Abstract
We previously identified a sequence-specific erythroid cell-enriched endoribonuclease (ErEN) activity involved in the turnover of the stable α-globin mRNA. We now demonstrate that ErEN activity is regulated by the poly(A) tail. The unadenylated α-globin 3′ untranslated region (3′UTR) was an efficient substrate for ErEN cleavage, while the polyadenylated 3′UTR was inefficiently cleaved in an in vitro decay assay. The influence of the poly(A) tail was mediated through the poly(A)-binding protein (PABP) bound to the poly(A) tail, which can inhibit ErEN activity. ErEN cleavage of an adenylated α-globin 3′UTR was accentuated upon depletion of PABP from the cytosolic extract, while addition of recombinant PABP reestablished the inhibition of endoribonuclease cleavage. PABP inhibited ErEN activity indirectly through an interaction with the αCP mRNA stability protein. Sequestration of αCP resulted in an increase of ErEN cleavage activity, regardless of the polyadenylation state of the RNA. Using electrophoretic mobility shift assays, PABP was shown to enhance the binding efficiency of αCP to the α-globin 3′UTR, which in turn protected the ErEN target sequence. Conversely, the binding of PABP to the poly(A) tail was also augmented by αCP, implying that a stable higher-order structural network is involved in stabilization of the α-globin mRNA. Upon deadenylation, the interaction of PABP with αCP would be disrupted, rendering the α-globin 3′UTR more susceptible to endoribonuclease cleavage. The data demonstrated a specific role for PABP in protecting the body of an mRNA in addition to demonstrating PABP's well-characterized effect of stabilizing the poly(A) tail.
The stability of mRNA is dictated by both general and specific stability determinants. Eukaryotic mRNAs have an m7G cap at the 5′ terminus and a poly(A) tail at the 3′ end. Both of these elements, along with the cap-binding proteins and the poly(A)-binding protein (PABP), are critical for mRNA stability and function. They provide a basal level of stability for an mRNA by preventing exoribonucleolytic degradation. Despite the presence of these elements on almost all RNA polymerase II transcripts, mRNA stabilities vary from several minutes to several days, indicating that elements inherent to a given mRNA also contribute to half-lives. Differential stability is determined by distinct cis elements which may either promote rapid degradation or confer increased stability onto an mRNA (40). These elements are thought to exert their influence through RNA-binding proteins that may either directly or indirectly influence the activities of ribonucleases.
A major regulatory component of eukaryotic mRNA turnover involves the interaction between PABP and the 3′ poly(A) tail (8, 41). Deadenylation has been most extensively characterized for Saccharomyces cerevisiae, where it appears to be the first rate-limiting step in the turnover of mRNA (2, 8, 15). Deadenylation also seems to be a critical first step in mammalian mRNA turnover as well (13, 45, 55). PABP functions to prevent access of a 3′-to-5′ poly(A)-specific exoribonuclease activity to an mRNA in mammalian cells (3, 17, 51) as well as to inhibit decapping in yeast (9). A gene encoding a poly(A)-specific exoribonuclease (PARN; referred to as deadenylating nuclease [25]) was recently cloned and may constitute the predominant deadenylating cellular enzyme. PABP consists of five domains, specifically, four highly conserved RNP motif RNA-binding domains at the amino half of the protein and a divergent carboxyl terminus (18). The first two RNP motifs of PABP are necessary and sufficient for specific poly(A) binding (7, 35). The recent cocrystalization of this domain with poly(A) RNA demonstrates that the RNA-binding domains are bound antiparallel to the RNA, in which the first RNP motif binds a segment of the poly(A) tail that is 3′ to the region bound by the second RNP motif (16).
Specific ribonucleases which target a particular mRNA for degradation by endoribonucleolytic cleavage are also involved in RNA turnover. Several mRNAs have been either implicated in targeting or directly demonstrated to be targeted by an endoribonuclease activity. These include mRNAs encoding albumin (37), apolipoprotein II (4), c-myc (39), Groα (46), insulin-like growth factor 2 (12, 42), transferrin receptor (5), Xlhbox 2B (6), 9E3 (47), β-globin (1, 30), and α-globin (52). Specific candidate nucleases which cleave the c-myc (27) and the Xlhbox 2B (6) mRNAs have been identified, and the gene encoding polysomal RNase 1, which cleaves the albumin mRNA, was recently cloned (10). Interestingly, the removal of the poly(A) tail does not seem to be a prerequisite for cleavage by the endoribonuclease activities characterized thus far (2, 40, 43). The significance of poly(A) tail-independent cleavage would be to allow a rapid response which circumvents the normal deadenylation pathway and initiates specific decay.
The current understanding of how specific mRNA stabilization is conferred to an mRNA is limited. One well-characterized example involves the cytosine-rich element (CRE) in the α-globin 3′ untranslated region (3′UTR). Mutations within the CRE were shown to decrease α-globin mRNA stability in mammalian cells (53) and prevent formation of an RNP complex termed the α-complex, which includes a poly(C)-binding protein, αCP (23, 24, 50). More recently, it was shown that αCP can directly interact with the CRE and constitute the α-complex (11). There are at least two αCP genes, αCP1 and αCP2 (also referred to as PCBP or hnRNP E), which are over 80% identical at the protein level and contain three hnRNP K homology RNA-binding domains (24, 28). The gene encoding αCP1 consists of an intronless structure, while the αCP2 gene contains a multiexon structure (31, 49). We have developed an in vitro decay assay system which faithfully recapitulates the differential stability of the α-globin mRNA in vitro with cytosolic S130 extract and demonstrated a functional role for the αCP proteins in stabilizing this mRNA (51). The α-globin 3′UTR containing a CRE deletion was less stable than the wild-type α-globin 3′UTR (αwt) in this system. Similarly, incubation of the αwt RNA with extract in which αCP had been sequestered, or extract depleted of αCP, rendered the RNA less stable. We have previously demonstrated that the αCP proteins confer a CRE-mediated mRNA stability by at least two mechanisms. First, αCP can interact with PABP to lower the rate of deadenylation (51), and second, it protects the αwt RNA from cleavage by a sequence specific erythroid cell-enriched endoribonuclease (ErEN) activity (52). We now report that unlike previously characterized mRNA-specific endoribonuclease activities which are poly(A) tail independent, ErEN activity is inhibited by the poly(A) tail and indirectly regulated by PABP.
