Abstract
Circular RNAs (circRNAs), covalently closed noncoding RNAs, are widely expressed in eukaryotes and viruses. They can function by regulating target gene expression, linear RNA transcription and protein generation. The phosphoinositide 3-kinase (PI3K)/AKT signaling pathway plays key roles in many biological and cellular processes, such as cell proliferation, growth, invasion, migration, and angiogenesis. It also plays a pivotal role in cancer progression. Emerging data suggest that the circRNA/PI3K/AKT axis modulates the expression of cancer-associated genes and thus regulates tumor progression. Aberrant regulation of the expression of circRNAs in the circRNA/PI3K/AKT axis is significantly associated with clinicopathological characteristics and plays an important role in the regulation of biological functions. In this review, we summarized the expression and biological functions of PI3K-AKT-related circRNAs in vitro and in vivo and assessed their associations with clinicopathological characteristics. We also further discussed the important role of circRNAs in the diagnosis, prognostication, and treatment of cancers.
Subject terms: Cancer therapy, Oncogenes
Introduction
The complexity of cancer and the variability of its clinical features are derived from its complex etiology, involving DNA, RNA, protein, and other factors.1–3 Cancer has become an important public health concern affecting people’s lives.4–6 In the past 10 years, the number of studies on cancer has increased rapidly, providing many novel clues for the treatment of cancer.7,8 The emergence of targeted therapy and immunotherapy has greatly improved the survival rate of cancer patients.9,10 However, cancer treatment remains a major scientific challenge.
Circular RNAs (circRNAs), a newly discovered type of noncoding RNA, have a covalently closed structure and high stability.11–13 CircRNAs are mainly formed by pre-mRNA a back-splicing and are widely expressed in eukaryotes and viruses.14,15 The regulatory role of circRNAs in physiological processes is still not very clear.16 However, accumulating evidence indicates that circRNAs are significantly associated with many diseases and play an important role in the occurrence and development of cancer. A common circRNA-mediated mechanism is that circRNAs act as competitive endogenous RNAs (ceRNAs) of microRNAs (miRNAs) in tumor progression. Circ101237 facilitates the expression of MAPK1 to suppress tumor progression by sponging miR-490-3p in non-small cell lung cancer (NSCLC).17 CircRNA also regulates cancer development and progression by interacting with protein. CircRNA cIARS suppresses cell autophagy via binding with RBP ALKBH5.18
Phosphoinositide 3-kinase (PI3K), a member of the lipid kinase family, is an important regulator of signaling and intracellular vesicular trafficking.19 Several studies have found that the PI3K/AKT pathway is aberrantly activated in cancer20–22 and controls core cellular functions, such as proliferation and survival.23,24 The PI3K/AKT pathway plays a pivotal role in the progression of cancer. Clinical trials targeting PI3K have also attracted increasing attention.25,26 Emerging evidence suggests that circRNAs interact with the PI3K/AKT pathway to regulate cancer progression. Importantly, circRNAs related to the PI3K/AKT pathway have become potential targets in the treatment of cancer. In this review, we summarized the current studies of the role of crosstalk between circRNAs and the PI3K/AKT pathway in the initiation and progression of cancer (Fig. 1). We also presented the clinical applications of PI3K/AKT-related circRNAs in patients with cancer.
Fig. 1.
CircRNAs interact with the PI3K/AKT pathway to regulate cancer progression. Image created with BioRender (https://biorender.com/)
The PI3K/AKT signaling pathway in tumorigenesis
PI3K
Phosphoinositide 3-kinase (PI3K), a member of the lipid kinase family,27,28 was first identified 3 decades ago.29 It can be divided into 3 types (class I–III) in mammals.19,30,31 Class I PI3Ks have gained much attention in the cancer-related field. PI3K is composed of one catalytic (p110) domain and one regulatory (p85) domain.32,33 p85, which contains the Src homology 2 (SH2) and SH3 protein-binding domains,34,35 can interact with target proteins with corresponding binding sites. The activation of PI3K mainly involves the binding of the substrate near the inner side of the plasma membrane.36,37 PI3K can be activated in two ways. One is that PI3K interacts with connexin or growth factor receptors with phosphorylated tyrosine residues, and then induces a conformational change of dimer.38–40 It also can be activated by the direct binding of p110 and Ras.41–43
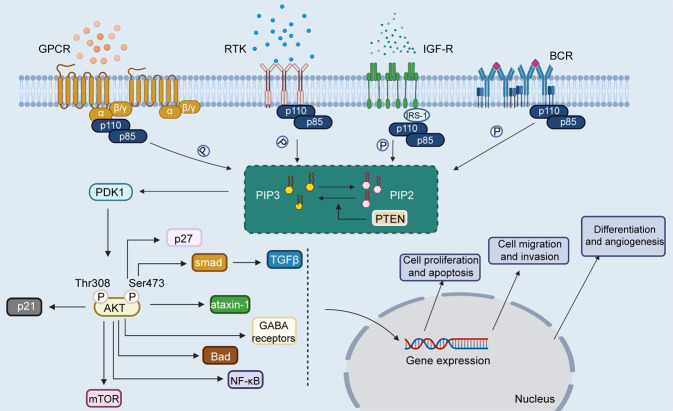
PI3K can be activated by multiple growth factors and signaling complexes, such as G-protein coupled receptors, B-cell receptors, vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), insulin and receptor tyrosine kinases (RTKs) (Fig. 2).20,44–48 These factors induce autophosphorylation through the activation of RTKs and then activate PI3K.49 The p85 subunit provides docking sites for autophosphorylation. In some cases, this process is mediated by the recruitment of adapter proteins. For example, the insulin receptor activates PI3K via insulin receptor substrate-1 (IRS-1).50,51 Activated PI3K increases the conversion of PIP2 to PIP3, which activates PDK1 and AKT.52,53 However, AKT is not the only target molecule of PI3K. PI3K regulates multiple signaling pathways by interacting with BTK, PDK1, and Rac.54
Fig. 2.
The activation process of PI3K/AKT signaling pathway. PI3K, composed of one catalytic (p110) domain and one regulatory (p85), can be activated by G-protein coupled receptor, RTK, IGF-R, and B-cell receptor. Activated PI3K facilitates the conversion of PIP2 to PIP3. PIP3 activates PDK1, and then PDK1 phosphorylates AKT at Thr308. AKT can be also phosphorylated, and activated by PDK2 at Ser473. Activated AKT can regulate mangy cellular biological functions by interacting with numerous downstream signaling molecules, such as p21, p27, TGFβ, ataxin-1, GABA receptors, Bad, NF-κB, and mTOR. Image created with BioRender (https://biorender.com/)
AKT
AKT, also called protein kinase B (PKB),55,56 is the cellular homolog of the oncogene v-Akt. AKT is a serine/threonine kinase that belongs to the AGC kinase family.57–59 There are three different AKT isoforms (AKT1, AKT2, and AKT3), which are widely expressed in most human tissues.60–62 AKT can link the interaction between receptors and PI3K to cellular anabolic pathways. AKT acts as a central regulator of cellular metabolism downstream of insulin signaling that is responsible for the regulation of glucose metabolism.63,64 In vivo experiments support that AKT2 plays a key role in the regulation of glucose metabolism.65,66 Researchers have found that germline mutations of AKT occur during the tumorigenesis and progression of some cancer.67,68
AKT plays a key role in multiple cellular processes, such as cell survival, proliferation, migration, apoptosis, and angiogenesis.69–72 AKT prevents TSC1/TSC2 complex formation and activates mTOR pathway, thereby regulating cell growth.73–75 It also regulates the expression of cyclin D1 and p53 to affect the cell cycle or the proliferation of various cell types through interacting with CDK inhibitors including p21 and p27.76 AKT boosts cell survival via inactivating the pro-apoptotic factors Bad and the transcription factor of the Forkhead (FKHR) family.77 The expression levels of GABA receptors and ataxin-1 are also regulated by AKT.78,79 Some studies observed that AKT regulates the TGFβ signaling pathway by binding with Smad.80 The present findings show that AKT is an important target for the treatment of cancer, diabetes, stroke, and neurodegenerative diseases.81–83
The activation of PI3K/AKT pathway
The PI3K/AKT signaling pathway plays key role in many biological and cellular functions.84,85 We have already elaborated on the activation of PI3K when introducing PI3K. The inositol ring of PI has five potential phosphorylation sites. PI3K activation could catalyze the phosphorylation of phosphatidylinositol (PI) at the 3′-position of the inositol ring.86 The phosphorylated products have a critical influence on cellular functions. PIP3 could enhance cell migration,87 and PI 3,4-bisphosphate regulates B cell activation and insulin sensitivity.88 AKT and PDK1, which contain PH domains can bind to PIP3. PIP3 activates PDK1,89 and then PDK1 phosphorylates AKT at Thr308.90,91 AKT can be also phosphorylated and activated by PDK2 at Ser473.92,93 Activated AKT regulates cell proliferation, differentiation, migration, and apoptosis by activating or inhibiting downstream target proteins, such as Bad,94 Caspase9,95 NF-κB,96,97 GSK-3,98 FKHR,99,100 p21,101 p53102 and FOXO1.103,104 Aberrant activation of PI3K/AKT pathway has been found in a variety of cancers,105 such as lung cancer,106 esophageal cancer,107 gastric cancer,108 breast cancer,109 laryngeal cancer,110 gallbladder cancer,111 and prostate cancer.112
PTEN is a widely mutated tumor suppressor gene that inhibits the oncogenic PI3K/AKT pathway.113–115 PTEN antagonizes the PI3K/Akt pathway by dephosphorylating PIP3 to PIP2,116,117 then induces changes in a variety of cellular biological functions.118,119 Carboxyl-terminal modulator protein (CTMP) could block the transmission of downstream signaling pathways by inhibiting AKT phosphorylation.120,121 PP2A has been found to dephosphorylate AKT-Thr308 and AKT-Ser473 to inhibit the activation of AKT.122,123
CircRNAs and cancer
CircRNAs were initially found in RNA viruses at the end of the 20th century and were considered transcriptional background noise.124–126 With the application of high-throughput RNA sequencing and bioinformatics approaches, circRNAs have attracted much attention from researchers.13,127,128 CircRNAs, covalently closed noncoding RNAs, are widely expressed in eukaryotes and viruses.11,129–131 Linear pre-mRNAs generate circRNAs through exon skipping or back-splicing events.132,133 The circular form of circRNAs protects them from degradation by exonucleases, causing them to show greater stability.11,12 CircRNAs can function by regulating target gene expression, linear RNA transcription, and protein generation.13,134,135 Moreover, circRNAs are involved in the occurrence and development of several cancers.129,136–139 Different circRNAs play distinct roles in diverse cancer types. The circRNA cSMARCA5 has tumor-suppressive properties in the progression of hepatocellular carcinoma.136 However, circMAPK4 suppresses cell apoptosis by regulating specific pathways in gliomas.140
There are mainly four mechanisms by which circRNAs can act in cancer progression: miRNA sponging, protein binding, regulation of gene transcription, and regulation of protein translation. CircRNAs function as natural miRNA sponges that regulate miRNA activity.141–143 miRNAs are essential players in almost all carcinogenic processes.144–146 Increasing evidence suggests that circRNAs modulate cancer progression by regulating the expression of miRNA targets.147–151 For example, cTFRC facilitates tumor progression by sponging miR-107 in bladder carcinoma.152 In addition, circRNAs regulate cancer development and progression by directly modifying the transcription of related genes. Zhang et al.153 reported a novel class of intron-derived circRNAs that is widely distributed throughout the nucleus. Intron-derived circRNAs can interact with RNA polymerase II to enhance the transcription of its target genes.154,155 CircRNAs could also act as protein decoys, and regulate RNA-binding proteins (RBPs) activity by combining with RBPs.156,157 The expression of circZKSCAN1 attenuates HCC cell stemness by targeting RBP fragile X mental retardation protein.158 Moreover, some circRNAs containing the AUG start codon and IRES can control gene expression at the translational level.159,160 However, this effect has not yet been fully elucidated in cancer.
The circRNA/PI3K/AKT axis in cancer
CircRNA plays a critical role in the initiation and development of human cancer.161–165 The studies on circRNA are changing our view of cancer genesis, progression, and treatment.166,167 CircRNAs alone may be insufficient for driving cancer progression. Similarly, traditional signaling pathways or signaling molecules alone may also be ineffective. Interestingly, studies have found that circRNAs are often interrelated with the PI3K/AKT signaling pathway. The PI3K/AKT signaling pathway plays key roles in many biological and cellular functions, such as cell proliferation, growth, invasion, migration, and angiogenesis.85,168 It also plays a pivotal role in the progression of cancer.27,169,170 Recently, a great deal of research regarding the interaction of circRNA and PI3K/AKT signaling pathways has attracted significant research interest. CircRNAs regulate cellular functions and control the occurrence and development of cancer via interactions with the PI3K/AKT pathway. Based on the current study, the mechanism/pattern of interaction between circRNA and PI3K/AKT pathway is primarily the ceRNA mechanism, which involves the activation or repression of downstream pathways by sponging miRNA. Research on the circRNA/PI3K/AKT axis is still in its infancy. With the deepening of research about the structure and function of circRNAs, the mechanism will add clarity regarding the circRNA/PI3K/AKT axis.
