Abstract
Background: Mitochondrial bioenergetic alterations are commonly observed metabolic adaptation in malignancies including acute myeloid leukemia (AML). Mitochondrial DNA alterations are well known in pediatric AML with possible prognostic significance; however, mitochondrial complex activity and its impact on disease outcome have not been previously explored. The aim of this study was to evaluate the mitochondrial complex II and complex V activity and its prognostic significance in pediatric AML patients. Methods: Consecutive 82 de novo pediatric (≤18 years) patients with AML were included in the study along with age and sex matched controls. Bone marrow mononuclear cells were isolated from baseline bone marrow samples from all patients and controls. DNA, RNA and proteins were extracted and relative expression of mitochondrial biogenesis genes TFAM, POLG, POLRMT were estimated along with mitochondrial DNA copy number. The mitochondrial complex II and V enzymes were immunocaptured and their activity was measured by substrate specific absorbance change by kinetic ELISA. The mitochondrial complex II and V activity was compared with controls and their association with clinico-pathological features and survival outcome were analysed. Complex activity was also correlated with relative expression of biogenesis genes. Results: The activity of mitochondrial complex II and V were found to be significantly enhanced (P = 0.010 and P = 0.0013 respectively) in pediatric AML patients compared to controls. The activity of mitochondrial complex II and V showed significant positive correlation with relative gene expression of mitochondrial biogenesis genes TFAM (P = 0.001 and P = 0.016 respectively) and POLG (P = 0.005 and P = 0.006 respectively). Neither of the two complex activities showed any significant association with baseline disease demographics or any clinico-pathological feature. Furthermore, the complex II and V activity did not show any impact on event free survival (P = 0.25 and P = 0.24 respectively) and overall survival (P = 0.14 and P = 0.17 respectively) in our cohort. Conclusion: The activity of both mitochondrial complex II and V are significantly elevated in bone marrow mononuclear cells of children with AML compared to controls. The enhanced activity may be related to upregulation of mitochondrial biogenesis genes TFAM and POLG. The enhanced activity of either of the complexes did not impact disease biology or survival outcomes in pediatric AML.
Keywords: Acute myeloid leukemia, children, mitochondrial complex activity, succinate dehydrogenase, ATP synthase, mitochondria, mitochondrial biogenesis, survival, outcome, pediatric
Introduction
Acute myeloid leukemia (AML) is a genetically heterogenous hematological malignancy where the impact of mitochondrial alterations in disease pathogenesis and outcomes are increasingly being recognised [1]. Mitochondrial alterations significantly affect cellular bioenergetics, which is exploited by tumor cells for energy needs and proliferation [2]. Mitochondrial energy production is predominantly dependent on oxidative phosphorylation (OXPHOS) complex, consisting of five protein complexes (complex I, II, III, IV and V) which are translated by mitochondrial and nuclear DNA except for complex II which is encoded exclusively by the nuclear genome [3]. Alterations in mitochondrial metabolism and OXPHOS is a common phenomenon in a multitude of malignancies [4].
There are only few studies which have explored mitochondrial OXPHOS complex variations in AML; it has been observed that about 8% of AML patients have mutations in one or more mitochondrial OXPHOS genes, which also contributes to poor survival outcome [5]. Previously, we have reported that the relative expression of mitochondrial complex I associated gene ND3 predicted worse survival [6].
Mitochondrial complex activity and its regulation have not been previously explored in AML. In a previous study in primary AML cell lines, mitochondrial complex II activity was observed to be significantly enhanced [7]. Complex II plays an important role in electron transport chain along with TCA cycle, while ATP is produced in the complex V of OXPHOS by utilizing protons of electron transport chain. Mitochondrial ATP is the main source of energy for all type of intracellular metabolic pathways [2]. Biogenesis of mitochondrial OXPHOS complexes depends on various transcription regulators especially mitochondrial biogenesis genes that regulates the expression of OXPHOS genes and hence mitochondrial bioenergetics [8,9]. In this study, we evaluated the activity of mitochondrial complex II and complex V in children with AML as well as explored their correlation with the relative expression of mitochondrial biogenesis genes. We also assessed the impact of these mitochondrial complex activities on baseline disease characteristics and survival outcome.
Methodology
Study design and patient recruitment
This study included consecutive de novo pediatric AML patients (≤18 years) who were registered at pediatric oncology clinic of our cancer centre during the period of July 2016 to July 2018. Patients of granulocytic sarcoma without marrow involvement, acute promyelocytic leukemia, and mixed phenotypic acute leukemia as well as those with insufficient samples were excluded from the study. Along with this, 30 age-matched cases of solid malignancies without any involvement of bone marrow were also enrolled in the study as controls. The study was approved from institute ethics committee (IEC/NP-336/2012, IECPG-79/22.03.2017). Informed consent was obtained from parents/legal guardians as well as assent was taken from all study participants (≥8 years).
