Abstract
All small mammalian hibernators periodically rewarm from torpor to high, euthermic body temperatures for brief intervals throughout the hibernating season. The functional significance of these arousal episodes is unknown, but one suggestion is that rewarming may be related to replacement of gene products lost during torpor due to degradation of mRNA. To assess the stability of mRNA as a function of the hibernation state, we examined the poly(A) tail lengths of liver mRNA from arctic ground squirrels sacrificed during four hibernation states (early and late during a torpor bout and early and late following arousal from torpor) and from active ground squirrels sacrificed in the summer. Poly(A) tail lengths were not altered during torpor, suggesting either that mRNA is stabilized or that transcription continues during torpor. In mRNA isolated from torpid ground squirrels, we observed a pattern of 12 poly(A) residues at greater densities approximately every 27 nucleotides along the poly(A) tail, which is a pattern consistent with binding of poly(A)-binding protein. The intensity of this pattern was significantly reduced following arousal from torpor and undetectable in mRNA obtained from summer ground squirrels. Analyses of polysome profiles revealed a significant reduction in polyribosomes in torpid animals, indicating that translation is depressed during torpor.
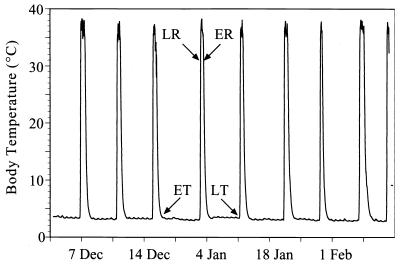
Hibernation in mammals is an energy-conserving strategy involving physiological and behavioral accommodations to low body temperature and low metabolic rate for extended, but not indefinite, periods (12). Hibernation involves a cyclic up- and down-regulation of metabolism (36). The arctic ground squirrel (Spermophilus parryii) illustrates an extreme example of hibernation under adverse conditions. Hibernation begins as early as August and continues until May (9), during which air temperatures drop to as low as −40°C and soil temperatures drop to as low as −18°C (9). Hibernating arctic ground squirrels maintain core body temperatures as low as −2.9°C for up to 3 weeks before spontaneously arousing (4). After arousing, ground squirrels maintain euthermic body temperatures for 15 to 24 h, most of which are spent sleeping (11, 35), before allowing their metabolism and body temperature to again decrease (Fig. 1) (5). The functional significance of these arousal episodes remains unknown (38).
FIG. 1.
Body temperature changes in a hibernating arctic ground squirrel housed at 5°C, illustrating periodic arousals from a low body temperature (torpor). Arrows indicate when animals were sacrificed during the hibernating season. ET, early torpor; LT, late torpor; LR, late rewarming; ER, early reentry.
Arousals from torpor are seen in all small hibernators. This periodicity of energetically costly returns to euthermic body temperatures when energy conservation is adaptive suggests that there is something critical about periodic arousal episodes (1, 36, 38). Of the many theories proposed for the functional significance of arousal episodes (8), one possibility is that replenishment of essential gene products can proceed only at euthermic body temperatures (25). This hypothesis predicts that at the low body temperatures of torpor, degradation of mRNA and/or protein pools proceeds faster than does their synthesis. For a wide range of gene products, the encoding mRNA is significantly less stable than the corresponding protein, with mRNAs exhibiting half-lives in the range of a few minutes to 12 h, approximately one-quarter the average half-life of proteins (17). In in vitro and in vivo studies, rates of transcription at temperatures close to 0°C are greatly reduced (7, 30). Given the instability of mRNA relative to protein, it would seem plausible that if gene products became limiting during torpor, it would first be noted in the mRNA pool. During active protein synthesis, mRNAs are associated with multiple ribosomes, forming polysomes. During the hibernation of ground squirrels, there is a marked loss of polysomes in liver and brain (13, 37). If arousal episodes allow protein synthesis to occur, thereby replenishing proteins depleted during torpor, then polysomes are expected to be depleted during torpor and then reappear during arousals.
Our goals were to compare mRNA stability and polysome profiles at different stages of hibernation and in summer-active animals. To address these goals, we took advantage of the fact that eukaryotic mRNA is polyadenylated on the 3′ end prior to translocation from the nucleus (31) and the length of the poly(A) tail can be estimated as a proxy for mRNA stability. In the cytoplasm, a full-length poly(A) tail is generally 200 to 250 adenosine residues in length. Although mammalian mRNA decay pathways have not been clearly identified, exonucleolytic removal of the poly(A) tail generally occurs prior to nucleolytic cleavage of the remainder of the mRNA (20). If total mRNA is degraded during torpor, there should be both a smaller pool of total mRNA and shorter poly(A) tails on those transcripts that remain. Our results demonstrate that poly(A) tail lengths are conserved during torpor, suggesting that mRNA is quite stable throughout a torpor bout. Stabilization of the poly(A) tail may be due in part to the selective binding of the poly(A)-binding protein (PABP) at low body temperatures.
