Abstract
Agrobacterium tumefaciens, a gram-negative soil bacterium, transfers DNA to many plant species. In the plant cell, the transferred DNA (T-DNA) is integrated into the genome. An in vitro ligation-integration assay has been designed to investigate the mechanism of T-DNA ligation and the factors involved in this process. The VirD2 protein, which is produced in Agrobacterium and is covalently attached to T-DNA, did not, under our assay conditions, ligate T-DNA to a model target sequence in vitro. We tested whether plant extracts could ligate T-DNA to target oligonucleotides in our test system. The in vitro ligation-integration reaction did indeed take place in the presence of plant extracts. This reaction was inhibited by dTTP, indicating involvement of a plant DNA ligase. We found that prokaryotic DNA ligases could substitute for plant extracts in this reaction. Ligation of the VirD2-bound oligonucleotide to the target sequence mediated by T4 DNA ligase was less efficient than ligation of a free oligonucleotide to the target. T-DNA ligation mediated by a plant enzyme(s) or T4 DNA ligase requires ATP.
The soil bacterium Agrobacterium tumefaciens is a plant pathogen responsible for tumor induction on dicotyledonous plants through its ability to transfer DNA carrying plant-active oncogenes to the plant cell (16, 25, 36). Because of this ability, Agrobacterium is widely used for plant transformation (12). The transferred DNA (T-DNA), which in Agrobacterium resides on a large tumor-inducing (Ti) plasmid, is processed within the bacterium and is exported to the plant, where it is integrated into the plant genome (28, 29, 30). Proteins encoded by the virulence (vir) region of the Ti plasmid regulate T-DNA processing and transfer. Virulence proteins recognize 24-bp-long imperfect direct repeats (border sequences) that define the T-DNA. In the presence of the VirD1 protein, VirD2 cleaves the border sequence in a site- and strand-specific manner and concomitantly becomes covalently attached to the 5′ end of the nicked DNA (9, 13, 33, 35). The nicked DNA is then displaced 5′ to 3′ from the plasmid, producing single-stranded T-DNA (8). The T-DNA–VirD2 complex and the VirE2 protein are believed to be transferred to the plant with the help of a pilus-like structure containing the VirB and VirD4 proteins (2, 4, 10, 37). In the plant cell, T-DNA is coated with the single-stranded DNA (ssDNA)-binding protein VirE2 (8, 25), forming a T-DNA–protein complex that is imported into the nucleus, where the T-DNA is integrated into the nuclear genome. The VirD2 protein is transferred into the nucleus in conjunction with the T-DNA; it presumably remains attached to it up to the integration step.
In higher eukaryotic organisms, such as plants, illegitimate recombination is the predominant mechanism of integration for naked DNA (20, 22, 24, 26). Likewise, the T-DNA is integrated into the plant genome by illegitimate recombination, a mechanism in which two DNA molecules that do not share extensive homology are joined (11, 18, 19, 31). It is not clear yet whether bacterial and/or plant factors mediate the integration of T-DNA. VirD2 has been shown not only to cleave the border sequence of ssDNA in vitro but also to rejoin the reaction partners (15, 23). However, although VirD2-mediated in vitro cutting and rejoining reactions are T-DNA border specific, T-DNA integration in vivo is sequence independent (11, 18, 19, 31). These findings suggest that the cutting and joining activity of VirD2 is likewise not involved in T-DNA ligation in vivo. On the other hand, it has been found that an R-to-G mutation in the H-R-Y integrase motif of VirD2 led to a decrease in the precision, but not in the efficiency, of the integration in vivo, suggesting involvement of VirD2 in T-DNA integration (31). Thus, the nature of the function of VirD2 in this process is still unclear.
To analyze the function, if any, of VirD2 in joining the 5′ terminus of the T-DNA to plant DNA, we designed an in vitro ligation-integration assay. We show that VirD2 does not possess general ligase activity. However, an enzyme(s) present in plant extracts, likely DNA ligase(s), was able to ligate the 5′ end of T-DNA from an artificial T-DNA–VirD2 complex to a partly double-stranded oligonucleotide. This reaction could be mimicked by other DNA ligases, e.g., T4 DNA ligase. Nevertheless, the reaction was less efficient than a standard ligation in the absence of VirD2. ATP was a cofactor for T-DNA ligation mediated by a plant enzyme(s) or T4 DNA ligase.
MATERIALS AND METHODS
DNA and proteins.