Baseline clinical characteristics, cytogenetics and molecular analysis
Baseline clinical details including demographic features, hematological parameters including complete blood count were recorded for all patients. Diagnosis was made by bone marrow aspiration morphology and immunophenotyping as per standard guidelines [10]. Cytogenetic analysis of baseline bone marrow sample of all the patients were evaluated by conventional karyotyping. Along with this, RNA was extracted from the bone marrow samples using TRIzol (Thermo) method. cDNA was used to identify translocation of RUNX1-RUNX1T1 (Runt-related transcription factor 1-RUNX1 partner transcriptional co-repressor 1 fusion transcript) and CBFB-MYH11 (Core binding factor beta-myosin heavy chain 11 fusion transcript) by RT-PCR. Furthermore, mutation of FLT3-ITD (Fms like tyrosine kinase 3-internal tandem duplication), and NPM1 (Nucleophosmin 1) gene were identified by DNA fragment length analysis [11,12]. Mutations as well as cytogenetic status were used for the risk stratification as per European LeukemiaNet (ELN) guidelines [13].
Treatment protocol
The uniform treatment protocol of 3+7 regimen including infusion of daunorubicin 60 mg/m2 day 1-3 and cytarabine 100 mg/m2 for 1-7 days, were used in all the patients for 7 days [14]. Induction therapy was followed by three cycles of high dose cytarabine at 18 g/m2 as consolidation therapy after achieving complete remission (CR) [15] whereas in refractory or relapse cases, repeat induction with ADE regimen (cytarabine: 100 mg/m2 twice daily, day 1-10; daunorubicin: 50 mg/m2, day 1-3; and etoposide: 100 mg/m2, day 1-5) was used [16]. Patients at CR2 underwent allogeneic hematopoietic stem cell transplantation with matched sibling donor if available [17].
Isolation of bone marrow mononuclear cells, protein extraction and estimation
Mononuclear cells were isolated from bone marrow aspirate samples by Ficoll-Hypaque (Sigma diagnostics, USA) density gradient centrifugation. Briefly, bone marrow sample (3 ml) with anticoagulant was diluted with 0.15 M NaCl in 1:1 ratio. Then the sample-NaCl mixture was layered onto a Ficoll column and centrifuged at 400 g for 30 minutes. The white interphase buffy layer was collected and washed twice with phosphate buffer saline 2000 rpm for 10 minutes at 25°C. The pellet was stored at -80°C and used for DNA, RNA and protein extraction.
For protein extraction, stored pellet of bone marrow mononuclear cells was treated with lysis buffer solution (200 μl per 2×106 cells) provided with the Abcam kit (Abcam, ab109908) and incubated on ice for 20 minutes for cell lysis. Samples were then centrifuged at 12000 g for 20 minutes at 4°C, the pellet was discarded and total protein concentration of supernatant was measured by the Pierce BCA Protein Assay (Thermo Fisher, Waltham, MA, USA).
Measurement of mitochondrial complex (II and V) activities
The activity of mitochondrial oxidative phosphorylation system complexes (complex II and complex V) was estimated by microplate coated ELISA based method as per manufacturer protocol (Abcam, ab109908 and Abcam, ab109714). Anti-Complex II monoclonal antibody (mAb) was immunocaptured into the 96 well plate which purifies the enzyme from a complex. Succinate was used as a substrate which was coupled with reduction of ubiquinol into ubiquinone and subsequently reduction of dye DCPIP (2,6-diclorophenolindophenol) and a decrease in its absorbance at OD600 nm in kinetics ELISA (BioTek, Gen5, Agilent). The rate of decrease in absorbance at 600 nm was monitored over time and calculated between two time points for all the samples in which the decrease in absorbance was the most linear. Rate (milli OD/min) was calculated as (Absorbance 1 - Absorbance 2)/Time (min), and the activity of immunocaptured complex-II as the mean of measurements obtained with immunocaptured enzyme minus the rate obtained without immunocaptured enzyme. Similarly for complex V, the production of ADP is coupled to the oxidation of NADH to NAD+, which was measured by absorbance at 340 nm (BioTek, Gen5, Agilent). The activity was calculated by expressing the enzymatic activity of succinate dehydrogenase for complex II and ATP synthase enzyme for complex V relative to the quantity of enzymes captured in each well.
Expression of mitochondrial biogenesis gene and estimation of mitochondrial DNA copy number
The relative mRNA expression of mitochondrial biogenesis genes such as Transcription factor A mitochondria (TFAM), DNA polymerase gamma (POLG) and RNA polymerase mitochondria (POLRMT) were estimated by quantitative real time PCR in all the samples along with relative mitochondrial DNA (mtDNA) copy number as per previously reported protocol [18].
