Abstract
Messenger RNA-based vaccines against COVID-19 induce a robust anti-SARS-CoV-2 antibody response with potent viral neutralization activity. Antibody effector functions are determined by their constant region subclasses and by their glycosylation patterns, but their role in vaccine efficacy is unclear. Moreover, whether vaccination induces antibodies similar to those in patients with COVID-19 remains unknown. We analyze BNT162b2 vaccine-induced IgG subclass distribution and Fc glycosylation patterns and their potential to drive effector function via Fcγ receptors and complement pathways. We identify unique and dynamic pro-inflammatory Fc compositions that are distinct from those in patients with COVID-19 and convalescents. Vaccine-induced anti-Spike IgG is characterized by distinct Fab- and Fc-mediated functions between different age groups and in comparison to antibodies generated during natural viral infection. These data highlight the heterogeneity of Fc responses to SARS-CoV-2 infection and vaccination and suggest that they support long-lasting protection differently.
Keywords: SARS-CoV-2, BNT162b2 mRNA vaccine, antibodies, IgG-Fc, Fcγ receptors, Fc-Glycosylation, complement, immunity, effector function, IgG glycosylation
Graphical abstract
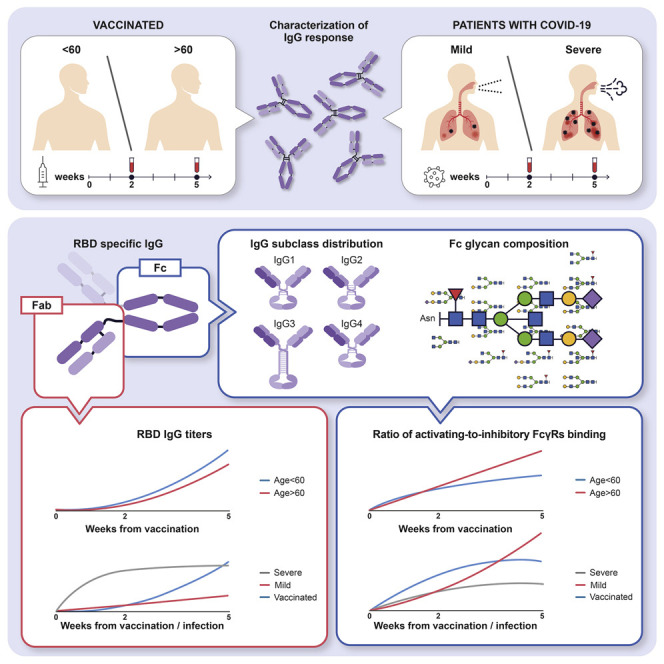
The Fc structures of IgGs produced during infection and vaccination have important roles in shaping the immune response. Farkash et al. show that vaccine- and infection-induced anti-SARS-CoV-2 IgGs differ in their Fc regions and in the engagement of complement and Fc receptors, implying distinct effector functions and immunity.
Introduction
The outbreak of the coronavirus disease 2019 (COVID-19) pandemic, with its associated tremendous health and economic consequences, has sparked a worldwide effort to design vaccines using different platforms. Messenger RNA (mRNA)-based vaccines have demonstrated high efficacy in reducing infection, symptomatic disease, and hospitalization caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) (McDonald et al., 2021). These vaccines elicit high titers of virus-neutralizing antibodies in the vast majority of vaccinated individuals (Goel et al., 2021; Wang et al., 2021). This response is dominated by the immunoglobulin G (IgG) isotype, and serum levels of neutralizing IgG induced by the vaccine were correlated with its efficacy in preventing SARS-CoV-2 infection (Baden et al., 2021; Polack et al., 2020). However, the generated IgG crystallizable fragment (Fc) structures of the vaccine-induced antibodies, their contribution to vaccine efficacy, and whether they induce similar effector function to viral-induced Ig remain unclear.
The response of IgG antibodies to infection and vaccination is elicited by two functional domains. Whereas the variable antigen-binding fragment (Fab) domain confers their antigen-binding specificity (Sela-Culang et al., 2013), the constant Fc domain determines their effector function. The latter is achieved by engagement of this domain with Fcγ receptor (FcγR) pathways to activate innate and adaptive immune responses, including cross-presentation of antigens for the activation of T cells, antibody-dependent cell-mediated phagocytosis (ADCP), antibody-dependent cellular cytotoxicity (ADCC), and complement-dependent cytotoxicity (CDC) (Bournazos and Ravetch, 2017; Lu et al., 2018). Fc function and, in particular, Fc-FcγR interactions are important for the activity of neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs), which have been developed to prevent or treat COVID-19 (Winkler et al., 2021; Yamin et al., 2021). However, studies on COVID-19 vaccine response have focused mostly on Fab-mediated viral binding and neutralization and, consequently, Fc characteristics and function in vaccine-induced IgG remain poorly known. In addition to the Fc-mediated elimination of infected cells and protective antiviral inflammation, modification of Fc structure and, particularly, of its glycan composition can affect IgG generation and the quality of Fab-mediated neutralization, as was previously shown for seasonal influenza vaccination (Wang et al., 2015a). It is unclear whether such Fc glycan modifications occur in response to the novel mRNA-based anti-COVID-19 vaccine and if so, how they affect the response to the vaccine.
Another question that remains open is how the IgG Fc response elicited by the mRNA vaccine compares to the immune response occurring in individuals who were naturally infected with SARS-CoV-2. Patients with severe, not mild, COVID-19 were reported to have a unique pro-inflammatory IgG signature during the early days post-infection, which was characterized by an elevation in afucosylation of Fc glycans in IgG1 antibodies. This Fc modification resulted in increased IgG1 binding to FcγRIIIA expressed on monocytes and macrophages and in subsequent release of inflammatory cytokines, which may have contributed to the development of pneumonia in these critically ill patients (Chakraborty et al., 2021a; Larsen et al., 2021). Nevertheless, the post-translational modifications occurring in IgG Fc domains of patients infected with SARS-CoV-2, convalescent, and vaccinated individuals and the role of this domain in the immune response against this disease have yet to be fully elucidated.
To address these questions, we performed an in-depth analysis of longitudinal IgG Fc response to the mRNA vaccine. To that end, we established a cohort of SARS-CoV-2-naive individuals aged 24–94 years who received 2 doses of SARS-CoV-2 mRNA BNT162b2 vaccine. We characterized the Fc structure and function of anti-Spike receptor binding domain (RBD) SARS-CoV-2-specific IgG generated by the 2-dose vaccination regime over a 5-week period and compared them to IgG from COVID-19 convalescents, mild, and severe patients. The results provide new insights into the IgG response to SARS-CoV-2 mRNA vaccine and infection.
Results
IgG Fab and Fc responses to BNT162b2 SARS-CoV-2 vaccine
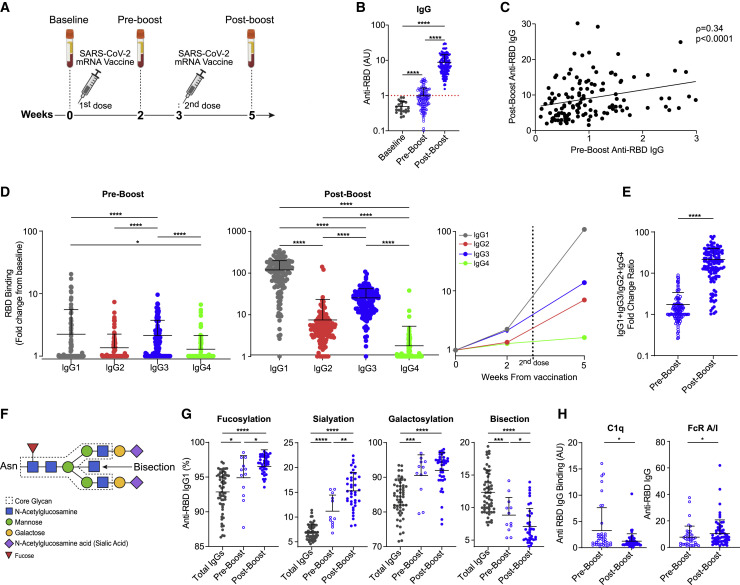
To explore the involvement of the Fc domain in IgG response to COVID-19 vaccine, we collected blood samples from 131 individuals with no evidence of prior SARS-CoV-2 infection 2 weeks after the first vaccine dose with BNT162b2 (“pre-boost”) and 2 weeks after the second vaccine dose (“post-boost”) (Figure 1 A; Table 1 ). Pre-vaccine samples were available from 23 individuals in this cohort. To characterize Fab-mediated IgG activity, we analyzed the serum titer of antibodies binding to the SARS-CoV-2 RBD, which correlates to and predicts the neutralization activity of the antibody (Wajnberg et al., 2020; Wu et al., 2020). Two weeks after the first vaccine dose, 50 of the 131 participants (38.2%) were found to be positive for the presence of RBD-binding IgGs. One participant was also positive for anti-N SARS-CoV-2 protein IgG, and therefore was suspected as COVID-19 convalescent and excluded from the study. In line with previous reports (Sahin et al., 2020), 2 weeks after the second vaccine dose, we observed a significant increase in IgG binding to RBD and 127 of the 130 individuals were positive for RBD binding (Figure 1B). The three participants who did not develop post-boost anti-RBD IgG response, presumably due to medical history associated with immunodeficiencies, were excluded from further analysis (for more details, see Method details). A strong correlation between pre-boost and post-boost anti-RBD IgG levels indicates that the early IgG response is predictive of the IgG levels induced by the two-dose vaccination (Figure 1C).