Synthetic oligonucleotides described in Table 1 were used. Oligonucleotides 3 and 6 were labeled at the 5′ end with [γ-32P]ATP using polynucleotide kinase (Boehringer Mannheim) according to the protocol provided by the supplier. Oligonucleotide 4 was phosphorylated (8-mer-P) at the 5′ end using polynucleotide kinase and ATP, as indicated in the protocol provided by the supplier. Oligonucleotide 4 was dephosphorylated (8-mer-OH) using calf intestine alkaline phosphatase (New England BioLabs) as advised by the supplier. VirD2 was purified as described previously (23). T4 DNA ligase, T4 RNA ligase, Taq DNA ligase, and Escherichia coli DNA ligase were from New England BioLabs. Trypsin was from Boehringer Mannheim.
TABLE 1.
Description of oligonucleotides used for in vitro T-DNA ligation studies
| Oligonucleotide | Length | Sequencea |
|---|---|---|
| 1 | 35-mer | 5′GCTCAAATTACAACGGTATATATCCTG∧CCAGTCAG3′ |
| 2 | 19-mer | 5′CTGACTGGGTACGTTTGGC3′ |
| 3 | 13-mer | 5′TAGCCAAACGTAC3′ |
| 4 | 8-mer | 5′CCAGTCAG3′ |
| 5 | 19-mer | 5′CTGACTGGCAGGATATATA3′ |
| 6 | 13-mer | 5′GGTATATATCCTG3′ |
The border sequence is underlined, and the cleavage site within it is marked (∧). Oligonucleotide 1 contained the complete border sequence, whereas oligonucleotides 5 and 6 contained only partial border sequences.
Plant extracts.
Nuclear extracts from synchronized tobacco BY2 suspension cultured cells (from S-phase) and extracts from pea shoot apices (soluble fractions) were prepared by following the protocols published previously (references 27 and 6, respectively).
Cleavage assay.
The activity of VirD2 was tested on 5′-radiolabeled oligonucleotide 1 in the assay described previously (23). The efficiency of VirD2-mediated cleavage of oligonucleotide 1 was 90 to 95%.
In vitro ligation assay.
Target DNA was produced by annealing 5′-radiolabeled oligonucleotide 3 (13-mer*) to oligonucleotide 2 (19-mer) at a 1:1 molar ratio, by first heating for 5 min at 95°C and then slowly cooling to room temperature. The T-DNA–VirD2 complex (8-mer–VirD2) was obtained by reaction of oligonucleotide 1 with VirD2 for 1 h at 37°C (1 μg of VirD2 per 3 pmol of oligonucleotide in TNM buffer [20 mM Tris-HCl, pH 8.8; 50 mM NaCl, 5 mM MgCl2]; for example, 33 μg of VirD2 and 100 pmol of oligonucleotide 1 in 80 μl of TNM buffer) as described previously (23). In some experiments the complex was treated in TNM buffer with trypsin (final concentration of 50 μg/μl) for 30 min at room temperature, followed by trypsin inactivation by incubation at 75°C for 10 min. The ligation test was performed for 15 min at room temperature in a final volume of 20 μl containing 20 mM Tris-HCl (pH 8.8), 50 mM NaCl, 5 mM MgCl2, either 5 mM ATP or 5 mM ATPγS (adenosine 5′-O-3-thiotriphosphate) or 5 mM AMPPNP (adenosine-5′-β,γ-imidotriphosphate) or 5 mM AMPCPP (adenosine-5′-α,β-methylentriphosphate), 4 pmol of target DNA, and various amounts (0 to 25 pmol) of T-DNA complex (8-mer–VirD2; standard reaction contained 8 pmol, calculated based on the efficiency of VirD2-mediated cleavage of oligonucleotide 1; see “Cleavage assay”) or 4 pmol of phosphorylated 8-mer (8-mer-P) with or without either plant extracts (6 μg of tobacco extracts or 20 μg of pea extracts) or T4 DNA ligase (4 U). Other ligases were used in the amounts indicated in the figure legends and under the buffer conditions recommended for each ligase. In some experiments the following inhibitors (Sigma) were added: aphidicolin (final concentration of 150 μM), ddTTP (final concentration of 5 μM), and dTTP (final concentration of 1 mM). The reactions were stopped by addition of formamide to the final concentration of 30%. The products were separated on 20% polyacrylamide–8 M urea gels run for 2 h at 40 V/cm and analyzed using a PhosphorImager (Molecular Dynamics). The efficiency of ligation was calculated as the percentage of the radioactivity of the ligation product (21-mer*) to the total radioactivity in the reaction (13-mer* plus 21-mer*).