MATERIALS AND METHODS
Plasmid constructs and protein preparations.
The human PABP expression plasmid pET28-PABP (51) and the human α2-globin expression plasmid pSV2Aneo-α2 (53) have previously been reported. The plasmid expressing the N-terminal segment of PABP (pET28-PABP-NT) was derived from pET28-PABP. The carboxyl-terminal (C-terminal) coding sequences were removed by deleting the cDNA from the MscI site to the HindIII site within the vector polylinker sequence. The resulting plasmid bears DNA that encodes a His tag at the amino terminus followed by the first 355 amino acids of PABP, which includes all four RNA-binding domains. The pET28-PABP-CT plasmid which expresses the C-terminal auxiliary domain of PABP (amino acids 352 to 631) was constructed by deleting the 5′ segment of PABP cDNA in pET28-PABP from the NdeI site to the MscI site. The NdeI site was filled in to reconstitute the translation start codon and ligated to the MscI-cut fragment, which maintains the correct frame. The human αCP1 expression plasmid (pET28a-αCP1) was constructed by inserting a PCR-generated αCP1 coding region with primers that introduce an EcoRI restriction site to both ends of the amplified DNA. The αCP1 fragment was inserted into the EcoRI site of pET28a (Novagen) such that the open reading frame was maintained with the histidine tag. Expression and purification of His-tagged PABP, PABP-NT, PABP-CT, and α-CP were conducted according to the instructions of the manufacturer (Novagen).
Extract preparation.
Mouse erythroleukemia (MEL) cell S130 extract was prepared as previously described (22). Cells were washed twice with phosphate-buffered saline, collected, and resuspended in buffer A (10 mM Tris [pH 7.5], 1 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol) at a density of 108 cells/ml of buffer. Cells were lysed by Dounce homogenization. Following removal of the nuclei by low-speed centrifugation (2,000 × g, 10 min), the supernatant was layered over a sucrose cushion (buffer A containing 30% [wt/vol] sucrose) and centrifuged at 130,000 × g for 1.5 h. The supernatant (S130 extract) was adjusted to 7 to 8 μg of protein/μl, supplemented with glycerol to a final concentration of 5% (vol/vol) and frozen in aliquots at −70°C. Poly(A)-depleted or poly(C)-depleted S130 extract was prepared as described by Wang et al. (51). Following depletion, the extract was repeatedly diluted and concentrated with a centricon filter (Amicon) to convert the buffer to buffer A and subsequently concentrated to the initial S130 protein concentration.
RNA substrate generation.
The α-globin 3′UTR template was PCR amplified from the pSV2Aneo-α2 plasmid with a T7 bacteriophage promoter added to the 5′ end as previously reported (52). RNAs for in vitro decay assays were generated with T7 RNA polymerase (Promega) using 200 ng of purified template and polyadenylated as described previously (51, 52). Uniformly labeled riboprobes were transcribed with [α-32P]UTP and the m7G(5′)ppp(5′)G cap analog according to the instructions of the manufacturer (Promega). 5′-end-labeled RNAs were generated by capping unlabeled and uncapped RNA with vaccinia virus capping enzyme and [α-32P]GTP as previously described (52). 3′-end-labeled RNA was generated with capped and unlabeled RNA ligated with [5′-32P]pCp using T4 RNA ligase for 16 h at 4°C. All labeled RNAs used in these studies were gel purified and resuspended as described by Wang et al. (51).
RNAs for electrophoretic mobility shift assays (EMSA) were produced as described by Kiledjian et al. (22) with slight modifications. The template for the αwt RNA containing 60 adenosine residues (αwtA60) was generated by PCR using a 3′ primer containing 60 thymidine nucleotides (nt) at the 5′ end. Uniformly labeled αwtA60 RNA was synthesized with T7 RNA polymerase as described above except that the nucleotide mixture consisted of 2 μl of [α-32P]UTP (3,000 Ci/mmol) and 0.4 mM (each) rATP, rGTP, and rCTP and 7 μM UTP. To generate RNAs containing a labeled poly(A) tail, approximately 20 pmol of unlabeled αwt RNA was polyadenylated with bovine poly(A) polymerase, 1.2 nmol of ATP, and 1 μl of [α-32P]ATP (3,000 Ci/mmol). All RNAs were gel purified prior to use in the assays.
In vitro mRNA decay assays.
All in vitro decay reactions were carried out at 25°C as described by Wang et al. (51). For each reaction, 0.1 pmol (2 × 104 cpm) of labeled RNA was incubated with 75 μg of MEL cell S130 extract or poly(A)-depleted S130 extract in a 20-μl total volume. Where indicated in the figures, 5 mM EDTA was added to inhibit exoribonuclease and deadenylase activity. Ten picomoles of a thioated oligonucleotide competitor consisting of 20 cytosine nucleotides, oligo(dC), was used to sequester αCP in the in vitro decay reactions as indicated in the figures. The use of a thioated oligonucleotide ensures minimal nonspecific degradation of the competitor and is an efficient competitor for αCP (51, 52).
EMSA.
