Abstract
Recent studies implicate the interferon (IFN) regulatory factors (IRF) IRF-3 and IRF-7 as key activators of the alpha/beta IFN (IFN-α/β) genes as well as the RANTES chemokine gene. Using coexpression analysis, the human IFNB, IFNA1, and RANTES promoters were stimulated by IRF-3 coexpression, whereas the IFNA4, IFNA7, and IFNA14 promoters were preferentially induced by IRF-7 only. Chimeric proteins containing combinations of different IRF-7 and IRF-3 domains were also tested, and the results provided evidence of distinct DNA binding properties of IRF-3 and IRF-7, as well as a preferential association of IRF-3 with the CREB binding protein (CBP) coactivator. Interestingly, some of these fusion proteins led to supraphysiological levels of IFN promoter activation. DNA binding site selection studies demonstrated that IRF-3 and IRF-7 bound to the 5′-GAAANNGAAANN-3′ consensus motif found in many virus-inducible genes; however, a single nucleotide substitution in either of the GAAA half-site motifs eliminated IRF-3 binding and transactivation activity but did not affect IRF-7 interaction or transactivation activity. These studies demonstrate that IRF-3 possesses a restricted DNA binding site specificity and interacts with CBP, whereas IRF-7 has a broader DNA binding specificity that contributes to its capacity to stimulate delayed-type IFN gene expression. These results provide an explanation for the differential regulation of IFN-α/β gene expression by IRF-3 and IRF-7 and suggest that these factors have complementary rather than redundant roles in the activation of the IFN-α/β genes.
Interferons (IFNs) are multifunctional secreted proteins involved in antiviral defense, cell growth regulation, and immune activation (44). Alpha/beta IFN (IFN-α/β) is produced by virus-infected host cells and constitutes the primary response against virus infection, while gamma IFN (IFN-γ), a TH1 cytokine produced by activated T cells and natural killer cells, is crucial in eliciting the proper immune response and pathogen clearance. Virus infection induces the transcription and synthesis of multiple IFN genes (16, 33, 44); newly synthesized IFN interacts with neighboring cells through cell surface receptors and the Janus-activated kinase (JAK)–STAT signaling pathway, resulting in the induction of over 30 new cellular proteins that mediate the diverse functions of the IFNs (6, 18, 21, 39). Among the many virus- and IFN-inducible proteins are members of the growing family of interferon regulatory factors (IRFs), which now consists of nine members, as well as several virus-encoded IRFs (4). The presence of IRF-like binding sites in the promoter regions of the IFNB and IFNA genes implicated the IRFs as direct regulators of IFN-α/β gene induction (11–14, 29). Within the IRF family, IRF-3 and IRF-7 have recently been identified as key regulators of the induction of IFNs (reviewed in reference 26).
IRF-3 is expressed constitutively in a variety of tissues and demonstrates a unique response to virus infection (1). Latent cytoplasmic IRF-3 is posttranslationally modified and activated through phosphorylation of specific serine residues located in its C-terminal end following virus infection or treatment with double-stranded RNA (24, 45–47). Overexpression of IRF-3 significantly enhances virus-mediated expression of IFN-α/β genes and results in the induction of an antiviral state (19). Other studies have demonstrated that transcription of the CC-chemokine RANTES is upregulated by virus infection, mediated through IRF-3 activation and binding to overlapping ISRE-like elements in the −100 region of the RANTES promoter (23).
Structure-function analysis has revealed that IRF-3 contains an N-terminal DNA binding domain (DBD); a strong but atypical transactivation domain, located between amino acids 134 and 394, a region that also contains a nuclear export sequence element; a proline-rich region; and an IRF association domain (IAD). Two autoinhibitory domains in IRF-3 form an intramolecular interaction that results in a closed conformation and masks the IAD and the DBD to prevent nuclear translocation and subsequent DNA binding (25). Following virus infection, inducible phosphorylation of IRF-3 at the carboxy terminus relieves the intramolecular association between the two autoinhibitory domains, unmasking the IAD and the DBD. The conformational change in IRF-3 results in the formation of homodimers through the IAD. IRF-3 dimerization leads to cytoplasmic to nuclear translocation, association with the CREB binding protein (CBP) coactivator, and stimulation of DNA binding and transcriptional activities (reviewed in references 17 and 26). IRF-3 phosphorylation ultimately results in its degradation via the ubiquitin-proteasome pathway (24, 34). These biological features implicate IRF-3 as an important component of the immediate-early response to virus infection (17, 26).
IRF-7 was first described to bind and repress the Qp promoter region of the Epstein-Barr virus (EBV) EBNA-1 gene, which contains an ISRE-like element (31, 48). Unlike IRF-3, IRF-7 is not expressed constitutively in cells; rather, expression is induced by IFN, lipopolysaccharide, and virus infection. As with IRF-3, virus infection appears to induce the phosphorylation of IRF-7 at its carboxy terminus, a region that is highly homologous to the IRF-3 C-terminal end (27, 37). IRF-7 also localizes to the cytoplasm in uninfected cells and translocates to the nucleus after phosphorylation (2, 37). Two groups have identified potential serine residues targeted for inducible phosphorylation by analogy to IRF-3. Marie et al. mutated Ser425 and Ser426 in murine IRF-7, based on homology to Ser385 and Ser386 in IRF-3. The mutant was not phosphorylated and did not activate IFN-α gene expression (27). Sato et al. generated a deletion mutant in which the region containing the potential sites of inducible phosphorylation between amino acids 411 to 453 was truncated. The mutant no longer translocated to the nucleus following virus infection, implicating inducible phosphorylation as a critical step for translocation (37).
Because of the common and distinct biological features of IRF-3 and IRF-7, we sought to identify the molecular basis for the differential activation of IFN-α/β genes by IRF-3 and IRF-7 in response to virus infection. Our results indicate that the distinct DNA binding specificities of IRF-3 and IRF-7—together with the different capacities of the IRF-3 and IRF-7 C-terminal domains to bind the CBP coactivator—provide an explanation for the differential regulation of IFN-α/β gene expression by these two transcription factors.
MATERIALS AND METHODS
Plasmid constructions and mutagenesis.
Plasmids expressing the wild-type and mutated forms of IRF-3 were described previously (23–25). IRF-7 expression plasmids were prepared by cloning the IRF-7A cDNA (PCR amplified from pcDNA-IRF-7A; a gift from L. Zhang and J. Pagano) into the pFlag-CMV-2 (pFlag-IRF-7) or 5′-myc-pcDNA3 (myc-IRF-7) vector. The point mutations of IRF-7 and the IRF-7 or IRF-3 chimeras were generated by overlap PCR mutagenesis with Vent DNA polymerase (New England Biolabs). Mutations were confirmed by sequencing. The deletion mutations of IRF-7 were generated by PCR. The IFNB-pGL3 luciferase reporter was generated by cloning the EcoRI-TaqI fragment (−280 to +20; filled in with the Klenow enzyme) from pUCβ26 into the NheI site (filled in with the Klenow enzyme) of the pGL3-basic vector (Promega). The RANTES-pGL3 luciferase reporter was prepared by cloning the BglII-SalI fragment (−397 to +5; filled in with the Klenow enzyme) from the RANTES-CAT reporter plasmid (25) into the NheI site (filled in with the Klenow enzyme) of the pGL3-basic vector. IFNA-pGL3 reporters (A1, −140 to +9; A2, −400 to +60; A4, −620 to +50; A7, −120 to +4; A14, −140 to +60) were generated by cloning the PCR products from 293 cell genomic DNA into the SmaI site of the pGL3-basic vector. For the construction of a minimum thymidine kinase (TK)-luciferase promoter containing one, two, or four copies of positive regulatory domain I (PRDI)-like and TG sites from the IFNA1, IFNA2, or IFNA14 promoters or two copies of the IRF-3 or IRF-7 binding sites, the double-stranded oligonucleotides were cloned into the SmaI site of the TK-pGL3 vector. IRF-3(4E)/pGEX-4T-2 (P10E/Q15E/N28E/K29E), IRF-3(+9)/pGEX-4T-2 (66-SSRG-69 and 75-AERAG-79), DBD-IRF-3-7/pGEX-4T-2 [IRF-3(1–67)–IRF-7(74–150)], and DBD-IRF-7-3/pGEX-4T-2 [IRF-7(1–73)–IRF-3(68–133)] were generated by PCR and cloned into the pGEX-4T-2 vector.
