are consistent with our immunofluorescence data in which we revealed significant alteration of NS1 and dsRNA positive DENV and ZIKV ROs in the absence of RACK1 expression. Collectively these results indicate that RACK1 is crucial for the biogenesis of both DENV and ZIKV (and most likely other flaviviruses) VPs to support robust viral replication. This raises the question as to how RACK1 contributes to VP formation. In the case of HCV, even though the ROs are morphologically distinct to those of DENV and ZIKV, the HCV NS5A protein induces autophagy and DMV formation through its interaction with RACK1 and ATG14L (15). Like HCV, we revealed that RACK1 resides and interacts with a viral NS protein, namely, NS1, using a number of experimental approaches. Interestingly, as for HCV NS5A protein, the flavivirus NS1 protein has recently been shown to be required for the biogenesis of DENV ROs independent of RNA genome replication (40). It is not inconceivable to envisage that RACK1 helps to stabilize NS1 at the ER membrane to facilitate VP formation or alternatively it provides a scaffold for the autophagy nucleation complex and formation of altered membrane vesicles as with HCV. Autophagy, the mechanism proposed for RACK1 assisted HCV DMV construction is also proposed to be induced during DENV and ZIKV infection (41), and it is possible that even though the ROs are morphologically different, similar mechanisms may be at play for flavivirus RO biogenesis and this warrants further investigation.
The dependency for RACK1 with regard to the flavivirus replication cycle most likely relates to its role in the biogenesis of ROs. Interestingly, the coronaviruses also initiate an extensive remodeling of intracellular ER membranes forming a three-dimensional structure referred to as the replication membranous web containing multiple interconnected vesicles with single or double membrane vesicles (DMVs) (42). DMV formation is not completely understood; however, non-structural viral proteins are critical for this process and hence it is not inconceivable to assume that RACK1 may also play a role in coronavirus DMV formation. While not a complete surprise, RACK1 depletion had a significant impact on SARS-CoV-2 genome replication, suggesting coronavirus cellular membrane rearrangements are RACK1 dependent and highlights the possibility of a shared mechanism by which positive stranded viruses remodel ROs. Given that both HCV and coronaviruses induce DMVs which are principal sites of virus replication (43), future experiments should investigate the role of RACK1 and autophagy as a common mechanism in the biogenesis of viral replication complex formation.
In conclusion, we have successfully used a genome-wide CRISPR KO screening approach to identify RACK1 as a host factor that is important for flavivirus infection. As we have demonstrated, RACK1 interacts with the flavivirus NS1 protein and colocalised to ROs containing dsRNA. Furthermore, depletion of RACK1 significantly impacts flavivirus RNA replication and the formation of ROs, highlighting the role of RACK1 as a key host factor essential for RO biogenesis. This study not only enhances our knowledge in the intricacies of the flavivirus life cycle and RO biogenesis, but also may guide novel approaches toward the design of pan-flavivirus and SARS-CoV-2 antiviral strategies.
MATERIALS AND METHODS
Cell and culture conditions.
All mammalian cell lines were maintained at 37°C in a 5% CO2 air atmosphere. Huh7 and Huh7.5 human hepatoma cells, HeLa human epithelial cells, HTR8/SVneo human trophoblast cells, HEK293T human embryonic kidney cells and Vero monkey kidney epithelial cells were maintained in DMEM (Gibco) containing 10% (vol/vol) FCS and 1% (vol/vol) penicillin and streptomycin. Huh7/Lunet-T7 cells stably expressing the bacteriophage T7 RNA polymerase have been described previously (44). A549-ACE2 cells were generated by transduction of lentiviruses encoding ACE2.
C6/36 Aedes albopictus cells were maintained in Basal Medium Eagle (BME) supplemented with l-glutamine, MEM non-essential amino acids, sodium pyruvate, 10% FBS and P/S and cultured at 28°C with 5% CO2.
Antibodies and Chemicals.
Mouse anti-envelope glycoprotein 4G2 (D1-4G2-4-15) was prepared from hybridoma cells purchased from ATCC. Mouse anti-dsRNA 3G1 and mouse anti-flavivirus NS1 4G4 was a generous gift from Roy Hall (University of Queensland). Human anti-flavivirus NS1 4G4 was a generous gift from Daniel Watterson (University of Queensland). Mouse anti-RACK1 was purchased from Santa Cruz Biotechnology (SC-17754) and mouse anti-FLAG and anti-HA were purchased from Sigma-Aldrich. Mouse anti-Myc and rabbit anti-FLAG were purchased from Cell Signaling Technologies. Mouse anti-SARS-CoV-2 spike antibody (1A9-GTX632604) was purchased from GeneTex. All secondary antibodies were purchased from Thermo Fisher Scientific.