Clinical features and cell biological functions related to the circRNA/PI3K/AKT axis
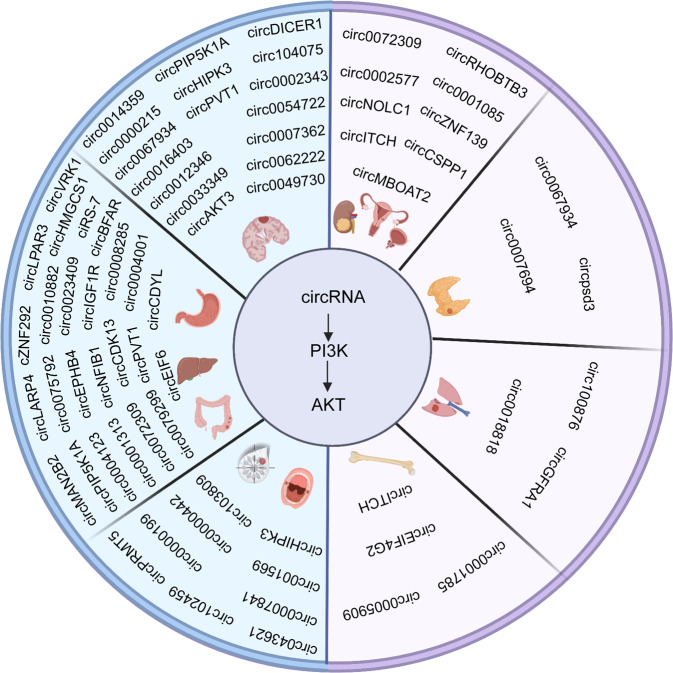
A large number of circRNAs have been found to be involved in the PI3K/AKT signaling pathway. The circRNA/PI3K/AKT axis modulates the expression of cancer-associated genes and thus regulates tumor progression. The circRNA/PI3K/AKT axis plays important role in the initiation and progression of several types of cancer. Current studies may lay the foundation for further research on the mechanisms of cancer progression and provide insights into circRNA-based clinical applications. In this section, we will summarize the expression, biological functions in vitro (Table 1), and associations with clinicopathological characteristics of circRNAs related to the PI3K/AKT signaling pathway (Table 2).
Table 1.
Role and biological functions of circRNA/PI3K/AKT axis in cancer progression in vitro
| Category | Type | CircRNA | Role | Function | Related genes; in vivo | Refs. |
|---|---|---|---|---|---|---|
| Digestive system neoplasms | Esophageal cancer | circLPAR3 | Oncogene | Cell migration and invasion | miR-198, MET, RAS, MAPK, PI3K, and AKT | 171 |
| Esophageal cancer | cZNF292 | Cell viability, migration, invasion, and apoptosis | miR-206, AMPK, PI3K, and AKT | 174 | ||
| Esophageal cancer | circVRK1 | Tumor suppressor | Cell proliferation, migration, EMT, and radioresistance | miR-624-3p, PTEN, PI3K, and AKT | 172 | |
| Esophageal cancer | circLARP4 | Tumor suppressor | Cell proliferation, migration, and apoptosis | miR-1323, PTEN, PI3K, and AKT | 173 | |
| Gastric cancer | circPIP5K1A | Oncogene | Cell proliferation, migration, invasion, and EMT | miR-671-5p, KRT80, PI3K, and AKT | 175 | |
| Gastric cancer | circ0010882 | Oncogene | Cell proliferation, migration, invasion, and apoptosis | PI3K, Akt, and mTOR | 176 | |
| Gastric cancer | circ0023409 | Oncogene | Cell viability, proliferation, migration, invasion, and apoptosis | miR-542-3p, IRS4, PI3K, and AKT | 177 | |
| Gastric cancer | ciRS-7 | Oncogene | miR-7, PTEN, PI3K, and AKT | 178 | ||
| Gastric cancer | circMAN2B2 | Oncogene | Cell viability, cell survival, migration, and apoptosis | miR-145, PI3K, AKT, and JNK | 179 | |
| Gastric cancer | circPVT1 | Oncogene | Cell viability, proliferation, apoptosis, and cisplatin sensitivity | miR-152-3p, HDGF, PI3K, and AKT | 180 | |
| Colorectal cancer | circ0001313 | Oncogene | Cell proliferation and apoptosis | miR-510-5p, PI3K, and AKT2 | 181 | |
| Colorectal cancer | circCDYL | Tumor suppressor | Cell viability, migration, invasion, and apoptosis | miR-105-5p, PTEN, PI3K, AKT, JAK2, and STAT5 | 182 | |
| Colorectal cancer | circ0008285 | Tumor suppressor | miR-382-5p, PTEN, PI3K, and AKT | 183 | ||
| Liver cancer | circCDK13 | Tumor suppressor | Cell migration, invasion, and cell cycle | JAK, STAT, PI3K, and AKT; tumor progression | 184 | |
| Liver cancer | circIGF1R | Oncogene | Cell proliferation, apoptosis, and cell cycle | PI3K, and AKT | 185 | |
| Liver cancer | circ0072309 | Tumor suppressor | Cell viability, colony formation, invasion, and migration | miR-665, PI3K, AKT, Wnt, and β-catenin | 186 | |
| Liver cancer | circ0079299 | Tumor suppressor | Tumor growth, cell cycle | PI3K, AKT, and mTOR; tumor size and tumor weight | 187 | |
| Liver cancer | circ0004001 | Oncogene | miRNAs, VEGF, VEGFR, PI3K, AKT, mTOR, and Wnt | 188 | ||
| Liver cancer | circ0004123 | Oncogene | miRNAs, VEGF, VEGFR, PI3K, AKT, mTOR, and Wnt | 188 | ||
| Liver cancer | circ0075792 | Oncogene | miRNAs, VEGF, VEGFR, PI3K, AKT, mTOR, and Wnt | 188 | ||
| Liver cancer | circEPHB4 | Tumor suppressor | Cell viability, apoptosis, migration, and invasion | HIF-1α, PI3K-AKT, and ZEB1; tumor weight, tumor size, and metastasis foci | 189 | |
| Liver cancer | circCDYL | Oncogene | miR-892a, miR-328-3p, HDGF, HIF1AN, NCL, PI3K, AKT, NOTCH2, C-MYC, and SURVIVIN | 190 | ||
| Hepatoblastoma | circHMGCS1 | Oncogene | Cell proliferation, apoptosis, and glutaminolysis | miR-503-5p, IGF2, IGF1R, PI3K, and AKT | 193 | |
| Pancreatic cancer | circNFIB1 | Tumor suppressor | miR-486-5p, PIK3R1, and VEGF-C | 194 | ||
| Pancreatic cancer | circEIF6 | Oncogene | Cell proliferation, migration, invasion, and apoptosis | miR-557, SLC7A11, PI3K, and AKT; tumor weight and volume | 195 | |
| Pancreatic cancer | circBFAR | Oncogene | miR-34b-5p, MET, and AKT; tumor weight and volume, Ki-67 level, MET inhibitor | 196 | ||
| Nervous system neoplasms | Glioma | circ0014359 | Oncogene | Cell viability, migration, invasion, and apoptosis | miR-153, PI3K, and AKT | 197 |
| Glioma | circDICER1 | Oncogene | Angiogenesis | MOV10, miR-103a-3p, miR-382-5p, ZIC4, Hsp90β, PI3K, and AKT | 198 | |
| Glioma | circHIPK3 | Oncogene | Cell proliferation, metastasis, apoptosis, and TMZ sensitivity | miR-524-5p, KIF2A, PI3K, and AKT; tumor growth | 199 | |
| Glioma | circPIP5K1A | Oncogene | Cell proliferation, invasion, apoptosis, and EMT | miR-515-5p, TCF12, PI3K, and AKT; tumor growth | 200 | |
| Glioma | circ104075 | Oncogene | Cell proliferation, apoptosis, and autophagy | Wnt, β-catenin, PI3K, and AKT, il-104075, and Bcl-9 | 201 | |
| Glioma | circ0000215 | Oncogene | Cell proliferation, invasion, apoptosis, and EMT | miR-495-3p, CXCR2, PI3K, and AKT | 202 | |
| Glioblastoma | circAKT3 | Tumor suppressor | Cell proliferation, and radiation resistance | PDK1, PI3K, and AKT; tumorigenicity | 62 | |
| Glioblastoma | circ0067934 | Oncogene | Cell proliferation, metastasis, apoptosis, and EMT | PI3K and AKT | 206 | |
| Glioblastoma | circPVT1 | Oncogene | Cell viability, migration, apoptosis, and EMT | miR-199a-5p, YAP1, PI3K, and AKT | 207 | |
| Neuroblastoma | circ0002343 | EMT | RAC1, PI3K, AKT, and mTOR | 211 | ||
| Genitourinary tumors | Kidney cancer | circ0072309 | Tumor suppressor | Cell proliferation, migration, invasion, and apoptosis | miR-100, PI3K, AKT, and mTOR | 218 |
| Kidney cancer | circC3P1 | Tumor suppressor | Cell viability, migration, invasion, and apoptosis | miR‐21, PTEN, PI3K, AKT, and NF‐κB | 219 | |
| Bladder cancer | circZNF139 | Oncogene | Cell proliferation, migration, invasion, and cell clones | 220 | ||
| Prostate cancer | circ0001085 | EMT | miR-196b-5p, miR-451a, PI3K, and AKT | 228 | ||
| Prostate cancer | circMBOAT2 | Oncogene | Cell proliferation, migration, and invasion | miR-1271-5p, mTOR, PI3K, and AKT; tumor volume, tumor weight, Ki-67 expression, and mTOR | 227 | |
| Prostate cancer | circITCH | Tumor suppressor | Cell proliferation, migration, and invasion | Wnt, β-catenin, PI3K, AKT, and mTOR | 226 | |
| Prostate cancer | circNOLC1 | Oncogene | Cell proliferation, and migration | NF-kappaB, miR-647, PAQR4, PI3K, and AKT | 225 | |
| Ovarian cancer | circRHOBTB3 | Tumor suppressor | Cell proliferation, metastasis, and glycolysis | PI3K and AKT | 231 | |
| Endometrial cancer | circ0002577 | Oncogene | Cell proliferation, migration, and invasion | miR-625-5P, IGF1R, PI3K, and AKT; tumor growth, and metastasis | 232 | |
| Cervical cancer | circCSPP1 | Oncogene | Cell proliferation and migration | miR-361-5p, ITGB1, PI3K, and AKT | 233 | |
| Tumors of the endocrine system | Thyroid cancer | circ0067934 | Oncogene | Cell proliferation, migration, invasion, apoptosis, and EMT | PI3K and AKT | 238 |
| Thyroid cancer | circ0007694 | Tumor suppressor | Cell proliferation, migration, invasion, and apoptosis | PI3K, AKT, mTOR, and Wnt; tumor growth | 239 | |
| Thyroid cancer | circpsd3 | Oncogene | Cell proliferation, metastasis, apoptosis, and cell cycle | miR-637, HEMGN, PI3K, and AKT | 240 | |
| Tumors of the respiratory system | Lung cancer | circGFRA1 | Oncogene | miR-188-3p, PI3K, and AKT; cell proliferation | 245 | |
| Lung cancer | circ100876 | Cell proliferation and apoptosis | miR-636, RET, PI3K, and AKT | 247 | ||
| Lung cancer | circ0018818 | Oncogene | Cell proliferation, invasion, apoptosis, and EMT | miR-767-3p, Nidogen 1(NID1), PI3K, and AKT | 246 | |
| Tumors of the musculoskeletal system | Osteosarcoma | circ0001785 | Oncogene | Cell proliferation and apoptosis | miR-1200, HOXB2, PI3K, AKT, and Bcl-2 | 250 |
| Osteosarcoma | circEIF4G2 | Oncogene | Cell proliferation, migration, and invasion | miR-218, PI3K, and AKT | 251 | |
| Osteosarcoma | circITCH | Tumor suppressor | Cell viability, proliferation, migration, invasion, and apoptosis | miR-22, PTEN, SP-1, PI3K, and AKT | 252 | |
| Osteosarcoma | circ0005909 | Oncogene | Cell viability and cell clones | miR-338-3p, HMGA1, MAPK-ERK, PI3K, and AKT | 253 | |
| Tumors of other systems | Oral squamous cell carcinoma | circ043621 | Oncogene | Cell proliferation, apoptosis, and cell cycle | MAPK, PI3K, AKT, and Bcl-2 | 257 |
| Oral squamous cell carcinoma | circ102459 | Tumor suppressor | Cell proliferation, apoptosis, and cell cycle | MAPK, PI3K, AKT, and Bcl-2 | 257 | |
| Multiple myeloma | circ0007841 | miR-338-3p, BRD4, PI3K, and AKT | 261 | |||
| Breast cancer | circ103809 | Oncogene | Cell proliferation, apoptosis, and cell cycle | PI3K and AKT | 262 | |
| Breast cancer | circPRMT5 | Oncogene | Cell proliferation, apoptosis, and angiogenesis | miR-509-3p, TCF7L2, PI3K, and AKT | 263 | |
| Breast cancer | circHIPK3 | Oncogene | Cell viability, proliferation, migration, and invasion | miR-193a, HMGB1, PI3K, and AKT | 264 | |
| Breast cancer | circ0000442 | Tumor suppressor | Cell viability, colony formation, and cell cycle | miR-148b-3p, PTEN, PI3K, and AKT | 265 | |
| Breast cancer | circ001569 | Oncogene | Cell growth and metastasis | PI3K and AKT | 266 | |
| Breast cancer | circ0000199 | Oncogene | Cell proliferation, migration, invasion, chemo-sensitivity, and autophagy | miR-206, miR-613, PI3K, AKT, and mTOR | 267 |
Table 2.