Cell cultures, transfections, and luciferase assays.
All transfections for luciferase assays were carried out with human embryonic kidney 293 cells grown in αMEM (GIBCO-BRL) supplemented with 10% fetal bovine serum, glutamine, and antibiotics. Subconfluent cells were transfected with 10 ng of the pRLTK reporter (Renilla luciferase for internal control), 100 ng of the pGL-3 reporter (firefly luciferase; experimental reporter), and 200 ng of expression plasmids by the calcium phosphate coprecipitation method. The reporter plasmids were RANTES-pGL3, IFNB-pGL3, IFNA1-pGL3, IFNA2-pGL3, IFNA4-pGL3, and IFNA14-pGL3 (38). The transfection procedures were previously described (22). At 24 h after transfection, the reporter gene activities were measured by a dual-luciferase reporter assay according to the manufacturer's instructions (Promega).
PCR-assisted DNA binding site selection from random oligonucleotides.
Binding site selection was performed as described previously but with slight modifications (41). The random double-stranded DNA oligonucleotides were synthesized by PCR using a random oligomer (5′-CCGACGCTCAGTGAATTCG[N]30TGGATCCGGTTCACATGGC-3′) and forward and reverse primers with the sequences 5′-CCGACGCTCAGTGAATTCG-3′ and 5′-GCCATGTGAACCGGATCCA-3′, respectively. The amplification reaction was carried out using 5 μg of random oligomer, 10 μg of forward primer, and 10 μg of reverse primer for three cycles, with each cycle consisting of 1.5 min at 95°C, 2 min at 55°C, and 2 min at 72°C. The binding mixture (25 μl) contained 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NaCl, 2 mM dithiothreitol [DTT], 5% glycerol, 0.5% Nonidet P-40 [NP-40], 10 μg of bovine serum albumin (BSA) per μl, 62.5 μg of poly(dI-dC) per ml, 100 ng of IRF-3–glutathione S-transferase (GST) or IRF-7–GST recombinant protein, and 1 μg of double-stranded N28. After incubation for 15 min, 10 μl of glutathione-Sepharose beads (Pharmacia) was added, and the mixture was incubated with constant rotation for 15 min at room temperature. Protein-DNA complexes were washed twice with 500 μl of cold binding buffer without BSA, and poly(dI-dC). The bound DNA was eluted at 50°C in 120 μl of elution buffer 1 (5 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 100 mM sodium acetate, 50 mM Tris-HCl [pH 7.6]). The DNA was then recovered by ethanol precipitation. The recovered DNA was amplified by PCR using 100 pmol of forward primer and 100 pmol of reverse primer for 15 cycles under the conditions described above. After five rounds of selection, the protein-DNA complexes were separated by electrophoresis with a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE). The bound DNA was excised from the dry gel and eluted in 400 μl of elution buffer 2 (0.5 M ammonium acetate, 1 mM EDTA [pH 8.0]). Bound DNA was recovered by ethanol precipitation and amplified by PCR. The products were then digested with EcoRI and BamHI, cloned into pBluescript KS(+), and subjected to sequence analysis.
Immunoblot analysis.
To confirm the expression of the transgenes, equivalent amounts of whole-cell extract (20 μg) were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) with a 10% polyacrylamide gel. After electrophoresis, proteins were transferred to a Hybond transfer membrane (Amersham) in a buffer containing 30 mM Tris, 200 mM glycine, and 20% methanol for 1 h. The membrane was blocked by incubation in phosphate-buffered saline (PBS) containing 5% dried milk for 1 h and then probed with anti-Flag antibody M2 (Sigma) in 5% milk–PBS at a dilution of 1:3,000. These incubations were done at 4°C overnight or at room temperature for 1 to 3 h. After four 10-min washes with PBS, the membrane was reacted with a peroxidase-conjugated secondary goat anti-mouse antibody (Amersham Corp.) at a dilution of 1:2,500. The reaction was then visualized with an enhanced chemiluminescence detection system as recommended by the manufacturer (Amersham).
Immunoprecipitation and immunoblot analysis of protein-protein interactions.
293 cells were cotransfected with expression plasmids encoding wild-type or mutated forms of IRF-3 or IRF-7. Whole-cell extracts (200 to 500 μg) were prepared from cotransfected cells and incubated with 2 μl of anti-myc antibody 9E10, anti-CBP antibody A-22, anti-CBP antibody N-15, anti-TAFIIp250 antibody 6B3, or anti-PCAF antibody (a gift from X. Yang) cross-linked to 30 μl of protein A-Sepharose beads for 1 h at 4°C. Precipitates were washed five times with lysis buffer (23) and eluted by boiling the beads for 3 min in SDS sample buffer. Eluted proteins were separated by SDS-PAGE, transferred to a Hybond transfer membrane, and incubated with anti-Flag, anti-CBP, anti-p300, anti-PCAF, anti-TAFIIp250, or anti-myc antibody (1:1,000 to 1:3,000). Immunocomplexes were detected by using a chemiluminescence-based system.
Protein expression and purification.
The IRF-3–GST and IRF-7–GST fusion proteins were expressed and isolated from Escherichia coli DH5α following 3 h of induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Pharmacia) at 37°C. Bacterial extracts in PBS containing 1% Triton X-100 were incubated with glutathione-Sepharose beads for 20 min at room temperature. After three washes with PBS, the fusion proteins were eluted with 15 mM glutathione in PBS.
Electromobility shift assay (EMSA).
Whole-cell extracts were prepared 48 h after transfection with 5 μg of expression plasmids. Cells were washed with PBS and lysed in 10 mM Tris-Cl (pH 8.0)–60 mM KCl–1 mM EDTA–1 mM DTT–0.5% NP-40–0.5 mM phenylmethylsulfonyl fluoride–10 μg of leupeptin per ml–10 μg of pepstatin per ml–10 μg of aprotinin per ml–0.5 ng of chymostatin per μl–0.25 μM microcystin. Equivalent amounts of whole-cell extract (20 μg) or various amounts of recombinant proteins were assayed for IRF-3 or IRF-7 binding by gel shift analysis using a 32P-labeled double-stranded oligonucleotide corresponding to the ISRE region of the RANTES promoter (5′-CTATTTCAGTTTTCTTTTCCGTTTTGTG-3′), the PRDI region of the IFNB promoter (5′-GAGAAGTGAAAGTG-3′), the PRDIII region of the IFNB promoter (5′-GAAAACTGAAAGGG-3′), the PRDI-PRDIII region of the IFNB promoter (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′), the PRDI-like and TG regions of the IFNA1 promoter (5′-GGAAAGCAAAAACAGAAATGGAAAGTGG-3′), the PRDI-like and TG regions of the IFNA2 promoter (5′-GAAAGCAAAAAGAGAAGTAGAAAGTAA-3′), the PRDI-like and TG regions of the IFNA14 promoter (5′-GGAAAGCCAAAAGAGAAGTAGAAAAAAA-3′), and selected IRF-3 and IRF-7 binding sites. Complexes were formed by incubating the probe with 20 μg of each whole-cell extract or various amounts of recombinant proteins. The binding mixture (20 μl) contained 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NaCl, 2 mM DTT, 5% glycerol, 0.5% NP-40, 10 μg of BSA per ml, and 62.5 μg of poly(dI-dC) per ml added to reduce nonspecific binding. After 20 min of incubation with the probe, extracts were loaded on a 5% polyacrylamide gel (60:1 cross-link) prepared in 0.5× TBE. After 2 h at 200 to 250 V, the gel was dried and exposed to Kodak film at −70°C overnight. To demonstrate the specificity of protein-DNA complex formation, a 200-fold molar excess of unlabeled oligonucleotide was added to the binding mixture before labeled probe was added.
RESULTS
Differential induction of IFNA and IFNB promoters by IRF-3 and IRF-7.