EMSA of uniformly labeled αwtA60 RNA and αCP1 protein were carried out as detailed by Kiledjian et al. (51). Briefly, 2 × 105 cpm of RNA (∼0.1 pmol per reaction mixture) was incubated in RNA-binding buffer with αCP1 protein at room temperature for 30 min, followed by RNase T1 (20 U) digestion for 10 min. The reaction mixture was incubated with heparin (5 mg/ml) for an additional 10 min at room temperature to minimize nonspecific RNA-protein interaction. The RNA-protein complex was resolved on a 5% native gel and visualized by autoradiography. The intensities of the bound complexes were quantitated using a Molecular Dynamics PhosphorImager with Image Quant software. For Fig. 4, the values are presented as the intensity of the αCP1 complex in the presence of PABP relative to the intensity of αCP1 alone. Similarly, the values in Fig. 5 are presented as the intensity of the PABP complex in the presence of αCP1 relative to the binding intensity of PABP alone. Values for both figures were derived from three independent experiments. EMSA of αwtA60 RNA containing a 32P-labeled poly(A) tail (2 × 105 cpm, 1 pmol) were carried out similarly except that 10 ng of RNase A instead of RNase T1 was used in each reaction mixture.
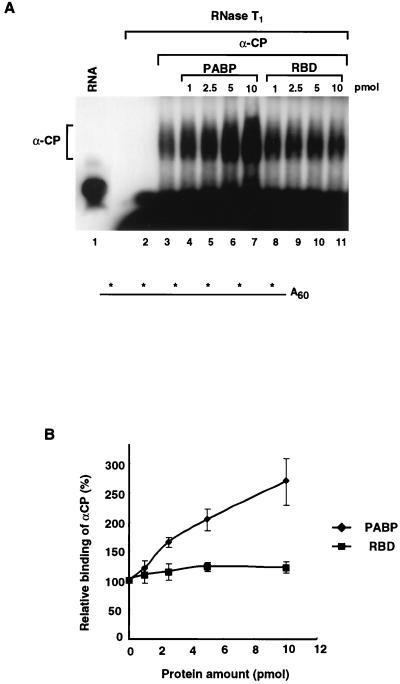
FIG. 4.
PABP enhances the binding of αCP1 to the αwtA+ RNA. EMSA were carried out to determine the effect of PABP on the binding of αCP1 to the αwtA+ RNA. (A) Binding of the αCP1 protein to uniformly labeled αwtA60 was carried out in the presence of increasing amounts of PABP as indicated. The RNase T1-resistant complex was resolved on a 5% native polyacrylamide gel. Migration of the bound αCP1 complex is shown on the left. Addition of PABP increases the binding of αCP1 (lanes 4 to 7) to the 3′UTR, while addition of an unrelated protein had no effect (lanes 8 to 11). A schematic of the uniformly 32P-labeled αwtA60 is shown at the bottom. (B) Quantitation of the results of the binding experiments presented in panel A are plotted as the relative levels of binding of αCP1 in the presence of PABP derived from three independent experiments. The vertical bars represent standard deviations. (C) An EMSA reaction mixture with uniformly labeled αwtA60 was incubated with 10 pmol of the indicated proteins. PABP-NT can also stimulate αCP1 binding, while PABP-CT or the hnRNP U RNA-binding domain (RBD) cannot. The schematic of the RNA is as described for panel A. (D) An EMSA similar to that described for panel C was carried out with uniformly 32P-labeled αwt RNA lacking a poly(A) tail. There was no detectable enhancement of αCP1 binding to the αwt RNA upon the addition of PABP when the RNA lacked a poly(A) tail. A schematic of uniformly 32P-labeled αwt is shown at the bottom.
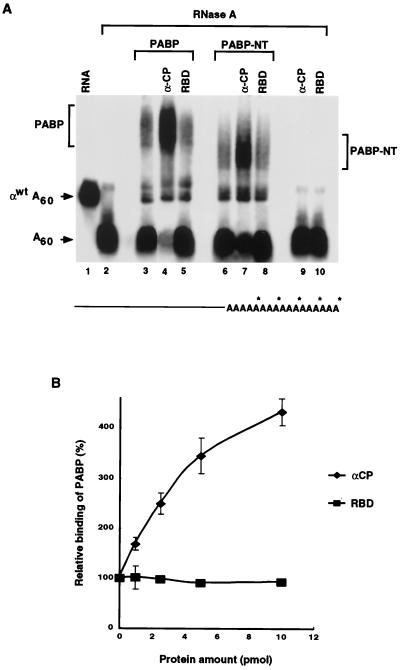
FIG. 5.
αCP1 enhances the binding of PABP to the poly(A) tail. (A) An αwtA+ RNA containing a 32P-labeled poly(A) tail was used in EMSA reaction mixtures to detect the binding of PABP to the poly(A) tail. Where indicated, 10 pmol of PABP or PABP-NT was used in the binding reaction mixtures. Lanes 4, 7, and 9 contain 10 pmol of αCP1, and lanes 5, 8, and 10 contain 10 pmol of the hnRNP U RNA-binding domain (RBD). The binding of both PABP and PABP-NT are enhanced by αCP1. The PABP-poly(A) tail and the PABP-NT–poly(A) tail complexes are indicated. The migrations of αwtA60 and the released poly(A) tail (A60) are indicated on the left of the figure. A schematic of αwtA60 labeled with 32P on the poly(A) tail is shown at the bottom. (B) The relative levels of binding of PABP to the poly(A) tail in the presence of increasing amounts of αCP1 derived from three independent experiments are plotted. The vertical bars denote standard deviations.
RESULTS
ErEN activity is influenced by the poly(A) tail.