Statistical analysis
Mann Whitney U test was used to compare median values of complex activities of patients with that of controls as well as for comparing across baseline patient characteristics. Spearman correlation was used for correlation of complex activity with relative gene expression and mtDNA copy number. Survival outcome of the cohort was analysed by Kaplan Meier statistics. Event free survival (EFS) was defined as time period from enrolment to relapse or death due to any cause, while time period from enrolment to death due to any cause was reported as overall survival (OS). Survival data was censored till 31st April 2021. Complex activity was categorized as high or low based on median value. Cox regression analysis was used to assess impact of complex activity on survival outcome of the patients. All the statistical analysis was carried out in Prism 6 (Graph Pad, La Jolla, CA, US) and SPSS software (version 23, IBM, NY, US) and P<0.05 was considered as statistically significant.
Results
Baseline patient characteristics
A total of 115 patients were evaluated for inclusion, of which 33 patients were excluded (3 patients were acute promyelocytic leukemia, 5 had granulocytic sarcoma without marrow involvement, and 25 patients had insufficient samples) from the study. A total of 82 patients and 30 controls were included for the final analysis. The baseline demographic and clinical characteristics of these patients are summarized in Table 1. The median age of the cohort was 12 years (range: 0.9-18 years) with slight male preponderance (Male:Female (1.73:1). Out of all, 21.95% patient had chloroma at presentation and more than half (53.25%) were good risk as per ELN risk stratification.
Table 1.
Baseline characteristic features of pediatric AML patients (n = 82)
| Characteristics | Number (%)/Median (Range) |
|---|---|
| Median age (years) | 12 (0.9-18) |
| Sex | |
| Male | 52 (63.4) |
| Female | 30 (36.58) |
| Haematological parameters | |
| Median haemoglobin, (g/dL) | 7.63 (2.3-14.6) |
| Median total leucocyte count, (×103/μL) | 25.30 (0.76-314.3) |
| Hyperleukocytosis (>50×103/μL) | 29 (35.37) |
| Median platelet count (×103/μL) | 40.0 (4.5-205) |
| Platelet count (<50×103/μL) | 48 (58.53) |
| Platelet count (≥50×103/μL) | 34 (41.46) |
| Clinical features at presentation | |
| Fever | 60 (73.17) |
| Chloroma | 18 (21.95) |
| Cytogenetics (n = 62)# | |
| Normal | 16 (25.81) |
| t(8;21) & inv (16)* | 34 (54.83) |
| Complex karyotype | 3 (4.84) |
| Other | 9 (14.52) |
| Failed cytogenetics | 12 (19.35) |
| Molecular analysis (n = 68) | |
| FLT3 ITD | 12 (17.64) |
| RUNX1-RUNX1T1 | 34 (50) |
| CBFB-MYH11 | 2 (2.94) |
| NPM1 | 1 (1.47) |
| Negative | 21 (30.88) |
| ELN risk stratification (n = 77)** | |
| Good | 41 (53.25) |
| Intermediate & poor | 36 (46.75) |
t(8;21) = t(8;21)(q22;q22) RUNX1-RUNX1T1; inv(16) = inv(16)(p13.1;q22) CBFB-MYH11; Median values were reported with range.
Cytogenetic analysis by conventional karyotyping was not available for remaining 20 patients.
ELN risk stratification was done using both cytogenetics and molecular markers in 61 patients. However, 9 patients risk stratification was done with only cytogenetics and in 7 patients, it was done by only molecular analysis.
n: number of patients; AML: Acute Myeloid Leukemia; Normal cytogenetics: 46,XX/46,XY; Failed cytogenetics: metaphases could not be isolated; FLT3 ITD: FMS-like tyrosine kinase internal tandem duplication; RUNX1-RUNX1T1: runt-related transcription factor 1-RUNX1 partner transcriptional co-repressor 1 fusion transcript; CBFB-MYH11: core binding factor beta-myosin heavy chain 11 fusion transcript; NPM1: nucleophosmin 1; ELN: European LeukemiaNet.
Mitochondrial OXPHOS complex activity in AML patients: comparison with controls and correlation with baseline demographics
The complex II activity was measured in 61 pediatric AML patients while complex V was evaluated in 71 patients. The median activity of mitochondrial OXPHOS complex II and complex V was significantly higher (P = 0.010 and P = 0.0013 respectively) in the bone marrow mononuclear cells of pediatric AML patients in comparison to age and sex matched controls (Figure 1).
Figure 1.
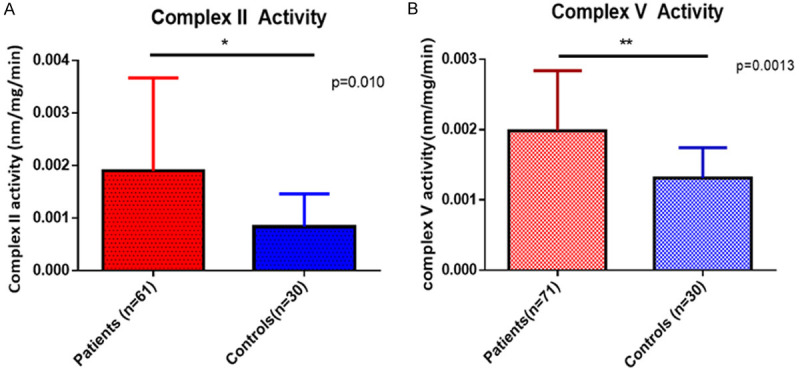
Comparison of mitochondrial complex II (A) and complex V (B) in pediatric AML patients compared to controls.