Figure 1.
IgG Fab and Fc responses to BNT162b2 SARS-CoV-2 vaccine
(A) Experimental scheme. Blood was collected before receiving any vaccine dose (baseline, n = 23), at 2 weeks after the first vaccine dose (pre-boost, n = 127), and at 5 weeks (post-boost, n = 127).
(B) RBD-binding IgG levels at baseline, pre-boost, and post-boost. Dotted red line indicates threshold for positivity.
(C) Correlation between anti-RBD IgG titers following the first vaccine dose and following the second dose (n = 127, non-parametric Spearman’s correlation).
(D) Anti-RBD IgG subclass distribution at the pre-boost and post-boost time points. Kruskal-Wallis test was used with Dunn’s post hoc test to correct for multiple comparisons.
(E) (IgG1 + IgG3):(IgG2 + IgG4) ratio of sera anti-RBD IgG subclasses. Pre-boost, n = 123; post-boost, n = 127. –
(F) Scheme of the IgG Fc glycan structure. The N-glycan is attached at the Asn297 position of each IgG heavy chain. The dashed line indicates the conserved heptasaccharide core, which may have the indicated saccharide extensions.
(G) Fc glycosylation patterns of IgG1 in vaccinated individuals, determined by mass spectrometry. Shown are the total IgGs produced at the pre-boost time point (n = 59) and anti-RBD IgGs of participants who had an IgG1 response at pre-boost (n = 12) and at post-boost (n = 39). Detected glycan structures are shown in Figure S2.
(H) Ratios between RBD-specific IgGs binding to activating (FcγRIIa + FcγRIIIa) versus inhibitory (FcγRIIb) receptors at each time point (pre-boost, n = 39; post-boost, n = 59; also see Figure S1).
Data are presented as scatterplots indicating individual measurements (dots); black line represents the mean; error bars represent standard deviations (SDs). Unless otherwise mentioned, unpaired 2-sided Mann-Whitney U test was used to evaluate the differences between groups. p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Table 1.
Baseline characteristics of BNT162b2 vaccinated cohort
| Overall | Age ≤60 | Age >60 | p | |
|---|---|---|---|---|
| n | 127 | 60 | 67 | – |
| Age, y, mean (SD) | 57.57 (17.99) | 40.88 (9.89) | 72.51 (7.10) | <0.001 |
| Male gender (%) | 56 (44.1) | 22 (36.7) | 34 (50.7) | 0.157 |
| Medical history | ||||
| Cardiovascular (%) | 9 (7.1) | 1 (1.7) | 8 (11.9) | 0.057 |
| Diabetes (%) | 12 (9.4) | 1 (1.7) | 11 (16.4) | 0.011 |
| Hypertension (%) | 29 (22.8) | 2 (3.3) | 27 (40.3) | <0.001 |
| Inflammatory/autoimmune (%) | 9 (7.1) | 3 (5.0) | 6 (9.0) | 0.602 |
| History ofcancer (%) | 10 (7.9) | 1 (1.7) | 9 (13.4) | 0.033 |
| Medications | ||||
| Statins (%) | 32 (25.2) | 2 (3.3) | 30 (44.8) | <0.001 |
| Aspirin (%) | 12 (9.4) | 0 (0.0) | 12 (17.9) | 0.002 |
To determine the IgG Fc response to the vaccine, we analyzed the subclass composition of serum anti-RBD IgGs (Figures 1D and S1A). The initial, pre-boost IgG response was mediated by significant elevation of IgG1 and IgG3 levels. At post-boost, serum binding levels of all IgG subclasses were elevated, as IgG1 became the predominant subclass, followed by IgG3 and IgG2 and a negligible IgG4 response. Pre-boost anti-RBD levels of both IgG1 and IgG3 were significant predictors of post-boost anti-RBD titers (Figure S1B). The overall dominance of IgG3 and IgG1 responses supports a strong Fc-mediated effector function. These two subclasses are characterized by high binding affinity to the activating FcγRs and the classic complement pathway, as compared to the lower affinity of IgG2 and IgG4 (Bruhns and Jönsson, 2015). Moreover, the ratio between higher affinity (IgG1 + IgG3) and lower affinity (IgG2 + IgG4) subclasses in the anti-RBD response increased from the pre-boost to post-boost time points (Figures. 1E and S1A). This indicates that the two-dose mRNA vaccine elicits an IgG subclass trajectory consistent with a pro-inflammatory Fc response.
The FcγR and complement-binding properties of a given IgG are dictated by the combination of its IgG subclass and the linked post-translational Fc glycosylation (Bournazos et al., 2017). We therefore analyzed Fc glycoform composition of vaccine-induced anti-SARS-CoV-2 IgGs. For that, we isolated SARS-CoV-2-specific IgGs from pre- and post-boost serum samples of 39 individuals who were positive for anti-RBD IgG response at pre-boost examination, as well as from additional post-boost samples of 19 age- and sex-matched individuals who were negative at pre-boost (Figure S1C). The IgG CH2 domain has a conserved glycan attached at the N297 position, which is composed of a core heptasaccharide structure that can be supplemented by additional saccharide units, namely fucose, N-acetylglucosamine (GlcNAc), up to two galactose units, and two sialic acid units, at conserved positions (Figure 1F). We detected dynamic changes in anti-SARS-CoV-2 IgG1 Fc glycan composition over time following vaccination as compared to Fc glycan structure in the total serum IgG (Figures 1G and S2). We observed a significant increase in IgG1 fucosylation and sialylation and a decrease in bisecting GlcNAc modification (Figure 1G) from the pre-boost to the post-boost time points. Previously described post-translational modifications in response to influenza and tetanus vaccines include increased galactosylation (Selman et al., 2012; Wang et al., 2015a). Therefore, the increased fucosylation and decreased bisection represent an antigen-specific Fc glycosylation kinetics that is unique to SARS-CoV-2 mRNA vaccination. Interestingly, increased sialyation seems to be a feature of both this mRNA-based vaccine and the mentioned tetanus and influenza vaccines. However, due to the temporally dynamic nature of the glycosylation modifications and the fact that different time points were analyzed (influenza, 0, 3, and 8 weeks post-vaccination; tetanus, 0, 2, and 4 weeks post-vaccination), the results of this comparison should be interpreted with care. In addition, whereas the influenza vaccine may elicit a recall response to a previous viral infection, the response to tetanus vaccine in children is more likely naive, similar to the response to SARS-CoV-2 vaccine. Fc sialyation was previously reported to play a role in affinity maturation of antigen-specific IgG1 generated by influenza vaccine (Wang et al., 2015a). Interestingly, we observed that the elevation in IgG1 sialylation, but also in fucosylation and galactosylation, correlated with higher anti-SARS-CoV-2 IgG titers (Figure S1F). This result supports a role for these post-translation modifications in driving a robust antiviral IgG response upon SARS-CoV-2 mRNA vaccination. In addition, we detected changes in glycosylation patterns in IgG2 and IgG3 that were not reported for other vaccines in humans (Figures S1 and S3)
While the functional significance of IgG Fc glycosylation is mostly known for the IgG1 subclass, due to the robust IgG3 response induced by the mRNA vaccine, we analyzed Fc glycosylation of all of the IgG subclasses (Figure S1D). Interestingly, the levels of vaccine-induced anti-SARS-CoV-2 IgG3 fucosylation decreased over time, as opposed to the increase in IgG1 fucosylation. This indicates a unique regulation process of fucosylation for each IgG subclass. In contrast, IgG3 sialylation increased in anti-SARS-CoV-2 antibodies over time and in comparison to total IgG, as observed in IgG1. The trends of the rest of the IgG3 glycan modifications, as well as of all glycan alterations in anti-RBD IgG2, which appeared only post-boost, were similar to what we observed in IgG1, but they did not reach statistical significance in our cohort. To our knowledge, previous studies of other vaccines in humans did not analyze Fc glycosylation patterns in non-IgG1 subclasses; thus, structural comparison of IgG2/3 Fc between SARS-CoV-2 mRNA and other vaccinations is lacking.