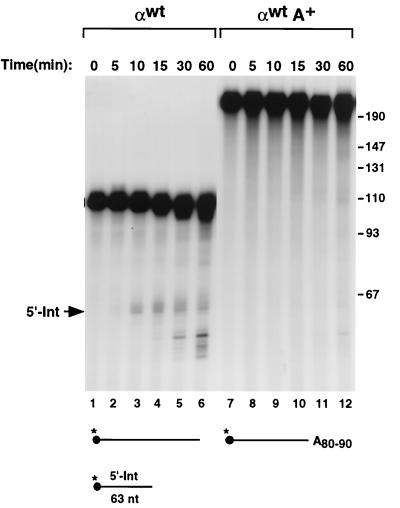
We have recently demonstrated that a sequence-specific erythroid cell-enriched endoribonuclease activity termed ErEN is involved in the turnover of the α-globin mRNA in vitro (52). The detection of the same ErEN-specific intermediate products in erythroid cells also suggested that this activity is involved in the natural turnover of α-globin mRNA (52). The majority of eukaryotic mRNA turnover pathways initiate with deadenylation. However, many of the mRNAs known to be targeted by endoribonucleases are cleaved independently of the polyadenylation state of the mRNA (2, 40, 43). To begin addressing whether ErEN is influenced by the poly(A) tail, we compared the ErEN activity on unadenylated αwt RNA to that on adenylated αwt (αwtA+) RNA. Cleavage of αwt by ErEN produces two intermediates, a 63-nt 5′ fragment and a 47-nt 3′ fragment, which are subsequently cleared by a 3′-to-5′ exoribonuclease(s) (52). Capped and 5′-end-labeled αwt RNAs which either contain or lack a poly(A) tail were incubated with S130 extract for up to 1 h. As seen in Fig. 1, the 5′ intermediate (5′-Int) (same as Int-1 in reference 52) was detected using the unadenylated RNA as well as subsequent smaller decay products which appeared with longer incubation times (lanes 2 to 6). Surprisingly, under identical conditions, ErEN's ability to cleave the αwtA+ RNA was reproducibly and significantly reduced (compare lanes 2 to 6 with lanes 8 to 12). These data demonstrate that the poly(A) tail can influence the cleavage of αwt by ErEN.
FIG. 1.
ErEN cleavage activity is poly(A) tail dependent. An in vitro decay assay was carried out with the 5′-end-32P-labeled and capped α-globin 3′UTR (αwt) and incubated with MEL cell S130 cytosolic extract. Reactions were carried out at room temperature for 5 to 60 min as indicated above the lanes with the unadenylated (αwt) or adenylated (αwtA+) 3′UTR. The location of the 5′ intermediate fragment (5′-Int) generated by the initial ErEN cleavage is shown. The smaller bands which accumulate with increasing incubation time in lanes 5 and 6 are a consequence of subsequent 3′-to-5′ exoribonuclease activities present within the extract (51). The RNA substrates used are shown schematically at the bottom of the figure. The filled circle denotes the 5′ m7G cap, the asterisks represent the position of the 32P labeling, and A80–90 signifies the 80 to 90 adenosine residues in the poly(A) tail. The relative position and size of the 5′-Int are shown. Single-stranded DNA size markers are shown on the right in nucleotides.
PABP inhibits ErEN activity.
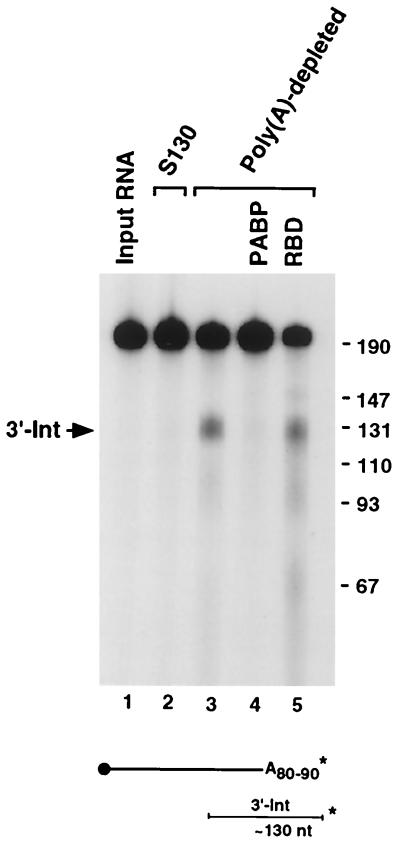
We next addressed whether the poly(A) tail effect on ErEN activity was a result of the adenosine residues or mediated through a trans-acting factor. To address this issue we used extract depleted of poly(A)-binding activity by incubation with poly(A) agarose beads. Incubation of a 3′-end-labeled αwtA+ RNA for 15 min with the poly(A)-depleted extract readily generated the ErEN cleavage product compared to what occurred in reactions with complete S130 extract (Fig. 2, compare lanes 3 to 2). Incubation of polyadenylated αwt with complete S130 extract even up to 1 h also failed to generate significant ErEN cleavage products (Fig. 1). PABP, which is most likely the predominant cytoplasmic poly(A)-binding activity, was tested to determine whether it can reconstitute the poly(A) tail-mediated inhibition of ErEN activity. Addition of recombinant PABP to the depleted extract restored resistance of the RNA to ErEN cleavage (compare lanes 4 to 3), while addition of a control RNA-binding protein consisting of the hnRNP U RNA-binding domain had no effect (lane 5). As expected, no difference in ErEN activity was observed between complete S130 extract and poly(A)-depleted S130 extract when the RNA substrate was not polyadenylated (data not shown). We conclude that the inhibitory role of the poly(A) tail on ErEN activity is mediated through PABP.
FIG. 2.
ErEN activity is inhibited by PABP bound to the poly(A) tail. 3′-end-labeled αwtA+ RNA containing 80 to 90 adenosines (A80–90) was incubated for 15 min with either MEL cell S130 extract (lane 2) or with S130 depleted with poly(A) agarose beads (lanes 3 to 5) in an in vitro decay assay. One microgram of PABP or the hnRNP U RNA-binding domain (RBD) was included as indicated (lane 4 or 5, respectively). The schematic of the RNA is shown at the bottom and is as described in the legend to Fig. 1, as are the size markers. The migration of the 3′ intermediate fragment is indicated on the left and shown schematically on the bottom.
The influence of PABP on ErEN activity is mediated through the binding of αCP.