Furthermore, there was no significant association of mitochondrial complex II and V activity with any of the clinicopathological parameters in pediatric AML patients (Table 2).
Table 2.
Association of clinico-pathological features with mitochondrial complex II and complex V activity in pediatric acute myeloid leukemia patients
| Variables | p value | Complex II Activity* (n = 61) | p value | Complex V Activity* (n = 71) |
|---|---|---|---|---|
| Age (years) | 0.86 | 0.26 | ||
| <10 (n1 = 23; n2 = 29) | 1.70E-03 (7.00E-04-4.60E-03) | 2.30E-03 (1.80E-03-2.90E-03) | ||
| ≥10 (n1 = 23; n2 = 45) | 1.90E-03 (6.00E-04-4.20E-03) | 1.90E-03 (1.30E-03-2.80E-03) | ||
| Gender | 0.92 | 0.88 | ||
| Male (n1 = 41; n2 = 45) | 1.90E-03 (6.00E-04-3.50E-03) | 2.00E-03 (1.20E-03-3.10E-03) | ||
| Female (n1 = 20; n2 = 26) | 1.60E-03 (6.70E-04-6.30E-03) | 2.20E-03 (1.80E-03-2.60E-03) | ||
| Chloroma | 0.26 | 0.38 | ||
| Present (n1 = 12; n2 = 15) | 2.10E-03 (6.00E-04-6.30E-03) | 2.00E-03 (1.30E-03-2.40E-03) | ||
| Absent (n1 = 49; n2 = 56) | 1.60E-03 (4.00E-04-2.90E-03) | 2.20E-03 (1.20E-03-2.70E-03) | ||
| Total leukocyte count (mm3) | 0.20 | 0.46 | ||
| <50000 (n1 = 35; n2 = 44) | 1.70E-03 (5.00E-04-3.00E-03) | 2.20E-03 (1.70E-03-2.90E-03) | ||
| ≥50000 (n1 = 26; n2 = 27) | 2.10E-03 (8.00E-04-7.20E-03) | 1.90E-03 (1.30E-03-2.60E-03) | ||
| Platelets | 0.096 | 0.69 | ||
| <50000 (n1 = 58; n2 = 45) | 1.43E-03 (5.33E-04-2.40E-03) | 2.10E-03 (1.28E-03-2.90E-03) | ||
| ≥50000 (n1 = 23; n2 = 27) | 2.67E-03 (7.94E-04-8.66E-03) | 2.90E-03 (1.80E-03-2.80E-03) | ||
| Haemoglobin (g/dl) | 0.81 | 0.51 | ||
| <8 (n1 = 39; n2 = 45) | 1.80E-03 (7.00E-04-4.70E-03) | 2.70E-03 (1.70E-03-2.70E-03) | ||
| ≥8 (n1 = 22; n2 = 26) | 2.10E-03 (5.00E-04-5.10E-03) | 1.90E-03 (1.00E-03-3.90E-03) | ||
| Cytogenetics | 0.059 | 0.29 | ||
| Good (n1 = 20; n2 = 29) | 2.40E-03 (1.40E-03-7.50E-03) | 2.60E-03 (1.70E-03-3.10E-03) | ||
| Others (n1 = 22; n2 = 23) | 8.00E-04 (5.00E-04-2.80E-03) | 1.90E-03 (1.30E-03-3.30E-03) | ||
| **ELN Risk group | 0.075 | 0.067 | ||
| Good (n1 = 25; n2 = 33) | 2.40E-03 (1.30E-03-5.10E-03) | 2.60E-03 (1.90E-03-3.20E-03) | ||
| Others (n1 = 30; n2 = 32) | 8.33E-04 (5.00E-04-2.80E-03) | 1.90E-03 (1.30E-03-2.60E-03) |
n1 = number of patients in complex II cohort; n2 = number of patients in complex V cohort.
The unit of complex II and complex V activity is nm/mg/min;
ELN = European LeukemiaNet.
Correlation of mitochondrial complex activity with mitochondrial biogenesis genes and mtDNA copy number
Mitochondrial complex II activity was found to have significant positive correlation with the relative expression of mitochondrial biogenesis genes TFAM (P = 0.001) and POLG (P = 0.005). Similarly, the activity of mitochondrial complex V also had significant positive correlation with relative expression of TFAM (P = 0.016) and POLG (P = 0.006). The relative mtDNA copy number as well as relative expression of POLRMT did not show any significant correlation with either mitochondrial complex II or complex V activity (Table 3).