Viruses and Plasmids.
The ZIKV strains MR766 (Uganda, 1947) and PRVABC59 (Puerto Rico, 2015) were propagated in C6/36 cells by infection at a multiplicity of infection (MOI) of 0.01 and supernatants harvested at 5–6 dpi. PRVABC59 is a contemporary strain that was isolated by CDC from the serum of a ZIKV infected patient who travelled to Puerto Rico in 2015. The complete genome sequence is published (Ref. Gene bank accession # KU501215). DENV infections utilised Mon601, a derivative of the New Guinea C strain that was produced from in vitro transcribed RNA, transfected into BHK-21 baby hamster kidney cells, amplified in C6/36 insect cells and titered in Vero cells. DENV 16681 infectious virus was generated as previously described (45) WNVKUN (Kunjin strain NSW2011) was generously provided by Karla J. Helbig (La Trobe University Melbourne, Australia). Other flaviviruses (46) and EBOV (47) have been described previously. SARS-CoV2 nCoV-WA1-2020 (MN985325.1) was provided by the Centers of Disease Control and Prevention (CDC) and propagated in Vero E6 cells in DMEM, supplemented with 2% (vol/vol) FCS, 1 mM l-glutamine and 1% (vol/vol) penicillin and streptomycin.
Plasmids were transfected with Lipofectamine 3000 (Thermo Fisher Scientific) following manufacturer’s recommendations. RACK1 tagged plasmids were cloned via PCR of cDNA from Huh7 cells and Gibson Assembly performed with restriction enzyme digested pcDNA6.2. N-EmGFP-DEST. pEGFP-N1-RACK1 was a gift from Anna Huttenlocher (Addgene plasmid # 41088). ZIKV NS tagged proteins were individually cloned via PCR of pZIKV-ICD (strain Paraiba_01/2015) and Gibson Assembly performed with restriction enzyme digested pcDNA6.2. N-EmGFP-DEST. LentiCRISPRv2 was a gift from Feng Zhang (Addgene plasmid# 52961). Plasmid encoding for RACK1 which was resistant to RACK1 siRNA (described below) was constructed by Gibson Assembly of restriction enzyme digested pcDNA6.2. N-EmGFP-DEST and a gBlock with silent mutations within RACK1 where the siRNA bound.
In vitro transcription of viral RNA.
Plasmid pFK-DVs containing the full-length DENV-2 genome (strain 16681), pFK-sgDVs-R2A and pFK-sgDVs-R2A-GND which are the sub-genomic DENV replicon and GND mutant based on the DENV-2 genome (strain 16681) have been previously described (48). To initiate virus replication, all DENV plasmid constructs were linearized with XbaI prior to in vitro RNA transcription with the mMessage Machine SP6 transcription kit (Thermo Fisher Scientific) and transfection of viral RNA into Huh7.5 cells by transfection with Lipofectamine 3000 (Thermo Fisher Scientific) following manufacturer’s recommendations.
CRISPR/Cas9 Genome-wide KO Screen.
The Human GeCKOv2 CRISPR knockout pooled library was a gift from Feng Zhang (Addgene #1000000048). Generation of the lentiviral CRISPR knockout pooled library and subsequent screening has been described previously (49, 50). Briefly, the library was amplified with the supplied protocol and lentivirus generated via transfection of 293FT cells with Lipofectamine 2000 (Thermo Fisher Scientific) and psPAX2 and PMD2.G lentiviral packaging plasmids. Following determination of the lentiviral titer, 300 million Huh7 cells were transduced with the lentiviral pooled library at an MOI of 0.3. 1 week postransduction, puromycin (3 μg/ml) was added for a week to select for cells with successful transduction. ZIKV (MR766) was added to the 300 million puromycin resistant transduced cells at an MOI = 5 and the screen terminated when the control un-transduced cells had died following ZIKV induced CPE. Surviving lentiCRISPRv2 transduced cells were harvested and genomic DNA isolated using a Blood and Cell Culture DNA Midi Kit (Qiagen). Genomic DNA from LentiCRISPRv2 transduced cells in the absence of ZIKV infection was also harvested to determine the representation of the library prior to ZIKV infection. Two rounds of PCR (Q5 - NEB) was performed on the genomic DNA to first amplify the guide region of both control and ZIKV-infected samples and second, introduce the Illumina primers to enable the samples to be run on next-generation sequencing. The NextSeq 2 × 75 bp kit (Illumina) was utilised and the data run through caRpools with the assistance of the Adelaide Bioinformatics Hub to identify enriched genes and therefore novel host factors required for ZIKV infection.