Relationship between circRNA/PI3K/AKT axis and clinical features in cancer
| Cancer type | CircRNA | Expression | Related features | Refs. |
|---|---|---|---|---|
| Bladder cancer | circZNF139 | Upregulated | Disease-free survival | 220 |
| Liver cancer | circIGF1R | Upregulated | Tumor size | 185 |
| Liver cancer | circRNA0072309 | Downregulated | 5-year survival | 186 |
| Liver cancer | circ0004001, circ0004123, and circ0075792 | Upregulated | TNM stage, and tumor size | 188 |
| Thyroid cancer | circ0067934 | Upregulated | Survival period and AJCC stage | 238 |
| Glioma | circPIP5K1A | Upregulated | Survival time, tumor volume, and tumor stage | 200 |
| Glioblastoma | circ0067934 | Upregulated | Disease-free survival and overall survival | 206 |
| Colorectal cancer | circ0008285 | Downregulated | Lymph node metastasis, TNM stage, and tumor size | 183 |
| Oral squamous cell carcinoma | circ043621 | Upregulated | Clinical stage, lymph node metastasis, and differentiation degree | 257 |
| Oral squamous cell carcinoma | circ102459 | Downregulated | Clinical stage, lymph node metastasis, and differentiation degree | 257 |
| Prostate cancer | circMBOAT2 | Upregulated | Gleason score, pathological T stage, and disease-free survival | 227 |
| Breast cancer | circPRMT5 | Upregulated | Overall survival | 263 |
| Breast cancer | cirCHIPK3 | Upregulated | Overall survival | 264 |
| Breast cancer | circ001569 | Upregulated | Lymph node metastasis, pathological stage, and overall survival | 266 |
| Breast cancer | circ0000199 | Upregulated | Tumor size, TNM stage, ki-67 level, and 3-year survival | 267 |
| Esophageal cancer | circLPAR3 | Upregulated | Lymph node metastasis and TNM stage | 171 |
| Esophageal cancer | circVRK1 | Downregulated | Overall survival | 172 |
| Gastric cancer | circ0010882 | Upregulated | Tumor size, histological grade, and overall survival | 176 |
| Gastric cancer | circ0023409 | Upregulated | Tumor size, histological grade, and lymph nodes metastasis | 177 |
| Gastric cancer | ciRS-7 | Upregulated | Overall survival | 178 |
| Pancreatic cancer | circNFIB1 | Downregulated | Lymph node metastasis | 194 |
| Pancreatic cancer | circBFAR | Upregulated | TNM stage, overall survival, and disease-free survival | 196 |
| Endometrial cancer | circ0002577 | Upregulated | Overall survival, histological grade, lymph node metastasis, and lymph vascular space invasion | 232 |
Digestive system neoplasms
Esophageal cancer
The expression of circVRK1 and circLARP4 is significantly downregulated and circLPAR3 levels are increased in esophageal squamous cell carcinoma (ESCC).171–173 Low circVRK1 expression predicts poor overall survival in patients with ESCC.172 Elevated circLPAR3 levels are markedly associated with lymph node metastasis (LNM) and advanced TNM stage.171 In addition, researchers have also observed alterations in biological functions of the circRNA/PI3K/AKT axis by in vitro functional assays. Silencing of the circRNA cZNF292 inhibits the activity of tumor cells and promotes cell apoptosis in ESCC.174 Upregulation of circVRK1 suppresses cell proliferation, increases the radiosensitivity of ESCC cells, and attenuates epithelial–mesenchymal transition (EMT).172 CircLARP4 inhibits cell apoptosis and promotes cell proliferation in ESCC.173 Furthermore, cZNF292, circVRK1, and circLARP4 all inhibit ESCC cell migration. Contrary to the aforementioned investigations, circLPAR3 functions as a tumor oncogene and enhances the malignant phenotype of ESCC tumors.171 Mechanistically, circLPAR3 increases the expression of the MET gene to enhance the RAS/MAPK and PI3K/Akt pathways by sponging miR-198 in ESCC. Knockdown of cZNF292 induces inactivation of the PI3K/AKT pathway and upregulation of AMPK signaling to exert effects in ESCC.174 CircVRK1 functions as a tumor suppressor gene by upregulating PTEN and inhibiting the PI3K/AKT axis.172 Similarly, circLARP4 promotes the expression of PTEN and inactivates the PI3K/AKT pathway to suppress the progression of ESCC.173
Gastric cancer
PI3K/AKT pathway-related circRNAs (circPIP5K1A, circ0010882, circ0023409, ciRS-7, circMAN2B2, and circPVT1) are all obviously upregulated in gastric cancer.175–180 The levels of circ0010882 and circ0023409 are positively associated with tumor size and histological grade in gastric cancer patients.176,177 In addition, higher expression of circ0010882 or ciRS-7 is associated with shorter overall survival. Circ0023409 promotes LNM in gastric cancer. In terms of biological function, increased circPIP5K1A, circ0010882, and circ0023409 expression reduces gastric cancer cell proliferation, migration, and invasion.175–177 High expression of circPVT1 may enhance the sensitivity of gastric cancer cells to cisplatin (DDP).180 We also found that circMAN2B2 upregulates cell viability and the surviving cell fraction by cell transfection experiments.179 Silencing of circ0010882 attenuated gastric cancer cell growth and motility in vitro.176 In terms of the mechanism, circPIP5K1A sponges miR-671-5p to facilitate tumor progression by upregulating the KRT80 and PI3K/AKT pathways in gastric cancer.175 Circ0010882 regulates biological functions by promoting PI3K/AKT/mTOR signaling.176 Further studies have demonstrated that circ0023409, ciRS-7, circMAN2B2, and circPVT1 regulate the PI3K/AKT pathway by acting as sponges of miRNAs in gastric cancer.177–180 For example, circ0023409 activates the PI3K/AKT pathway by sponging miR-542-3p to increase IRS4 levels.177 In addition, researchers have established in vivo xenograft nude mouse models to further explore the relationship between gastric cancer and the circRNA/PI3K/AKT axis. The expression of circPIP5K1A facilitates tumor growth in gastric cancer in vivo.175
Colorectal cancer (CRC)
The expression level of circ0001313 is dramatically upregulated while the levels of circCDYL and circ0008285 are decreased in CRC.181–183 Circ0008285 expression is positively associated with LNM, tumor-node-metastasis (TNM) stage, and tumor size in patients with CRC.183 Functionally, circCDYL inhibits CRC cell migration and invasion.182 Circ0001313 and circCDYL significantly reduce cell apoptosis in CRC.181,182 Silencing the expression of circ0008285 enhances cell proliferation and migration in CRC.183 The expression of circ0001313 increases the level of AKT2, thus contributing to CRC progression by downregulating miR-510-5p expression.181 CircCDYL inactivates PI3K/AKT and JAK/STAT signaling by decreasing miR-150-5p levels in colon cancer.182 Circ0008285 expression reduces migration and proliferation via regulation of the miR-382-5p/PTEN/PI3K/AKT axis in CRC.183
Liver cancer
A series of circRNAs related to the circRNA/PI3K/AKT axis has been found to be closely related to the occurrence and progression of hepatocellular carcinoma (HCC). These circRNAs with aberrant expression are listed in Table 1.184–190 Tumor size positively correlates with the expression of circIGF1R, circ0004001, circ0004123, and circ0075792 in HCC.185,188 High expression of circ0072309 is related to better 5-year survival in patients with HCC.186 Decreased circCDK13 levels enhance cell motility while low levels of circIGF1R inhibit cell growth in HCC.184,185 High expression of circ0072309 impairs cell growth and motility, affecting cell viability, colony formation, invasion, and migration.186 Mechanistically, circCDK13 inhibits HCC progression by regulating the PI3K/AKT and JAK/STAT pathways (Table 1).184 Circ0072309 functions as a sponge of miR-665 to negatively regulate the PI3K/AKT and Wnt/β-catenin pathways in the pathophysiologic processes of HCC.186 The expression of circEPHB4 impedes HCC progression by negatively regulating the HIF-1α/PI3K/AKT axis and HIF-1α/ZEB1 pathway.189 Hepatoblastoma is the most common primary malignant hepatic tumor in children.191,192 The expression of circHMGCS1 is significantly upregulated in hepatoblastoma cell lines compared to normal hepatocyte cells and HCC cells.193 circHMGCS1 also promotes cell proliferation and inhibits apoptosis in hepatoblastoma cell lines. CircHMGCS1 markedly upregulates the IGF2/IGF1R/PI3K/AKT axis to regulate proliferation by sponging miR-503-5p.193 The expression of circEPHB4 was negatively associated with tumor weight, size, and metastatic foci in vivo.189 A higher level of circ0079929 predicted decreased tumor size and weight in nude mouse models.187 CircCDK13 is an important negative regulator in the development and progression of HCC.184
Pancreatic cancer
The level of circNFIB1 is markedly decreased while circEIF6 and circBFAR expression levels are elevated in pancreatic cancer.194–196 High expression of circNFIB1 restrains lymphatic metastasis of pancreatic cancer.194 Upregulated levels of circBFAR predict high TNM stage and poor prognosis.196 Functionally, we found that the expression of circEIF6 promotes cell proliferation, increases cell migration and invasion, and inhibits cell apoptosis by performing siRNA-mediated knockdown experiments in pancreatic cancer cells.195 Mechanistically, circNFIB1 induces VEGF-C inhibition and attenuates LNM by sponging miR-486-5p and inhibiting the PI3K/AKT pathway in pancreatic ductal adenocarcinoma.194 CircEIF6 regulates biological functions by upregulating miR-557 expression, downregulating SLC7A11 levels, and inactivating the PI3K/AKT pathway in pancreatic cancer.195 CircBFAR facilitates mesenchymal–epithelial transition by sponging miR-34b-5p and upregulating the MET/PI3K/AKT axis in pancreatic ductal adenocarcinoma.196 In vivo experiments showed that downregulation of circBFAR or circEIF6 expression can lead to lower tumor weight and volume in pancreatic ductal adenocarcinoma.195
Nervous system neoplasms
Glioma
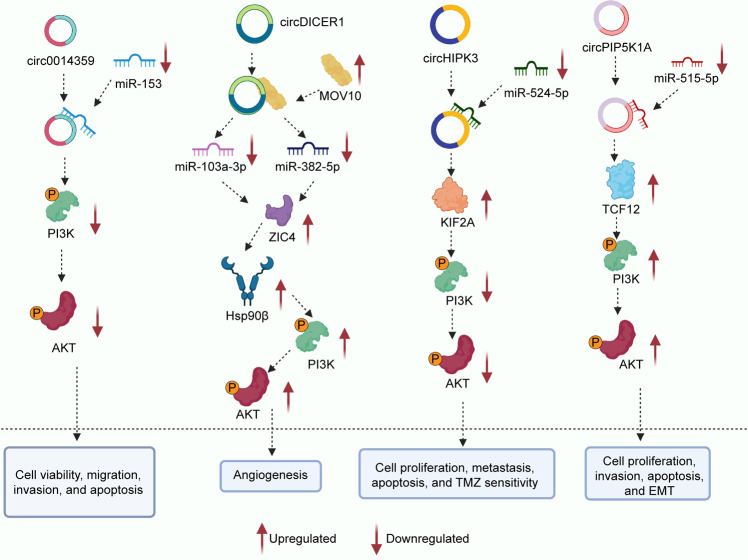
PI3K/AKT axis-associated circRNAs are significantly upregulated in glioma (Table 1).197–202 Elevated circPIP5K1A expression is positively correlated with shorter survival time, larger tumor volume, and higher tumor stage in patients with glioma.200 CircHIPK3, circPIP5K1A, circ104075, and circ0000215 increase glioma cell proliferation in vitro.197,199,200,202 Cic0014359, circHIPK3, circPIP5K1A, and circ0000215 facilitate cell motility in glioma.197,199,200,202 Furthermore, circDICER1 markedly attenuates the angiogenesis of glioma-exposed endothelial cells.198 Downregulated expression of circHIPK3 induces a significant upregulation of temozolomide sensitivity in glioma.199 Mechanistic studies have revealed that circ0014359 exerts its effects by inhibiting the level of miR-153 and regulating the PI3K axis in glioma197 (Fig. 3). CircDICER1 in combination with MOV10 plays a critical role in glioma angiogenesis via regulation of miR-103a-3p (miR-382-5p)/ZIC4.198 CircHIPK3 regulates biological functions to improve sensitivity to temozolomide through suppression of the miR-524-5p/KIF2A-mediated PI3K/AKT pathway.199
Fig. 3.
The specific mechanism of glioma progression between circRNAs and PI3K/AKT pathway. Circ0014359 exerts its effects by inhibiting the level of miR-153 and regulating the PI3K/AKT axis. CircDICER1 in combination with MOV10 plays a critical role in glioma angiogenesis via regulation of miR-103a-3p (miR-382-5p)/ZIC4. CircHIPK3 regulates biological functions to improve sensitivity to temozolomide through suppression of the miR-524-5p/KIF2A-mediated PI3K/AKT pathway. circRNAs can also facilitate glioma tumorigenesis and progression by regulating the circPIP5K1A/miR-515-5p/TCF12/PI3K/AKT axis in glioma. Image created with BioRender (https://biorender.com/)
A series of studies have shown that circRNAs can facilitate glioma tumorigenesis and progression by regulating the circPIP5K1A/miR-515-5p/TCF12/PI3K/AKT and circ0000215/miR-495-3p/CXCR2/PI3K/AKT pathways200,202 (Fig. 3). Glioblastoma (GBM) is the most malignant glioma and has an extremely poor prognosis.203–205 CircAKT3 is overexpressed while circ0067934 and circPVT1 expression are significantly downregulated in GBM.62,206,207 A higher level of circ0067934 portends shorter disease-free survival and decreased overall survival rates in GBM.206 Inhibition of circ0067934 expression may be a promising strategy for improving GBM prognosis. The upregulation of circAKT3 suppresses GBM cell proliferation and increases sensitivity to radiation.62 The expression of circ0067934 facilitates cell proliferation and metastasis and inhibits cell apoptosis in GBM by upregulating the PI3K-AKT pathway.206
Neuroblastoma (NB) and pituitary tumor
NB is the most common extracranial solid tumor in childhood.208–210 The expression of circ0002343 was found to be involved in the regulation of EMT in NB.211 circ0002343 significantly affects EMT by regulating the RAC1/PI3K/AKT/mTOR axis. Pituitary tumors are some of the most common benign neoplasms of the central nervous system.212,213 The levels of circ0054722, circ0012346, and circ0007362 are significantly increased while the expression of some circRNAs (circ0062222, circ0016403, circ0033349, and circ0049730) is downregulated in invasive nonfunctioning pituitary adenomas compared with the levels in noninvasive nonfunctioning pituitary adenomas.214
Genitourinary tumors
Kidney cancer and bladder cancer
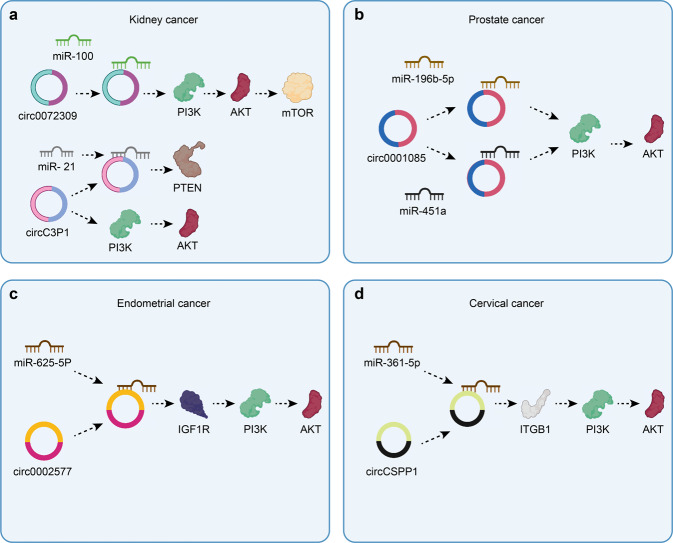
Kidney cancer is not a single disease but comprises different types of cancer that occur in the kidney.215–217 Renal carcinoma-associated transcripts (circ0072309 and circC3P1) are significantly downregulated in renal carcinoma tissues compared to corresponding normal tissues.218,219 These circRNAs significantly suppresses cell proliferation, migration, and invasion and promote cell apoptosis in kidney cancer. Circ-0072309 sponges miR-100 to inhibit the PI3K/AKT and mTOR pathways in kidney cancer.218 CircC3P1 exerts diverse biological functions by inhibiting the PI3K/AKT and NF-κB pathways by regulating the miR-21/PTEN axis219 (Fig. 4a). The overexpression of circZNF139 is markedly associated with disease-free survival in bladder cancer.220 circZNF139 overexpression also attenuates bladder cancer cell proliferation, colony formation, migration, and invasion by regulating the PI3K/AKT pathway.