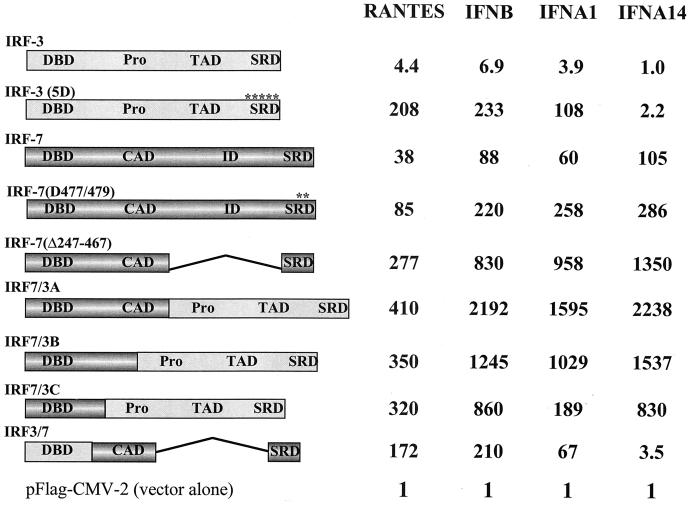
The IRF-7 transcription factor shares many structural features with IRF-3, including a structurally conserved DBD and a serine-rich C-terminal region that is the target of virus-inducible phosphorylation (27, 37). Because of the involvement of IRF-3 in the activation of immediate-early IFNB and IFNA genes and the role of IRF-7 in the induction of delayed-type IFNA genes (27), we compared the activation of IFN-α/β and RANTES promoters by different forms of IRF-3 and IRF-7. During studies on the structure and function of IRF-7, two constitutively active forms of IRF-7 were generated (R. Lin, Y. Mamane, and J. Hiscott, submitted for publication), one with a substitution of Ser477 and Ser479 with the phosphomimetic Asp [IRF-7(D477/479)] and the other with a deletion of a portion of the C-terminal domain [IRF-7(Δ247–467)]. Plasmids expressing wild-type IRF-3, IRF-3(5D) (25), IRF-7, IRF-7(D477/479), and IRF-7(Δ247–467) were cotransfected into 293 cells together with the IFNB, RANTES, and different IFNA promoter constructs and examined for their ability to stimulate reporter gene activity. As shown in Fig. 1, both IFNB and RANTES promoters were activated by IRF-3 or IRF-7. The constitutively active form of IRF-3 activated IFNB and RANTES luciferase reporter gene activities 200- and 150-fold, respectively, while IRF-7 resulted in 88- and 38-fold stimulation of IFNB and RANTES promoter activities, respectively. Substitution of Ser477 and Ser479 with the phosphomimetic Asp resulted in a form of IRF-7 that activated the IFNB and RANTES promoters up to 220- and 85-fold, respectively, while IRF-7(Δ247–467) activated the IFNB and RANTES promoters up to 830- and 277-fold, respectively.
FIG. 1.
Activities of IRF-7 and IRF-3 fusion proteins. The structures of the fusion proteins are illustrated schematically (Pro, proline-rich domain; TAO, transactivation domain; SRD, signal response domain; CAD, constitutive activation domain; ID, inhibitory domain). IRF-7/3A contains 541 amino acids, 246 from IRF-7 (1 to 246) and 295 from IRF-3(5D) (133 to 427). IRF-7/3B consists of 495 amino acids, 200 from IRF-7 (1 to 200) and 295 from IRF-3(5D) (133 to 427). IRF-7/3C consists of 445 amino acids, 150 from IRF-7 (1 to 150) and 295 from IRF-3(5D) (133 to 427). IRF-3/7 contains 265 amino acids, 132 from IRF-3 and 133 from IRF-7(Δ247–467). For transactivation assays, 293 cells were transfected with the pRLTK control plasmid, the RANTES-pGL3, IFNB-pGL3, IFNA1-pGL3, or IFNA4-pGL3 reporter plasmid, and the expression plasmids encoding IRF-3, IRF-3(5D), IRF-7, IRF-7(D477/479), IRF-7(Δ247–467), IRF-3/7, or IRF-7/3, as indicated. Luciferase activity was analyzed at 24 h posttransfection by the dual-luciferase reporter assay as described by the manufacturer (Promega). Relative luciferase activity was measured as fold activation (relative to the basal level for the reporter gene in the presence of the pFlag-CMV-2 vector after normalization to cotransfected relative light unit activity); the values represent the average of three experiments performed in duplicate, with variability of 10 to 25%.
The IFNA1 and IFNA2 promoters were activated 108-fold (Fig. 1) and 38-fold (data not shown), respectively, by the constitutively active form of IRF-3, demonstrating that both promoters are recognized and activated by IRF-3. However, the IFNA14 promoter was not induced by IRF-3(5D) coexpression (Fig. 1). The IFNA4 and IFNA7 promoters responded in the same manner as the IFNA14 promoter (data not shown), not being induced by IRF-3(5D). When IRF-7 was used to induce the IFNA promoters, all five IFNA promoters were strongly transactivated by wild-type IRF-7 (50-fold induction of IFNA7 to 100-fold induction of IFNA4) or by the two constitutively active forms of IRF-7. In all cases, the induction of these IFNA reporter gene constructs by IRF-7 or IRF-7(D477/479) was enhanced four- to eightfold by concomitant Sendai virus infection (data not shown). This experiment confirms the differential responsiveness of IFNA and IFNB genes to induction by IRF-3 or IRF-7.
Chimeric IRF-7 and IRF-3 proteins produce supraphysiological induction of RANTES and IFN promoters.
The possibility that these two factors possess distinct DNA binding and/or activation properties was next evaluated by generating chimeric proteins containing the IRF-7 or IRF-3 N-terminal DBD fused to the IRF-3(5D) or IRF-7 C-terminal transactivation domain (Fig. 1). Chimeric proteins containing the IRF-7 DBD and the IRF-3(5D) transactivation domain (IRF-7/3A, IRF-7/3B, and IRF-7/3C) strongly activated RANTES, IFNB, IFNA1, and IFNA4 promoter activities (Fig. 1) as well as IFNA7 and IFNA14 promoter activities (data not shown). In most cases, the levels of transactivation were 10- to 20-fold higher than those observed with the constitutively active forms of IRF-3 or IRF-7. Although the chimeric protein containing the IRF-3 DBD and the IRF-7 transactivation domain (IRF-3/7) was a strong transactivator for the RANTES, IFNB, and IFNA1 promoters, IRF-3/7 only weakly stimulated expression from the IFNA4, IFNA7, and IFNA14 promoters (Fig. 1 and data not shown). This result suggests that the DBDs of IRF-3 and IRF-7 recognized different DNA binding sites or possessed distinct affinities for the same DNA site, with this selectivity contributing to the differential regulation of IFN-α/β gene expression.
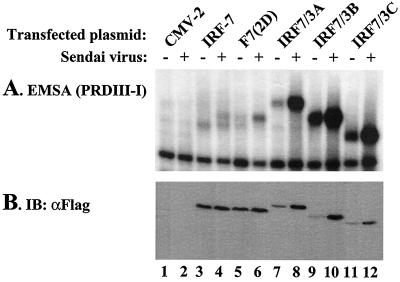
Expression of the IRF-7/3A, IRF-7/3B, or IRF-7/3C chimeric proteins was therefore analyzed by an EMSA to correlate transactivation with DNA binding activity. In uninfected cells, an IRF-7–PRDIII-PRDI complex was identified (Fig. 2, lane 3), whereas after virus infection, a slower-migrating form of the IRF-7–PRDIII-PRDI complex was detected (Fig. 2, lane 4). The higher-molecular-weight IRF-7–PRDIII-PRDI complex was also detected in IRF-7(D477/479)-expressing cells before and after virus infection (Fig. 2, lanes 5 and 6), reminiscent of the effect of IRF-3(5D) (25). The IRF-7/3 chimeric proteins all possessed constitutive DNA binding activity that appeared to be enhanced about fourfold by virus infection, although increased DNA binding appeared to be the result of higher protein levels in the infected cell extracts, as detected by immunoblotting (Fig. 2B).
FIG. 2.