To examine how the dynamic changes in the IgG Fc structures translate to potential effector functions, we examined the FcγRs and complement component 1q (C1q) binding profiles of vaccine-induced IgG (Figures 1H and S1E). To determine pure Fc binding properties without the effect of the IgG titer, we corrected for the number of RBD-binding IgGs and characterized a similar quantity of IgG samples. The effector function potency of a given IgG response is dictated by the balance between activating and inhibitory FcγR signaling it induces and is directly correlated with the activating/inhibitory (A/I) FcγR binding ratio (Anthony and Nimmerjahn, 2011; Nimmerjahn et al., 2015). We therefore analyzed the A/I binding ratio of vaccine-induced anti-SARS-CoV-2 RBD IgG and identified significant increase from pre- to post-boost time points (Figure 1H). This kinetic change in FcγR binding intensity over time was not specific to any individual FcγR (Figure S1E), but seemed to be dominated by decreased binding to the inhibitory FcγRIIB post-boost. This highlights the complexity of IgG Fc-FcγR interactions, which can be dictated by the heterogeneous combination of subclasses and Fc glycoforms. For instance, FcγRIIIA binding, which is expected to increase due to the robust post-boost increase in IgG1 and IgG3 response, can also be attenuated due to the relative decreased portion of afucosylated IgG1 and relative levels of IgG3 in the post-boost IgG pool. These opposite effects on IgG binding intensity to FcγRIIIA can neutralize one another, resulting in similar pre-boost versus post-boost binding levels. Moreover, there was decreased binding to C1q in the post-boost time point. Overall, our data reveal unique kinetics of Fc structure generation over the 5 weeks following vaccination with BNT162b2. Moreover, these results suggest that the generated anti-RBD SARS-CoV-2 antibodies acquire increased capability to engage FcγR pathways as a potential mechanism to eliminate infection, in addition to their Fab-mediated neutralization activity.
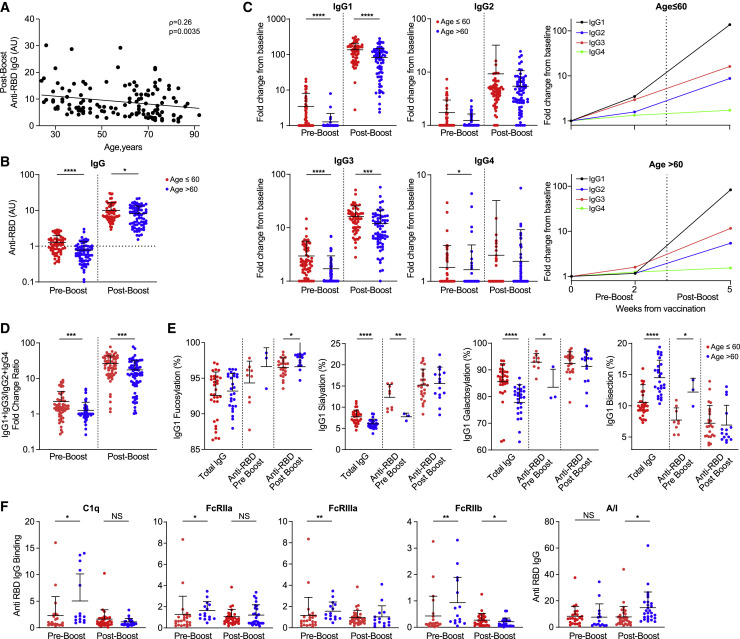
Age, but not sex, affects the BNT162b2-induced IgG Fc structures and function
Immune aging is associated with a diminished ability to mount an effective response against pathogens (Ciabattini et al., 2018), and age is a significant risk factor for severe disease and mortality from COVID-19 (Shahid et al., 2020; Williamson et al., 2020). To determine how age affects the IgG response to BNT162b2 vaccine, we compared the Fab and Fc responses, namely subclass trajectory, Fc glycosylation, and engagement of immune receptors, in young versus elderly individuals. In line with previous reports (Bates et al., 2021; Jalkanen et al., 2021), we found a strong correlation between age and vaccine-induced anti-RBD titers (Figure 2 A). Because individuals older than 60 years are at increased risk for severe outcomes from COVID-19 (Shahid et al., 2020) and were given priority for COVID-19 vaccination, we used this age as a cutoff in analyzing the vaccine response. Individuals older than 60 years had lower anti-RBD SARS-CoV-2 titers in both pre- and post-boost time points, more so in the pre-boost time point (Figure 2B). Whereas in individuals younger than 60 years, a combined IgG1-IgG3 response was observed 2 weeks after the first vaccine dose, the response in the older population was significantly dominated by IgG3, with negligible involvement of other subclasses, implying a delayed switch from IgG3 to IgG1 subclass (Figures 2C and S3A). However, at the post-boost time point, IgG1 became the predominant subclass in both age groups. At both time points, the individuals in the >60 years old group displayed decreased (IgG1+ IgG3):(IgG2+IgG4) ratios as compared to the < 60 years old individuals, due to lower IgG1 and IgG3 levels but similar IgG2 response (Figures 2D and S3A).
Figure 2.
Age affects the BNT162b2-induced IgG Fc structures and function
(A) Correlation of post-boost anti-RBD IgG titers and age. Non-parametric Spearman’s correlation was used, n = 127.
(B) Pre- and post-boost anti-RBD IgG titers when using a cutoff of 60 years of age (age ≤60, n = 60; age >60, n = 67). Dotted line depicts threshold for positivity.
(C) Pre- and post-boost anti-RBD IgG subclass distributions by age. Fold change from baseline anti-RBD IgG levels were determined as described above (age ≤60, n = 60; age >60, n = 67).
(D) Age-dependent pre- and post-boost (IgG1 + IgG3):(IgG2 + IgG4) ratios of anti-RBD IgG (age ≤60, n = 60, red; age >60, n = 67, blue).
(E) Age-dependent Fc glycosylation patterns of IgG1 among vaccinated individuals. Levels are compared for the total IgGs produced at the pre-boost time point (age ≤60, n = 27; age >60, n = 32), for individuals who had an IgG1 response at pre-boost (age ≤60, n = 9; age >60, n = 3), and for individuals who had an IgG1 response at post-boost (age ≤60, n = 24; age >60, n = 15).
(F) Age-dependent binding activity of RBD-specific IgGs to FcγRs and C1q. Binding to each receptor was determined at pre-boost (age ≤60, n = 22, red; age >60, n = 15, blue) and post-boost (age ≤60, n = 32, age >60, n = 27). Ratios between RBD-specific IgG binding to activating inhibitory receptors were determined as described above. See also Figure S3.
Data are presented as scatterplots indicating individual measurements (dots); the black line represents the mean; error bars represent SDs. Unpaired 2-sided Mann-Whitney U test was used to evaluate differences between groups. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Aging is associated with an inherent alteration in Fc glycosylation patterns (Chen et al., 2012), which was observed when we compared total IgG glycosylation patterns between age groups (Figure 2E). Decreased IgG1 sialyation and galactosylation and elevated IgG1 bisection were found in total IgG as well as anti-RBD IgG1 in the pre-boost examination among individuals who had an IgG1 response at that time point. At the post-boost time point, these typical age-dependent differences were attenuated and elevated IgG1 RBD fucosylation was the only significant Fc glycosylation pattern unique to the older population. IgG3 Fc glycosylation displayed similar trends in which the initial IgG response resembled that of the inherent age-related differences, which was subsequently attenuated in the post-boost time point. No age dependence in differences in IgG2 Fc glycosylation were observed at any time point (Figure S3B).
Given the differences in subclass trajectories and Fc glycosylation that we observed between age groups, we proceeded to compare the binding profiles to FcγRs and C1q (Figure 2F). At pre-boost, the elderly population displayed elevated binding to all of the FcγRs and C1q. This finding could be explained by the predominance of IgG3 at this time point in the elderly (Vidarsson et al., 2014). In the post-boost time point, decreased binding to FcγRIIb was the only significant difference, causing an overall increase in the A/I ratio of RBD-specific IgGs in the elderly population. To determine whether this response was unique to the newly formed IgGs or to an overall trend in elderly individuals, we compared the FcγR-binding capacity of total IgGs (Figure S3C) and found no baseline differences between the populations. Overall, these results reveal distinct Fab and Fc functional properties of anti-RBD SARS-CoV-2-elicited IgG among vaccinated adults aged >60 years. Despite their reduced Fab-mediated anti-RBD titer and neutralization activity (Bates et al., 2021), the effectiveness of mRNA vaccination in this age group seems to be similar to the effectiveness in younger adult populations (Baden et al., 2021; Polack et al., 2020; Tenforde et al., 2021). Our data suggest that this could be explained by increased pre-boost Fc engagement of complement and post-boost activation of FcγR pathways, which may represent distinct levels of the IgG-mediated protective mechanism of older adults. However, this aspect needs to be characterized further by functional studies, such as ADCP/ADCC assays.
There are known differences between the sexes in the frequency and intensity of inflammation and immune-related diseases. Furthermore, male patients with COVID-19 tend to develop more severe disease outcomes (Takahashi et al., 2020) and display lower anti-RBD titers compared to female patients (Peckham et al., 2020). Hence, we proceeded to examine gender-related IgG response to the vaccine. We did not find significant differences in anti-RBD IgG titers, IgG subclass distribution or Fc glycosylation patterns, or in Fc engagement of downstream receptors (Figure S4).