Generation of EMC1/EMC6/RACK1 CRISPR KO polyclonal cell lines.
Benchling was utilised to generate 20 nt guide sequences targeting EMC1, EMC6 and RACK1. 20 nt oligos were synthesized and annealed prior to insertion into BsmBI digested LentiCRISPRv2. Following the generation of lentiviruses encoding LentiCRISPRv2 with EMC1, EMC6 or RACK1, Huh7.5 were transduced and puromycin selected 5 days postransduction for 1 week. Surviving cells were infected with ZIKV (PRVABC59) and qRT-PCR performed to quantify mRNA levels of EMC1, EMC6 or RACK1 and ZIKV RNA levels. sgRNA sequences are listed below: EMC1= TCCTGGGAGACTAACATCGG, EMC6 = CCGGCAATAATCCAGGACGG and RACK1 = CGCCATTTTGCATGGTCGAG.
RACK1 knockdown and pIRO plasmid transfection.
RACK1 knockdown for use in infection, immunofluorescence and pIRO assays was achieved using a RACK1 siRNA pool (Dharmacon: L-006876-00-0005, 5 nmol). Concentrations of reagents were modified dependent on vessel size. For a well of a 6-well plate, 3 μl of either RACK1 or non-targeting control (NTC) (Dharmacon: D-001810-10) siRNA pool from a stock solution of 20 μM was added to 100 μl of Opti-MEM (ThermoFisher: 31985070) and 5 μl of Lipofectamine RNAi-MAX was added to another tube containing 100 μl of OPTI-MEM. After 5 min incubation at room temperature (RT), Opti-MEM solutions containing the siRNA and transfection reagent were mixed and incubated for 15 min at RT. siRNA transfection mix was added to Huh-7 cells (seeded 24 h prior) and 24 h posttransfection cells were either infected with virus or transfected with plasmids/RNA. Studies using the pIRO system are described eleswhere (19). In brief, Huh7/Lunet T7 cells (3 × 105) were seeded per well of a 6-well plate well, together with either RACK1 or NTC siRNA transfection mixture. After 24 h cells, were trypsinized and 5 × 104 cells were seeded per well of a 24-well plate containing a glass coverslip. For expression of viral proteins (ZIKV or DENV) in these cells, either plasmid DNA (500 ng) pIRO-Z:Δ 5’SLAB or pIRO-D:Δ 5’SLAB was added to 100 μl of Opti-MEM media, mixed briefly and 1.5 μl of Trans-IT-LT1 transfection reagent (Mirus: MIR2304) was added to the DNA - Opti-MEM mixture and mixed briefly. After 20 min incubation at RT, the transfection mixture was added onto the cells. Following a 4 h incubation period, medium was changed, and cells were fixed 16 to 20 h after transfection for transmission electron microscopy (EM) or immunofluorescence (IF) analysis. TEM was performed as has been recently described (19).
Immunofluorescence microscopy, Immunoprecipitation and Immunoblot.