Fig. 4.
The specific mechanism of circRNAs and PI3K/AKT pathway in different cancers. a Circ-0072309 sponges miR-100 to inhibit the PI3K/AKT/mTOR pathway in kidney cancer. CircC3P1 inhibits kidney cancer progression via regulation of miR/PTEN pathways and the PI3K/AKT pathway. b Circ0001085 regulates prostate cancer progression through the PI3K/AKT pathway by sponging miR-196b-5p and miR-451a. c Overexpression of circ0002577 enhances the IGF1R/PI3K/AKT axis to increase the migration, invasion, and proliferation of endometrial cancer cells. d CircCSPP1 expression inhibits cervical cancer cell apoptosis and promotes cell proliferation and migration via the miR-361-5p/ITGB1/PI3K/AKT axis in cervical cancer. Image created with BioRender (https://biorender.com/)
Prostate cancer (PCa)
PCa is a major cause of male cancer-related mortality worldwide.221–224 The level of circNOLC1 is increased while circITCH expression is obviously downregulated in PCa.225,226 CircMBOAT2 is overexpressed in PCa and contributes to poor prognosis.227 Moreover, increased circMBOAT2 levels are positively correlated with Gleason score and pathological T stage. Functionally, circNOLC1, circITCH, and circMBOAT2 govern multiple cellular processes, such as cell proliferation, migration, and invasion, via the circRNA/PI3K/AKT axis in PCa.225–227 Circ0001085 induces EMT in PCa cells in vitro.228 Circ0001085 regulates PCa progression through the PI3K/AKT pathway by sponging miR-196b-5p and miR-451a (Fig. 4b). CircMBOAT2 clearly promotes tumorigenesis and metastasis in PCa in vivo.227
Female reproductive system cancers
Ovarian, endometrial, and cervical cancer are three major malignant tumors causing a severe threat to women’s health.229,230 The downregulation of circRHOBTB3 not only attenuates cell proliferation and metastasis but also inhibits glycolysis by suppressing the PI3K/AKT pathway in ovarian cancer.231 Circ0002577 expression is markedly increased in endometrial cancer.232 Circ0002577 expression is positively correlated with the histological grade of the tumor, LNM, and lymph vascular space invasion. Studies have revealed that patients with high expression of circ0002577 have a poor prognosis. The overexpression of circ0002577 enhances the IGF1R/PI3K/AKT axis to increase the migration, invasion, and proliferation of endometrial cancer cells (Fig. 4c). Silencing of circ0002577 expression significantly inhibits the growth and metastasis of tumors in nude mouse models of endometrial cancer.232 The expression of circCSPP1 is markedly upregulated in cervical cancer tissues.233 CircCSPP1 expression inhibits cervical cancer cell apoptosis and promotes cell proliferation and migration via the miR-361-5p/ITGB1/PI3K/AKT axis in cervical cancer (Fig. 4d).
Tumors of the endocrine system
Thyroid cancer is the most common malignancy occurring in the endocrine system.234–237 The expression of circ0067934 and circpsd3 is upregulated whereas circ0007694 expression is downregulated in thyroid tumors.238–240 High circ0067934 expression is associated with a shorter survival period of thyroid cancer patients.238 The expression of circ0067934 and circ0007694 affects diverse cell biological functions, such as cell proliferation, migration, invasion, and apoptosis, in thyroid cancer via the PI3K/AKT signaling pathway.238,239 During the regulation of different cellular biological processes, circ0067934 acts as an oncogene, but circ0007694 may function as a tumor suppressor gene in the progression of thyroid cancer. Increased circ0007694 expression effectively suppresses the growth of papillary thyroid carcinoma in vivo.239
Tumors of the respiratory and musculoskeletal systems
Lung cancer
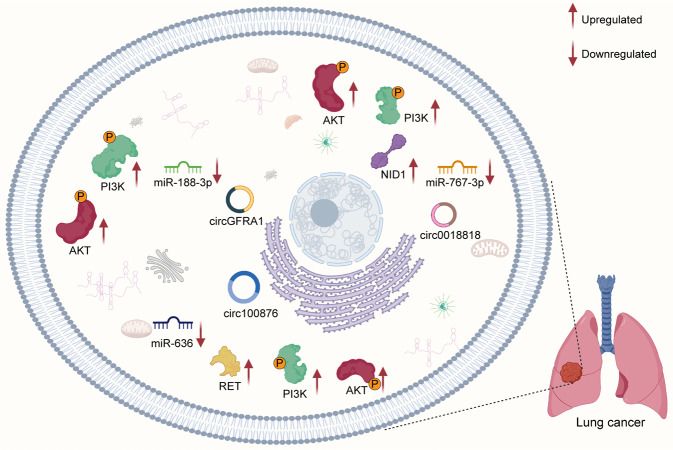
Lung cancer is one of the leading causes of cancer-related death worldwide, with NSCLC accounting for 85% of all lung cancers.241–244 The expression of circGFRA1 and circ0018818 is significantly upregulated in NSCLC tissues compared to normal counterparts.245,246 Silencing of circ0018818 expression inhibits proliferation, invasion, and EMT and promotes cell apoptosis.246 In addition, circGFRA1 activates the PI3K/AKT pathway by downregulating the expression of miR-188-3p in lung cancer. Knockdown of circ100876 reduces cell proliferation, migration, and invasion and facilitates NSCLC cell apoptosis by regulating the miR-636/RET axis and PI3K/AKT signaling.247 The circ0018818/miR-767-3p/NID1/PI3K/AKT axis also plays a key role in the progression of lung cancer (Fig. 5).
Fig. 5.
The mechanism of circRNAs and PI3K/AKT pathway in lung cancer. CircGFRA1 activates the PI3K/AKT pathway by downregulating the expression of miR-188-3p in lung cancer. Circ100876 affects biological functions via PI3K/AKT signaling by regulating the miR-636/RET axis. The circ0018818/miR-767-3p/NID1/PI3K/AKT axis also plays a key role in the progression of lung cancer. Image created with BioRender (https://biorender.com/)
Osteosarcoma (OS)
OS is the most common primary malignant bone tumor in children and adolescents.248,249 The expression of circRNAs associated with the PI3K/AKT axis is listed in Table 1.250–253 The expression of circEIF4G2 and circITCH affects cell biological functions, such as cell proliferation, migration, and invasion, in OS.251,252 Silencing of circ0005909 obviously decreases cell viability and cell clone capacity in OS cell lines.253 Decreased expression of circ0001785 reduces cell proliferation and facilitates cell apoptosis in OS.250 Mechanistically, the expression of circ-ITCH attenuates cell biological functions because circ-ITCH acts as a competing endogenous RNA (ceRNA) for miR-22 to inactivate the PTEN/PI3K/AKT and SP-1 pathways in OS.252 Circ0005909 expression enhances OS malignant progression by upregulating the MAPK-ERK and PI3K-Akt signaling pathways by sponging miR-338-3p to inhibit the level of HGMA1.253
Tumors of other systems
Oral squamous cell carcinoma (OSCC) is a malignant type of head and neck squamous cell carcinoma.254–256 Circ043621 expression is remarkably elevated and circ102459 levels are dramatically decreased in OSCC tissues.256 CircPARD3 and circ043621 expression levels are relatively associated with clinical stage, LNM, and differentiation degree in OSCC. In vitro assays have revealed that increased circ043621 levels and decreased circ102459 expression can induce arrest in the G0 and/or G1 phase, apoptosis, and inhibition of cell proliferation by activating the MAPK and PI3K/AKT pathways.257 Multiple myeloma (MM) is a plasma cell malignancy.258–260 The expression of circ0007841 is significantly upregulated in MM cell lines and bone marrow-derived cells.261 High circ0007841 expression enhances the malignant behaviors of MM cells, for example, promoting cell proliferation, cell cycle progression, and metastasis, by activating the PI3K/AKT pathway.
PI3K/AKT axis-associated circRNAs are aberrantly regulated in breast cancer262–267 (Table 1). The overexpression of circ0000199 is significantly associated with tumor size, TNM stage, and Ki-67 level in patients with breast cancer.267 Higher levels of circPRMT5, circHIPK3, circ001569, and circ0000199 predict poor prognosis in breast cancer.263,264,266,267 circ0000199 can affect tumor cell tolerance of chemotherapy via suppression of the PI3K/AKT/mTOR pathway and activation of the miR-206/miR-613 axis.267 circ0000199 also enhances cell proliferation, migration, and invasion in breast cancer. Silencing of circPRMT5 expression attenuates angiogenesis and proliferation and induces apoptosis.263 CircPRMT5 contributes to malignant phenotypes by activating the PI3K/AKT/mTOR axis via regulation of the miR-509-3p/TCF7L2 pathway. High expression of cirCHIPK3 significantly promotes cell migration, invasion, viability, and proliferation by targeting the miR-193a/HMGB1/PI3K/AKT axis.264 High circ0000442 expression induces suppression of cell viability and cell cycle arrest at the G1 phase and decreases colony formation in breast cancer.265 circ0000442 knockdown experiments have further confirmed this result. circ0000442 acts as a sponge of miR-148b-3p to downregulate the PTEN/PI3K/AKT pathway to impede tumor progression. Moreover, the knockdown of cirCHIPK3 attenuates breast cancer growth in vivo.264
CircRNAs related to the PI3K/AKT pathway as biomarkers
In recent years, researchers have focused on identifying effective molecular biomarkers to improve the early detection, monitoring, and prediction of therapy response in cancer patients.268–270 Technological advances have contributed to an up-to-date understanding of the roles of circRNAs in the initiation and progression of cancer. A growing number of circRNAs related to the PI3K/AKT pathway have been found to be potential biomarkers for the diagnosis, treatment, and prognostication of many cancers. In this section, we will further discuss the important role of circRNAs in clinical applications.
Diagnostic biomarkers
The diagnosis of cancer at an early stage is critical for effective treatment and monitoring.271,272 A critical factor of early diagnosis is the identification of diagnostic biomarkers.273–275 Many circRNAs in the PI3K/AKT pathway have been identified as aberrantly expressed during the progression of different cancers (Table 1). For example, the expression of circCSPP1 is markedly upregulated in cervical cancer tissues.233 The expression of circGFRA1 and circ0018818 is significantly upregulated in NSCLC tissues compared to normal tissues.245,246 CircRNAs with significantly abnormal expression have diagnostic potential in many cancers. In addition, the levels of circ0004001, circ0004123, and circ0075792 in serum are markedly upregulated in patients with HCC.188 The expression of circ0010882 in serum is obviously elevated in gastric cancer patients.176
The expression of circ0007841 in serum is significantly increased in MM patients.261 These results suggest that early diagnosis based on circRNAs is practical. More studies about the diagnostic roles of circRNAs in serum are needed.
Prognosis prediction
Emerging evidence suggests that many circRNAs are reliable for predicting the prognosis of patients with cancer.196,276,277 which provides important guidance for cancer therapy. A significant number of circRNAs have been found to be markedly associated with survival parameters, such as overall survival, disease-free survival, and the 5-year survival rate (Table 2). Low circVRK1 expression predicts poor overall survival in patients with ESCC.172 A higher level of circ0067934 portends shorter disease-free survival and decreased overall survival rates in GBM.206 The expression of circ0072309 is positively correlated with the 5-year survival rate in patients with liver cancer.186 In addition, some circRNAs have been found to be significantly associated with other clinical features in cancer. Elevated circLPAR3 levels are markedly associated with LNM and advanced TNM stage in esophageal cancer.171 The levels of circ0010882 and circ0023409 are positively associated with tumor size and histological grade in gastric cancer patients.176,177 The elevated expression of circMBOAT2 has positively correlated with the Gleason score and pathological T stage in PCa.227 These results provide an important reference for cancer treatment.
Targeted therapies
Targeted therapy, a recent trend in cancer therapy, is emerging as a novel therapeutic strategy.278–280 Targeted therapies significantly enhance the efficiency of cancer therapy.281,282 CircRNAs can positively or negatively modulate biological functions and cancer progression through multiple signaling pathways. CircLARP4 promotes the expression of PTEN and inactivates the PI3K/AKT pathway to suppress the progression of ESCC.173 CircPIP5K1A sponges miR-671-5p to facilitate tumor progression by upregulating KRT80 and the PI3K/AKT pathway in gastric cancer.175 Circ0067934 facilitates cell proliferation and metastasis and inhibits cell apoptosis in GBM by upregulating the PI3K-AKT pathway.206 CircEPHB4 impedes HCC progression by negatively regulating the HIF-1α/PI3K/AKT axis and the HIF-1α/ZEB1 pathway in HCC.189 Upregulating or downregulating the expression of circRNAs may be a feasible way to regulate tumor progression. Silencing of circ0010882 attenuates gastric cancer cell growth and motility in vitro.176 Knockdown of circ100876 reduces cell proliferation, migration, and invasion and facilitates NSCLC cell apoptosis.247 In addition, a miR-671-5p inhibitor was able to significantly reduce the level of circPIP5K1A to inhibit the progression of gastric cancer.175 Rapamycin, an mTOR inhibitor, blocks the circMBOAT2/PI3K/AKT/mTOR pathway to suppress PCa progression.227 CircHIPK3 regulates biological functions to improve sensitivity to temozolomide through suppression of the miR-524-5p/KIF2A-mediated PI3K/AKT pathway in glioma.199 Circ0000199 can make tumor cells sensitive to chemotherapy via suppression of the PI3K/AKT/mTOR pathway and activation of the miR-206/miR-613 axis in breast cancer.267 High expression of circPVT1 enhances the sensitivity of gastric cancer cells to cisplatin (DDP).180 These results provide important information for the clinical treatment of cancers.