Site-specific in vitro ligation assay.
The assay was performed as described above, with the exception that a sequence-specific target DNA (produced by annealing oligonucleotide 5 and 5′-radiolabeled oligonucleotide 6) was used in the absence of DNA ligase or plant extract.
RESULTS
T-DNA ligation is performed by plant enzymes, likely a DNA ligase.
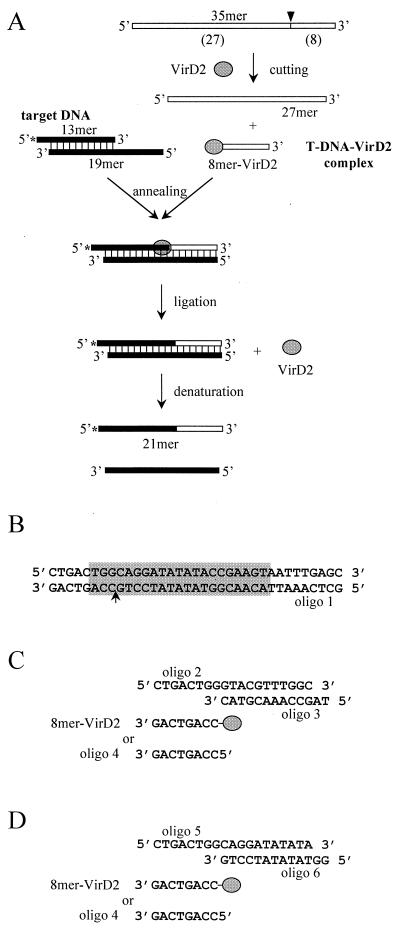
Ligation of the 5′ end of T-DNA to the 3′ end of plant DNA is an important step in the T-DNA integration process. To mimic it, we designed an in vitro ligation assay using a T-DNA–VirD2 complex formed in vitro and an artificial target DNA (Fig. 1). The T-DNA–VirD2 complex (8-mer–VirD2) was produced by VirD2-dependent endonucleolytic cleavage of an oligonucleotide containing the border sequence (oligonucleotide 1 [Fig. 1A]). During this reaction, VirD2 became covalently attached to the 5′ end of the DNA via a phosphotyrosine bond (not shown; see reference 23). The artificial target DNA was composed of annealed oligonucleotides 2 and 3 (Fig. 1C). Its 5′ part (8 nucleotides long) was complementary to the T-DNA sequence (8-mer–VirD2 or oligonucleotide 4) in order to facilitate duplex formation, although in vivo complementarity between the 5′ end of T-DNA and plant DNA is very limited and usually does not extend beyond one base pair (31). The sequence of the 3′ part of the target DNA was not specific for VirD2 and mimicked plant genomic DNA (Fig. 1C). In fact, the target DNA chosen to mimic the in planta ligation reaction was a fragment of a preinsertion sequence isolated from Arabidopsis thaliana (11). Ligation of the T-DNA–VirD2 complex (8-mer–VirD2) to the target DNA was monitored by the appearance of a radiolabeled 21-mer oligonucleotide (Fig. 1A). For comparison, a site-specific target DNA (composed of annealed oligonucleotides 5 and 6) was used in which the border sequence was restored (Fig. 1D).
FIG. 1.
T-DNA in vitro ligation assay. See text for details. (A) Scheme of the assay. (B) Fragment of pTi DNA containing the border sequence (grey box) with the cleavage site (arrow). The lower strand represents oligonucleotide 1. (C) Substrates for T-DNA in vitro ligation. (D) Substrates for site-specific in vitro ligation.