Immunofluorescent labeling was performed as described previously (51). Cells were examined using either a Nikon Eclipse Ti microscope or an Olympus FV3000 confocal microscope. To quantify positioning of red and green fluorescence within Huh7.5 cells transfected with either NTC or RACK1 siRNA, we made use of a previously validated approach based on Imaris Coloc Suite (v9.3.0, Bitplane Inc., Switzerland) (52). Briefly, this approach provides an automated quantitation of signals’ co-occurrence based on correlation coefficients that measure the strength of the linear relationship between two variables, i.e., the gray values of fluorescence intensity voxels of green and red image pairs. Pearson’s correlation coefficient (PCC) calculates the relationship between intensities in two images by linear regression and the slope of the fitted line provides the rate of association of two fluorophores. Its value can range from +1 to -1, with 1 standing for complete positive correlation and -1 for negative correlation, with zero indicating no correlation (52, 53). To perform immunoprecipitation, cell lysates were harvested from cells using a lysis buffer containing 1% Triton X-100, 50 mM Tris pH 8, 150 mM NaCl and 1x Protease Inhibitor cocktail (Sigma) and centrifuged at 21,000 × g to remove cell debris. Following overnight incubation of the antibody of interest (0,5 μg/sample at 4°C), Protein A MagBeads (Genscript) was added to each sample and the manufacturers recommended protocol followed. To isolate bound protein from the beads during elution, 2x sample buffer (Bio-Rad) was added to each sample and boiled at 100°C for 10 min. After separation from the MagBeads, the boiled lysate was utilised for Western blotting. Western blotting was performed as described elsewhere (51). Membrane-bound protein was detected by chemiluminescence using SuperSignal West Femto (Thermo Fisher Scientific) and imaged using a ChemiDoc MP imaging system (Bio-Rad).
Nano Luciferase Assays.
Luciferase assays to quantify nano-luciferase (NLuc) were performed as described previously (54). Briefly, Huh7.5 cells were seeded into 48-well plates prior to transfection with NTC/RACK1 siRNA or infection with DENV2-NS1-Nluc at the indicated MOI. Samples were harvested with 1x passive lysis buffer before measurement of NLuc activity using the Nano-Glo luciferase assay system (Promega) with the GloMax-96 luminometer (Promega).
Proximity Ligation Assay.
Proximity ligation assay was performed as described previously (55). Briefly, proximity ligation kits (Duolink PLA) were purchased from Sigma-Aldrich and the manufacturer’s protocol was followed. Briefly, cells were fixed with acetone/methanol, blocked with 5% BSA and incubated with primary antibodies of interest raised in divergent species. Cells were then incubated with mouse PLA probe PLUS and rabbit PLA probe MINUS for 1 h in a humidity chamber at 37°C. The ligation mix containing the ligase enzyme was incubated with the cells for 30 min in the humidity chamber at 37°C, followed by the amplification mix (90 min in the humidity chamber at 37°C) containing the polymerase which facilitates rolling circle amplification if the PLA probes are sufficient proximity in the cells. Cells were then incubated with DAPI for 5 min at room temperature and visualized under the immunofluorescence microscope as outlined above.
Plaque Assay.
Virus infectivity was determined by plaque assay as described previously (51). Briefly, Vero cells in 24-well plates were infected with 200 μl of serially-diluted virus-containing supernatants for 1 h at 37°C before the addition of 1 ml overlay of complete media containing 1.5% (wt/vol) carboxymethylcellulose (CMC) (Sigma) and cells returned to culture for various days dependent on the virus. Following fixation with 10% formation for 1 h, the CMC overlay was removed and plaques visualized via crystal violet stain. Plaques were counted and virus infectivity expressed as PFU per ml.
Quantitative Real-Time PCR.
Cells were seeded in 24-well plates for 24 h prior to transfection or infection. Total RNA was extracted from cells using NucleoZOL (Macherey-Nagel) per manufacturers recommendation. Both cDNA synthesis and qRT-PCR were performed simultaneously with the Luna Universal One-Step RT-qPCR Kit (NEB) with the Quantstudio 7 Flex (Life Technologies) to quantitate relative levels of ZIKV RNA, RACK1 mRNA and housekeeping gene RPLPO. All primer sequences used are outlined below: EMC1 FP: 5’CGGCCTGAGCGGCTGTATATC3’, EMC1 RP: 5’CTCCACAGCACCACCTTCCC3’, EMC6 FP: 5’TTCTACCTGCTCGCCTCCGT3’, EMC6 RP: 5′ CCCGATGAGGCCTCCTGTAA3’, RACK1 FP: 5′ TAACCGCTACTGGCTGTGTG3’ and RACK1 RP: 5′ GCCTTGCTGCTGGTACTGAT3’.
Statistics.
Data was graphed and analyzed with Prism 9 (GraphPad Software) using Student's t test, with P < 0.05 considered to be significant.
ACKNOWLEDGMENTS
This work was supported by the NHMRC of Australia (APP1145613). We thank Blagojce Jovcevski for technical assistance.