MATERIALS AND METHODS
Animals.
Arctic ground squirrels were born in captivity at the University of Alaska Fairbanks from wild caught adults trapped in May along the Denali Highway, east of Cantwell, Alaska. Weaned young were housed individually at an ambient temperature of either 4°C (hibernating groups) or 22°C (summer-euthermic group). The photoperiod mimicked that of the natural photoperiod for the trapping area (decreasing from 12 h of light and 12 h of darkness on 21 September to 4 h of light and 20 h of darkness on 21 December). Animals were fed Mazuri rodent pellets and water ad lib, supplemented with carrots. Animals were sacrificed at several time points in the hibernation cycle (Fig. 1) by intracardiac injection of pentobarbital sodium. Body temperatures of animals in the late-rewarming and early-reentry groups were monitored by temperature-sensitive radio transmitters (Mini-Mitter Inc., Sun River, Oreg.) implanted abdominally under halothane anesthesia prior to the hibernating season. Animals sacrificed at early torpor and late torpor were monitored daily to determine the arousal state by placing wood shavings on their back. Disturbance of the shavings indicated that the animals had aroused.
Animals were allowed to go through several torpor bouts until a predictable bout length could be estimated (Fig. 1). Animals in the late rewarming phase of an arousal episode were sacrificed within 2 h after the abdominal temperature reached 30°C (n = 3; mean body temperature = 32.7°C standard error [SE] = 0.5°C). Early-reentry animals were sacrificed at the end of the euthermic phase, within 1 h of body temperature decreasing below 35°C as animals reentered torpor (n = 3; mean body temperature = 32.5°C, SE = 1.4°C; length of time body temperature was above 35°C = 8.5 ± 0.5 h). Late-torpor animals were sacrificed after completing 80 to 90% of a torpor bout, which was determined by averaging previous torpor bout lengths (n = 4; mean body temperature = 4.5°C, SE = 0.6°C, after 8 to 11 days of torpor). Early-torpor animals were sacrificed less than 24 h into a torpor bout (n = 3; mean body temperature = 7.1°C, SE = 1.2°C). Summer-euthermic animals had not been hibernating and were housed at an ambient temperature of approximately 20°C (n = 2; mean body temperature = 36.6°C, SE = 0.5°C). Body temperature at sacrifice was measured rectally by thermocouple thermometer for all animals. Samples of the liver were quickly removed at sacrifice, flash-frozen in liquid nitrogen, and stored at −70°C until they were used for polysome and Northern blot analysis. All protocols for animal care and use were approved by the University of Alaska Institutional Animal Care and Use Committee.
Analysis of poly(A) tail length.
Total RNA was extracted from approximately 100 mg of liver tissue using TriReagent (Sigma). Poly(A) RNA was isolated from 200 μg of total RNA using an oligo(dT) cellulose (Gibco BRL) batch preparation method, as described previously (19). Isolated poly(A) RNA was then end labeled using T4 RNA ligase and 32P-pCp. Unincorporated label was removed by passing the reaction mixture through a Sephadex G-50 spin column. The radioactivity of the product was assayed by liquid scintillation. A constant amount of radioactivity (50,000 cpm) was RNase A digested to isolate the poly(A) tail (RNase A cleaves 3′ of U and C residues). The RNase digestion reaction was stopped by the addition of phenol-chloroform-isoamyl alcohol (25:24:1). Poly(A) RNA was ethanol precipitated and separated on a 6% polyacrylamide gel (0.5× Long Ranger Tris-borate-EDTA with 7 M urea). Century RNA size markers (Ambion Inc.) were similarly labeled and run alongside the samples. The gel was run at 1,250 V until the bromophenol blue dye migrated 50 cm. Autoradiography was performed at −70°C with an intensifying screen for 28 h. The autoradiograph was analyzed by light densitometry on an Alpha Innotech Corporation IS-1000 digital imaging system to quantitate changes in the poly(A) tail lengths of the pool of poly(A) mRNA in each animal.
Polysome analysis.