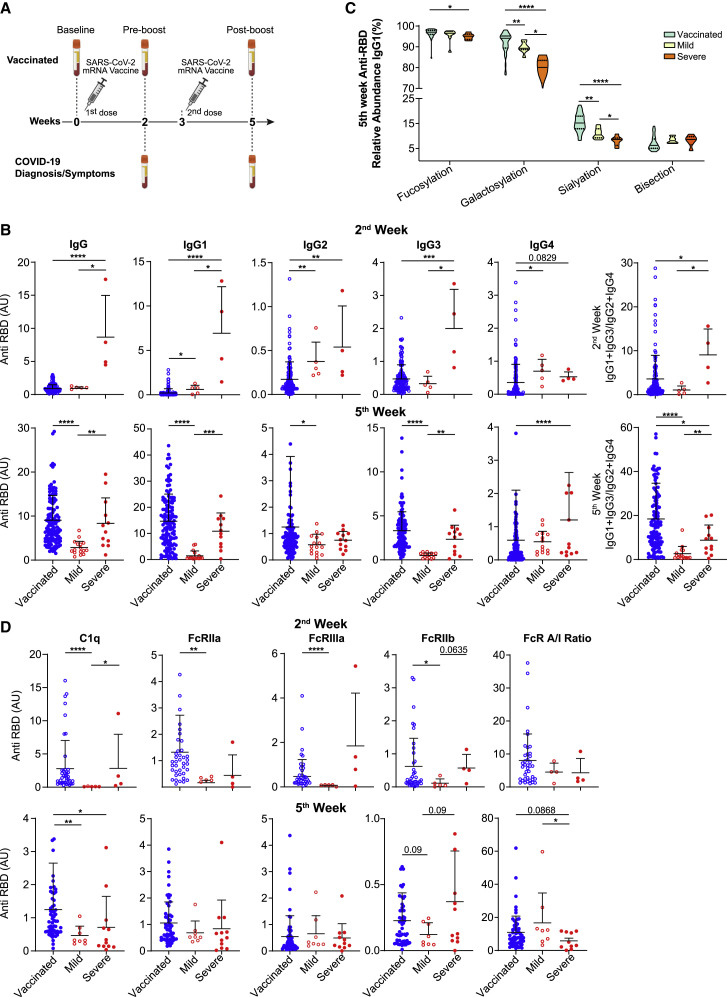
Structural and functional properties of anti-RBD IgG response are distinct in vaccinated individuals versus patients with COVID-19
To compare IgG trajectories between vaccination-induced response and natural SARS-CoV-2 infection, we analyzed the responses in our vaccinated cohort as well as those of convalescent, mild, and severe patients with COVID-19 at similar time points, as measured from either symptom onset or diagnosis (Figure 3 A; Table 2 ). At the 2-week time point, patients with severe COVID-19 had higher titers of RBD-binding IgGs as compared to both vaccinated individuals and patients with mild COVID-19. Differences between severe patients and vaccinated individuals were observed in levels of all IgG subclasses, whereas differences between severe and mild patients were mainly at the IgG1 and IgG3 levels (Figure 3B). Unlike that in vaccinated individuals, in patients with both mild and severe COVID-19, an initial IgG2 response was evident at the early time point and then was elevated. At the 2-week time point, the (IgG1 + IgG3):(IgG2 + IgG4) ratio was higher in severe patients as compared to both mild patients and vaccinated individuals. However, following the second vaccine dose, this ratio increased significantly over patients with both severe and mild COVID-19 at an equivalent time point from disease onset. These data indicate that the anti-RBD IgG titer and Fab-mediated neutralization potential following the two vaccine doses are higher than in COVID-19 convalescents and similar to patients suffering from a severe disease, while the vaccine-induced subclass composition is enriched in pro-inflammatory IgGs, as compared to both convalescent and severe patients.
Figure 3.
Structural and functional properties of anti-RBD IgG response are distinct in vaccinated individuals versus patients with COVID-19
(A) Experimental scheme of the COVID-19 and vaccine cohorts.
(B) Anti-RBD total IgG and subclass responses 2 weeks (vaccinated, n = 127; mild, n = 5; severe, n = 4) and 5 weeks (vaccinated, n = 127; mild, n = 16; severe, n = 12) after vaccine or COVID-19 diagnosis. IgG levels were determined by ELISA.
(C) Patterns of Fc glycosylation in anti-RBD IgGs produced at 5 weeks from vaccine or COVID-19 diagnosis. RBD-specific IgGs were isolated and Fc glycan structure was determined for each subclass by mass spectrometry as described above (vaccinated, n = 39; mild, n = 8; severe, n = 6). Data are presented as violin plots, with solid lines representing median and dotted lines representing upper and lower quartiles.
(D) Dynamic of FcγR and C1q binding properties of anti-RBD IgGs from vaccinated individuals and patients with COVID-19. FcγR binding was determined at 2 weeks (vaccinated, n = 39; mild, n = 5; severe, n = 4) and 5 weeks (vaccinated, n = 59; mild, n = 8; severe, n = 12) from vaccine or diagnosis. See also Figure S5.
Unless otherwise mentioned, data are presented as scatterplots indicating individual measurements (dots); the black line represents the mean; error bars represent SDs. Unpaired 2-sided Mann-Whitney U test was used to evaluate differences between groups. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Table 2.
Baseline characteristics of patients with COVID-19
| 2nd week | 5th week | |
|---|---|---|
| n | 9 | 28 |
| Age, y, mean (SD) | 52.11 (28.42) | 44.14 (18.80) |
| Male gender (%) | 6 (66.7) | 15 (60.0) |
| Age >60 y (%) | 3 (33.3) | 8 (28.6) |
| Fatalities (%) | 3 (33.3) | 3 (10.7) |
| Severe patients (%) | 4 (44.4) | 12 (42.9) |
| Days from diagnosis, mean (SD) | 12.33 (2.40) | 34.29 (5.02) |
Next, we compared glycosylation patterns of RBD-specific and total IgGs in the vaccinated versus patient cohort at 5 weeks from diagnosis. In RBD-specific IgGs of patients with COVID-19, IgG1 Fc fucosylation was decreased as compared to vaccinated individuals, a trend that was correlated with disease severity (Figure 3C). This gradient between mild and severe patients is consistent with previous reports on the association of increased afucosylated anti-SARS-CoV-2 IgG early after infection and severe disease outcome (Chakraborty et al., 2021a; Larsen et al., 2021). Similar trends distinguishing between vaccination and disease severities were observed in anti-RBD IgG1 sialyation and galactosylation. Together with distinct Fc glycan profiles of IgG2 and IgG3/4 (Figure S5A), these unique subclass-specific Fc glycosylation changes support the notion of a different regulatory process in response to vaccine versus acute infection.
We then compared the binding capacity of anti-RBD IgGs of vaccinated and patients with COVID-19 to FcγRs and C1q. At 2 weeks, patients with mild COVID-19 displayed attenuated binding of RBD-specific IgGs compared to vaccinated individuals. High variability and a small cohort size prevented us from determining whether at that time point severe COVID-19 patients had an increased potential to engage effector function, as would be expected given their subclass composition and enrichment of afucosylated IgG1. Post-boost vaccine-elicited RBD-specific IgGs were characterized by a trend toward increased engagement of effector function relative to COVID-19 patients, as indicated by both C1q binding capacities and FcγR A/I ratio.
While the mRNA-based vaccine elicits a response only against the viral spike protein, the immune response during SARS-CoV-2 infection is broader, targeting multiple viral antigens (Chakraborty et al., 2021b). To evaluate the IgG response of non-RBD-binding antibodies in the sera of patients with COVID-19, we analyzed the Fc structure and function of their total IgG. At 5 weeks post-diagnosis, there was a similar gradient of Fc glycan modifications in the total IgG from severe patients through to vaccinated individuals, as observed in RBD-specific IgGs. For instance, decreased galactosylation was observed in all IgG subclasses of severe but not in mild/convalescent patients, as compared with vaccinated individuals (Figure S5B). Moreover, total IgGs of severe patients displayed elevated binding capabilities to FcγRs and C1q as compared to mild patients at all time points. As opposed to anti-RBD IgGs, total IgGs of the vaccinated cohort displayed lower binding capacities compared to patients, as well as lower A/I FcγR ratio. This indicates an increased systemic inflammatory state and a more comprehensive IgG response against a multitude of SARS-CoV-2 epitopes in patients with COVID-19, as opposed to the specific anti-Spike response that is generated by the mRNA-based vaccine.