Author contributions: Byron Shue, Abhilash I. Chiramel, Berati Cerikan, Sonja Frölich, Emily N. Kirby, and Michael R. Beard conducted experiments and analyzed the results, Thu-Hien To, and Stephen M. Pederson provided bioinformatics analysis while Nicholas S. Eyre, Stephen M. Pederson, Ralf F. W. Bartenschlager, and Sonja M. Best provided experimental design input and reagents. Byron Shue and Michael R. Beard conceived the study and wrote the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael R. Beard, Email: michael.beard@adelaide.edu.au.
Julie K. Pfeiffer, University of Texas Southwestern Medical Center
REFERENCES
- 1.Daep CA, Muñoz-Jordán JL, Eugenin EA. 2014. Flaviviruses, an expanding threat in public health: focus on dengue, West Nile, and Japanese encephalitis virus. J Neurovirol 20:539–560. 10.1007/s13365-014-0285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deen J. 2016. The dengue vaccine dilemma: balancing the individual and population risks and benefits. PLoS Med 13:e1002182. 10.1371/journal.pmed.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawes BE, Smalley CA, Tiner BL, Beasley DW, Milligan GN, Reece LM, Hombach J, Barrett AD. 2016. Research and development of Zika virus vaccines. NPJ Vaccines 1:16007–16007. 10.1038/npjvaccines.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatel-Chaix L, Bartenschlager R. 2014. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside–caught in the web. J Virol 88:5907–5911. 10.1128/JVI.03404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aktepe TE, Mackenzie JM. 2018. Shaping the flavivirus replication complex: It is curvaceous!. Cell Microbiol 20:e12884. 10.1111/cmi.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puschnik AS, Majzoub K, Ooi YS, Carette JE. 2017. A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol 15:351–364. 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi J-G, Shi G, Qi L, Manjunath N, Wu H. 2015. A CRISPR-Based screen identifies genes essential for West-Nile-Virus-Induced Cell Death. Cell Rep 12:673–683. 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, Guo Z, Green S, Kowalik TF, Brass AL. 2016. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep 16:232–246. 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 10.Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, Diamond MS. 2016. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535:164–168. 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Muffat J, Omer Javed A, Keys HR, Lungjangwa T, Bosch I, Khan M, Virgilio MC, Gehrke L, Sabatini DM, Jaenisch R. 2019. Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc Natl Acad Sci USA 116:9527–9532. 10.1073/pnas.1900867116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams DR, Ron D, Kiely PA. 2011. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun Signal 9:22–24. 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winter J, Breinig M, Heigwer F, Brügemann D, Leible S, Pelz O, Zhan T, Boutros M. 2016. caRpools: an R package for exploratory data analysis and documentation of pooled CRISPR/Cas9 screens. Bioinformatics 32:632–634. 10.1093/bioinformatics/btv617. [DOI] [PubMed] [Google Scholar]
- 14.Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, Baumert TF, Schuster C, Imler J-L. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159:1086–1095. 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JS, Tabata K, Twu W-I, Rahman MS, Kim HS, Yu JB, Jee MH, Bartenschlager R, Jang SK. 2019. RACK1 mediates rewiring of intracellular networks induced by hepatitis C virus infection. PLoS Pathog 15:e1008021. 10.1371/journal.ppat.1008021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, Bonnet-Madin L, Kümmerer BM, Delaugerre C, Roingeard P, Vidalain P-O, Amara A. 2018. A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell Rep 22:1364. 10.1016/j.celrep.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 17.Shao Q, Herrlinger S, Zhu Y-N, Yang M, Goodfellow F, Stice SL, Qi X-P, Brindley MA, Chen J-F. 2017. The African Zika virus MR-766 is more virulent and causes more severe brain damage than current Asian lineage and dengue virus. Development 144:4114–4124. 10.1242/dev.156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 19.Cerikan B, Goellner S, Neufeldt CJ, Haselmann U, Mulder K, Chatel-Chaix L, Cortese M, Bartenschlager R. 2020. A non-replicative role of the 3' terminal sequence of the dengue virus genome in membranous replication organelle formation. Cell Rep 32:107859. 