Conclusions and future perspectives
CircRNAs are emerging biomarkers in cancer diagnosis and treatment. Complex circRNA regulatory networks have important implications in cancer research and have revolutionized our views on cancer genesis, progression, and treatment. In terms of circRNA-mediated cellular signaling studies, the most exciting finding is that circRNAs can function through molecular associations with the components of classical signaling pathways. The PI3K/AKT pathway is closely associated with the pathogenesis and development of cancer. It can regulate cell survival and proliferation and plays an essential role in cell migration, invasion, and angiogenesis. The circRNA/PI3K/AKT axis has recently attracted increasing attention. The modulating effect of tumor cellular biological functions is of interest for researchers studying the circRNA/PI3K/AKT axis. In terms of the circRNA/PI3K/AKT axis, plenty of circRNAs have been extensively studied. The ubiquitous expression and tumor specificity of circRNAs have ushered in new opportunities for cancer diagnosis. The expression of circRNAs is significantly associated with the clinical phenotype and survival time, indicating that it has important guiding significance for cancer prognostic evaluation. However, the expression level and expression stability of circRNAs in circulating body fluids need further study. Assessment of the expression stability of circRNAs in circulating body fluids, including urine and blood, has vast prospects in terms of clinical applications. In addition, considering the aberrant expression of a large number of cancer-related circRNAs, it is crucial to identify circRNAs related to certain types of cancer.
CircRNAs positively or negatively regulate biological functions in cancer development and progression via the PI3K/AKT signaling pathway. Thus, we may control the cancer process by regulating circRNAs in the circRNA/PI3K/AKT axis. The implementation of this idea relies on in-depth research of pharmacologic therapies. A promising drug must stably regulate circRNA activity and efficiently transduce the effect, thus controlling cancer progression. This necessitates a deeper understanding of the functions and mechanisms of circRNA related to the PI3K/AKT pathway under physiological and pathophysiological conditions. At present, research on the circRNA/PI3K/AKT axis is still in its infancy. Structural and functional data for circRNAs related to PI3K/AKT pathway remain limited. The mechanism of interactions between circRNAs and the PI3K/AKT pathway has yet to be established. Without detailed information on the structure and function of circRNAs, therapeutic options based on PI3K/AKT pathway are difficult to identify.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (81790631), the Zhejiang University Academic Award for Outstanding Doctoral Candidates (2020055), the Independent Project Fund of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the National Key Research and Development Program of China (2016YFC1101404/3), and Zhejiang Basic Public Welfare Research Program of China (LQ20H030012).
Author contributions
Lanjuan Li designed the study and reviewed and edited the paper; Chen Xue and Ganglei Li participated in original draft preparation; Juan Lu collected the references and help with reviewing the paper; all authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Chen Xue, Ganglei Li, Juan Lu
References
- 1.Andrei L, et al. Advanced technological tools to study multidrug resistance in cancer. Drug Resist. Updat. 2020;48:100658. doi: 10.1016/j.drup.2019.100658. [DOI] [PubMed] [Google Scholar]
- 2.Gapstur SM, et al. A blueprint for the primary prevention of cancer: targeting established, modifiable risk factors. CA Cancer J. Clin. 2018;68:446–470. doi: 10.3322/caac.21496. [DOI] [PubMed] [Google Scholar]
- 3.Martínez-Jiménez F, et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer. 2020;20:555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 4.Alcaraz KI, et al. Understanding and addressing social determinants to advance cancer health equity in the United States: a blueprint for practice, research, and policy. CA Cancer J. Clin. 2020;70:31–46. doi: 10.3322/caac.21586. [DOI] [PubMed] [Google Scholar]
- 5.Yabroff KR, Gansler T, Wender RC, Cullen KJ, Brawley OW. Minimizing the burden of cancer in the United States: goals for a high-performing health care system. CA Cancer J. Clin. 2019;69:166–183. doi: 10.3322/caac.21556. [DOI] [PubMed] [Google Scholar]
- 6.Bright CJ, et al. Risk of subsequent primary neoplasms in survivors of adolescent and young adult cancer (Teenage and Young Adult Cancer Survivor Study): a population-based, cohort study. Lancet Oncol. 2019;20:531–545. doi: 10.1016/S1470-2045(18)30903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, Xue W, Anderson DG. CRISPR-Cas: a tool for cancer research and therapeutics. Nat. Rev. Clin. Oncol. 2019;16:281–295. doi: 10.1038/s41571-019-0166-8. [DOI] [PubMed] [Google Scholar]
- 9.Akhave NS, Biter AB, Hong DS. Mechanisms of resistance to KRAS(G12C)-targeted therapy. Cancer Discov. 2021;11:1345–1352. doi: 10.1158/2159-8290.CD-20-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans ER, Bugga P, Asthana V, Drezek R. Metallic nanoparticles for cancer immunotherapy. Mater. Today (Kidlington) 2018;21:673–685. doi: 10.1016/j.mattod.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 12.Vo JN, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e813. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng Z, et al. Circular RNA CircMAP3K5 acts as a microRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation. Circulation. 2021;143:354–371. doi: 10.1161/CIRCULATIONAHA.120.049715. [DOI] [PubMed] [Google Scholar]
- 14.Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2020;5:1–36. doi: 10.1007/s13238-020-00799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YG, et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e109. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legnini I, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang ZY, et al. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci. Rep. 2020;10:9024. doi: 10.1038/s41598-020-65920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, et al. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72. doi: 10.1038/s41420-020-00306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilanges B, Posor Y, Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nat. Rev. Mol. Cell Biol. 2019;20:515–534. doi: 10.1038/s41580-019-0129-z. [DOI] [PubMed] [Google Scholar]
- 20.Molinaro A, et al. Insulin-driven PI3K-AKT signaling in the hepatocyte is mediated by redundant PI3Kα and PI3Kβ activities and is promoted by RAS. Cell Metab. 2019;29:1400–1409.e1405. doi: 10.1016/j.cmet.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins BD, Goncalves MD, Cantley LC. Insulin-PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 2020;16:276–283. doi: 10.1038/s41574-020-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fruman DA, et al. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanker AB, Kaklamani V, Arteaga CL. Challenges for the clinical development of PI3K inhibitors: strategies to improve their impact in solid tumors. Cancer Discov. 2019;9:482–491. doi: 10.1158/2159-8290.CD-18-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okkenhaug K, Graupera M, Vanhaesebroeck B. Targeting PI3K in cancer: impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov. 2016;6:1090–1105. doi: 10.1158/2159-8290.CD-16-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le X, et al. Systematic functional characterization of resistance to PI3K inhibition in breast cancer. Cancer Discov. 2016;6:1134–1147. doi: 10.1158/2159-8290.CD-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fresno Vara JA, et al. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004;30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Yuan TL, Cantley LC. Introduction. Curr. Top. Microbiol. Immunol. 2010;346:1–7. doi: 10.1007/82_2010_55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arafeh R, Samuels Y. PIK3CA in cancer: the past 30 years. Semin. Cancer Biol. 2019;59:36–49. doi: 10.1016/j.semcancer.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Gulluni F, De Santis MC, Margaria JP, Martini M, Hirsch E. Class II PI3K functions in cell biology and disease. Trends Cell Biol. 2019;29:339–359. doi: 10.1016/j.tcb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 32.El Motiam A, et al. SUMOylation modulates the stability and function of PI3K-p110β. Cell Mol. Life Sci. 2021;78:4053–4065. doi: 10.1007/s00018-021-03826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Cruz-Herrera CF, et al. Conjugation of SUMO to p85 leads to a novel mechanism of PI3K regulation. Oncogene. 2016;35:2873–2880. doi: 10.1038/onc.2015.356. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann BT, Jücker M. Activation of PI3K/Akt signaling by n-terminal SH2 domain mutants of the p85α regulatory subunit of PI3K is enhanced by deletion of its c-terminal SH2 domain. Cell Signal. 2012;24:1950–1954. doi: 10.1016/j.cellsig.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 2007;88:13–18. doi: 10.1099/vir.0.82419-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Jang H, Nussinov R. PI3K driver mutations: a biophysical membrane-centric perspective. Cancer Res. 2021;81:237–247. doi: 10.1158/0008-5472.CAN-20-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katan M, Cockcroft S. Phosphatidylinositol(4,5)bisphosphate: diverse functions at the plasma membrane. Essays Biochem. 2020;64:513–531. doi: 10.1042/EBC20200041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa S, et al. Role of connexin-43 in protective PI3K-Akt-GSK-3β signaling in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H2536–H2544. doi: 10.1152/ajpheart.00940.2011. [DOI] [PubMed] [Google Scholar]
- 39.Dey JH, et al. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Nagle AM, Wang YF, Boone DN, Lee AV. Controlled dimerization of insulin-like growth factor-1 and insulin receptors reveals shared and distinct activities of holo and hybrid receptors. J. Biol. Chem. 2018;293:3700–3709. doi: 10.1074/jbc.M117.789503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murillo MM, et al. RAS interaction with PI3K p110α is required for tumor-induced angiogenesis. J. Clin. Investig. 2014;124:3601–3611. doi: 10.1172/JCI74134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaskovic R, et al. Intermolecular biparatopic trapping of ErbB2 prevents compensatory activation of PI3K/AKT via RAS-p110 crosstalk. Nat. Commun. 2016;7:11672. doi: 10.1038/ncomms11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson C, Chun-Jen Lin C, Stern M. Ras-dependent and Ras-independent effects of PI3K in Drosophila motor neurons. Genes Brain Behav. 2012;11:848–858. doi: 10.1111/j.1601-183X.2012.00822.x. [DOI] [PubMed] [Google Scholar]
- 44.Bresnick AR, Backer JM. PI3Kβ-A versatile transducer for GPCR, RTK, and small GTPase signaling. Endocrinology. 2019;160:536–555. doi: 10.1210/en.2018-00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oudit GY, et al. The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J. Mol. Cell Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Houslay DM, et al. Coincident signals from GPCRs and receptor tyrosine kinases are uniquely transduced by PI3Kβ in myeloid cells. Sci. Signal. 2016;9:ra82. doi: 10.1126/scisignal.aae0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu F, et al. PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia. 2019;33:2898–2911. doi: 10.1038/s41375-019-0489-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Liu WX, Huang XG. MicroRNA-199a-3p inhibits angiogenesis by targeting the VEGF/PI3K/AKT signalling pathway in an in vitro model of diabetic retinopathy. Exp. Mol. Pathol. 2020;116:104488. doi: 10.1016/j.yexmp.2020.104488. [DOI] [PubMed] [Google Scholar]
- 49.Duan Y, Haybaeck J, Yang Z. Therapeutic potential of PI3K/AKT/mTOR pathway in gastrointestinal stromal tumors: rationale and progress. Cancers. 2020;12:2972. doi: 10.3390/cancers12102972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Damayanti NP, et al. Therapeutic targeting of TFE3/IRS-1/PI3K/mTOR axis in translocation renal cell carcinoma. Clin. Cancer Res. 2018;24:5977–5989. doi: 10.1158/1078-0432.CCR-18-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talbot K, et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Investig. 2012;122:1316–1338. doi: 10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo H, et al. The PI3K/AKT pathway and renal cell carcinoma. J. Genet. Genomics. 2015;42:343–353. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Q, Jiang W, Hou P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin. Cancer Biol. 2019;59:112–124. doi: 10.1016/j.semcancer.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: beyond AKT. Curr. Opin. Cell Biol. 2017;45:62–71. doi: 10.1016/j.ceb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 58.Revathidevi S, Munirajan AK. Akt in cancer: mediator and more. Semin. Cancer Biol. 2019;59:80–91. doi: 10.1016/j.semcancer.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Leroux AE, Schulze JO, Biondi RM. AGC kinases, mechanisms of regulation and innovative drug development. Semin. Cancer Biol. 2018;48:1–17. doi: 10.1016/j.semcancer.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Palumbo S, Paterson C, Yang F, Hood VL, Law AJ. PKBβ/AKT2 deficiency impacts brain mTOR signaling, prefrontal cortical physiology, hippocampal plasticity and select murine behaviors. Mol. Psychiatry. 2021;26:411–428. doi: 10.1038/s41380-020-00964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hinz N, Jücker M. Distinct functions of AKT isoforms in breast cancer: a comprehensive review. Cell Commun. Signal. 2019;17:154. doi: 10.1186/s12964-019-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia X, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Häggblad Sahlberg S, et al. Different functions of AKT1 and AKT2 in molecular pathways, cell migration and metabolism in colon cancer cells. Int J. Oncol. 2017;50:5–14. doi: 10.3892/ijo.2016.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S, et al. The flavonoid baicalin improves glucose metabolism by targeting the PH domain of AKT and activating AKT/GSK3β phosphorylation. FEBS Lett. 2019;593:175–186. doi: 10.1002/1873-3468.13305. [DOI] [PubMed] [Google Scholar]
- 65.Takenaka N, Nakao M, Matsui S, Satoh T. A crucial role for the small GTPase Rac1 downstream of the protein kinase Akt2 in insulin signaling that regulates glucose uptake in mouse adipocytes. Int. J. Mol. Sci. 2019;20:5443. doi: 10.3390/ijms20215443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang D, Wang J, Zhou C, Xiao W. Zebrafish akt2 is essential for survival, growth, bone development, and glucose homeostasis. Mech. Dev. 2017;143:42–52. doi: 10.1016/j.mod.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 67.Wen L, et al. Characterization of AKT somatic mutations in Chinese breast cancer patients. Cancer Manag. Res. 2021;13:3055–3065. doi: 10.2147/CMAR.S299624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S, et al. Spectrum of PIK3CA/AKT mutations across molecular subtypes of triple-negative breast cancer. Breast Cancer Res. Treat. 2021;187:625–633. doi: 10.1007/s10549-021-06242-3. [DOI] [PubMed] [Google Scholar]
- 69.Conduit SE, et al. AKT signaling promotes DNA damage accumulation and proliferation in polycystic kidney disease. Hum. Mol. Genet. 2020;29:31–48. doi: 10.1093/hmg/ddz232. [DOI] [PubMed] [Google Scholar]
