Abstract
Coronavirus disease (COVID-19) is a severe infectious disease that causes respiratory illness and has had devastating medical and economic consequences globally. Therefore, early, and precise diagnosis is critical to control disease progression and management. Compared to the very popular RT-PCR (reverse-transcription polymerase chain reaction) method, chest CT imaging is a more consistent, sensible, and fast approach for identifying and managing infected COVID-19 patients, specifically in the epidemic area. CT images use computational methods to combine 2D X-ray images and transform them into 3D images. One major drawback of CT scans in diagnosing COVID-19 is creating false-negative effects, especially early infection. This article aims to combine novel CT imaging tools and Virtual Reality (VR) technology and generate an automatize system for accurately screening COVID-19 disease and navigating 3D visualizations of medical scenes. The key benefits of this system are a) it offers stereoscopic depth perception, b) give better insights and comprehension into the overall imaging data, c) it allows doctors to visualize the 3D models, manipulate them, study the inside 3D data, and do several kinds of measurements, and finally d) it has the capacity of real-time interactivity and accurately visualizes dynamic 3D volumetric data. The tool provides novel visualizations for medical practitioners to identify and analyze the change in the shape of COVID-19 infectious. The second objective of this work is to generate, the first time, the CT African patient COVID-19 scan datasets containing 224 patients positive for an infection and 70 regular patients CT-scan images. Computer simulations demonstrate that the proposed method’s effectiveness comparing with state-of-the-art baselines methods. The results have also been evaluated with medical professionals. The developed system could be used for medical education professional training and a telehealth VR platform.
Keywords: COrona-VIrus Disease (COVID-19), COVID lesion segmentation, VR visualization, Lesion measurements
1. Introduction
The number of people killed by the coronavirus pandemic continues to grow, where over (112,461) confirmed cases were reported in Algeria by the end of February 2021. When we refer to the testing categories, the polymerize chain reaction (PCR) tests are the most accurate form available today [1]. However, it takes around 48 h before the results are known. On the other hand, CT scan images are preferred by an experienced doctor to detect the infected lesions in the lungs. CT scan benefits comprise less cost, provide valuable data, and wide availability [2].
Image segmentation has proven a fundamental part of many processing pipelines in the biomedical imaging field [3], [4], [5], [6]. However, when using clinical, low-resolution lung data [7], image segmentation can be challenging. E.g., in acute respiratory syndrome (COVID-19). Many of the existing algorithms were developed to deal with different stages for facing the COVID-19 outbreak, as well as segmentation techniques [8]. These methods can be categorized into two main types: Classical approaches [9], [10] and AI-based techniques [11]. Ulhaq et al in [12], show a broad survey of computer vision methods for screening the COVID-19 lesions. Most recently, methods based on Deep learning have been developed [13]; yet, just a few of them are sufficiently mature to present efficient detection and accurate segmentation of the lesion [14]. Furthermore, these methods require a higher: duration of development, a huge amount of data, and they are computationally expensive [15]. Moreover, there is a distinct lack of labeled medical images of COVID-19 lesions [16]. Some examples of image segmentation and classification methods in COVID-19 applications are summarized in Table 1 along with results obtained of each approach. The abbreviations used in this table are as follow: AUC = Area Under the Curve, SEN = Sensetivity, ACC = Accuracy, Dice = Sørensen–Dice Coefficient, SPE = Specificity, ROC = Receiver Operating Characteristic, PC = Precision-Recall, CT-IM-D = CT-Scan images Data.
Table 1.
Related works on COVID-19 segmentation approache.
| Literature | Segmentation approach | CT-IM | Patients | Performance |
|---|---|---|---|---|
| Zhou et al. [17] | U-Net and Focal Tversky | 473 | – | Dice = 83.1%, SEN = 86.7%, SPE = 99.3% |
| Cao et al. [18] | U-Net architecture | – | 2 | – |
| Gozes et al. [19] | Deep Learning CT Image | – | 157 | AUC = 0.99, SPE = 92.2% |
| Qiu et al. [20] | MiniSeg | 100 | – | SPE = 97.4%, Dice = 77.2% |
| Jin et al. [21] | UNet+, CNN | – | 723 | AUC = 0.99, SEN = 0.97, SPE = 0.92 |
| Fei Shan et al. [22] | VB-Net | 249 | 249 | Dice = 91.6% |
| Xie et al. [23] | Contextual two stage U-Net | 204 | – | IOU = 0.91, HD95 = 6.44 |
| Shen et al. [24] | Region growing | – | 44 | R = 0.7679, P < 0.05 |
| Yan et al. [25] | COVID-SegNet | 861 | – | Dice = 72.60%, SEN = 75.10%. |
| Jun et al. [26] | UNet++ | 46,096 | 106 | SPE = 93.5%, ACC = 95.2% |
| Zheng et al. [27] | Pretrained UNet | – | 540 | ROC-AUC = 0.959 |
| Xiaowei et al. [28] | VNET | 618 | 106 | ACC = 86.7% |
| Fan et al. [29] | Semi-Inf-Net | 100 | – | Dice = 73.9%, SEN = 72.5% |
| Cheng et al. [30] | 2D-CNN | 970 | 496 | ACC = 94.9%, AUC = 97.9% |
| Chen et al. [31] | U-Net Res | 110 | – | Dice = 94.0%, ACC = 89.0% |
| Ophir et al. [19] | U-net architecture | – | 56 | AUC = 0.99, SEN = 98.2%, SPE = 92.2% |
| Lin Li [32] | U-Net | 4356 | 3,322 | SEN = 90%, SPE = 96% |
In terms of VR visualization, great progress performances were achieved [33]. Particularly, the entertainment industry (gaming) where great progress was achieved in the last decade, and these developments are expected to radically transform healthcare services [34]. Furthermore, VR technology could improve the ergonomic environments of clinicians in such a way as to improve training process, treatment plan, and support clinical decision [35], [36].
VR could be considered as an effective solution for 3D visualization of medical images since it could provide efficient disease analysis and diagnosis regarding the classic approaches. Virtual Reality (VR) gives an opportunity to immerse users in a fully artificial digital medical environment that involves the human anatomy described in 3D models. Today VR systems overcome classical medical imagery problems with novel 3D imagery visualization techniques [37].
In the ongoing pandemic COVID-19, it has been shown that VR-developed techniques help healthcare-related applications [38], [39]. Another advantage of VR is the possibility to offer valuable learnings for medical students and learners for COVID-19 management situations in hospitals and clinics.
Thus, the primary goal will be to develop an advanced VR visualization platform for automated COVID-19 lesion segmentation and infection measurements, analysis, and diagnosis. In summary, our contributions are:
-
•
An advanced Virtual Reality-based 3D Visualization system for COVID-19 lesion recognition, measurements, and analysis.
-
•
Novel optimized unsupervised segmentation procedure through a new cost function maximization.
-
•
Comparison’s study and validation result by the medical staff of the infected patient by different COVID-19 methods.
-
•
New African COVID-19 patient VR visualization database.
The remainder of the paper is organized as follows. Section 1 reviews the related works related to COVID-19 segmentation. Section 2 outlines the proposed scheme along with a comparison both on objective and subjective perspectives. Meanwhile medical validation is presented in Section 3 and in terms of the VR visualization in Section 4. We conclude by highlighting the outcome achieved (in the last Section 6).