DNA binding activity of IRF-7/3 chimeric proteins. (A) An EMSA was performed with whole-cell extracts (20 μg) derived from 293 cells transfected with various Flag-tagged IRF-7 or IRF-7/3 expression plasmids. At 24 h posttransfection, cells were infected with Sendai virus for 6 h (+) or left uninfected (−), as indicated. The 32P-labeled probe corresponded to the PRDI-PRDIII (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′) motif of the IFNB promoter. (B) Twenty micrograms of whole-cell extracts from panel A was analyzed by immunoblotting (IB) with anti-Flag antibody. F7(2D), F7 D477/479.
IRF-7 does not interact with the CBP coactivator.
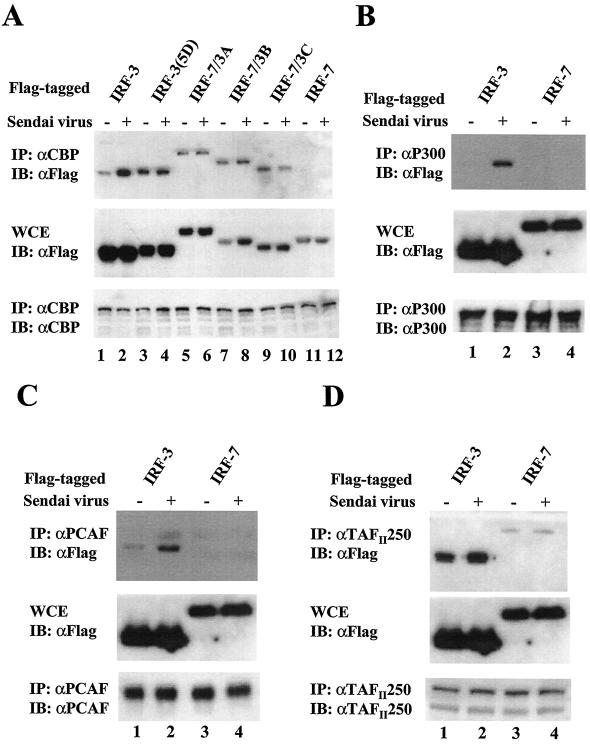
IRF-3(5D) was previously shown to associate with the CBP coactivator (25); therefore, we next examined the interaction of CBP with IRF-7 or with IRF-7/3 chimeric proteins by immunoprecipitation of Flag-tagged IRF forms. After immunoprecipitation of endogenous CBP with anti-CBP antibody A-22 (Fig. 3A), immunoblot analysis with anti-Flag antibody revealed that IRF-7/3A, IRF-7/3B, or IRF-7/3C associated with CBP in both unstimulated and virus-infected cells (Fig. 3A, lanes 5 to 10), while IRF-7 did not interact with CBP (Fig. 3A, lanes 11 and 12). These data indicate that the strong transactivation activity displayed by the IRF-7/3 chimeric proteins was due to the DNA binding activity of the IRF-7 domain, coupled with the capacity of the IRF-3(5D) C-terminal domain to associate with the CBP coactivator. Two other histone acetyltransferase coactivators (p300 and PCAF) were also examined, and both p300 and PCAF associated with IRF-3 in virus-infected cells but failed to interact with IRF-7 (Fig. 3B and C).
FIG. 3.
Activated forms of IRF-3 and IRF-7/3 chimeric proteins but not IRF-7 are associated with histone acetyltransferases. 293 cells were transfected with Flag-tagged IRF-3, IRF-7, or IRF-7/3 expression plasmids, as indicated above the lanes. At 24 h posttransfection, cells were infected with Sendai virus for 12 h (+) or left uninfected (−), as indicated. Whole-cell extracts (WCE) (200 μg for CBP and 500 μg for p300, PCAF, and TAFIII250) were immunoprecipitated with anti-CBP antibody A-22 (A), anti-p300 antibody N-15 (B), anti-PCAF antibody (C), or anti-TAFIIp250 antibody 6B3 (D). Immunoprecipitated complexes (upper panel) or 20 μg of whole-cell extracts (middle panel) was analyzed by SDS–8% PAGE and subsequently probed with anti-Flag antibody M2. The membranes shown in the upper panel were reprobed with anti-CBP antibody A-22 (A), anti-p300 antibody N-15 (B), anti-PCAF antibody (C), or anti-TAFIIp250 antibody 6B3 (D) (lower panel). IP, immunoprecipitation; IB, immunoblotting.
The transcription initiation factor TFIID is a multimeric protein complex composed of TATA-binding protein (TBP) and many TBP-associated factors (TAFIIs). TAFIIs are important cofactors that mediate activated transcription by providing interaction sites for distinct transcriptional activators. TAFII250 serves as the core subunit of TFIID and interacts with a variety of other TAFIIs as well as TBP. TAFII250 is required for the activation of particular genes and associates with components of the basal transcriptional machinery, such as TFIIA, TFIIE, and TFIIF (7). In addition, TAFII250 functions as both a protein kinase and a histone acetyltransferase (7, 30). Therefore, we also examined the interaction of TAFII250 with IRF-3 and IRF-7. IRF-3 constitutively associated with TAFII250, and this interaction was further enhanced by virus infection (Fig. 3D, lanes 1 and 2); IRF-7 also weakly interacted with TAFII250 in unstimulated or virus-infected cells (Fig. 3D, lanes 3 and 4). These results suggest that IRF-3 and IRF-7 may recruit TAFII250 to target promoters and activate transcription.
Distinct DNA binding specificities of IRF-3 and IRF-7.
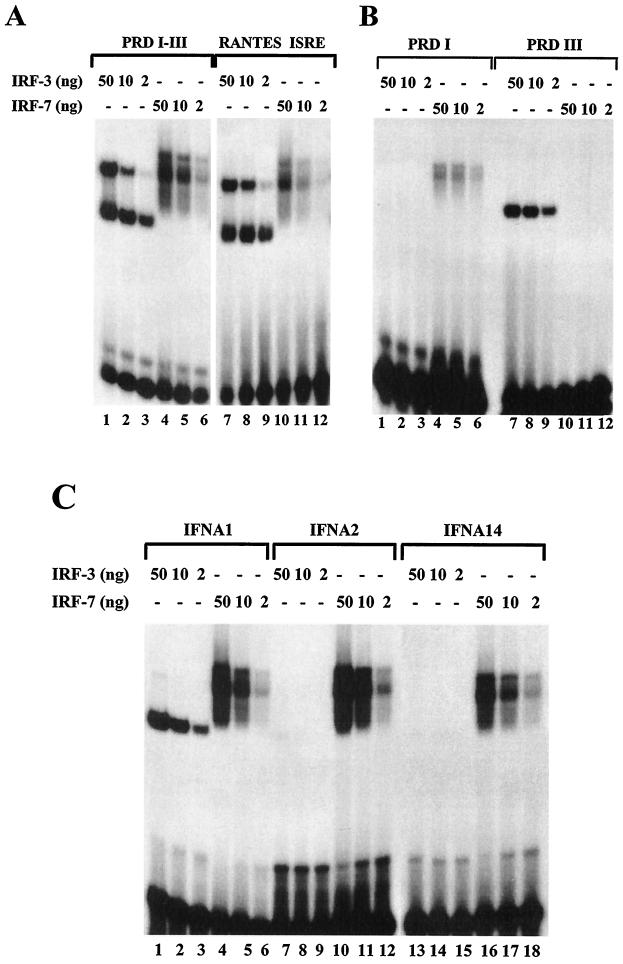
Recombinant IRF-3 and IRF-7 proteins were next used to measure protein-DNA interactions within the ISRE domain of the RANTES promoter and the PRDI-PRDIII region of the IFNB promoter. In an EMSA analysis, IRF-3 bound strongly to both PRDI-PRDIII and RANTES ISRE probes (Fig. 4A, lanes 1 to 3 and 7 to 9), whereas IRF-7 bound more strongly to PRDI-PRDIII than to the RANTES ISRE (Fig. 4A, lanes 4 to 6 and 10 to 12). The PRDI and PRDIII regions of the IFNB promoter were separately evaluated to determine whether IRF-3 and IRF-7 binding could be localized to an individual domain of the IFNB promoter. IRF-3 strongly bound to the PRDIII probe (Fig. 4B, lanes 7 to 9) but failed to bind to the PRDI probe (Fig. 4B, lanes 1 to 3); strikingly, IRF-7 bound to the PRDI probe (Fig. 4B, lanes 4 to 6) but did not bind to the PRDIII probe (Fig. 4B, lanes 10 to 12).