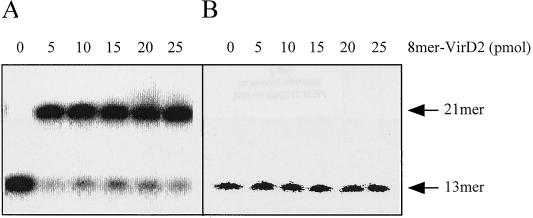
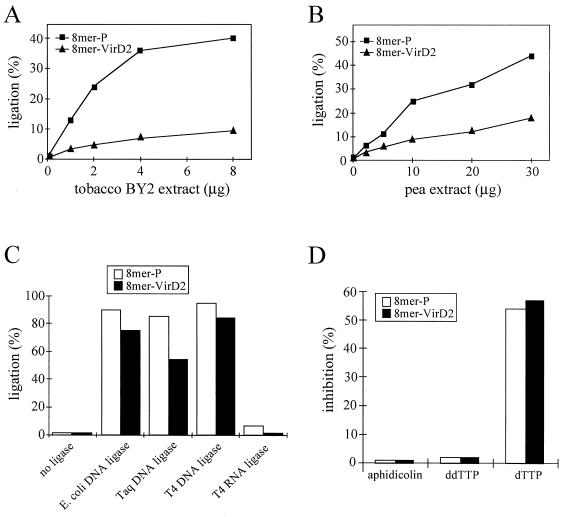
The site-specific ligation (rejoining) of the 8-mer–VirD2 complex occurred with high efficiency (Fig. 2A). This is consistent with the previously shown activity of VirD2 in rejoining oligonucleotides containing complementary parts of the border sequence (23). However, VirD2 bound to the oligonucleotide was unable to ligate it to the non-sequence-specific target DNA (Fig. 2B). These results suggested that plant-derived ligases and possibly other factors are required for T-DNA integration. Since a purified plant DNA ligase was not yet available, plant extracts were tested for their ability to ligate T-DNA to plant DNA. Indeed, a low but reproducible ligase activity could be detected in extracts from tobacco BY2 cells and from pea shoot apices (Fig. 3A and B). The activities of these extracts for ligation of the 8-mer–VirD2 complex to target DNA were low in comparison to ligation of VirD2-free oligonucleotides of the same sequence, containing a phosphoryl group at the 5′ terminus (Fig. 3A and B). In order to improve the efficiency of ligation we tested different assay conditions, including pH (6.8, 7.0, 7.5, and 8.0), salt concentration (25, 100, 150, 200, 250, 300, 400, and 500 mM NaCl), Mg2+ concentration (0, 1, 2.5, 10, and 25 mM MgCl2), addition of polyethylene glycol (final concentration in the range of 0 to 30%) or spermidine (final concentration in the range of 0 to 300 mM), and temperature (16, 24, and 30°C), as well as different combinations of these parameters. The 8-mer–VirD2 complex ligation efficiencies could not be improved by the tested modifications of the reaction conditions.
FIG. 2.
VirD2 by itself is able to perform site-specific rejoining but not T-DNA ligation in vitro. (A) Site-specific ligation by VirD2; (B) T-DNA ligation without any ligase added. 8-mer–VirD2 was used as the ligation substrate.
FIG. 3.
T-DNA ligation in vitro is performed by plant enzymes, likely by a DNA ligase. In vitro ligation was performed with nuclear extract from tobacco BY2 cells (A), with extract from pea shoot apices (B), and with the following purified prokaryotic ligases: E. coli DNA ligase (10 U), Taq DNA ligase (40 U), T4 DNA ligase (40 U), and T4 RNA ligase (20 U) (C). (D) Effect of inhibitors (150 μM aphidicolin, 5 μM ddTTP, and 1 mM dTTP) on T-DNA in vitro ligation. Inhibition values represent comparisons of the ligation efficiencies in the presence and absence of the inhibitor. 8-mer–VirD2 (8 pmol) and 8-mer-P (4 pmol) were used as ligation substrates.
The activity of plant extracts responsible for T-DNA ligation is likely provided by a plant DNA ligase, although the detected labeled 21-mer oligonucleotide could also be a product of a DNA polymerase activity present in these extracts. In order to settle this dilemma, the effects of known inhibitors of in vitro ligation were tested. Aphidicolin (inhibitor of DNA polymerases α, δ, and ɛ [5, 14, 34]) and ddTTP (inhibitor of DNA polymerases β and γ [17]) did not inhibit the reaction (Fig. 3D). On the other hand, an inhibitor of plant DNA ligases (dTTP [7]) showed an inhibitory effect (Fig. 3D). In addition, the labeled 21-mer product was not detected in the standard in vitro T-DNA ligation assay when 8-mer–VirD2 (or 8-mer-P) was omitted (data not shown). These results indicate that a plant DNA ligase is indeed responsible for ligation of the 8-mer–VirD2 complex to the target DNA. However, our efforts to purify the enzyme were unsuccessful due to loss of activity already in early steps of purification (data not shown).