2. Proposed lung segmentation on CT-images
In this section, the proposed COVID-19 lesion segmentation and VR visualization used are presented in detail. CT-scan images considered one of the best sensing approaches since it allows the physicians and radiologists to identify internal structures and see their shape, size, density and texture [40], [41] in a better settings. Our contribution in the context of COVID-19 includes segmentation and VR visualization of COVID-19 lesions so the radiologists will have a clear idea to provide preliminary analysis and interpretation. Fundamentally, digital image processing is an exciting field of research [4], [42], especially in biomedical and medical research [43], [44], [45]. On the other hand, the segmentation process is the approach to detecting and screening images of diseases such as COVID-19 lesions. The image segmentation then is the focal point of the present work, including VR visualization.
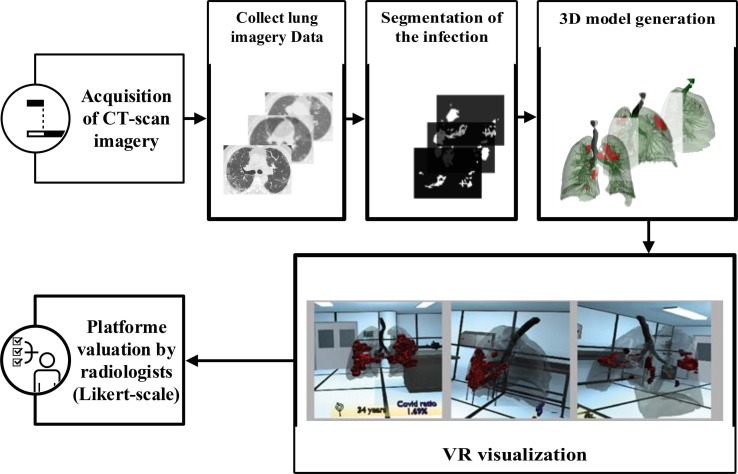
Deciding on COVID-19 infected area in the lung is one of the essential for the correct diagnosis. One issue is to identify which regions are infected and which are healthy precisely. Therefore, radiologists are required to view the 3D extent of the lesion to determine the infected area. Fig. 1 presents the process of 3D visualization of COVID-19 from CT-scan images.
Fig. 1.
The general architecture of our advanced VR COVID-19 visualization process.
Firstly, complete DICOM dataset imagery is collected by a CT-scanner. Secondly, the proposed segmentation approach is applied to segment the lesions of giving CT-scan imagery efficiently. A 3D model is generated where the lungs, bronchi, and COVID lesions are separated; this model will be integrated into the VR platform through Unity 3D software. Finally, six radiologists independently evaluate the quality of the platform using a Likert scale (seven-point scale).
Since CT-images illustrate a low-contrast image and contain some noises. The use of the original CT-images without pre-processing affects the accuracy of image segmentation. Therefore, the original image needs to be: i) denoised with informative preservation; and ii) normalized the local contrast. To deal with this issue, the below equation explains the phenomenon of enhancement adopted.
Let Xi,j be an original image, consisting of L-th luminance levels, i and j represent the pixel location of an image. and Ri,j be the lung regions, containing two luminance levels, Ri,j = {x 0 , x 1 |∀x ⊂ I}
| (1) |
where Fu,v,δ represents a directional filter. α represents the gamma correction of the linear luminance normalization. u, v and δ represent the location of a 3-D filter. u is position in a row, v is position in a column, and δ is position in a level. ⊗ denotes a 2D convolution operator. β represents the gamma correction of the non-linear luminance normalization. α can improve the details under the bright regions while β is very sensitive and impacts to dark regions. log(.) represents a common logarithm to base 10, log 10(.) = log(.). To avoid all ambiguity, it is best to explicitly specify log (·) when the logarithm to base 10 is intended.
2.1. Threshold based local mean condition
The accuracy of the proposed segmentation algorithm depends on a segmentation condition. In this sub-section, we defined the mean for local-region segmentation. It can be used to exit the iterative segmentation process or control the accuracy of the segmentation, simultaneously. The mean of regions will be repeatedly changed when the size of regions also gradually changed. To generalize the threshold-based local mean method, we combined two threshold-based local mean functions as a single function. It can be applied for wide-range segmentation-based applications. It can be described as:
| µ = ωKµK + ωCµC | (2) |
where µK and µC represent the Kapur’s entropy [46] mean value and a cross-mean [47] value, respectively. The function balance two thresholds by using weights, ωK and ωC.
Kapur et al. [46], [47] introduced the single signal-based entropy of segmented classes to generate optimal threshold values. This thresholding technique is extensively improved and illustrates remarkable performance in many image
segmentation problems. First of all, the signal must be separated into two components: a dark component, Yi,j ≤ xt, and a bright component, Yi,j > xt. The local Kapur’s entropy can be calculated as:
| (3) |
| (4) |
where (x) and (x) represent a probability density function of a dark component and a bright component, respectively. and denote a local Kapur’s entropy number of a dark component and a bright component, respectively.
Another local mean is a cross-mean thresholding technique. It was first introduced by Trongtirakul et al. [47] in 2020 for segmenting backlit regions and non-backlit regions. The concept of this technique contains the fundamental of enhancement measure by entropy (EME) method. It optimally separates the entire component into several local components by taking the maximum relationship between the local mean of each component. Also, this technique can be applied for a grayscale-based imaging application like CT-scanned, thermal, night-vision applications including multi-spectral, hyper-spectral, and terahertz imaging applications. The cross-mean thresholding can be calculated as:
| (5) |
| (6) |
Due to the local mean values on a dark component, Yi,j xt, and the local mean value on a bright component, Yi,j > xt, contain different initialized position. In general, the mean of the bright component is higher than another component. Therefore, it needs to be normalized their whole luminance range. written as:
| (7) |
| (8) |
where and represent a region-based local mean of a dark component and a bright component, respectively.
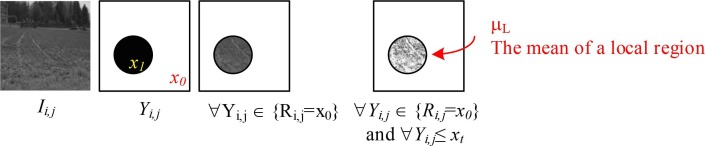
To calculate a region-based local mean, let Yi,j denote the pre-segmented image. X 0 and X 1 represent a binary value of Yi,j. S denotes the size of a local region, Ri,j, and xL − 1 represents the maximum luminance level of an image. The illustrative example and the calculation of region-based local mean can be illustrated in Fig. 2 and written as:
Fig. 2.
Illustrative example of the region-based local mean calculation.
where ωK and ωC represents the weight of cross-entropy [46] threshold-based local mean number and cross-mean [47] threshold-based local mean number, respectively. argmaxn(•) represents the n − largest search operator to return the elements of entropy functions in which the function numbers are maximized. Yi,j represents the pre-segmented image. xt represents a threshold luminance level, starting from t = 2, 3, …, L − 2. M and N represent the size of an image, and xL−1 represents the total number of original luminance levels.
After that, we designed the fast segmentation on luminance levels. Most local segmentation need to be defined the criteria to exit an iterative segmentation process. We used the local mean number as aforementioned in the previous sub-section. Another problem for CT-image segmentation is to avoid small, segmented regions. We set this parameter to control the accuracy of the segmentation process. Occasionally, missing significant parts of the desired region to segment, sometimes due to i) the wrong setting in a CT-scanning machine; or ii) the inappropriate processes of the pre-segmentation, for example, unsuitable filters applied for a denoising process, over-brightness or under-exposure for an image enhancement process. The proposed segmentation process contributes the segmentation performance controlled by two conditions: i) threshold-based local mean; and ii) the minimum pixel number in each luminance level. The proposed segmentation function can be described as:
| Bi,j = { x0, Mi,j ≥ µ, card(Mi,j) > cx1, otherwise | (11) |
where Mi,j represents a morphologically open image. The morphologically open image function, imopen, is also available in MATLAB. µ represents a threshold-based local mean. Card (•) represents a cardinality operator. c represents the minimum pixel number in each luminance level. x 0 and x 1 represent a non-segmented region and a segmented region, e.g., x 0 = 0 and x 1 = 1 illustrate that segmented regions become white and other regions are presented by black. For satisfied segmentation results, we set some parameter in the proposed segmentation algorithm as: α = 2.0, β = 2.0, ωK = 1.0, c = 10.