FIG. 4.
Binding of IRF-3 and IRF-7 to IRF binding sites of the IFN and RANTES promoters. (A) Recombinant N-terminal IRF-3 and IRF-7 bind to PRDI-PRDIII and RANTES ISRE probes. An EMSA was performed with the indicated amounts of recombinant protein; the 32P-labeled probe corresponded to the PRDI-PRDIII region (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′) or the ISRE of the RANTES gene (5′-CTATTTCAGTTTTCTTTTCCGTTTTGTG-3′). (B) Recombinant N-terminal IRF-3 and IRF-7 bind to PRDI and PRDIII probes. An EMSA was performed with the indicated amounts of recombinant protein; the 32P-labeled probe corresponded to PRDI (5′-GAGAAGTGAAAGTG-3′) or PRDIII (5′-GAAAACTGAAAGGG-3′). (C) Recombinant N-terminal IRF-3 and IRF-7 bind to the PRDI-like and TG sites from the IFNA1, IFNA2, and IFNA14 promoters. An EMSA was performed with the indicated amounts of IRF-3 or IRF-7 and 32P-labeled probes corresponding to the following PRDI-like sites: IFNA1, 5′-GGAAAGCAAAAACAGAAATGGAAAGTGG-3′; IFNA2, 5′-GAAAGCAAAAAGAGAAGTAGAAAGTAA-3′; and IFNA14, 5′-GGAAAGCCAAAAGAGAAGTAGAAAAAAA-3′.
Binding of recombinant IRF-3 and IRF-7 to the regulatory regions of different IFNA promoters (IFNA1, IFNA2, and IFNA14) which contained PRDI-like binding sites was also analyzed (Fig. 4C). Both IRF-3 and IRF-7 bound strongly to the IFNA1 probe (Fig. 4C, lanes 1 to 6). Interestingly, probes from the equivalent regions of IFNA2 and IFNA14 associated with IRF-7 (Fig. 4C, lanes 10 to 12 and 16 to 18) but failed to bind detectable IRF-3 protein (Fig. 4C, lanes 7 to 9 and 13 to 15).
Identification of consensus DNA binding sites for IRF-3 and IRF-7.
All of the above studies suggested that IRF-3 and IRF-7 recognized different binding sites. To clarify IRF-3 and IRF-7 DNA binding site specificities, a PCR-based DNA binding site selection strategy (41) was used to select a panel of oligonucleotides recognized by IRF-3 and/or IRF-7. Candidate binding sites generated from a pool of radiolabeled oligonucleotides containing 28 random base pairs were incubated with recombinant IRF-3–GST or IRF-7–GST; the IRF-bound oligonucleotides were eluted, PCR amplified, and used as input for subsequent rounds of selection and purification. The PCR-amplified oligonucleotides recovered after each round of selection were subjected to gel shift analysis using recombinant IRF-3–GST (Fig. 5A) or IRF-7–GST (Fig. 5B). Oligonucleotides selected with IRF-3 consisted of enriched sequences that bound to IRF-3 (Fig. 5A, lanes 1 to 5) but interacted weakly with IRF-7 (Fig. 5A, lanes 6 to 10), while oligonucleotides selected with IRF-7 bound weakly to both IRF-7 (Fig. 5B, lanes 6 to 10) and IRF-3 (Fig. 5B, lanes 1 to 5). After five rounds of selection, bound oligonucleotides were purified from the gel, PCR amplified, and prepared for sequencing. The sequences recovered from 16 cloned IRF-3 binding sites and the consensus sequence are shown in Fig. 6. The sequences selected with IRF-3 were virtually identical to the consensus sequence recognized by IRF-1 and IRF-2 (5′-GAAANNGAAANN-3′) (41).
FIG. 5.
Gel shift analysis of selected IRF-3 and IRF-7 binding sites. Oligonucleotides selected with recombinant IRF-3–GST (lanes 1 to 5) or IRF-7–GST (lanes 6 to 10) at each round were amplified by PCR using 32P-labeled primers and subsequently used as probes in a gel shift analysis. Fifty nanograms of IRF-3–GST (A) or IRF-7–GST (B) was used in each binding reaction. The number of selection cycles is shown above each lane. Arrows indicate protein-DNA complexes.
FIG. 6.
Consensus sequences binding to IRF-3. Sequences of 16 cloned IRF-3 binding sites derived by five rounds of binding site selection are shown. Each sequence was aligned with respect to its homologous sequence (in boldface type). The frequency of each nucleotide at each position of the homologous sequence and the consensus sequence are shown at the bottom.
Interestingly, the sequences recovered from 28 cloned IRF-7 binding sites were closely related to the IRF-3 consensus sequence, but upon closer examination it became clear that IRF-7 bound with greater flexibility than IRF-3 to the consensus binding site (Fig. 7). For example, from the sequences selected with IRF-3, 12 out of a total 16 sequences contained the tandem repeat GAAANNGAAANN motif, while from the sequences selected with IRF-7, only 5 of 28 sequences contained the GAAANNGAAANN motif. The majority of the IRF-7 binding sites had a minimum of one nucleotide replacement in either the 5′-GAAA or the 3′-GAAA motif of the half-site. Thus, for IRF-7, either the 5′ or the 3′ half-site consisted of GAAANN and the other half-site contained at least one nucleotide substitution. This result indicates that a single nucleotide alteration in the GAAANNGAAANN consensus site precluded the binding of IRF-3 to ISRE or PRDI-like sites, whereas the nucleotide replacement(s) did not affect IRF-7 DNA binding.
FIG. 7.
Consensus sequences binding to IRF-7. Sequences of 28 cloned IRF-7 binding sites derived by five rounds of binding site selection are shown. Each sequence was aligned with respect to its homologous sequence (in boldface type). The frequency of each nucleotide at each position of the homologous sequence and the consensus sequence are shown at the bottom.
Binding of IRF-3 and IRF-7 to selected sequences.
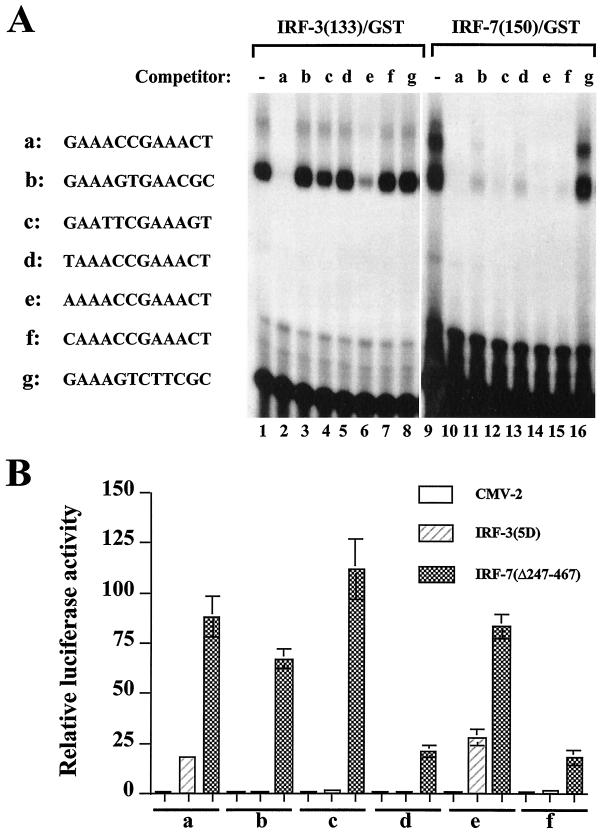
The difference between the DNA binding sites selected with IRF-3 and IRF-7 is minimally a single nucleotide replacement in one of the GAAA core sequences of the GAAANNGAAANN motif. Oligonucleotides from the binding site selection were subjected to a DNA gel shift binding competition analysis to determine the relative binding affinities of IRF-3 and IRF-7 for different sites. IRF-3 binding to the GAAACCGAAACT oligonucleotide (Fig. 8A, a) was competed by the homologous GAAACCGAAACT motif (a) or the AAAACCGAAACT motif (e) (Fig. 8A, lanes 2 and 6) but not by oligonucleotide b, c, d, f, or g (Fig. 8A, lanes 3 to 5, 7, and 8), which were altered by one nucleotide in one of the GAAA core motifs. With IRF-7, all oligonucleotides (a to f) (Fig. 8A, lanes 10 to 15) were able to compete for the binding of IRF-7 to the GAAACCGAAACT motif. The competition for binding was specific, since oligonucleotide g, which contained one intact GAAANN half-site, failed to compete for the binding of either IRF-3 or IRF-7 (Fig. 8A, lanes 8 and 16).