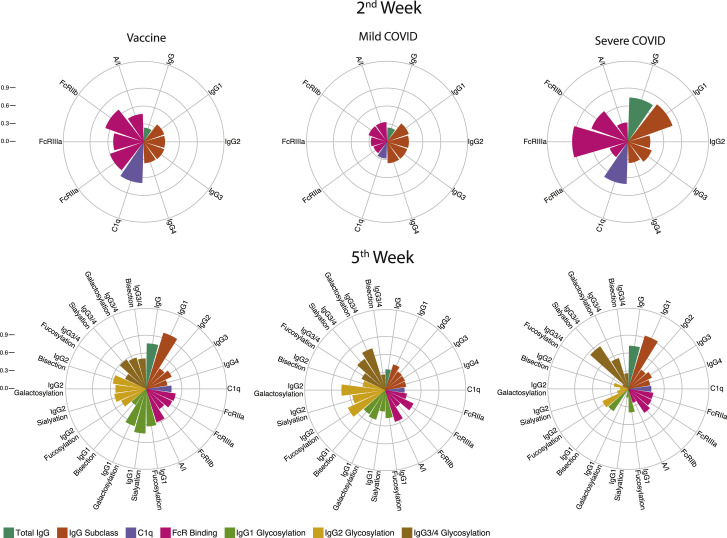
Overall, these results indicate that 2 weeks after the first vaccine dose or disease onset, patients with COVID-19 had higher anti-RBD IgG responses and increased binding to both activating and inhibitory FcγRs and to C1q in comparison to the vaccinated individuals, more evidently in severe patients. However, at the later time point, IgG subclass composition in vaccinated individuals demonstrated an increased inflammatory response, and Fc glycosylation patterns differed between vaccinated individuals and patients with COVID-19. Our data indicate that mRNA vaccine and COVID-19 infections induce distinct IgG trajectories. These unique RBD-specific IgG responses and associated characteristics of effector function potential are graphically summarized in Figure 4 .
Figure 4.
Anti-RBD IgG response in vaccinated individuals versus patients with COVID-19
The circular bar plots depict the mean percentile of each feature ranging from 0 to 1.
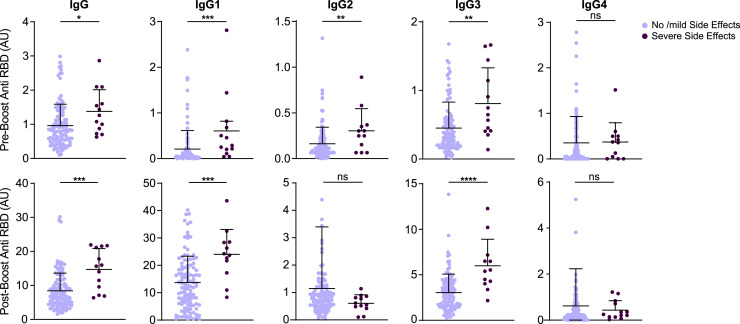
Elevated anti-RBD IgG titers in individuals with severe side effects after the second vaccine dose
Lastly, we examined whether severe side effects to the vaccine were predictors of IgG response. For that, we questioned each participant for the presence of localized and systemic side effects and their severity (for more details, see Method details).Whereas after the first vaccine dose, severe side effect reports were very rare, following the second dose, 13 individuals reported severe systemic symptoms. These individuals were mostly younger than 60 years of age (mean age, 42.7 years) and female (10/13). The presence of severe systemic side effects after the second dose correlated with elevated anti-RBD binding titers following both the first and second doses. The elevated IgG titer was mainly in IgG1 and IgG3 subclasses, while IgG2 levels were also elevated in vaccinated individual who suffered from systemic side effects following the first but not the second dose of the vaccine (Figure 5 ). The same results were obtained after correcting the groups for matched age and sex (Figure S6). Thus, BNT16b2 mRNA vaccine-associated systemic side effects after the second dose were correlated with elevated pro-inflammatory IgG subclass response.
Figure 5.
Elevated anti-RBD IgG titers in individuals with severe side effects after the second vaccine dose
Anti-RBD total IgG and subclass composition were determined for the indicated serum samples. Symptom severity was determined by the participants and was assigned as no/mild side effects and severe side effects groups. No side effects, n = 114; severe side effects, n = 13. See also Figure S6.
Data are presented as scatterplots indicating individual measurements (dots); the black line represents the mean; error bars represent SDs. Unpaired 2-sided Mann-Whitney U test was used to evaluate differences between groups. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Discussion
While neutralization levels are correlated with vaccine efficacy against breakthrough infections, less is known about the effect of vaccine-induced Fc properties. Here, we demonstrate that the IgG response elicited by BNT162b2 mRNA vaccine involves dynamic modulation of the Fc structures and increased binding to activating FcγRs after the second dose. In addition to the increased antibody neutralization activity that is generated from vaccine prime to boost, we report here an increased FcγR engagement potential of the Fc domain of generated antibodies. Upon post-vaccine exposure to SARS-CoV-2, these Fc structures in the generated antibody-virus immune complexes (ICs) can affect the phagocytic activity of these complexes. Thus, even when neutralizing IgG levels are highly effective, the overall FcγR engagement potential will dictate the intensity of IC clearance and of the sequential antiviral immune responses, including cross-presentation to prime and activate T cell response and elimination of infected host cells.
We observed age differences in vaccine-induced IgG Fab and Fc structures and receptor engagement properties. The vaccine-primed IgG subclass trajectory we identified was characterized by an initial IgG3 response, mainly in individuals older than 60 years, whereas in younger individuals, IgG1 response was also generated at the pre-boost time point. The delay in IgG3 to IgG1 switch, with associated increased binding capacities to FcγRs and C1q that we observed in older participants, could be related to immune aging, which could also be relevant in the context of inflammatory-mediated acute illnesses such as COVID-19. Moreover, reduced RBD binding and neutralization potential, but increased C1q binding pre-boost and A/I FcγR binding ratio post-boost, characterized the vaccine-generated antibodies in older adults. These age-dependent differences in IgG characteristics may result in the activation of distinct mechanisms of vaccine efficacy. Interestingly, a slight reduction in vaccine efficacy for asymptomatic infection, but not for symptomatic and severe COVID-19, was reported in older adults (Haas et al., 2021; Jalkanen et al., 2021). While the reduced neutralization activity in older adults could result in increased asymptomatic infection, increased Fc engagement may enhance SARS-CoV-2 clearance and compensate for the reduced IgG titer, resulting in similar levels of protection from symptomatic infection as in younger individuals with higher neutralizing IgG levels.
While the A/I FcγRs binding ratio was elevated from vaccine prime to boost, this kinetic change was not significant for any individual FcγR and seemed to be dominated by decreased binding to the inhibitory FcγRIIB post-boost. This observation highlights the complexity of IgG Fc-FcγR interactions, which can be dictated by numerous combinations of IgG subclasses and Fc glycoforms. For instance, FcγRIIIA binding, which would be expected to increase due to the strong post-boost increase in IgG1 and IgG3 response, could also be attenuated due to the decreased portion of afucosylated IgG1 and reduced portion of IgG3 in the post-boost IgG pool. Moreover, the high levels of sialylation on core-fucosylated IgG1 in the post-boost samples can further reduce the A/I binding ratio and antibody-mediated killing (Li et al., 2017). Thus, despite having robust IgG1 and IgG3 responses post-boost, complex regulatory mechanisms may affect the Fc engagement of immune receptors. Additional modifications to the Fc scaffold, including allotypic differences and IgG2 and IgG3 Fc glycosylations, may further contribute to IgG function and binding to FcγRs and C1q. However, the roles of these modifications are currently poorly understood.
We characterized here the dynamics of the IgG response over a 5-week period from the first vaccine dose, during which a boost injection was administered after 3 weeks. Due to this uniform vaccination regime, we could not distinguish between the contribution of the vaccine boost or merely the time that passed on the distinct IgG Fc response we identified 2 or 5 weeks post-vaccination. The effect of vaccine boost on IgG Fc structures may have important implications for the planning of long-term vaccination strategies, particularly in the context of the third vaccine boost. This additional dose is being delivered in several countries and being considered worldwide in an attempt to fight breakthrough post-vaccine infections, mainly by the B.1.617.2 (Delta) variant, and to prolong the efficacy period of COVID-19 vaccines.
IgG1 and IgG3 were the predominant subclasses following both COVID-19 vaccination and natural infection. However, in patients, but not in healthy vaccinated individuals, the early post-infection IgG response included enhanced IgG2 and IgG4 levels. The differences in subclass distribution and Fc glycoforms during IgG response to infection could be attributed to the presence of a multitude of antigens, including glycosylated epitopes, which elicit dominant IgG2 responses (Vidarsson et al., 2014). In our cohort, the ratios of IgG1 and IgG3 to IgG2 and IgG4 in patients with severe COVID-19 were significantly higher than in mild patients and vaccinated individuals at the early time point. However, following the vaccine boost, these ratios in the vaccinated surpassed those of the severe patients, supporting the notion of vaccine-induced increased A/I FcγR engagement by IgG.
In summary, we identified unique and dynamic modifications in the IgG scaffold during antibody response to the BNT162b2 vaccine. These age- but not sex-dependent alterations in IgG subclasses, Fc glycosylation, and binding to immune receptors were distinct from those we detected in COVID-19 convalescents and patients and from previously reported responses to other types of vaccines. We suggest that alongside the serum levels and neutralization activity of the antibodies, these Fc features may affect the quantity, quality, and mode of protection achieved by the COVID-19 mRNA vaccine.