2.2. Hyper-parameter tuning
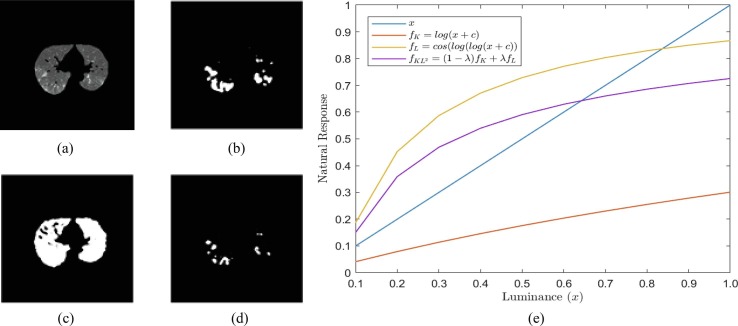
The Otsu method is one of the most well-known methods for automatic image thresholding (segmentation and binarization). It is minimizing the inter-class variance between two foreground and background image components information to choose the threshold value. However, the Otsu has a limitation; for instance, if the class variance is larger, it can miss the weak objects in the image see Fig. 3 (b). Also, it performs purely if the input image has low contrast,
Fig. 3.
Parameter influence of the proposed segmentation method a) Input Image; b) Otsu’s Threshold, xt = 105; c) Kapur’s Entropy, xt = 80, d) KL2-Entropy, xt = 122, λ = 0.75, and e) natural response functions.
salt-and-pepper noise corrupted, or uneven illumination. To solve the problem mentioned above, entropy-based segmentation methods are used. Those existing state-of-the-art entropy-based methods also have some limitations, see Fig. 3. They generate a natural response with a slight difference among various luminance levels. Therefore, these methods, including Kapur’s and Tsaills’ approaches, fail to segment in some cases, as shown in Fig. 3(c-d). The proposed KL2-Entropy method enhances the natural response function on the dark image component (x ≤ 0.5), as shown in Fig. 3. The proposed process generates a notable response when the input image is low-contrast like a CT-scanned image. The (λ) parameter is set up into the equation to control the output segmented regions, as shown in Fig. 3(d).
3. Results
This section compares the proposed segmentation method with the COVID-19 lesion segmentation method on new CT-images from a local hospital containing 224 patients in Algeria who tested positive for the coronavirus. These data are provided with a manual labeled ground truth manually labeled by a medical expert. Then, we show the VR visualization and the effect of the COVID-lesion
on the patient lungs. We pick active contour [49], GraphCut [50], ResNet [51], robust threshold [52], [53], Tunable Weka [54] methods to quantitatively compare the results against the proposed in terms of segmentation quality.
3.1. COVID-SVR dataset
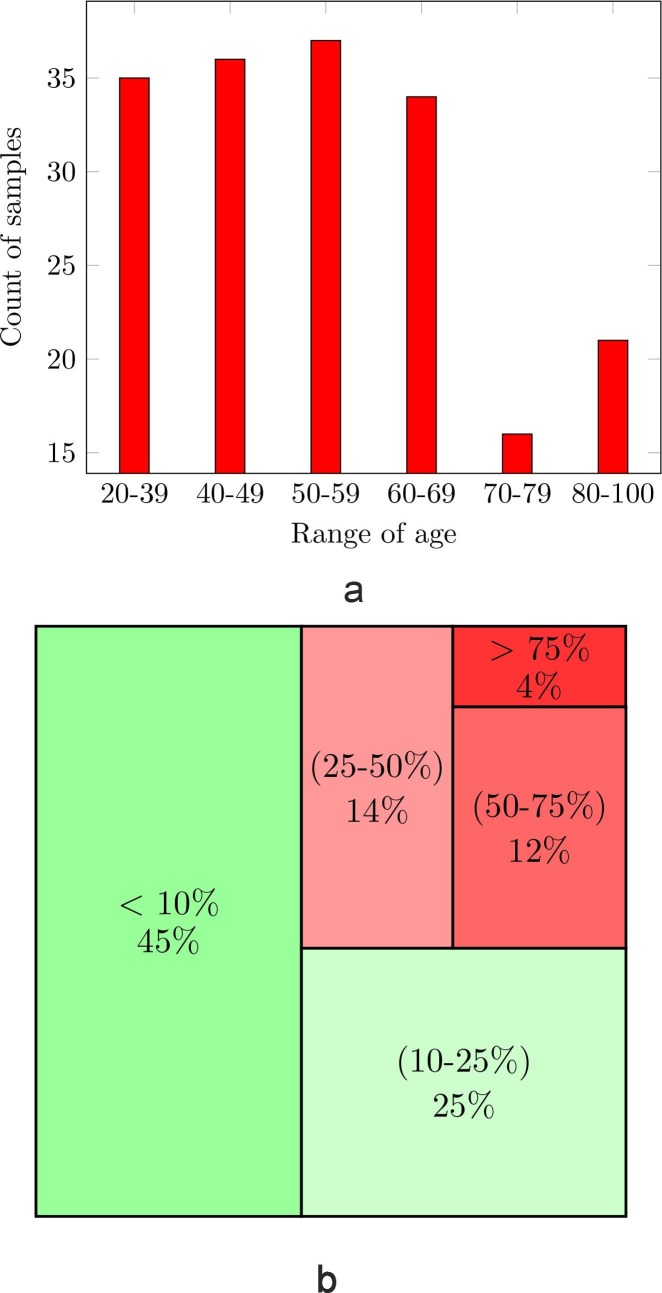
Ct-scan data acts as a vital stage in the VR segmentation and diagnosis platform. Presently, there are many COVID-19 public datasets available but few data from African countries are available. We collect a new COVID-19 segmentation and Virtual Reality (COVID-SVR) dataset to fill in this gap. In this subsection, we introduce the data collection, the professional medical labeling, along with significant statistics of our database as shown in Fig. 4 .
Fig. 4.
COVID-SVR data statistics, (a) The ages; (b) severity level of infection in %
To generate high-quality labeling, we first invited two radiologists to mark as many lesions as possible from infected CT-images based on their clinical experience. As can be seen in the second row of Fig. 4 the COVID-SVR data covers different levels of cases.