To our surprise, prokaryotic DNA ligases efficiently established the link between the 8-mer–VirD2 complex and target DNA (Fig. 3C). RNA ligase, as expected, was not able to ligate VirD2-bound or VirD2-free substrate to target DNA. However, in all analyzed cases the efficiency of ligation of the complex was lower than that of the free DNA. The fact that this difference was particularly pronounced in the extracts used (Fig. 3A and B) points to an interesting deficiency and/or inhibitor present in them. A similar inhibitory effect, although caused by other factors, could be responsible for the difference in the efficiency of 8-mer–VirD2 ligation by purified enzymes and those present in plant extracts (Fig. 3C versus A and B). These factors could include nucleases and/or proteases destroying reaction substrates or enzymes and thereby decreasing the reaction efficiency.
The following experiments, designed to analyze this unique ligation reaction of a DNA-protein complex to target DNA, were carried out with plant extracts and T4 DNA ligase whenever possible. Particularly attractive was the fact that with T4 DNA ligase, a well-characterized enzyme could be employed.
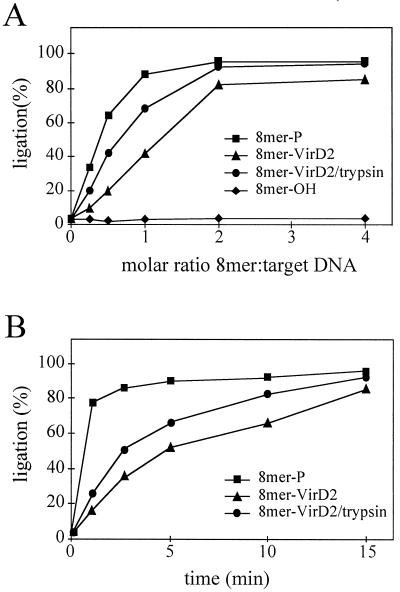
The T-DNA–VirD2 complex is ligated to a target sequence less efficiently than a VirD2-free substrate.
As already shown for plant extracts (see above), ligation of the 8-mer–VirD2 complex was less efficient than ligation of a phosphorylated substrate (8-mer-P), as indicated by tests of reaction substrate dependence and reaction rate (Fig. 4). It reached a plateau at a molar ratio of 8-mer–VirD2 complex to target DNA of 2:1 (Fig. 4A). In contrast, a substrate with a free phosphoryl group at the 5′ terminus was ligated efficiently with a 1:1 molar ratio. This difference was also reflected in the ligation kinetics; ligation of the 8-mer–VirD2 complex was much slower than that of the phosphorylated 8-mer-P substrate, although both reactions were essentially completed upon 15 min of incubation (Fig. 4B). Limited accessibility of the phosphoryl group due to the attached VirD2 protein may be the cause of this delay. To test this idea, a preformed 8-mer–VirD2 complex was digested with trypsin, and the dependence of the molar ratio of the substrates and the kinetics of the reactions were tested. Digestion with trypsin of VirD2 complexed with an oligonucleotide resulted in the oligonucleotide-peptide (14 amino acids) complex (for details see reference 23). Trypsin treatment led to an increase in the ligation efficiency in both molar ratio dependence and kinetics tests (Fig. 4). Similar results were obtained when pea extracts were used (data not shown). These findings indicated that ligase-dependent ligation of T-DNA–VirD2 complex is independent of any enzymatic activity of VirD2.
FIG. 4.
T4 DNA ligase-mediated ligation to the target sequence is more efficient for a phosphorylated than a VirD2-bound substrate. (A) Dependence on T-DNA to target DNA molar ratio. (B) Kinetics of the ligation reaction at a 2:1 molar ratio of T-DNA to target DNA. 8-mer-P, 8-mer–VirD2, trypsin-treated 8-mer–VirD2, and 8-mer-OH were used as ligation substrates.
T-DNA ligation requires ATP.
Upon ligation mediated by T4 DNA ligase or other ATP-dependent DNA ligases, the AMP moiety is transferred first to the enzyme and then to the 5′ end of the 5′-ligation partner (21, 32). Ligation of the 8-mer–VirD2 complex mediated by T4 DNA ligase or enzymes in plant extracts was also ATP dependent (Table 2). Thus, ATP could be replaced by ATP analogues that can be hydrolyzed to AMP, such as ATPγS or AMPPNP. In contrast, in the presence of AMPCPP the ligation could not proceed, thus demonstrating the need for ATP as a source of AMP also for 8-mer–VirD2 complex ligation. This is also true for enzymes derived from plant extracts isolated from pea (Table 2) and tobacco (data not shown) cells.