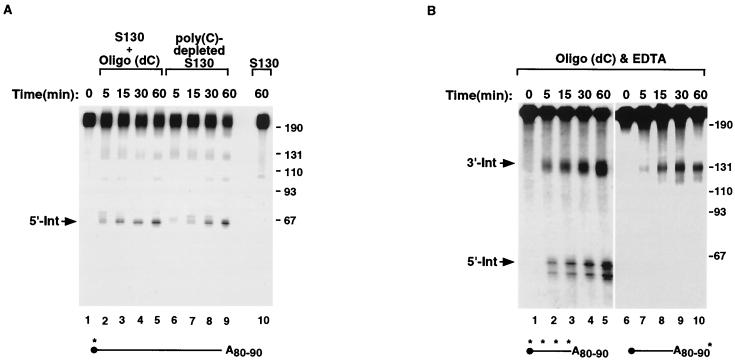
We next addressed how PABP could function to regulate ErEN activity. In our initial identification of ErEN we demonstrated that the binding of αCP to unadenylated αwt RNA prevented access of ErEN to the RNA (52). Sequestration of the αCP proteins with an oligo(dC) oligonucleotide competitor exposed the ErEN target sequence within the 3′UTR and enabled more efficient cleavage (51, 52). The effect of αCP on αwtA+ was tested in an in vitro decay reaction identical to that described for Fig. 1, except that oligo(dC) was included as a competitor to sequester αCP. As seen in Fig. 3A, ErEN activity was detected on the αwtA+ RNA when oligo(dC) was included despite the fact that the 5′-end-labeled RNA was polyadenylated (lanes 2 to 5). Similar results were obtained when poly(C)-depleted extract, which removes the αCP proteins (51), was used (lanes 6 to 9). Incubation of complete S130 extract in the absence of competitor resulted in inappreciable ErEN activity (lane 10) similar to that observed in Fig. 1. Therefore, it appears that upon the removal of the αCP proteins, ErEN was able to cleave even a polyadenylated RNA. However, the formal possibility existed that the RNA was rapidly deadenylated and subsequently cleaved by ErEN. To rule out this possibility, the reactions were repeated with uniformly labeled or 3′-end-labeled αwtA+ RNAs containing approximately 90 adenosine residues. EDTA was included in the reaction mixtures to minimize both deadenylation and exonucleolytic degradation to more readily detect the ErEN intermediates. ErEN activity is not sensitive to, or altered by, low levels of EDTA (52). As shown in Fig. 3B, both the 5′-Int as well as the ∼130-nt polyadenylated 3′-Int were detected with uniformly labeled αwtA+ (lanes 2 to 5). Further confirmation that the 3′-Int is indeed polyadenylated was provided with the use of a 3′-end-labeled αwtA+ RNA containing ∼90 adenosines. Again, the ∼130-nt 3′-Int was detected (lanes 6 to 10), illustrating that the RNA can remain adenylated and be cleaved by ErEN. These data demonstrate that influence of the poly(A) tail on ErEN activity is mediated through the αCP protein and that sequestration of αCP off of the α-globin 3′UTR bypasses the inhibitory role of the poly(A) tail.
FIG. 3.
ErEN can cleave polyadenylated αwt upon sequestration of αCP. ErEN activity on polyadenylated αwt RNA was determined in the in vitro decay assays by using oligo(dC), which is an efficient competitor for the sequestration of αCP, or by using poly(C)-depleted extract, which is devoid of αCP (51). (A) An in vitro RNA decay reaction was carried out with 5′-end-labeled αwt containing a poly(A) tail of approximately 80 to 90 residues as described in the legend to Fig. 1, except that 10 pmol of an oligo(dC) competitor was included in lanes 2 to 5 or poly(C)-depleted extract was used in lanes 6 to 9 for the indicated times. The reaction with complete S130 extract at the 60-min time point is shown in lane 10. (B) In vitro decay reactions were carried out as described for panel A except 5 mM EDTA was included in the reaction mixtures to minimize deadenylation and exoribonuclease activity. αwtA+ uniformly labeled with 32P was used in lanes 1 to 5, and 3′-end-labeled αwtA+ was used in lanes 6 to 10. Labeling is as described in the legend to Fig. 1.
The observations that both αCP and PABP can influence ErEN activity and that removal of αCP circumvents the significance of the poly(A) tail suggests that PABP exerts its effect through αCP. One way PABP may influence αCP is by increasing its binding affinity to the αwtA+ RNA, which in turn would protect the RNA from degradation by ErEN. The αCP proteins were recently shown to bind directly to the αwt RNA 3′UTR (11). Therefore, we were able to test this hypothesis with an EMSA using a reconstituted system with recombinant αCP1 and PABP proteins in the absence of extract. An uniformly labeled αwtA+ RNA containing an unlabeled poly(A) tail was incubated with recombinant αCP1 in the presence of increasing amounts of PABP. Following a 20-min incubation, the reaction mixtures were treated with RNase T1 to resolve the RNase-resistant αCP-RNA ribonucleoprotein complex (Fig. 4A, lane 3). Addition of recombinant PABP to the reaction mixture reproducibly increased the formation of the αCP1 complex (Fig. 4A, lanes 4 to 7). Similar results were obtained when αCP2 was used instead of αCP1 (data not shown). An increase in the binding of αCP1 was not detected with an unrelated RNA-binding domain (lanes 8 to 11). Relative αCP1 binding intensities in the presence of PABP or the control protein are plotted in Fig. 4B. The increase in the intensity of the αCP complex signal was not due to an increase in the size of the labeled RNA within the bound complex when PABP was included, since the RNA fragments in both complexes are the same size (data not shown). These results are consistent with our previous demonstration that αCP and PABP can interact with one another both in vitro and in vivo (51).