10.1016/j.celrep.2020.107859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rastogi M, Sharma N, Singh SK. 2016. Flavivirus NS1: a multifaceted enigmatic viral protein. Virol J 13:131–10.. 10.1186/s12985-016-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothan HA, Kumar M. 2019. Role of endoplasmic reticulum-associated proteins in flavivirus replication and assembly complexes. Pathogens 8:148. 10.3390/pathogens8030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ci Y, Liu Z-Y, Zhang N-N, Niu Y, Yang Y, Xu C, Yang W, Qin C-F, Shi L. 2020. Zika NS1-induced ER remodeling is essential for viral replication. J Cell Biol 219:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holden P, Horton WA. 2009. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res Notes 2:243–250. 10.1186/1756-0500-2-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDougall WM, Perreira JM, Reynolds EC, Brass AL. 2018. CRISPR genetic screens to discover host-virus interactions. Curr Opin Virol 29:87–100. 10.1016/j.coviro.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Lin DL, Inoue T, Chen Y-J, Chang A, Tsai B, Tai AW. 2019. The ER membrane protein complex promotes biogenesis of dengue and Zika virus non-structural multi-pass transmembrane proteins to support infection. Cell Rep 27:1666–1674.e4. 10.1016/j.celrep.2019.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki T, Okamoto T, Katoh H, Sugiyama Y, Kusakabe S, Tokunaga M, Hirano J, Miyata Y, Fukuhara T, Ikawa M, Satoh T, Yoshio S, Suzuki R, Saijo M, Huang DCS, Kanto T, Akira S, Matsuura Y. 2018. Infection with flaviviruses requires BCLXL for cell survival. PLoS Pathog 14:e1007299. 10.1371/journal.ppat.1007299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallina A, Rossi F, Milanesi G. 2001. Rack1 binds HIV-1 Nef and can act as a Nef-protein kinase C adaptor. Virology 283:7–18. 10.1006/viro.2001.0855. [DOI] [PubMed] [Google Scholar]
- 28.Demirov D, Gabriel G, Schneider C, Hohenberg H, Ludwig S. 2012. Interaction of influenza A virus matrix protein with RACK1 is required for virus release. Cell Microbiol 14:774–789. 10.1111/j.1462-5822.2012.01759.x. [DOI] [PubMed] [Google Scholar]
- 29.Bi J, Zhao Q, Zhu L, Li X, Yang G, Liu J, Yin G. 2018. RACK1 is indispensable for porcine reproductive and respiratory syndrome virus replication and NF-κB activation in Marc-145 cells. Sci Rep 8:2985–2992. 10.1038/s41598-018-21460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaFontaine E, Miller CM, Permaul N, Martin ET, Fuchs G. 2020. Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 545:53–62. 10.1016/j.virol.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González-Calixto C, Cázares-Raga FE, Cortés-Martínez L, Del Ángel RM, Medina-Ramírez F, Mosso C, Ocádiz-Ruiz R, Valenzuela JG, Rodríguez MH, Hernández-Hernández F de LC. 2015. AealRACK1 expression and localization in response to stress in C6/36 HT mosquito cells. J Proteomics 119:45–60. 10.1016/j.jprot.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sall AA, Faye O, Diallo M, Firth C, Kitchen A, Holmes EC. 2010. Yellow fever virus exhibits slower evolutionary dynamics than dengue virus. J Virol 84:765–772. 10.1128/JVI.01738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck A, Tesh RB, Wood TG, Widen SG, Ryman KD, Barrett ADT. 2014. Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing. J Infect Dis 209:334–344. 10.1093/infdis/jit546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Garcia MD, Meertens L, Chazal M, Hafirassou ML, Dejarnac O, Zamborlini A, Desprès P, Sauvonnet N, Arenzana-Seisdedos F, Jouvenet N, Amara A. 2016. Vaccine and wild-type strains of yellow fever virus engage distinct entry mechanisms and differentially stimulate antiviral immune responses. mBio 7:e01956-15–e01915. 10.1128/mBio.01956-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447. 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet M-S, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM. 2017. The host protein reticulon 3.1A is utilized by flaviviruses to facilitate membrane remodelling. Cell Rep 21:1639–1654. 10.1016/j.celrep.2017.10.055. [DOI] [PubMed] [Google Scholar]