Table 3.
Correlation of mitochondrial complex activity with mitochondrial biogenesis gene expression and mitochondrial DNA copy number
| Gene name | Complex II Activity (n = 61) | Complex V activity (n = 71) | ||
|---|---|---|---|---|
|
|
|
|||
| Correlation coefficient (r) | p value | Correlation coefficient (r) | p value | |
| TFAM | 0.42 | 0.001 | 0.28 | 0.016 |
| POLG | 0.36 | 0.005 | 0.32 | 0.006 |
| POLRMT | -0.16 | 0.21 | -0.17 | 0.16 |
| mtDNA copy number | -0.01 | 0.94 | 0.04 | 0.78 |
mtDNA: Mitochondrial DNA. r = Spearman correlation coefficient.
Impact of mitochondrial complex activity on survival outcome of pediatric AML patients
At a median follow-up of 47.07 months (38.44 months-55.69 months), the median EFS of the cohort was 10.17 months (5.85 months-14.49 months), while the median OS of the cohort was 24.03 months (9.38 months-31.22 months). The activity of mitochondrial complex II and complex V did not show any impact on OS (P = 0.14 and P = 0.17 respectively) (Figure 2A, 2B; Table 4) as well as on EFS (P = 0.25 and P = 0.24 respectively) (Figure 2C, 2D; Table 4).
Figure 2.
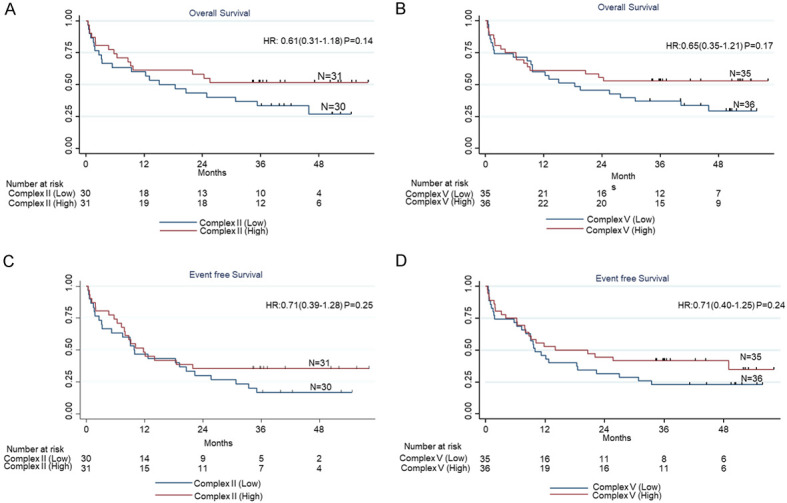
Impact of complex II and complex V on overall survival (A, B) and event free survival (C, D).
Table 4.
Impact of mitochondrial complex activity on event free survival and overall survival of pediatric AML patients
| Complex activity | Event Free Survival | Overall survival | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Median (Months) | Hazard Ratio (95% CI) | p value | Median (Months) | Hazard ratio (95% CI) | p value | ||
| Complex II Activity (n = 61) | Low (n = 30) | 9.87 | 0.71 (0.39-1.28) | 0.25 | 15.07 | 0.61 (0.31-1.18) | 0.14 |
| High (n = 31) | 11.80 | Not reached | |||||
| Complex V Activity (n = 71) | Low (n = 35) | 9.97 | 0.71 (0.40-1.25) | 0.24 | 18.37 | 0.65 (0.35-1.21) | 0.17 |
| High (n = 36) | 14.07 | Not reached | |||||
Categorisation of mitochondrial complex activity into low and high is based on the median values of complex activity. CI = Confidence interval; AML: Acute myeloid leukemia.
Discussion
It is postulated as per Warburg hypothesis that cancer cells predominantly depend on glycolytic pathways for their energy needs. However, it is now becoming increasingly evident that mitochondrial energy metabolism plays a key role in tumor initiation and progression [19-21]. A meta-analysis suggested that ATP production by OXPHOS is significantly higher in cancerous cells as compared to normal cells [22]. Furthermore, expression of OXPHOS proteins are also known to be upregulated in multiple malignancies including leukemias [21,23-25]. In our study, we observed that mitochondrial complex II and complex V activity were significantly upregulated in the bone marrow mononuclear cells of children with AML compared to controls, suggesting their putative role in tumor biology and energy metabolism. A previous study by Sriskanthadevan et al, which evaluated the activity of OXPHOS complexes in AML primary culture and cell lines, suggested that there was an increase in complex I and complex II activity in AML cells compared to normal cells. Another study reported enriched expression of OXPHOS proteins in AML leukemic stem cells (LSCs) in comparison to normal hematopoietic stem cells suggesting heterogeneity in metabolic phenotype of AML cells [26]. These corroborative findings suggest that enhanced mitochondrial complex activity is a common metabolic adaptation in AML blasts.