The isolation and display of polysomes were performed as described previously (16, 29), with some modifications. Briefly, 300 mg of frozen tissue was pulverized with a mortar and pestle under liquid N2 and then homogenized in 3 ml of buffer (25 mM Tris-HCl [pH 7.6], 25 mM NaCl, 10 mM MgCl2, 250 mM sucrose, 1 mg of heparin per ml, 100 μg of cycloheximide per ml) using a Dounce homogenizer. Six strokes with a loose pestle were followed by six strokes with a tight pestle, and the homogenate was centrifuged at 16,000 × g at 4°C for 15 min. The supernatant was mixed with a 1/10 volume of detergent (5% sodium deoxycholate, 5% Triton X-100). Aliquots (0.3 ml) of the supernatant were layered over a continuous 10-ml 0.5 to 1.5 M sucrose gradient with a 1-ml pad of 2 M sucrose (sucrose contained 300 mM NaCl, 10 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 100 μg of heparin per ml, and 100 μg of cycloheximide per ml). The gradients were centrifuged at 4°C for 2.5 h at 37,000 rpm in an SW41 rotor. The UV absorbance of the resulting gradients was monitored at 254 nm (UA-5 absorbance/fluorescence detector; ISCO Inc.). The gradients were collected into 11 fractions of approximately 1 ml each for RNA isolation, immediately frozen on dry ice, and then stored at −70°C.
Northern analysis.
RNA was isolated from 250 μl of each polysome fraction using TriReagent (Sigma) and loaded onto a 1.25% agarose formaldehyde gel for Northern blot analysis. Gels were blotted to a Hybond N+ membrane (Amersham) and UV cross-linked. Blots were then probed with a 316-bp probe for the mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (Ambion) and labeled using a Strip-EZ RNA probe synthesis kit (Ambion). Hybridization and washing of the blots were done using a NorthernMax kit (Ambion). Blots were exposed to film at −70°C for 24 to 48 h, in order to obtain an exposure within the linear range of the film. Blots were analyzed by light densitometry on an Alpha Innotech Corporation IS-1000 digital imaging system to determine the distribution of GAPDH mRNA within the polysome profile. For statistical analysis, fractions 2 through 5 were classified as the monosome region and fractions 6 through 11 were classified as the polysome region, based upon the appearance of the spectrophotometric analysis of the polysome profile. The first discernible ribosomal peak generally fell in fraction 4 or 5. Earlier fractions were included, since in some animals a substantial amount of RNA was apparently unassociated with any ribosomal units. Optical densities of these regions for all animals in a group were averaged and analyzed by the Tukey-Kramer multiple-comparison test to determine differences in RNA distribution between groups.
RESULTS
Stability of the total mRNA pool.
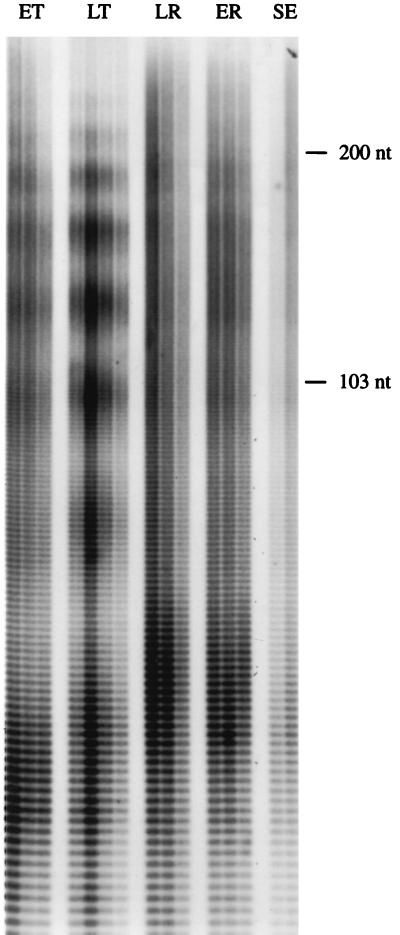
To estimate the stability of mRNA during a torpor bout, we compared poly(A) tail lengths of the total mRNA pool obtained from the livers of animals at different stages of hibernation: early torpor, late torpor, late rewarming, early reentry, and summer euthermy. The length of the poly(A) tracts was assessed by RNase A digestion of end-labeled mRNA. There was no evidence of differential stability of mRNA during hibernation. Poly(A) tail lengths isolated from liver mRNA ranged from approximately 30 to more than 200 nucleotides (nt) for all ground squirrels, regardless of the stage of hibernation or arousal episode when sampled (Fig. 2). In addition, distributions of poly(A) tail lengths were similar among groups, and shorter poly(A) tails, suggestive of degradation, were not more abundant in one stage of hibernation than another.