TABLE 2.
Use of ATP and its analogues in T-DNA ligationa
| Nucleoside triphosphate | Efficiency of ligation (%)b:
|
|||
|---|---|---|---|---|
| T4 DNA ligase
|
Plant extract
|
|||
| 8-mer–VirD2 | 8-mer-P | 8-mer–VirD2 | 8-mer-P | |
| None | 5.1 ± 0.9 | 5.9 ± 1.2 | 2.2 ± 1.7 | 4.2 ± 0.8 |
| ATP | 80.8 ± 2.8 | 92.3 ± 3.9 | 12.4 ± 1.6 | 32.9 ± 2.2 |
| AMPCPP | 5.8 ± 0.6 | 7.2 ± 0.3 | 3.4 ± 1.3 | 3.9 ± 1.5 |
| AMPPNP | 69.2 ± 4.3 | 80.7 ± 2.5 | 11.8 ± 1.5 | 32.1 ± 1.2 |
| ATPγS | 78.3 ± 2.7 | 90.8 ± 3.6 | 17.4 ± 1.0 | 34.9 ± 1.5 |
The in vitro ligation assays were done without or with ATP or different analogues and with T4 DNA ligase (4 U) or pea extract (20 μg). 8-mer–VirD2 and 8-mer-P were used as ligation substrates.
The numbers are average values from three independent experiments.
DISCUSSION
Our understanding of the early events of T-DNA transfer has increased considerably during recent years, but the mechanism of the T-DNA integration remains largely unknown. Previous analyses of several T-DNA insertions and their respective preinsertion sites (insertional target sites) isolated from A. thaliana and tobacco transformants (11, 18, 19, 31) not only suggested a possible role of VirD2 in T-DNA integration but also provided new details of the mechanism of this process that led to a model for T-DNA integration (31).
According to this model, the 3′ end of the T-DNA (or adjacent sequence) finds a short complementary region in the upper strand of plant DNA and anneals, with concomitant displacement of the bottom strand. The displaced plant DNA and 3′ overhang of T-DNA are digested away by nucleases. Simultaneously, the nucleotide(s) of the 5′ end of T-DNA, in particular the one directly attached to VirD2, anneals to its partner nucleotide of the upper strand of plant DNA and is subsequently ligated to the 3′ end of the bottom strand. Finally, the upper strand of the integrating T-DNA is produced by the plant DNA repair machinery. As a consequence, (i) a short fragment of plant DNA is lost at the insertion site, and (ii) the 5′ end of T-DNA is preserved (precision of integration), while the 3′ end of T-DNA is truncated.
Based on the following findings, the bacterial VirD2 protein has been suggested to function actively, as an integrase-ligase, in T-DNA integration. First, the influence of a mutation in the H-R-Y integrase-like motif of VirD2 on T-DNA integration was determined (31). The H-R-Y motif is perfectly conserved within a family of site-specific recombinases from bacteriophages λ, φ80, P22, P2, 186, P4, and P1, as well as from the yeast 2μm plasmid (1). These residues were suggested to contribute to the active site of this family of recombinases. As a consequence of the R-to-G mutation in the H-R-Y motif, precision of integration was lost without any change in its efficiency (31). Replacement of VirD2 by the MobA protein caused a similar effect (3). The unchanged efficiency argues against a function of VirD2 as an integrase and suggests that other factors may be involved in T-DNA integration. However, loss of precision of integration (defined as a lack of conservation of the 5′-end nucleotide attached to VirD2 in the integrated T-DNA) suggests the importance of VirD2 for the T-DNA integration process. Second, VirD2 was found to be able not only to cleave ssDNA at the border sequence in vitro but also to ligate cleaved ssDNA to the 3′ preformed end of another ssDNA molecule (23), suggesting a ligase function of VirD2 in T-DNA integration. However, both cleavage and ligation reactions were sequence specific, while in vivo T-DNA integration shows limited requirements for sequence homology. We decided to test the potential function of VirD2 as a ligase for T-DNA integration directly.