In this study, we observed that the altered OXPHOS complex activity had no association with disease phenotype or demographics. This is in contrast to previous studies in various malignancies which observed that mitochondrial OXPHOS alterations have an impact on tumor biology [27]. In solid tumors like pancreatic cancer, it has been observed that cancer stem cells, which are more dependent on OXPHOS, had high metastatic and tumorigenic potential [28]. Cancers like melanoma and diffuse large B-cell lymphoma have also shown to have distinct metabolic fingerprints and associated survival mechanisms based on OXPHOS activity [5,24]. This suggests that there is significant heterogeneity in metabolic adaptations across different malignancies, wherein the role of mitochondrial OXPHOS alterations may be variable.
Studies which have evaluated mutations in OXPHOS complexes have shown mutations in mitochondrial OXPHOS complex I and IV to be associated with TP53 mutation and also predictive of inferior OS in AML patients [5]. In vivo studies on AML cell line also reported that increased expression of OXPHOS is a protective mechanism for AML blasts during chemotherapy and may contribute to chemoresistance [29,30]. A study by Lagadinou et al suggested that LSCs are sensitive to OXPHOS inhibition and targeting OXPHOS may promote selective eradication of AML LSCs [31]. We have also previously observed that elevated expression of OXPHOS complex I gene ND3 was associated with ELN poor risk group in pediatric cohort of AMLL as well as its inferior EFS and OS [6]. However, none of the above studies have explored mitochondrial complex activity in AML patient samples. Our current study did not observe any impact of mitochondrial complex activity on survival outcome in the cohort of children with AML. This suggests that while enhanced mitochondrial complex activity is a metabolic adaptation of AML blast cells for energy needs for survival, this enhanced activity may not necessarily predict disease aggressiveness or survival outcomes. Also, alterations in gene expression or mutations may not equate to enhanced functionality of the complexes [24].
Altered expression of mitochondrial biogenesis and subsequent change in OXPHOS has been reported in lung cancer, breast cancer, lymphomas and cholangiocarcinoma [32-34]. Furthermore, studies have reported that AML blast cells rely on mitochondria for energy production driven by upregulation of mitochondrial biogenesis genes [35-37]. We have previously reported upregulation of mitochondrial biogenesis genes in pediatric AML patients [18]. In the current study, we observed a significant positive correlation of mRNA expression of mitochondrial biogenesis genes i.e., TFAM and POLG with mitochondrial complex II and V activity, suggesting that enhanced biogenesis of mitochondria may result in increased mitochondrial OXPHOS activity. The finding of our current study in patient samples supports the previous finding in AML cell line wherein inhibition of mitochondrial biogenesis gene POLG led to selective decrease in OXPHOS activity with targeting of leukemia cells [38]. This metabolic adaptation may be crucial for supporting higher energy requirement of rapidly proliferating cancer cells and can be a potential therapeutic target in future [39].
The limitations of our study are lack of evaluation of relative expression of various mitochondrial complex related genes, which could have helped in correlating complex activity with up-regulation at genetic level. Additionally, the overall functionality of mitochondria was also not assessed.
In conclusion, our study suggests that pediatric AML patients had significantly elevated mitochondrial complex II and V activity in their bone marrow mononuclear cells which is possibly driven by enhanced mitochondrial biogenesis, as part of metabolic adaptation of blasts for survival. This adaptation does not impact disease phenotype or survival outcome in our cohort, but this needs to be validated in further studies. Furthermore, targeting mitochondrial OXPHOS complex by selectively inhibiting regulators of OXPHOS and exploiting this vulnerable metabolic alteration is an exciting area for developing targeted therapeutics for AML.
Acknowledgements
The authors acknowledge the funding support from DST-SERB (Department of Science and Technology-Science and Engineering Research Board), Government of India (Extramural Research grant: EMR/2016/006376) and ICMR (Indian council of medical research) funding of senior research fellowship (Project no. 2019-6059/CMB/BMS) for this work. The authors also acknowledge every member of pediatric oncology team of our centre including research staff, nurses and dietician for their exemplary clinical services.
Disclosure of conflict of interest
None.