FIG. 2.
Autoradiograph of a 6% polyacrylamide gel showing the range of poly(A) tail lengths isolated from mRNA of animals at different stages of hibernation and different body temperatures. RNA molecular size markers correspond to 200 and 103 nt. ET, early torpor; LT, late torpor; LR, late rewarming; ER, early reentry; SE, summer euthermy.
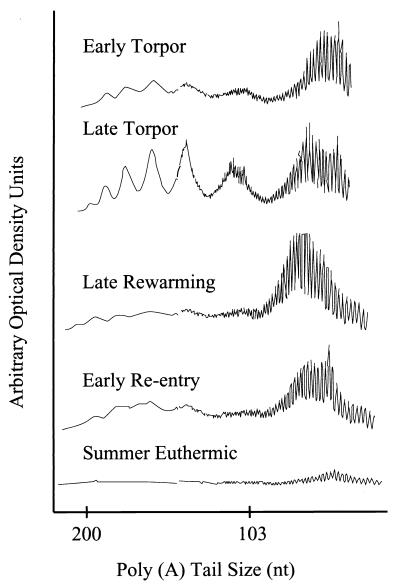
A reproducible pattern of dense bands spanning 12 to 15 nt separated by approximately 27 nt was observed in poly(A) tail lengths isolated from liver taken from torpid animals. To further characterize the temporal appearance and unique size distribution pattern of poly(A) tail lengths, densitometric analysis of the poly(A) tracts was conducted. Peaks representing overabundant poly(A) tail lengths begin to appear in animals during early torpor, increase in magnitude during late torpor, and considerably diminish during the late-rewarming and early-reentry phases of an arousal episode. Summer-euthermic animals did not display a poly(A) size distribution pattern (Fig. 3).
FIG. 3.
Densitometry scan of poly(A) tail sizing gel, illustrating the distribution of poly(A) tails into discrete size classes as a torpor bout progresses.
Translational inhibition during torpor.
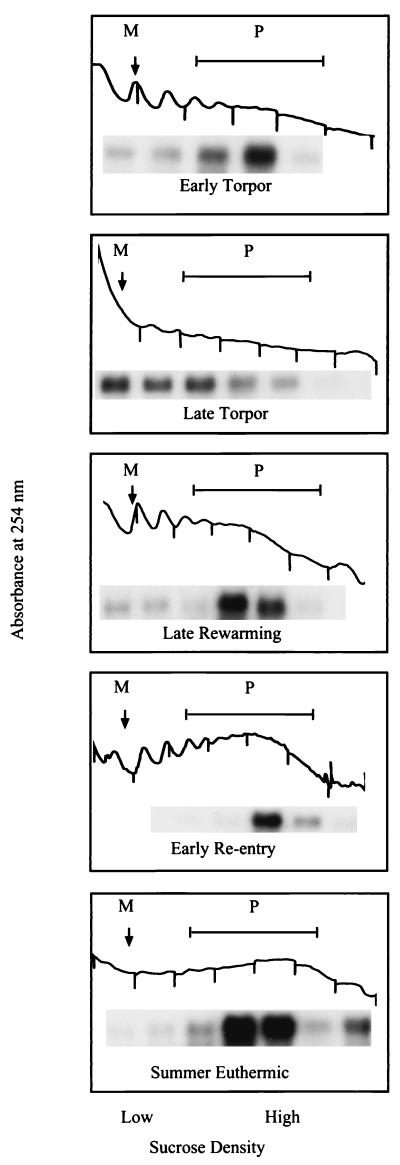
The size distribution of poly(A) tails, protected every 27 nt, suggested that differential binding of PABP may be responsible for the pattern of protected poly(A) tracts during torpor. Since PABP is also thought to influence translatability of poly(A) mRNA, we measured the degree of ribosomal disaggregation in liver homogenates to gauge the activity of the translational machinery as a function of hibernation state. Polysome profiles from summer-active animals displayed a pronounced peak in the polysomal region, which is consistent with active translation. Profiles from animals in early torpor begin to show a disaggregation of polysomes, which continues into late torpor, where we observed an enhanced monosome peak with little or no evidence of a polysome peak. Profiles from the late-rewarming-phase animals show a shift back towards polysomes, and profiles from early-reentry-phase animals appear similar to those from summer-active animals (Fig. 4).
FIG. 4.