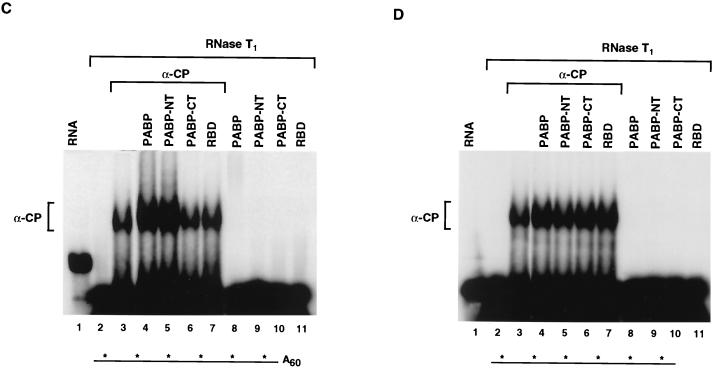
To determine what region of PABP was responsible for enhancing αCP1 binding activity, the N-terminal and C-terminal halves of PABP were individually tested. Similar to the full-length PABP, the N-terminal segment of PABP stimulated binding of αCP1 (Fig. 4C, lane 5). This truncated form of PABP contains the first 355 amino acids of the protein and includes the four RNA-binding domains. Addition of the C-terminal auxiliary domain had no effect (lane 6). As shown in lanes 8 through 11, neither PABP, its truncated derivatives, nor the control protein can form an RNase-resistant complex with the labeled RNA on its own. Any binding of PABP to the unlabeled poly(A) tail would not be detected, since the RNA is cleaved with RNase T1 following complex formation, which separates the unlabeled poly(A) tail from the labeled 3′UTR. The observed increase in αCP1 binding by PABP required a polyadenylated substrate since αCP1 binding to the 3′UTR lacking a poly(A) tail was not stimulated by PABP in trans (Fig. 4D). These data show that PABP once bound to the poly(A) tail can then stimulate the binding of αCP1 to the α-globin 3′UTR. Furthermore, the effector domain is contained within the first four RNA-binding domains of PABP.
αCP enhances the binding of PABP to the poly(A) tail.
Our findings that PABP can stabilize the binding of αCP to αwtA+ RNA (Fig. 4) and that αCP can interact with PABP to impede deadenylation (51) suggests that αCP may also stabilize the binding of PABP to the poly(A) tail. Such an interaction might provide a mutual stabilization of αCP to the CRE and of PABP to the poly(A) tail and in turn constitute a higher-order complex to protect the mRNA. EMSA were used to test whether the binding of PABP is enhanced by αCP1. An αwtA+ RNA containing an unlabeled 3′ UTR and a 32P-labeled poly(A) tail was used to enable the detection of PABP binding. Treatment of the RNA with RNase A, which selectively cleaves after pyrimidines, degrades the 3′UTR but leaves the poly(A) tail intact (Fig. 5A, lane 2). It therefore can be used to specifically detect the binding of PABP to the poly(A) tail. Incubation of recombinant PABP with the αwtA+ probe generated an RNase A-resistant PABP complex as shown in Fig. 5A (lane 3). As shown in Fig. 5B, formation of the PABP complex increased upon addition of an increasing amount of recombinant αCP1 but not upon addition of an unrelated RNA-binding domain. Similar results were also detected with αCP2 (data not shown). Consistent with the interaction domain of PABP to αCP1 being contained in its amino terminus, Fig. 5A demonstrates that the binding of the N-terminal segment of PABP to the poly(A) tail (lane 6) was also increased with the addition of αCP1 (lane 7). We conclude that there is a synergistic interaction between αCP and PABP which increases the binding affinity of each protein to its respective substrate, thus providing a higher-order network which stabilizes the α-globin mRNA.
DISCUSSION
This report demonstrates that the poly(A) tail-PABP complex can influence the fate of mRNA turnover by regulating a process distinct from its well-characterized role of preventing deadenylation. PABP can interact with a protein bound to the body of the mRNA and influence the activity of an endoribonuclease. This role of PABP is indirect and exerted through αCP. Binding of αCP to the αwt RNA prevents cleavage of this RNA by ErEN (52). Therefore, by increasing the binding affinity of αCP to the 3′UTR, the RNA becomes further refractory to ErEN cleavage. Collectively, this study along with previous reports (33, 51, 52) supports the following model for α-globin mRNA stability and turnover. αwtA+ RNA contains αCP bound to the CRE and PABP bound to the poly(A) tail. The αCP proteins by an interaction with PABP stabilize the PABP-poly(A) tail interaction and lower the rate of deadenylation. At the same time, this interaction also improves the αCP-CRE interaction and more efficiently prevents ErEN from accessing the mRNA. Upon eventual deadenylation, the αCP-PABP interaction is disrupted and ErEN is more likely to displace αCP and cleave the RNA, thus accounting for the influence of the poly(A) tail on ErEN activity. Since the role of PABP is exerted through the binding of αCP, sequestration of αCP should eliminate the poly(A) tail dependence, and this is indeed what is observed (Fig. 3). The requirement of PABP to function indirectly through the binding of αCP may provide an erythroid cell with an override mechanism that is able to bypass the need for prior deadenylation. The α-globin mRNA can therefore be degraded by two pathways, one initiated by, and dependent on, deadenylation and the other being a deadenylation-independent pathway initiated by the removal of αCP, which exposes an endoribonuclease cleavage site. Furthermore, the αCP-PABP interaction appears to be synergistic and improves the binding of PABP to the poly(A) tail, which may account for the observed influence of αCP on deadenylation (33, 51). It is possible that this is not a unique feature to αCP and that it is a more general strategy utilized by other mRNA-stabilizing proteins.
The control of α-globin transcript stability by a regulated endoribonuclease activity may be critical during two stages of erythropoiesis. First, it may provide a mechanism to ensure that excess toxic α-globin does not accumulate relative to the level of β-globin, which leads to ineffective erythropoiesis and hemolysis (34, 44). ErEN activity might be influenced by excess α-globin protein levels in a differentiating erythrocyte and specifically initiate degradation of a fraction of α-globin mRNA regardless of its polyadenylation state. A second role for ErEN activity may be during terminal erythroid differentiation, when αCP levels decrease (33) and all mRNAs, including α-globin, are cleared from the erythrocyte (36). At this point ErEN would be able to access its target site within the 3′UTR and initiate the demise of the α-globin mRNA irrespective of its polyadenylation state. Identification of the ErEN protein and the gene encoding this activity will more thoroughly address its biological role.