- 70.Lei N, Peng B, Zhang JY. CIP2A regulates cell proliferation via the AKT signaling pathway in human lung cancer. Oncol. Rep. 2014;32:1689–1694. doi: 10.3892/or.2014.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JH, et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat. Commun. 2017;8:949. doi: 10.1038/s41467-017-00906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimmeler S, Zeiher AM. Akt takes center stage in angiogenesis signaling. Circ. Res. 2000;86:4–5. doi: 10.1161/01.res.86.1.4. [DOI] [PubMed] [Google Scholar]
- 73.Montaner S. Akt/TSC/mTOR activation by the KSHV G protein-coupled receptor: emerging insights into the molecular oncogenesis and treatment of Kaposi’s sarcoma. Cell Cycle. 2007;6:438–443. doi: 10.4161/cc.6.4.3843. [DOI] [PubMed] [Google Scholar]
- 74.Huang H, et al. Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (mTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy. 2013;9:175–195. doi: 10.4161/auto.22791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar P, Raman T, Swain MM, Mishra R, Pal A. Hyperglycemia-induced oxidative-nitrosative stress induces inflammation and neurodegeneration via augmented tuberous sclerosis complex-2 (TSC-2) activation in neuronal cells. Mol. Neurobiol. 2017;54:238–254. doi: 10.1007/s12035-015-9667-3. [DOI] [PubMed] [Google Scholar]
- 76.Li T, et al. P21 and P27 promote tumorigenesis and progression via cell cycle acceleration in seminal vesicles of TRAMP mice. Int. J. Biol. Sci. 2019;15:2198–2210. doi: 10.7150/ijbs.35092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yip WK, Leong VC, Abdullah MA, Yusoff S, Seow HF. Overexpression of phospho-Akt correlates with phosphorylation of EGF receptor, FKHR and BAD in nasopharyngeal carcinoma. Oncol. Rep. 2008;19:319–328. [PubMed] [Google Scholar]
- 78.Barati MT, Scherzer J, Wu R, Rane MJ, Klein JB. Cytoskeletal rearrangement and Src and PI-3K-dependent Akt activation control GABA(B)R-mediated chemotaxis. Cell Signal. 2015;27:1178–1185. doi: 10.1016/j.cellsig.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calandria JM, et al. Ataxin-1 poly(Q)-induced proteotoxic stress and apoptosis are attenuated in neural cells by docosahexaenoic acid-derived neuroprotectin D1. J. Biol. Chem. 2012;287:23726–23739. doi: 10.1074/jbc.M111.287078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu S, et al. Human adipose‑derived mesenchymal stem cells promote breast cancer MCF7 cell epithelial‑mesenchymal transition by cross interacting with the TGF‑β/Smad and PI3K/AKT signaling pathways. Mol. Med. Rep. 2019;19:177–186. doi: 10.3892/mmr.2018.9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeong SH, Yang MJ, Choi S, Kim J, Koh GY. Refractoriness of STING therapy is relieved by AKT inhibitor through effective vascular disruption in tumour. Nat. Commun. 2021;12:4405. doi: 10.1038/s41467-021-24603-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang K, Liang Y, Su Y, Wang L. DhHP-6 ameliorates hepatic oxidative stress and insulin resistance in type 2 diabetes mellitus through the PI3K/AKT and AMPK pathway. Biochem. J. 2020;477:2363–2381. doi: 10.1042/BCJ20200402. [DOI] [PubMed] [Google Scholar]
- 83.Lv H, Li J, Che YQ. CXCL8 gene silencing promotes neuroglial cells activation while inhibiting neuroinflammation through the PI3K/Akt/NF-κB-signaling pathway in mice with ischemic stroke. J. Cell Physiol. 2019;234:7341–7355. doi: 10.1002/jcp.27493. [DOI] [PubMed] [Google Scholar]
- 84.Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol. Biosyst. 2015;11:1946–1954. doi: 10.1039/c5mb00101c. [DOI] [PubMed] [Google Scholar]
- 85.Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tessier M, Woodgett JR. Serum and glucocorticoid-regulated protein kinases: variations on a theme. J. Cell Biochem. 2006;98:1391–1407. doi: 10.1002/jcb.20894. [DOI] [PubMed] [Google Scholar]
- 87.Yang Y, et al. Leep1 interacts with PIP3 and the Scar/WAVE complex to regulate cell migration and macropinocytosis. J. Cell Biol. 2021;220:e202010096. doi: 10.1083/jcb.202010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, Marshall AJ. Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: a distinct branch of PI3K signaling. Cell Signal. 2015;27:1789–1798. doi: 10.1016/j.cellsig.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 89.Dieterle AM, et al. PDK1 controls upstream PI3K expression and PIP3 generation. Oncogene. 2014;33:3043–3053. doi: 10.1038/onc.2013.266. [DOI] [PubMed] [Google Scholar]
- 90.Misra UK, Pizzo SV. Activated α2-macroglobulin binding to cell surface GRP78 induces T-loop phosphorylation of Akt1 by PDK1 in association with Raptor. PLoS ONE. 2014;9:e88373. doi: 10.1371/journal.pone.0088373. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Dangelmaier C, et al. PDK1 selectively phosphorylates Thr(308) on Akt and contributes to human platelet functional responses. Thromb. Haemost. 2014;111:508–517. doi: 10.1160/TH13-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawakami Y, et al. Protein kinase C betaII regulates Akt phosphorylation on Ser-473 in a cell type- and stimulus-specific fashion. J. Biol. Chem. 2004;279:47720–47725. doi: 10.1074/jbc.M408797200. [DOI] [PubMed] [Google Scholar]
- 93.Hresko RC, Murata H, Mueckler M. Phosphoinositide-dependent kinase-2 is a distinct protein kinase enriched in a novel cytoskeletal fraction associated with adipocyte plasma membranes. J. Biol. Chem. 2003;278:21615–21622. doi: 10.1074/jbc.M302937200. [DOI] [PubMed] [Google Scholar]
- 94.Zhao J, et al. 7,8-Dihydroxyflavone suppresses proliferation and induces apoptosis of human osteosarcoma cells. Acta Biochim. Biophys. Sin. 2021;53:903–911. doi: 10.1093/abbs/gmab060. [DOI] [PubMed] [Google Scholar]
- 95.Zhu S, et al. 20(S)-ginsenoside Rh2 induces caspase-dependent promyelocytic leukemia-retinoic acid receptor A degradation in NB4 cells via Akt/Bax/caspase9 and TNF-α/caspase8 signaling cascades. J. Ginseng Res. 2021;45:295–304. doi: 10.1016/j.jgr.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang B, et al. MiR-217 inhibits apoptosis of atherosclerotic endothelial cells via the TLR4/PI3K/Akt/NF-κB pathway. Eur. Rev. Med. Pharm. Sci. 2020;24:12867–12877. doi: 10.26355/eurrev_202012_24190. [DOI] [PubMed] [Google Scholar]
- 97.Liu YC, et al. Lenvatinib inhibits AKT/NF-κB signaling and induces apoptosis through extrinsic/intrinsic pathways in non-small cell lung cancer. Anticancer Res. 2021;41:123–130. doi: 10.21873/anticanres.14757. [DOI] [PubMed] [Google Scholar]
- 98.Duda P, et al. GSK-3 and miRs: master regulators of therapeutic sensitivity of cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2020;1867:118770. doi: 10.1016/j.bbamcr.2020.118770. [DOI] [PubMed] [Google Scholar]
- 99.Shang J, Gao ZY, Zhang LY, Wang CY. Over-expression of JAZF1 promotes cardiac microvascular endothelial cell proliferation and angiogenesis via activation of the Akt signaling pathway in rats with myocardial ischemia-reperfusion. Cell Cycle. 2019;18:1619–1634. doi: 10.1080/15384101.2019.1629774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, et al. MiR-4787-5p regulates vascular smooth muscle cell apoptosis by targeting PKD1 and inhibiting the PI3K/Akt/FKHR pathway. J. Cardiovasc. Pharmacol. 2021;78:288–296. doi: 10.1097/FJC.0000000000001051. [DOI] [PubMed] [Google Scholar]
- 101.Mu M, Niu W, Zhang X, Hu S, Niu C. LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene. 2020;39:6879–6892. doi: 10.1038/s41388-020-01466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chibaya L, Karim B, Zhang H, Jones SN. Mdm2 phosphorylation by Akt regulates the p53 response to oxidative stress to promote cell proliferation and tumorigenesis. Proc Natl Acad Sci USA. 2021;118:e2003193118. doi: 10.1073/pnas.2003193118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li K, et al. High cholesterol induces apoptosis and autophagy through the ROS-activated AKT/FOXO1 pathway in tendon-derived stem cells. Stem Cell Res. Ther. 2020;11:131. doi: 10.1186/s13287-020-01643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang Y, et al. Effect of miR-182 on hepatic fibrosis induced by Schistosomiasis japonica by targeting FOXO1 through PI3K/AKT signaling pathway. J. Cell Physiol. 2018;233:6693–6704. doi: 10.1002/jcp.26469. [DOI] [PubMed] [Google Scholar]
- 105.Yu L, Wei J, Liu P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin. Cancer Biol. 2021;S1044-579X:00188–7. doi: 10.1016/j.semcancer.2021.06.019. [DOI] [PubMed] [Google Scholar]
- 106.Chen J, Alduais Y, Zhang K, Zhu X, Chen B. CCAT1/FABP5 promotes tumour progression through mediating fatty acid metabolism and stabilizing PI3K/AKT/mTOR signalling in lung adenocarcinoma. J. Cell. Mol. Med. 2021;25:9199–9213. doi: 10.1111/jcmm.16815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Du W, et al. Methylation of NRN1 is a novel synthetic lethal marker of PI3K-Akt-mTOR and ATR inhibitors in esophageal cancer. Cancer Sci. 2021;112:2870–2883. doi: 10.1111/cas.14917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu L, et al. SLC1A3 promotes gastric cancer progression via the PI3K/AKT signalling pathway. J. Cell Mol. Med. 2020;24:14392–14404. doi: 10.1111/jcmm.16060. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Qu J, et al. AKR1B10 promotes breast cancer cell proliferation and migration via the PI3K/AKT/NF-κB signaling pathway. Cell Biosci. 2021;11:163. doi: 10.1186/s13578-021-00677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Starska K, et al. Fibroblast growth factor receptor 1 and 3 expression is associated with regulatory PI3K/AKT kinase activity, as well as invasion and prognosis, in human laryngeal cancer. Cell Oncol. (Dordr.) 2018;41:253–268. doi: 10.1007/s13402-017-0367-z. [DOI] [PubMed] [Google Scholar]
- 111.Wang L, Yang M, Jin H. PI3K/AKT phosphorylation activates ERRà by upregulating PGC-1à and PGC-1á in gallbladder cancer. Mol. Med. Rep. 2021;24:613. doi: 10.3892/mmr.2021.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hou Y, Li H, Huo W. THBS4 silencing regulates the cancer stem cell-like properties in prostate cancer via blocking the PI3K/Akt pathway. Prostate. 2020;80:753–763. doi: 10.1002/pros.23989. [DOI] [PubMed] [Google Scholar]
- 113.Haddadi N, et al. PTEN/PTENP1: ‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol. Cancer. 2018;17:37. doi: 10.1186/s12943-018-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim GW, et al. HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology. 2021;73:533–547. doi: 10.1002/hep.31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Y, et al. Hydrazinocurcumin and 5-fluorouracil enhance apoptosis and restrain tumorigenicity of HepG2 cells via disrupting the PTEN-mediated PI3K/Akt signaling pathway. Biomed. Pharmacother. 2020;129:109851. doi: 10.1016/j.biopha.2020.109851. [DOI] [PubMed] [Google Scholar]
- 116.Nguyen Huu T, et al. Redox regulation of PTEN by peroxiredoxins. Antioxidants. 2021;10:302. doi: 10.3390/antiox10020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Henager SH, Henriquez S, Dempsey DR, Cole PA. Analysis of site-specific phosphorylation of PTEN by using enzyme-catalyzed expressed protein ligation. Chembiochem. 2020;21:64–68. doi: 10.1002/cbic.201900316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li J, Tanhehco EJ, Russell B. Actin dynamics is rapidly regulated by the PTEN and PIP2 signaling pathways leading to myocyte hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2014;307:H1618–H1625. doi: 10.1152/ajpheart.00393.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang X, et al. IMP3 accelerates the progression of prostate cancer through inhibiting PTEN expression in a SMURF1-dependent way. J. Exp. Clin. Cancer Res. 2020;39:190. doi: 10.1186/s13046-020-01657-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Wang J, Fry CME, Walker CL. Carboxyl-terminal modulator protein regulates Akt signaling during skeletal muscle atrophy in vitro and a mouse model of amyotrophic lateral sclerosis. Sci. Rep. 2019;9:3920. doi: 10.1038/s41598-019-40553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li J, Shan W, Zuo Z. Age-related upregulation of carboxyl terminal modulator protein contributes to the decreased brain ischemic tolerance in older rats. Mol. Neurobiol. 2018;55:6145–6154. doi: 10.1007/s12035-017-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu T, et al. HEATR1 negatively regulates Akt to Help sensitize pancreatic cancer cells to chemotherapy. Cancer Res. 2016;76:572–581. doi: 10.1158/0008-5472.CAN-15-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu L, et al. Inhibition of protein phosphatase 2A sensitizes mucoepidermoid carcinoma to chemotherapy via the PI3K-AKT pathway in response to insulin stimulus. Cell Physiol. Biochem. 2018;50:317–331. doi: 10.1159/000494008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang G, et al. Identification and characterization of circular RNAs during the sea buckthorn fruit development. RNA Biol. 2019;16:354–361. doi: 10.1080/15476286.2019.1574162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tomar D, Yadav AS, Kumar D, Bhadauriya G, Kundu GC. Non-coding RNAs as potential therapeutic targets in breast cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2020;1863:194378. doi: 10.1016/j.bbagrm.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 127.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 130.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 131.Toptan T, et al. Circular DNA tumor viruses make circular RNAs. Proc. Natl Acad. Sci. USA. 2018;115:E8737–e8745. doi: 10.1073/pnas.1811728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guarnerio J, et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29:628–640. doi: 10.1038/s41422-019-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li S, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods. 2021;18:51–59. doi: 10.1038/s41592-020-01011-4. [DOI] [PubMed] [Google Scholar]
- 134.Li Y, Ge YZ, Xu L, Jia R. Circular RNA ITCH: a novel tumor suppressor in multiple cancers. Life Sci. 2020;254:117176. doi: 10.1016/j.lfs.2019.117176. [DOI] [PubMed] [Google Scholar]
- 135.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 136.Yu J, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 137.Gao X, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol. 2021;23:278–291. doi: 10.1038/s41556-021-00639-4. [DOI] [PubMed] [Google Scholar]
- 138.Tan S, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28:693–695. doi: 10.1038/s41422-018-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat. Rev. Cancer. 2021;21:22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 140.He J, et al. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer. 2020;19:17. doi: 10.1186/s12943-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hall IF, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 142.Wang R, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen L, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol. Cancer. 2019;18:13. doi: 10.1186/s12943-019-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hill M, Tran N. MicroRNAs regulating microRNAs in cancer. Trends Cancer. 2018;4:465–468. doi: 10.1016/j.trecan.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 145.Chen X, et al. TDP-43 regulates cancer-associated microRNAs. Protein Cell. 2018;9:848–866. doi: 10.1007/s13238-017-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee TJ, et al. Strategies to modulate MicroRNA functions for the treatment of cancer or organ injury. Pharm. Rev. 2020;72:639–667. doi: 10.1124/pr.119.019026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen Y, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang X, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen LY, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol. Cancer. 2020;19:164. doi: 10.1186/s12943-020-01272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xie F, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol. Cancer. 2018;17:144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lu Q, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Su H, et al. Circular RNA cTFRC acts as the sponge of microRNA-107 to promote bladder carcinoma progression. Mol. Cancer. 2019;18:27. doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Y, et al. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 154.Ma N, et al. circTulp4 functions in Alzheimer’s disease pathogenesis by regulating its parental gene, Tulp4. Mol. Ther. 2021;29:2167–2181. doi: 10.1016/j.ymthe.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 155.Li Z, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 156.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J. Neurosci. Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 157.Wang Z, Lei X. Matrix factorization with neural network for predicting circRNA-RBP interactions. BMC Bioinforma. 2020;21:229. doi: 10.1186/s12859-020-3514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhu YJ, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526–3540. doi: 10.7150/thno.32796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Prats AC, et al. Circular RNA, the key for translation. Int. J. Mol. Sci. 2020;21:8591. doi: 10.3390/ijms21228591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zheng SL, Li L, Zhang HP. Progress on translation ability of circular RNA. Yi Chuan. 2020;42:423–434. doi: 10.16288/j.yczz.19-354. [DOI] [PubMed] [Google Scholar]