Limitations of the study
This study focuses on the structure of IgGs produced against the RBD domain of the SARS-CoV-2 virus. We did not address the humoral response mediated by non-IgG antibody isotypes. Moreover, we did not address the response by non-RBD binding IgGs, such as those that bind other epitopes of the Spike protein (in the vaccinated cohort) or other proteins of the virus (in infected individuals). Additional limitations arise from the methods used in this study. While we tried to provide a comprehensive characterization of the Fc glycoforms in all IgG subclasses, the mass spectrometry-based method used to analyze IgG Fc glycans cannot distinguish quantitatively between glycans attached to IgG3 and IgG4 subclasses. Therefore, glycan analysis results refer to these two subclasses together. However, when we assessed the RBD-IgG subclass response with ELISA, we did not detect a significant IgG4 response. Thus, we assumed negligible IgG4 levels in our samples, and throughout the text, we refer to the glycosylation data of IgG3/4 as IgG3 glycosylations. When we assessed antibody responses by ELISA, we could not distinguish between quantities of IgGs produced as opposed to higher binding affinities of IgGs. Another limitation was the relatively small cohort of patients, especially in the early time point, which reduced the power of the statistical analysis and our ability to draw conclusions from it. Lastly, while we were able to provide structural information on the produced IgGs and determine their Fc receptor binding capabilities, given the low affinity of monomeric IgGs to FcRs and the low sensitivity of the assay, it is possible that the effects were underestimated. A functional assay such as ADCC/ADCP may add another aspect to the importance of the presented findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Horseradish peroxidase-conjugated anti-human IgG | Jackson ImmunoResearch | Cat# 109-035-088; RRID: AB_2337584 |
| Mouse anti-human IgG1 | Southern biotech | Cat# SB-9054-05; RRID: AB_2796627 |
| Mouse anti-human IgG2 | Southern biotech | Cat# SB-9060-05; RRID: AB_2796633 |
| Mouse anti-human IgG3 | ThermoFisher | Cat# MH1732; RRID: AB_2539713 |
| Mouse anti-human gG4 | Southern biotech | Cat# SB-9200-05; RRID: AB_2796691 |
| Biological samples | ||
| Human plasma from COVID19 patients | Isolated from patients in Ichilov and Sheba medical centers | Helsinky: #0262-20-TLV, #7287-20), WIS IRB #1280 |
| Human plasma from Pfizer–BioNTech COVID-19 (Tozinameran) Vaccinated donors | In house, Isolated from volunteers | WIS IRB #1280 |
| Chemicals, peptides, and recombinant proteins | ||
| TMB One Component Substrate | Bethyl laboratories, INC | Cat# TOA-48156708 |
| Pierce NHS-Activated Magnetic Beads | ThermoFisher | Cat# TS88832 |
| protein G Sepharose 4 Fast Flow beads | GE Healthcare | Cat# 17-0618-05 |
| IgGs Elution Buffer | ThermoFisher | Cat# 21009 |
| The SARS-CoV-2 (2019-nCoV) Nucleoprotein His-tag protein | Sino biological | Cat# 40588-V08B |
| HisPur Ni-NT resin | ThermoFisher | Cat# TS-88222 |
| imidazole | Sigma-Aldrich | Cat# I5513 |
| the human recombinant receptors FcγRIIA | Sino Biological | Cat# 10374-H08H |
| the human recombinant receptors FcγRIIB | Sino Biological | Cat# 10259-H08H |
| the human recombinant receptors FcγR | Sino Biological | Cat# 10389-H08H |
| C1q | Sigma Aldrich | Cat# C1740 |
| Nonident P40 Substitute (GEPAL) | Sigma Aldrich | Cat# CA-630 |
| TopTip Borate p10 | Glygen Corp | https://www.glysci.com/TopTip/ |
| Critical commercial assays | ||
| Pierce Direct magnetic IP/Co-IP kit | ThermoFisher | Cat# 88828 |
| Deposited data | ||
| Mass spectrometry data | This paper | https://www.ebi.ac.uk/ Project accession: PXD029321 |
| Experimental models: cell lines | ||
| Expi293 Cells | ThermoFisher | Cat# A14527 |
| Recombinant DNA | ||
| hexahistidine-tagged SARS-CoV-2 RBD construct | F. Krammer | Icahn School of Medicine at Mount Sinai |
| The SARS-CoV-2 (2019-nCoV) Nucleoprotein His-tag plasmid | Sino Biological | VG40588-CH |
| Software and algorithms | ||
| Graphpad Prism Prism 9.2.0 | GraphPad Prism | RRID: SCR_002798 |
| Rstudio (version 1.3.1093) | R studio | https://www.rstudio.com/ |
| Byonic | Protein metrics | https://proteinmetrics.com/byos/ |
| Skyline | McCoss Lab,Univeristy of Washington | https://skyline.ms/project/home/software/Skyline/begin.view |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rony Dahan (rony.dahan@weizmann.ac.il).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Clinical cohorts and samples
Study design
This study was designed to investigate the overall IgG response, IgG subclass distribution, subclass-specific Fc glycosylation patterns and effector functions of IgGs produced following administration of the SARS-CoV-2 mRNA-based BNT162b2 vaccine and to compare these to the response to natural infection with SARS-CoV-2. For this purpose, we analyzed plasma samples that had been collected independently from adults (aged ≥ 18 years) at four centers. Samples were obtained at the Sheba and Tel Aviv Sourasky medical centers and at a motel designated for patients with mild COVID-19 in quarantine. Blood from the vaccine cohort was collected at the Weizmann Institute of Science. Samples were coded and blinded and the relevant review boards in each institute (see below) approved this study.
The vaccine
The Pfizer–BioNTech COVID-19 vaccine, also known as tozinameran or BNT162b2, co-developed by BioNTech and Pfizer, is based on mRNA products that encode a genetically modified SARS-CoV-2 spike protein. The vaccination campaign in Israel started in December 2020.
Research subjects
Vaccine cohort
Recruitment of participants was performed according to and following the approval of the Weizmann Institute of Science Institutional Review Board. 131 volunteers signed an informed consent form prior to the first blood collection. In some cases, plasma samples were collected from 1 week up to 2 hours before first vaccine dose. Samples from all participants were collected 2 weeks after each vaccine dose (Figure 2A). Inclusion criteria were age > 18 and ability to sign informed consent. Exclusion criteria were previous SARS-CoV-2 infection as determined by history or presence of N binding IgGs at the pre-boost time point or presence of an immune deficiency causing aberrant IgG response. Overall 131 individuals volunteered of them 127 were included in the study according to these criteria.
We performed screening for the presence of N-binding IgGs and found one volunteer with high binding titers, as well as high RBD-binding IgGs following the first vaccine. This volunteer was excluded from further analysis due to potential past infection with SARS-CoV-2. Of the remaining 130 volunteers, 127 were positive for the presence of anti-RBD antibodies at 5 weeks. Of the three that were negative, one had a history of chronic lymphocytic leukemia with known low immunoglobulin levels, one was on chronic replacement IVIG therapy following rituximab treatment for lymphoma 9 years earlier, and the third individual was an 86-year-old male with no known underlying medical condition. These individuals were excluded from further analysis.
Patient cohort
We obtained blood samples from patients who had recovered from COVID-19 as well as de-identified samples from active patients at diverse clinical stages of the disease (asymptomatic, mild to severe). The samples and relevant clinical data were obtained from Tel Aviv Sourasky and Sheba Medical Centers (The Israeli Central Virology Laboratory (iCVL) and the hospital blood bank; as well as from hotels hosting mild COVID-19 patients in quarantine in accordance with the relevant institutional committees. Diagnosis of COVID-19 was confirmed by qRT-PCR performed on RNA extracted from nasopharyngeal swab samples. We included samples from patients at 2 and 5 weeks from diagnosis; each sample was obtained from a different individual. Blood samples were handled in an enhanced biosafety level facility (BSL2+) in accordance with the Weizmann Institute of Science standard operating procedure.
Method details
Cloning, expression and protein purification
The hexahistidine-tagged SARS-CoV-2 RBD construct was a kindly provided by gift from F. Krammer (Icahn School of Medicine at Mount Sinai). The SARS-CoV-2 (2019-nCoV) Nucleoprotein His-tag protein and plasmid were purchased from Sino Biological. To produce RBD and N proteins, plasmids were transfected transiently into Expi293 cells (ThermoFisher) as per manufacturer’s instructions. For RBD production, the secreted protein was collected from the supernatant and affinity purified on HisPur Ni-NT resin (ThermoFisher). For the production of N protein, cells were transfected as above, and the cell palette was frozen overnight at −20°C. Frozen pellets were thawed on ice for 15-20 minutes and re-suspended in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.5% Nonident P 40 Substitute (IGEPAL Sigma) and EDTA-free protease inhibitors) at 4°C on dry ice for 20-25 minutes and further incubated on a rotator at 4°C for 30 minutes. Next, the lysate was centrifuged at 13,000 x g, 4°C for 15 minutes to pellet debris and incubated with the HisPur Ni-NT resin. Polypropylene columns (Bio-Rad Laboratories) were loaded with the supernatant-resin mixtures and then washed with wash buffer (50 mM NaH2PO4H2O, 30 mM NaCl, 20 mM imidazole) four times. Columns were then eluted using elution buffer, which contained a high (235 mM) concentration of imidazole (Sigma-Aldrich). Four fractions were collected from each column by incubating the resin in the column with 3 mL of elution buffer for each fraction. Eluate was collected directly in a 50 mL falcon tube placed on ice. Purified proteins were dialyzed against PBS and sterile filtered (0.22 μm). Purity was assessed by SDS-PAGE and Coomassie staining (Amanat et al., 2020).