Previous studies by us and others (23, 24, 50) were unable to detect direct binding of αCP to the αwt RNA; however, Chkheidze et al. (11) recently detected direct binding of recombinant αCP to this RNA. The apparent discrepancy appears to be due to the assay conditions employed. The earlier studies used an RNase cocktail containing both RNase A and RNase T1 to degrade the αwt RNA following formation of a RNP complex (23, 24, 50), while the later study used only RNase T1 (11). We were unable to detect direct binding of αCP1 or αCP2 to αwt RNA when RNase A was included in the assays (unpublished observations), yet complex formation was readily detected when RNase A was omitted (Fig. 4). Perhaps efficient RNase A-resistant binding of αCP to αwt requires additional proteins that can associate with the 3′UTR as previously proposed (23). This suggests that the interaction of αCP with PABP is only part of a larger network of protein-protein interactions that are involved in α-globin mRNA stability.
Several regions of PABP have been mapped as protein-protein interaction domains. In the yeast PABP (Pab1), the second RNP motif is required for the interaction with eIF4G within the cap-binding complex (21). The carboxyl terminus of PABP appears to mediate protein-protein interactions with multiple proteins, including the translational termination release factor eRF3 (19) and the yeast αCP-like hnRNP K homology domain-containing protein Pbp2 (32), as well as mediating PABP homotypic interactions for poly(A) binding (26). We had initially reported that the region required for an interaction of PABP with αCP resides within amino acids 199 to 631 (51). The present study demonstrates that amino acids 1 to 355 contains the region of PABP which can interact with and enhance the binding of αCP to the CRE. This region does not include the C-terminal auxiliary domain and eliminates this region as the potential interaction domain. Taken together, these data indicate that the interaction domain is contained within amino acids 199 to 355, which includes part of the third RNP motif and all of the fourth. Considering that the highly structured third RNP motif is truncated and is unable to form the native RNP motif structure, we predict that the interaction domain for PABP with αCP is contained in the fourth RNP motif. Further deletional studies will address this hypothesis.
The interaction of PABP with the 5′ end of an mRNA has been demonstrated for yeast and mammalian systems (14, 20, 38, 48, 54). The fact that PABP could also interact with proteins bound to the α-globin 3′UTR implies a link between the 3′UTR and the 5′ cap. Such an interaction may produce a “pretzel” type of structure where PABP may juxtapose αCP and other α-globin 3′UTR-binding proteins with the 5′ cap. We are currently testing whether αCP, or other proteins which associate with the α-globin 3′UTR (23), can interact with the 5′ cap and/or cap-binding proteins to influence events at the 5′ end.
Endoribonucleases characterized to date which specifically target mRNA seem to function in a deadenylation-independent manner (2, 40, 43). For example, a specific endoribonuclease cleaves the transferrin receptor 3′UTR, releasing a polyadenylated 3′ fragment (5). This is also the case with the 9E3 growth factor mRNA, where a polyadenylated 3′ endoribonuclease product is detected (47). Similarly, the Xlhbox 2 mRNA is targeted by a specific endoribonuclease that cleaves the mRNA independently of the state of the poly(A) tail in vitro (6). In these cases it appears that an endoribonuclease provides a means to bypass deadenylation and respond quickly to an environmental stimulus. Endoribonuclease cleavage of polyadenylated β-globin mRNA has also been reported (30). In a particular β0 thalassemia variant which contains a premature translational stop codon, three distinct aberrant β-globin transcripts that are cleaved within exon 1 or 2 and are missing the 5′ segment of the mRNA are detected (29, 30).
The inhibition of ErEN cleavage by the poly(A) tail is surprising and indicative of a more complex regulatory mechanism involving two modes, one that is poly(A) tail dependent and one that is not. The ErEN cleavage site has been mapped to 47 nt upstream of the poly(A) addition site in the α-globin 3′UTR (52). A requirement for an endoribonucleolytic cleavage 47 nt from the end of the mRNA after deadenylation seems redundant since the deadenylated mRNA should already be a target for a 3′-to-5′ exoribonuclease(s). We therefore propose that the terminal 47 nt of the α-globin mRNA also contribute to the stability of this mRNA. The endoribonuclease would function to remove this region and expose the mRNA to 3′-to-5′ exonucleolytic degradation. Detection of the short 47-nt ErEN cleavage fragment in cells expressing the α-globin mRNA (52) further underscores the unusual stability of this RNA segment. Consistent with a role for the terminal 47 nt in mRNA stability, preliminary data indicate that this region is required to hinder a 3′-to-5′ exoribonuclease activity in vitro (N. Rodgers and M. Kiledjian, unpublished observations). The stability of the α-globin mRNA appears to be regulated by a complex network of interactions, only one of which involves the interaction of αCP with PABP to regulate deadenylation and cleavage by ErEN. Further studies will address the significance of additional α-globin 3′UTR-binding proteins and the terminal 47 nt of this mRNA in the overall stability of the α-globin message.
ACKNOWLEDGMENTS
We thank N. D. Rodgers and P. Trifillis for helpful discussions and critical reading of the manuscript and N. Shanmugam for providing the hnRNP U RNA-binding domain protein.
This work was supported by funds from the National Institutes of Health (grant DK51611) to M.K.