References
- 1.Longo DL, Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;12:1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Luo Y, Ma J, Lu W. The significance of mitochondrial dysfunction in cancer. Int J Mol Sci. 2020;21:1–24. doi: 10.3390/ijms21165598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirches E. Mitochondrial and nuclear genes of mitochondrial components in cancer. Curr Genomics. 2009;10:281–293. doi: 10.2174/138920209788488517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S, Akhtari M, Alachkar H. Characterization of mutations in the mitochondrial encoded electron transport chain complexes in acute myeloid leukemia. Sci Rep. 2018;8:13301. doi: 10.1038/s41598-018-31489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyagi A, Pramanik R, Bakhshi R, Singh A, Vishnubhatla S, Bakhshi S. Expression of mitochondrial genes predicts survival in pediatric acute myeloid leukemia. Int J Hematol. 2019;110:205–212. doi: 10.1007/s12185-019-02666-2. [DOI] [PubMed] [Google Scholar]
- 7.Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, Jhas B, Hurren R, Gronda M, Wang X, Jitkova Y, Sukhai MA, Lin FH, Maclean N, Laister R, Goard CA, Mullen PJ, Xie S, Penn LZ, Rogers IM, Dick JE, Minden MD, Schimmer AD. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125:2120–2130. doi: 10.1182/blood-2014-08-594408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez-Enríquez S, Marín-Hernández Á, Gallardo-Pérez JC, Pacheco-Velázquez SC, Belmont-Díaz JA, Robledo-Cadena DX, Vargas-Navarro JL, Corona de la Peña NA, Saavedra E, Moreno-Sánchez R. Transcriptional regulation of energy metabolism in cancer cells. Cells. 2019;8:1225. doi: 10.3390/cells8101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M, Hirpara JL, Eu JQ, Sethi G, Wang L, Goh BC, Wong AL. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2019;25:101073. doi: 10.1016/j.redox.2018.101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Béné MC, Nebe T, Bettelheim P, Buldini B, Bumbea H, Kern W, Lacombe F, Lemez P, Marinov I, Matutes E, Maynadié M, Oelschlagel U, Orfao A, Schabath R, Solenthaler M, Tschurtschenthaler G, Vladareanu AM, Zini G, Faure GC, Porwit A. Immunophenotyping of acute leukemia and lymphoproliferative disorders: a consensus proposal of the European LeukemiaNet Work Package 10. Leukemia. 2011;25:567–574. doi: 10.1038/leu.2010.312. [DOI] [PubMed] [Google Scholar]
- 11.Sharawat SK, Raina V, Kumar L, Sharma A, Bakhshi R, Vishnubhatla S, Gupta R, Bakhshi S. High FMS-like tyrosine kinase-3 (FLT3) receptor surface expression predicts poor outcome in FLT3 internal tandem duplication (ITD) negative patients in adult acute myeloid leukaemia: a prospective pilot study from India. Indian J Med Res Suppl. 2016;143:11–16. doi: 10.4103/0971-5916.191740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chopra A, Soni S, Pati H, Kumar D, Diwedi R, Verma D, Vishwakama G, Bakhshi S, Kumar S, Gogia A, Kumar R. Nucleophosmin mutation analysis in acute myeloid leukaemia: immunohistochemistry as a surrogate for molecular techniques. Indian J Med Res. 2016;143:763–768. doi: 10.4103/0971-5916.192027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz M, Sierra J, Tallman MS, Tien HF, Wei AH, Löwenberg B, Bloomfield CD. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi A, Pramanik R, Chaudhary S, Chopra A, Bakhshi S. Cytogenetic profiles of 472 Indian children with acute myeloid leukemia. Indian Pediatr. 2018;55:469–473. [PubMed] [Google Scholar]
- 15.Bakhshi S, Singh P, Swaroop C. Outpatient consolidation chemotherapy in pediatric acute myeloid leukemia: a retrospective analysis. Hematology. 2009;14:255–260. doi: 10.1179/102453309X446144. [DOI] [PubMed] [Google Scholar]
- 16.Garg A, Ganguly S, Vishnubhatla S, Chopra A, Bakhshi S. Outpatient ADE (cytarabine, daunorubicin, and etoposide) is feasible and effective for the first relapse of pediatric acute myeloid leukemia: a prospective, phase II study. Pediatr Blood Cancer. 2020;67:e28404. doi: 10.1002/pbc.28404. [DOI] [PubMed] [Google Scholar]
- 17.Arora S, Pushpam D, Tiwari A, Choudhary P, Chopra A, Gupta R, Kumar R, Bakhshi S. Allogeneic hematopoietic stem cell transplant in pediatric acute myeloid leukemia: lessons learnt from a tertiary care center in India. Pediatr Transplant. 2021;25:e13918. doi: 10.1111/petr.13918. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhary S, Ganguly S, Palanichamy JK, Singh A, Bakhshi R, Jain A, Chopra A, Bakhshi S. PGC1A driven enhanced mitochondrial DNA copy number predicts outcome in pediatric acute myeloid leukemia. Mitochondrion. 2021;58:246–254. doi: 10.1016/j.mito.2021.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 22.Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 23.