Purification of total IgG from plasma
5-10 mL of blood were collected from volunteers/patients into EDTA-containing tubes, (BD Biosciences, San Jose, CA) and then centrifuged at 1200 rpm for 10 minutes. Plasma was collected and kept for further analysis. Samples originating from patients underwent an additional heat inactivation step and was heat-inactivated for 30 minutes at 56°C to avoid potential viral spread. Antibodies were purified from the obtained plasma by protein G Sepharose 4 Fast Flow beads (GE Healthcare) according to the manufacturer’s protocol. In short, 2 mL of beads were washed in PBS, re-suspended with up to 15 mL of plasma and rotated overnight at 4°C. Next, the plasma-bead mix was transferred onto a column (Bio-Rad). Beads were then washed by several bead volumes until run-through was clear of protein, as determined by a 260/280 ratio measured by a Nanodrop spectrophotometer. Elution of IgGs was obtained with Elution Buffer (ThermoFisher) into 10% 1 M Tris (pH 9.0) in tubes. Purified antibodies were concentrated, quantified and dialyzed in PBS and sterile filtered (0.22 μm) and kept at 4°C for further analysis. These IgGs were further analyzed via either ELISA and/or mass spectrometry (see below).
Isolation of RBD-specific IgG
Pierce NHS-Activated Magnetic Beads (Thermo fisher) were used to purify antigen (RBD) specific IgG from total IgG. For column RBD immobilization, 10 μg of RBD were bound to 25 μL NHS magnetic bead slurry according to the manufacturer’s protocol. Each column was incubated with up to 6 mg of total IgGs from each sample and subsequently eluted according to the manufacturer’s protocol. RBD-specificity and purity of the eluted IgG was verified by RBD ELISA as described below. These RBD specific IgGs were further analyzed via either ELISA and/or mass spectrometry (see below).
ELISA screening for binding to RBD and N proteins
Plasma samples were screened for binding to both RBD and N proteins as in Amanat et al. (2020), with small modifications. 96-well half-area ELISA microplates (Greiner Bio-One) were coated with 20 μl of RBD or N protein at 1 μg/ml/well in PBS and incubated at 37°C for 1 hour. All sequential steps were performed at room temperature. Plates were washed (PBS/Tween 0.05%) and 100 μL blocking solution (2% FCS/PBS) was added for 2 h. Then, samples from either vaccinated individuals or heat-inactivated plasma samples from patients with COVID-19 were diluted, first at 1:100 and then serially in blocking solution 1:4. 20 μl of the serially diluted samples were added to each well and incubated for 2 h. After washing, plates were incubated for 1 hour with horseradish 1:2500 peroxidase-conjugated anti-human IgG (Jackson ImmunoResearch). Detection was performed using a one-component substrate solution (TMB, Bethyl laboratories, INC) and reactions were stopped by addition of 0.18 M sulphuric acid. Absorbance at 450 nm was immediately recorded using a SpectraMax Plus spectrophotometer (Molecular Devices), and background absorbance from negative control samples was subtracted. Serum samples taken before 2020 were used as negative controls. The mean of these negative controls plus 3 times the standard deviation was assigned a value of 1. Patient samples varying in their anti-RBD/N titer were used as controls to standardize the reaction in all tested plates. Additional modified ELISA protocols, which are described in the following subsections, were all based on the above protocol.
Isotyping of plasma samples by ELISA
To assess the serum distribution of RBD binding antibody subclasses, plate coating was done with RBD 2 μg/ml/well, and 2% FCS/PBS was used as blocking solution. The following mouse anti-human IgG secondary antibodies were used: IgG1 1:4000 (Southern biotech) and IgG3 1:500 (ThermoFisher). Serum was initially diluted 1:25 and then serially diluted 1:2. For IgG2 1:500 (Southern biotech,) and IgG4 1:500 (Southern biotech), serum was initially diluted 1:10 and then serially diluted 1:2.Trajectories of IgG subclass distribution over time (Figures 1, 2, and S3) are presented as fold change from baseline. To calculate the fold change, we determined for each subclass the pre-vaccine baseline mean level.
ELISA of IgG samples for FcγRs binding
To determine the binding of total IgGs and anti-RBD IgGs to the different FcγRs, the human recombinant receptors FcγRIIA, IIB, and IIIA (Sino Biological) were immobilized to half area plates at 2 μg\ml in PBS. Blocking solution contained 10% BSA. Initial concentrations of 50 μg/ml of total IgG and 10 μg/ml of anti-RBD IgG and were serially diluted 1:2. HRP-conjugated anti-human IgG was used at 1:2500. For each FcR assay, the optimal concentration that fulfilled linearity was determined and these values were used for plotting.
ELISA of IgG samples for C1q binding
C1q (Sigma) was immobilized to half area plates (5 μg/ml/well) and 5% BSA/PBS was used as blocking solution. Initial concentrations of 50 μg/ml of total IgG and 10 μg/ml of anti-RBD IgG were serially diluted 1:2. HRP-conjugated anti-human IgG was used at 1:2500.
Preparation of IgG samples for mass spectrometry
SDS in 50 mM Tris-HCl was added to 95 μg from each total IgG sample, to a final concentration of 5% SDS. Samples were reduced with 5 mM dithiothreitol (Sigma) for 1 h at 56°C, and alkylated with 10 mM iodoacetamide (Sigma) in the dark for 45 min at room temperature. Next, samples were acidified with 1% phosphoric acid (Sigma). Each sample was loaded onto 96-well S-Trap plate (Protifi, USA) according to the manufacturer’s instructions. In brief, after loading, samples were washed with 90:10 methanol/50 mM ammonium bicarbonate. Samples were then digested with 2 μg trypsin for 1 h at 47°C. The digested peptides were eluted using 50 mM ammonium bicarbonate; 2 μg trypsin was added to this fraction and incubated overnight at 37°C. Two more elutions were made using 0.2% formic acid and 0.2% formic acid in 50% acetonitrile. The three eluates were pooled together and vacuum-centrifuged to dry. Samples were kept at −20°C until analysis.
RBD Speicifc IgGs were processed similarly with the following modifications. SDS in 50 mM Tris-HCl was added to the samples to a final concentration of 5% SDS. 0.3 μg of trypsin was used for sample digestion.
Identification of Fc-attached glycopeptides by mass spectrometry
Borate TopTips (Glygen, USA) were equilibrated with loading buffer (0.5 M triethylammonium acetate, pH 10.5, 50% acetonitrile). Samples were dissolved in 20 μl loading buffer, loaded onto the tips, washed with loading buffer and eluted with 5% formic acid, 50% acetonitrile. Samples were then vacuum-centrifuged to dry and kept at −20°C until analysis. Samples were analyzed using a nanoAcquity nano-liquid chromatograph (Waters, MA, USA) coupled to a Q-Exactive Exploris mass spectrometer (Thermo Fisher Scientific). Samples were injected and concentrated on a nanoEase Symmetry C18 trap column (180 μm X 20 mm, 5-μm particles, Waters), and separated on a nanoEase HSS C18 T3 analytical column (75 μm X 250 mm, 1.8-μm particles, Waters). Samples were separated using a gradient of 4%–25% buffer “B” (0.1% formic acid, 99.9% MeCN) for 48 min at 45°C, at 350 nl/min flow rate. Mass spectra were acquired in parallel reaction monitoring (PRM) with MS1 acquisition. MS1 was performed using 375-1800 m/z survey scans at 120,000 resolution (at 200 m/z) with AGC set at Standard and maximum injection time at Auto. PRM was performed using 1.3 m/z isolation window. Precursors were chosen from a list for stepped HCD fragmentation 32 and 15 normalized collision energy (NCE). AGC was set to Standard and maximum injection time to Auto. Fragments were acquired from 140 m/z at 15,000 resolution (at 200 m/z).
Glycopeptide data analysis
Data were searched against the IgG glycan-carrying peptide sequences allowing for deamidation and Fc glycan database using Byonic (v4.0.1.) search engine, and quantified using Skyline (v20.2.0.343). The following peptide sequences were used for subclass determination: IgG1, EEQYNSTYR; IgG2, EEQFNSTFR; IgG3, EEQYNSTFR; and IgG4, EEQFNSTYR. However, due to an inability to differentiate the readouts of the IgG3 and IgG4 peptides, these are shown together as IgG3/IgG4 in all the figures.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier ProteomeXchange: PXD02932.