- 161.Li R, et al. CircRNA: a rising star in gastric cancer. Cell Mol. Life Sci. 2020;77:1661–1680. doi: 10.1007/s00018-019-03345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. 2019;13:669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu T, et al. CircRNAs in cancer metabolism: a review. J. Hematol. Oncol. 2019;12:90. doi: 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 166.Zhan W, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J. Cell Physiol. 2020;235:1733–1745. doi: 10.1002/jcp.29092. [DOI] [PubMed] [Google Scholar]
- 167.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tewari D, Patni P, Bishayee A, Sah AN, Bishayee A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: a novel therapeutic strategy. Semin. Cancer Biol. 2019;S1044-579X:30405–5. doi: 10.1016/j.semcancer.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 169.Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–132. doi: 10.1016/j.semcancer.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 170.O’Donnell JS, Massi D, Teng MWL, Mandala M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin. Cancer Biol. 2018;48:91–103. doi: 10.1016/j.semcancer.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 171.Shi Y, et al. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111:2824–2836. doi: 10.1111/cas.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He Y, et al. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int. J. Biol. Macromol. 2019;125:116–123. doi: 10.1016/j.ijbiomac.2018.11.273. [DOI] [PubMed] [Google Scholar]
- 173.Chen Z, et al. Circular RNA_LARP4 sponges miR-1323 and hampers progression of esophageal squamous cell carcinoma through modulating PTEN/PI3K/AKT pathway. Dig. Dis. Sci. 2020;65:2272–2283. doi: 10.1007/s10620-019-05973-0. [DOI] [PubMed] [Google Scholar]
- 174.Liu Z, et al. Silence of cZNF292 suppresses the growth, migration, and invasion of human esophageal cancer Eca-109 cells via upregulating miR-206. J. Cell Biochem. 2020;121:2354–2362. doi: 10.1002/jcb.29458. [DOI] [PubMed] [Google Scholar]
- 175.Song H, et al. CircPIP5K1A activates KRT80 and PI3K/AKT pathway to promote gastric cancer development through sponging miR-671-5p. Biomed. Pharmacother. 2020;126:109941. doi: 10.1016/j.biopha.2020.109941. [DOI] [PubMed] [Google Scholar]
- 176.Peng YK, et al. Circular RNA hsa_circ_0010882 promotes the progression of gastric cancer via regulation of the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharm. Sci. 2020;24:1142–1151. doi: 10.26355/eurrev_202002_20165. [DOI] [PubMed] [Google Scholar]
- 177.Li J, Yang Y, Xu D, Cao L. hsa_circ_0023409 accelerates gastric cancer cell growth and metastasis through regulating the IRS4/PI3K/AKT Pathway. Cell Transpl. 2021;30:963689720975390. doi: 10.1177/0963689720975390. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 178.Pan H, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 179.Sun B, et al. Circular RNA circMAN2B2 promotes growth and migration of gastric cancer cells by down-regulation of miR-145. J. Clin. Lab. Anal. 2020;34:e23215. doi: 10.1002/jcla.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang X, Zhang Y, Li W, Liu X. Knockdown of cir_RNA PVT1 elevates gastric cancer cisplatin sensitivity via sponging miR-152-3p. J. Surg. Res. 2021;261:185–195. doi: 10.1016/j.jss.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 181.Tu FL, et al. Circ-0001313/miRNA-510-5p/AKT2 axis promotes the development and progression of colon cancer. Am. J. Transl. Res. 2020;12:281–291. [PMC free article] [PubMed] [Google Scholar]
- 182.Cui W, Dai J, Ma J, Gu H. circCDYL/microRNA-105-5p participates in modulating growth and migration of colon cancer cells. Gen. Physiol. Biophys. 2019;38:485–495. doi: 10.4149/gpb2019037. [DOI] [PubMed] [Google Scholar]
- 183.Wang J, Luo J, Liu G, Li X. Circular RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation and migration via the miR-382-5p/PTEN axis. Biochem. Biophys. Res. Commun. 2020;527:503–510. doi: 10.1016/j.bbrc.2020.03.165. [DOI] [PubMed] [Google Scholar]
- 184.Lin Q, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int. J. Oncol. 2018;53:246–256. doi: 10.3892/ijo.2018.4371. [DOI] [PubMed] [Google Scholar]
- 185.Fu HW, et al. Circ-IGF1R has pro-proliferative and anti-apoptotic effects in HCC by activating the PI3K/AKT pathway. Gene. 2019;716:144031. doi: 10.1016/j.gene.2019.144031. [DOI] [PubMed] [Google Scholar]
- 186.Yu Q, Dai J, Shu M. Circular RNA-0072309 has antitumor influences in Hep3B cell line by targeting microRNA-665. Biofactors. 2020;46:1618–1629. doi: 10.1002/biof.1618. [DOI] [PubMed] [Google Scholar]
- 187.Zheng H, et al. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag. Res. 2019;11:443–454. doi: 10.2147/CMAR.S189338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:226. doi: 10.1186/s12935-020-01302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tan Y, et al. Antitumor effects of circ-EPHB4 in hepatocellular carcinoma via inhibition of HIF-1α. Mol. Carcinog. 2019;58:875–886. doi: 10.1002/mc.22976. [DOI] [PubMed] [Google Scholar]
- 190.Wei Y, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71:130–147. doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 191.Eichenmüller M, et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J. Hepatol. 2014;61:1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 192.Rougemont AL, McLin VA, Toso C, Wildhaber BE. Adult hepatoblastoma: learning from children. J. Hepatol. 2012;56:1392–1403. doi: 10.1016/j.jhep.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 193.Zhen N, et al. CircHMGCS1 promotes hepatoblastoma cell proliferation by regulating the IGF signaling pathway and glutaminolysis. Theranostics. 2019;9:900–919. doi: 10.7150/thno.29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kong Y, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol. Cancer. 2020;19:82. doi: 10.1186/s12943-020-01205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang T, Li M, Lu H, Peng T. Up-regulation of circEIF6 contributes to pancreatic cancer development through targeting miR-557/SLC7A11/PI3K/AKT signaling. Cancer Manag. Res. 2021;13:247–258. doi: 10.2147/CMAR.S280307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Guo X, et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol. Cancer. 2020;19:83. doi: 10.1186/s12943-020-01196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shi F, Shi Z, Zhao Y, Tian J. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem. Biophys. Res. Commun. 2019;510:614–620. doi: 10.1016/j.bbrc.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 198.He Q, et al. MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4 expression change. J. Exp. Clin. Cancer Res. 2019;38:9. doi: 10.1186/s13046-018-0990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yin H, Cui X. Knockdown of circHIPK3 facilitates temozolomide sensitivity in glioma by regulating cellular behaviors through miR-524-5p/KIF2A-mediated PI3K/AKT pathway. Cancer Biother. Radiopharm. 2021;36:556–567. doi: 10.1089/cbr.2020.3575. [DOI] [PubMed] [Google Scholar]
- 200.Zheng K, et al. CircRNA PIP5K1A promotes the progression of glioma through upregulation of the TCF12/PI3K/AKT pathway by sponging miR-515-5p. Cancer Cell Int. 2021;21:27. doi: 10.1186/s12935-020-01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chi G, Xu D, Zhang B, Yang F. Matrine induces apoptosis and autophagy of glioma cell line U251 by regulation of circRNA-104075/BCL-9. Chem. Biol. Interact. 2019;308:198–205. doi: 10.1016/j.cbi.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 202.Mutalifu N, et al. Circ_0000215 increases the expression of CXCR2 and promoted the progression of glioma cells by sponging miR-495-3p. Technol. Cancer Res. Treat. 2020;19:1533033820957026. doi: 10.1177/1533033820957026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tan AC, et al. Management of glioblastoma: state of the art and future directions. CA Cancer J. Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 204.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 205.Gan HK, van den Bent M, Lassman AB, Reardon DA, Scott AM. Antibody-drug conjugates in glioblastoma therapy: the right drugs to the right cells. Nat. Rev. Clin. Oncol. 2017;14:695–707. doi: 10.1038/nrclinonc.2017.95. [DOI] [PubMed] [Google Scholar]
- 206.Xin J, Zhang XY, Sun DK, Tian LQ, Xu P. Up-regulated circular RNA hsa_circ_0067934 contributes to glioblastoma progression through activating PI3K-AKT pathway. Eur. Rev. Med. Pharm. Sci. 2019;23:3447–3454. doi: 10.26355/eurrev_201904_17709. [DOI] [PubMed] [Google Scholar]
- 207.Chi G, Yang F, Xu D, Liu W. Silencing hsa_circ_PVT1 (circPVT1) suppresses the growth and metastasis of glioblastoma multiforme cells by up-regulation of miR-199a-5p. Artif. Cells Nanomed. Biotechnol. 2020;48:188–196. doi: 10.1080/21691401.2019.1699825. [DOI] [PubMed] [Google Scholar]
- 208.Cohen MA, et al. Formation of human neuroblastoma in mouse-human neural crest chimeras. Cell Stem Cell. 2020;26:579–592.e576. doi: 10.1016/j.stem.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Delloye-Bourgeois C, et al. Microenvironment-driven shift of cohesion/detachment balance within tumors induces a switch toward metastasis in neuroblastoma. Cancer Cell. 2017;32:427–443.e428. doi: 10.1016/j.ccell.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 210.Fletcher JI, et al. Too many targets, not enough patients: rethinking neuroblastoma clinical trials. Nat. Rev. Cancer. 2018;18:389–400. doi: 10.1038/s41568-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 211.Zhang L, et al. Comprehensive characterization of circular RNAs in neuroblastoma cell lines. Technol. Cancer Res. Treat. 2020;19:1533033820957622. doi: 10.1177/1533033820957622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lecoq AL, Kamenický P, Guiochon-Mantel A, Chanson P. Genetic mutations in sporadic pituitary adenomas—what to screen for? Nat. Rev. Endocrinol. 2015;11:43–54. doi: 10.1038/nrendo.2014.181. [DOI] [PubMed] [Google Scholar]
- 213.Elston MS, McDonald KL, Clifton-Bligh RJ, Robinson BG. Familial pituitary tumor syndromes. Nat. Rev. Endocrinol. 2009;5:453–461. doi: 10.1038/nrendo.2009.126. [DOI] [PubMed] [Google Scholar]
- 214.Wang J, et al. Circular RNA in invasive and recurrent clinical nonfunctioning pituitary adenomas: expression profiles and bioinformatic analysis. World Neurosurg. 2018;117:e371–e386. doi: 10.1016/j.wneu.2018.06.038. [DOI] [PubMed] [Google Scholar]
- 215.Linehan WM, et al. The metabolic basis of kidney cancer. Cancer Discov. 2019;9:1006–1021. doi: 10.1158/2159-8290.CD-18-1354. [DOI] [PubMed] [Google Scholar]
- 216.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089–2100. doi: 10.1101/gr.131110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Junker K, et al. Potential role of genetic markers in the management of kidney cancer. Eur. Urol. 2013;63:333–340. doi: 10.1016/j.eururo.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 218.Chen T, Shao S, Li W, Liu Y, Cao Y. The circular RNA hsa-circ-0072309 plays anti-tumour roles by sponging miR-100 through the deactivation of PI3K/AKT and mTOR pathways in the renal carcinoma cell lines. Artif. Cells Nanomed. Biotechnol. 2019;47:3638–3648. doi: 10.1080/21691401.2019.1657873. [DOI] [PubMed] [Google Scholar]
- 219.Chen T, Yu Q, Xin L, Guo L. Circular RNA circC3P1 restrains kidney cancer cell activity by regulating miR-21/PTEN axis and inactivating PI3K/AKT and NF- kB pathways. J. Cell Physiol. 2020;235:4001–4010. doi: 10.1002/jcp.29296. [DOI] [PubMed] [Google Scholar]
- 220.Yao J, et al. ZNF139/circZNF139 promotes cell proliferation, migration and invasion via activation of PI3K/AKT pathway in bladder cancer. Aging (Albany NY) 2020;12:9915–9934. doi: 10.18632/aging.103256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Siegel DA, O’Neil ME, Richards TB, Dowling NF, Weir HK. Prostate cancer incidence and survival, by stage and race/ethnicity—United States, 2001-2017. MMWR Morb. Mortal. Wkly Rep. 2020;69:1473–1480. doi: 10.15585/mmwr.mm6941a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gandaglia G, et al. Structured population-based prostate-specific antigen screening for prostate cancer: the European Association of Urology Position in 2019. Eur. Urol. 2019;76:142–150. doi: 10.1016/j.eururo.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 223.Zhang L, et al. The first integrins β3-mediated cellular and nuclear targeting therapeutics for prostate cancer. Biomaterials. 2019;223:119471. doi: 10.1016/j.biomaterials.2019.119471. [DOI] [PubMed] [Google Scholar]
- 224.Wong MC, et al. Global incidence and mortality for prostate cancer: analysis of temporal patterns and trends in 36 countries. Eur. Urol. 2016;70:862–874. doi: 10.1016/j.eururo.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 225.Chen W, et al. Circular RNA CircNOLC1, upregulated by NF-KappaB, promotes the progression of prostate cancer via miR-647/PAQR4 axis. Front. Cell Dev. Biol. 2020;8:624764. doi: 10.3389/fcell.2020.624764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li S, et al. Circular RNA cir-ITCH is a potential therapeutic target for the treatment of castration-resistant prostate cancer. Biomed. Res. Int. 2020;2020:7586521. doi: 10.1155/2020/7586521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shi J, et al. Circular RNA circMBOAT2 promotes prostate cancer progression via a miR-1271-5p/mTOR axis. Aging (Albany NY) 2020;12:13255–13280. doi: 10.18632/aging.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yan Z, Xiao Y, Chen Y, Luo G. Screening and identification of epithelial-to-mesenchymal transition-related circRNA and miRNA in prostate cancer. Pathol. Res. Pr. 2020;216:152784. doi: 10.1016/j.prp.2019.152784. [DOI] [PubMed] [Google Scholar]
- 229.Ventriglia J, et al. Immunotherapy in ovarian, endometrial and cervical cancer: state of the art and future perspectives. Cancer Treat. Rev. 2017;59:109–116. doi: 10.1016/j.ctrv.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 230.Mu AK, Lim BK, Aminudin N, Hashim OH, Shuib AS. Application of SELDI-TOF in N-glycopeptides profiling of the urine from patients with endometrial, ovarian and cervical cancer. Arch. Physiol. Biochem. 2016;122:111–116. doi: 10.3109/13813455.2016.1151441. [DOI] [PubMed] [Google Scholar]
- 231.Yalan, S., Yanfang, L., He, C., Yujie, T. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. Preprint at http://www.medrxiv.org/content/10.23736/S0031-0808.20.03957-9 (2020).