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, Broda P, Minetti C, Lisanti MP, Sotgia F. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. 2011;10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, Zhou F, Green MR, Chen L, Monti S, Marto JA, Shipp MA, Danial NN. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashton TM, Gillies McKenna W, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 26.Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, Bullinger L, Poschet G, Nonnenmacher Y, Barnert A, Bahr C, Zeisberger P, Przybylla A, Sohn M, Tönjes M, Erez A, Adler L, Jensen P, Scholl C, Fröhling S, Cocciardi S, Wuchter P, Thiede C, Flörcken A, Westermann J, Ehninger G, Lichter P, Hiller K, Hell R, Herrmann C, Ho AD, Krijgsveld J, Radlwimmer B, Trumpp A. BCAT1 restricts αkG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature. 2017;551:384–388. doi: 10.1038/nature24294. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, Bosc C, Sugita M, Stuani L, Fraisse M, Scotland S, Larrue C, Boutzen H, Féliu V, Nicolau-Travers ML, Cassant-Sourdy S, Broin N, David M, Serhan N, Sarry A, Tavitian S, Kaoma T, Vallar L, Iacovoni J, Linares LK, Montersino C, Castellano R, Griessinger E, Collette Y, Duchamp O, Barreira Y, Hirsch P, Palama T, Gales L, Delhommeau F, Garmy-Susini BH, Portais JC, Vergez F, Selak M, Danet-Desnoyers G, Carroll M, Récher C, Sarry JE. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Dis. 2017;7:716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moschoi R, Imbert V, Nebout M, Chiche J, Mary D, Prebet T, Saland E, Castellano R, Pouyet L, Collette Y, Vey N, Chabannon C, Recher C, Sarry JE, Alcor D, Peyron JF, Griessinger E. Protective mitochondrial transfer from bone marrow stromal cells to acute myeloid leukemic cells during chemotherapy. Blood. 2016;128:253–264. doi: 10.1182/blood-2015-07-655860. [DOI] [PubMed] [Google Scholar]
- 31.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O’Dwyer KM, Liesveld JL, Brookes PS, Becker MW, Jordan CT. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samper E, Morgado L, Estrada JC, Bernad A, Hubbard A, Cadenas S, Melov S. Increase in mitochondrial biogenesis, oxidative stress, and glycolysis in murine lymphomas. Free Radic Biol Med. 2009;46:387–396. doi: 10.1016/j.freeradbiomed.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellance N, Benard G, Furt F, Begueret H, Smolková K, Passerieux E, Delage JP, Baste JM, Moreau P, Rossignol R. Bioenergetics of lung tumors: alteration of mitochondrial biogenesis and respiratory capacity. Int J Biochem Cell Biol. 2009;41:2566–2577. doi: 10.1016/j.biocel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Raggi C, Taddei ML, Sacco E, Navari N, Correnti M, Piombanti B, Pastore M, Campani C, Pranzini E, Iorio J, Lori G, Lottini T, Peano C, Cibella J, Lewinska M, Andersen JB, di Tommaso L, Viganò L, Di Maira G, Madiai S, Ramazzotti M, Orlandi I, Arcangeli A, Chiarugi P, Marra F. Mitochondrial oxidative metabolism contributes to a cancer stem cell phenotype in cholangiocarcinoma. J Hepatol. 2021;74:1373–1385. doi: 10.1016/j.jhep.2020.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Panina SB, Pei J, Kirienko NV. Mitochondrial metabolism as a target for acute myeloid leukemia treatment. Cancer Metab. 2021;9:17. doi: 10.1186/s40170-021-00253-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marlein C, Zaitseva L, Piddock R, Shafat M, Collins A, Bowles K, Rushworth S. PGC1α driven mitochondrial biogenesis within the bone marrow stromal cells of the acute myeloid leukemia micro-environment is a pre-requisite for mitochondrial transfer to leukemic blasts. Blood. 2017;130:3927–3927. [Google Scholar]
- 37.Bralha FN, Liyanage SU, Hurren R, Wang X, Son MH, Fung TA, Chingcuanco FB, Tung AY, Andreazza AC, Psarianos P, Schimmer AD, Salmena L, Laposa RR. Targeting mitochondrial RNA polymerase in acute myeloid leukemia. Oncotarget. 2015;6:37216–37228. doi: 10.18632/oncotarget.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liyanage SU, Hurren R, Voisin V, Bridon G, Wang X, Xu C, MacLean N, Siriwardena TP, Gronda M, Yehudai D, Sriskanthadevan S, Avizonis D, Shamas-Din A, Minden MD, Bader GD, Laposa R, Schimmer AD. Leveraging increased cytoplasmic nucleoside kinase activity to target mtDNA and oxidative phosphorylation in AML. Blood. 2017;129:2657–2666. doi: 10.1182/blood-2016-10-741207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Kalpage HA, Wang D, Edwards H, Hüttemann M, Ma J, Su Y, Carter J, Li X, Polin L, Kushner J, Dzinic SH, White K, Wang G, Taub JW, Ge Y. Cotargeting of mitochondrial complex I and Bcl-2 shows antileukemic activity against acute myeloid leukemia cells reliant on oxidative phosphorylation. Cancers (Basel) 2020;12:1–19. doi: 10.3390/cancers12092400. [DOI] [PMC free article] [PubMed] [Google Scholar]