3.2. Visual evaluation
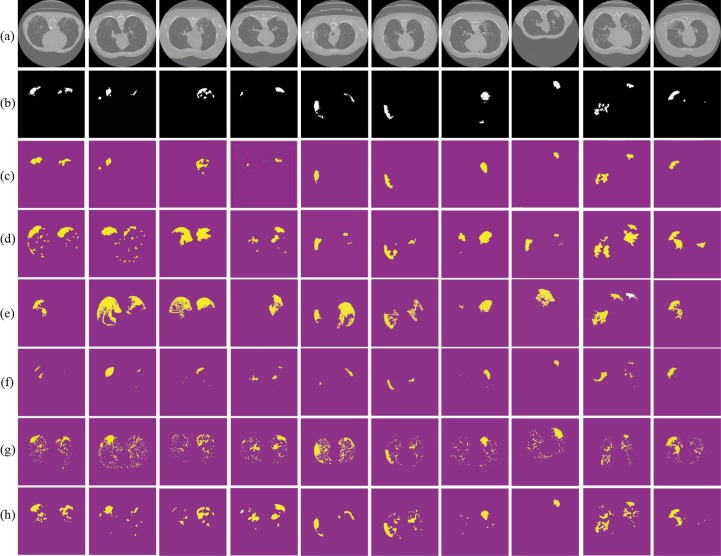
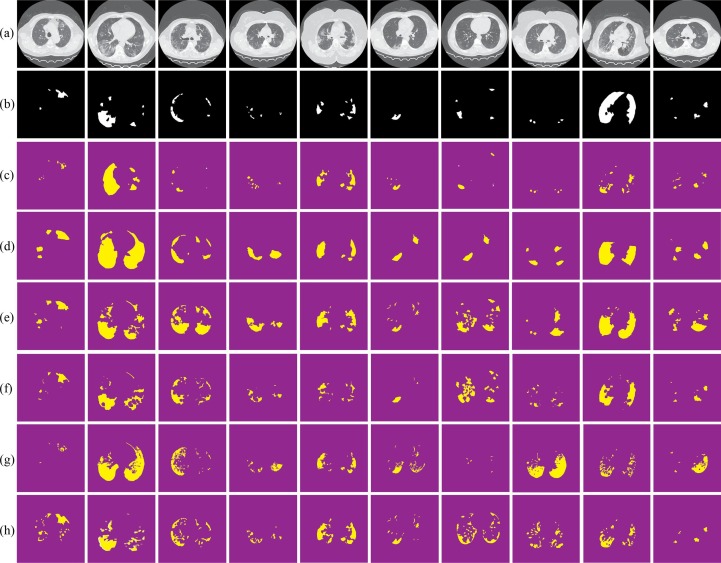
Fig. 6 summarizes the usefulness of the proposed COVID-19 approach. Regardless of the size or quality of the input CT scan image, by enhancing the lungs tissue following by applying our segmentation, it is proven visually (see Fig. 6(c) (third row)) that the proposed provides accurate extraction of the COVID lesion comparing to state-of-the-art segmentation methods (Fig. 6(d–h) (forth-to-eighth).
Fig. 6.
Visual comparisons of different methods using MosMedData against our segmentation test set (c); (a) original images; (b) ground truths; (d) Active contour; (e) GraphCut; (f) ResNet; (g) Robust Threshold and (h) tunable Weka.
3.3. Statistical evaluation metrics
The metrics used to measure the segmentation performance include the Dice coefficient (SDC), Intersection-Over-Union (Jaccard Index), and Hausdorff metric. The reason for choosing the Hausdorff metrics is: (i) it measures the distance between sets of points, (ii) it commonly used metrics over geometric objects, (iii) it operates as a measure of dissimilarity between binary images, for which it is well suited both for theoretical and for intuitive reasons, (iv) it gives a good idea of the difference in the optical impressions a human would get from two images, and (v) it uses extensively in computer vision, pattern recognition, medicine, and computational chemistry [56]. Mathematically, the Hausdorff (HD) [57] metric is defined as the similarity between two sets A and B.
| (12) |
The justification for choosing the Jaccard index (J) is: it measures the similarity between sets of patterns by converting each pattern into a single element within the set; it is conceptually and computationally simple, it produces easily interpretable results, and it is an appropriate measure in a wide variety of domain [57]. Mathematically, the Jaccard Index (the similarity between two sets A and B) is defined as the intersection (∩) size divided by the size of the union (∪) of the two sets A and B: where we say J(A, B) = 1 if |A ∪ B| = 0
| (13) |
Dice Index is a similarity measure related to the Jaccard index [59]. It defined as:
| (14) |
Dice Coefficient. The dice similarity coefficient is a measure of positive overlap
between two binary images. It defines as:
| DC = 2·TP 2·TP + FP + FN | (15) |
Where TP is the number of actual positive pixels in the images, FP is the number of false-positive pixels, and FN is the number of false negatives. It is a similarity measure ranging from zero to one–one meaning the two images are identical. The primary benefit of the is that it does not depend on actual negative values. Mean IoU measure: it calculates the ratio of overlapping area between two images with the intersected area subtracted from their union as shown in:
4. meanIoU
| A,B = |A ∩ B| |A| + |B| − |A ∩ B| | (16) |
A perfect segmentation yields an SDC and J score of 1. On the flip side, the Hausdorff metric measures the degree of mismatch between two sets by measuring the distance of the point of A that is farthest from any point of B and vice versa. Small values of H indicate better segmentation, (h) is the distance between point A and the closest point in set B. The resultant segmentation for a set of samples COVID images is shown in Table 2 . From the mean values listed in the aforementioned table, it has been seen that the means of SDC and Jaccard metrics are significantly higher using the proposed, except in terms of SDC where tunable Weka competes (see Table 3 ).
Table 2.
Statistical segmentation quality measurement by using COVID-SVR Data of the proposed against active contour [49], GraphCut [50], ResNet [51], Gated-UNet [60], Dense UNet [61], Robust Threshold [52], [53], and Tunable Weka [54]. The best results are shown in red.
| Segmentation methods | Quality measurements |
||
|---|---|---|---|
| Dice (SDC) ↑ | Jaccard ↑ | Hausdorff ↓ | |
| Proposed | 0.49 | 0.46 | 5.62 |
| Active Contour [49] | 0.30 | 0.32 | 17.77 |
| GraphCut [50] | 0.29 | 0.29 | 18.64 |
| ResNet [51] | 0.33 | 0.31 | 27 |
| Robust Threshold [52], [53] | 0.28 | 0.34 | 15.81 |
| Gated-UNet [60] | 0.44 | – | – |
| Dense-UNet [61] | 0.41 | – | – |
| Tunable Weka [54] | 0.51 | 0.43 | 9.68 |
Table 3.
Statistical segmentation quality measurement by using MosMedData[63] of the proposed against to state-of-the-art segmentation methods. The best results are shown in red.
| Methods | Dice (SDC) ↑ | Jaccard ↑ | Hausdorff |
|---|---|---|---|
| Proposed | 0.74 | 0.61 | 1.62 |
| Active Contour [49] | 0.32 | 0.29 | 3.23 |
| GraphCut [50] | 0.36 | 0.39 | 3.62 |
| ResNet [51] | 0.43 | 0.41 | 3 |
| Robust Threshold [52], [53] | 0.53 | 0.44 | 2.1 |
| Gated-UNet [60] | 0.44 | – | – |
| Dense-UNet [61] | 0.41 | – | – |
| Tunable Weka [54] | 0.54 | 0.53 | 3.68 |
| BiSe-Net[62] | 0.7 | – | – |
| A-COVID LIRS&M [8] | 0.71 | 0.57 | – |
Visual assessments for the lung segmentation are given in Fig. 5 and Fig. 6 respectively on the COVID-SVR and MosMedData. These examples illustrate those segmented regions using state-of-the-art methods make different mistakes. Furthermore, some areas are wrongly predicted to be a lesion or healthy tissue by most of the approaches or even all of them. Concerning clinical practice, the near to ground truth segmentation of lesion is the most important that showcases the severity of the disease. Thus, from the qualitative results provided by the proposed (see Fig. 5, Fig. 6) we notice that the propose (Figs. 5 and 6c) provide a near segmentation to ground truth.
Fig. 5.
Visual comparisons of different methods using COVID-SVR Dataset against our segmentation method (c); (a) original images; (b) ground truths; (d) Active contour; (e) GraphCut; (f) ResNet; (g) Robust Threshold and (h) tunable Weka.