FIG. 8.
Characterization of selected binding sites. (A) An EMSA was performed with 20 ng of recombinant IRF-3–GST (lanes 1 to 8) or IRF-7–GST (lanes 9 to 16), 32P-labeled oligonucleotide a (5′-GAAACCGAAACTGAAACCGAAACT-3′), and a 1,000-fold molar excess of competitor DNA. Selected binding sites (two copies, indicated beside the gel as a to g) were used as competitors. (B) Activation of selected promoters by IRF-3 and IRF-7. 293 cells were transfected with the pRLTK control plasmid, reporter constructs containing the minimum TK-luciferase promoter and two copies of selected binding sites (designated a to f), and the active forms of IRF-3(5D) and IRF-7(Δ247–467) expression plasmids, and luciferase activity was analyzed at 24 h posttransfection. Relative luciferase activity was measured as fold activation (relative to the basal level for the reporter gene in the presence of the pFlag-CMV-2 vector after normalization to cotransfected relative light unit activity); the values represent the average of three experiments performed in duplicate, with variability of 10 to 25%.
The ability of IRF-3 and IRF-7 to activate transcription from minimal TK promoter constructs linked to two copies of oligonucleotides a to f was next examined (Fig. 8B). Cotransfection of the constitutively active form of IRF-3 led to an 18- and 28-fold induction of reporter genes containing two copies of GAAACCGAAACT (Fig. 8B, a) and AAAACCGAAACT (e) motifs, respectively. However, no induction was observed from promoter constructs containing two copies of TAAACCGAAACT, CAAACCGAAACT, GAATTCGAAAGT, or GAAAGTGAACGC (Fig. 8B, b, c, d, and f). Interestingly, coexpression of IRF-7(Δ247–467) stimulated expression from all reporter genes to different extents, consistent with the DNA binding competition analysis results.
Differential gene activation by the IRF-like and TG motifs of IFNA promoters.
To determine whether the PRDI-like and TG sites (−98 to −71) of the different IFNA promoters were sufficient to mediate virus-induced or IRF-3- or IRF-7-activated expression, the same sequences that were used as probes in the EMSA were cloned upstream of a minimal TK promoter in the pGL3 luciferase reporter construct. As shown in Table 1, row 1, a single copy of PRDI-like and TG sites from the IFNA1 promoter was not responsive to Sendai virus infection and was only weakly activated by the constitutively active form of IRF-3 (5-fold); this element was moderately induced by coexpression of either IRF-7 (6-fold) or the constitutively active form of IRF-7 (18-fold). Sendai virus infection together with IRF coexpression further augmented (two- to threefold) the induction of the PRDI-like and TG sites from the IFNA1 promoter. Two copies of the PRDI-like and TG sites from the IFNA1 promoter were generally more responsive to IRF-3 and IRF-7 stimulation (Table 1, row 2). In particular, IFNA1 was strongly activated by the constitutively active forms of IRF-3 and IRF-7 (300- and 270-fold induction, respectively). Two copies of the PRDI-like and TG sites from the IFNA1 promoter were also responsive to Sendai virus infection (12-fold induction). Coexpression of different forms of IRF-3 or IRF-7 generally had the effect of increasing the virus-induced activation of the IFNA1 promoter (Table 1, row 2).
TABLE 1.
Activation of PRDI-like and TG motifs from distinct IFNA promoters by IRF-3 and IRF-7a
| Promoter (no. of copies) | Relative luciferase activity for the following construct in the presence (+) or absence (−) of Sendai virus infection:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CMV-2
|
IRF-3
|
IRF-3 (5D)
|
IRF-7
|
IRF-7 (D477/479)
|
||||||
| − | + | − | + | − | + | − | + | − | + | |
| IFNA1 (1) | 1 | 1.4 | 1.0 | 3.6 | 4.8 | 7.3 | 6.5 | 22 | 18 | 46 |
| IFNA1 (2) | 1 | 12 | 9.3 | 25 | 302 | 267 | 57 | 329 | 271 | 592 |
| IFNA2 (1) | 1 | 1.1 | 0.9 | 1.7 | 0.8 | 1.6 | 4.7 | 12.4 | 7.8 | 21 |
| IFNA2 (2) | 1 | 2.6 | 1.1 | 4.7 | 2.0 | 8.2 | 31 | 212 | 123 | 362 |
| IFNA14 (2) | 1 | 1.1 | 0.8 | 1.9 | 0.8 | 1.3 | 3.3 | 6.3 | 5.2 | 9.6 |
| IFNA14 (4) | 1 | 1.6 | 0.9 | 2.9 | 0.8 | 2.2 | 24 | 47 | 32 | 86 |
293 cells were transfected with the pRLTK control plasmid, IFNA TK-pGL3 reporter constructs (minimum TK-luciferase promoter containing one, two, or four copies of PRDI-like and TG sites from the IFNA1, IFNA2, or IFNA14 promoters), and various IRF-3 and IRF-7 expression plasmids, as indicated. Eight hours after transfection, cells were infected with Sendai virus (+) or left uninfected (−), as indicated. Luciferase activity was analyzed at 24 h posttransfection. Relative luciferase activity was measured as fold activation (relative to the basal level for the reporter gene in the presence of the pFlag-CMV-2 vector after normalization to cotransfected relative light unit activity); the values represent the average of three experiments performed in duplicate, with variability of 10 to 25%.
The construct carrying a single copy of the PRDI-like and TG regions of IFNA2 was similarly weakly responsive to IRF coexpression and/or virus activation (Table 1, row 3). However, two copies of the PRDI-like and TG sites from the IFNA2 promoter were strongly activated by the constitutively active form of IRF-7 (120-fold induction) but were not activated by IRF-3(5D) (Table 1, row 4), consistent with the differential binding of IRF-7 and IRF-3 to the IFNA2 promoter (Fig. 4C). Two copies of the PRDI-like and TG sites from IFNA14 were activated modestly by IRF-7 (about 3- to 10-fold) but were not stimulated by IRF-3(5D) (Table 1, row 5). Even with four copies of the PRDI-like and TG sites from IFNA14, the activation of this promoter by IRF-7 and/or Sendai virus infection was in the range of 20- to 80-fold (Table 1, row 6). These results support the observations of Fig. 1 and 4 demonstrating that the PRDI-like and TG sites of the IFNA promoter are differentially regulated by the IRF-3 and IRF-7 transcription factors. The requirement for multimerization of these domains further suggests that these elements likely cooperate with other cis regulatory elements to control IFNA gene expression.
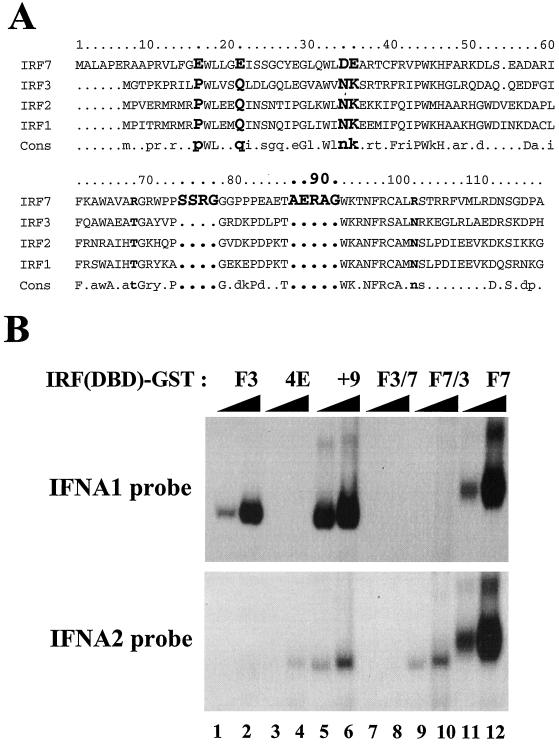
Primary sequence differences in the DBD of IRF-7 contribute to DNA binding affinity.