Polar plots
Each feature in the polar plots is drawn as a wedge indicating the mean percentile of that group ranging from 0-1. Mean group percentiles were calculated by determination the groups’ Z-score based on the mean and standard deviation of the entire feature based on all the relevant pre-boost, post-boost and COVID-19 time-points (2nd week and 5th week) values. For IgG subclass distribution, fold change values were used.
Quantification and statistical analysis
Statistical data were evaluated using GraphPad Prism (v.9) software. The confidence level was 95%, with a significance level of 5% (α = 0.05). Results are expressed as the arithmetic means ± SD. Data comparison with P values of % 0.05 was considered statistically significant. Based on normality testing (Anderson-Darling, D’Agostino-Pearson and Shapiro-Wilk tests) non-parametric tests were used for statistical group comparison. Mann-Whitney U test was used to compare between 2 groups and Kruskal-Wallis test was used to compare 3 or more groups, followed by Dunn’s post hoc test. Statistical details of the assays applied can be found in Figure legends. Tables and circular bar plots were produced in Rstudio (version 1.3.1093 © 2009-2020 RStudio, PBC) using the tableone and ggplot packages, respectively.
Acknowledgments
We thank all of the volunteers who donated blood and participated in the study. We thank Dahan lab members Jasmine Balanga, Yuval Shapir, Ella Herzog, and Avia Habshush for technical work and fruitful discussion on the data presented in the manuscript. We thank F. Krammer (Icahn School of Medicine at Mount Sinai) for providing the RBD expression construct. This study was supported by the Ben B. and Joyce E. Eisenberg Foundation, Miel de Botton, and the CoronaVirus Weizmann Institute of Science Fund. R.D. is incumbent of the Rina Gudinski Career Development Chair. He is supported by the Rising Tide Translation Cancer Research Fund, the Melanoma Research Alliance (MRA), the Israel Cancer Research Fund (ICRF), the Israel Cancer Association (ICA), the Israel Science Foundation (ISF), Teva Pharmaceuticals, the Moross Integrated Cancer Center (MICC), the Dwek Institute for Cancer Therapy Research, Yeda Research and Development Co., the Mizutani Foundation for Glycoscience, the Harry J. Lloyd Charitable Trust (HJLCT), the Emerson Collective Cancer Research Fund, the Flight Attendant Medical Research Institute (FAMRI), the Garvan-Weizmann Program, the Ira and Diana Riklis Fund for CAR-T Therapy, the Enoch Foundation, the Pearl Welinsky Merlo Foundation, the Benoziyo Fund for the Advancement of Science, the fund honoring Gerty Schwarz Schaier, a research grant from Max Saad, and the Mexican Association of Friends of the Weizmann Institute.
Author contributions
Conceptualization, R.D.; methodology, I.F., T.F., Y. Levin, and D.M.; investigation, I.F., T.F., D.M., N.C.-S., Y.A., D.M., G.M. and N.B.; writing – original draft, R.D., T.F., and I.F.; funding acquisition, R.D.; resources, I.F., Y. Lustig, A.B., L.M., I.K., and D.S.; supervision, R.D.
Declaration of interests
R.D. serves as a consultant for Teva Pharmaceutical Industries and NucleAi and receives research grant support from Teva Pharmaceutical Industries. The other authors declare no competing interests.
Published: December 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110114.
Supplemental information
Data and code availability
-
•
De-identified human raw mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and accession numbers are listed in the key resources table.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony R.M., Nimmerjahn F. The role of differential IgG glycosylation in the interaction of antibodies with FcγRs in vivo. Curr. Opin. Organ Transplant. 2011;16:7–14. doi: 10.1097/MOT.0b013e328342538f. [DOI] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T.A., Leier H.C., Lyski Z.L., Goodman J.R., Curlin M.E., Messer W.B., Tafesse F.G. Age-Dependent Neutralization of SARS-CoV-2 and P.1 Variant by Vaccine Immune Serum Samples. JAMA. 2021;326:868–869. doi: 10.1001/jama.2021.11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Ravetch J.V. Diversification of IgG effector functions. Int. Immunol. 2017;29:303–310. doi: 10.1093/intimm/dxx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Wang T.T., Dahan R., Maamary J., Ravetch J.V. Signaling by Antibodies: Recent Progress. Annu. Rev. Immunol. 2017;35:285–311. doi: 10.1146/annurev-immunol-051116-052433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns P., Jönsson F. Mouse and human FcR effector functions. Immunol. Rev. 2015;268:25–51. doi: 10.1111/imr.12350. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Gonzalez J.C., Sievers B.L., Mallajosyula V., Dubey M., Cheng B.Y.-L., Tran K.Q.T., Chakraborty S., Cassidy A., Chen S.T., et al. Divergent early antibody responses define COVID-19 disease trajectories. bioRxiv. 2021 doi: 10.1101/2021.05.25.445649. [DOI] [Google Scholar]
- Chen G., Wang Y., Qiu L., Qin X., Liu H., Wang X., Wang Y., Song G., Li F., Guo Y., et al. Human IgG Fc-glycosylation profiling reveals associations with age, sex, female sex hormones and thyroid cancer. J. Proteomics. 2012;75:2824–2834. doi: 10.1016/j.jprot.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Ciabattini A., Nardini C., Santoro F., Garagnani P., Franceschi C., Medaglini D. Vaccination in the elderly: the challenge of immune changes with aging. Semin. Immunol. 2018;40:83–94. doi: 10.1016/j.smim.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:1–19. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K., et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397:1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen P., Kolehmainen P., Häkkinen H.K., Huttunen M., Tähtinen P.A., Lundberg R., Maljanen S., Reinholm A., Tauriainen S., Pakkanen S.H., et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat. Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Nouta J., Hoepel W., Chen H.-J., Linty F., Visser R., Brinkhaus M., et al. Amsterdam UMC COVID-19. Biobank Study Group Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371:eabc8378. doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., DiLillo D.J., Bournazos S., Giddens J.P., Ravetch J.V., Wang L.X. Modulating IgG effector function by Fc glycan engineering. Proc. Natl. Acad. Sci. USA. 2017;114:3485–3490. doi: 10.1073/pnas.1702173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald I., Murray S.M., Reynolds C.J., Altmann D.M., Boyton R.J. Comparative systematic review and meta-analysis of reactogenicity, immunogenicity and efficacy of vaccines against SARS-CoV-2. NPJ Vaccines. 2021;6:74. doi: 10.1038/s41541-021-00336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Gordan S., Lux A. FcγR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol. 2015;36:325–336. doi: 10.1016/j.it.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., Rosser E.C., Webb K., Deakin C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I., Kunik V., Ofran Y. The structural basis of antibody-antigen recognition. Front. Immunol. 2013;4:302. doi: 10.3389/fimmu.2013.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M.H.J., De Jong S.E., Soonawala D., Kroon F.P., Adegnika A.A., Deelder A.M., Hokke C.H., Yazdanbakhsh M., Wuhrer M. Changes in antigen-specific IgG1 Fc N-glycosylation upon influenza and tetanus vaccination. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014563. M111.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid Z., Kalayanamitra R., McClafferty B., Kepko D., Ramgobin D., Patel R., Aggarwal C.S., Vunnam R., Sahu N., Bhatt D., et al. COVID-19 and Older Adults: What We Know. J. Am. Geriatr. Soc. 2020;68:926–929. doi: 10.1111/jgs.16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Yale IMPACT Research Team Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenforde M.W., Olson S.M., Self W.H., Talbot H.K., Lindsell C.J., Steingrub J.S., Shapiro N.I., Ginde A.A., Douin D.J., Prekker M.E., et al. IVY Network. HAIVEN Investigators Effectiveness of Pfizer-BioNTech and Moderna Vaccines Against COVID-19 Among Hospitalized Adults Aged ≥65 Years - United States, January-March 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:674–679. doi: 10.15585/mmwr.mm7018e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidarsson G., Dekkers G., Rispens T. IgG Subclasses and Allotypes: From Structure to Effector Functions. Front. Immunol. 2014;20:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.T., Maamary J., Tan G.S., Bournazos S., Davis C.W., Krammer F., Schlesinger S.J., Palese P., Ahmed R., Ravetch J.V. Anti-HA Glycoforms Drive B Cell Affinity Selection and Determine Influenza Vaccine Efficacy. Cell. 2015;162:160–169. doi: 10.1016/j.cell.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L., et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. Nat. Commun. 2020;12:2670. [Google Scholar]
- Yamin R., Jones A.T., Hoffmann H.-H., Kao K.S., Francis R.L., Sheahan T.P., Baric R.S., Rice C.M., Ravetch J.V., Bournazos S. Fc-engineered antibody therapeutics with improved efficacy against COVID-19. Res. Sq. 2021 doi: 10.21203/rs.3.rs-555612/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
De-identified human raw mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and accession numbers are listed in the key resources table.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.