Virtual Reality was considered with CT-scan imaging to provide a comprehensive display of lungs with COVID-19 detailed lesions. The results obtained from the segmentation of CT-scan images are used as processed data to generate a 3D COVID-19 lesions visualization system. Although our segmentation method provides a reasonable segmentation, the method proposed is less time-consuming and doesn’t require a high-performance graphical card. This makes
it possible to perform segmentation and 3D visualization faster. Our approach could be suitable for practical situations. Radiologists, in hospitals and clinics, should have the results as quickly as possible in order to deal with a high number of patients, provide adapted treatment, facilitate management of disease and reduce coma situation.
5. VR system design
In this section, we dealt with improving the medical diagnostic proposed method. For that purpose, we provide for radiologists an immersive and interactive VR application to visualize and interact with 3D infected COVID-19 lungs of actual patients. To measure the proposed COVID-based VR application performance, we conducted experiments with doctors and radiologists on a set of patients with COVID-19, including those with mild, moderate, severe, and critical diseases.
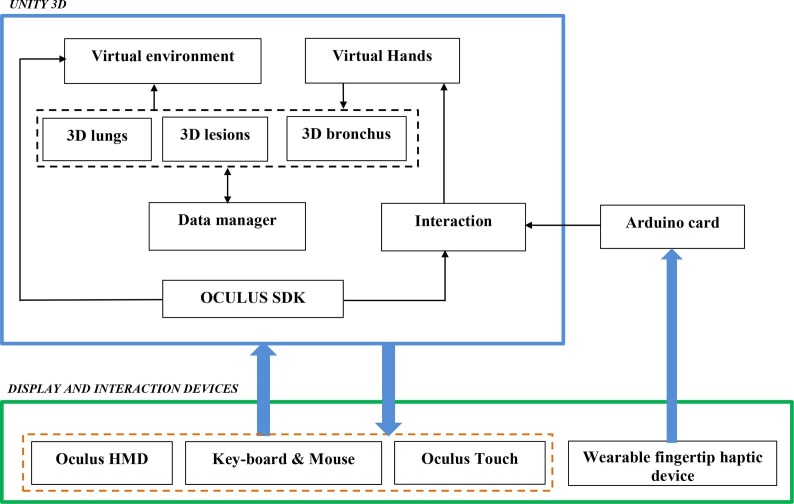
Fig. 7 shows the general diagram developed in the Unity game engine, showing the components of the VR developed application. The software part was considered by Unity 3D. In this case, we integrated seven packages: the virtual environment package which integrated optional 3D objects of the scene such as walls, tables, lamps, paintings, etc., three packages for 3D lungs design (3D lung models, 3D bronchus models, and 3D covid-lesion models), interaction package for human–computer interaction management, data manager package for data exchange between packages and 3D scene updates.
Fig. 7.
Packages of the VR application.
The hardware part is composed of Oculus Rift S Head Mounted Display (HMD) (version updated of Oculus Rift [56]) connected to a computer monitor. It integrates sensors to recognize the user’s head movements in the space. Oculus Touch, mouse, and keyboard, manage the 3D interaction between the user and the VR application. The user manipulates can manipulate the 3D infected lungs and go inside the lungs to view the 3D COVID-lesion in more detail (which is possible with 2D CT-scan images). Oculus SDK [64] supports 3D visualization and Hand interaction management.
6. VR application results
We developed a VR application that allows 3D data generation, visualization, and manipulation from medical imaging resources (in our case, we used scanner images). In fact, a DICOM imagery stack was converted to [obj] file and/or [slt]. In our configuration, we used Blender software framework [65] to import [obj] files and generates FBX format [66] directly used to provide 3 lung visualizations.
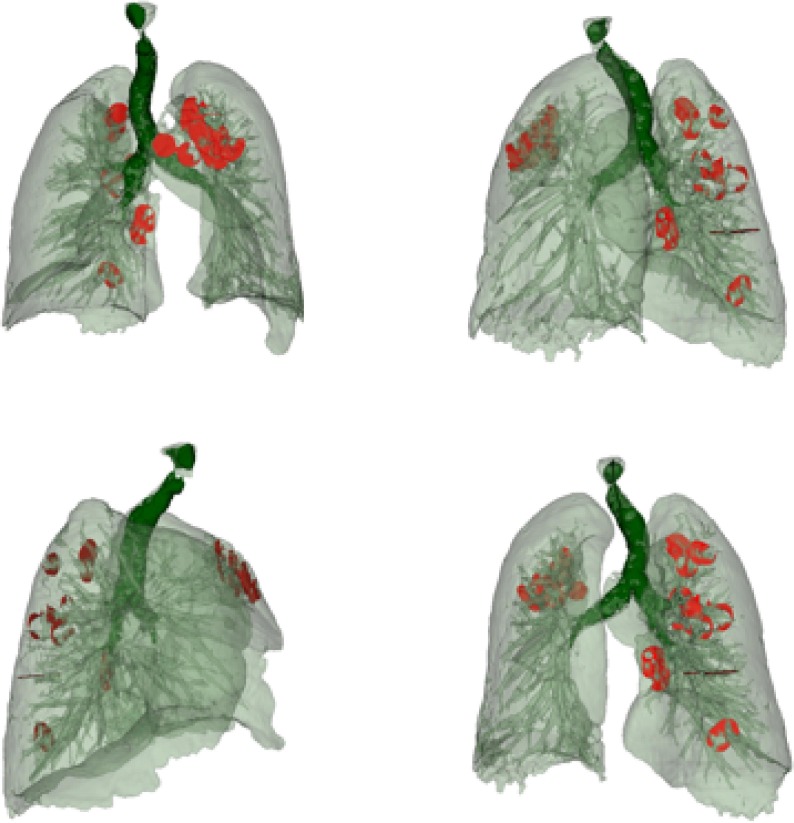
Fig. 8 shows 3D images obtained using a 3D slicer, with a segmented area of the bronchi, lung with veins, and COVID lesions. The color map that transforms the intensities of the RGB scale into color was manually adjusted so that bronchi, veins, and lesion that contains different views of the object. We used the Unity 3D game engine [67] to import FBX formats and design VR infected lungs where COVID-19 lesions are well-segmented, and 3D described as shown in Fig. 8. Table 4 presents a comparison of the statistics of the progression and growth of the COVID lesion over time of the same person. From the fourth row, we can see that the SDC coefficient is equal to (0.057). This value SDC proves that the lesion’s severity is proportional to the amount of lung tissue destruction, and the lesion grows drastically in 10 days only (as shown in the third row). The lesion’s effect is also higher since the volume (in cc) of the lesion comparison in voxels in CT scans is on the order of (4289.33).
Fig. 8.
VR volume visualization of the automatic segmentation for lung with blood vessels along with COVID-19 lesion.
Table 4.
Comparison and calculation the volume of the lungs and lesion of the effect of COVID-19 on a patient who tested twice by COVID-19
| Segmentation statistics | Number of Voxels |
||
|---|---|---|---|
| Lung | Lesion | ||
| Day 1 | 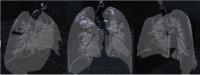 |
8,010,730 | 16,077 |
| Day 10 | 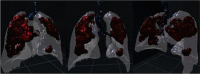 |
6,621,017 | 1,306,384 |
| Comparison | |||
| SDC Coefficient | 0.057 | ||
| Compare volume (cc) | 4289.33 | ||
7. Participants
A medical staff composed of six (06) participants (radiologists, doctors, students, and collaborators) volunteered to explore our COVID-19 VR Visualization system and provide their opinions through user experience in a subjective survey. The latter was elaborated with VR experts to measure doctors’ interaction with 3D realistic infected lungs in immersed virtual environments, representing an instant volume-rendered segmented data. In our case, we used Oculus Rift S Head Mounted Display (HMD), to track the user’s head and hands movement. This experimentation could be performed using tablets and smartphones as well.