Polysome profiles prepared from animals in each of five different temperature states. Below each fraction collected is the corresponding Northern blot lane probed for GAPDH. Vertical lines on the polysome profile indicate separations between fractions collected. M, monosome peak; P, polysome region.
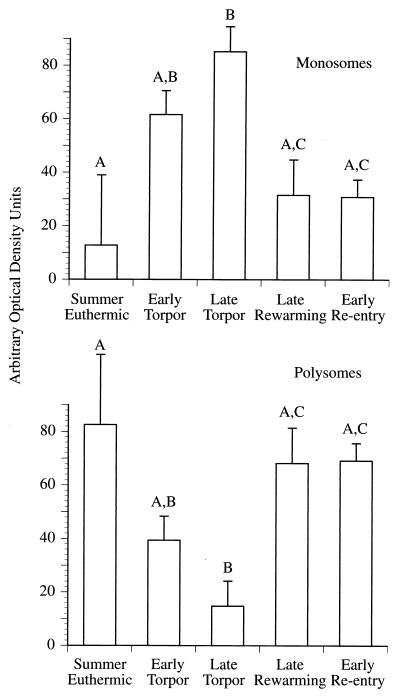
To verify that mRNA present in the polysome profiles was intact, and to test whether mRNA had shifted from polysome to monosome fractions, we isolated RNA from 1-ml fractions of the polysome profile and assayed for the presence of a ubiquitously expressed gene (the GAPDH gene) by Northern analysis. The GAPDH probe hybridized to the total RNA as a single band in all cases, indicating that the observed diminution of polysomes in early torpor, late torpor, and late rewarming was not due to degradation of the samples (Fig. 4). Densitometric analysis of these blots was conducted to quantify the distribution of GAPDH mRNA levels in the monosome and polysome fractions of the polysome profiles as a function of the hibernation state (Fig. 5). Animals in late torpor have significantly more GAPDH mRNA in the monosome region and less in the polysome region of the polysome profiles than summer-euthermic animals or animals in either stage of an interbout euthermic arousal (late rewarming or early reentry, P = 0.05). Early-torpor animals also displayed significantly more GAPDH mRNA in the monosome region and significantly less in the polysome region of the polysome profiles than summer-euthermic animals (P = 0.05).
FIG. 5.
Densitometry analysis of Northern blots prepared from individual fractions of polysome profiles. Values were obtained by scanning individual lanes and summing the values for the fractions that correspond to either the monosome (fractions 2 through 5) or polysome (fractions 6 through 11) region of the polysome profile. Different letters indicate significantly different fractions between groups (P ≤ 0.05).
DISCUSSION
Poly(A) tail length is stable during the torpor phase of hibernation.
There was no evidence of depletion of mRNA during torpor based on analysis of poly(A) tail lengths of total mRNA isolated from hibernating arctic ground squirrels at different stages of torpor and arousal episodes. The ranges of poly(A) tail sizes observed were equivalent among groups, with no apparent depletion of the longest poly(A) tail lengths during torpor. This is consistent with recent studies of specific mRNAs by Northern analysis (13, 27). Northern blots probed for GAPDH revealed discrete bands of the appropriate size for intact GAPDH, indicating that, for at least this message, there was no evidence of degradation of the body of the message. Persistence of mRNA levels throughout a torpor bout can be accomplished only if rates of synthesis and degradation remain matched. Since transcription of mRNA is low or absent at low temperatures (7, 30), persistence of mRNA is likely due to stabilization of transcripts.
Potential role of PABP in the stabilization of mRNA during torpor.
Animals sacrificed during torpor showed a poly(A) tail size distribution pattern consistent with the binding of PABP (2, 3). PABP bound to the poly(A) tail inhibits deadenylation under physiological conditions in mammals (20). In both in vitro reconstitution assays and cell extract assays (6, 20, 26), the presence of PABP results in a distribution of poly(A) tail size abundance similar to what we observed. Depletion of PABP from cell extract assays results in mRNA degradation as much as 10 times faster than in the presence of PABP (6). While it is not surprising that PABP might be involved in producing this pattern of poly(A) tail sizes, the temporal pattern of poly(A) tail length distribution throughout the arousal-state changes of hibernation is striking and suggests that PABP could function to stabilize mRNA throughout a torpor bout.
There is growing evidence that PABP can enhance translation through an interaction with proteins bound to the 5′ cap structure (14, 18, 28). PABP bound to the poly(A) tail appears to interact with eukaryotic initiation factor 4G or PABP-interacting protein 1, which in turn promotes the binding of additional initiation factors that facilitate ribosome binding and start-codon recognition (10, 28). This suggests a second potential role for PABP in hibernating animals; the PABP interaction that stabilizes mRNA during torpor could also serve to facilitate initiation of translation upon arousal.