The results from PCR-mediated DNA binding site selection indicated that IRF-3 binds to the consensus site recognized by IRF-1 and IRF-2 (5′-GAAANNGAAANN-3′) (41), while IRF-7 binds with greater flexibility than IRF-3 to a related sequence (Fig. 6 and 7). In an attempt to identify amino acid differences that may contribute to differential DNA binding specificity, the amino acid sequences within the DBDs of IRF-1, IRF-2, IRF-3, and IRF-7 were compared by sequence alignment. Two major differences are present in IRF-7: (i) IRF-7 contains four acidic residues in the N-terminal region of the DBD (E16E21 in helix α1 and D34E35 in antiparallel β sheet β1), compared with P10Q15N28K29 in IRF-1, IRF-2, and IRF-3); and (ii) IRF-7 contains a four-amino-acid insert (SSRG) in loop L2 and a five-amino-acid insert (AERAG) in helix α3 (9) (Fig. 9A). To determine whether these amino acid differences could alter DNA binding specificity, the construct IRF-3(4E) (P10E/Q15E/N28E/K29E) was generated; in this construct, the PQNK residues of IRF-3 were converted to glutamic acid (E). Also, IRF-3 was modified to include nine additional amino acids [IRF-3(+9)], which included the SSRG amino acids after position 66 and the AERAG amino acids after position 75. Hybrid DBD chimeric proteins IRF-3-7 [IRF-3(1–67)–IRF-7(74–150)] and DBD-IRF-7-3 [IRF-7(1–73)–IRF-3(68–133)] were also generated by PCR and cloned into the pGEX-4T-2 vector. As shown in Fig. 9B, the IRF-3(4E) (lanes 3 and 4), IRF-3(+9) (lanes 5 and 6), and DBD-IRF-7-3 (lanes 9 and 10) fusion proteins were able to bind to the IFNA2 probe, suggesting that these amino acid sequence differences may contribute to differential DNA binding specificity. However, all of the mutated or chimeric proteins were characterized by a decrease in DNA binding affinity of more than 100-fold compared with the affinity of wild-type IRF-3 (Fig. 9B, lanes 1 and 2) or IRF-7 (lanes 11 and 12). These results indicate that primary sequence information does not predict crucial amino acid residues involved in differential specificity. Rather, the intact three-dimensional structure of IRF-3 and IRF-7 is required to convey subtle differences in DNA binding specificity.
FIG. 9.
Amino acids of IRF-7 involved in different DNA binding specificities. (A) Sequence alignment of the DBDs of IRF-7, IRF-3, IRF-2, and IRF-1. The sequential numbering of IRF-7 is shown at the top. Identical residues in IRF-1, IRF-2, and IRF-3 but not IRF-7 are shown in boldface type. (B) Binding of mutated forms of IRF-3 or IRF3 and IRF7 chimeric recombinant proteins to the PRDI-like- and TG sites from the IFNA1 and IFNA2 promoters. An EMSA was performed with the indicated amounts of recombinant GST fusion proteins and 32P-labeled probes corresponding to the following PRDI-like sites: IFNA1, 5′-GGAAAGCAAAAACAGAAATGGAAAGTGG-3′; and IFNA2, 5′-GAAAGCAAAAAGAGAAGTAGAAAGTAA-3′.
DISCUSSION
In the present study, we sought to examine the molecular basis for the differential regulation of several members of the IFN-α/β gene family (IFNA and IFNB) by IRF-3 and IRF-7. The IFNB, IFNA1, IFNA2, and RANTES promoters were activated by coexpression of either IRF-3 or IRF-7, whereas the IFNA4, IFNA7, and IFNA14 promoters were exclusively activated by IRF-7 and not by IRF-3. Analysis of protein-DNA interactions revealed that recombinant IRF-3 and IRF-7 selectively bound to different regions of the IFNB promoter; IRF-3 bound preferentially to the PRDIII domain of the IFNB promoter, while IRF-7 interacted exclusively with the PRDI domain. PCR-mediated DNA binding site selection results demonstrated that IRF-3 recognized the IRF consensus element 5′-GAAANNGAAANN-3′. Replacement of a single nucleotide within the GAAA core half-site was sufficient to preclude IRF-3 DNA binding. IRF-7 bound to a related sequence motif but with greater flexibility than IRF-3; a single nucleotide replacement did not decrease IRF-7 DNA binding. These results demonstrate that the DNA binding site specificities of IRF-3 and IRF-7, together with the different capacities of the IRF-3 and IRF-7 C-terminal domains to bind the CBP coactivator, provide a partial explanation for the differential regulation of IFN-α/β gene expression by these two transcription factors.
The chimeric forms of IRF-7 and IRF-3 generated during the present study combined the DNA binding specificity of the IRF-7 DBD with the strong transactivation capacity of the IRF-3(5D) C-terminal domain. When tested with the IFNA and IFNB promoters, 10- to 20-fold-higher levels of reporter gene activity were observed with the chimeric proteins than with the constitutively active forms of either protein alone. Furthermore, the strong transcriptional activity of the IRF-7/3 chimera was not matched by the IRF-3/7 fusion protein. It is possible that the more restricted DNA binding specificity of the IRF-3 DBD and the apparent lack of interaction of IRF-7 with the histone acetyltransferase coactivators account for the difference between these chimeric forms. The failure of wild-type IRF-7, constitutively active IRF-7, or virus-activated IRF-7 to associate with CBP, p300, or PCAF was surprising, particularly since IRF-7 was able to stimulate IFN gene expression (Fig. 1). It is possible that IRF-7 interacts in a restricted manner with a distinct histone acetyltransferase coactivator or recruits coactivators only when bound to DNA and in association with other factors. The activity of IRF-7/3 in human cells stably transfected with IRF-7/3-expressing constructs has been difficult to characterize, since it appears that the expression of the chimeric protein is proapoptotic. Inducible regulation of the IRF-7/3-expressing constructs may provide an interesting system for examining downstream IRF-regulated genes.
TFIID has been identified as a potential target for transcriptional regulation (40). TFIID is a multimeric protein complex consisting of TBP and many TAFIIs. Although TBP alone is able to bind core promoters containing TATA elements and can support basal transcription, TAFIIs are required for activated transcription. Some TAFIIs have been shown to serve as coactivators that directly contact enhancer-bound activators to modulate gene-specific transcription (43). TAFII250 (CCG1), a cell cycle regulatory protein thought to be important for progression through G1 phase, is one of the TAF subunits of TFIID (36). It binds directly to TBP as well as several other TAFs, including TAFII32 and TAFII70 (5). TAFII250 has also been shown to possess both histone acetyltransferase activity (30) and a protein kinase activity (7). The histone acetyltransferase activity of TAFII250 is conserved in yeasts, flies, and humans and may play an important role in controlling access of the transcription machinery to nucleosome-bound promoter sequences. TAFII250 is a bipartite protein kinase, consisting of N- and C-terminal kinase domains, and directly interacts with and phosphorylates RAP74, the large subunit of TFIIF (7, 35). The association of TAFII250 with IRF-3 and IRF-7 may target histone acetylation to IRF target promoters and allow TFIID to gain access to transcriptionally repressed chromatin.