8. Procedure
The medical staff wears the HMD and visualizes the 3D infected lungs of patients for seven minutes but can be extended if needed. Immersion time was
recorded. Then, no training, nor other explanation, for the VR-naive examiner was performed. After the experience, participants were asked to complete a subjective questionnaire without being disturbed or influenced.
Fig. 9 shows a user using the VR visualization system, which depicts the volume-rendered COVID-19 lesions inside 3D lungs in a virtual room. Furthermore, we provided a VR background surrounding the 3D lung so that participants could be familiar with the VR application.
Fig. 9.
VR visualization of COVID-19 lesions inside 3D lungs.
In the evaluation part, subjects were asked to compare VR infected lung models with scanner images to identify the lesion properties (volume, extension, distribution, location). After the completion of trials, the subjects were asked to answer a short survey about their feelings and fill a questionnaire rated on a 7-point Likert scale.
9. Analysis
Participants were asked to state to the extent to which they agreed or disagreed (a 7-point scale) with 09 statements related to the VR diagnostic COVID-19 platform. The data were also analyzed to explore whether medical staff volunteers who were new to VR experiences had a high comfort score. In this case, it is possible to study the usefulness of the VR application regarding radiologists in their daily work and in extreme conditions. Finally, participants were asked to state what they believed to be the best features. The parameters to evaluate for the experimentation are (parameters from 5 to 9 inspired from [68]). Meanwhile, parameters from 1 to 4 were proposed by authors:
-
1.
Immersion is perceived.
-
2.
Lesion is well observed.
-
3.
Easy to use.
-
4.
Could be used for another lesion detection.
-
5.
Improve knowledge of disease.
-
6.
An enjoyable experience.
-
7.
Could help reduce error.
-
8.
Enhance understanding.
-
9.
Provide a realistic view of clinical case.
10. Discussion
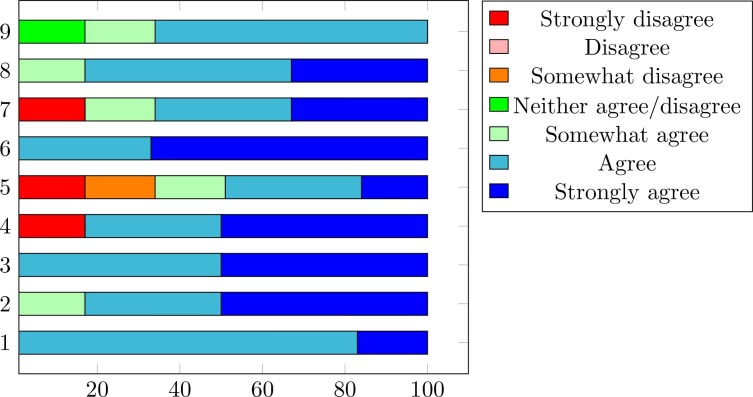
According to question 1, 83% of participants felt Immersed in the virtual world when they firstly manipulate the 3D lung. Among them, one participant was surprised and provided a strongly agreed opinion. More than 80% of experimenters found the 3D COVID-19 lesion clear, visible, and realistic regarding the second question. Simultaneously, all participants found the system easy to use (Question 3). In question 4, 17% of experimenters judged that the application could not be used for another disease detection, where others estimate that the application could be suitable to be applied for other forms of pathology diagnosis. In terms of question 5, 66% believed that 3D technology improves disease knowledge and provides appropriate medical treatment. Similarly, most participants denote those diagnostic errors (Question 6) could be reduced significantly, they also notice that they perceive a realistic 3D view of the COVID-19 lesion and its volume and distribution within the lung (Question 8). Finally, all participants found these experiences enjoyable and improve their understanding of the disease. Moreover, they wish to repeat this experience if they are asked again. Besides, they express their ability to recommend these applications to their colleagues. The summary of the agreement level provided by doctors is illustrated in Fig. 10 .
Fig. 10.
Agreement level provided by medical stuff using 9 statements.
11. Conclusions
The accurate visualization and segmentation of the COVID-19 lesion is a significant stage to help the radiologist in the interpretation of the lesion in a faster way. In this paper, we designed and evaluated an automatic tool for automatic COVID-19 lung infection segmentation and VR visualization using chest CT-scan images. The computer simulations using the MosMedData and COVID-SVR dataset containing (2 2 4) collected from EL-BAYANI center for radiology show better efficiency comparing to the state-of-the-art segmentation approaches, including Active contour, GraphCut, ResNet, Robust threshold, Gated-UNet, Dense-UNet, tunable Weka, BiSe-Net, and A-COVID LIRSM. The proposed algorithm’s performance is measured using the following assessment scores: SDC, Jacquard index, and Hausdorff distance. These contributions prove the prospect of improving diagnosis and visualization for COVID-19 and could support the medical professional to evaluate a possible lesion and abnormalities inside the lungs. After that, the cutting-edge technology using VR is performed to develop a platform of COVID-19 VR viewer, where it was
experimented with by medical staff. As different radiologists with doctors conclude after using our system, the visualization of CT-scan images using Virtual Reality provides a better interpretation of the radiological results and can be a breakthrough for medical treatment planning. According to participants’ opinions, the proposed application provides a more detailed and more realistic view of the COVID-19 lesion’s location, volume evolution, and distribution within the lung. According to a doctor taking part in the trials, the drawback of the proposed approach is that it only gives credible results when the doctor has preliminary information that patients got COVID-19. The doctor said that a lung could have various complex pathologies that differ from the COVID-19 disease. Suppose we provide this application to a doctor without knowing that COVID-19 affected the concerned patient. In that case, he cannot identify the type of pathology even if he uses our VR visualization tool. A medical staff composed of six (06) participants volunteered to explore our COVID-19 VR Visualization system and provide their opinions through a subjective user experience survey. We believe that medical professionals could use this tool to visualize radiological results.
Funding
This research was funded by the national research fund (DGRSDT/MESRS) of Algeria and the Centre de Développement des Technologies Avancées (CDTA).
Author Contributions
S.A. and A.O. conceived the project; methodology, A.O. and T.T; validation, S.B. and A.O, N.Z. formal analysis; investigation, S.A.; data creation, A.O. and M-L.A; Data analysis and preparation, A.O.; writing—review and editing, S.A.; visualization, D.A.; project administration, N.Z.;. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank the staff of Centre Médico-Social (CMS-CDTA) Amira Hamou, Ait Saada Kamal, and Dr. Yeddou to perform the data annotation task which is a costly and time-consuming process. Moussa Abdou for all his hard work and assistance in video editing. Last, we would like to pay our gratitude and our respects to Prof. Ghouti MERAD died after a battle with COVID-19 who, although no longer with us who always believed in our ability to be successful in the academic arena.