Additional RNA-protein interactions have been observed in hibernating animals.
Ultrastructural examination of tissue from hibernating and aroused hazel dormice (Muscardinus avellanarius) revealed several novel structures present only during torpor in the nuclei of brown adipose tissue, liver, pancreas, and adrenal cortex (21–24, 34, 39). Cytochemical and immunocytochemical examination of these structures revealed that they are complexes of small nuclear RNPs, heterogeneous nuclear RNAs, and protein, suggesting that these constituents may continue to combine to form spliceosomes at low body temperatures. Perichromatin fibers were also detected in nuclei of torpid dormice, indicating that transcription may also progress at a low rate during deep torpor. Malatesta et al. (23) and Tamburini et al. (34) note that these structures in torpid dormice resemble previously described structures observed in cells treated with either temperature or chemicals to inhibit both RNA and protein synthesis and that perhaps these structures represent an accumulation of splicing factors which can be brought into use as soon as metabolic activity increases during the rewarming phase of an arousal episode. Although these interactions take place in the nucleus, they are temporally correlated with the cytoplasmic mRNA-PABP interactions we describe in this paper. Future mechanistic studies will be necessary to elucidate whether RNA-protein interactions unique to the torpor phase of the hibernation cycle play a physiological role in stabilization of mRNA during torpor or preparation for translation during arousal episodes.
Inhibition of the translational machinery during torpor.
Analysis of polysome profiles prepared from the livers of animals sacrificed at different stages of torpor and arousal episodes suggests a reduction in translation during torpor. Animals early in a torpor bout displayed a reduction in polysomes, and polysomes were undetectable in tissue homogenates obtained from animals sacrificed after several days of torpor. Tissue homogenates obtained from animals during the rewarming phase of an arousal episode displayed a polysome profile indicating a reassembly of polysomes, and animals sacrificed at the end of the euthermic phase of an arousal episode had a polysome profile which resembled that obtained from summer-euthermic animals. All Northern blots prepared with RNA isolated from fractions of each polysome profile and probed for GAPDH revealed a distinct band of the appropriate size, indicating that changes in the appearance of the polysome profiles were not due to general degradation of mRNA. It should be noted that previous studies have shown that GAPDH levels (protein and mRNA) in the liver do not vary between the active and hibernating states in hibernating jerboas (Jaculus orientalis) (32).
These findings agree with previous work demonstrating a disaggregation of polysomes in torpid ground squirrels; moreover, extracts prepared from hibernating animals, when warmed and assayed at 37°C, still display an approximately threefold reduction in translation (13, 37). Frerichs and coworkers further demonstrated in vivo that as an animal is entering torpor but is still at a relatively high body temperature (body temperature decreased from 19.0 to 7.5°C during a 2-h experiment), protein synthesis is still suppressed, as measured by [14C]leucine administration (13). They suggest that this suppression is not due solely to temperature effects and attribute at least part of this active suppression to a change in the phosphorylation state of eukaryotic initiation factor 2α. Since it is unlikely that the detected degree of change in the phosphorylation state is sufficient to fully account for the drastic reduction in translation, modification of other aspects of the translation process cannot be ruled out (13). The reduction in polysomes suggests that the suppression of protein synthesis is due largely to an asynchrony between the rates of initiation and elongation, where initiation is most severely affected. It is possible that the reduction of initiation is mediated by modifying the mRNA template into a translationally inert state. This process, referred to as mRNA “masking,” has been well described for oocytes, where maternal mRNA is both stabilized and translationally repressed until fertilization occurs by an array of proteins, including PABP (15, 33).
Although our evidence of stabilization of mRNA through the presence of PABP and the inhibition of translation through the disassembly of polysomes in torpid arctic ground squirrels is indirect, restricting protein synthesis so that it occurs only during arousal episodes and with preexisting mRNAs would be advantageous to a hibernating mammal. If the many enzymatic processes involved in translation have differing temperature sensitivities, as body temperature decreases from euthermic to near-freezing temperatures, a mismatch of kinetics could occur, making regulation and integration of translation difficult. If one of the functions of arousal episodes is to allow protein synthesis to occur, having translatable mRNA already associated with PABP upon arousal could minimize the duration of the euthermic phase of arousal episodes. Because thermoregulatory costs during the euthermic phase are a significant portion of the total energetic cost of hibernating, minimizing the intervals of euthermy would be consistent with the energy-saving strategy of hibernation.