The ligation of the 5′ end of T-DNA to the 3′ end of plant DNA is one of the crucial steps in T-DNA integration. We designed an in vitro ligation assay mimicking T-DNA ligation. A T-DNA–VirD2 complex was made in vitro from endonucleolytic cleavage by VirD2 of an oligonucleotide containing a border sequence, and its ligation to target DNA (plant DNA sequence) was tested. We showed that VirD2 by itself was not able to ligate T-DNA in vitro, at least under the conditions optimal for efficient site-specific ligation. This result indicates that the function of VirD2 in T-DNA integration does not involve the site-specific ligation activity of the protein. The ligation, therefore, must be performed by plant enzymes, most probably by a DNA ligase. A ligase activity responsible for T-DNA ligation was indeed found in plant extracts, but prokaryotic DNA ligases could substitute for these specific plant activities.
The T-DNA–VirD2 complex is certainly an interesting substrate for ligation to plant DNA. To study this ligation in detail, we used the well-characterized T4 DNA ligase and compared it, whenever possible, to activities present in plant extracts. Ligation of DNA bound to VirD2 was less efficient than that of DNA with a free phosphoryl group at the 5′ end. In our in vitro assay, enzymatic activity of VirD2 was not required for ligation to proceed; a trypsinized complex was ligated even more efficiently than a complete one. This points to steric problems that need to be resolved for the two involved proteins, VirD2 and DNA ligase. In light of these findings one may wonder why this reaction takes place at all. Plant-specific enzymes, factors, or nucleosomal substructures at the target locus may, possibly in combination, improve this process. Moreover, nucleoprotein complexes seem to be the dominant form of DNA that could serve as the ligation substrate(s), as has been directly demonstrated for repair of chromosomal breaks (26). Alternatively, or in addition, the presence of VirD2 attached to the DNA may ensure the integrity of the 5′ end of integrating T-DNA by protecting the DNA from 5′→3′ exonucleases, as has been shown previously in in vitro experiments (9).
Although Agrobacterium-mediated DNA transfer is a powerful tool for plant transformation, it still requires some improvement for biotechnological use, for example, in directing the transgene to a defined locus in the plant genome. Although a lot of effort went into the targeting of T-DNA to a wild-type or introduced locus in the plant genome, the frequency of gene targeting remained low (11, 22, 24), suggesting a very efficient system of illegitimate recombination. Identification of plant factors involved in T-DNA integration not only will help to answer why higher eukaryotes have such an efficient illegitimate recombination system but also may lead to the development of plant lines with high frequencies of gene targeting.
ACKNOWLEDGMENTS
We thank Witold Filipowicz for helpful discussions and critical reading of the manuscript and Wen-Hui Shen (Strasbourg, France) for providing nuclear extracts from synchronized tobacco BY2 suspension cultured cells. We also thank Jean P. Jost, Ulrich Hübscher, and Jan Lucht for critical reading of the manuscript.
Collaboration with J.B. was started by a short-term EMBO fellowship to A.Z. (grant no. ASTF 8505).
REFERENCES
- 1.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V L, Pierson III L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron C, Zambryski P C. Plant transformation: a pilus in Agrobacterium T-DNA transfer. Curr Biol. 1996;6:1567–1569. doi: 10.1016/s0960-9822(02)70773-2. [DOI] [PubMed] [Google Scholar]
- 3.Bravo-Angel A M, Gloeckler V, Hohn B, Tinland B. Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J Bacteriol. 1999;181:5758–5765. doi: 10.1128/jb.181.18.5758-5765.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coello P, Rodrigez R, Garcia E, Vazquez-Ramos J M. A DNA polymerase from maize axes: its purification and possible role. Plant Mol Biol. 1992;20:1159–1168. doi: 10.1007/BF00028902. [DOI] [PubMed] [Google Scholar]
- 6.Daniel P P, Bryant J A. DNA ligase activity in pea seedlings (Pisum sativum L.): development of a sensitive assay system and partial characterization of soluble and chromatin-bound DNA ligases. Biochem Int. 1985;11:645–652. [Google Scholar]
- 7.Daniel P P, Bryant J A. DNA ligase in pea (Pisum sativum L.) seedlings: changes in activity during germination and effects of deoxyribonucleotides. J Exp Bot. 1988;39:481–486. [Google Scholar]
- 8.de la Cruz F, Lanka E. Function of the Ti-plasmid Vir proteins: T-complex formation and transfer to the plant cell. In: Spaink H, Hooykaas P, Kondorosi A, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 281–301. [Google Scholar]