References
- 1.Zitek T. The appropriate use of testing for covid-19. West. J. Emerg. Med. 2020;21(3):470. doi: 10.5811/westjem.2020.4.47370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diwakar M., Kumar M. A review on ct image noise and its denoising. Biomed. Signal Process. Control. 2018;42:73–88. [Google Scholar]
- 3.Trongtirakul T., Oulefki A., Agaian S., Chiracharit W. Mobile Multimedia/Image Processing, Security, and Applications 2020. International Society for Optics and Photonics; 2020. Enhancement and segmentation of breast thermograms. 113990F. [Google Scholar]
- 4.Oulefki A., Trongtirakul T., Agaian S., Chiracharit W. Mobile Multimedia/Image Processing, Security, and Applications 2020. International Society for Optics and Photonics; 2020. Detection and visualization of oil spill using thermal images. 113990L. [Google Scholar]
- 5.Pandey S., Singh P.R., Tian J. An image augmentation approach using two-stage generative adversarial network for nuclei image segmentation. Biomed. Signal Process. Control. 2020;57 [Google Scholar]
- 6.Agaian S., Madhukar M., Chronopoulos A.T. Automated screening system for acute myelogenous leukemia detection in blood microscopic images. IEEE Syst. J. 2014;8(3):995–1004. [Google Scholar]
- 7.Zhao C., Wang Z., Li H., Wu X., Qiao S., Sun J. A new approach for medical image enhancement based on luminance-level modulation and gradient modulation. Biomed. Signal Process. Control. 2019;48:189–196. [Google Scholar]
- 8.Oulefki A., Agaian S., Trongtirakul T., Kassah Laouar A. Automatic covid-19 lung infected region segmentation and measurement using ct-scans images. Pattern Recogn. 2021;114:107747. doi: 10.1016/j.patcog.2020.107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayesh M., Mohammad K., Qaroush A., Agaian S., Washha M. A robust line segmentation algorithm for arabic printed text with diacritics. Electron. Imaging. 2017;2017(13):42–47. [Google Scholar]
- 10.Almuntashri A., Finol E., Agaian S. 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC) IEEE; 2012. Automatic lumen segmentation in ct and pc-mr images of abdominal aortic aneurysm; pp. 2891–2896. [Google Scholar]
- 11.Civit-Masot J., Luna-Perejón F., Domínguez Morales M., Civit A. Deep learning system for covid-19 diagnosis aid using x-ray pulmonary images. Appl. Sci. 2020;10(13):4640. [Google Scholar]
- 12.Ulhaq A., Born J., Khan A., Gomes D.P.S., Chakraborty S., Paul M. Covid-19 control by computer vision approaches: a survey. IEEE Access. 2020;8:179437–179456. doi: 10.1109/ACCESS.2020.3027685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. Bullock, K. H. Pham, C. S. N. Lam, M. Luengo-Oroz, et al., Mapping the landscape of artificial intelligence applications against covid-19, arXiv preprint arXiv:2003.11336.
- 14.L. Wang, A. Wong, Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images, arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- 15.Rajab M.I., Woolfson M.S., Morgan S.P. Application of region-based segmentation and neural network edge detection to skin lesions. Comput. Med. Imaging Graph. 2004;28(1-2):61–68. doi: 10.1016/s0895-6111(03)00054-5. [DOI] [PubMed] [Google Scholar]
- 16.F. Shi, J. Wang, J. Shi, Z. Wu, Q. Wang, Z. Tang, K. He, Y. Shi, D. Shen, Review of artificial intelligence techniques in imaging data acquisition, seg-mentation and diagnosis for covid-19, IEEE reviews in biomedical engineering. [DOI] [PubMed]
- 17.T. Zhou, S. Canu, S. Ruan, An automatic covid-19 ct segmentation network using spatial and channel attention mechanism, arXiv preprint arXiv:2004.06673. [DOI] [PMC free article] [PubMed]
- 18.Cao Y., Xu Z., Feng J., Jin C., Han X., Wu H., Shi H. Longitudinal assessment of covid-19 using a deep learning–based quantitative ct pipeline: Illustration of two cases. Radiol.: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200082. e200082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O. Gozes, M. Frid-Adar, H. Greenspan, P. D. Browning, H. Zhang, W. Ji, A. Bernheim, E. Siegel, Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis, arXiv preprint arXiv:2003.05037.
- 20.Y. Qiu, Y. Liu, J. Xu, Miniseg: An extremely minimum network for efficient covid-19 segmentation, arXiv preprint arXiv:2004.09750. [DOI] [PubMed]
- 21.S. Jin, B. Wang, H. Xu, C. Luo, L. Wei, W. Zhao, X. Hou, W. Ma, Z. Xu, Z. Zheng, et al., Ai-assisted ct imaging analysis for covid-19 screening: Building and deploying a medical ai system in four weeks, MedRxiv. [DOI] [PMC free article] [PubMed]
- 22.F. Shan, Y. Gao, J. Wang, W. Shi, N. Shi, M. Han, Z. Xue, Y. Shi, Lung infection quantification of covid-19 in ct images with deep learning, arXiv preprint arXiv:2003.04655.
- 23.W. Xie, C. Jacobs, J.-P. Charbonnier, B. van Ginneken, Contextual two- stage u-nets for robust pulmonary lobe segmentation in ct scans of covid-19 and copd patients, arXiv preprint arXiv:2004.07443.
- 24.Shen C., Yu N., Cai S., Zhou J., Sheng J., Liu K., Zhou H., Guo Y., Niu G. Quantitative computed tomography analysis for stratifying the severity of coronavirus disease 2019. J. Pharm. Anal. 2020;10(2):123–129. doi: 10.1016/j.jpha.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Q. Yan, B. Wang, D. Gong, C. Luo, W. Zhao, J. Shen, Q. Shi, S. Jin, L. Zhang, Z. You, Covid-19 chest ct image segmentation–a deep convolutional neural network solution, arXiv preprint arXiv:2004.10987.
- 26.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., Hu S., Wang Y., Hu X., Zheng B., Zhang K., Wu H., Dong Z., Xu Y., Zhu Y., Chen X.i., Zhang M., Yu L., Cheng F., Yu H. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speidel M.A., Wilfley B.P., Star-Lack J.M., Heanue J.A., Van Lysel M.S. Scanning-beam digital x-ray (sbdx) technology for interventional and diagnostic cardiac angiography. Med. Phys. 2006;33(8):2714–2727. doi: 10.1118/1.2208736. [DOI] [PubMed] [Google Scholar]
- 28.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., Lang G., Li Y., Zhao H., Liu J., Xu K., Ruan L., Sheng J., Qiu Y., Wu W., Liang T., Li L. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 30.C. Jin, W. Chen, Y. Cao, Z. Xu, X. Zhang, L. Deng, C. Zheng, J. Zhou, H. Shi, J. Feng, Development and evaluation of an ai system for covid-19 diagnosis. medrxiv 2020, preprint https://doi. org/10.1101/2020.03.20.20039834. [DOI] [PMC free article] [PubMed]
- 31.X. Chen, L. Yao, Y. Zhang, Residual attention u-net for automated multi-class segmentation of covid-19 chest ct images, arXiv preprint arXiv:2004.05645.
- 32.L. Li, L. Qin, Z. Xu, Y. Yin, X. Wang, B. Kong, J. Bai, Y. Lu, Z. Fang, Q. Song, et al., Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct, Radiology. [DOI] [PMC free article] [PubMed]
- 33.González Izard S., Sánchez Torres R., Alonso Plaza Ó., Juanes Méndez J.A., García-Peñalvo F.J. Nextmed: Automatic imaging segmentation, 3d reconstruction, and 3d model visualization platform using augmented and virtual reality. Sensors. 2020;20(10):2962. doi: 10.3390/s20102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.W. Greenleaf, How vr technology will transform healthcare, in: ACM SIGGRAPH 2016 VR Village, 2016, pp. 1–2.