ACKNOWLEDGMENTS
This work was done under the tenure of a graduate research fellowship from the American Heart Association, Alaska Affiliate, Inc., to J.E.K. and with the support of ARO grant DAAG559810234 to S.L.M., NIH grant GM27757 to A.J., NSF OPP grant 9819540 to B.M.B., and an NSF CAREER Award to B.B.B.
We thank David Mangus for invaluable methodological advice.
REFERENCES
- 1.Anufriev A I, Akhremenko A K. Energetic cost of hibernation of the arctic ground squirrel. Ekologiya. 1990;5:68–72. [Google Scholar]
- 2.Baer B W, Kornberg R D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer B W, Kornberg R D. Repeating structures of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes B M. Freeze avoidance in a mammal: body temperatures below 0°C in an arctic hibernator. Science. 1989;244:1593–1595. doi: 10.1126/science.2740905. [DOI] [PubMed] [Google Scholar]
- 5.Barnes B M, Ritter D. Patterns of body temperature change in hibernating arctic ground squirrels. In: Carey C, Florant G L, Wunder B A, Horwitz B, editors. Life in the cold: ecological, physiological and molecular mechanisms. Boulder, Colo: Westview Press; 1993. pp. 119–130. [Google Scholar]
- 6.Bernstein P, Peltz S W, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bocharova L S, Gordon R Y, Arkhipov V I. Uridine uptake and RNA synthesis in the brain of torpid and awakened ground squirrels. Comp Biochem Physiol. 1992;101B:189–192. doi: 10.1016/0305-0491(92)90177-s. [DOI] [PubMed] [Google Scholar]
- 8.Boyer B B, Barnes B M. Molecular and metabolic aspects of mammalian hibernation. BioScience. 1999;49:713–724. [Google Scholar]
- 9.Buck C L, Barnes B M. Annual cycle of body composition and hibernation in free-living arctic ground squirrels. J Mammal. 1999;80:430–442. [Google Scholar]
- 10.Craig A W, Haghighat A, Yu A T, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 11.Daan S, Barnes B M, Strijkstra A M. Warming up for sleep? Ground squirrels sleep during arousals from hibernation. Neurosci Lett. 1991;128:265–268. doi: 10.1016/0304-3940(91)90276-y. [DOI] [PubMed] [Google Scholar]
- 12.French A R. The patterns of mammalian hibernation. Am Sci. 1988;76:569–575. [Google Scholar]
- 13.Frerichs K U, Smith C B, Brenner M, DeGracia D J, Krause G S, Marrone L, Dever T E, Hallenbeck J M. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc Natl Acad Sci USA. 1998;95:14511–14516. doi: 10.1073/pnas.95.24.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Tekur S, Reinbold R, Eppig J J, Choi Y C, Zheng J Z, Murray M T, Hecht N B. Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol Reprod. 1998;59:1266–1274. doi: 10.1095/biolreprod59.5.1266. [DOI] [PubMed] [Google Scholar]
- 16.Hammond M L, Bowman L H. Insulin stimulates the translation of ribosomal proteins and the transcription of rDNA in mouse myoblasts. J Biol Chem. 1988;263:17785–17791. [PubMed] [Google Scholar]
- 17.Hargrove J L, Schmidt F H. The role of mRNA and protein stability in gene expression. FASEB J. 1989;3:2360–2370. doi: 10.1096/fasebj.3.12.2676679. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 19.Jacobson A. Purification and fractionation of poly(A)+RNA. Methods Enzymol. 1987;152:254–261. doi: 10.1016/0076-6879(87)52028-6. [DOI] [PubMed] [Google Scholar]
- 20.Korner C G, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 21.Malatesta M, Cardinali A, Battistelli S, Zancanaro C, Martin T E, Fakan S, Gazzanelli G. Nuclear bodies are usual constituents in tissues of hibernating dormice. Anat Rec. 1999;254:389–395. doi: 10.1002/(SICI)1097-0185(19990301)254:3<389::AID-AR10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22.Malatesta M, Zancanaro C, Marcheggiani F, Cardinali A, Rocchi M B L, Capizzi D, Vogel P, Fakan S, Gazzanelli G. Ultrastructural, morphometrical and immunocytochemical analyses of the exocrine pancreas in a hibernating dormouse. Cell Tissue Res. 1998;292:531–541. doi: 10.1007/s004410051082. [DOI] [PubMed] [Google Scholar]