In virus-infected human and murine cells, IFNA and IFNB genes are coordinately induced, and their individual mRNAs are expressed at different levels. Human IFNA1, IFNA2, and IFNA4 are highly expressed in virus-infected peripheral blood mononuclear and lymphoblastoid Namalwa cells; their respective mRNA levels are 5- to 20-fold higher than those of IFNA5, IFNA7, IFNA8, and IFNA14 in the same cells (15). Since the virus-responsive elements within the IFNA gene promoters contain multiple GAAANN sequences similar to the PRDI and PRDIII domains of IFNB, many studies have shown that the PRDI-like sites of the human IFNA gene promoters are essential for IFNA-induced expression (reviewed in reference 3). In this study, we show that IRF-7 specifically binds to the PRDI site of the IFNB gene promoter. Activation of the IFNA gene promoters by IRF-7 likewise occurs through the binding of IRF-7 to the PRDI-like sites of the IFNA gene promoters, based on the fact that recombinant IRF-7 binds to the PRDI-like sites of the IFNA1, IFNA2, and IFNA14 promoters.
Recent molecular and biological results have also suggested a temporal regulation of IFN gene activation by IRF-3 and IRF-7 (reviewed in reference 26). IFN-α/β genes can be subdivided into two groups: (i) immediate-early genes activated in response to virus infection by a protein synthesis-independent pathway (IFNB and murine IFNA4, which is equivalent to human IFNA1); and (ii) delayed-type genes (which include the other IFNA subtypes), whose expression is dependent on de novo protein synthesis (27). Following virus infection, IRF-3, NF-κB, and ATF-2–c-Jun are posttranslationally activated by inducer-mediated phosphorylation. These proteins cooperate to form a transcriptionally active enhanceosome at the IFNB promoter, together with the CBP transcriptional coactivator and the chromatin-associated high-mobility-group protein (9, 20, 28, 32, 42). IRF-3 also upregulates murine IFNA4 expression (27). Secreted IFN produced from a subset of initially infected cells acts through an autocrine and paracrine loop which requires intact IFN receptors and JAK-STAT pathways. IFN activation of the IFN-stimulated gene factor 3 complex results in the transcriptional upregulation of IRF-7 (27, 37). Virus infection activates IRF-7 through inducible phosphorylation, and phosphorylated IRF-7 participates together with IRF-3 in the transcriptional induction of immediate-early and delayed-type IFN genes (27, 37). In mice with a targeted disruption of either STAT-1, p48, or the IFN-α/β receptor, IRF-7 is not upregulated; hence, the amplification loop of IFN induction is not observed. Finally, the formation of distinct homo- or heterodimers between activated IRF-3 and activated IRF-7 may lead to differential regulation of target IFNA genes (27, 37).
Recently, the crystal structure of the DBDs of IRF-1 and IRF-2 bound to DNA demonstrated that AANNGAAA is the sequence physically recognized by IRF-1 and IRF-2 (8, 10). Our DNA binding site selection demonstrated that DNA sequences recognized by IRF-3 are identical to the IRF-E consensus element G(A)AAANNGAAANN, the consensus sequence for IRF-1 and IRF-2 (41). IRF-7 binds to related sequence elements but with greater flexibility in binding site specificity. From the sequences selected with IRF-3, 12 out of a total 16 sequences contained a tandem repeat of GAAA, while with IRF-7, only 5 out of 28 sequences contained the GAAANNGAAANN consensus motif. Most of the sequences selected with IRF-7 had at least one nucleotide replacement in either the 5′-GAAA or the 3′-GAAA core motif. Furthermore, the oligonucleotides containing two copies of selected IRF-7 binding sites with one nucleotide replacement in either 5′-GAAA (GAATTCGAAAGT) or 3′-GAAA (GAAAGTGAACGC) were able to compete for IRF-7 binding but not for IRF-3 binding to IRF-E. Consistent with their DNA binding activities, reporter constructs carrying two copies of these two binding sites were activated by a constitutively active form of IRF-7 but not by a constitutively active form of IRF-3.
As shown in Table 1, several PRDI-like sequences from different IFNA promoters were also analyzed for IRF-3 and/or IRF-7 transactivation. IRF-7 bound to the PRDI-like motif of the IFNA1, IFNA2, and IFNA14 promoters and activated transcription from the reporter gene promoters containing these motifs. In contrast, IRF-3 bound to the PRDI-like motif of IFNA1 and activated the expression of a reporter gene containing the PRDI-like motif from IFNA1 but did not bind to or activate related sequences from IFNA2 or IFNA14.
The flexibility in the binding site specificity of IRF-7 indicates that more target genes may be recognized by IRF-7 than by IRF-3; for example, genes containing the sequence GAANNGAAANN (interleukin 4, HLA-B7, major histocompatibility complex class I, H-2Dd, immunoglobulin λB, 2′,5′-oligoadenylate synthase, and Mx) or the sequence GAAATGGAAGAG (interleukin 7 receptor) may be preferentially activated by IRF-7 but not by IRF-3. The restricted binding site specificity of IRF-3 is nonetheless consistent with its role as a specific inducer of immediate-early IFN genes. In conclusion, we have demonstrated that despite an overall similarity in structure between IRF-3 and IRF-7, both transcription factors possess unique functional characteristics and share complementary rather than redundant roles in the activation of the IFN-α/β genes.
ACKNOWLEDGMENTS
We thank Paula Pitha, Luwen Zhang, Joseph Pagano, Xiang-Jiao Yang, and Illka Julkunen for reagents used in this study and members of the Molecular Oncology Group, Lady Davis Institute for Medical Research, for helpful discussions.
This research was supported by grants from the Cancer Research Society Inc. and the Medical Research Council of Canada. R.L. was supported in part by a Fraser Monat McPherson fellowship from McGill University, P.G. was supported by an FRSQ postdoctoral fellowship, Y.M. was supported by an MRC studentship, and J.H. was supported by an MRC senior scientist award.
REFERENCES
- 1.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au W C, Moore P A, LaFleur D W, Tombal B, Pitha P M. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 3.Braganca J, Civas A. Type I interferon gene expression: differential expression of IFN-A genes induced by viruses and double-stranded RNA. Biochimie. 1998;80:673–687. doi: 10.1016/s0300-9084(99)80021-2. [DOI] [PubMed] [Google Scholar]
- 4.Burysek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 5.Chen J, Attardi L, Verrijzer P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 6.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 7.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 8.Escalante C R, Yie J, Thanos D, Aggarwal A K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 9.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 10.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel recognition of and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita T, Kimura Y, Miyamoto M, Barsoumian E L, Taniguchi T. Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 12.Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-β gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 14.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:903–913. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 15.Hiscott J, Cantell K, Weissmann C. Differential expression of human interferon genes. Nucleic Acids Res. 1984;12:3727–3746. doi: 10.1093/nar/12.9.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiscott J, Nguyen H, Lin R. Molecular mechanisms of interferon beta gene induction. Semin Virol. 1995;6:161–173. [Google Scholar]
- 17.Hiscott J, Pitha P, Génin P, Nguyen H, Heylbroeck C, Mamane Y, Algarté M, Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 18.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 19.Juang Y T, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factory-3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1998;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 21.Levy D E. Interferon induction of gene expression through the Jak-Stat pathway. Semin Virol. 1995;6:181–190. [Google Scholar]
- 22.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin R, Heylbroeck C, Genin P, Pitha P, Hiscott J. Essential role of IRF-3 in direct activation of RANTES gene transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant M, Lepage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 27.Marie I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to the IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 30.Mizzen C, Yang X, Kokubo T, Brownell J, Bannister A, Owen-Hughes T, Workman J, Wang L, Berger S, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 31.Nonkwello C, Ruf I K, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 33.Pitha P M, Au W-C. Induction of interferon alpha gene expression. Semin Virol. 1995;6:151–159. [Google Scholar]
- 34.Ronco L, Karpova A, Vidal M, Howley P. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruppert S, Tjian R. Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev. 1995;9:2747–2755. doi: 10.1101/gad.9.22.2747. [DOI] [PubMed] [Google Scholar]
- 36.Ruppert S, Wang E, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 37.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 38.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type 1 interferon gene expression by interferon regulatory factor 3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 39.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 40.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka N, Kawakami T, Taniguchi T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 43.Verrijzer C, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 44.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 45.Wathelet M G, Lin C H, Parakh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 46.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneyama M, Suhara W, Fukuhara Y, Fukada M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]