- 35.W. S. Khor, B. Baker, K. Amin, A. Chan, K. Patel, J. Wong, Augmented and virtual reality in surgery—the digital surgical environment: applications, limitations and legal pitfalls, Ann. Transl. Med. 4 (23). [DOI] [PMC free article] [PubMed]
- 36.Østergaard M.L., Konge L., Kahr N., Albrecht-Beste E., Nielsen M.B., Nielsen K.R. Four virtual-reality simulators for diagnostic abdominal ultrasound training in radiology. Diagnostics. 2019;9(2):50. doi: 10.3390/diagnostics9020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D. Aouam, N. Zenati-Henda, S. Benbelkacem, C. Hamitouche, An interactive vr system for anatomy training, in: Mixed Reality and Three-Dimensional Computer Graphics, IntechOpen, 2020.
- 38.Ueda M., Martins R., Hendrie P.C., McDonnell T., Crews J.R., Wong T.L., McCreery B., Jagels B., Crane A., Byrd D.R., et al. Managing cancer care during the covid-19 pandemic: agility and collaboration toward a common goal. J. Natl. Compr. Canc. Netw. 2020;18(4):366–369. doi: 10.6004/jnccn.2020.7560. [DOI] [PubMed] [Google Scholar]
- 39.Haleem A., Javaid M., Vaishya R., Deshmukh S.G. Areas of academic research with the impact of covid-19. Am. J. Emerg. Medicine. 2020;38(7):1524–1526. doi: 10.1016/j.ajem.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva F., Pereira T., Frade J., Mendes J., Freitas C., Hespanhol V., Costa J.L., Cunha A., Oliveira H.P. Pre-training autoencoder for lung nodule malignancy assessment using ct images. Appl. Sci. 2020;10(21):7837. [Google Scholar]
- 41.Li C., Chen W., Tan Y. Point-sampling method based on 3d u-net architecture to reduce the influence of false positive and solve boundary blur problem in 3d ct image segmentation. Appl. Sci. 2020;10(19):6838. [Google Scholar]
- 42.Oulefki A., Aouache M., Bengherabi M. Iberian Conference on Pattern Recognition and Image Analysis. Springer; 2019. Low-light face image enhancement based on dynamic face part selection; pp. 86–97. [Google Scholar]
- 43.Artzi M., Aizenstein O., Jonas-Kimchi T., Myers V., Hallevi H., Bashat D.B. Flair lesion segmentation: application in patients with brain tu mors and acute ischemic stroke. Eur. J. Radiol. 2013;82(9):1512–1518. doi: 10.1016/j.ejrad.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 44.Chen C.L.P., Li H., Wei Y., Xia T., Tang Y.Y. A local contrast method for small infrared target detection. IEEE Trans. Geosci. Remote Sens. 2014;52(1):574–581. [Google Scholar]
- 45.Trongtirakul T., Chiracharit W., Imberman S., Agaian S. Fractional contrast stretching for image enhancement of aerial and satellite images. Electron. Imaging. 2020;2020(10):60411-1–60411-11. [Google Scholar]
- 46.Kapur T., Grimson W.E.L., Wells W.M., III, Kikinis R. Segmentation of brain tissue from magnetic resonance images. Med. Image Anal. 1996;1(2):109–127. doi: 10.1016/S1361-8415(96)80008-9. [DOI] [PubMed] [Google Scholar]
- 47.Trongtirakul T., Chiracharit W., Agaian S.S. Single backlit image enhancement. IEEE Access. 2020;8:71940–71950. [Google Scholar]
- 49.Zhang K., Zhang L., Song H., Zhou W. Active contours with selective local or global segmentation: a new formulation and level set method. Image Vis. Comput. 2010;28(4):668–676. [Google Scholar]
- 50.Boykov Y., Jolly M.-P. International Conference on Medical Image Computing and Computer-assisted Intervention. Springer; 2000. Interactive organ segmentation using graph cuts; pp. 276–286. [Google Scholar]
- 51.Xia K.-J., Yin H.-S., Zhang Y.-D. Deep semantic segmentation of kidney and space-occupying lesion area based on scnn and resnet models combined with sift-flow algorithm. J. Med. Syst. 2019;43(1):2. doi: 10.1007/s10916-018-1116-1. [DOI] [PubMed] [Google Scholar]
- 52.Li Z., Liu G., Zhang D., Xu Y. Robust single-object image segmentation based on salient transition region. Pattern Recogn. 2016;52:317–331. [Google Scholar]
- 53.Stauffer W., Sheng H., Lim H.N. Ezcolocalization: an imagej plugin for visualizing and measuring colocalization in cells and organisms. Sci. Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-33592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K.W., Schindelin J., Cardona A., Sebastian Seung H. Trainable weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33(15):2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 56.Schuhmacher D., Vo B.-T., Vo B.-N. A consistent metric for performance evaluation of multi-object filters. IEEE Trans. Signal Process. 2008;56(8):3447–3457. [Google Scholar]
- 57.Anastasopoulos C., Weikert T., Yang S., Abdulkadir A., Schmülling L., Bühler F., Paciolla R., Sexauer J., Cyriac I.N., et al. Development and clinical implementation of tailored image analysis tools for covid-19 in the midst of the pandemic: the synergetic effect of an open, clinically embedded software development platform and machine learning. Eur. J. Radiol. 2020;131 doi: 10.1016/j.ejrad.2020.109233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakagawa H., Nagatani Y., Takahashi M., Ogawa E., Van Tho N., Ryujin Y., Nagao T., Nakano Y. Quantitative ct analysis of honeycombing area in idiopathic pulmonary fibrosis: correlations with pulmonary function tests. Eur. J. Radiol. 2016;85(1):125–130. doi: 10.1016/j.ejrad.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 60.Schlemper J., Oktay O., Schaap M., Heinrich M., Kainz B., Glocker B. Rueckert, Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 2019;53:197–207. doi: 10.1016/j.media.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X., Chen H., Qi X., Dou Q., Fu C.-W., Heng P.-A. H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes. IEEE Trans. Med. Imaging. 2018;37(12):2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 62.C. Yu, J. Wang, C. Peng, C. Gao, G. Yu, N. Sang, Bisenet: Bilateral segmentation network for real-time semantic segmentation, in: Proceedings of the European conference on computer vision (ECCV), 2018, pp. 325–341.
- 63.Morozov S.P., Andreychenko A.E., Blokhin I.A., Gelezhe P.B., Gonchar A.P., Nikolaev A.E., Pavlov N.A., Chernina V.Y., Gombolevskiy V.A. Mosmeddata: data set of 1110 chest ct scans performed during the covid-19 epidemic. Digital Diagn. 2020;1(1):49–59. [Google Scholar]
- 64.<https://developer.oculus.com/downloads/package/oculus-platform-sdk/>.
- 65.Flaischlen S., Wehinger G.D. Synthetic packed-bed generation for cfd simulations: Blender vs. star-ccm+ ChemEngineering. 2019;3(2):52. [Google Scholar]
- 66.Hoffman M., Provance J. AMIA Summits on Translational Science Proceedings. 2017. Visualization of molecular structures using hololens-based augmented reality; p. 68. [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S.L., Suk H.J., Kang J.H., Jung J.M., Laine T.H., Westlin J. 2014 IEEE World Forum on Internet of Things (WF-IoT) IEEE; 2014. Using unity 3d to facilitate mobile augmented reality game development; pp. 21–26. [Google Scholar]
- 68.Maloca P.M., de Carvalho J.E.R., Heeren T., Hasler P.W., Mushtaq F., Mon-Williams M., Scholl H.P.N., Balaskas K., Egan C., Tufail A., Witthauer L., Cattin P.C. High-performance virtual reality volume rendering of original optical coherence tomography point-cloud data enhanced with real-time ray casting. Transl. Vision Sci. Technol. 2018;7(4):2. doi: 10.1167/tvst.7.4.2. [DOI] [PMC free article] [PubMed] [Google Scholar]