REFERENCES
- 1.Albrecht G, Krowczynska A, Brawerman G. Configuration of beta-globin messenger RNA in rabbit reticulocytes. Identification of sites exposed to endogenous and exogenous nucleases. J Mol Biol. 1984;178:881–896. doi: 10.1016/0022-2836(84)90317-6. [DOI] [PubMed] [Google Scholar]
- 2.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein P, Peltz S W, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binder R, Gordon D A, Hwang S P, Williams D L. Estrogen-induced destabilization and associated degradation intermediates of apolipoprotein II mRNA. Prog Clin Biol Res. 1990;322:227–240. [PubMed] [Google Scholar]
- 5.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown B D, Zipkin I D, Harland R M. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 1993;7:1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- 7.Burd C G, Matunis E L, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caponigro G, Parker R. mRNA turnover in yeast promoted by the MATalpha1 instability element. Nucleic Acids Res. 1996;24:4304–4312. doi: 10.1093/nar/24.21.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 10.Chernokalskaya E, Dubell A N, Cunningham K S, Hanson M N, Dompenciel R E, Schoenberg D R. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA. 1998;4:1537–1548. doi: 10.1017/s1355838298980451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chkheidze A N, Lyakhov D L, Makeyev A V, Morales J, Kong J, Liebhaber S A. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol Cell Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christiansen J, Kofod M, Nielsen F C. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig A W, Haghighat A, Yu A T, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 15.Decker C J, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 16.Deo R C, Bonanno J B, Sonenberg N, Burley S K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 17.Ford L P, Bagga P S, Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol Cell Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grange T, de Sa C M, Oddos J, Pictet R. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 20.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler S H, Sachs A B. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiledjian M, Day N, Trifillis P. Purification and RNA binding properties of the polycytidylate-binding proteins αCP1 and αCP2. Methods. 1999;17:84–91. doi: 10.1006/meth.1998.0710. [DOI] [PubMed] [Google Scholar]
- 23.Kiledjian M, DeMaria C T, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Körner C G, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kühn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 27.Lee C H, Leeds P, Ross J. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. J Biol Chem. 1998;273:25261–25271. doi: 10.1074/jbc.273.39.25261. [DOI] [PubMed] [Google Scholar]
- 28.Leffers H, Dejgaard K, Celis J E. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 29.Lim S, Mullins J J, Chen C M, Gross K W, Maquat L E. Novel metabolism of several beta zero-thalassemic beta-globin mRNAs in the erythroid tissues of transgenic mice. EMBO J. 1989;8:2613–2619. doi: 10.1002/j.1460-2075.1989.tb08401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim S K, Maquat L E. Human beta-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5′ termini. EMBO J. 1992;11:3271–3278. doi: 10.1002/j.1460-2075.1992.tb05405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makeyev A V, Chkheidze A N, Liebhaber S A. A set of highly conserved RNA-binding proteins, alphaCP-1 and alphaCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J Biol Chem. 1999;274:24849–24857. doi: 10.1074/jbc.274.35.24849. [DOI] [PubMed] [Google Scholar]
- 32.Mangus D A, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales J, Russell J E, Liebhaber S A. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J Biol Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 34.Nathan D G, Gunn R B. Thalassemia: the consequences of unbalanced hemoglobin synthesis. Am J Med. 1966;41:815–830. doi: 10.1016/0002-9343(66)90039-8. [DOI] [PubMed] [Google Scholar]
- 35.Nietfeld W, Mentzel H, Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papayannopoulou T, Abkowitz J, D'Andrea A. Biology of erythropoiesis, erythroid differentiation, and maturation. In: Hoffman R, Benz E J, Shattil S J, Furie B, Cohen H J, Silberstein L E, McGlave P, editors. Hematology: basic principles and practice. 3rd ed. New York, N.Y: Churchill Livingstone; 2000. pp. 202–219. [Google Scholar]
- 37.Pastori R L, Moskaitis J E, Schoenberg D R. Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry. 1991;30:10490–10498. doi: 10.1021/bi00107a018. [DOI] [PubMed] [Google Scholar]
- 38.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prokipcak R D, Herrick D J, Ross J. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J Biol Chem. 1994;269:9261–9269. [PubMed] [Google Scholar]
- 40.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 42.Scheper W, Holthuizen P E, Sussenbach J S. The cis-acting elements involved in endonucleolytic cleavage of the 3′ UTR of human IGF-II mRNAs bind a 50 kDa protein. Nucleic Acids Res. 1996;24:1000–1007. doi: 10.1093/nar/24.6.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoenberg D R, Chernokalskaya E. Ribonucleases involved in eukaryotic mRNA turnover. In: Harford J B, Morris D R, editors. mRNA metabolism and post-transcriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. pp. 217–240. [Google Scholar]
- 44.Schwartz E, Benz E J. The thalassemia syndromes. In: Hoffman R, Benz E J, Shatill S, Furie B, Cohen H J, editors. Hematology: basic principles and practice. New York, N.Y: Churchill Livingston; 1991. pp. 368–392. [Google Scholar]
- 45.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 46.Stoeckle M Y. Removal of a 3′ non-coding sequence is an initial step in degradation of gro alpha mRNA and is regulated by interleukin-1. Nucleic Acids Res. 1992;20:1123–1127. doi: 10.1093/nar/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoeckle M Y, Hanafusa H. Processing of 9E3 mRNA and regulation of its stability in normal and Rous sarcoma virus-transformed cells. Mol Cell Biol. 1989;9:4738–4745. doi: 10.1128/mcb.9.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 49.Tommerup N, Leffers H. Assignment of human KH-box-containing genes by in situ hybridization: HNRNPK maps to 9q21.32–q21.33, PCBP1 to 2p12–p13, and PCBP2 to 12q13.12–q13.13, distal to FRA12A. Genomics. 1996;32:297–298. doi: 10.1006/geno.1996.0121. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol Cell Biol. 1995;15:1769–77. doi: 10.1128/mcb.15.3.1769. . (Erratum, 15:2331.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Kiledjian M. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J. 2000;19:295–305. doi: 10.1093/emboj/19.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss I M, Liebhaber S A. Erythroid cell-specific mRNA stability elements in the α2-globin 3′ nontranslated region. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 55.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]