- 232.Wang Y, Yin L, Sun X. CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2020;39:169. doi: 10.1186/s13046-020-01679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yang W, Xie T. Hsa_circ_CSPP1/MiR-361-5p/ITGB1 regulates proliferation and migration of cervical cancer (CC) by modulating the PI3K-Akt signaling pathway. Reprod. Sci. 2020;27:132–144. doi: 10.1007/s43032-019-00008-5. [DOI] [PubMed] [Google Scholar]
- 234.Kim J, Gosnell JE, Roman SA. Geographic influences in the global rise of thyroid cancer. Nat. Rev. Endocrinol. 2020;16:17–29. doi: 10.1038/s41574-019-0263-x. [DOI] [PubMed] [Google Scholar]
- 235.Ito Y, Nikiforov YE, Schlumberger M, Vigneri R. Increasing incidence of thyroid cancer: controversies explored. Nat. Rev. Endocrinol. 2013;9:178–184. doi: 10.1038/nrendo.2012.257. [DOI] [PubMed] [Google Scholar]
- 236.Lin RY. Thyroid cancer stem cells. Nat. Rev. Endocrinol. 2011;7:609–616. doi: 10.1038/nrendo.2011.127. [DOI] [PubMed] [Google Scholar]
- 237.Mazeh H, Chen H. Advances in surgical therapy for thyroid cancer. Nat. Rev. Endocrinol. 2011;7:581–588. doi: 10.1038/nrendo.2011.140. [DOI] [PubMed] [Google Scholar]
- 238.Wang H, Yan X, Zhang H, Zhan X. CircRNA circ_0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med. Sci. Monit. 2019;25:1342–1349. doi: 10.12659/MSM.913463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Long MY, et al. Comprehensive circular RNA profiling reveals the regulatory role of circRNA_0007694 in papillary thyroid carcinoma. Am. J. Transl. Res. 2020;12:1362–1378. [PMC free article] [PubMed] [Google Scholar]
- 240.Li Z, et al. Circ_PSD3 promotes the progression of papillary thyroid carcinoma via the miR-637/HEMGN axis. Life Sci. 2021;264:118622. doi: 10.1016/j.lfs.2020.118622. [DOI] [PubMed] [Google Scholar]
- 241.Jonas DE, et al. Screening for lung cancer with low-dose computed tomography: updated evidence report and systematic review for the US preventive services task force. J. Am. Med. Assoc. USA. 2021;325:971–987. doi: 10.1001/jama.2021.0377. [DOI] [PubMed] [Google Scholar]
- 242.Duruisseaux M, Esteller M. Lung cancer epigenetics: from knowledge to applications. Semin. Cancer Biol. 2018;51:116–128. doi: 10.1016/j.semcancer.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 243.Gridelli C, et al. Non-small-cell lung cancer. Nat. Rev. Dis. Prim. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 244.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–e172. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yao J, Xu G, Zhu L, Zheng H. circGFRA1 enhances NSCLC progression by sponging miR-188-3p. Onco Targets Ther. 2020;13:549–558. doi: 10.2147/OTT.S230795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Xu X, Zhou X, Gao C, Cui Y. Hsa_circ_0018818 knockdown suppresses tumorigenesis in non-small cell lung cancer by sponging miR-767-3p. Aging (Albany NY) 2020;12:7774–7785. doi: 10.18632/aging.103089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Song J, Shi W, Gao Z, Liu X, Wang W. Downregulation of circRNA_100876 inhibited progression of NSCLC in vitro via targeting miR-636. Technol. Cancer Res. Treat. 2020;19:1533033820951817. doi: 10.1177/1533033820951817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Angulo P, et al. Natural compounds targeting major cell signaling pathways: a novel paradigm for osteosarcoma therapy. J. Hematol. Oncol. 2017;10:10. doi: 10.1186/s13045-016-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Li S, et al. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle. 2019;18:1281–1291. doi: 10.1080/15384101.2019.1618127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lin E, Liu S, Xiang W, Zhang H, Xie C. CircEIF4G2 promotes tumorigenesis and progression of osteosarcoma by sponging miR-218. Biomed. Res Int. 2020;2020:8386936. doi: 10.1155/2020/8386936. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 252.Ren C, et al. The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artif. Cells Nanomed. Biotechnol. 2019;47:3359–3367. doi: 10.1080/21691401.2019.1649273. [DOI] [PubMed] [Google Scholar]
- 253.Zhang C, Na N, Liu L, Qiu Y. CircRNA hsa_circ_0005909 promotes cell proliferation of osteosarcoma cells by targeting miR-338-3p/HMGA1 Axis. Cancer Manag. Res. 2021;13:795–803. doi: 10.2147/CMAR.S285118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Omori H, et al. YAP1 is a potent driver of the onset and progression of oral squamous cell carcinoma. Sci. Adv. 2020;6:eaay3324. doi: 10.1126/sciadv.aay3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Peng QS, et al. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway. Cell Death Dis. 2020;11:112. doi: 10.1038/s41419-020-2273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pai S, et al. CD47-SIRPα signaling induces epithelial-mesenchymal transition and cancer stemness and links to a poor prognosis in patients with oral squamous cell carcinoma. Cells. 2019;8:1658. doi: 10.3390/cells8121658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Deng W, et al. Microarray profile of circular RNAs identifies hsa_circRNA_102459 and hsa_circRNA_043621 as important regulators in oral squamous cell carcinoma. Oncol. Rep. 2019;42:2738–2749. doi: 10.3892/or.2019.7369. [DOI] [PubMed] [Google Scholar]
- 258.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat. Rev. Cancer. 2012;12:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 259.Ledergor G, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 2018;24:1867–1876. doi: 10.1038/s41591-018-0269-2. [DOI] [PubMed] [Google Scholar]
- 260.Kumar SK, et al. Multiple myeloma. Nat. Rev. Dis. Prim. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 261.Wang Y, Lin Q, Song C, Ma R, Li X. Circ_0007841 promotes the progression of multiple myeloma through targeting miR-338-3p/BRD4 signaling cascade. Cancer Cell Int. 2020;20:383. doi: 10.1186/s12935-020-01475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Qiu X, Wang Q, Song H, Shao D, Xue J. circ_103809 promotes breast cancer progression by regulating the PI3K/AKT signaling pathway. Oncol. Lett. 2020;19:3725–3730. doi: 10.3892/ol.2020.11507. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 263.Wu D, Jia H, Zhang Z, Li S. Circ-PRMT5 promotes breast cancer by the miR-509-3p/TCF7L2 axis activating the PI3K/AKT pathway. J. Gene Med. 2021;23:e3300. doi: 10.1002/jgm.3300. [DOI] [PubMed] [Google Scholar]
- 264.Chen ZG, et al. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac. Cancer. 2020;11:2660–2671. doi: 10.1111/1759-7714.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Zhang XY, Mao L. Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene. 2021;766:145113. doi: 10.1016/j.gene.2020.145113. [DOI] [PubMed] [Google Scholar]
- 266.Xu JH, Wang Y, Xu D. Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomark. 2019;25:193–201. doi: 10.3233/CBM-182293. [DOI] [PubMed] [Google Scholar]
- 267.Li H, et al. Hsa_circ_0000199 facilitates chemo-tolerance of triple-negative breast cancer by interfering with miR-206/613-led PI3K/Akt/mTOR signaling. Aging (Albany NY) 2021;13:4522–4551. doi: 10.18632/aging.202415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Redman MW, et al. Biomarker-driven therapies for previously treated squamous non-small-cell lung cancer (Lung-MAP SWOG S1400): a biomarker-driven master protocol. Lancet Oncol. 2020;21:1589–1601. doi: 10.1016/S1470-2045(20)30475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat. Rev. Clin. Oncol. 2020;17:11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Whiteside TL. Validation of plasma-derived small extracellular vesicles as cancer biomarkers. Nat. Rev. Clin. Oncol. 2020;17:719–720. doi: 10.1038/s41571-020-00433-5. [DOI] [PubMed] [Google Scholar]
- 271.Rebbeck TR, et al. Precision prevention and early detection of cancer: fundamental principles. Cancer Discov. 2018;8:803–811. doi: 10.1158/2159-8290.CD-17-1415. [DOI] [PubMed] [Google Scholar]
- 272.Wender RC, Brawley OW, Fedewa SA, Gansler T, Smith RA. A blueprint for cancer screening and early detection: advancing screening’s contribution to cancer control. CA Cancer J. Clin. 2019;69:50–79. doi: 10.3322/caac.21550. [DOI] [PubMed] [Google Scholar]
- 273.Borrebaeck CA. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer. 2017;17:199–204. doi: 10.1038/nrc.2016.153. [DOI] [PubMed] [Google Scholar]
- 274.Lin XJ, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16:804–815. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 275.Wu L, Qu X. Cancer biomarker detection: recent achievements and challenges. Chem. Soc. Rev. 2015;44:2963–2997. doi: 10.1039/c4cs00370e. [DOI] [PubMed] [Google Scholar]
- 276.Fan L, Cao Q, Liu J, Zhang J, Li B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol. Cancer. 2019;18:16. doi: 10.1186/s12943-018-0936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ding L, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Cooper BM, Iegre J, Donovan DanielHO', Ölwegård Halvarsson M, Spring DR. Peptides as a platform for targeted therapeutics for cancer: peptide-drug conjugates (PDCs) Chem. Soc. Rev. 2021;50:1480–1494. doi: 10.1039/d0cs00556h. [DOI] [PubMed] [Google Scholar]
- 279.Oh DY, Bang YJ. HER2-targeted therapies—a role beyond breast cancer. Nat. Rev. Clin. Oncol. 2020;17:33–48. doi: 10.1038/s41571-019-0268-3. [DOI] [PubMed] [Google Scholar]
- 280.Comoglio PM, Trusolino L, Boccaccio C. Known and novel roles of the MET oncogene in cancer: a coherent approach to targeted therapy. Nat. Rev. Cancer. 2018;18:341–358. doi: 10.1038/s41568-018-0002-y. [DOI] [PubMed] [Google Scholar]
- 281.Srinivasarao M, Galliford CV, Low PS. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015;14:203–219. doi: 10.1038/nrd4519. [DOI] [PubMed] [Google Scholar]
- 282.Schmitt MW, Loeb LA, Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nat. Rev. Clin. Oncol. 2016;13:335–347. doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]