- 38.Neufeldt CJ, Cortese M, Scaturro P, Cerikan B, Wideman JG, Tabata K, Moraes T, Oleksiuk O, Pichlmair A, Bartenschlager R. 2019. ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat Microbiol 4:2416–2429. 10.1038/s41564-019-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, La Cruz-Rivera De PC, Douglas C, Konopka G, Xing C, Schoggins JW. 2018. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol 3:1214–1223. 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Płaszczyca A, Scaturro P, Neufeldt CJ, Cortese M, Cerikan B, Ferla S, Brancale A, Pichlmair A, Bartenschlager R. 2019. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog 15:e1007736. 10.1371/journal.ppat.1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiramel AI, Best SM. 2018. Role of autophagy in Zika virus infection and pathogenesis. Virus Res 254:34–40. 10.1016/j.virusres.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopfer H, Herzig MC, Gosert R, Menter T, Hench J, Tzankov A, Hirsch HH, Miller SE. 2020. Hunting coronavirus by transmission electron microscopy: a guide to SARS-CoV-2-associated ultrastructural pathology in COVID-19 tissues. Histopathology 150:w20203. 10.1111/his.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff G, Melia CE, Snijder EJ, Bárcena M. 2020. Double-membrane vesicles as platforms for viral replication. Trends Microbiol 28:1022–1033. 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appel N, Pietschmann T, Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol 79:3187–3194. 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyre NS, Johnson SM, Eltahla AA, Aloi M, Aloia AL, McDevitt CA, Bull RA, Beard MR. 2017. Genome-wide mutagenesis of dengue virus reveals plasticity of the NS1 Protein and enables generation of infectious tagged reporter viruses. J Virol 91:504. 10.1128/JVI.01455-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiramel AI, Meyerson NR, McNally KL, Broeckel RM, Montoya VR, Méndez-Solís O, Robertson SJ, Sturdevant GL, Lubick KJ, Nair V, Youseff BH, Ireland RM, Bosio CM, Kim K, Luban J, Hirsch VM, Taylor RT, Bouamr F, Sawyer SL, Best SM. 2019. TRIM5α restricts flavivirus replication by targeting the viral protease for proteasomal degradation. Cell Rep 27:3269–3283.e6. 10.1016/j.celrep.2019.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kondoh T, Letko M, Munster VJ, Manzoor R, Maruyama J, Furuyama W, Miyamoto H, Shigeno A, Fujikura D, Takadate Y, Yoshida R, Igarashi M, Feldmann H, Marzi A, Takada A. 2018. Single-nucleotide polymorphisms in human NPC1 influence filovirus entry into cells. J Infect Dis 218:S397–S402. 10.1093/infdis/jiy248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischl W, Bartenschlager R. 2013. High-throughput screening using dengue virus reporter genomes. Methods Mol Biol 1030:205–219. 10.1007/978-1-62703-484-5_17. [DOI] [PubMed] [Google Scholar]
- 49.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12:828–863. 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Hoek KH, Eyre NS, Shue B, Khantisitthiporn O, Glab-Ampi K, Carr JM, Gartner MJ, Jolly LA, Thomas PQ, Adikusuma F, Jankovic-Karasoulos T, Roberts CT, Helbig KJ, Beard MR. 2017. Viperin is an important host restriction factor in control of Zika virus infection. Sci Rep 7:4475. 10.1038/s41598-017-04138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liffner B, Frölich S, Heinemann GK, Liu B, Ralph SA, Dixon MWA, Gilberger T-W, Wilson DW. 2020. PfCERLI1 is a conserved rhoptry associated protein essential for Plasmodium falciparum merozoite invasion of erythrocytes. Nat Commun 11:1411–1414. 10.1038/s41467-020-15127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frölich S, Robker R, Russell D. 2020. Development of automated microscopy-assisted high-content multiparametric assays for cell cycle staging and foci quantitation. Cytometry A 97:378–393. 10.1002/cyto.a.23988. [DOI] [PubMed] [Google Scholar]
- 54.Eyre NS, Aloia AL, Joyce MA, Chulanetra M, Tyrrell DL, Beard MR. 2017. Sensitive luminescent reporter viruses reveal appreciable release of hepatitis C virus NS5A protein into the extracellular environment. Virology 507:20–31. 10.1016/j.virol.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Khantisitthiporn O, Shue B, Eyre NS, Nash CW, Turnbull L, Whitchurch CB, Van der Hoek KH, Helbig KJ, Beard MR. 2021. Viperin interacts with PEX19 to mediate peroxisomal augmentation of the innate antiviral response. Life Sci Alliance 4 10.26508/lsa.202000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1 (CRISPR screen data set.)<br>. Download jvi.00596-21-s0001.xls, XLS file, 2.5 MB (2.5MB, xls)