- 9.Dürrenberger F, Crameri A, Hohn B, Koukolikova-Nicola Z. Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci USA. 1989;86:9154–9158. doi: 10.1073/pnas.86.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fullner K J, Cano L, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 11.Gheysen G, Villarroel R, Van Montagu M. Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- 12.Hansen G, Chilton M D. Lessons in gene transfer to plants by a gifted microbe. Curr Top Microbiol Immunol. 1999;240:21–57. doi: 10.1007/978-3-642-60234-4_2. [DOI] [PubMed] [Google Scholar]
- 13.Herrera-Estrella A, Chen Z M, Van Montagu M, Wang K. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5′ terminus of T-strand molecules. EMBO J. 1988;7:4055–4062. doi: 10.1002/j.1460-2075.1988.tb03299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huberman J. New views of the biochemistry of eukaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase α. Cell. 1981;23:647–648. doi: 10.1016/0092-8674(81)90426-8. [DOI] [PubMed] [Google Scholar]
- 15.Jasper F, Koncz C, Schell J, Steinbiss H H. Agrobacterium T-strand production in vitro: sequence-specific cleavage and 5′ protection of single-stranded DNA templates by purified VirD2 protein. Proc Natl Acad Sci USA. 1994;91:694–698. doi: 10.1073/pnas.91.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lartey R, Citovsky V. Nucleic acid transport in plant-pathogen interactions. Genet Eng. 1997;19:201–214. doi: 10.1007/978-1-4615-5925-2_11. [DOI] [PubMed] [Google Scholar]
- 17.Litvak S, Castroviejo M. DNA polymerases from plant cells. Mutat Res. 1987;181:81–91. [Google Scholar]
- 18.Matsumoto S, Ito Y, Hosoi T, Takahashi Y, Machida Y. Integration of Agrobacterium T-DNA into a tobacco chromosome: possible involvement of DNA homology between T-DNA and plant DNA. Mol Gen Genet. 1990;224:309–316. doi: 10.1007/BF00262423. [DOI] [PubMed] [Google Scholar]
- 19.Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei G P, Schell J, Hohn B, Koncz C. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrihew R V, Marburger K, Pennington S L, Roth D B, Wilson J H. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol Cell Biol. 1996;16:10–18. doi: 10.1128/mcb.16.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nash R, Lindahl T. DNA ligases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 575–586. [Google Scholar]
- 22.Offringa R, de Groot M J, Haagsman H J, Does M P, van den Elzen P J, Hooykaas P J. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 1990;9:3077–3084. doi: 10.1002/j.1460-2075.1990.tb07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paszkowski J, Baur M, Bogucki A, Potrykus I. Gene targeting in plants. EMBO J. 1988;7:4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi L, Tinland B, Hohn B. Role of virulence proteins of Agrobacterium in the plant. In: Spaink H, Hooykaas P, Kondorosi A, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 302–330. [Google Scholar]
- 26.Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen W H, Gigot C. Protein complexes binding to cis elements of the plant histone gene promoters: multiplicity, phosphorylation and cell cycle alteration. Plant Mol Biol. 1997;33:367–379. doi: 10.1023/a:1005797104536. [DOI] [PubMed] [Google Scholar]
- 28.Sheng J, Citovsky V. Agrobacterium-plant cell DNA transport: have virulence proteins, will travel. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tinland B. The integration of T-DNA into plant genomes. Trends Plant Sci. 1996;1:178–184. [Google Scholar]
- 30.Tinland B, Hohn B. Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. Genet Eng. 1995;17:209–229. [PubMed] [Google Scholar]
- 31.Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel A M, Hohn B. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomkinson A E, Levin D S. Mammalian DNA ligases. Bioessays. 1997;19:893–901. doi: 10.1002/bies.950191009. [DOI] [PubMed] [Google Scholar]
- 33.Ward E R, Barnes W M. VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science. 1988;242:927–930. [Google Scholar]
- 34.Weisser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hübscher U. Biochemical and functional comparison of DNA polymerases α, δ and ɛ from calf thymus. J Biol Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 35.Young C, Nester E W. Association of the VirD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J Bacteriol. 1988;170:3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zupan J, Zambryski P. The Agrobacterium DNA transfer complex. Crit Rev Plant Sci. 1997;16:279–295. [Google Scholar]
- 37.Zupan J, Ward D, Zambryski P. Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells. Curr Biol. 1998;1:649–655. doi: 10.1016/s1369-5274(98)80110-0. [DOI] [PubMed] [Google Scholar]