- 23.Malatesta M, Zancanaro C, Martin T E, Chan E K L, Amalric F, Lührmann R, Vogel P, Fakan S. Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur J Cell Biol. 1994;65:82–93. [PubMed] [Google Scholar]
- 24.Malatesta M, Zancanaro C, Tamburini M, Martin T E, Fu X, Vogel P, Fakan S. Novel nuclear ribonucleoprotein structural components in the dormouse adrenal cortex during hibernation. Chromosoma. 1995;104:121–128. doi: 10.1007/BF00347694. [DOI] [PubMed] [Google Scholar]
- 25.Martin S L, Srere H K, Belke D, Wang L C H, Carey H V. Differential gene expression in the liver during hibernation in ground squirrels. In: Carey C, Florant G L, Wunder B A, Horwitz B, editors. Life in the cold: ecological, physiological and molecular mechanisms. Boulder, Colo: Westview Press; 1993. pp. 443–453. [Google Scholar]
- 26.Morales J, Russell J E, Liebhaber S A. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J Biol Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 27.O'Hara B F, Watson F L, Srere H K, Kumar H, Wiler S W, Welch S K, Bitting L, Heller H C, Kilduff T S. Gene expression in the brain across the hibernation cycle. J Neurosci. 1999;19:3781–3790. doi: 10.1523/JNEUROSCI.19-10-03781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otero L J, Ashe M P, Sachs A B. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios R, Palmiter R D, Schimke R T. Identification and isolation of ovalbumin-synthesizing polysomes. J Biol Chem. 1972;247:2316–2321. [PubMed] [Google Scholar]
- 30.Scholtissek C. Nucleotide metabolism in tissue culture cells at low temperatures. I. Phosphorylation of nucleosides and deoxynucleosides in vivo. Biochim Biophys Acta. 1967;145:228–237. doi: 10.1016/0005-2787(67)90041-x. [DOI] [PubMed] [Google Scholar]
- 31.Sheiness D, Darnell J E. Polyadenylic acid segment in mRNA becomes shorter with age. Nat New Biol. 1973;241:265–268. doi: 10.1038/newbio241265a0. [DOI] [PubMed] [Google Scholar]
- 32.Soukri A, Valverde F, Hafid N, Elkebbaj M S, Serrano A. Occurrence of a differential expression of the glyceraldehyde-3-phosphate dehydrogenase gene in muscle and liver from euthermic and induced hibernating jerboa (Jaculus orientalis) Gene. 1996;181:139–145. doi: 10.1016/s0378-1119(96)00494-5. [DOI] [PubMed] [Google Scholar]
- 33.Spirin A S. Storage of messenger RNA in eukaryotes: envelopment with protein, translational barrier at 5′ side, or conformational masking by 3′ side? Mol Reprod Dev. 1994;38:107–117. doi: 10.1002/mrd.1080380117. [DOI] [PubMed] [Google Scholar]
- 34.Tamburini M, Malatesta M, Zancanaro C, Martin T E, Fu X, Vogel P, Fakan S. Dense granular bodies: a novel nucleoplasmic structure in hibernating dormice. Histochem Cell Biol. 1996;106:581–586. doi: 10.1007/BF02473273. [DOI] [PubMed] [Google Scholar]
- 35.Trachsel L, Edgar D, Heller H. Are ground squirrels sleep deprived during hibernation? Am J Physiol. 1991;260:R1123–R1129. doi: 10.1152/ajpregu.1991.260.6.R1123. [DOI] [PubMed] [Google Scholar]
- 36.Wang L C H. Time patterns and metabolic rates of natural torpor in the Richardson's ground squirrel. Can J Zool. 1979;57:149–155. [Google Scholar]
- 37.Whitten B K, Schrader L E, Huston R L, Honold G R. Hepatic polyribosomes and protein synthesis: seasonal changes in a hibernator. Int J Biochem. 1970;1:406–408. [Google Scholar]
- 38.Willis J S. The mystery of the periodic arousal. In: Lyman C, Willis J, Malan A, Wang L, editors. Hibernation and torpor in mammals and birds. New York, N.Y: Academic Press; 1982. pp. 92–103. [Google Scholar]
- 39.Zancanaro C, Malatesta M, Vogel P, Fakan S. Ultrastructure of the adrenal cortex of hibernating, arousing, and euthermic dormouse, Muscardinus avellanarius. Anat Rec. 1997;249:359–364. doi: 10.1002/(SICI)1097-0185(199711)249:3<359::AID-AR6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]